Abstract
The World Health Organization (WHO) has recognised all Cronobacter species as human pathogens. Among premature neonates and immunocompromised infants, these infections can be life-threatening, with clinical presentations of septicaemia, meningitis and necrotising enterocolitis. The neurological sequelae can be permanent and the mortality rate as high as 40–80 %. Despite the highlighted issues of neonatal infections, the majority of Cronobacter infections are in the elderly population suffering from serious underlying disease or malignancy and include wound and urinary tract infections, osteomyelitis, bacteraemia and septicaemia. However, no age profiling studies have speciated or genotyped the Cronobacter isolates. A clinical collection of 51 Cronobacter strains from two hospitals were speciated and genotyped using 7-loci multilocus sequence typing (MLST), rpoB gene sequence analysis, O-antigen typing and pulsed-field gel electrophoresis (PFGE). The isolates were predominated by C. sakazakii sequence type 4 (63 %, 32/51) and C. malonaticus sequence type 7 (33 %, 17/51). These had been isolated from throat and sputum samples of all age groups, as well as recal and faecal swabs. There was no apparent relatedness between the age of the patient and the Cronobacter species isolated. Despite the high clonality of Cronobacter, PFGE profiles differentiated strains across the sequence types into 15 pulsotypes. There was almost complete agreement between O-antigen typing and rpoB gene sequence analysis and MLST profiling. This study shows the value of applying MLST to bacterial population studies with strains from two patient cohorts, combined with PFGE for further discrimination of strains.
Introduction
The Cronobacter genus belongs to the family Enterobacteriaceae and consists of seven species: C. sakazakii, C. malonaticus, C. muytjensii, C. turicensis, C. dublinensis, C. universalis and C. condimenti [1, 2]. In 2002, the International Commission on Microbiological Specifications for Foods (ICMSF) classified Cronobacter as pathogenic organisms to a restricted population, endangering their lives and causing serious long-term consequences [3]. The World Health Organization (WHO) has recognised all Cronobacter species as microorganisms pathogenic for human beings [4]. Among premature neonates and immunocompromised infants, these infections can be life-threatening, with clinical presentations of septicaemia, meningitis and necrotising enterocolitis. The neurological sequelae can be permanent and the mortality rate can be as high as 40–80 % [5]. Despite the highlighted issues of neonatal infections, the majority of Cronobacter infections are in the adult population, especially those suffering from serious underlying disease or malignancy [6]. Cronobacter species are also part of the normal flora carriage [7–9].
The first reported age-profiled data was for 819 Cronobacter bacteraemia cases in England and Wales between 1992 and 2007 [4]. The majority (91 %) of bacteraemia cases were patients >15 years in age. Holý et al. reported the age profile of Cronobacter carriage from a survey of >45,000 patients from two hospitals sampled from 2005 to 2011 [9]. The organism was isolated from every age group, with a higher frequency in children less than 14 years of age. The majority of Cronobacter spp. isolates were from throat swabs, followed by urine, tracheal aspirates, bronchoalveolar lavage, cannulae and sputum samples. Patrick et al. also reported an age profile for Cronobacter infections from an earlier period (2003–2009), which confirmed its prominence in the adult population, especially in urinary tract infections (UTIs) [6]. However, none of these age profiling studies speciated or genotyped the Cronobacter isolates. To date, over 1000 Cronobacter strains have been genotyped according to a 7-loci multilocus sequence typing (MLST) scheme [10]. This genotyping has revealed a prevalence of C. sakazakii clonal complex 4 with neonatal meningitis cases and C. malonaticus clonal complex 7 with adult infections [10–12]. Whole genome phylogenetic analysis (164 genomes) has confirmed the use of fusA for Cronobacter speciation [10, 13].
This study aimed to address this lack of knowledge using the collection of 51 clinical Cronobacter strains, which included those from the study by Holý et al. [9]. These strains were speciated and genotyped using 7-loci MLST, rpoB gene sequence analysis, O-antigen typing and pulsed-field gel electrophoresis (PFGE).
Materials and methods
Bacterial strains and cultivation
Fifty-one clinical Cronobacter strains were used in this study. The strains had been collected during a survey of Cronobacter carriage by patients from two hospitals, during a 6-year period from May 2007 to August 2013. This includes strains isolated in the previous study by Holý et al. [9]. Patient information such as age, sex, clinical presentation, isolated site and date of isolation are given in Table 1. Bacterial strains were routinely cultivated on tryptone soya agar (Fluka, UK) at 37 °C overnight.
Table 1.
