Abstract
Background: Since more and more evidences support that NUMB orchestrates many cell physiological and pathological processes of diseases including cancer, based on our previous work, we studied deeply the function of NUMB in endometrial cancer (EC) and tried to understand the mechanism of NUMB's nucleus translocation which might be relative to the occurrence of EC and will contribute to find a new targeting therapeutic strategy for EC.
Methods: Immunohistochemistry was employed to test NUMB and HDM2 expression in endometrial cancer tissue from clinical patients. CCK-8 assay, cell cycle tested by Flow cytometer and PCNA determined by RT-PCR were employed to test the effects of NUMB on cell proliferation and apoptosis. In order to investigate the mechanism how NUMB, HDM2 and p53 interact in EC cell, western blot, Co-IP and immunofluorescent were used to observe the combination and location of NUMB, HDM2 and p53 as well as the interaction among them.
Results: Both NUMB and HDM2 expressed greater in endometrial cancer tissues than in normal endometrial tissues. Overexpression of NUMB induced apoptosis in Ishikawa cell while inhibition of NUMB increased cell proliferation. NUMB could combine HDM2 and p53, moreover the PTB domain of NUMB is the main site combining with p53. The effects of NUMB in cell was closely associated with p53. Not only NUMB regulated P53 expression level but also NUMB acts depending on P53, in turn p53 impacted the NUMB level as a feedback. Overexpression of NUMB could not bring itself into nuclear. Both siHDM2 and siP53 didn't bring NUMB into nucleus, However overexpression of HDM2 and p53 increased the NUMB level in nucleus, and the NUMB nuclear location induced by overexpression of HDM2 was stronger than that of p53 overexpression.
Conclusions: Based on present data, we think NUMB acts as an anti-oncogene role and could regulate p53 level and function in endometrial cancer like in other cancers, meanwhile, the function of NUMB depend on P53. On the other hand, the location of NUMB could be regulated mainly by HDM2. So far we are not able to explain why endometrial cancer patients had high NUMB expression level since NUMB was regarded as a tumor suppressor, which is worthy studying further to explore underlying mechanism.
Keywords: NUMB, endometrium cancer, nuclear translocation, HDM2, p53
Introduction
NUMB was first described as a cell fate determiner in neuron system, and consequently was reported to be responsible for the anti-oncogenic properties in cancers1. In breast cancer, low expression of NUMB was proven to be related with the poor prognosis. However, there were other evidences showing NUMB has oncogene characters in some context instead of anti-oncogenic properties. For example, in experimental gliomas, NUMB overexpression did not exert a tumor suppressor function with no inhibition of proliferation and failing to induce the differentiation of neural or glial cells2. The other two new isoforms of NUMB (NUMB 5, NUMB 6) was proven to play the role of oncogene3. So, what is the role of NUMB in endometrial cancer, which is one of the most common women malignant genital cancers worldwide? However, it has not yet been reported so far. In our previous study, it showed overexpression of NUMB and the accumulation of NUMB in nucleus in endometrial cancer4. This nuclear translocation was accordance with the findings in pilocytic astrocytomas5. Therefore, we wanted to go further to understand which role NUMB acts as in endometrial cancer, oncogenetic or anti-oncogenic role? Who regulated the nuclear location of NUMB occurred since it was regarded as locating in cytoplasm mainly6.
Material and methods
1. Antibodies, plasmids, and other materials
Main antibodys: Anti-NUMB antibody (Abcam, ab14140, Hong Kong Ltd.). HDM2 antibody (EPITOMICS, Cat.#S1357, Epitomics Inc.). P53 Rabbit Monocloal antibody (EPITOMICS, Cat.#1026-1, Epitomics Inc.). Main reagents: Dulbecco's modified Eagle medium- high glucose (GIBCO, 11965-092). Fetal bovine serum (GIBCO, 10099-141), 0.25% trypsin (GIBCO, 15050), SYBR Green Realtime PCR Master Mix (TOYOBO, QRT-101), First Strand cDNA Synthesis Kit (TOYOBO, FSK-101), Trizol (Invitrogen, 66012), Lipofectamine® 2000 Reagent (invitrogen, cat#11618-019), Opti-MEM (invitrogen, cat#31985-062).
2. Cell culture and clinic samples
HEC-1A, HEC-1B, Ishikawa cell lines were obtained from Shanghai HengYuan Biological Technology Co. Ltd. All of the cell lines were cultured in Dulbecco's modified Eagle medium- high glucose (DMEM-HG) supplemented with 10% fetal bovine serum and penicillin streptomycin (1:100), at 37 °C in a humidified incubator with 5% CO2. Upon reaching 90% confluence, cells were dissociated with 0.25% trypsin and split.
From October, 2010 to November, 2011, 33 patients with EC and 17 women without EC, aged from 37 to 78 years, were enrolled. 33 patients were treated at the Gynaecology & Obstetrics Hospital of Fudan University. Hysterectomy, bilateral salpingo-oophorectomy, and cytological examination of peritoneal fluid, with or without lymphadenectomy were performed. Patients without macroscopic lesion during the process of operation and with non-EC pathologic diagnosis (e.g. Cervical cancer) post operation were ruled out. For every patient, the tissues of normal endometrium and EC were collected and stored in 4% paraformaldehyde (PFA) promptly after obtained. Patients without normal endometrium area were ruled out. This study was approved by the Medicine Ethics Committee of Gynaecology & Obstetrics Hospital of Fudan University. All patient-derived tissues were obtained with oral informed consent. Because we need the patient's tissue not patient's basic information except age, such as name, vocation, married or not, and so on. In addition, the results were analyzed anonymously. So, we did not design a written informed consent table to be signed. We document this process with the following sentence " Do you agree we collect a very small part of your sectioned tissue to do some research works after you have a hysterectomy, we promise we will never let your personal basic information open, thank you for supporting our studies" when we talk with patients for the consent of surgery. All of patients agreed. This document was approved by our ethic committee.
