The zinc-finger antiviral protein prevents replication of Sindbis virus
Keywords: host defense, IFN, innate immunity, pattern-recognition receptor, RNA degradation
Abstract
Accumulating evidence indicates that type I interferon (IFN) mediates the host protective response to RNA viruses. However, the anti-viral effector molecules involved in this response have not been fully identified. Here, we show that zinc-finger anti-viral protein (ZAP), an IFN-inducible gene, plays a critical role in the elimination of Sindbis virus (SINV) in vitro and in vivo. The loss of ZAP greatly enhances the replication of SINV but does not inhibit type I IFN production in primary mouse embryonic fibroblasts (MEFs). ZAP binds and destabilizes SINV RNA, thereby suppressing the replication of SINV. Type I IFN fails to suppress SINV replication in ZAP-deficient MEFs, whereas the ectopic expression of ZAP is sufficient to suppress the replication of SINV in MEFs lacking the expression of type I IFN and the IFN-inducible genes. ZAP-deficient mice are highly susceptible to SINV infection, although they produce sufficient amounts of type I IFN. Therefore, ZAP is an RNA-sensing anti-viral effector molecule that mediates the type-I-IFN-dependent host defense against SINV.
Introduction
Pattern-recognition receptors (PRRs) detect viral nucleic acids and induce an anti-viral innate immune response (1, 2). Retinoic acid-inducible gene I (RIG-I), also called DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58), and melanoma differentiation-associated gene 5 (MDA5), also called interferon (IFN)-induced with helicase C domain 1 (IFIH1), are RNA helicases that detect cytosolic viral double-stranded (ds) RNAs (3, 4), whereas cyclic GMP-AMP synthase (cGAS) detects cytosolic viral dsDNA (5, 6). After cytosolic viral nucleic acids are detected, RIG-I-like receptors (RLRs) and cGAS trigger the activation of IFN regulatory factor 3 (IRF3), a transcription factor that induces the expression of type I IFN and IFN-inducible genes. Endosomal Toll-like receptor 7 (TLR7) and TLR9 detect extracellular viral single-stranded (ss) RNA and ssDNA, respectively, and activate IRF7 to induce the expression of type I IFN and the IFN-inducible genes (7–10). Thus, nucleic-acid-sensing PRRs are essential to the establishment of an anti-viral state by mediating the expression of type I IFN and IFN-inducible genes.
The anti-viral effector molecules responsible for the type-I-IFN-mediated host defense against viruses are not fully understood. Among the IFN-inducible proteins, zinc-finger anti-viral protein (ZAP), also called zinc-finger CCCH type, anti-viral 1 (ZC3HAV1), is a well-characterized anti-viral effector molecule (11). ZAP was originally identified as the anti-viral effector molecule that inhibits murine leukemia virus (MLV) infection (12). ZAP binds to MLV transcripts via its CCCH-type zinc-finger (C3H ZF) domains and reduces the level of MLV transcripts (13). After it detects the MLV transcripts, ZAP associates with the exosome complex, an RNA degradation structure, and destabilizes the MLV transcripts (14). Endogenous ZAP is involved in the anti-MLV response in mouse embryonic fibroblasts (MEFs) (15). Recent studies have shown that Sindbis virus (SINV), Ebola virus, Marburg virus, MLV, HIV-1 and hepatitis B virus (HBV) are also targeted by ZAP (12, 16–19). However, the roles of ZAP in type-I-IFN-mediated virus elimination and in anti-viral responses in vivo are unclear. In this study, we examined these issues and show the essential involvement of ZAP in the host defense against viruses.
Methods
Reagents
High-molecular-weight poly rI:rC, LPS, R848, oligodeoxynucleotide (ODN) 1585, ODN 1668, 5′ppp-dsRNA and IFN stimulatory DNA (ISD) were purchased from InvivoGen. Anti-VSV-G (A190-131A) antibody was purchased from Bethyl Laboratories. Anti-Myc antibody (2276S) and mouse IgG1 isotype control antibody (5415S) were purchased from Cell Signaling Technology. Horseradish-peroxidase-conjugated anti-β-actin antibody (sc-1615) was purchased from Santa Cruz Biotechnology. Recombinant mouse IFN-β was purchased from ProSpec. Recombinant mouse granulocyte macrophage colony-stimulating factor (GM-CSF) and human Fms-like tyrosine kinase 3 ligand (FLT3L) were purchased from Peprotech. Lipofectamine™ 2000 was purchased from Invitrogen. Anti-SINV E2 antibody was originally developed by D. E. Griffin (Johns Hopkins University School of Medicine, USA) and obtained from Y. Yoshinaka (Tokyo Medical and Dental University, Japan).