Source of Cronobacter strains used in this study
| Strain number | Hospital | Department | Patient age (years) | Patient sex | Isolation date | Isolation site |
|---|---|---|---|---|---|---|
| 1830 | Olomouc | Paediatrics | <1 | Male | 09/05/2007 | Throat swab |
| 1829 | Olomouc | Paediatrics | 1 | Male | 04/06/2007 | Throat swab |
| 1828 | Olomouc | Paediatrics | 2 | Male | 12/10/2007 | Nose swab |
| 1831 | Olomouc | Paediatrics | 3 | Male | 06/06/2007 | Throat swab |
| 1832 | Olomouc | Paediatrics | 3 | Female | 27/03/2009 | Throat swab |
| 1999 | Olomouc | Paediatrics | 3 | Male | 30/01/2013 | Throat swab |
| 2020 | Olomouc | Paediatrics | 5 | Female | 26/05/2013 | Stool |
| 1835 | Olomouc | Paediatrics | 6 | Male | 30/03/2012 | Throat swab |
| 2015 | Olomouc | Paediatrics | 7 | Female | 16/08/2013 | Throat swab |
| 2014 | Olomouc | Paediatrics | 8 | Male | 08/04/2013 | Throat swab |
| 1917 | Olomouc | Paediatrics | 15 | Male | 28/10/2012 | Throat swab |
| 1834 | Olomouc | Paediatrics | 16 | Male | 31/05/2010 | Throat swab |
| 2004 | Olomouc | Paediatrics | 17 | Female | 02/03/2013 | Throat swab |
| 1827 | Olomouc | Internal Medicine III | 76 | Female | 09/10/2007 | Cannula |
| 1833 | Olomouc | CMPa | 5 | Male | 11/01/2010 | Stool |
| 1838 | Olomouc | AICUb | 63 | Female | 10/04/2012 | Sputum |
| 1998 | Prostějov | Internal Medicine (A) | 49 | Female | 22/01/2013 | Sputum |
| 2008 | Prostějov | Internal Medicine (A) | 68 | Male | 12/03/2013 | Sputum |
| 2011 | Prostějov | Internal Medicine (A) | 68 | Male | 31/03/2013 | USCd |
| 2006 | Prostějov | Internal Medicine (A) | 70 | Female | 28/02/2013 | Sputum |
| 2007 | Prostějov | Internal Medicine (A) | 70 | Female | 06/03/2013 | Sputum |
| 2022 | Prostějov | Internal Medicine (A) | 70 | Female | 06/03/2013 | Sputum |
| 1842 | Prostějov | Internal Medicine (A) | 72 | Female | 27/06/2012 | Sputum |
| 2005 | Prostějov | Internal Medicine (A) | 73 | Female | 24/02/2013 | Sputum |
| 2021 | Prostějov | Internal Medicine (A) | 76 | Female | 07/04/2013 | Sputum |
| 1841 | Prostějov | Internal Medicine (A) | 79 | Female | 18/06/2012 | Sputum |
| 2003 | Prostějov | Internal Medicine (A) | 83 | Male | 20/02/2013 | Sputum |
| 1915 | Prostějov | Internal Medicine (A) | 84 | Female | 18/10/2012 | Sputum |
| 1996 | Prostějov | Internal Medicine (A) | 84 | Female | 14/01/2013 | Sputum |
| 2010 | Prostějov- | Internal Medicine (A) | 84 | Female | 12/03/2013 | Throat swab |
| 2019 | Prostějov | Internal Medicine (A) | 87 | Male | 10/05/2013 | Sputum |
| 2001 | Prostějov | Internal Medicine (B) | 68 | Male | 29/01/2013 | SOCe |
| 2000 | Prostějov | Internal Medicine (B) | 71 | Male | 03/02/2013 | Rectal Swab |
| 2002 | Prostějov | Internal Medicine (B) | 77 | Male | 19/02/2013 | Sputum |
| 1916 | Prostějov | Internal Medicine (B) | 84 | Male | 06/11/2012 | Sputum |
| 2013 | Prostějov | Internal Medicine (B) | 91 | Female | 04/04/2013 | Sputum |
| 2012 | Prostějov | Internal Medicine (C) | 70 | Male | 04/04/2013 | Sputum |
| 2009 | Prostějov | Internal Medicine (C) | 77 | Female | 16/03/2013 | Tongue swab |
| 1903 | Prostějov | Internal Medicine—ICU | 59 | Male | 24/08/2012 | Sputum |
| 1902 | Prostějov | Internal Medicine—ICU | 69 | Male | 21/08/2012 | Sputum |
| 1901 | Prostějov | Internal Medicine—ICU | 82 | Male | 15/08/2012 | Sputum |
| 1997 | Prostějov | ICUc | 65 | Male | 21/01/2013 | Sputum |
| 1839 | Prostějov | ICU | 73 | Female | 12/06/2012 | SPEGf |
| 1840 | Prostějov | ICU | 80 | Female | 19/06/2012 | Sputum |
| 1836 | Prostějov | Surgery | 63 | Male | 23/05/2012 | Wound swab |
| 1837 | Prostějov | Surgery | 85 | Female | 25/05/2012 | Wound swab |
| 1914 | Prostějov | Infectious Diseases | 69 | Male | 02/10/2012 | Sputum |
| 2018 | Prostějov | Infectious Diseases | 72 | Male | 05/05/2013 | Sputum |
| 2016 | Prostějov | AICU | 27 | Male | 18/04/2013 | Sputum |
| 2017 | Prostějov | AICU | 27 | Male | 22/04/2013 | Sputum |
| 1995 | Prostějov | Outpatient | 50 | Male | 10/01/2013 | Sputum |
a CMP Clinical and Molecular Pathology
b AICU Anaesthesiology and Intensive Care Unit
c ICU Intensive Care Unit
d USC Urine suction catheter
e SOC Swab of the oral cavity
f SPEG Smear from area of percutaneous endoscopic gastrostomy
Phenotyping
Cronobacter isolates were phenotyped using the ID 32E kit (bioMérieux), according to the manufacturer’s instructions. The resultant phenotypic profiles were compared to the bioMérieux online database at https://apiweb.biomerieux.com.