3. Plasmid, siRNA, RT-PCR and transfection
Constructed plasmid with pEGFP-N1 (CLONTECH) vector, PCR primers as follows:
PTB F: 5'-GGAATTCATGAACAAATTACGG -3',
PTB R: 5'- TCCCCGCGGTCCTTCTCTTGTAAA -3',
Delt PTB F: 5'-GGAATTCATGGGATCATTCCGTGTC-3',
Delt PTB R: 5'-TCCCCGCGGAAGTTCAATTTCAAA -3'
All constructed plasmids were sent to be sequenced.
siRNA sequence:
NUMB siRNA: 5'- CAG CCA CUG AAC AAG CAG A-3'.
HDM2 siRNA: 5'-GCC ACA AAU CUG AUA GUA U-3'.
P53 siRNA pool: 5'- CUA CUU CCU GAA AAC AAC GTT-3',
5'- GAC UCC AGU GGU AAU CUA CTT-3',
5'- UGG UUC ACU GAA GAC CCA GTT-3'.
Quantitative RT-PCR was performed on a Roche Lightcycler 480 (Roche, UK) using SYBR Green qPCR. qRT-PCR primers were designed using the Primer premier 5, Real-time PCR primers as follows (H refers to human):
HNUMB F AGGCCAGTCGTCCACATCA
HNUMB R GGTACTTAACCGGGAAGCTACAT
HHDM2 F CAGTAGCAGTGAATCTACAGGGA
HHDM2 R CTGATCCAACCAATCACCTGAAT
P53 F ACAGCTTTGAGGTGCGTGTTT
P53 R CCCTTTCTTGCGGAGATTCTCT
HGAPDH F GGAGCGAGATCCCTCCAAAAT
HGAPDH R GGCTGTTGTCATACTTCTCATGG
Lentiviral particles were prepared by transiently transfecting HEK293T cells with lentiviral vectors together with packaging vectors (pMD2-VSVG and psPAX2) using the standard calcium-phosphate method. Retroviral packaging was made by calcium phosphate transfection of 293-GP packaging cells with the appropriate plasmids in combination with pMD2ENV coding for envelope proteins, and collected 48 h later. Infected cells were selected with 2mg/ml puromycin.
Ishikawa cells with or without Lentivirus containing appropriate plasmids were transfected using Lipofectamine® 2000 Reagent (invitrogen) and 166pmol siRNA per 60mm dish, 10 ug plasmid per 60mm dish.
4. Immunohistochemistry
To test the expression level of NUMB and HDM2 in endometrial cancer tissues and normal endometrial tissues, immunohistochemistry was employed. Tissue was fixed 4% PFA and then embedded with paraffin. A standard immunohistochemistry staining procedure was performed. Briefly, a series of deparaffinization was done with xylene and ethanol alcohol. Antigen retrieval was performed by boiling tissue slides with 0.01 mol/L citric buffer. Hydrogen peroxide was used to quench the endogenous peroxidase activity. After blocking, the sections were incubated with primary antibodies rabbit against human NUMB (Abcam ab14140) with work concentration 1:1000, overnight at 4°C. Corresponding biotin goat anti-rabbit IgG secondary antibodies (Jackson 111-065-003) with work concentration 1:3000, incubated 1hr at room temperature and peroxidase-conjugated streptavidn (Jackson 016-030-084), incubated 30min at room temperature. In final, coloration with DAB. Stained slides were counterstained with formulation hematoxylin and then dehydrated with alcohol and xylene and mounted in resinous mounting media. Tissue sections stained with isotype IgG were used as controls. All slides stained with the same antibody were processed at the same time. Stained tissue slides were reviewed under an Olympus microscope (OLYMPUS CKX41, Tokyo, Japan), and images were captured by a digital camera and recorded into a microscope-linked PC computer. Average intensity quantification was performed with image pro plus 6.0 software. Intensity was measured in five equally divided regions. Average intensity per area was determined by dividing the sum of all pixel intensities by the measured area. All compared images were acquired under identical parameters. Data represent the mean and SEM.
5. Cell apoptosis analysis by Flow Cytometry
Cell apoptosis was analyzed by flow cytometry after 3 days of transfection with plasmid-NUMB, plasmid-PTB, plasmid-delta-PTB and plasmid-GFP. The fluorescent probe annexin V was used for the quantification of apoptosis. Mix 100 μl of the cell suspension (1-5x106/ml.) and 5 μl of fluorochrome-conjugated Annexin V (eBioscience. cat. 88-8007). The samples were incubated at RT for 15 min, Wash cells in 1X Binding Buffer and resuspend in 200 μl of 1X Binding Buffer. Add 5 μl of Propidium Iodide Staining Solution (eBioscience,cat. cat. 00-6990). Analyze by flow cytometry using the PE channel for Propidium Iodide. A flow-cytometer (FACS Calibular, B-D) with 488 nm excitation was used. A 515 nm band pass filter was utilized to examine SA-FLOUS and a 560 nm band pass filter was used to test for PI.
6. CCK8 assay
After 24 of transfection, seeding 96-well plate with 1×104/well of cell density. At selected time course including 0h, 24h, 48h, 72h, 96h after cell attachment, added 10ul CCK-8 into each well containing cells and incubated 30min before measuring with 450nm wave length. Did every steps According to protocol of the CCK8 assay (Lot#EX783, Dojindo Laboratories, Tokyo, Japan). Results are presented as mean values from three replicate experiments with different passages enclosing ten wells each.
7. PCNA assay
QPCR was used to test PCNA, which steps was same as described above. Real-time PCR primers as follows: F:5'-CCTGCTGGGATATTAGCTCCA-3'; R:5'-CAGCGGTAGGTGTCGAAGC-3'.
8. Immunoprecipitation
After protein extraction, mixed 1mg protein and 2ug rabbit IgG (Santa Cruz, Cat.# H270), 40ul Protein G Plus-Agarose (Santa Cruz, cat.# sc-2002), 4℃, reversed for 30 min, and supernatant incubated at 4℃ overnight with 2ug GFP- antibody (Abmart,cat.#M20004). The beads were then pelleted and washed five times with lysis buffer. After a final centrifugation, the pellet was suspended in 2×SDS-PAGE loading buffer (SIGMA, S3401-1VL) and boiled for 5min. Then did the steps of immunoblotting (see immunoblotting).