Plasmids
The retroviral expression vector pMRX–ires–puro was kindly donated by S. Yamaoka (Tokyo Medical and Dental University, Japan). The cytomegalovirus-promoter-driven expression vector pcDNA3.1(+) was purchased from Invitrogen. A synthesized cDNA fragment encoding the Myc-tag sequence was cloned into pMRX–ires–puro and pcDNA3.1(+), generating pMRX–Myc–ires–puro and pcDNA3–Myc, respectively. cDNA fragments encoding ZAP-L, ZAP-S, ZAP-N and ZAP-C were amplified from an MEF cDNA library with PCR and cloned into pMRX–Myc–ires–puro, generating pMRX–ZAP-L–Myc–ires–puro, pMRX–ZAP-S–Myc–ires–puro, pMRX–ZAP-N–Myc–ires–puro and pMRX–ZAP-C–Myc–ires–puro, respectively. A cDNA fragment encoding ZAP-L was cloned into pcDNA3–Myc, generating pcDNA–ZAP-L–Myc. pEGFP-N1 was purchased from Clontech. pdsTE12Q was kindly provided by B. Levine (University of Texas Southwestern Medical Center, Dallas, TX, USA).
Mice and cells
The Zc3hav1 –/– mice (C57BL/6 and 129 mixed background) have been described previously (15) and were backcrossed to C57BL/6 mice over eight generations. Age-matched C57BL/6 mice were used as the control Zc3hav1 +/+ mice. The Ddx58 –/–/Ifih1 –/– mice have been described previously (20). The Irf3 –/–/Irf7 –/– mice were kindly donated by T. Taniguchi (The University of Tokyo, Japan). All animal experiments were performed in accordance with the guidelines of Osaka University and were approved by the Ethics Committee of the Research Institute for Microbial Diseases of Osaka University.
The Zc3hav1 +/–, Ddx58 +/–/Ifih1 +/– and Irf3 +/–/Irf7 +/– mice were crossed separately, and primary MEFs were prepared from pregnant female mice on embryonic day 13.5, as described previously (15). Spontaneously immortalized wild-type MEFs have been described previously (21). Primary MEFs were used in most in vitro experiments. Because the transfection efficiency of the plasmid is quite low in primary MEFs, immortalized wild-type MEFs, which are competent for plasmid transfection, were used in the experiment shown in Fig. 5(A). Plate-E packaging cells were kindly donated by T. Kitamura (The University of Tokyo). Vero cells have been described previously (22). The MEFs, Plat-E cells and vero cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, penicillin (100U ml−1) and streptomycin (100U ml−1). To prepare the primary dendritic cells, mouse bone-marrow cells were treated for 6 days with GM-CSF (10ng ml−1) or FLT3L (20ng ml−1) in RPMI 1640 medium supplemented with 10% fetal bovine serum, 50mM 2-mercaptoethanol, penicillin (100U ml−1) and streptomycin (100U ml−1).
Fig. 5.
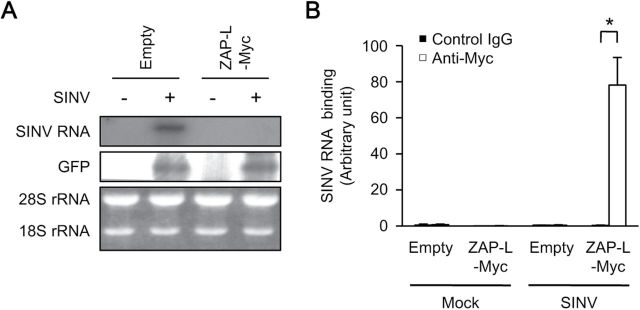
ZAP binds to and reduces SINV RNA. (A) Immortalized wild-type MEFs were transiently transfected with SINV RNA and an enhanced green fluorescent protein (EGFP)-expressing plasmid with or without the ZAP-L–Myc-expressing plasmid for 24h. The total cellular RNA was subjected to a northern blotting analysis for SINV RNA. (B) Immortalized wild-type MEFs retrovirally transduced with or without ZAP-L–Myc were transiently transfected with SINV RNA for 24h. The level of SINV RNA binding to ZAP-L–Myc was measured with RNA immunoprecipitation coupled to quantitative RT–PCR. The level of immunoprecipitated SINV RNA was normalized to that of the input SINV RNA to determine the SINV RNA binding activity. The experiments were performed at least three times and representative data are shown (means ± SD of three independent samples). *P < 0.05.
Viruses
SINV and VSV have been described previously (4, 23). SINV RNA was synthesized from pdsTE12Q using SP6 RNA polymerase (Takara Bio), according to the manufacturer’s protocol. In a mouse model of SINV infection, 10-day-old pups were subcutaneously inoculated with 100 plaque-forming units (pfu) of SINV. The viral titers were determined with 50% tissue culture infectious dose (TCID50) assays, as described previously (22). A retroviral vector was produced by Plat-E cells transfected with pMRX–ires–puro, as described previously (24).