PFGE of Cronobacter isolates
PFGE analysis of Cronobacter isolates was as previously described by Caubilla-Barron et al. [14] using the two restriction enzymes Xbal and Spel (Promega, UK). The bands were separated using a CHEF-DR II System (Bio-Rad, Belgium) at 14 °C, 6 V for 20 h with initial and final switch of 5 and 50 s, respectively. The DNA band profiles were analysed using BioNumerics software version 7.1 (Applied Maths, Belgium). The banding patterns obtained from the PFGE for both XbaI and SpeI were combined within the Bionumerics software and analysed by the unweighted pair-group method using arithmetic averages (UPGMA). Isolates with band similarity values of less than 95 % were considered to be non-clonal [15].
Molecular serotyping of Cronobacter O-antigens
Cronobacter serotypes were determined using the multiplex polymerase chain reaction (PCR) assay as described by Jarvis et al. and Sun et al. [16, 17]. The allocated serotypes were uploaded to the Cronobacter PubMLST database for open access; http://PubMLST.org/cronobacter/.
DNA extraction
DNA was extracted from the target strains using the GenElute™ kit (Sigma, UK), according to the manufacturer’s instructions. The DNA concentration was confirmed using a NanoDrop® ND-2000 UV–vis spectrometer (Thermo Scientific, UK), and the DNA was stored at −20 °C for 6 months.
rpoB allele sequence analysis
rpoB allele profiling was performed as described by Brady et al. [18]. PCR products were visualised on a 1 % agarose gel stained with SYBR Safe. The PCR product (637 bp) was sequenced and aligned with additional sequences from the Cronobacter PubMLST database in MEGA (Molecular Evolutionary Genetics Analysis) software version 5.2 [19] using the ClustalW algorithm. rpoB alleles were allocated numbered profiles according to the PubMLST database and were uploaded for open access.
MLST
MLST was performed as previously described by Baldwin et al. [20] and as given on the Cronobacter PubMLST open access database (http://www.pubmlst.org/cronobacter/). The seven housekeeping genes amplified were ATP synthase beta chain (atpD), elongation factor G (fusA), glutaminyl-tRNA synthetase (glnS), glutamate synthase large subunit (gltB), DNA gyrase subunit B (gyrB), translation initiation factor IF-2 (infB) and phosphoenolpyruvate synthase (ppsA). For multilocus sequence analysis (MLSA), concatenated sequences (3036 bp total length) were aligned in MEGA version 5.2 using the ClustalW algorithm.
Results
A total of 51 Cronobacter strains were characterised by several phenotyping and genotyping methods. Presumptive identification using ID 32E phenotyping identified 49 isolates as Enterobacter sakazakii, one strain (1838) as Pantoea spp. and the remaining strain (1841) as E. cloacae. Since the bioMérieux ID 32E online database does not recognise the Cronobacter genus, the strains could not be further identified using this method.
Using the fusA sequence analysis and comparison with the Cronobacter PubMLST database identified the 51 strains as primarily C. sakazakii (33/51), followed by C. malonaticus (17/51) and one C. muytjensii strain. The strains were then further genotyped using the 7-loci MLST scheme. This supported the species identification-based fusA sequence analysis, and further subtyped the isolates (Table 2). The C. sakazakii strains were from two sequence types; ST4 (32/51, 63 %) and ST64 (1/51, 2 %). All the C. malonaticus strains were ST7 (17/51, 33 %) and the single C. muytjensii isolate was ST28 (2 %).
Table 2.
Number of isolated Cronobacter strains from various hospital departments
| Hospital | Department | Number of Cronobacter strains isolated |
|---|---|---|
| Olomouc | Paediatrics | 13 |
| Internal Medicine | 1 | |
| AICUa | 1 | |
| Pathology | 1 | |
| Prostějov | Internal Medicine | 22 |
| Internal Medicine—ICUb | 3 | |
| Surgery | 2 | |
| ICU | 3 | |
| Infectious Diseases | 2 | |
| AICU | 2 | |
| Outpatient | 1 | |
| Total | 51 |
a AICU Anaesthesiology and Intensive Care Unit
b ICU Intensive Care Unit
The identification of strains using rpoB sequence analysis [18] and comparison with rpoB sequences in the Cronobacter PubMLST database agreed with species designation using fusA allele sequence analysis (Table 2). There were four different rpoB profiles, 1, 18, 35 and 36, which correlated with their 7-loci sequences types. All C. sakazakii ST4 and ST64 strains were rpoB profiles 1 and 35, respectively. The C. malonaticus ST7 strains were rpoB profile 18 and C. muytjensii ST28 was rpoB profile 36. See Table 2 for more information.
