Abstract
The plasmid is a very well-known mobile genetic element that participates in the acquisition of virulence genes, such as staphylococcal enterotoxins (SEs), via horizontal transfer. SEs are emetic toxins and causative agents in staphylococcal food poisoning (SFP). We herein identified the types of plasmids harbored by seven SFP isolates and examined their production of plasmid-related SE/SEl to determine whether the new types of plasmid-related SE or SE-like (SEl) toxins (i.e. SElJ and SER) were involved in SFP. These isolates harbored pIB485-like plasmids, and all, except for one isolate, produced SElJ and SER. The amount of SER produced by each isolate accounted for the highest or second highest percentage of the total amount of SE/SEl produced. These new types of plasmid-related SE/SEls as well as classical SE may play a role in SFP. The seven isolates were classified into two SED-production types; a high SED-production type (>500 ng/ml) and no SED-production type. A nucleotide sequencing analysis revealed that three plasmids harbored by the SED-non-producing isolates had a single-base deletion in the sed gene with a resulting stop codon (from 233 amino acids of the intact SED to 154 amino acids of the mutant SED (mSED)). A real-time reverse transcription-PCR analysis showed that the mRNA of the msed gene was transcribed in the isolates. If the msed gene was translated as a protein, mSED may act as an emetic toxin instead of intact SED.
Keywords: mutant SED, plasmid, staphylococcal enterotoxin, staphylococcal food poisoning, Staphylococcus aureus
Staphylococcal food poisoning (SFP) is one of the most prevalent food-borne intoxications following the ingestion of staphylococcal enterotoxins (SEs), which are produced by Staphylococcus aureus (S. aureus) [9, 27]. SEs belong to a broad family of superantigens (SAGs). Most SAGs induce emesis and/or toxic shock syndrome [2, 6, 7, 20]. The mechanisms responsible for toxic shock syndrome have been examined, with various molecular evidence being obtained, including the identification of toxic shock syndrome toxin 1 (TSST-1) as a superantigen and the activation of T cell receptors [1, 10]. However, the mechanisms underlying emetic activity have not yet been elucidated in detail [10, 14, 19].
SEs have been divided into five serological types (classical SEs; SEA to SEE) based on their antigenicity. New types of SEs and SE-like toxins (SEls) have recently been reported (SEG to SElX) [2, 9, 14, 20, 26, 28]. Many SEs are encoded in accessory genetic elements that have been identified as mobile elements (mobile genetic elements; MGEs) [2, 13]. A previous study demonstrated that MGEs incorporated prophages, plasmids, transposons, the highly variable genomic region νSaβ and S. aureus pathogenicity islands [13]. This finding implies that SE/SEl genes are transferred among staphylococcal strains by horizontal transfer; therefore, MGEs may play a role in the evolution of pathogens [13, 14].
The plasmid is one of the best known MGEs and participates in the acquisition of beneficial genes for bacteria, such as antibiotic resistance genes or virulence genes, via horizontal transfer. At least five types of SE/SEl genes (i.e. sed, selj, ser, ses and set) are known to be encoded and transferred by plasmids [2, 14, 20, 24]. For example, previous studies reported that sed, selj and ser were encoded adjoiningly by the same plasmid, such as the pIB485-like plasmid, pUO-Sa-SED1 and pUO-Sa-SED2 [2, 14]. selj, ser, ses and set were also shown to be encoded by pF5 [20]. However, the types of plasmids harbored in SFP isolates collected during at least seven SFP outbreaks in Tokyo between 1992 and 2013 have not yet been identified, and it currently remains unknown whether new types of plasmid-related SEs (beyond the classical type of SED) are involved in these SFP outbreaks.
Although classical SEs were identified as the main factor in these outbreaks in Tokyo, Japan, we suspected that other plasmid-related SE/SEls were also involved. We analyzed the plasmids harbored by the representative S. aureus isolates of each SFP and examined the production of plasmid-related SEs in these isolates using molecular and protein analyses. We then attempted to confirm whether a new factor was involved in SFP.
MATERIALS AND METHODS
Bacterial strains and culture conditions: The bacterial strains used in this study are listed in Table 1. S. aureus strains were isolated from the feces and vomitus of patients or the suspected foods with SFP in Tokyo, Japan. The genotypes of SEs of these strains were determined using multiplex PCR techniques that have been reported previously [15]. S. aureus strains were cultured overnight in brain heart infusion (BHI) broth (Becton Dickinson, Sparks, MD, U.S.A.) at 37°C with shaking (100 rpm) to isolate plasmid DNA. The S. aureus cultures were grown in BHI broth supplemented with 1% yeast extract (Becton Dickinson) at 37°C for 48 hr with shaking (100 rpm) in order to assess SE production or at 37°C for 24 hr with shaking (100 rpm) to extract total RNA. Escherichia coli (E. coli) strain DH5α was purchased from Promega (Madison, WI, U.S.A.). The E. coli strains were cultured overnight in Luria-Bertani (LB) broth (Sigma, St. Louis, MO, U.S.A.) containing 50 µg/ml of ampicillin (Wako Pure Chemicals, Osaka, Japan) at 37°C with shaking (100 rpm) for plasmid isolation.
