Abstract
Stage-specific embryonic antigen-4 (SSEA-4) is a glycosphingolipid, which is overexpressed in some cancers and has been linked to disease progression. However, little is known about the functions of SSEA-4 and the characteristics of SSEA-4 expressing tumor cells. Our studies identified SSEA-4 expression on a subpopulation of cells in many solid tumor cell lines but not in leukemic cell lines. Fluorescence-activated cell sorting-sorted SSEA-4+ prostate cancer cells formed fibroblast-like colonies with limited cell–cell contacts, whereas SSEA-4− cells formed cobblestone-like epithelial colonies. Only colonies derived from SSEA-4+ cells were enriched for pluripotent embryonic stem cell markers. Moreover, major epithelial cell-associated markers Claudin-7, E-cadherin, ESRP1 and GRHL2 were down-regulated in the SSEA-4+ fraction of DU145 and HCT-116 cells. Similar to cell lines, SSEA-4+ primary prostate tumor cells also showed down-regulation of epithelial cell-associated markers. In addition, they showed up-regulation of epithelial-to-mesenchymal transition as well as mesenchymal markers. Furthermore, SSEA-4+ cells escape from adhesive colonies spontaneously and form invadopodia-like migratory structures, in which SSEA-4, cortactin as well as active pPI3K, pAkt and pSrc are enriched and colocalized. Finally, SSEA-4+ cells displayed strong tumorigenic ability and stable knockdown of SSEA-4 synthesis resulted in decreased cellular adhesion to different extracellular matrices. In conclusion, we introduce SSEA-4 as a novel marker to identify heterogeneous, invasive subpopulations of tumor cells. Moreover, increased cell-surface SSEA-4 expression is associated with the loss of cell–cell interactions and the gain of a migratory phenotype, suggesting an important role of SSEA-4 in cancer invasion by influencing cellular adhesion to the extracellular matrix.
Keywords: adhesion, epithelial-to-mesenchymal transition, invadopodia, invasion, SSEA-4
Introduction
Tumor heterogeneity refers to the existence of subpopulations of cells, with distinct genotypes and phenotypes that may harbor divergent biological behaviors, within a primary tumor or between a primary tumor and its metastases (intra- and inter-tumor, respectively). Intra-tumoral heterogeneity may give rise to distinct cellular phenotypes based on antigen expression and can lead to important consequences like differential response to chemotherapy and metastatic proclivity (Valeriote and van Putten 1975; Liu et al. 2013). A variety of cell surface-specific markers have been employed to prospectively isolate heterogeneous cancer cells using immunomagnetic or fluorescence-activated cell sorting (FACS) techniques. These markers include CD271, CD166, CD133, CD47, CD44, CD24, EpCAM, GD2, Tra-1-60 and several other molecules (Xia 2014), which appear to be expressed not only in tumors but also in normal epithelial stem cells, progenitor cells and mesenchymal stem cells (Sivasubramaniyan et al. 2012; Verga Falzacappa et al. 2012; Harichandan et al. 2013).
Cell surface antigens that have been used to characterize human embryonic stem cells (hESCs) include stage-specific embryonic antigen (SSEA)-3 and -4, as well as keratin sulfate-associated antigens TRA-1-60 and TRA-1-81 (Fong et al. 2009). Interestingly, all of these markers are carbohydrates carried on glycolipids or glycoproteins. SSEA-4 is a ganglioside which consists of a glycosphingolipid (GSL) containing a terminal sialic acid residue (N-acetylneuraminic acid). It is synthesized from SSEA-3 by the enzyme ST3 beta-galactoside alpha-2,3-sialyltransferase 2 (ST3GAL2; Saito et al. 2003). The expression of SSEA-4 undergoes qualitative and quantitative changes during development, differentiation and tumorigenesis. During human preimplantation development, SSEA-3 and -4 are first observed on the pluripotent cells of the inner cell mass and are lost upon differentiation (Tondeur et al. 2008). In addition, human germ stem cells in the testis and ovary express SSEA-4 (Harichandan et al. 2013; Virant-Klun et al. 2013).
Promotion or inhibition of tumorigenesis through glycosylation is of crucial importance in cancer research. Some studies show that GSL SSEA-4 is implicated in the malignancy of cancers, such as invasion and metastasis (Katagiri et al. 2001; Hung et al. 2013). The expression of SSEA-4 is associated with the increase in metastatic potential and poor prognosis of lung, renal, breast and oral cancer (Katagiri et al. 2001; Gottschling et al. 2013; Hung et al. 2013; Noto et al. 2013). Katagiri et al. (2005) reported that SSEA-4 is involved in laminin binding through interactions with the laminin receptor, laminin-binding protein 34/67.
So far, little is known about the functions of SSEA-4 in cancer and the characteristics of SSEA-4 expressing tumor cells. In this study, SSEA-4 was identified as a promising antigenic target of the de novo generated monoclonal antibody (mAb) IPS-K-4A2B8, which recognized distinct subpopulations of solid cancer cell lines. In addition, the role of SSEA-4 expression in regulation of different properties of cancer cells including adhesion, migration and tumorigenicity was investigated. We could demonstrate that SSEA-4 identifies tumor cells that undergo spontaneous loss of epithelial phenotype and might play a role in tumor progression by influencing cellular adhesion to extracellular matrix (ECM).
Results
Generation of mAbs reactive with subsets of tumor cells
This study was aimed to identify novel mAbs that recognize highly tumorigenic subpopulations of human cancer cells. For this purpose, we screened a large panel of in-house generated mAbs against cell surface antigens for their reactivity with different human solid cancer and leukemic-derived cell lines. In addition, novel mAbs with specific reactivity against cell surface molecules expressed on human induced pluripotent stem cell line 122 (iPS 122) were generated. In an initial screening effort, the reactivity analysis of selected mAbs with several cell lines revealed that most of antibody-defined antigens were homogenously present or absent on the majority of the tested cell lines. As shown in Supplementary Tables S1 and S2, most antibodies were unable to discriminate between distinct subpopulations in multiple cell lines. In contrast, mAbs IPS-K-1A6G5 and IPS-K-3C4A6 reacted with subpopulations of the testis cancer cell lines TCAM2, NT2, NCCIT and 2102Ep, whereas mAb IPS-K-4A2B8 (immunoglobulin class IgM) additionally reacted with subpopulations of cancer cell lines derived from other tissues including the breast, colon and prostate.
The heterogeneous reactivity profile of mAb IPS-K-4A2B8 prompted us to analyze its reactivity on a large number of solid tumor and leukemic cell lines. Interestingly, the mAb reacted with many solid tumor cell lines (Figure 1) but not with any of the screened leukemic cell lines (Supplementary Figure S1A).
Fig. 1.
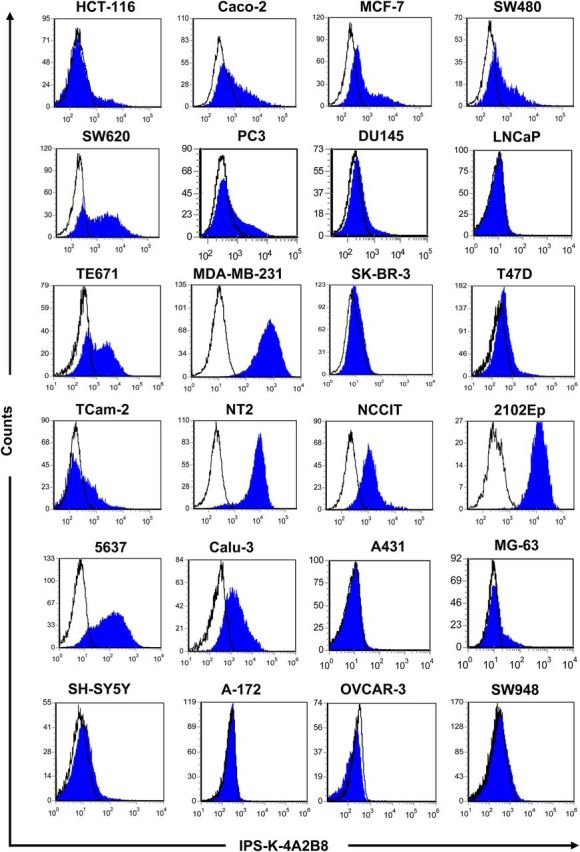
Reactivity profiles of mAb IPS-K-4A2B8 on solid tumor cell lines. Cells were labeled with mAb IPS-K-4A2B8 using indirect immunofluorescence staining as described in Methods. Cells were analyzed on a FACSCanto flow cytometer. Data were processed using the FCS Express software. Note that antibody IPS-K-4A2B8 reacts with most cells of embryonal carcinoma-derived cell lines (NT2, NCCIT, 2102Ep) and only with a subpopulation of cells in many other cancer cell lines (HCT-116, Caco-2, MCF-7, SW480, SW620, PC3, DU145, TE671, TCam-2, MG-63).
Target identification of IPS-K-4A2B8 antibody
The interesting reactivity profile of mAb IPS-K-4A2B8 with several solid tumor-derived cell lines prompted us to identify the target molecule. For this purpose, we applied several strategies.