Enzyme-linked immunosorbent assays
The ELISA kits for IFN-α and IFN-β were purchased from PBL Interferon Source. The ELISA kits for CXCL10 and IL-6 were purchased from R&D Systems. The levels of IFN-α, IFN-β, IL-6 and CXCL10 were measured with ELISAs, according to the manufacturers’ instructions.
RNA immunoprecipitation assay
The interaction between SINV RNA and ZAP was detected with the Ribocluster Profiler™/RIP-Assay kit (MBL), according to the manufacturer’s instructions.
Immunoblotting
Total cell lysates were separated by SDS–PAGE and immunoblotted with the indicated antibodies, as described previously (24).
Northern blot analysis
Total RNA was isolated from cells with an RNeasy Kit (Qiagen), according to the manufacturer’s instructions. The RNA samples (8 µg each) were separated by electrophoresis, transferred to a nylon membrane and hybridized for 12h with the indicated 32P-labeled probes, prepared with the MegaPrime™ DNA Labelling System (GE Healthcare). The nylon membrane was washed with 2 × saline sodium citrate (SSC) and 0.2 × SSC and exposed to x-ray film to quantify the relative RNA levels. The probe was designed to target SINV RNA nucleotides 4048–4259.
Quantitative reverse transcription–PCR
Total RNA was isolated from cultured cells with the ZR RNA MicroPrep™ kit (Zymo Research). Total RNA was isolated from the brain with an RNeasy Lipid Tissue Mini Kit (Qiagen). Reverse transcription (RT) was performed with ReverTra Ace® reverse transcriptase (Toyobo). For the quantitative PCR, cDNA fragments were amplified from the RT products with Realtime PCR Master Mix (Toyobo). The fluorescence from the TaqMan probe was detected with the StepOnePlus™ Real-Time PCR System (Applied Biosystems). The expression of Ifnb1, Cxcl10, IL-1b and IL-6 mRNAs was normalized to that of β-actin mRNA. The experiments were performed in accordance with the manufacturer’s instructions.
Statistical analysis
The Student’s t-test was used to analyze all data. A P value of <0.05 was considered significant.
Results
ZAP suppresses SINV replication in primary MEFs
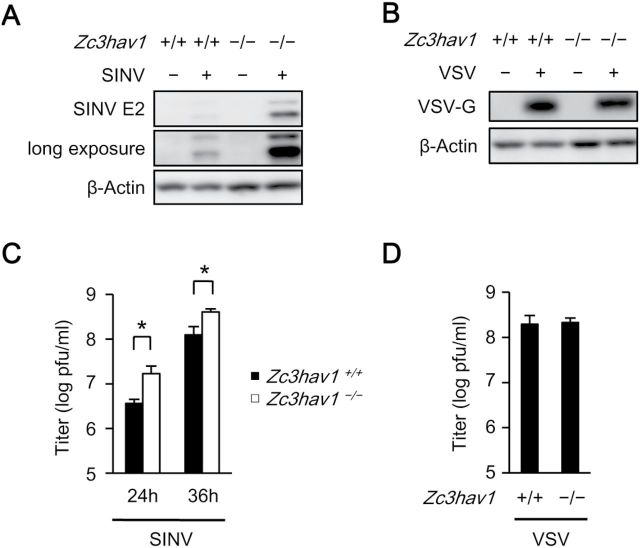
SINV is a positive-sense RNA virus belonging to the genus Alphavirus in the family Togaviridae. SINV causes fever, rash, arthralgia and encephalopathy in humans and animals (25). Type I IFN is critically involved in the host defense against SINV (26). Therefore, we used SINV as the model pathogen in the present study. We first examined the involvement of endogenous ZAP in the anti-viral response to SINV in primary MEFs. The replication of SINV was greatly enhanced in primary Zc3hav1 –/– MEFs (Fig. 1A and C), although the replication of vesicular stomatitis virus (VSV) was normal (Fig. 1B and D). These findings indicate that endogenous ZAP contributes to the suppression of SINV replication in primary MEFs.
Fig. 1.
Loss of ZAP enhances the replication of SINV in primary MEFs. (A and B) Primary Zc3hav1 +/+ and Zc3hav1 –/– MEFs were infected with SINV [multiplicity of infection (MOI) = 0.1] (A) or VSV (MOI = 0.1) (B) for 24h. The levels of viral proteins and β-actin in the cell lysates were measured with immunoblotting. (C and D) Primary Zc3hav1 +/+ and Zc3hav1 –/– MEFs were infected with SINV (MOI = 0.1) (C) for the indicated periods or VSV (MOI = 0.1) (D) for 24h. The viral titers in the culture supernatants were determined with TCID50 assays. The experiments were performed three times and representative data are shown (means ± SD of three independent samples). *P < 0.05.