9. Immunoblotting
Equal loading was run on a SDS polyacrylamide gel and transferred to PVDF membranes. After blocking 1hr in 5% w/v nofat milk, 1×TBS, 0.05% Tween-20, membranes were incubated with primary antibodies rabbit against human NUMB (abcam,ab14140,1ug/ml) with work concentration 1:1000, rabbit against human p53(EPITOMICS, Cat.#1026-1) with work concentration 1:10000, rabbit against human HDM2 (EPITOMICS, Cat.#S1357) with work concentration 1:3000, overnight at 4 °C. Primary antibody bound membranes were washed 3 times in TBS-T and incubated for 1 h at RT with secondary antibody conjugated with horseradish peroxidase (santa cruz,sc2004) with work concentration 1:100000. Immunosignals were visualized, in accordance to manufacturer's instructions, using the ECL(GE Healthcare,9510090). Experiment repeated three times.
10. immunofluorence
After 24h of transfection or not, Ishikawa cells were cultured on coverslips to appropriate density. Cells were fixed with 100% ethanol for 15 min and then Permeabilized with 0.2% Triton X-100 10 min. After blocking in 10% goat serum for 30 min, slides were incubated with the first antibody diluted in 10% goat serum for 1 h. After washing with PBS, slides were incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies (1:1000 dilution) for 1 h. After washing with PBS, DAPI (SIGMA, 1:1000 dilution) incubated 10min, then sealed to a cover glass plate with VECTASHIELD (Vector Laboratories, Inc. Cat.#H-1000).
11. Statistical analysis
Statistical analysis was performed using Prism 6.0. Expression of NUMB and HDM2 in patients' samples and corresponding normal tissues (IHC) were compared with t tests. The PCR, WB, PCNA and PI (proliferation Index) assays were compared with one-way ANOVA and dunnett's multiple comparisons test. The CCK8 assay was compared with two-way ANOVA and Tukey's multiple comparisons test. P<0.05 was considered to be statistically significant.
Results
1. The expression level of NUMB and HDM2 in endometrium cancer tissue and cell lines
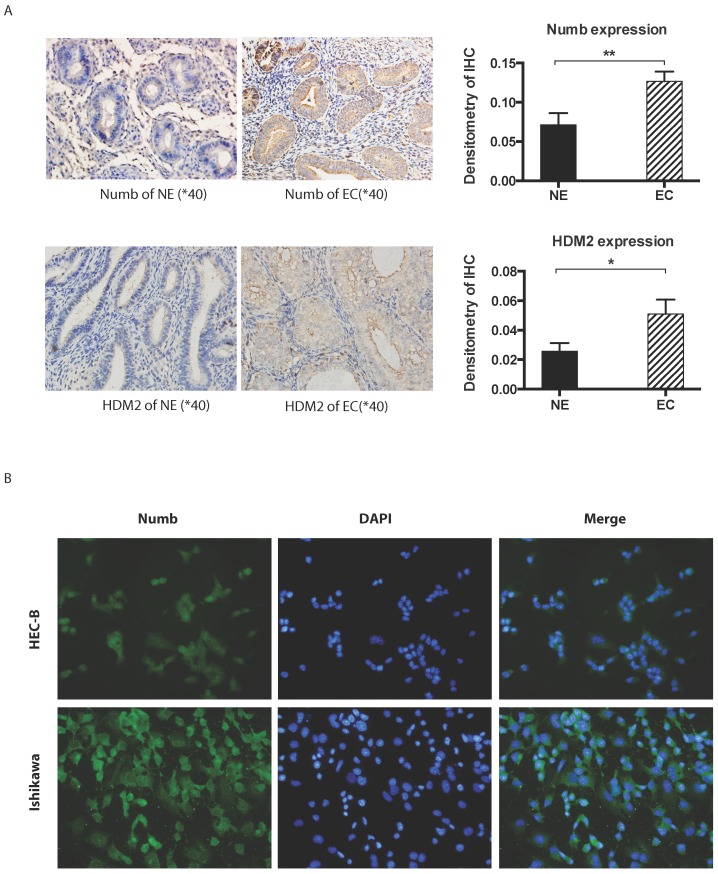
Immunohistochemistory was employed to compare the difference of NUMB and HDM2 expression level between cancer tissue and normal tissue. As shown in Fig. 1A, we found both NUMB and HDM2 in cancer tissue was higher than that of normal tissue with statistical significance (NUMB of NE and EC were 0.07134 ± 0.01476 and 0.1266 ± 0.01247 (Mean ± SEM) respectively, P value is 0.0093, HDM2 of NE and EC were 0.02578 ± 0.00546 and 0.05103 ± 0.009720 (Mean ± SEM) respectively P value is 0.0283). We used three endometrial cancer cell lines to test the location of NUMB in cell. As shown in Fig. 1B, NUMB expressed in all tested cell lines (pictures of HEC-1A has not be shown), and it could be seen that NUMB located in both cytoplasm and nucleus of Ishikawa cell and HEC-1B cell. The level of NUMB in Ishikawa appears to be higher than that of HEC-1A and 1B by visual observation.
Fig 1.
The expression of NUMB and HDM2 in endometrial cancer(EC). 1A. shows NUMB and HDM2 expression in EC with immunohistochemistry. The brown color presented the NUMB and HDM2. In the bar graphs of NUMB and HDM2 expression in normal endometrium (NE) and endometrial cancer (EC) tissue, the P Values were 0.0093 and 0.0283 respectively. 1B. is NUMB expression in endometrial cancer cell lines, it showed NUMB could locate at the cytoplasm and nucleolus of both two cell lines.