Table 1. Bacterial strains used in the present study.
| Strains | SEa)/SElb) genotypes | Source | |
|---|---|---|---|
| S. aureus | |||
| Tokyo10539 | sed seg sei selj sem sen seo ser | Food poisoning outbreak | |
| Tokyo11726 | sea sed selj ser | Food poisoning outbreak | |
| Tokyo12261 | sea sed selj ser | Food poisoning outbreak | |
| Tokyo12804 | sea sed selj ser | Food poisoning outbreak | |
| Tokyo12902 | sea sed selj ser | Food poisoning outbreak | |
| Tokyo13057 | sed seg sei selj sem sen seo ser | Food poisoning outbreak | |
| Tokyo13231 | sea sed selj ser | Food poisoning outbreak | |
| E. coli | |||
| DH5α | SE negative | Promega | |
Boldface type indicates staphylococcal enterotoxins or staphylococcal enterotoxin-like toxins harbored by plasmids. a) SE: Staphylococcal enterotoxin. b) SEl: Staphylococcal enterotoxin-like toxin.
Extraction of Staphylococcus aureus plasmid DNA and analysis of restriction fragment length polymorphism: The extraction of plasmid DNA from S. aureus was performed according to a previous method with a slight modification [4]. In brief, cultured S. aureus cells were pelleted by centrifugation. The pellets were then dissolved with 200 µl of lysozyme solution (5 µg/ml lysostaphin, 50 mM glucose, 10 mM EDTA and 25 mM Tris-HCl, pH 8.0) and then incubated at 37°C for 1 hr. After being incubated, 400 µl of alkaline SDS solution (0.2 N NaOH, 1% SDS) was added and gently mixed by inversion for a few sec, and 300 µl of high salt solution (3M sodium acetate, pH 4.8) was then added and gently mixed by inversion for a few sec. The samples are maintained on ice for 10 min and then centrifuged at 12,000 × g for 15 min at 4°C. The supernatant containing plasmid DNA was transferred into new tubes and then purified by a phenol-chloroform treatment, isopropanol precipitation and ethanol precipitation. After purification, each plasmid DNA was dissolved with TE buffer. The yield and purity of the extracted plasmid DNA were quantified using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, U.S.A.). Plasmid DNA was digested with EcoRI (2 U, Takara, Otsu, Japan) at 37°C for 1 hr, and the enzyme was then inactivated by the addition of a 10 ×loading buffer (Takara) to perform a restriction fragment length polymorphism (RFLP) analysis. The digested plasmid DNA was electrophoresed on 0.7% SeaKem GTG Agarose gels (Lonza, Rockland, ME, U.S.A.).
Plasmid constructs: All plasmids used in this study are listed in Table 2. The primers employed to construct the plasmids are indicated in Table 3. The plasmid pG10539F was constructed by cloning approximately 4.9 kbp of the EcoRI-fragment of pTokyo10539 into the plasmid vector pGEM-3Zf (+) (Promega). The plasmids pG11726F, pG12261F, pG12804F, pG12902F, pG13057F and pG13231F were also constructed by cloning approximately 4.9 kbp of the EcoRI-fragment obtained from the appropriate S. aureus plasmids into the plasmid vector pGEM-3Zf (+) (Table 2). The plasmids pGTsed, pGTftsZ, pGTsodA and pGTrpoB were constructed by cloning the PCR products into the pGEM-T easy TA cloning vector (Promega). Subcloning plasmids were transformed into E. coli DH5α (Promega).
Table 2. Plasmids used in the present study.
| Plasmid | Relevant characteristic | Source |
|---|---|---|
| pGEM-3Zf (+) | Apr, cloning vector | Promega |
| pGEM-T easy | Apr, TA cloning vector | Promega |
| pTokyo10539 | S. aureus plasmid purified from strain Tokyo10539 | This study |
| pTokyo11726 | S. aureus plasmid purified from strain Tokyo11726 | This study |
| pTokyo12261 | S. aureus plasmid purified from strain Tokyo12261 | This study |
| pTokyo12804 | S. aureus plasmid purified from strain Tokyo12804 | This study |
| pTokyo12902 | S. aureus plasmid purified from strain Tokyo12902 | This study |
| pTokyo13057 | S. aureus plasmid purified from strain Tokyo13057 | This study |
| pTokyo13231 | S. aureus plasmid purified from strain Tokyo13231 | This study |
| pG10539F | Apr, pGEM-3Zf (+) with cloned 4916 bp EcoRI-fragment of pTokyo10539 | This study |
| pG11726F | Apr, pGEM-3Zf (+) with cloned 4915 bp EcoRI-fragment of pTokyo11726 | This study |
| pG12261F | Apr, pGEM-3Zf (+) with cloned 4916 bp EcoRI-fragment of pTokyo12261 | This study |
| pG12804F | Apr, pGEM-3Zf (+) with cloned 4915 bp EcoRI-fragment of pTokyo12804 | This study |
| pG12902F | Apr, pGEM-3Zf (+) with cloned 4917 bp EcoRI-fragment of pTokyo12902 | This study |
| pG13057F | Apr, pGEM-3Zf (+) with cloned 4916 bp EcoRI-fragment of pTokyo13057 | This study |
| pG13231F | Apr, pGEM-3Zf (+) with cloned 4915 bp EcoRI-fragment of pTokyo13231 | This study |
| pGTsed | Apr, pGEM-T easy with cloned PCR product containing sed (msed) sequence (41 bp–425 bp) | This study |
| pGTftsZ | Apr, pGEM-T easy with cloned PCR product containing ftsZ sequence (53 bp–448 bp) | This study |
| pGTsodA | Apr, pGEM-T easy with cloned PCR product containing sodA sequence (71 bp–539 bp) | This study |
| pGTrpoB | Apr, pGEM-T easy with cloned PCR product containing rpoB sequence (965 bp–1576 bp) | This study |
Table 3. Nucleotide sequences used in the present study.