IPS-K-4A2B8 antigen is associated with lipid rafts
To gain more insight into the distribution of IPS-K-4A2B8 antigen on the cell surface and to design effective antigen extraction procedures for target identification, the potential localization of the antigen in lipid rafts was studied. To assess the potential distribution of IPS-K-4A2B8 antigen in lipid rafts, the reactivity of the antibody was analyzed by flow cytometry on 1% Triton X-100 (TX-100) treated as well as untreated human iPS122 and DU145 cells. CD44, which is known to be resistant to TX-100 treatment (Filatov et al. 2003), was used as a control antigen. As shown in Figure 2A and B, IPS-K-4A2B8 epitope expression did not decrease after TX-100 treatment in both iPS122 and DU145 cells. This indicated that IPS-K-4A2B8 antigen is localized in the TX-100 resistant lipid raft membrane fraction of both iPS122 and DU145 cells.
Fig. 2.
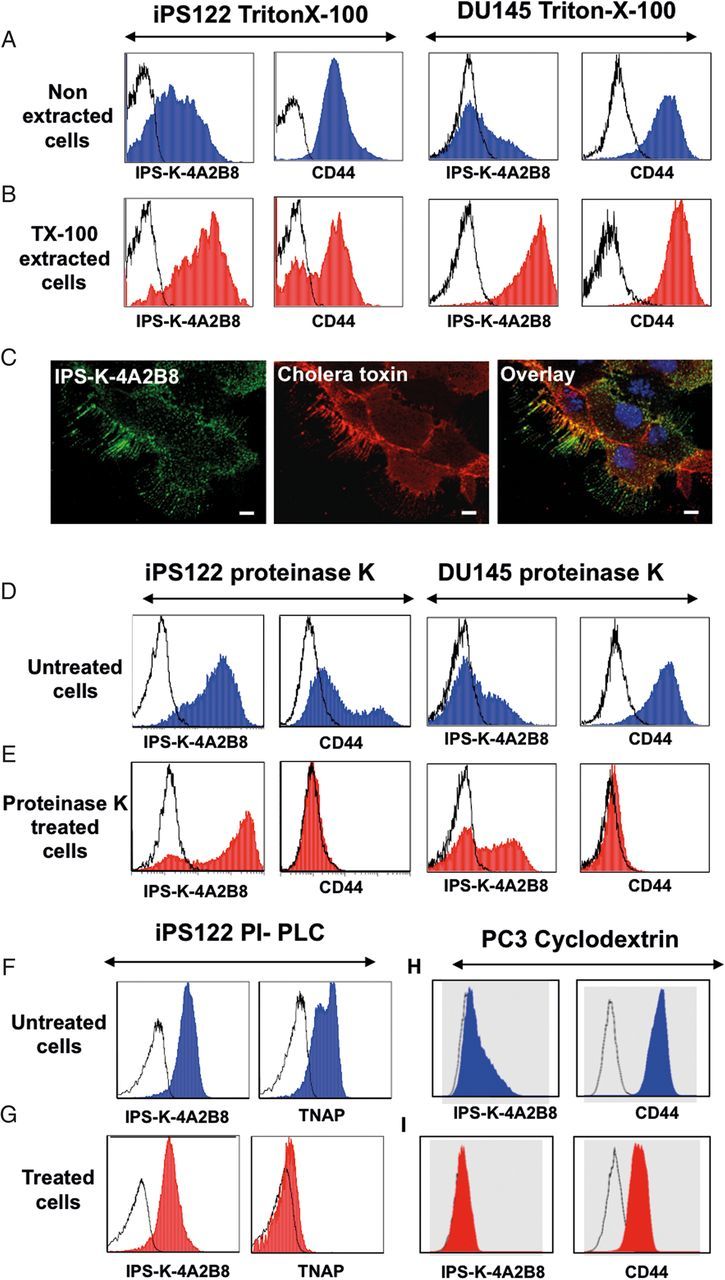
Antibody IPS-K-4A2B8 recognizes a GSL in lipid rafts. Open, blue and red histograms represent controls, untreated and treated cell staining, respectively. (A, B) Resistance of IPS-K-4A2B8 antigen to solubilization with TX-100. Native (A) and TX-100 (B) extracted iPS122 or DU145 cells were stained with IPS-K-4A2B8 or CD44 antibody followed by PE-labeled secondary antibody. Note that both CD44 and IPS-K-4A2B8 antigen are resistant to solubilization with TX-100. (C) IPS-K-4A2B8 antigen colocalizes with the lipid raft marker CTB in DU145 cells. DU145 cells were preincubated with mAb IPS-K-4A2B8 and then with the lipid raft marker Alexa Fluor 555-CTB at 4°C as described in Methods. IPS-K-4A2B8 antigen and CTB strongly colocalize at the plasma membrane. (D and E) Resistance of IPS-K-4A2B8 antigen to proteinase K. iPS122 and DU145 cells were treated with proteinase K to strip off the membrane proteins. Untreated (D) or treated (E) cells fixed with 1% paraformaldehyde and stained with IPS-K-4A2B8 or CD44 antibody. Note that the reactivity of mAb IPS-K-4A2B8 is retained, whereas CD44 expression is completely lost after proteinase K digestion. Untreated (F) and PI-PLC-treated (G) iPS122 cells were stained with IPS-K-4A2B8 or tissue non-specific alkaline phosphatase antibody (clone: IPS-K-4A2B8). (H and I) MβCD treatment abrogates binding of IPS-K-4A2B8 antibody. PC3 cells were incubated with (I) and without (H) 5 mM MβCD at 37°C for 1 h. The cells were then stained with anti-CD44 or IPS-K-4A2B8 antibody. Note that the binding of mAb IPS-K-4A2B8 is completely lost after MβCD treatment.
To confirm the localization of the IPS-K-4A2B8 antigen to lipid rafts, we used Alexa Fluor 555 conjugated cholera toxin B (CTB) subunit, which binds to GM1 ganglioside, a component of lipid raft membrane microdomains in the plasma membrane. As illustrated in Figure 2C, IPS-K-4A2B8 antigen colocalized with CTB in DU145 cells, thus supporting the view that IPS-K-4A2B8 antigen is expressed in lipid rafts.
Antibody IPS-K-4A2B8 recognizes a GSL in lipid rafts
Lipid rafts are highly enriched for GSLs, glycophosphatidylinositol (GPI)-anchored proteins and glycoproteins (Varma and Mayor 1998). To determine which of these components is recognized by mAb IPS-K-4A2B8, the effect of proteinase K, phosphoinositol phospholipase C (PI-PLC) and methyl-beta-cyclodextrin (MβCD) treatment was studied.
To analyze whether the recognized epitope of mAb IPS-K-4A2B8 is a surface protein, the reactivity of mAb IPS-K-4A2B8 with proteinase K treated and non-treated DU145 cells as well as iPS122 cells was compared. CD44 was used as a control antigen (sensitive to proteinase K treatment). As shown in Figure 2D and E, the reactivity of anti-CD44 mAb was lost after proteinase K treatment. In contrast, an increase in the reactivity of IPS-K-4A2B8 mAb after proteinase K treatment indicates that the recognized epitope of mAb IPS-K-4A2B8 is not a cell surface protein. Similarly, removal of GPI-anchored proteins after PI-PLC treatment did not inhibit the binding of mAb IPS-K-4A2B8, whereas it completely abrogated the binding of antibody IPS-K-4A4F2 which recognizes the PI-linked molecule tissue non-specific alkaline phosphatase (TNAP; control) (Figure 2F and G). To analyze whether the target of mAb IPS-K-4A2B8 is a glycolipid, DU145 cells were treated with MβCD, which is known to deplete cholesterol from the cell membrane (Ostermeyer et al. 1999) and affects the cell surface localization of GSLs. As shown in Figure 2H and I, treatment of cells with MβCD resulted in complete abrogation of the binding of mAb IPS-K-4A2B8. Collectively, these data indicate that IPS-K-4A2B8 antigen is a GSL in the lipid rafts.
Antibody IPS-K-4A2B8 recognizes the extracellular region of GSL SSEA-4
To shed further light on the target specificity of mAb IPS-K-4A2B8, we performed glycan-binding analysis of Alexa fluor 555 conjugated IPS-K-4A2B8 on a glycan array comprising of 611 glycan targets. As shown in Supplementary Figure S1B, IPS-K-4A2B8 bound with very high affinity (30,000 RFU) to a single glycan Neu5Aca2-3Galb1-3GalNAcb1-3Gala1-4Galb1-4Glcb-Sp0, which corresponds to SSEA-4. To further confirm the specificity of antibody IPS-K-4A2B8 for SSEA-4, glycan array analysis was performed with the commercially available SSEA-4 antibody MC-813-70. As expected, MC-813-70 also bound to SSEA-4 glycan but with a 50-fold lower affinity (600 RFU) compared with mAb IPS-K-4A2B8 (Supplementary Figure S1C). Both antibodies did not bind to any other glycan in the array demonstrating their high specificity for SSEA-4. Comparison of reactivity profiles of IPS-K-4A2B8 and the commercially available mAb MC-813-70 showed that IPS-K-4A2B8 gave rise to a superior separation capacity (Supplementary Figure S2A–D).
To further prove the specificity of IPS-K-4A2B8 mAb, blocking experiments with SSEA-4 molecule, GM1b glycan and structurally related SSEA-3 molecule were performed. As shown in Supplementary Figure S2E, preincubation of SSEA-4 with mAb IPS-K-4A2B8 showed a significant inhibition (87.94%) of mAb IPS-K-4A2B8 binding to NT-2 cells when compared with the control. In contrast, preincubation of SSEA-3 or GM1b with mAb IPS-K-4A2B8 did not significantly influence the binding of mAb IPS-K-4A2B8 to cells (Supplementary Figure S2E and Table S3). These results verify the specificity of IPS-K-4A2B8 antibody for SSEA-4.