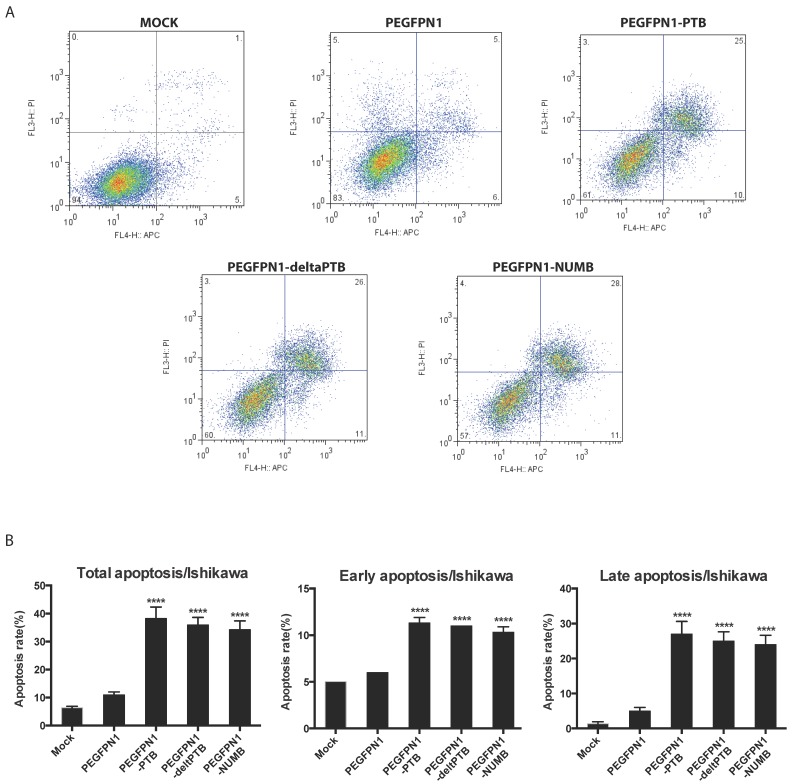
2. Overexpression of NUMB prompted cell apoptosis
We explored the function of NUMB in regulating cell biological behaviors. We constructed successfully three plasmids which were NUMB-PEGFP-N1, ΔPTB-PEGFP-N1 and PTB-PEGFP-N1. After transfecting them into Ishikawa cell, we determined the apoptosis of cell by flow cytometer. As shown in Fig. 2A and 2B, whatever overexpression of NUMB, ΔPTB or PTB, all increased the apoptosis rate including the total apoptosis rate, early apoptosis rate or late apoptosis (P<0.0001), which indicated, at a certain point, the PTB domain does not affect the function of NUMB in prompting cell apoptosis.
Fig 2.
Overexpression of NUMB prompt apoptosis of Ishikawa cell(ISIC). 2A. shows the graph of apoptosis assay. 2B. the bar graph showing the statistical significance. overexpression of NUMB, ΔPTB or PTB all increased the apoptosis rate of cell, including the total apoptosis rate, early apoptosis rate or late apoptosis (P<0.0001)
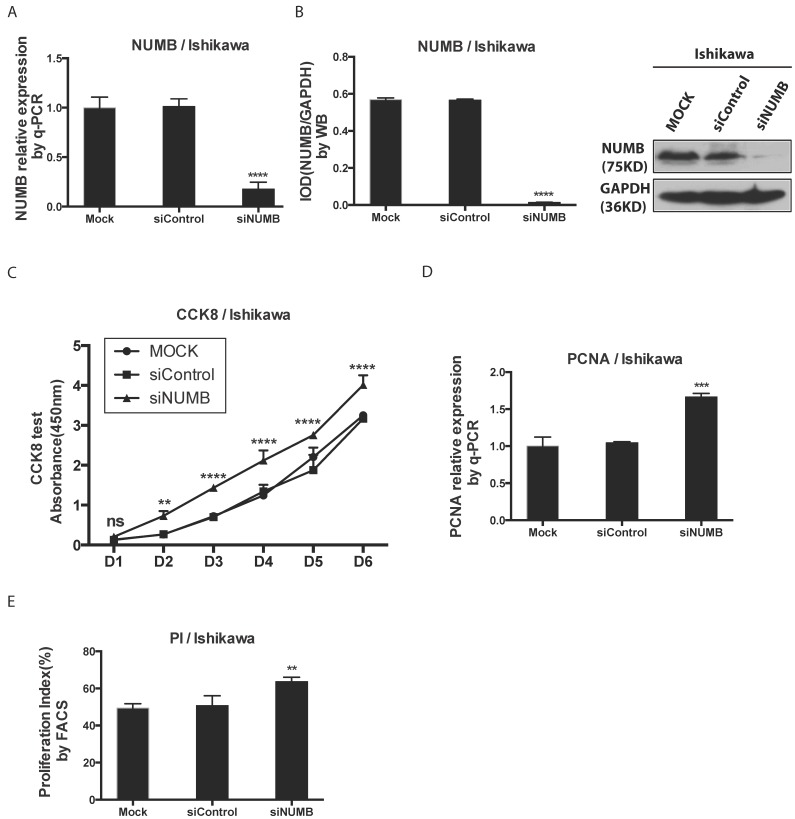
3. The knockdown of NUMB increased the proliferation of Ishikawa cell
We investigated the impact of knockdown NUMB on the proliferation of Ishikawa cell with the CCK8 test, PCNA assay as well as cell cycle by flow cytometer. First, as shown in Fig. 3A-3B, the mRNA level and protein level of NUMB were obviously decreased by siNUMB (P<0.0001), which suggested the success of siRNA targeting to NUMB. The proliferation capacity of Ishikawa cell was increased when NUMB was knocked down since day 2 (P<0.001)(Fig. 3C). As shown in Fig. 3D-3E, we also got both the proliferation index measured by flow cytometer and PCNA relative expression measured by RT-PCR. In siNUMB group they were higher than those of control group with a statistical significance (p=0.0014 for flow cytometer, P<0.0001 for RT-PCR). All of them supported the conclusion that inhibition of NUMB expression increased the proliferation capacity of Ishikawa cell.
Fig 3.
The knockdown of NUMB increased the proliferation of Ishikawa cell. 3A. shows the success of siNUMB method tested by q-PCR. 3B. shows the success of siNUMB method tested by WB. 3C. shows the increased proliferation of Ishikawa cell with knock down of NUMB tested by CCK8 assay. 3D. shows the bar graph of PCNA assay through QPCR method. 3E. shows the proliferation index by determined cell cycle with flow cytometer assay.