| Purpose | Gene | Name | Oligonucleotide sequence (5′→3′) |
|---|---|---|---|
| TA-Cloning | sed or msed | SED41F | CTAGTTTGGTAATATCTCCTTTAAACG |
| SED425R | TTACCTTCGTGTGGAGTGACA | ||
| ftsZ | FTSZ53F | GTGTAGGTGGTGGCGGTAAC | |
| FTSZ448R | CAGCAGCAGCTTGAGTTTGA | ||
| sodA | SODA71F | TGGAAATTCACCATGACAGA | |
| SODA539R | CCAATGTAGTCAGGGCGTTT | ||
| rpoB | RPOB965F | TTGAATCAAACGCAAACAGC | |
| RPOB1576R | ATCCAATGTTTGGTCCCTCA | ||
| Quantitative RT-PCR | sed or msed | SED291F | TTCAAAAGAAATGGCTCAACA |
| SED425R | TTACCTTCGTGTGGAGTGACA | ||
| ftsZ | FTSZ315F | GGGTGGCGGAACTGGTACT | |
| FTSZ376R | CGCCCATTTCTTTTGCAATT | ||
| sodA | SODA373F | GGTTCAGGTTGGGCTTGGT | |
| SODA440R | TCTTGGTTTGGTGTAGTCACAATTTC | ||
| rpoB | RPOB1482F | ACGTGAACGTGCTCAAATGG | |
| RPOB1563R | TCCCTCAGGCGTTTCAATTG |
Sandwich ELISA: In order to determine the concentrations of SEs in cultures, sandwich ELISA was performed according to our and Omoe’s previously reported method with slight modifications [16, 18, 21, 22]. In brief, the culture supernatant of the S. aureus strain was treated overnight with 50% (v/v) normal rabbit serum (Thermo Fisher Scientific) at 4°C to eliminate any nonspecific reactions caused by Protein A, and the treated culture supernatant was then diluted 10- to 10,000-fold in Can Get Signal Immunoreactions Enhancer Solution 1 (TOYOBO, Osaka, Japan). Each anti-SE antibody labeled with HRP was diluted at an appropriate ratio in Can Get Signal Immunoreactions Enhancer Solution 2 (TOYOBO). The luminescence signal was determined using a SpectraMax Paradigm (Molecular Devices, Sunnyvale, CA, U.S.A.) and SuperSignal ELISA Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). The concentration of each SE in the culture supernatants was determined by converting the luminescence values to the corresponding concentrations by use of a standard curve.
Sequencing of EcoRI-fragments including sed, selj and ser genes: Omoe et al. previously reported that the sed, selj and ser genes were encoded by 4.5–5.0 kbp of the EcoRI-fragments of pIB485-like plasmids [14]. Therefore, we cloned 4.5–5.0 kbp of the EcoRI-fragments of seven S. aureus plasmids into pGEM-3Zf (+) and determined the inserted sequences using the primer walking method with an ABI3130 capillary sequencer (Thermo Fisher Scientific) and BigDye Terminator Ver. 3.1 Cycle Sequencing kit (Thermo Fisher Scientific). Sequence data were assembled by MEGA5 software [25]. The extraction of open reading frames (ORFs) was performed using GeneMark (http://exon.gatech.edu/) and ORF Finder software (http://www.ncbi.nlm.nih.gov/projects/gorf/). ORFs were annotated by the Basic Local Alignment Search Tool (BLAST) (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Comparisons of the sequences of each ORF were performed using Genetyx software program, ver. 8 (GENETYX CORPORATION, Tokyo, Japan).
Total RNA extraction and reverse transcription reaction: Total RNA (tRNA) was extracted from seven S. aureus isolates. These isolates were cultured in BHI broth supplemented with 1% yeast extract, the cell bodies were harvested by centrifugation at 5,000 × g for 10 min, and the tRNA of the cells was then extracted using a RiboPure-Bacteria Kit (Thermo Fisher Scientific) according to the manufacturer’s instruction. The amounts of tRNA were measured using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific). The quality of tRNA was confirmed by measuring the A260/A280 ratio and 1% agarose gel electrophoresis with no degradation of 23S or 16S ribosomal RNA. The extracted tRNA (2 µg) was reversely transcribed using random primers with the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instruction. The reverse transcription (RT) cycle comprised 25°C for 10 min, 37°C for 120 min and 85°C for 5 min in a thermal cycler, and the resultant cDNA was stored at −20°C.