SSEA-4+ cells in human solid cancer cell lines are enriched for novel pluripotent stem cell markers
To determine the phenotype of SSEA-4+ and SSEA-4− cells, DU145, HCT-116, PC3 and MCF-7 cells were fractionated by FACS into SSEA-4+ and SSEA-4− cells (Figure 3A, Supplementary Figure S3) and plated into culture flasks at defined cell numbers (50,000 cells per T25). Strikingly, SSEA-4+ DU145 cells gave rise to spindle-shaped, fibroblast like colonies, with limited cell–cell contacts. Conversely, SSEA-4− cells gave rise to colonies with cobblestone-like epithelial morphology (Figure 3B). After 4–11 days, adherent cells were trypsinized and stained with markers indicated in Figure 3C. Flow cytometric analysis revealed that SSEA-4+ DU145 and HCT-116 cells were highly enriched for the markers of pluripotent embryonic stem cells Tra-1-60 and Tra-1-81 compared with SSEA-4− cells or unfractionated cells (Figure 3C and Supplementary Figure S3A). Similarly, SSEA-4+ MCF-7 cells were highly enriched for the pluripotent embryonic stem cell marker SSEA-3 (Supplementary Figure S3B). In contrast to DU145, HCT-116 and MCF-7 cells, the embryonic markers Tra-1-60, Tra-1-81 or SSEA-3 were not enriched in SSEA-4+ PC3 cells as the parental PC3 cells do not express these markers (Supplementary Figure S3C). Notably, SSEA-4− cells from all cell lines spontaneously generated SSEA-4+ cells and the size of the positive population was almost identical to that of parental cells. Similarly, SSEA-4+ cells also generated SSEA-4− cells.
Fig. 3.
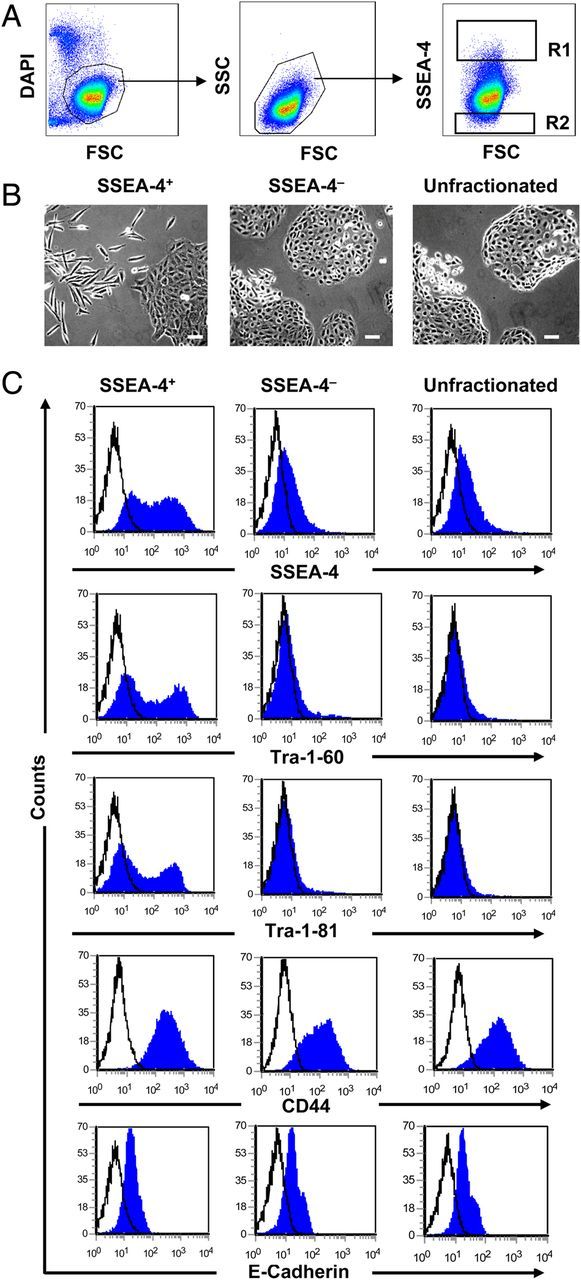
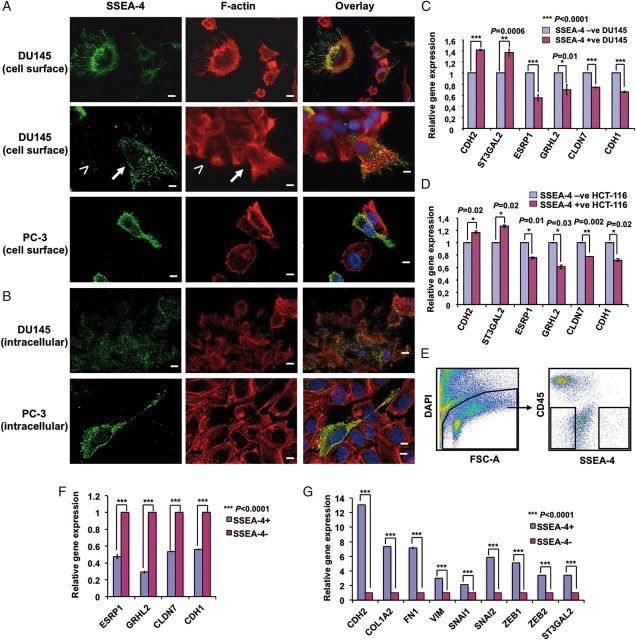
SSEA-4 defines a distinct subpopulation of DU145 cells. (A) Display of SSEA-4 expression on DU145 cells. Sort windows R1 and R2 were set and cells were sorted on a FACS Aria cell sorter. (B) Morphology of SSEA-4+ and SSEA-4− cells. When cultured, SSEA-4+ DU145 cells gave rise to fibroblast-like colonies with limited cell–cell contacts. Conversely, the SSEA-4− cells gave rise to cobblestone like epithelial colonies. Scale bars: 20 µm. (C) Phenotypic characterization of SSEA-4+ and SSEA-4− DU145 cells after 11 days of culture. Note that SSEA-4+ cells are highly enriched for pluripotent stem cell markers Tra-1-60 and Tra-1-81.
Localization of SSEA-4 to polarized cells with long filopodia like structures
To further confirm that SSEA-4 is enriched in fibroblast-like spindle-shaped cells, the subcellular localization of SSEA-4 along with F-actin was analyzed on DU145 and PC3 cells by immunofluorescence microscopy. Cell surface staining using IPS-K-4A2B8 mAb and F-actin staining with phalloidin demonstrated that SSEA-4 accumulated predominantly in polarized cells with long filopodia-like structures in both cell lines (Figure 4A). However, some non-polarized cells also expressed SSEA-4. Strikingly, a few peripheral cells in epithelial islands of DU145 change shape and show a similar morphology as migratory fibroblast-like cells, which expressed SSEA-4 on the cell surface (Figure 4A). To determine the intracellular distribution of SSEA-4 as well, cells were permeabilized with TX-100 prior to incubation with antibodies. Weak staining of SSEA-4 was detected in the cytoplasm of all DU145 cells. In contrast to DU145 cells, most of cell surface SSEA-4-negative PC3 cells were negative for cytoplasmic SSEA-4 (Figure 4B).
Fig. 4.
Characterization of SSEA-4 expressing cells. (A and B) Localization of SSEA-4 on DU145 and PC3 cells. (A) Adherent DU145 and PC3 cells were surface labeled with SSEA-4, fixed and then permeabilized, followed by staining with Alexa Fluor 488-labeled secondary antibody and Rhodamine-labeled phalloidin. Cells were mounted in DAPI containing mounting media as described in Methods. Note the conversion of cells from a colonial epithelial morphology (less than symbol) into a polarized, migratory mesenchymal morphology (up-arrow) and the localization of SSEA-4 to the plasma membrane of polarized cells. (B) Adherent cells were fixed, permeabilized, labeled with IPS-K-4A2B8 followed by staining with Alexa Fluor 488-labeled secondary anti-mouse IgM antibody and Rhodamine-labeled phalloidin. Note weak staining of SSEA-4 in the cytoplasm of all DU145 cells. In contrast to DU145 cells, most PC3 cells that were negative for cell surface SSEA-4 did not express cytoplasmic SSEA-4. Scale bars: 10 µm. (C and D) Expression of several EMT-related genes are modulated in SSEA-4+ and SSEA-4− DU145 as well as HCT-116 cells. Relative gene expression of EMT markers on SSEA-4+ and SSEA-4− (C) DU145 as well as (D) HCT-116 cells shows higher expression of mesenchymal marker (CDH2) in SSEA-4+ subset and epithelial markers (ESRP1, GRHL2, CLDN7 and CDH1) in SSEA-4− cells. Also note the increase in the expression of ST3GAL2 in SSEA-4+ cells. Shown are data from one representative biological experiment performed in technical replicates, quantified relatively to SSEA-4− cells and normalized to GAPDH. Error bars denote mean ± standard error. p-values were calculated using Student's t-test. (E) Display of SSEA-4 expression on primary prostate cancer cells. Sort windows were set and cells were sorted on a FACS Aria cell sorter. (F and G) Differential expression of EMT-related genes in SSEA-4+ and SSEA-4− primary prostate tumor cells. (F) Relative gene expression of EMT-related markers in SSEA-4+ and SSEA-4− cells shows higher expression of epithelial markers in SSEA-4− cells. (G) In contrast, SSEA-4+ cells show an increased expression of mesenchymal markers (CDH2, COL1A2, FN1 and VIM), EMT markers (SNAI1, SNAI2, ZEB1 and ZEB2) and SSEA-4 synthesizing enzyme ST3GAL2. Data are shown from one patient sample performed in technical replicates, quantified relatively to SSEA-4− cells and normalized to GAPDH. Error bars denote mean ± standard error. p-values were calculated using Student's t-test.