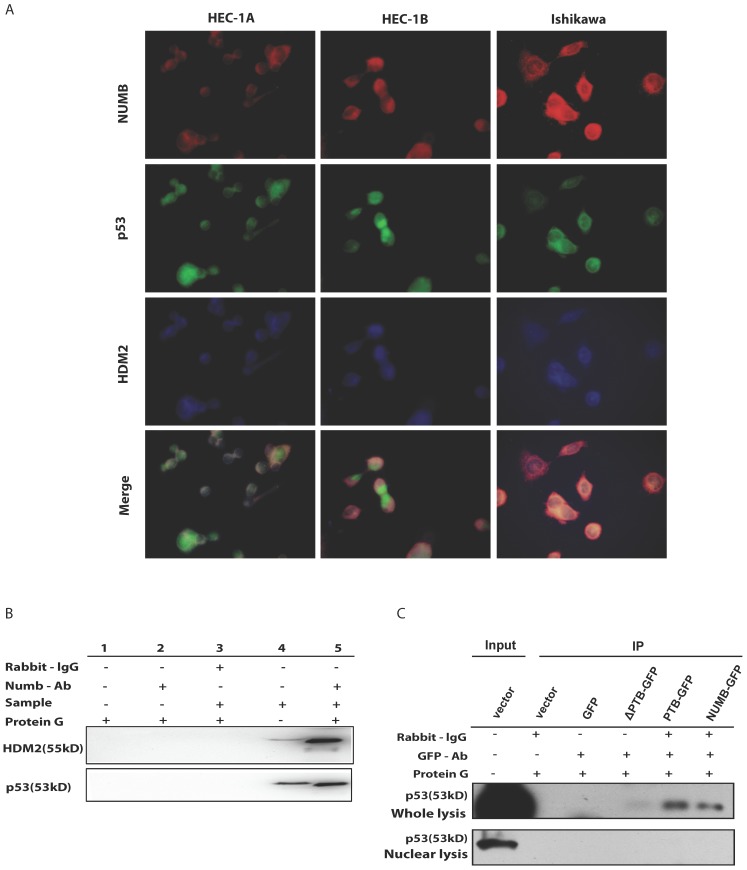
4. The formation of NUMB-HDM2-p53 tri-complex and the regulation of p53 expression and function by NUMB in Ishikawa cell
Since NUMB induced cell apoptosis in Ishikawa cell, to our best knowledge, p53 controls the process of cell apoptosis and HDM2 regulates the p53 level. In breast cancer, NUMB protect p53 against HDM2 by formation of the tri-complex NUMB-HDM2-P537, we next sought to explore whether this tri-complex exists in endometrial cancer through immunofluorescent and immunoprecipitation assay. As shown in Fig. 4A, in three endometrial cancer cell lines, we observed there really existed three color overlapping, however HEC-1A and HEC-1B cell had more two color overlapping (red and blue, i.e. NUMB and HDM2) in cytoplasm rather than three color overlapping, which nearly happened in Ishikawa cell. So, we think the formation of tri-complex of NUMB-p53-HDM2 more important to Ishikawa than HEC-1A or HEC-1B. Meanwhile, the IP result showed NUMB was able to combine p53 and HDM2 in Ishikawa cell (Fig. 4B).
Fig 4.
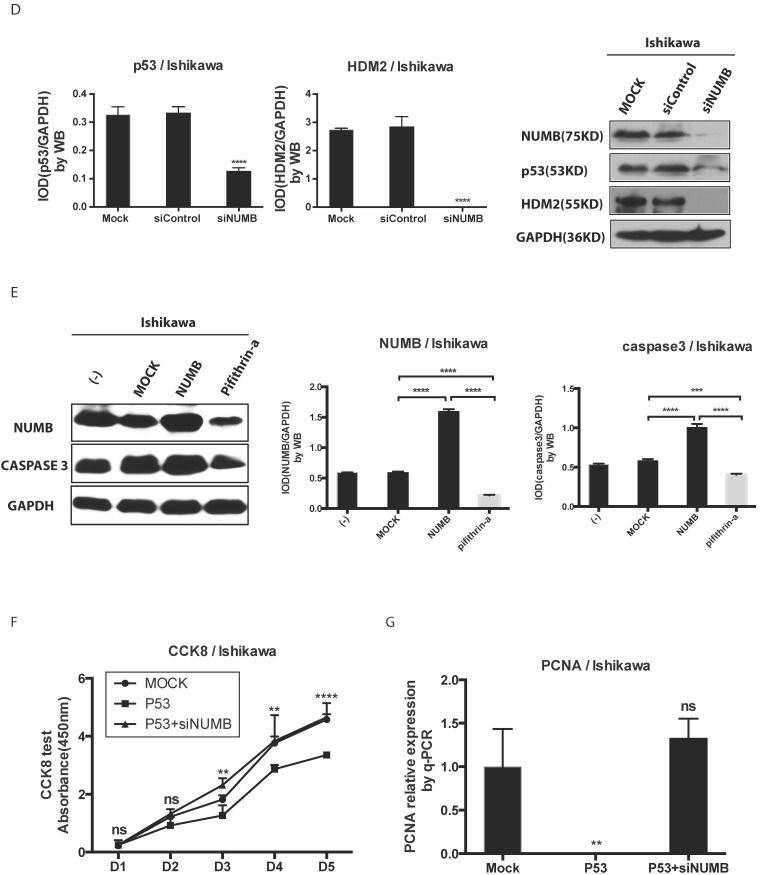
The formation of NUMB-HDM2-p53 tri-complex and the regulation of p53 expression and function by NUMB in Ishikawa cell. 4A. shows the existing NUMB-HDM2-p53 tri-complex in the endometrial cancer cell lines (HEC-1A, HEC-1B, Ishikawa) with immunofluorescent. Red color refers to the location of NUMB inside of the cell, green color refers to p53, and blue color refers to the HDM2. We observed three color overlapping in cytoplasm or nuclear or both. Meanwhile, in Ishikawa cell, the area of color overlap was the largest. 4B. shows the NUMB-HDM2-p53 tri-compound with immunoprecipitation. Endogenous NUMB-HDM2 interaction and NUMB-p53 interaction in Ishikawa cells. NUMB was immunoprecipitated from cell lysates of Ishikawa cells. HDM2 and p53 was detected by a specific antibody. Lane 1 was used as a vector control, lane 2 as a negative control, lane 3 as a non-related antibody control, lane 4 as a positive control. 4C. Immunoprecipitation was employed to determine which domain of NUMB could combine p53. using the whole cell lysis and GFP antibody to pull down the overexpressed NUMB protein. 4D. WB was employed to explore the regulation of P53 and HDM2 by NUMB at the protein level. The knock down of NUMB caused the decrease of p53 and HDM2. 4E. WB results shows overexpression of NUMB induced the increase of caspase3, a downstream target of p53 while the inhibitor of HDM2-p53 interaction induced the decrease of caspase3. 4F. CCK8 assay proved siNUMB albated the inhibition of p53 on cell proliferation. 4G. PCNA determination by q-PCR also supported siNUMB antagonized the effects of p53.