Quantitative real-time RT-PCR: The expression levels of sed or mutant sed (msed) in the S. aureus isolates were confirmed by quantitative real-time RT-PCR (qRT-PCR) using the Power SYBR Green PCR Master Mix (Thermo Fisher Scientific) and ABI7500 Real Time PCR System (Thermo Fisher Scientific) according to the manufacturer’s instructions. According to a previous study [5], we selected three housekeeping genes (i.e. cell division protein (ftsZ), superoxide dismutase (sodA) and RNA polymerase beta subunit (rpoB)) for normalization. The primer sets for sed, ftsZ, sodA and rpoB were used as shown in Table 3. The standard curves for each gene were generated by the serial dilution of the above-constructed plasmids containing PCR products for one of these genes in order to quantify their mRNA concentrations. A melting curve was used to confirm that the SYBR green-based amplicons were accurate because SYBR green also detects double-stranded DNA, including primer dimers, contaminating DNA and PCR products from misannealed primers. The expression ratio of sed or msed to that of ftsZ, sodA or rpoB was calculated to adjust for variations among the qRT-PCR reactions.
Statistical analysis: Statistical analyses were performed using Statcel2 software (The Publisher OMS Ltd., Saitama, Japan). SE production data and gene expression data were analyzed initially by ANOVA and then the Tukey-Kramer multiple comparison. P values of <0.05 were considered significant.
RESULTS
Determination of SE/SEl genotypes of S. aureus isolates from seven outbreaks: Seven SFP isolates were subjected to a multiplex PCR analysis in order to detect the SE/SEl genes. All isolates harbored two plasmid-related SE/SEl genes (selj and ser) in addition to the sed gene (Table 1). Furthermore, five out of seven isolates harbored the sea gene, while two harbored the seg, sei, sem, sen and seo genes (Table 1).
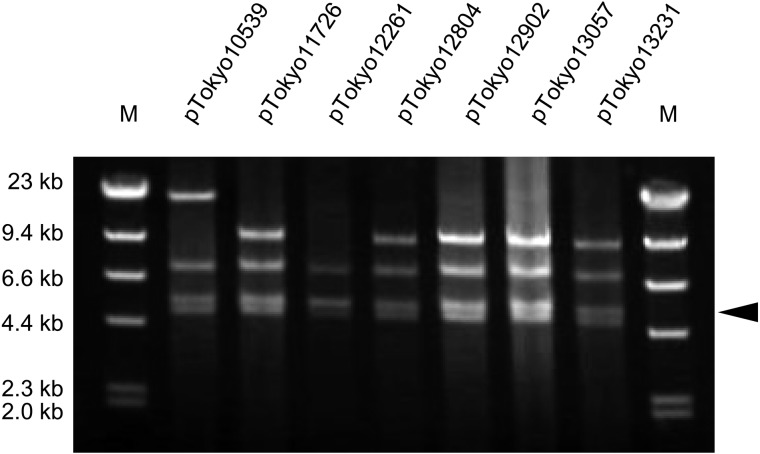
Identification of plasmids harbored by seven S. aureus isolates: Since three plasmid-related SE/SEl genes were commonly detected among all seven S. aureus isolates, the plasmids were purified from these isolates and identified by RFLP. The EcoRI-treated plasmids revealed respective agarose-gel electrophoretic patterns (Fig. 1). The RFLP patterns of the digested plasmids were identical to or very similar to those of the pIB485-like plasmids (originally extracted from Omoe’s laboratory strains [14]). The same RFLP patterns were commonly detected in pTokyo11726, pTokyo12804, pTokyo12902, pTokyo13057 and pTokyo13231. The RFLP patterns of these plasmids were identical to those of p196E and p361 analyzed by Omoe et al. [14]. The RFLP pattern of pTokyo12261 was identical to those of p1151 and pI7 [14]. The p196E, p361, p1151 and pI7 plasmids were each a type of pIB485-like plasmid [14]. On the other hand, the RFLP pattern of pTokyo10539 was not identical to those of the other plasmids, but was very similar to those of the above pIB485-like plasmids.
Fig. 1.
Restriction fragment length polymorphism analysis of purified Staphylococcus aureus plasmids. Plasmids were purified from sed-, selj- and ser-positive S. aureus isolates originating from staphylococcal food poisoning outbreaks that occurred in Tokyo. These plasmids were digested by EcoRI. Lanes: M, size marker λ-HindIII digest. The arrowhead indicates the EcoRI fragment (approximately 4.9 kbp), which is assumed to involve the sed, selj and ser genes.