Markers defining epithelial phenotype are enriched in SSEA-4− cells
To gain molecular insight into the expression profiles of SSEA-4+ and SSEA-4− cells, whole-genome microarray analysis of sorted PC3 cells was performed. The positive fraction contained 3% of cells expressing the highest levels of SSEA-4, whereas the negative fraction was completely negative for SSEA-4. Gene chip analysis of sorted SSEA-4− cells revealed increased expression of epithelial cell markers including CDH1 (also known as E-cadherin), epithelial-specific splicing factor 1 (ESRP1), grainyhead transcription factor 2 (GRHL2) and tight junction protein CLDN7 when compared with cells of the SSEA-4+ fraction (data not shown). The microarray data were further verified by quantitative real-time polymerase chain reaction (PCR) analysis. Consistent with the microarray data, sorted SSEA-4− cells from DU145 and HCT-116 showed an increased expression of CDH1, ESRP1, GRHL2 and CLDN7 compared with SSEA-4+ cells (Figure 4C and D). We also included the mesenchymal marker CDH2 (also known as N-cadherin) as well as SSEA-4 synthesizing enzyme ST3GAL2 (EC 2.4.997) in our analysis. As expected, both markers were up-regulated in the SSEA-4+ cells compared with SSEA-4− cells (Figure 4C and D). Collectively, these data indicate that SSEA-4 is selectively enriched in cells that undergo spontaneous epithelial to mesenchymal transition (EMT).
To investigate whether a fraction of SSEA-4+ cells is also observed in primary tumor cells and whether the expression profile of this population correlates with that of cell lines, primary prostate tumor cells were stained with anti-SSEA-4 antibody and analyzed by multiparameter flow cytometry. After exclusion of CD45+ hematopoietic cells from the dissociated tumor samples, the remaining cells were fractionated into SSEA-4+ and SSEA-4− cells (Figure 4E) and analyzed for gene expression. Quantitative real-time PCR analysis revealed an increased expression of epithelial phenotype-related genes CDH1, ESRP1, GRHL2 and CLDN7 in sorted SSEA-4− cells compared with SSEA-4+ cells (Figure 4F). In contrast, mesenchymal phenotype-related genes CDH2, COL1A2, FN1 and VIM and EMT-related genes SNAI1, SNAI2, ZEB1, ZEB2 and SSEA-4 synthesizing enzyme ST3GAL2 were up-regulated in SSEA-4+ cells compared with SSEA-4− cells (Figure 4G).
Accumulation of SSEA-4 and integrins in F-actin-rich cellular structures
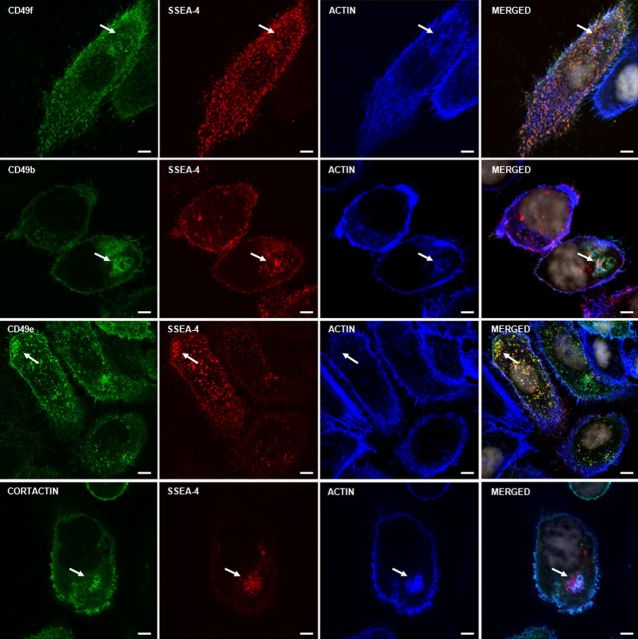
During the course of our study, we noticed strong colocalization of SSEA-4 with F-actin to cell motility structures and some unknown cellular compartments. This raised our interest in evaluating the role of SSEA-4 in cell adhesion and migration. One mechanism by which GSLs could affect cell adhesion is via their interaction with integrins (Zheng et al. 1993; Sharma et al. 2005). We therefore examined colocalization of SSEA-4 or F-actin with integrins on PC3 cells. Figure 5 and Supplementary Figure S4 shows that integrins CD49b, CD49e and CD49f accumulated in SSEA-4 and F-actin-rich puncta. These results indicate that F-actin accumulating cellular structures are highly enriched for SSEA-4 and integrins.
Fig. 5.
Localization of SSEA-4, F-actin and integrins on PC3 cells. Three color staining of adherent PC3 cells with anti-integrins, phalloidin (F-actin) and mAb IPS-K-4A2B8 shows that integrins CD49f, CD49e and CD49b colocalize with SSEA-4 and F-actin accumulating spots. Arrows point to accumulation of integrins at F-actin and SSEA-4 accumulating structures. Scale bars: 5 µm.
Identification of the cellular compartment enriched for SSEA-4
Next, we were interested to identify the cellular compartment in which SSEA-4, F-actin and integrins accumulate. For this purpose, we studied the potential distribution of SSEA-4 as well as F-actin in various intra and subcellular compartments in PC3 cells using compartment-specific antibodies. As GSLs are frequently internalized via caveolae to the Golgi apparatus (Le and Nabi 2003), we examined the accumulation of SSEA-4 as well as F-actin in caveolae and Golgi apparatus by using the cis-Golgi marker GM130 and the caveolae-specific marker caveolin-1. GM130 and caveolin-1 showed no overlap with SSEA-4 and F-actin. These data demonstrate that SSEA-4 and F-actin do not accumulate in the Golgi apparatus and in caveolae or caveolin-1 enriched early endosomes. However, examination at higher magnification showed GM130 to be present adjacent to SSEA-4 and F-actin accumulating structures (Figure 6 and Supplementary Figure S5).
Fig. 6.
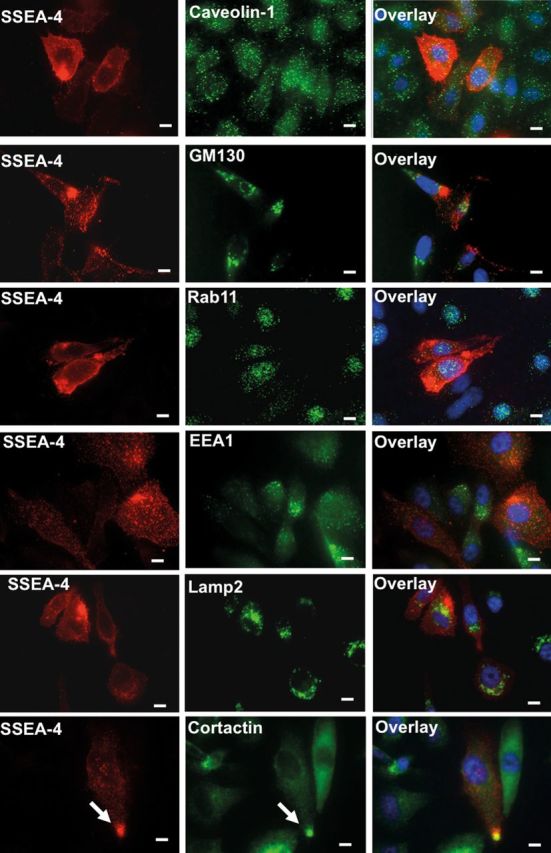
Localization of SSEA-4 and organelle-specific markers on PC3 cells. Adherent PC3 cells were stained for organelle-specific markers including caveolin-1 (caveolae), GM130 (Golgi apparatus), Rab11 (recycling endosomes), EEA1 (early endosomes), Lamp2 (lysosomes) or cortactin (invadopodia) and SSEA-4. SSEA-4 accumulating structures showed no colocalization with caveolin-1, GM130, Rab11, EEA1 and Lamp2, suggesting that SSEA-4 accumulating puncta does not correspond to caveolae, Golgi apparatus, recycling endosomes, early endosomes and lysosomes. On the other hand, SSEA-4 strongly colocalized with cortactin (marked by up-arrow), indicating that SSEA-4 accumulating puncta correspond to invadopodia. Scale bars: 10 µm.
In subsequent experiments, accumulation of SSEA-4 and F-actin in recycling endosomes, early endosomes, lysosomes and invadopodia were tested. For this purpose, the recycling endosomal marker Rab11, the early endosomal marker EEA-1, the lysosomal marker LAMP-2 and the invadopodia specific marker cortactin were employed. SSEA-4 and F-actin showed no colocalization with Rab11, EEA-1 or LAMP-2 (Figure 6 and Supplementary Figure S5), suggesting that SSEA-4 and F-actin accumulating puncta do not correspond to recycling endosomes, early endosomes or lysosomes. In contrast, SSEA-4 and F-actin accumulating puncta strongly overlapped with cortactin, suggesting that SSEA-4 and F-actin-rich cellular structures correspond to invadopodia (Figure 6, Supplementary Figures S4 and S5).