The ZAP long isoform has zinc-finger domains at its N-terminus and a poly(ADP-ribose) polymerases (PARP) domain at its C-terminus, whereas the ZAP short isoform has zinc-finger domains at the N-terminus but lacks the PARP domain (Fig. 2A). Previous studies have shown that the zinc-finger domains of ZAP are sufficient to trigger an anti-viral response, but another study has shown that both the zinc-finger and PARP domains of ZAP are involved in the anti-viral response in the 293 cell line (27, 28). Therefore, the role of the PARP domain of ZAP in the anti-viral response is still unclear. We assessed the involvement of the zinc-finger and PARP domains of ZAP in the anti-viral response to SINV. The complementation of the ZAP N-terminal portion, but not that of ZAP C-terminal portion restored the anti-viral activity against SINV in Zc3hav1 –/– MEFs (Fig. 2B), confirming the importance of the zinc-finger domains of ZAP in the suppression of SINV replication. Complementation of both the ZAP long isoform and the ZAP short isoform in Zc3hav1 –/– MEFs restored the anti-viral activity against SINV (Fig. 2B), indicating that the PARP domain of ZAP is not essential for the suppression of SINV replication in primary MEFs.
Fig. 2.
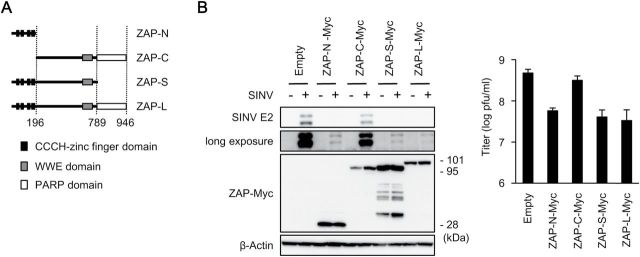
N-terminal zinc-finger domains of ZAP mediate the anti-viral response to SINV. (A) Schematic of ZAP. (B) Primary Zc3hav1 –/– MEFs retrovirally transduced with ZAP-N–Myc, ZAP-C–Myc, ZAP-S–Myc and ZAP-L–Myc were infected with SINV (MOI = 0.05) for 24h. The levels of viral proteins, β-actin and Myc-tagged proteins in the cell lysates were measured with immunoblotting. The viral titers in the culture supernatants were determined with TCID50 assays. The experiments were performed at least three times and representative data are shown (means ± SD of three independent samples).
ZAP reduces SINV RNA levels independently of type I IFN production
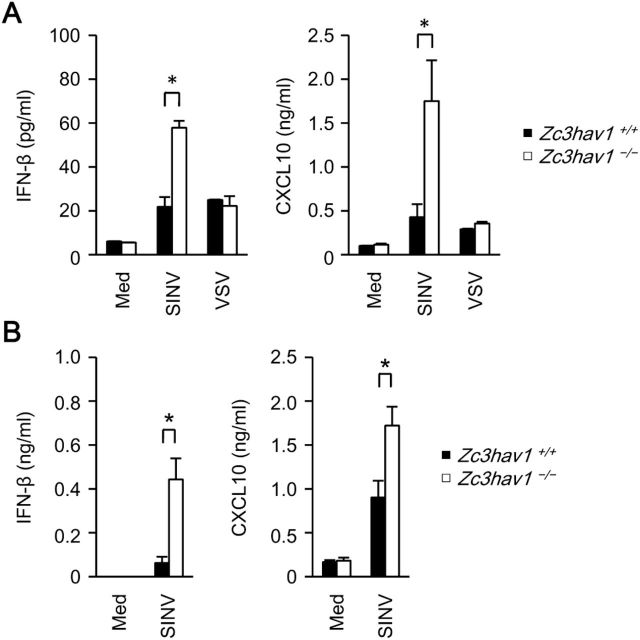
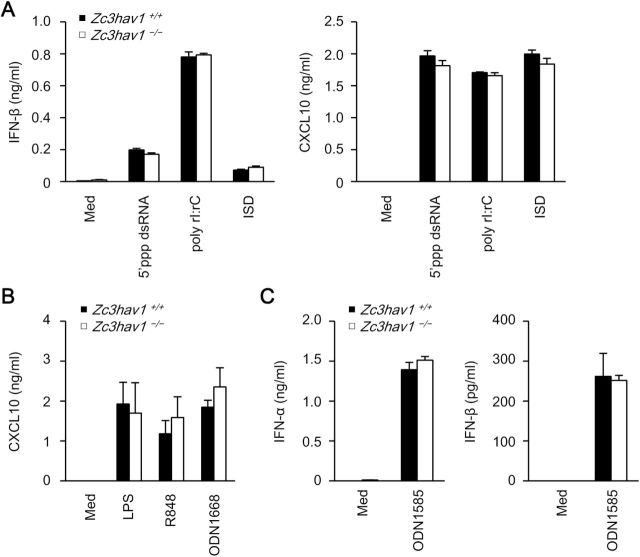
Type I IFN and chemokines are critically involved in anti-viral host defenses (29). ZAP is reported to be involved in the regulation of RIG-I signaling in the 293 cell line (30). Therefore, we assessed the involvement of ZAP in the production of these factors. The SINV-induced production of IFN-β, a type I IFN, and chemokine (C-X-C motif) ligand 10 (CXCL10), an IFN-inducible chemokine, was enhanced in Zc3hav1 –/– MEFs and Zc3hav1 –/– conventional dendritic cells (Fig. 3A and B). However, the VSV-induced production of IFN-β and CXCL10 was normal in Zc3hav1 –/– MEFs (Fig. 3A). Furthermore, the production of IFN-β and CXCL10 was normal in Zc3hav1 –/– MEFs stimulated with the RLR and cGAS ligands (Fig. 4A). These findings indicate that ZAP is not essential for the RLR- or cGAS-mediated expression of type I IFN or IFN-inducible genes in primary mouse cells and suggests that an increase in SINV causes the enhanced activation of RLR in primary Zc3hav1 –/– MEFs. Zc3hav1 –/– conventional dendritic cells normally produce CXCL10 after they are stimulated with the TLR4, TLR7 and TLR9 ligands (Fig. 4B). Zc3hav1 –/– plasmacytoid dendritic cells normally produce IFN-α and IFN-β after they are stimulated with the TLR9 ligand (Fig. 4C). These findings also indicate that ZAP is not essential for the TLR-mediated expression of type I IFN or IFN-inducible genes.