Comparison with serotyping profiling showed a strong correlation between some sequence types and serotypes. O-serotype C. sakazakii O:2 corresponded with C. sakazakii ST4. The association was not exclusive however, as C. sakazakii ST64 (strain 1995) was also serotype C. sakazakii O:2. In addition, the serotype of all (n = 17) C. malonaticus ST7 strains corresponded with the two designated serotypes C. malonaticus O:2 and C. sakazakii O:6 according to the schemes of Jarvis et al. and Sun et al., respectively [16, 17]. Based on fusA speciation, C. malonaticus O:2 was given as the serotype for these strains (Table 3). No serotype could be determined for the C. muytjensii strain as no PCR products were obtained with either PCR serotyping scheme.
Table 3.
Speciation and genotyping of Cronobacter spp. from two hospitals
| Strain | Hospital | Species | Pulsotype | rpoB allele | fusA allele | Serotype | Sequence type |
|---|---|---|---|---|---|---|---|
| 2021 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2022 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1901 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1915 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1996 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1837 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1841 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1842 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2003 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2005 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2007 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2010 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2016 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2019 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 2017 | Prostějov | C. sakazakii | 12 | 1 | 1 | CS O:2 | ST4 |
| 1916 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 1840 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 2000 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 2001 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 2002 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 2009 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 2011 | Prostějov | C. sakazakii | 7 | 1 | 1 | CS O:2 | ST4 |
| 1917 | Olomouc | C. malonaticus | 4 | 18 | 7 | CMal O:2 | ST7 |
| 1999 | Olomouc | C. malonaticus | 4 | 18 | 7 | CMal O:2 | ST7 |
| 2004 | Olomouc | C. malonaticus | 4 | 18 | 7 | CMal O:2 | ST7 |
| 2015 | Olomouc | C. malonaticus | 4 | 18 | 7 | CMal O:2 | ST7 |
| 2014 | Olomouc | C. malonaticus | 4 | 18 | 7 | CMal O:2 | ST7 |
| 2020 | Olomouc | C. malonaticus | 4 | 18 | 7 | CMal O:2 | ST7 |
| 1828 | Olomouc | C. malonaticus | 5 | 18 | 7 | CMal O:2 | ST7 |
| 1829 | Olomouc | C. malonaticus | 5 | 18 | 7 | CMal O:2 | ST7 |
| 1830 | Olomouc | C. malonaticus | 5 | 18 | 7 | CMal O:2 | ST7 |
| 1831 | Olomouc | C. malonaticus | 5 | 18 | 7 | CMal O:2 | ST7 |
| 1832 | Olomouc | C. malonaticus | 5 | 18 | 7 | CMal O:2 | ST7 |
| 1903 | Prostějov | C. sakazakii | 10 | 1 | 1 | CS O:2 | ST4 |
| 1998 | Prostějov | C. sakazakii | 10 | 1 | 1 | CS O:2 | ST4 |
| 2006 | Prostějov | C. sakazakii | 10 | 1 | 1 | CS O:2 | ST4 |
| 2008 | Prostějov | C. sakazakii | 10 | 1 | 1 | CS O:2 | ST4 |
| 1833 | Olomouc | C. malonaticus | 3 | 18 | 7 | CMal O:2 | ST7 |
| 1834 | Olomouc | C. malonaticus | 3 | 18 | 7 | CMal O:2 | ST7 |
| 1835 | Olomouc | C. malonaticus | 3 | 18 | 7 | CMal O:2 | ST7 |
| 1902 | Prostějov | C. sakazakii | 11 | 1 | 1 | CS O:2 | ST4 |
| 1997 | Prostějov | C. sakazakii | 11 | 1 | 1 | CS O:2 | ST4 |
| 1914 | Prostějov | C. malonaticus | 1 | 18 | 7 | CMal O:2 | ST7 |
| 2018 | Prostějov | C. malonaticus | 1 | 18 | 7 | CMal O:2 | ST7 |
| 1827 | Olomouc | C. malonaticus | 2 | 18 | 7 | CMal O:2 | ST7 |
| 2013 | Prostějov | C. sakazakii | 8 | 1 | 1 | CS O:2 | ST4 |
| 2012 | Prostějov | C. sakazakii | 9 | 1 | 1 | CS O:2 | ST4 |
| 1839 | Prostějov | C. sakazakii | 13 | 1 | 1 | CS O:2 | ST4 |
| 1836 | Prostějov | C. sakazakii | 14 | 1 | 1 | CS O:2 | ST4 |
| 1995 | Prostějov | C. sakazakii | 15 | 35 | 8 | CS O:2 | ST64 |
| 1838 | Olomouc | C. muytjensii | 6 | 36 | 24 | No PCR product | ST28 |
PFGE was used to ascertain whether the strains in each sequence type (i.e. C. sakazakii ST4 and C. malonaticus ST7) could be further distinguished and whether this could be used to profile the strains from the two hospitals. Using the restriction enzyme XbaI, C. sakazakii strains gave 12 to 17 DNA fragments per strain, whereas C. malonaticus strains gave 8 to 10 bands (Fig. 1). Comparable numbers of fragments were obtained using SpeI: 14 to 17 bands for C. sakazakii strains and 14 to 16 bands for C. malonaticus strains. The XbaI restriction enzyme separated the collection into 16 pulsotypes: ten for C. sakazakii, five for C. malonaticus and one for C. muytjensii, while the SpeI restriction enzyme divided the collection into 14 pulsotypes: eight for C. sakazakii, five for C. malonaticus and one for C. muytjensii. Combining the PFGE profiles generated with the restriction enzymes XbaI and SpeI grouped the 51 strains into a total of 15 pulsotypes: nine for C. sakazakii, five for C. malonaticus and one for C. muytjensii. Strains of the same sequence type from different hospital departments were distinguishable by PFGE and are considered in more detail below.