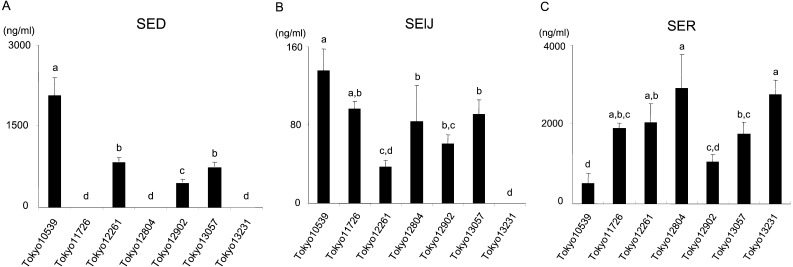
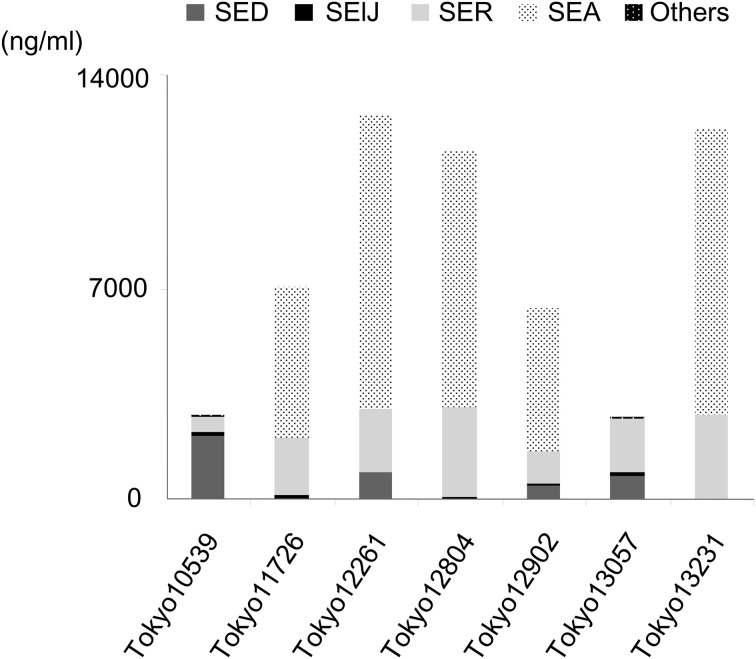
Detection of SE production in seven S. aureus isolates: The production of plasmid-related SE/SEls was measured in the seven isolates by sandwich ELISA. SElJ and SER were produced by most of the isolates; however, the intensity of their production was dependent on each isolate (SElJ: 37–135 ng/ml, except for Tokyo13231, SER: 523–2925 ng/ml; Fig. 2B and 2C). On the other hand, the seven isolates were classified into two SED-production types: a high SED-production type (higher than 500 ng/ml) and no SED-production type (lower than the detection limit (<0.1 ng/ml)). The former included Tokyo10539, Tokyo12261, Tokyo12902 and Tokyo13057, while the latter included Tokyo11726, Tokyo12804 and Tokyo13231 (Fig. 2A). SED accounted for 0–76% of the total amount of SE/SEls among the seven isolates (Fig. 3), while SElJ and SER accounted for 0–5% and 16–66%, respectively. SEA accounted for 71–78% of the total amount of SE/SEl produced in sea-positive isolates (Tokyo11726, Tokyo12261, Tokyo12804, Tokyo12902 and Tokyo13231). The production of SEG, SEI, SEM, SEN and SEO was very weak (<0.1%) among these SE genes-positive isolates (Tokyo10539 and Tokyo13057).
Fig. 2.
Amounts of plasmid-related staphylococcal enterotoxins (SEs) produced by seven isolates originating from staphylococcal food poisoning outbreaks. (A) SED, (B) SElJ and (C) SER production levels are shown. Experiments were performed in triplicate, and data are presented as the mean value ± standard deviation. Data labeled with different letters are significantly different from each other (P<0.05).
Fig. 3.
Total amount of staphylococcal enterotoxins (SEs) produced by seven isolates originating from staphylococcal food poisoning outbreaks. Experiments were performed in triplicate, and data are presented as the mean value. ‘Others’ in the figure include the production of SE, such as SEG, SEI, SEM, SEN and SEO.
Cloning and sequencing analyses of EcoRI-fragments harboring sed or the shortened sed-like (mutant sed) sequence: Since SED was not detected in three isolates, we speculated that these isolates may have a different sequence. We cloned 4.5–5.0 kbp of the EcoRI-fragments of seven S. aureus plasmids into pGEM-3Zf (+) (see the part indicated by the arrowhead in Fig. 1) and determined their sequences. Three different nucleotide sequences, 4,915 bp, 4,916 bp, and 4,917 bp, were detected: 4,915 bp originated from the EcoRI-fragments of pTokyo11726, pTokyo12804 and pTokyo13231, 4,916 bp originated from the EcoRI-fragments of pTokyo10539, pTokyo12261 and pTokyo13057, and 4,917 bp originated from the EcoRI-fragments of pTokyo12902. All EcoRI-fragments had three open reading frames (ORFs) potentially encoding proteins over 100 amino acids in length. These three ORFs corresponded to the sed, selj and ser genes with high homology. However, compared with sed harbored by these EcoRI-fragments, the sed genes encoded by pTokyo11726, pTokyo12804 and pTokyo13231 were 99% identical to those encoded by other plasmids, but had a deletion in the gene at the 514th base “A” from the start codon (temporally named mutant sed; msed). On the other hand, an intact sequence was detected in pTokyo10539, pTokyo12261, pTokyo12902 and pTokyo13057 (Fig. 4).