Fig. 3.
ZAP is not essential for SINV-induced type I IFN production in primary MEFs or dendritic cells. (A) Primary Zc3hav1 +/+ and Zc3hav1 –/– MEFs were infected with SINV (MOI = 1) or VSV (MOI = 1) for 24h. (B) Zc3hav1 +/+ and Zc3hav1 –/– GM-CSF-induced bone-marrow dendritic cells were infected with SINV (MOI = 5) for 24h. The levels of IFN-β and CXCL10 in the culture supernatants were measured with ELISAs. The experiments were performed three times (means ± SD of three independent samples). *P < 0.05.
Fig. 4.
ZAP is not essential for nucleic-acid-induced type I IFN production in primary MEFs or dendritic cells. (A) Primary Zc3hav1 +/+ and Zc3hav1 –/– MEFs were stimulated with 5′ppp-dsRNA (1 µg ml−1), poly rI:rC (1 µg ml−1) and ISD (1 µg ml−1), together with Lipofectamine™ 2000 for 24h. The levels of IFN-β and CXCL10 in the culture supernatants were measured with ELISAs. (B) Zc3hav1 +/+ and Zc3hav1 –/– GM-CSF-induced bone-marrow dendritic cells were stimulated with LPS (TLR4 ligand, 100ng ml−1), R848 (TLR7 ligand, 10 µg ml−1) and ODN 1668 (TLR9 ligand, 1 µM) for 24h. The levels of CXCL10 in the culture supernatants were measured by an ELISA. (C) Zc3hav1 +/+ and Zc3hav1 –/– FLT3L-induced bone-marrow dendritic cells were stimulated with ODN 1585 (TLR9 ligand, 5 µM) for 24h. The levels of IFN-α and IFN-β in the culture supernatants were measured with ELISAs. The experiments were performed three times (means ± SD of three independent samples).
We next examined whether ZAP directly affects SINV replication. Although a previous study showed that ZAP reduced the level of luciferase mRNA fused with fragmented SINV RNA (13), there is as yet no direct evidence to support the destabilization of SINV RNA by ZAP or to confirm that ZAP binds to SINV RNA. Ectopically expressed ZAP greatly reduced the level of transfected SINV RNA in wild-type MEFs (Fig. 5A). Stably expressed ZAP was also immunoprecipitated with transfected SINV RNA in wild-type MEFs (Fig. 5A and B). The interaction between ectopically expressed ZAP and transfected SINV RNA was barely detectable, probably because the level of SINV RNA was extremely low (data not shown). These findings indicate that ZAP detects SINV RNA to induce its degradation.
Type I IFN requires ZAP to suppress SINV replication in MEFs
Type I IFN suppresses SINV replication (26). Therefore, we examined the involvement of ZAP in the type-I-IFN-mediated suppression of SINV replication. Treatment with IFN-β induced the expression of ZAP mRNA and suppressed SINV replication in wild-type MEFs (Fig. 6A). However, treatment with IFN-β failed to suppress SINV replication in Zc3hav1 –/– MEFs (Fig. 6B). Although SINV replication was enhanced in Ddx58 –/–/Ifih1 –/– and Irf3 –/–/Irf7 –/– MEFs, which are deficient in the expression of type I IFN and IFN-inducible genes, the ectopic expression of ZAP was sufficient to suppress SINV replication in these MEFs (Fig. 6C). These findings indicate that ZAP is an anti-viral effector molecule responsible for the type-I-IFN-mediated anti-viral response to SINV.