Then, we transfected whole NUMB, PTB or ΔPTB and then used immunoprecipitation to determine which domain of NUMB could combine p53. As shown in Fig. 4C, using the whole cell lysis and GFP antibody to pull down the overexpressed NUMB protein, in lane 1, 5, 6 emerged p53 bands proved again the combination of NUMB with p53 and showed the PTB domain of NUMB is the place which p53 protein could combine. But, using the lysis extracted from nucleus, there is no p53 band in lane 5 and 6. It also indicated that overexpression of NUMB could not bring NUMB into nucleus.
Deeply, we found siNUMB induced the decrease of P53 and HDM2 at the protein level (Fig. 4D). To our best knowledge, HDM2 could inhibit p53 function and increase p53 ubiquination, and p53 could induce proteins of caspase family to increase cell apoptosis. We used pifithrin-a, a p53 inhibitor, to observe the effect of NUMB on the HDM2-p53 pathway. We found overexpression of NUMB increased caspase 3 level while, pifithrin-a decreased caspase3 level, which implied NUMB might induce cell apoptosis through regulating p53 and downstream proteins of p53 (Fig. 4E). In addition, overexpression of p53 inhibited the cell proliferation, however siNUMB alleviated the inhibition of p53 on cell proliferation through CCK8 assay and PCNA test (Fig. 4F-4G).
5. The regulation of NUMB location by HDM2 in Ishikawa cell
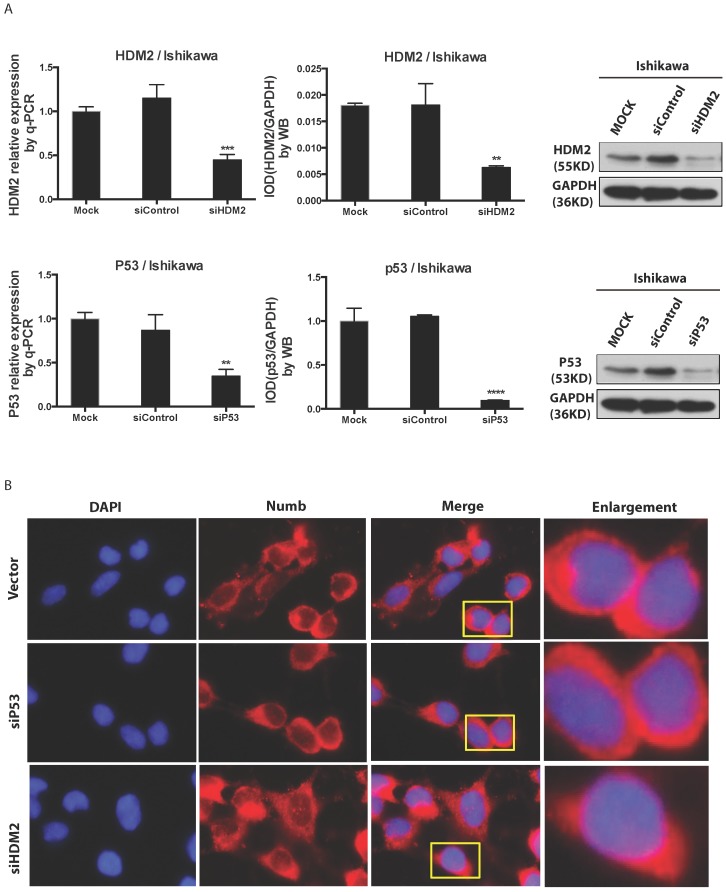
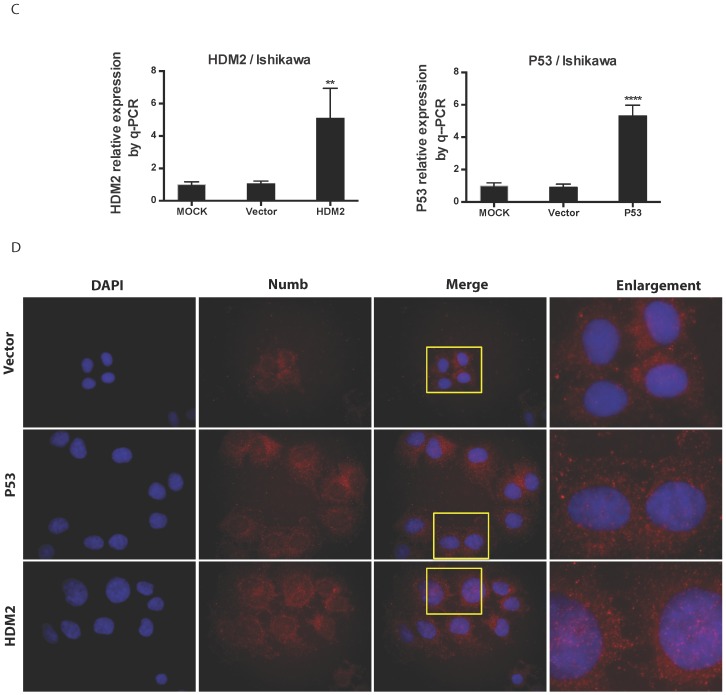
We found NUMB transferred into nucleus in our previous work, in this study we wanted to explore whether or not p53 and HDM2 could change the location of NUMB in cell compartment? We used siRNA to knockdown HDM2 and P53 separately. The data of RT-PCR and Western blot shown the success of siHDM2 and siP53 (Fig. 5A). We didn't observe a obvious change in location of NUMB (Fig. 5A). However, when exogenous HDM2 and P53 were transfected into cell, high HDM2 and p53 level induced the increase of NUMB in nuclear, especially in high HDM2 group (Fig. 5C-5D).