Invadopodia are enriched in expression of SSEA-4 and signaling molecules
The formation of invadopodia in human cancer cells requires the activity of PI3K, Akt and Src (Ayala et al. 2008; Yamaguchi et al. 2011). Hence, it is expected that active signaling molecules including phosphorylated PI3K, Akt and Src (pPI3K, pAkt and pSrc) accumulate in invadopodia. Therefore, we examined the potential colocalization of SSEA-4 and active signaling molecules. As shown in Figure 7A, SSEA-4 expression colocalized with the expression of pSrc, pPI3K and pAkt. Although pSrc accumulated to some extent in most SSEA-4-rich structures, it also accumulated in SSEA-4− compartments. However, pPI3K expression mostly overlapped with SSEA-4 expression. Taken together, these data confirm that SSEA-4 accumulates in invadopodia.
Fig. 7.
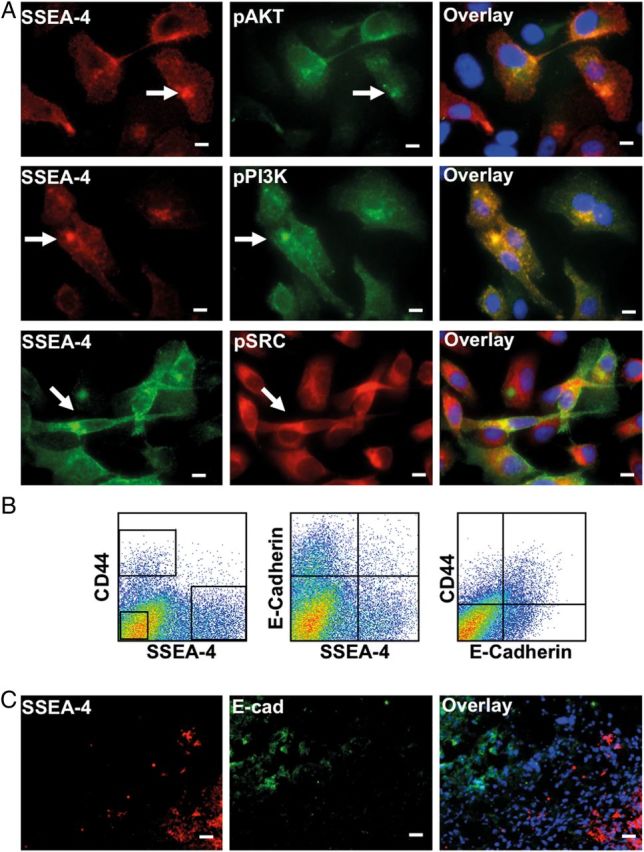
(A) Invadopodia are enriched in SSEA-4 and active signaling molecules. Adherent PC3 cells were stained for SSEA-4 and active signaling molecules. Note the colocalization of pPI3K, pAkt and pSrc at SSEA-4 accumulating structures (marked by up-arrow). Scale bars: 10 µm. (B and C) SSEA-4+ cells show increased tumorigenicity in vivo in immunopermissive NSG mice. (B) Flow cytometric analysis of the xenografts derived from unfractionated DU145 cells revealed that most SSEA-4 expressing cells did not coexpress E-cadherin. In contrast, most CD44 expressing cells coexpressed E-cadherin. (C) Immunohistochemical staining of the xenografts derived from unfractionated DU145 cells confirmed that SSEA-4 and E-cadherin do not colocalize. SSEA-4 was mainly expressed by the cells on the periphery of the tumor, whereas E-cadherin was expressed by the cells located in the core of the tumor. Scale bars: 10 µm.
SSEA-4+ cells show increased tumorigenicity in vivo
To explore the relevance of SSEA-4 expression in vivo, we performed xenotransplantation of SSEA-4+ and SSEA-4− DU145 cells in immunopermissive NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. The same numbers of unfractionated, SSEA-4+ and SSEA-4− cells were implanted subcutaneously in the flanks of 4–5 months old male mice and tumor induction was monitored every week by palpation of the injection sites. When 150,000 or 50,000 cells were injected per flank, all populations including unfractionated, SSEA-4+ and SSEA-4− cells generated tumors after 3–4 weeks post-injection in all animals. Reducing the number of transplanted cells to 3000 cells per animal reduced the tumor formation from SSEA-4− cells and tumors appeared only in two of six injection sites. In contrast, SSEA-4+ cells robustly induced tumors in all transplanted animals (Supplementary Table S4). The xenografts derived from unfractionated DU145 cells were analyzed for the expression of SSEA-4 in combination with markers known to associate with prostate cancer cells including E-cadherin and CD44 (Gao et al. 1997; Kallakury et al. 2001). Flow cytometric analysis revealed that the majority of SSEA-4+ cells are not positive for CD44 and E-cadherin (Figure 7B). To further confirm that E-cadherin and SSEA-4 do not coexpress, the xenograft was stained for these markers by immunohistochemistry (IHC). As expected, IHC staining revealed that SSEA-4 and E-cadherin do not show coexpression (Figure 7C). Notably, SSEA-4 expression was enriched in the tumor periphery but not in the tumor core.
In subsequent experiments, we compared the tumorigenic ability of SSEA-4+ and CD44+ cells. For this purpose, xenografts derived from unfractionated DU145 cells were harvested and stained with anti-SSEA-4 and CD44 antibody. Three thousand cells of unfractionated, SSEA-4+CD44−, SSEA-4−CD44+ and SSEA-4−CD44− cells were sorted by FACS and injected subcutaneously. Recording of tumor size and number after 80 days of transplantation showed that 5/6, 3/6 and 2/6 mice injected with SSEA-4+CD44−, SSEA-4−CD44− and SSEA-4−CD44+ cells gave rise to tumors, respectively (Supplementary Figure S6A). Furthermore, tumors derived from SSEA-4+ cells were comparatively larger than the tumors derived from CD44+ cells (Supplementary Figure S6B). These results show that SSEA-4+CD44− cells display higher tumorigenic propensity than SSEA-4−CD44+ cells. However, further studies are needed to analyze the tumorigenic ability of SSEA-4+CD44+ cells. Since we observed only a 2- to 3-fold difference in tumor initiation between SSEA-4+ and SSEA-4− populations, we investigated whether this was due to the generation of SSEA-4+ cells from SSEA-4− cells and vice versa. For this purpose, we harvested the tumors generated from different groups of mice injected with unfractionated, SSEA-4+CD44− cells or SSEA-4−CD44+ cells, after 80 days of transplantation, and triple stained the single-cell suspension for SSEA-4, CD44 and E-cadherin. Flow cytometric analysis revealed that SSEA-4− cells spontaneously achieve a SSEA-4+ phenotype and vice versa (Supplementary Figure S6C). Also, the expression profiles of CD44 and E-cadherin were almost similar among the different groups.
Down-regulation of SSEA-4 alters cell adhesion
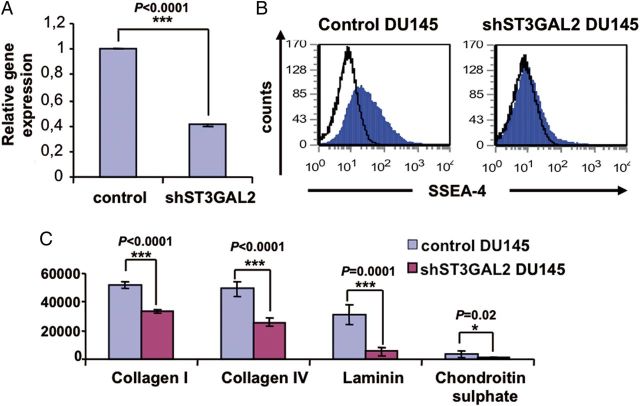
To investigate the functional role of SSEA-4 in cancer, we stably suppressed the expression of ST3GAL2, the critical enzyme involved in the biosynthesis of SSEA-4. Suppression of ST3GAL2 was achieved by using a lentiviral-based small hairpin RNA (shRNA) expression vector in DU145 cells. As shown in Figure 8A, an about 60% knockdown in gene expression was achieved. Cells transduced with empty lentiviruses (construct of vector PLKO.1-Puro without shST3GAL2) were used as controls. As expected, flow cytometric analysis revealed that ST3GAL2 knockdown reduced the percentage of SSEA-4+ cells from 21.5 to 4% in DU145 cells (Figure 8B).
Fig. 8.
ST3GAL2 knockdown impairs SSEA-4 expression and cell adhesion. (A and B) Modulation of ST3GAL2 mRNA in DU145 cells by lentiviral ST3GAL2 knockdown impairs SSEA-4 expression. Panel (A) shows relative gene expression levels of ST3GAL2 normalized to internal standard GAPDH of three biological experiments each performed in triplicates. Error bars denote mean ± standard error. p-values were calculated using the Student's t-test. Panel (B) shows representative flow cytometry analysis of SSEA-4 expression in the control and knockdown DU145 cells. (C) Adhesion assays show that the number of adherent cells was significantly higher in the control compared with the knockdown DU145 cells under all matrix conditions. Error bars denote mean ± standard error. p-values were calculated using the Student's t-test; n = 5 per group.