Fig. 6.
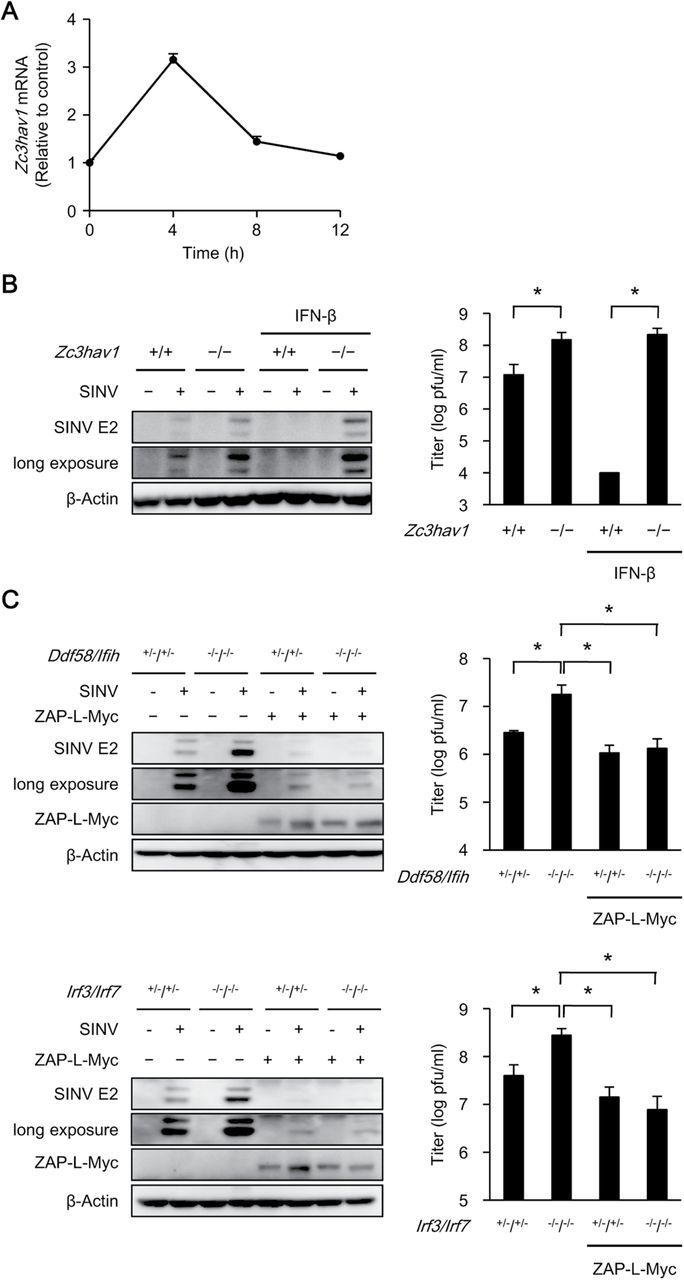
ZAP is essential for the type-I-IFN-induced anti-viral response to SINV. (A) Primary wild-type MEFs were stimulated with IFN-β (50 pg ml−1) for the indicated periods. The level of Zc3hav1 mRNA was measured with quantitative RT–PCR. (B) Primary Zc3hav1 +/+ and Zc3hav1 –/– MEFs were treated with or without IFN-β (50 pg ml−1) for 12h and then infected with SINV (MOI = 0.05) for 24h. The levels of SINV E2 and β-actin in the cell lysates were measured with immunoblotting. The viral titers were determined with TCID50 assays after 24h. (C) Primary Ddx58 +/–/Ifih1 +/–, Ddx58 –/–/Ifih1 –/–, Irf3 +/–/Irf7 +/– and Irf3 –/–/Irf7 –/– MEFs retrovirally transduced with or without ZAP-L–Myc were infected with SINV (MOI = 0.05) for 24h. The levels of SINV E2, β-actin and Myc-tagged proteins in the cell lysates were measured with immunoblotting. The viral titers were determined with TCID50 assays after 24h. The experiments were performed three times and representative data are shown (means ± SD of three independent samples). *P < 0.05.