Fig 5.
The regulation of NUMB location by HDM2 in Ishikawa cell. 5A. shows the success of siNUMB method tested by q-PCR and WB. 5B. shows no obvious change of NUMB location when siHDM2 or siP53 compared with vector group through immunoflurorescence staining. “Enlargement” pictures refer to the enlargement of yellow frame in “merge” pictures. 5C. shows the success of exogenous HDM2 and p53 transfection tested by q-PCR. 5D. shows the increase of NUMB nuclear translocation upon siHDM2 compared with vector group through immunoflurorescence staining.
Discussion
Accumulating evidence suggests a potential tumor suppressor role for NUMB due to the stabilization of p53 and inhibition of Notch signaling 7, 8, 9, 10. This tumor suppressive function of Numb had been proven in many cancers, such as breast cancer, non-small cell lung cancer, pancreatic cancer, salivary gland carcinomas and so on. Our findings confirmed that endometrial cancer tissue had higher NUMB level than that of normal endometrium, which is consistent with our previous work4, although it is opposite to results from other studies that reported the low expression of NUMB in cancer7, 11, 12. It seems to be a paradox that endometrial cancer tissue has higher NUMB level than that of normal endometrium tissue when we believe NUMB is a tumor suppressor in EC. However, it should be noted that the role of Numb might not be explained thoroughly with its expression level, the function of Numb impacted by a lot of factors. Therefore, it is still controversial about the role of NUMB so far. For example, Euskirchen et al tried to prove the role of the potential tumor suppressor NUMB in malignant gliomas, but they could not detect loss of NUMB neither at transcriptional nor protein level, even no significant antiproliferative effect of NUMB overexpression in the adherent U87 glioma cell line2. We supposed several possible reasons should be considered to explain the discrepancy of manifestations between in vitro and in vivo.
First, NUMB protein expressed highly in cancer tissue, which is compensatory behavior, could not contribute to prevent the tumorigenesis. Wang et al proposed Numb plays a key role in cell epithelial polarity and is a critical and dynamic link between the TJ and the AJ under physiological and pathological conditions through the formation of Numb-aPKC-Par6 complex and interaction with E-cad and Par313. Weiwei Sheng, et al presented the differential expression of NUMB between well-differentiated pancreatic cancer (PC) and paired normal tissues. The decrease of cell epithelial polarity and adhesion in well-differentiated PC may stimulate NUMB expression for maintaining epithelial integrity. So, they found NUMB expression was slightly increased in PC compared to the paired adjacent normal tissues14. In our previous work, all samples of endometrial cancer were from the patients with early stage (stage I)4, and in this study, most of 33 samples also were from early stage, which might explain partly the divergence. Being accordance with PC The decrease of cell epithelial polarity and adhesion in well-differentiated EC may stimulate NUMB expression for maintaining epithelial integrity while extensive loss of NUMB may be a feature of cancers of epithelial origin.
Second, NUMB was stimulated as an adaptive response to HDM2 overexpression to balance the effects of HDM2, which is p53 can be inhibited by HDM2 and NUMB can protect p53 from the inhibition of HDM215, 16. It was proved the high expression of HDM2 in cancer tissue in this study. So, NUMB could increase to prevent the degradation of p53 by HDM2.
In addition, various NUMB isoforms have different effects on the proliferation or differentiation. NUMB Isoform with PRRL (long proline-rich region) promotes proliferation while NUMB Isoform with PRRS promotes differentiation. Chen H, et al found in cervical cancer there was a significantly higher expression of mRNA for the NUMB-PRRL isoform but not for the NUMB-PRRS isoform17. However, in our study, our antibody does not distinguish between the different isoforms. We also think this discrepancy might be related to cancer type which indicated the different mechanism of tumor occurrence. Even in same type of cancer, we also observed this discrepancy among different cell lines, for example, Ishikawa cell had higher NUMB level than HEC-1A and HEC-1B.
Based on what we found in our in vitro study, we observed the overexpression of exogenous Numb induced the apoptosis via flow cytometry assay. A recent literature showed the NUMB promotes apoptosis in a p53-dependent manner, the apoptotic function is abolished when Numb is methylated by Set8 or the Lys methylation sites in Numb are mutated18. At the mean time, we found the knockdown of Numb inhibited the proliferation of tumor cells via CKK8, PCNA and PI (Proliferation Index) assays. Kang et al also proved the overexpression of NUMB in NCI-H2452 cells significantly inhibited proliferation and promoted apoptosis, even enhanced sensitivity to sisplatin19. Sima et al also provided overexpression of Numb could induce G0/G1 phase arrest and inhibit cell proliferation, migration and invasion20. These above data supported the fact Numb should act as a tumor suppressor, which is consistent with the conclusion that Numb acts as a tumor suppressor presented by most of literatures about other cancers, like breast cancer, pancreatic cancer, lung cancer and so on.
NUMB can combine with HDM2 and p53 in breast cancer7, we also proved NUMB combined with HDM2 and p53 as well as PTB domain is the main site combining with p53 in Ishikawa cell. Data of immunohistochemistry and immunofluorescent from endometrial cancer tissue and Ishikawa cell showed the expression of NUMB both in cytoplasm and nucleus, which suggested NUMB could enter into nucleus. Overexpression of NUMB didn't contribute to its nuclear translocation.
To our knowledge, HDM2 could bring some protein into nucleus21 and HDM2 really could regulate the subcellular localization of NUMB through a direct interaction with PTB of NUMB in other cancer cell line9, The author did not know why the transmitting function of MDM2 just occurred in some cells not all of them. In our study, as shown in immunofluorescent data, we found NUMB entered into nucleus increased upon overexpression of HDM2 and p53 while it didn't happen upon siHDM2 and siP53. We thought the nucleus translocation of NUMB upon overexpression of HDM2 could be a response to rescue the inhibition of high HDM2 on p53.