We next analyzed the role of SSEA-4 in cell adhesion. The effect of ST3GAL2 knockdown on DU145 cell adherence to different ECM components including collagen I, collagen IV, chondroitin sulfate and laminin was assessed using the fluorometric cell adhesion assay. The results show that the efficiency of adhesion to collagen I, collagen IV, laminin and chondroitin sulfate was 1.5, 1.9, 5.9 and 3.9 times higher in the control compared with the knockdown DU145 cells (Figure 8C). These results show that SSEA-4 is involved in cellular adhesion.
Discussion
In this study, we identified the ganglioside SSEA-4 as a marker for detecting intra-tumor heterogeneity. Among the tested cell lines, SSEA-4 expression was exclusively found in cells derived from solid tumors but not from leukemic blasts, independent of the fact that all cell lines expressed ST3GAL2, an enzyme involved in SSEA-4 synthesis. In most cases, SSEA-4+ tumor cells displayed high levels of embryonic stem cell-specific markers including SSEA-3, Tra-1-60 and Tra-1-81. SSEA-4 was found to be expressed predominantly in the E-cadherin−CD44− fraction isolated from xenografts induced by DU145 prostate cancer cells. In addition, the gene expression signature derived from SSEA-4+ cells strikingly correlated with the loss of epithelial cell-specific markers and gain of mesenchymal cell-specific markers. Moreover, inhibition of SSEA-4 synthesis using shRNA against ST3GAL2 resulted in a significant reduction in cell adhesion of DU145 cells to different ECM molecules. Of note, SSEA-4 predominantly accumulated in the filopodia and invadopodia of PC3 prostate cancer cells. This is of particular significance because the ability to form invadopodia is known to closely correlate with the invasive and metastatic properties of tumor cells (Yamaguchi et al. 2005, 2009). Finally, in vivo experiments demonstrated high tumorigenic capacity of SSEA-4+ DU145 cells compared with their negative counterparts. Collectively, our results indicate that SSEA-4 is a marker to identify invasive tumorigenic cells, which may be responsible for the development of metastasis.
Of note, among the prostate cancer cell lines used in this study, PC3 cells displayed a higher percentage of SSEA-4+ cells than DU145 cells, whereas LNCaP cells were completely negative for SSEA-4. This observation is in line with the finding that PC3 cells show a higher metastatic potential than DU145 cells or LNCaP cells, which are characterized by a moderate or very low metastatic potential, respectively (Pulukuri et al. 2005).
Many investigators showed that tumorigenic cells undergo reversible changes in the expression of many markers. Chaffer et al. (2011) reported that FACS selected CD44loCD24hi transformed human mammary epithelial cells could give rise to stem-like CD44hiCD24lo cells and vice versa. In addition, recent reports proposed that more differentiated cancer cells spontaneously acquired stem cell-like properties through dedifferentiation (Quintana et al. 2010; Battula et al. 2012). In line with these findings, we detected a spontaneous generation of SSEA-4+ cells from SSEA-4− DU145, HCT-116, MCF-7 or PC3 cells and vice versa. The fact that the difference of tumor initiation capacity in SSEA-4+ cells was only 2–3-fold higher than that in SSEA-4− DU145 cells can be explained by the dynamic phenotypic plasticity allowing resynthesis of SSEA-4 in SSEA-4− cells. In order to prevent switching on of SSEA-4 synthesis in SSEA-4− DU145 cells, we generated a stable ST3GAL2 knockdown. Our preliminary data indicate that the tumorigenic potential of SSEA-4+ DU145 cells is much more pronounced than that of SSEA-4− shST3GAL2 DU145 cells.
Among the cell surface markers differentially expressed on SSEA-4+ and SSEA-4− DU145, HCT-116 and MCF-7 cells, we found that Tra-1-60, Tra-1-81 or SSEA-3 were highly enriched in SSEA-4+ cells. In line with this finding, Rajasekhar et al. (2011) introduced Tra-1-60 as a key marker to isolate tumor initiating cells from primary prostate tumors and prostate cancer cell lines. In addition, Chang et al. (2008) reported that SSEA-3 is absent in normal breast tissue but highly expressed in breast cancer stem cells derived from primary tumors. Moreover, as cancer cells are known to acquire characteristics of stem cells such as self-renewal, differentiation (Malecki et al. 2013) and expression of embryonic antigens (Fong et al. 2009), it is likely that SSEA-4+ cancer cells consist of more stem cell-like cells.
Xiang et al. showed that GRHL2 is an epithelial-specific transcription factor that determines the epithelial phenotype of breast cancer cells. The loss of its expression during tumor progression contributes to loss of the epithelial phenotype leading to EMT (Xiang et al. 2012). By using multiple functional assays, they confirmed that GRHL2 plays a dominant role in maintaining the epithelial phenotype, and reduced or increased GRHL2 levels in cancer cells lead to EMT or MET induction, respectively. In addition, GRHL2 regulates a broad range of epithelial genes including epithelial-specific splicing factor (ESRP1), cell adhesion and tight junction (CDH1, TJP2, CLDN4 and CLDN7) and Wnt ligands (WNT7A, WNT7B). These data are in line with our finding that SSEA-4+ cells show reduced expression of epithelial genes CDH1, CLDN7, GRHL2 and ESRP1. Hence, the down-regulation of epithelial markers in SSEA-4+ cells may contribute to loss of the epithelial phenotype and promote EMT.
In our study, we observed that most of SSEA-4+ DU145 cells derived from xenografts did not coexpress CD44 and E-cadherin. However, most of CD44+ cells did coexpress E-cadherin. This is of particular significance because a reduction in cell–cell adhesion due to down-regulation of E-cadherin is known to associate with the invasive and metastatic phenotype of many carcinomas (Hirohashi 1998). In addition, the loss of CD44 is reported to associate with transformation, particularly in Burkitt's lymphoma, neuroblastoma and prostate cancer (Gao et al. 1997; Kallakury et al. 2001; Kauffman et al. 2003).
Katagiri et al. (2005) demonstrated that SSEA-4 is involved in laminin binding through interactions with the laminin receptor, laminin-binding protein 34/67. Moreover, several investigators proposed that an increase in invasiveness of tumor cells is associated with the ability of these cells to attach to laminin (Loeber and Runyan 1990; Yudoh et al. 1995; Brenner et al. 2000). In addition, YIGSR, a pentapeptide which is known to inhibit the laminin-mediated cell attachment, also inhibited the cell migration induced by the cell attachment to laminin (Loeber and Runyan 1990; Yudoh et al. 1995; Brenner et al. 2000). In line with this, we found that cell adhesion to laminin was significantly reduced by knockdown of ST3GAL2, suggesting that SSEA-4 participates in tumor invasion by assisting tumor cells to adhere to laminin.
A number of studies demonstrated a decrease in the expression of GSL SSEA-4 during differentiation of hESCs (Park et al. 2010; Stelling et al. 2013). So far, little is known about the role of SSEA-4 in both embryonic stem cells and cancer. Although each type of GSL has the potential for a specific biological function, GSLs, in general, have been suggested to be associated with signal transducers (Src family kinases and small G-proteins), tetraspanins, growth factor receptors and integrins. Such organizational framework, defining GSL modulated or dependent cell adhesion, motility and growth, is termed glycosynapse (Hakomori Si 2002). Glycosynapses are known to participate in (i) modulation of growth factor receptor activities through intrinsic tyrosine kinases (Mutoh et al. 1995; Kaucic et al. 2006), (ii) modulation of integrin function (Mitsuzuka et al. 2005; Sharma et al. 2005) and (iii) interaction with cytoplasmic signal transducers such as Src family kinases and small G proteins (Iwabuchi et al. 1998; Kasahara and Sanai 2000). In agreement with the above mentioned functions of glycosynapses, we propose that SSEA-4 modulates cell adhesion and migration through its interactions with integrins, F-actin and signaling molecules. This view is based on the following observations: (i) decreased attachment of DU145 cells to laminin after ST3GAL2 knockdown, (ii) colocalization of SSEA-4 with F-actin and integrins, (iii) colocalization of SSEA-4 with active signaling molecules including pSrc, pPI3K and pAkt and (iv) accumulation of SSEA-4 in invadopodia.
In conclusion, we introduce SSEA-4 as a marker to identify tumor cells that undergo spontaneous partial loss of the epithelial phenotype and gain of the mesenchymal-like migratory phenotype. Future studies may corroborate a variety of aspects of the role of SSEA-4 in the regulation of adhesive and migratory properties of invasive cancer cells. The elucidation of the mechanism underlying SSEA-4 mediated regulation of cancer cell adhesion and migration will provide new insights into the mechanism of cancer metastasis and also provide new targets for cancer therapy. Because of its very high affinity and specificity for SSEA-4, the IPS-K-4A2B8 antibody may be a very promising candidate for cancer therapy.