ZAP mediates host defense against SINV
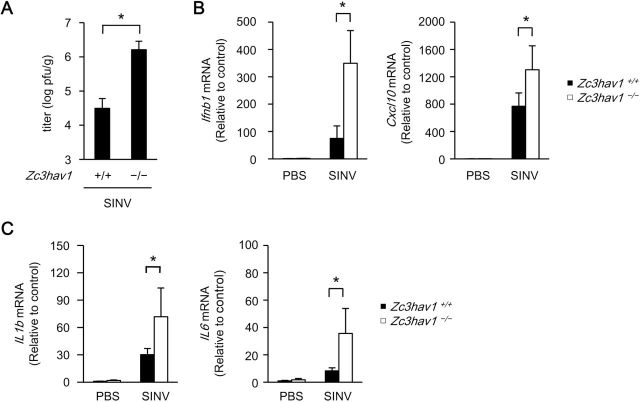
A number of studies have shown that ZAP suppresses viral replication in immortalized and cancer cells (31). Therefore, we examined the involvement of ZAP in the anti-viral response in vivo using Zc3hav1 –/– mice. SINV replicated efficiently in the brain in infant Zc3hav1 –/– mice (Fig. 7A). The expression of Ifnb1 and Cxcl10 was elevated in infant Zc3hav1 –/– mice infected with SINV (Fig. 7B). The expression of Il1b and Il6, which are associated with the severity of SINV-induced encephalopathy (32, 33), were also elevated in infant Zc3hav1 –/– mice infected with SINV (Fig. 7C). These findings indicate that ZAP protects infant mice from SINV infection.
Fig. 7.
Loss of ZAP renders mice susceptible to SINV infection. (A–C) Ten-day-old Zc3hav1 +/+ and Zc3hav1 –/– mice (n = 5 each) were subcutaneously inoculated with SINV (100 pfu per mouse). The brains of the SINV-infected Zc3hav1 +/+ and Zc3hav1 –/– mice were isolated on day 5. The viral titers (pfu per tissue weight) in the brains were determined with TCID50 assays (A). The levels of Ifnb1, Cxcl10, Il1b and Il6 mRNAs in the brains were measured with quantitative RT–PCR (B and C). The experiments were performed five times (means ± SD of five independent samples). *P < 0.05.
Discussion
Although we and others have demonstrated the importance of ZAP in anti-viral responses, the molecular mechanisms underlying the elimination of viruses by ZAP are still unclear. ZAP suppresses the replication of SINV, Ebola virus, Marburg virus, MLV, HIV-1 and HBV (12, 16–19), indicating that ZAP targets positive-sense RNA viruses, negative-sense RNA viruses, retroviruses and DNA viruses. However, ZAP does not suppress the replication of VSV, a negative-sense RNA virus. ZAP inhibits viral protein translation and viral RNA splicing and destabilizes viral RNA (34). These findings raise important questions. How does ZAP sense a wide variety of viruses? How does ZAP specifically identify its target viruses? How does ZAP distinguish the different modes of anti-viral actions? In future studies, we will address these questions to better understand the anti-viral innate immune response.
Members of the C3H ZF protein family can bind to RNA and are conserved from yeasts to mammals. In mammals, >50 members of the C3H ZF protein family have been identified (35). This family is involved in a variety of cellular events, including RNA synthesis, mRNA splicing, mRNA degradation and protein translation, and thus regulates the innate and acquired immunity of the host (36–38). ZAP is a member of the C3H ZF protein family and acts as an RNA-sensing anti-viral effector molecule. In addition to ZAP, ZFP36 and regnase-1, other C3H ZF protein family members, sense viral RNA and are involved in the regulation of viral replication (39–41). Therefore, the C3H ZF protein family seems to play a major role in the direct elimination of viruses. However, the anti-viral activities of the majority of C3H ZF protein family members have not yet been investigated. It would be interesting to identify the C3H ZF protein family members involved in host defenses against viruses in a future study.
Funding
National Institutes of Health (grant PO1-AI070167), the Japan Society for the Promotion of Science (JSPS) Funding Program for World-Leading Innovative R&D on Science and Technology ‘FIRST Program’, JSPS KAKENHI (grant nos. 26713005, 25117717, 23659231), the Research Grant of the Naito Foundation and the Visionary Research Grant of the Takeda Science Foundation. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Acknowledgements
We thank T. Kitamura for the Plat-E cells; S. Yamaoka for pMRX–ires–puro; D. E. Griffin and Y. Yoshinaka for the anti-SINV E2 antibody; B. Levine for pdsTE12Q and the members of the Laboratory of Host Defense for their assistance. T.K. performed most of the experiments and analyzed the data; M.T., T.M., Y.Y. and Y.M. helped with the experiments; S.A. supervised the overall research project; T.S. designed and helped with the experiments and wrote the manuscript. The authors confirm that all data underlying the findings are fully available without restriction.
Conflict of interest statement: The authors declared no conflict of interests.