Fig. 1.
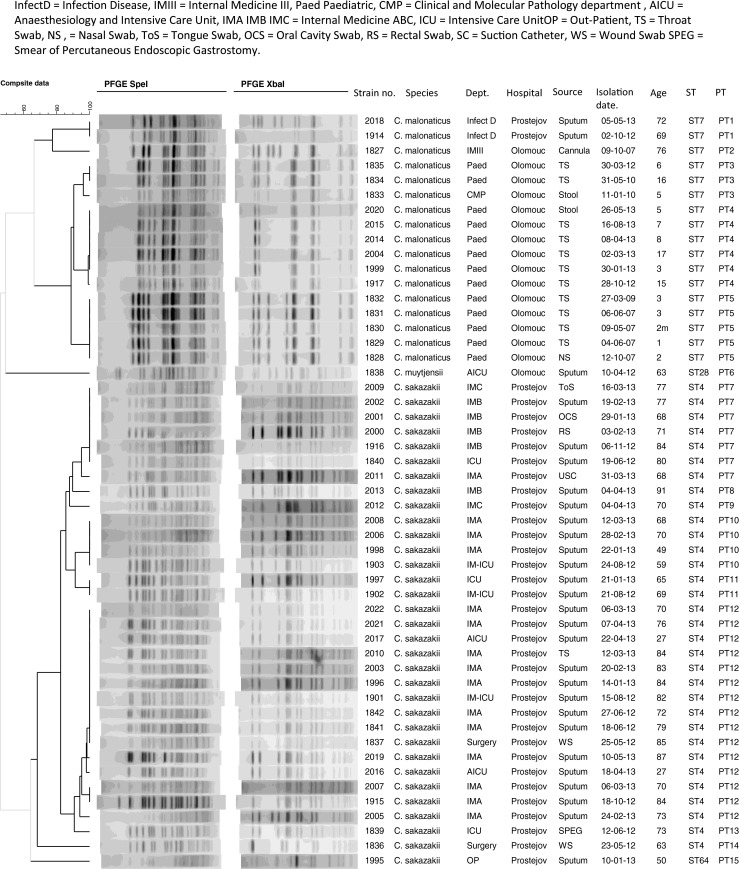
Combined XbaI and SpeI pulsed-field gel electrophoresis (PFGE) profiles of 51 Cronobacter strains. Infect D Infectious Diseases, IMIII Internal Medicine III, Paed Paediatric, CMP Clinical and Molecular Pathology, AICU Anaesthesiology and Intensive Care Unit, IMA IMB IMC Internal Medicine A, B, C, respectively, ICU Intensive Care Unit, OP Outpatient, TS throat swab, NS nasal swab, ToS tongue swab, OCS oral cavity swab, RS rectal swab, USC urine suction catheter, WS wound swab, SPEG smear from area of percutaneous endoscopic gastrostomy
The isolates from Olomouc hospital formed four distinguishable C. malonaticus pulsotypes (PT2 to 5) and one C. muytjensii pulsotype (PT6), which were recovered from different age groups of patients from four hospital departments. PT2 was one C. malonaticus ST7 strain (1827) isolated in the Internal Medicine Department from the intravenous cannula of a 76-year-old patient in 2007. PT3 was composed of three C. malonaticus ST7 strains (1834, 1835, 1833), two of which were isolated from the Paediatric Department and one was from the Clinical and Molecular Pathology Department. The three PT3 strains had been isolated over a 2-year period from throat and stool samples of patients under 16 years of age. The six isolates in PT4 were all C. malonaticus ST7 strains. Five had been isolated from the Paediatric Department over a 10-month period from throat swabs and one from a stool sample. The patient ages ranged from 3 to 17 years old. The majority (4/5) of PT5 strains were isolated from the throat and one from nose from the same Paediatric Department. These strains were also C. malonaticus ST7 and had been collected over a period of 2 years. The patient ages ranged from 2 months to 3 years. C. muytjensii ST28 strain 1838 was in a unique pulsotype (PT6). This strain was isolated in 2012 at the Anaesthesiology and Intensive Care Unit, from the sputum of a 63-year-old patient.