Methods
In vitro cell culture of cell lines and induced pluripotent cells
In this study, 27 solid cancer cell lines, 15 leukemic cell lines and induced pluripotent cell line iPS122 (kindly provided by Dr Andras Nagy, Lunenfeld-Tanenbaum Research Institute, Toronto, Ontario, Canada) were used. Solid cancer and leukemic cell lines (listed in Supplementary Figure S1A and Tables S1 and S2) were plated and expanded in vitro in media containing RPMI 1640 supplemented with 1% penicillin–streptomycin, 1% l-glutamine, 1% amino acids, 0.005% 1-thioglycerol and 10% fetal bovine serum (all reagents from PAA, Freiburg, Germany). In the case of iPS122, cells were plated and expanded on a monolayer of mouse embryonic fibroblasts plated at a density of 2400 cells/mm2. The iPS colonies were cultured in embryonic stem cell media containing KnockOut dulbecco's modified Eagle's medium supplemented with 20% KnockOut serum replacement (Invitrogen, Bleiswijk, Netherlands), 1% penicillin–streptomycin, 1% l-glutamine (PAA), 1% non-essential amino acids (Invitrogen, Bleiswijk, Netherlands), 6 µM β-mercaptoethanol (Sigma-Aldrich, Munich, Germany) and 5 ng/mL of recombinant human basic fibroblast growth factor (Peprotech, Hamburg, Germany).
Generation of mAbs using induced pluripotent stem cell line iPS122
Novel mAbs against cell surface molecules expressed on human iPS122 cells were raised by immunization of 6–8-week-old female BALB/c mice with 107 iPS122 cells as described previously (Buhring et al. 2007).
Flow cytometric analysis and cell sorting
Fluorescein isothiocyanate (FITC)-conjugated antibodies against CD44, SSEA-3, SSEA-4, Tra 1-60 and Tra 1-81 were purchased from Becton Dickinson (Heidelberg, Germany). Allophycocyanin-conjugated CD324 (E-cadherin) was obtained from Miltenyi Biotec (Bergisch, Gladbach, Germany). In-house generated and commercial monoclonal antibodies used in the study are listed in Supplementary Tables S1, S2 and S5. After blocking of non-specific binding with 10 mg/mL polyglobin (10 min, 4°C), cells were labeled with antibodies as described previously (Sivasubramaniyan et al. 2013). In brief, cells were stained with proprietary antibodies or fluorochrome-conjugated antibodies. Cells stained with conjugates were washed twice in FACS buffer containing phosphate buffered saline (PBS) supplemented with 0.1% bovine serum albumin (BSA) and 0.001% of sodium azide and used for flow cytometry. Cells labeled with proprietary antibodies were stained with 10 µL of 1:25 diluted F (ab) 2 fragment of an R-phycoerythrin (PE)-conjugated goat anti-mouse antibody (Dako Cytomation, Hamburg, Germany) for 15 min and washed twice. Cells were sorted on a FACSAria cell sorter in the high-purity single-cell mode (Becton Dickinson) or analyzed on a FACS Canto II flow cytometer (Becton Dickinson) and FCS express software (De Novo Software, Ontario, Canada) or FlowJo software (Tree Star Inc., Ashland, USA).
TX-100 treatment
To determine the potential resistance of the IPS-K-4A2B8 antigen to solubilization with non-ionic detergents and its association with lipid rafts, TX-100 treatment was performed on iPS122 and DU145 cells as described previously (Filatov et al. 2003). In the next step, both treated and untreated cells were stained with proprietary antibodies or fluorochrome-conjugated antibodies for flow cytometric analysis.
Immunocytochemical staining with CTB conjugates
DU145 cell line cultured to 60% confluence was incubated with 5 µg/mL of purified mAb IPS-K-4A2B8 for 60 min. The monolayer was fixed with 4% paraformaldehyde (PFA) for 15 min on ice and stained with 0.5 µg/mL of Alexa Fluor 555 conjugated CTB (Invitrogen) in PBS for 60 min at 4°C. Cells were permeabilized with 0.5% TX-100 for 5 min at room temperature and blocked in 5% BSA (Sigma-Aldrich) for 30 min at room temperature. After blocking, cells were incubated with Alexa Fluor 488 conjugated anti-mouse IgM antibody (1:500) (Life Technologies) for 50 min at room temperature. Cells were mounted with 4′6′-diamidino-2 phenylindole (DAPI) containing Vectashield mounting medium (Vector Labs, Lörrach, Germany) and photographed with Zeiss Observer.Z1 AX10 microscope with ApoTome and AxioVision 4.8 imaging software 488 (Carl Zeiss MicroImaging, Jena, Germany).
Proteinase K treatment
For proteinase K treatment, we used iPS122 and DU145 cells. Single-cell suspension of 2 × 106 cells were treated with PBS containing 100 µg/mL proteinase K enzyme (Qiagen, Düsseldorf, Germany) for 1 h at 37°C with intermittent shaking. The control cells were resuspended at the same cell density in PBS without proteinase K. The enzymatic reaction was stopped by adding phenylmethylsulfonyl fluoride at a concentration of 1 mM/mL and incubated on ice for 10 min. In the next step, cells were fixed with 1% PFA on ice for 15 min and washed twice with 5 mL PBS and pelleted at 1200 rpm (241 × g) for 7 min at 4°C. Finally, fixed cells were stained with IPS-K-4A2B8 antibody or anti-human CD44 antibody for flow cytometric analysis.
Treatment of cells with PI-PLC
iPS122 cells were treated with or without 3 units of PI-PLC isolated from Bacillus cereus (Invitrogen) and incubated for 1 h in PBS at 37°C. Cells were pelleted by centrifugation at 10,000 × g for 3 min. Treated and untreated cells were stained with anti-TNAP and IPS-K-4A2B8 antibody for flow cytometric analysis.
MβCD treatment
Sixty percent of confluent PC3 cells were incubated in serum-free complete media supplemented with or without 5 mM MβCD (Sigma-Aldrich) for 1 h at 37°C. In the next step, single-cell suspension was prepared by using 1% trypsin-ethylenediaminetetraacetic acid. The cells were pelleted at 1200 rpm (241 × g) for 7 min at 4°C and stained with IPS-K-4A2B8 antibody or anti-human CD44 antibody for flow cytometric analysis.
Glycan array
Cy3-labeled IPS-K-4A2B8 and FITC-labeled SSEA-4 (clone MC813-70, Becton Dickinson) were sent to the Consortium for Functional Glycomics Core H, where they were screened against versions 5.0 (611 glycans) of the printed array. Both mAbs were used at three different concentrations for screening analysis on the array in replicates of six.
Blocking analysis with SSEA-4 glycan
For blocking studies, mAb IPS-K-4A2B8 was incubated with purified SSEA-3, SSEA-4 or GM1b glycan (ELICITYL OligoTech, Crolles, France) at concentrations of 0, 15, 30, 45, 60 and 75 µM/mL for 30 min on ice. For control, IPS-K-4A2B8 was incubated with PBS. In the next step, NT-2 cells (0.5 × 106 cells/mL) were stained with IPS-K-4A2B8 antibody. After washing, cells were analyzed on a FACSCanto II flow cytometer. The percent blocking was calculated as follows: 100 − [(MFI of cells stained with IPS-K-4A2B8 incubated with test glycan/MFI of cells stained with IPS-K-4A2B8 incubated with PBS) × 100].
Immunocytochemical staining
Purified mouse anti-human antibodies against Caveolin and rat anti-human antibodies against CD49f were purchased from Becton Dickinson. Purified mouse anti- human antibodies against CD29, CD49b, CD49e, CD49f, CD51/61, CD104 and CD107b were purchased from BioLegend (San Diego, CA, USA). Purified mouse anti-human antibodies against EEA1 and Rab11 were purchased from Abcam. Alexa Fluor 488-conjugated isotype-specific goat anti-mouse antibodies (IgG1, IgG2a, IgG2b, IgM), Cy3-conjugated goat anti-mouse IgG, Alexa Fluor 555 goat anti-mouse IgM antibodies and Rhodamine Phalloidin were purchased from Life Technologies.
Cells cultured to 60–80% confluency on a 22 mm glass cover slip were fixed with 4% paraformaldehyde for 15 min at 4°C, permeabilized with 0.2% TX-100 in PBS for 5 min and blocked in 5% BSA in PBS for 1 h at room temperature. After blocking, cells were incubated with purified antibodies diluted in staining buffer containing PBS supplemented with 5% BSA, overnight at 4°C. In the next step, cells were incubated with fluorochrome-conjugated isotype-specific secondary antibody in staining buffer for 1 h at room temperature. In order to perform cell surface immunocytochemical staining, the permeabilization with TX-100 was avoided. Cells were mounted with DAPI containing Vectashield mounting medium (Vector Labs) and photographed with Zeiss Observer.Z1 AX10 microscope with ApoTome and AxioVision 4.8 imaging software 488 (Carl Zeiss MicroImaging) or with Zeiss LSM510 META confocal laser scanning microscope equipped with an argon laser (at 488 nm) and two HeNe lasers (at 543 and 633 nm) (Carl Zeiss MicroImaging) and the images were analyzed using ZEN 2012 software.
Xenotransplantation model
All animal experimental procedures were approved by the University of Tübingen Institutional Animal Care and Use Committee. Immunopermissive NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (also termed NOD/SCID/IL2Rγnull, abbreviated as NSG) mice (Shultz et al. 2005) used in this study were maintained under pathogen-free conditions. FACS sorted or unfractionated DU145 cells were implanted subcutaneously in individual flanks of the 4–5-month-old male mice. Tumor growth was monitored by palpation of the injection site and quantification of metric traits. Mice were sacrificed 4–20 weeks after implantation.