Fig. 4.
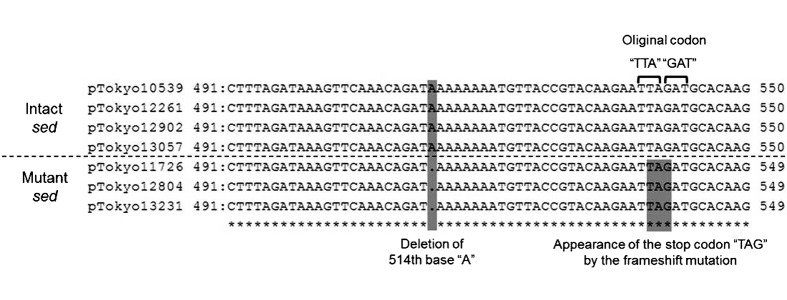
Comparison of sed or mutant sed sequences harbored by each of the seven plasmids that originated from staphylococcal food poisoning isolates. Two shades show the deletion point (left) or the stop codon by the frameshift mutation (right). The square brackets show the original codons of intact sed.
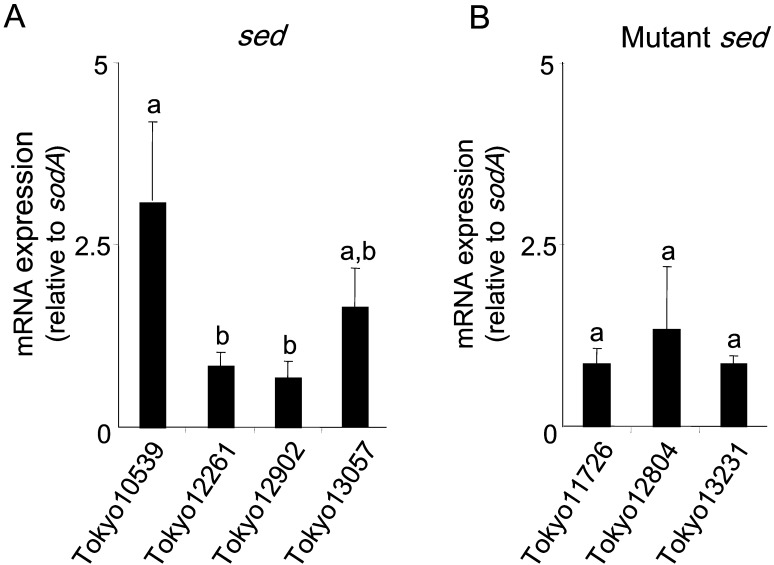
Detection of sed or mutant sed mRNA by quantitative RT-PCR: The sed gene was expressed by all S. aureus isolates harboring intact sed, with the intensity of expression depending on each isolate (Fig. 5). These results coincided with the protein expression profiles (Fig. 2A). On the other hand, the transcription of the msed was detected in the SED-non-producing isolates. No significant differences were observed among these isolates (P>0.05, Fig. 5) after normalization using sodA. Similar results were also confirmed using ftsZ and rpoB as internal controls (data not shown).
Fig. 5.
Gene expression profiles in each of the seven isolates that originated from staphylococcal food poisoning outbreaks. (A)sed and (B) mutant sed are shown. The amount of sed or msed mRNA was normalized to that of sodA mRNA as an internal control. Data labeled with different letters were significantly different from each other (P<0.05).
DISCUSSION
Classical SEs are known to be the toxin causatives of SFP [6]. SED, which is one of the classical SEs, is the second or third most common SE associated with SFP [3]. Some S. aureus strains produce SED through the transfer of plasmids encoding the sed gene. Such sed-harboring plasmids also encode other SE/SEl genes (e.g. selj and ser), similar to pIB485-like plasmids [2, 14]. In the present study, we purified seven plasmids from SFP isolates that were sed-, selj- and ser-positive. All of these plasmids were identical to or very similar to pIB485-like plasmids [14]. These results suggested that the SFP isolates examined in the present study acquired these SE/SEl genes through the transfer of pIB485-like plasmids from other S. aureus strains.
In the present study, the production of SElJ and SER was confirmed among most of the S. aureus isolates tested (Fig. 2B and 2C). These isolates produced large amounts of SER (from 523 to 2,925 ng/ml) (Fig. 2C). A recent study demonstrated that SER induced emesis in a primate model following its oral administration [20]. On the other hand, SElJ has not been tested for its emetic activity in a primate model. However, SElJ may indeed exhibit emetic activity, because its amino acid (AA) sequence is similar to that of SEA (64.6% homology) and SEE (63.4% homology) in all SE/SEls. Omoe et al. also reported that the symptoms of SFP may be induced by multiple classical and new SEs produced from a single S. aureus strain [17]. In the present study, the amount of SER produced by each of the seven isolates accounted for the highest or second highest percentage of the total amount of SE/SEl produced (Fig. 3). These results suggested that SElJ and SER participated in SFP, even though classical SEs may be the main cause of SFP.