The isolates from Prostějov hospital were recovered from seven departments and were clustered in ten distinguishable Cronobacter pulsotypes (Table 3). PT1 was the only C. malonaticus pulsotype (strains 1914 and 2018). These were both C. malonaticus ST7 strains which were isolated from patients’ sputum at the Infectious Disease Department. The collection was over a 7-month period, and the patients were 69 and 72 years in age. All the remaining isolates were strains of C. sakazakii, which formed nine pulsotypes (PT7 to 15). Eight of these pulsotypes (PT7 to 14) were composed of 32 strains of C. sakazakii ST4. PT15 was composed of one C. sakazakii ST64 strain (1995). Most of the 15 C. sakazakii ST4 strains in PT12 were isolated from sputum except strains 1837 and 2010, which were isolated from a wound swab and throat swab, respectively. This pulsotype was collected over period of about 1 year and the patients ages ranged from 27 to 87 years. In PT12, 12 isolates were collected from the Internal Medicine Department, two from the Anaesthesiology and Intensive Care Unit and one from the Surgery Department. PT13 and PT14 each contained single C. sakazakii ST4 strains; 1839 and 1836, respectively. PT15 contained a single C. sakazakii ST64 strain (1995). These strains were isolated from a percutaneous endoscopic gastrostomy smear ICU, wound surgery and the sputum of an outpatient, respectively. The isolations were over a 7-month period and the patient ages ranged from 50 to 73 years. PT7 consisted of seven C. sakazakii ST4; strains 1840, 1916 and 2002 were isolated from sputum, strain 2000 from rectal swab, strain 2001 from oral cavity swab, strain 2009 from tongue swab and strain 2011 from section catheter. Six of the isolates were collected from the Internal Medicine department, and strain 1840 was isolated from an Intensive Care Unit patient. The collection was over a 7-month period and all patients were over 68 years of age. PT8, 9, 10 and 11 consisted of eight C. sakazakii ST4 strains. All these strains except one (1997) were isolated from sputum at the Internal Medicine Department, whereas strain 1997 was collected from the Intensive Care Unit. The PT8 strain was isolated in 2013 from a 91-year-old patient. PT9 was isolated in 2013 from a 70-year-old patient. PT10 was collected over a roughly 8-month period and the patient ages were between 49 and 70 years old. The two strains in PT11 were collected in 2012 and 2013 and the mean patient age was 67 years (Table 4).
Table 4.
Distribution of Cronobacter species and genotype according to hospital and patient details
| Cronobacter species | Sequence type | No. of isolates (%) | Pulsotype (n) | Hospital | Period of isolation | Age (years) | Sex | Source (n) | |
|---|---|---|---|---|---|---|---|---|---|
| Male | Female | ||||||||
| C. sakazakii | ST4 | 32 (63) | 12 (15), 7 (7), 10 (4), 11 (2), 8 (1), 9 (1), 13 (1), 14 (1) | Prostějov | 12/06/12–10/05/13 | >27 | 16 | 16 | Sputum (24), wound swab (2), section catheter (1), tongue swab (1), throat swab (1), oral cavity (1), rectal swab (1), SPEGa (1) |
| C. sakazakii | ST64 | 1 (2) | 15 (1) | Prostějov | 10/01/2013 | 50 | 1 | 0 | Sputum (1) |
| C. malonaticus | ST7 | 17 (33) | 4 (6), 5 (5), 3 (3), 2 (1) | Olomouc | 06/05/07–16/08/13 | <1 to 76 | 12 | 5 | Throat swab (11), faecal material (2), cannula (1), nasal swab (1) |
| 1 (2) | Prostějov | 2/10/2012 & 5/05/2013 | 69 and 72 | 2 | 0 | Sputum (2) | |||
| C. muytjensii | ST28 | 1 (2) | 6 (1) | Olomouc | 10/04/2012 | 63 | 0 | 1 | Sputum (1) |
| Total | 51 | 6 years | 29 | 22 | |||||
a SPEG Smear from area of percutaneous endoscopic gastrostomy
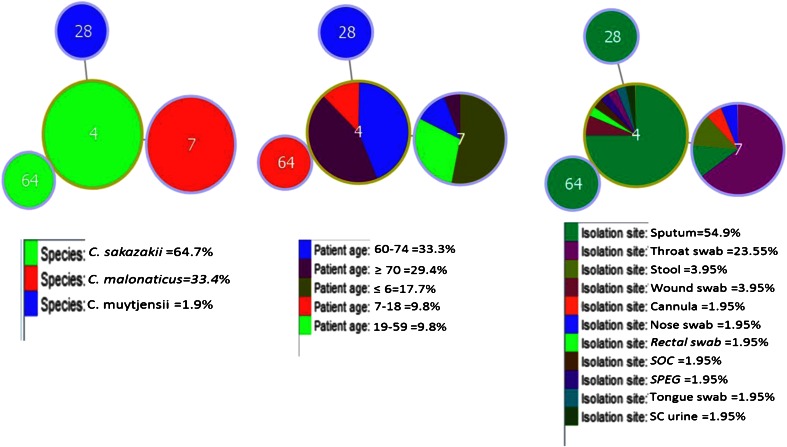
goeBURST analysis showed the range of patient ages and sources with Cronobacter species (Fig. 2). C. sakazakii ST4 strains were predominantly sputum samples from adults >70 years in age, whereas C. malonaticus ST7 were from throat swabs of children <6 years old.