References
- 1. Kawai T., Akira S. 2009. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int. Immunol. 21:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Iwasaki A. 2012. A virological view of innate immune recognition. Annu. Rev. Microbiol. 66:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yoneyama M., Kikuchi M., Natsukawa T., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730. [DOI] [PubMed] [Google Scholar]
- 4. Kato H., Takeuchi O., Sato S., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101. [DOI] [PubMed] [Google Scholar]
- 5. Sun L., Wu J., Du F., Chen X., Chen Z. J. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu J., Sun L., Chen X., et al. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heil F., Hemmi H., Hochrein H., et al. 2004. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303:1526. [DOI] [PubMed] [Google Scholar]
- 8. Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303:1529. [DOI] [PubMed] [Google Scholar]
- 9. Hemmi H., Kaisho T., Takeda K., Akira S. 2003. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J. Immunol. 170:3059. [DOI] [PubMed] [Google Scholar]
- 10. Honda K., Yanai H., Negishi H., et al. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772. [DOI] [PubMed] [Google Scholar]
- 11. Zhu Y., Gao G. 2008. ZAP-mediated mRNA degradation. RNA Biol. 5:65. [DOI] [PubMed] [Google Scholar]
- 12. Gao G., Guo X., Goff S. P. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703. [DOI] [PubMed] [Google Scholar]
- 13. Guo X., Carroll J. W., Macdonald M. R., Goff S. P., Gao G. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78:12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo X., Ma J., Sun J., Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl Acad. Sci. USA 104:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee H., Komano J., Saitoh Y., et al. 2013. Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc. Natl Acad. Sci. USA 110:12379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bick M. J., Carroll J. W., Gao G., Goff S. P., Rice C. M., MacDonald M. R. 2003. Expression of the zinc-finger antiviral protein inhibits Alphavirus replication. J. Virol. 77:11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Müller S., Möller P., Bick M. J., et al. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 81:2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhu Y., Chen G., Lv F., et al. 2011. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc. Natl Acad. Sci. USA 108:15834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mao R., Nie H., Cai D., et al. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog. 9:e1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kato H., Takeuchi O., Mikamo-Satoh E., et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saitoh T., Tun-Kyi A., Ryo A., et al. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598. [DOI] [PubMed] [Google Scholar]
- 22. Saitoh T., Fujita N., Hayashi T., et al. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc. Natl Acad. Sci. USA 106:20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moriishi K., Koura M., Matsuura Y. 2002. Induction of Bad-mediated apoptosis by Sindbis virus infection: involvement of pro-survival members of the Bcl-2 family. Virology 292:258. [DOI] [PubMed] [Google Scholar]
- 24. Saitoh T., Fujita N., Jang M. H., et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 456:264. [DOI] [PubMed] [Google Scholar]
- 25. Ryman K. D., Klimstra W. B. 2008. Host responses to Alphavirus infection. Immunol. Rev. 225:27. [DOI] [PubMed] [Google Scholar]
- 26. Ryman K. D., Klimstra W. B., Nguyen K. B., Biron C. A., Johnston R. E. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gläsker S., Töller M., Kümmerer B. M. 2014. The alternate triad motif of the poly(ADP-ribose) polymerase-like domain of the human zinc finger antiviral protein is essential for its antiviral activity. J. Gen. Virol. 95(Pt 4):816. [DOI] [PubMed] [Google Scholar]
- 28. Kerns J. A., Emerman M., Malik H. S. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sadler A. J., Williams B. R. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayakawa S., Shiratori S., Yamato H., et al. 2011. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat. Immunol. 12:37. [DOI] [PubMed] [Google Scholar]
- 31. Lin Y., Zhang H., Liang J., et al. 2014. Identification and characterization of Alphavirus M1 as a selective oncolytic virus targeting ZAP-defective human cancers. Proc. Natl Acad. Sci. USA 111:E4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang X. H., Goldman J. E., Jiang H. H., Levine B. 1999. Resistance of interleukin-1beta-deficient mice to fatal Sindbis virus encephalitis. J. Virol. 73:2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klimstra W. B., Ryman K. D., Bernard K. A., Nguyen K. B., Biron C. A., Johnston R. E. 1999. Infection of neonatal mice with sindbis virus results in a systemic inflammatory response syndrome. J. Virol. 73:10387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu Y., Wang X., Goff S. P., Gao G. 2012. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J. 31:4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liang J., Song W., Tromp G., Kolattukudy P. E., Fu M. 2008. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS One 3:e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hurt J. A., Obar R. A., Zhai B., Farny N. G., Gygi S. P., Silver P. A. 2009. A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. J. Cell Biol. 185:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen C. Y., Gherzi R., Ong S. E., et al. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451. [DOI] [PubMed] [Google Scholar]
- 38. Matsushita K., Takeuchi O., Standley D. M., et al. 2009. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature 458:1185. [DOI] [PubMed] [Google Scholar]
- 39. Maeda M., Sawa H., Tobiume M., et al. 2006. Tristetraprolin inhibits HIV-1 production by binding to genomic RNA. Microbes Infect. 8:2647. [DOI] [PubMed] [Google Scholar]
- 40. Lin R. J., Chien H. L., Lin S. Y., et al. 2013. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 41:3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lin R. J., Chu J. S., Chien H. L., et al. 2014. MCPIP1 suppresses hepatitis C virus replication and negatively regulates virus-induced proinflammatory cytokine responses. J. Immunol. 193:4159. [DOI] [PubMed] [Google Scholar]