A nucleotide sequencing analysis detected two distinctly different types of sequence structures in the sed gene; intact and aberrant. Four plasmids: pTokyo10539, pTokyo12261, pTokyo12902 and pTokyo13057, were intact, while the other three: pTokyo11726, pTokyo12804 and pTokyo13231, had a single-base deletion in sed (514th base “A” from the start codon). The frameshift mutation due to this single-base deletion resulted in a stop codon in the gene (msed gene) (Fig. 4). This result suggested that msed transcription was stopped in the middle of intact sed. By converting the nucleotide sequence of msed into an AA sequence, the length of the mSED protein (154 AA) was found to be shorter than that of the intact SED protein (233 AA). This is the first study to have identified such a plasmid harboring the msed gene that originated from SFP isolates.
The functional regions of SEs were previously shown to be responsible for emetic and superantigenic activities [10, 12, 23]; for example, the residues of 21–50 and 81–100 in SEA were found to be important for emetic and superantigenic activities, whereas the residues of 161–180 were only responsible for superantigenic activity [12]. Several residues (H187, H225 and D227) for binding to major histocompatibility complex (MHC) class II have been identified by mutagenesis [10, 23], and the major MHC binding sites, which were located on the C-terminal side, were marked out for superantigenic activity. SED has a similar type of binding site to SEA [10]. mSED then conserves the region of the N-terminal side responsible for emetic activity. These findings suggest the mSED protein retains emetic activity. On the other hand, it has also been reported that a one-point mutant of SE (in the N-terminal or C-terminal site) may result in the loss of emetic and/or superantigenic activities [8, 11]. Therefore, we intend to examine the precise function of mSED in a future study.
A significant amount of SED (more than 500 ng/ml) was detected in each S. aureus isolate harboring the plasmids encoding intact sed (Fig. 2A). On the other hand, the production of mSED was not detected in any S. aureus isolate harboring the plasmids encoding msed by conventional sandwich ELISA using an anti-SED antibody (Fig. 2A). However, the msed gene was expressed in a similar manner to the intact sed gene (Fig. 5). If the msed gene was translated as a protein, the mSED protein may possess slightly different properties (e.g. three-dimensional structure and epitopes) from the intact SED protein and act as an emetic toxin instead of intact SED. The amount of the mSED protein produced by SFP isolates as well as its emetic activity needs to be directly examined in future studies.
In summary, we herein demonstrated that plasmid-related SE/SEls (SElJ and SER) and mutant SED may have contributed to SFP outbreaks. All SFP isolates harbored pIB485-like plasmids and produced the plasmid-related SE/SEls encoded by these plasmids. The new types of plasmid-related SE/SEls as well as classical SEs play a role in SFP. In addition, we found that mutant SED was encoded by three pIB485-like plasmids, which were harbored by SED-non-detecting isolates. Since the mRNA of msed was transcribed in these isolates, mSED may act as an emetic toxin instead of intact SED.
Acknowledgments
We thank members of the food-poisoning laboratory (at the Tokyo Metropolitan Institute of Public Health) for kindly providing the S. aureus strains used in this study. We thank Prof. Kazuyoshi Hashizume (at Iwate University) for his comments and suggestions, which helped us to considerably improve our manuscript. We would like to express our sincere gratitude to Prof. Katsuhiko Omoe (at Iwate University).
REFERENCES
- 1.Alouf J. E., Müller-Alouf H.2003. Staphylococcal and streptococcal superantigens: molecular, biological and clinical aspects. Int. J. Med. Microbiol. 292: 429–440. doi: 10.1078/1438-4221-00232 [DOI] [PubMed] [Google Scholar]
- 2.Argudín M. Á., Mendoza M. C., Rodicio M. R.2010. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2: 1751–1773. doi: 10.3390/toxins2071751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argudín M. Á., Mendoza M. C., González-Hevia M. A., Bances M., Guerra B., Rodicio M. R.2012. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl. Environ. Microbiol. 78: 2930–2935. doi: 10.1128/AEM.07487-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnboim H. C., Doly J.1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7: 1513–1523. doi: 10.1093/nar/7.6.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derzelle S., Dilasser F., Duquenne M., Deperrois V.2009. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol. 26: 896–904. doi: 10.1016/j.fm.2009.06.007 [DOI] [PubMed] [Google Scholar]
- 6.Dinges M. M., Orwin P. M., Schlievert P. M.2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13: 16–34. doi: 10.1128/CMR.13.1.16-34.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser J. D., Proft T.2008. The bacterial superantigen and superantigen-like proteins. Immunol. Rev. 225: 226–243. doi: 10.1111/j.1600-065X.2008.00681.x [DOI] [PubMed] [Google Scholar]