Besides what we mentioned above, Numb gene has been proven it is an interactive partner of TRPV6 and PTEN is a novel regulator of this complex in prostate cancer cells22. Numb also interacts with microRNA. miR-146a directly targets the 3'UTR of NUMB and a region within the NUMB coding sequence when suppressing NUMB expression, oncogenic enhancement modulated by miR-146a expression is attenuated by exogenous NUMB expression in oral squamous cell Carcinoma23. Although it is controversial that NUMB is a tumor suppressor, we believe Numb is a novel and promising target in cancer therapy. In this study, we believe NUMB played a role of anti-oncogene in endometrial cancer. HDM2 contributed to the location of NUMB. So, NUMB is closed to the occurrence of EC and it will be a new promising target of EC treatment in the future with understanding of NUMB step by step.
Acknowledgments
This work was supported by National Natural Science Foundation of China (81001149) and Shanghai Outstanding Youth Training Plan of China, (XYQ2011062).
References
- 1.Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Experimental cell research. 2010;316:900–6. doi: 10.1016/j.yexcr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 2.Euskirchen P, Skaftnesmo KO, Huszthy PC, Brekkå N, Bjerkvig R, Jacobs AH. et al. NUMB does not impair growth and differentiation status of experimental gliomas. Exp Cell Res. 2011;317:2864–73. doi: 10.1016/j.yexcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Karaczyn A, Bani-Yaghoub M, Tremblay R, Kubu C, Cowling R, Adams TL. et al. Two novel human NUMB isoforms provide a potential link between development and cancer. Neural development. 2010;5:31. doi: 10.1186/1749-8104-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C, Cui T, Feng W, Li H, Hu L. Role of Numb expression and nuclear translocation in endometrial cancer. Oncology letters. 2015;9:1531–6. doi: 10.3892/ol.2015.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan B, Omar FM, Das K, Ng WH, Lim C, Shiuan K. et al. Characterization of Numb expression in astrocytomas. Neuropathology. 2008;28:479–84. doi: 10.1111/j.1440-1789.2008.00907.x. [DOI] [PubMed] [Google Scholar]
- 6.Dho SE, French MB, Woods SA, McGlade CJ. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem. 1999:33097–104. doi: 10.1074/jbc.274.46.33097. [DOI] [PubMed] [Google Scholar]
- 7.Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G. et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- 8.Adams SJ, Aydin IT, Celebi JT. GAB2--a scaffolding protein in cancer. Molecular cancer research: MCR. 2012;10:1265–70. doi: 10.1158/1541-7786.MCR-12-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juven-Gershon T, Shifman O, Unger T, Elkeles A, Haupt Y, Oren M. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Molecular and cellular biology. 1998;18:3974–82. doi: 10.1128/mcb.18.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGill MA, Dho SE, Weinmaster G, McGlade CJ. Numb regulates post-endocytic trafficking and degradation of Notch1. The Journal of biological chemistry. 2009;284:26427–38. doi: 10.1074/jbc.M109.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rennstam K, McMichael N, Berglund P, Honeth G, Hegardt C, Ryden L. et al. Numb protein expression correlates with a basal-like phenotype and cancer stem cell markers in primary breast cancer. Breast Cancer Res Tr. 2010;122:315–24. doi: 10.1007/s10549-009-0568-x. [DOI] [PubMed] [Google Scholar]
- 12.Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. The Journal of cell biology. 2004;167:215–21. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Z, Sandiford S, Wu C, Li SS. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 2009;28:2360–73. doi: 10.1038/emboj.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheng W, Dong M, Zhou J, Li X, Liu Q, Dong Q. et al. Cooperation among Numb, MDM2 and p53 in the development and progression of pancreatic cancer. Cell and tissue research. 2013;354:521–32. doi: 10.1007/s00441-013-1679-6. [DOI] [PubMed] [Google Scholar]
- 15.Boyd MT, Vlatkovic N, Rubbi CP. The nucleolus directly regulates p53 export and degradation. The Journal of cell biology. 2011;194:689–703. doi: 10.1083/jcb.201105143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. Journal of biomedical research. 2013;27:254–71. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Chen X, Ye F, Lu W, Xie X. Symmetric division and expression of its regulatory gene Numb in human cervical squamous carcinoma cells. Pathobiology. 2009;76:149–54. doi: 10.1159/000209393. [DOI] [PubMed] [Google Scholar]
- 18.Dhami GK, Liu H, Galka M, Voss C, Wei R, Muranko K. et al. Dynamic methylation of Numb by Set8 regulates its binding to p53 and apoptosis. Molecular cell. 2013;50:565–76. doi: 10.1016/j.molcel.2013.04.028. [DOI] [PubMed] [Google Scholar]
- 19.Kang Y, Ding M, Tian G, Guo H, Wan Y, Yao Z. et al. Overexpression of Numb suppresses tumor cell growth and enhances sensitivity to cisplatin in epithelioid malignant pleural mesothelioma. Oncology reports. 2013;30:313–9. doi: 10.3892/or.2013.2429. [DOI] [PubMed] [Google Scholar]
- 20.Sima J, Zhang B, Yu Y, Sima X, Mao Y. Overexpression of Numb suppresses growth, migration, and invasion of human clear cell renal cell carcinoma cells. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2885–92. doi: 10.1007/s13277-014-2918-5. [DOI] [PubMed] [Google Scholar]
- 21.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cellular and molecular life sciences: CMLS. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SY, Hong C, Wie J, Kim E, Kim BJ, Ha K. et al. Reciprocal positive regulation between TRPV6 and NUMB in PTEN-deficient prostate cancer cells. Biochemical and biophysical research communications. 2014;447:192–6. doi: 10.1016/j.bbrc.2014.03.123. [DOI] [PubMed] [Google Scholar]
- 23.Hung PS, Liu CJ, Chou CS, Kao SY, Yang CC, Chang KW. et al. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PloS one. 2013;8:e79926. doi: 10.1371/journal.pone.0079926. [DOI] [PMC free article] [PubMed] [Google Scholar]