Fig. 2.
goeBURST analysis of Cronobacter strains
Discussion
Reported Cronobacter infections have primarily concerned infants, especially premature neonates with clinical presentations of necrotising enterocolitis and invasive meningitis [21, 22]. Although many of these cases have been linked to contaminated reconstituted infant formula [23], other routes appear to exist, as infections occur in breast-fed infants as well [22, 24, 25]. The carriage of the organism by adults [9] and the high incidence of UTIs [6] indicate that the exposure routes to this bacterium still require further elucidation. In order to have a wider perspective on the exposure to Cronobacter, this study speciated and genotyped Cronobacter strains from age-profiled clinical isolates, and extended the previous study by Holý et al., who reported the incidence of Cronobacter from >45,000 patients [9].
Of the 51 strains, the majority were C. sakazakii (65 %) and C. malonaticus (33 %) (Table 3). The prominence of these two species in clinical isolates has been previously reported in a review of the international Cronobacter PubMLST database with >1000 strains (Forsythe et al. 2014) [10]. C. sakazakii ST4 was the predominant sequence type (32/51 strains) and composed all isolates from Prostějov hospital during a 1-year period. Seventeen C. malonaticus ST7 strains were isolated from two hospitals, Olomouc and Prostějov, during the 6-year period from 2007 to 2013. Two further strains were identified as ST64 and ST28, which are C. sakazakii and C. muytjensii, respectively.
PFGE analysis of isolates revealed that the 35 strains isolated at Prostějov hospital could be divided into three groups. The majority (32/35) of strains belonged to C. sakazakii ST4 and were serotype C. sakazakii O:2. These strains were isolated from various hospital departments during 2012–2013. Two other group isolates were also recovered from patients in this hospital. These were two strains of C. malonaticus ST7 and were serotype C. malonaticus O:2, and were the only strains isolated from the Department of Infectious Diseases. The remaining strain was C. sakazakii ST64 serotype O:2, which was isolated from an outpatient (50 years old, sputum).
In contrast, all but one of the 16 Cronobacter strains isolated from patients at Olomouc hospital were C. malonaticus ST7 ; the other isolate was C. muytjensii. The C. malonaticus strains belonged to the identical sequence type 7 and identical serotype C. malonaticus O:2. With two exceptions, all these strains were from patients at the Department of Paediatrics and had an age range of 0–18 years. There were two strains from adults, one C. malonaticus from an intravenous cannula and another which was C. muytjensii from sputum.
Despite the greater discrimination of strains using PFGE than MLST, isolates from patients for whom there were no known links could not be further differentiated. For example, the C. sakazakii ST4, pulsotype 12 strains were isolated from 15 adults (aged 27–85 years) between May 2012 and May 2013. This could be due to the reported high clonality of sequence types within C. sakazakii and C. malonaticus limiting the discriminatory power of PFGE [1, 10].
In summary, these clinical isolates were predominated by C. sakazakii ST4 (63 %, 32/51) and C. malonaticus ST7 (33 %, 17/51). These had been isolated from throat and sputum samples of all age groups, as well as recal and faecal swabs. There was no apparent relatedness between the age or sex of the patient and the Cronobacter species isolated. Despite the high clonality of Cronobacter, PFGE profiles differentiated strains within each sequence type into 15 pulsotypes. There was almost complete agreement between O-antigen typing and rpoB gene sequence analysis and MLST profiling. The majority (43/51) of strains were from the upper respiratory system (i.e. throat swabs and sputum samples) and only three were from faeces and one from urine; two being C. sakazakii ST4 and the remaining two C. malonaticus ST7. Hence, it is plausible that this small sampling of the lower intestinal tract and UTIs does not reflect the diversity of Cronobacter in those samples. Given the high incidence of Cronobacter in UTI, this area needs further consideration [6].
This study shows the value of applying MLST to bacterial population studies with strains from two patient cohorts, combined with PFGE for further discrimination of strains.
Acknowledgements
This work was supported by Research Support Foundation, Vaduz (801100021/39). We also thank the Libyan Embassy for their funding of Abdlrhman Alsonosi and Umm Al-Qura University for funding Sumyya Hariri.
Ethical statement
Collection of material: We used only laboratory samples and we had no contact with patients, so no informed consent was required.
Submission of manuscript: All authors have contributed sufficiently to the scientific work presented in the manuscript and, therefore, share collective responsibility and accountability for the results. All authors agree with the final version of the manuscript under submission. The manuscript has not previously been submitted to any journal and is not under consideration by any other journal. No parts of the data have been previously submitted for publication.
Conflict of interest
The authors have no declared conflicts of interests.