- 8.Harris T. O., Betley M. J.1995. Biological activities of staphylococcal enterotoxin type A mutants with N-terminal substitutions. Infect. Immun. 63: 2133–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennekinne J. A., De Buyser M. L., Dragacci S.2012. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev. 36: 815–836. doi: 10.1111/j.1574-6976.2011.00311.x [DOI] [PubMed] [Google Scholar]
- 10.Hu D. L., Nakane A.2014. Mechanisms of staphylococcal enterotoxin-induced emesis. Eur. J. Pharmacol. 722: 95–107. doi: 10.1016/j.ejphar.2013.08.050 [DOI] [PubMed] [Google Scholar]
- 11.Hu D. L., Omoe K., Sashinami H., Shinagawa K., Nakane A.2009. Immunization with a nontoxic mutant of staphylococcal enterotoxin A, SEAD227A, protects against enterotoxin-induced emesis in house musk shrews. J. Infect. Dis. 199: 302–310. doi: 10.1086/596065 [DOI] [PubMed] [Google Scholar]
- 12.Maina E. K., Hu D. L., Asano K., Nakane A.2012. Inhibition of emetic and superantigenic activities of staphylococcal enterotoxin A by synthetic peptides. Peptides 38: 1–7. doi: 10.1016/j.peptides.2012.08.013 [DOI] [PubMed] [Google Scholar]
- 13.Malachowa N., DeLeo F. R.2010. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67: 3057–3071. doi: 10.1007/s00018-010-0389-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omoe K., Hu D. L., Takahashi-Omoe H., Nakane A., Shinagawa K.2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun. 71: 6088–6094. doi: 10.1128/IAI.71.10.6088-6094.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omoe K., Hu D. L., Takahashi-Omoe H., Nakane A., Shinagawa K.2005. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol. Lett. 246: 191–198. doi: 10.1016/j.femsle.2005.04.007 [DOI] [PubMed] [Google Scholar]
- 16.Omoe K., Ishikawa M., Shimoda Y., Hu D. L., Ueda S., Shinagawa K.2002. Detection of seg, seh, and sei genes in Staphylococcus aureus isolates and determination of the enterotoxin productivities of S. aureus isolates Harboring seg, seh, or sei genes. J. Clin. Microbiol. 40: 857–862. doi: 10.1128/JCM.40.3.857-862.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Omoe K., Hu D. L., Ono H. K., Shimizu S., Takahashi-Omoe H., Nakane A., Uchiyama T., Shinagawa K., Imanishi K.2013. Emetic potentials of newly identified staphylococcal enterotoxin-like toxins. Infect. Immun. 81: 3627–3631. doi: 10.1128/IAI.00550-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omoe K., Imanishi K., Hu D. L., Kato H., Fugane Y., Abe Y., Hamaoka S., Watanabe Y., Nakane A., Uchiyama T., Shinagawa K.2005. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect. Immun. 73: 5540–5546. doi: 10.1128/IAI.73.9.5540-5546.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono H. K., Nishizawa M., Yamamoto Y., Hu D. L., Nakane A., Shinagawa K., Omoe K.2012. Submucosal mast cells in the gastrointestinal tract are a target of staphylococcal enterotoxin type A. FEMS Immunol. Med. Microbiol. 64: 392–402. doi: 10.1111/j.1574-695X.2011.00924.x [DOI] [PubMed] [Google Scholar]
- 20.Ono H. K., Omoe K., Imanishi K., Iwakabe Y., Hu D. L., Kato H., Saito N., Nakane A., Uchiyama T., Shinagawa K.2008. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect. Immun. 76: 4999–5005. doi: 10.1128/IAI.00045-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato’o Y., Omoe K., Naito I., Ono H. K., Nakane A., Sugai M., Yamagishi N., Hu D. L.2014. Molecular epidemiology and identification of a Staphylococcus aureus clone causing food poisoning outbreaks in Japan. J. Clin. Microbiol. 52: 2637–2640. doi: 10.1128/JCM.00661-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato’o Y., Omoe K., Ono H. K., Nakane A., Hu D. L.2013. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol. Immunol. 57: 91–99. doi: 10.1111/1348-0421.12007 [DOI] [PubMed] [Google Scholar]
- 23.Schad E. M., Zaitseva I., Zaitsev V. N., Dohlsten M., Kalland T., Schlievert P. M., Ohlendorf D. H., Svensson L. A.1995. Crystal structure of the superantigen staphylococcal enterotoxin type A. EMBO J. 14: 3292–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schelin J., Wallin-Carlquist N., Cohn M. T., Lindqvist R., Barker G. C., Rådström P.2011. The formation of Staphylococcus aureus enterotoxin in food environments and advances in risk assessment. Virulence 2: 580–592. doi: 10.4161/viru.2.6.18122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas D. Y., Jarraud S., Lemercier B., Cozon G., Echasserieau K., Etienne J., Gougeon M. L., Lina G., Vandenesch F.2006. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 74: 4724–4734. doi: 10.1128/IAI.00132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tranter H. S.1990. Foodborne staphylococcal illness. Lancet 336: 1044–1046. doi: 10.1016/0140-6736(90)92500-H [DOI] [PubMed] [Google Scholar]
- 28.Wilson G. J., Seo K. S., Cartwright R. A., Connelley T., Chuang-Smith O. N., Merriman J. A., Guinane C. M., Park J. Y., Bohach G. A., Schlievert P. M., Morrison W. I., Fitzgerald J. R.2011. A novel core genome-encoded superantigen contributes to lethality of community-associated MRSA necrotizing pneumonia. PLoS Pathog. 7: e1002271. doi: 10.1371/journal.ppat.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]