References
- 1.Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, Forsythe SJ. Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J Clin Microbiol. 2012;50:3031–3039. doi: 10.1128/JCM.00905-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holý O, Forsythe S. Cronobacter spp. as emerging causes of healthcare-associated infection. J Hosp Infect. 2014;86:169–177. doi: 10.1016/j.jhin.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 3.International Commission on Microbiological Specifications for Foods (ICMSF) Microbiological testing in food safety management. New York: Kluwer Academic/Plenum Publishers; 2002. [Google Scholar]
- 4.Food and Agriculture Organization/World Health Organization (FAO/WHO) (2008) Enterobacter sakazakii (Cronobacter spp.) in powdered follow-up formulae. In: Microbiological Risk Assessment Series No. 15. Rome. 90 pp. Available online at: http://www.who.int/foodsafety/publications/mra_followup/en/
- 5.Lai KK. Enterobacter sakazakii infections among neonates, infants, children, and adults. Case reports and a review of the literature. Medicine (Baltimore) 2001;80:113–122. doi: 10.1097/00005792-200103000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Patrick ME, Mahon BE, Greene SA, Rounds J, Cronquist A, Wymore K, Boothe E, Lathrop S, Palmer A, Bowen A. Incidence of Cronobacter spp. Infections, United States, 2003–2009. Emerg Infect Dis. 2014;20:1520–1523. doi: 10.3201/eid2009.140545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gosney MA, Martin MV, Wright AE, Gallagher M. Enterobacter sakazakii in the mouths of stroke patients and its association with aspiration pneumonia. Eur J Intern Med. 2006;17:185–188. doi: 10.1016/j.ejim.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Cui JH, Cui ZG, Hu GC, Yang YL, Li J, Shi YW. Cronobacter carriage in neonate and adult intestinal tracts. Biomed Environ Sci. 2013;26:861–864. doi: 10.3967/bes2013.011. [DOI] [PubMed] [Google Scholar]
- 9.Holý O, Petrželová J, Hanulík V, Chromá M, Matoušková I, Forsythe SJ. Epidemiology of Cronobacter spp. isolates from patients admitted to the Olomouc University Hospital (Czech Republic) Epidemiol Mikrobiol Imunol. 2014;63:69–72. [PubMed] [Google Scholar]
- 10.Forsythe SJ, Dickins B, Jolley KA. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics. 2014;15:1121. doi: 10.1186/1471-2164-15-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph S, Forsythe SJ. Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg Infect Dis. 2011;17:1713–1715. doi: 10.3201/eid1709.110260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hariri S, Joseph S, Forsythe SJ. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg Infect Dis. 2013;19:175–177. doi: 10.3201/eid1901.120649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, Rico A, Brzoska P, Hamby SE, Masood N, Hariri S, Sonbol H, Chuzhanova N, McClelland M, Furtado MR, Forsythe SJ. Comparative analysis of genome sequences covering the seven Cronobacter species. PLoS One. 2012;7:e49455. doi: 10.1371/journal.pone.0049455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caubilla-Barron J, Hurrell E, Townsend S, Cheetham P, Loc-Carrillo C, Fayet O, Prère M-F, Forsythe SJ. Genotypic and phenotypic analysis of Enterobacter sakazakii strains from an outbreak resulting in fatalities in a neonatal intensive care unit in France. J Clin Microbiol. 2007;45:3979–3985. doi: 10.1128/JCM.01075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis KG, Grim CJ, Franco AA, Gopinath G, Sathyamoorthy V, Hu L, Sadowski JA, Lee CS, Tall BD. Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl Environ Microbiol. 2011;77:4017–4026. doi: 10.1128/AEM.00162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Wang M, Wang Q, Cao B, He X, Li K, Feng L, Wang L. Genetic analysis of the Cronobacter sakazakii O4 to O7 O-antigen gene clusters and development of a PCR assay for identification of all C. sakazakii O serotypes. Appl Environ Microbiol. 2012;78:3966–3974. doi: 10.1128/AEM.07825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol. 2013;36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, Forsythe S. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol. 2009;9:223. doi: 10.1186/1471-2180-9-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Acker J, de Smet F, Muyldermans G, Bougatef A, Naessens A, Lauwers S. Outbreak of necrotizing enterocolitis associated with Enterobacter sakazakii in powdered milk formula. J Clin Microbiol. 2001;39:293–297. doi: 10.1128/JCM.39.1.293-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowen AB, Braden CR. Invasive Enterobacter sakazakii disease in infants. Emerg Infect Dis. 2006;12:1185–1189. doi: 10.3201/eid1208.051509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.[No authors listed] From the Centers for Disease Control and Prevention. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. JAMA. 2002;287:2204–2205. doi: 10.1001/jama.287.17.2204. [DOI] [PubMed] [Google Scholar]
- 24.Stoll BJ, Hansen N, Fanaroff AA, Lemons JA. Enterobacter sakazakii is a rare cause of neonatal septicemia or meningitis in VLBW infants. J Pediatr. 2004;144:821–823. doi: 10.1016/j.jpeds.2004.02.045. [DOI] [PubMed] [Google Scholar]
- 25.Ravisankar S, Syed SS, Garg P, Higginson J. Is Cronobacter sakazakii infection possible in an exclusively breastfed premature neonate in the neonatal intensive care unit? J Perinatol. 2014;34:408–409. doi: 10.1038/jp.2014.14. [DOI] [PubMed] [Google Scholar]