Tissue preparation and cell isolation from xenografts and primary prostate cancer
Xenografts were surgically excised after euthanizing the mice by CO2 inhalation. Human prostate cancer tissue from one patient with advanced prostate cancer and inguinal metastasis (Gleason score 5 + 4 = 9) was obtained following cystectomy after written informed consent and approval of the Ethics Committee (No. 379/2010BO2) of the University of Tübingen.
The tissue was extensively rinsed in sterile PBS, mechanically minced with a tissue chopper and enzymatically digested for 60–90 min at 37°C in hank's balanced salt solution (HBSS) with Ca2+ and Mg2+ containing 250 µg/mL of DNAse I (Sigma-Aldrich), 250 µg/mL of Dispase II (Roche, Mannheim, Germany) and 750 FALGPA units/mL of Collagenase XI (Sigma-Aldrich). The digest was filtered through 100 and 40 µm sieves to remove undigested tissue. The cells were then centrifuged at 1000 rpm (168 × g) for 10 min and pellets were incubated in ammonium chloride (Stemcell Technologies, Köln, Germany) for 10 min on ice to selectively lyse the erythrocytes. The cells were washed twice with PBS and kept on ice until further use.
Immunohistochemical staining
The tissue was fixed overnight in 4% paraformaldehyde, equilibrated the following night in 20% sucrose and frozen in Tissue Tek OCT compound. Immunohistochemical staining was performed on 5 µm cryosections. The sections were washed three times in Tris-buffered saline (TBS) for 5 min and blocked for 30 min with TBS containing 0.1% TX-100, 10% goat serum, 0.1% sodium azide, 0.1% cold fish skin gelatin (CFSG). The sections were stained with purified antibodies diluted in staining buffer containing TBS supplemented with 5% goat serum, 0.1% sodium azide and 0.1% CFSG for overnight at 4°C. The sections were washed three times for 5 min with TBS containing 0.05% Tween-20 (TBST), followed by staining with fluorophore-conjugated secondary antibody diluted in staining buffer. The sections were washed three times for 5 min with TBST and were mounted with DAPI containing Vectashield hard set mounting medium. The slides were visualized using a Zeiss Observer.Z1 AX10 microscope with ApoTome and AxioVision 4.8 imaging software.
Lentiviral transduction
Lentivirus carrying ST3GAL2 small hairpin RNA (cat#NM_006927) or control construct of vector PLKO.1-Puro (cat#promSHC001) was purchased from Sigma-Aldrich. Lentiviral production and cell transduction was performed using standard protocols as described previously (Konantz et al. 2013). Lentiviruses were concentrated using Vivaspin 20 mL centrifugal concentrators (Sartorius Stedim Biotech, Goettingen, Germany) at 3000 × g for 50 min. Transduced cells harboring the ST3GAL2 or control shRNA cassettes were selected in the presence of 2 µg/mL puromycin.
Gene expression analysis
RNA isolation and cDNA preparation was performed as described previously (Sivasubramaniyan et al. 2013). One microgram of RNA was reverse transcribed using the ImProm-II Reverse Transcription system (Promega, Mannheim, Germany) as described by the manufacturer's protocol. Real-time quantitative PCR was performed using the SYBR Green reagent (Eurogentec, Cologne, Germany) on a Light Cycler480 Real-time PCR instrument (Roche Applied Science, Mannheim, Germany). Primer sequences (intron spanning) are listed in Supplementary Table S6. All primers were used at annealing temperature of 58°C.
Adhesion assay
Fluorimetric-based cell adhesion assays of ST3GAL2 knockdown DU145 cells or DU145 cells containing control construct were performed. Polystyrene 96 well clear tissue culture plates were coated (replicates of 5) with 100 µL of prediluted ECM for 24 h at 4°C. ECM used were collagen I (5 µg/cm2), collagen IV (5 µg/cm2), laminin (1 µg/cm2) and chondroitin sulfate (100 µg/cm2). After wash with PBS supplemented with 1× ion mix (1 mM CaCl2, 1 mM MgCl2 and 50 µM MnCl2 in RPMI1640), non-specific cell binding to ECM was prevented by pre-incubation with 1% BSA for 60 min at room temperature. For quantitative determination of the cell adhesion, 1 × 106 of control and ST3GAL2 knockdown cells were incubated with 8.9 µM carboxyfluorescein diacetate succinimidyl ester (Sigma-Aldrich) in HBSS (without Ca2+ and Mg2+) for 15 min at 37°C and 1 × 105 cells were plated and incubated for 45–60 min at 37°C under 5% CO2. The non-adherent cells were removed by gently rinsing the wells with PBS supplemented with 1× ion mix and resuspended in 100 µL of PBS. The mean fluorescence intensity from adherent cells in each well was measured using GloMaxR – Multi Detection System (Promega).
Statistics
Analysis of variance with two factors (tumor weight and frequency) was performed to determine the difference in the tumorigenic potential of the sorted populations isolated from the tumor explants (Supplementary Figure S6B). The acceptable level of significance for the analyses was set at p ≤ 0.05. For all other experiments, graphical data are presented as mean values ± standard error. p-values were derived via the application of a two-tailed, unpaired Student's t-test. Statistically significant differences are indicated by asterisks.
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
This work was supported by research collaboration with Roche Diagnostics GmbH, Penzberg, Germany, entitled: Functional profiling of novel antibodies.
Conflict of interest
None declared.
Abbreviations
BSA, bovine serum albumin; CFSG, cold fish skin gelatin; CTB, cholera toxin B; DAPI, 4′6′-diamidino-2 phenylindole; ECM, extracellular matrix; EMT, epithelial to mesenchymal transition; FACS, fluorescence-activated cell sorting; FITC, fluorescein isothiocyanate; GSL, glycosphingolipid; hESC, human embryonic stem cell; IHC, immunohistochemistry; mAb, monoclonal antibody; MβCD, methyl-beta-cyclodextrin; PE, R-phycoerythrin; PI-PLC, phosphoinositol phospholipase C; SSEA-4, stage-specific embryonic antigen-4; TBS, Tris-buffered saline; TBST, TBS containing 0.05% Tween 20.
Supplementary Material
Acknowledgement
The authors would like to acknowledge the Consortium for Functional Glycomics funded by the NIGMS GM62116 for services provided by the Glycan Array Synthesis Core (Core D) at The Scripps Research Institute in La Jolla, CA, USA. They produced the mammalian glycan microarray and the Protein–Glycan Interaction Core (H) at Emory University School of Medicine in Atlanta, GA, USA, assisted with analysis of samples on the array.
References
- Ayala I, Baldassarre M, Giacchetti G, Caldieri G, Tete S, Luini A, Buccione R. 2008. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J Cell Sci. 121:369–378. [DOI] [PubMed] [Google Scholar]
- Battula VL, Shi Y, Evans KW, Wang RY, Spaeth EL, Jacamo RO, Guerra R, Sahin AA, Marini FC, Hortobagyi G, et al. 2012. Ganglioside GD2 identifies breast cancer stem cells and promotes tumorigenesis. J Clin Invest. 122:2066–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner W, Gross S, Steinbach F, Horn S, Hohenfellner R, Thuroff JW. 2000. Differential inhibition of renal cancer cell invasion mediated by fibronectin, collagen IV and laminin. Cancer Lett. 155:199–205. [DOI] [PubMed] [Google Scholar]
- Buhring HJ, Battula VL, Treml S, Schewe B, Kanz L, Vogel W. 2007. Novel markers for the prospective isolation of human MSC. Ann NY Acad Sci. 1106:262–271. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. 2011. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 108:7950–7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WW, Lee CH, Lee P, Lin J, Hsu CW, Hung JT, Lin JJ, Yu JC, Shao LE, Yu J, et al. 2008. Expression of Globo H and SSEA3 in breast cancer stem cells and the involvement of fucosyl transferases 1 and 2 in Globo H synthesis. Proc Natl Acad Sci USA. 105:11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov AV, Shmigol IB, Kuzin II, Sharonov GV, Feofanov AV. 2003. Resistance of cellular membrane antigens to solubilization with Triton X-100 as a marker of their association with lipid rafts–analysis by flow cytometry. J Immunol Methods. 278:211–219. [DOI] [PubMed] [Google Scholar]
- Fong CY, Peh GS, Gauthaman K, Bongso A. 2009. Separation of SSEA-4 and TRA-1–60 labelled undifferentiated human embryonic stem cells from a heterogeneous cell population using magnetic-activated cell sorting (MACS) and fluorescence-activated cell sorting (FACS). Stem Cell Rev. 5:72–80. [DOI] [PubMed] [Google Scholar]
- Gao AC, Lou W, Dong JT, Isaacs JT. 1997. CD44 is a metastasis suppressor gene for prostatic cancer located on human chromosome 11p13. Cancer Res. 57:846–849. [PubMed] [Google Scholar]
- Gottschling S, Jensen K, Warth A, Herth FJ, Thomas M, Schnabel PA, Herpel E. 2013. Stage-specific embryonic antigen-4 is expressed in basaloid lung cancer and associated with poor prognosis. Eur Respir J. 41:656–663. [DOI] [PubMed] [Google Scholar]
- Hakomori Si SI. 2002. The glycosynapse. Proc Natl Acad Sci USA. 99:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harichandan A, Sivasubramaniyan K, Buhring HJ. 2013. Prospective isolation and characterization of human bone marrow-derived MSCs. Adv Biochem Eng Biotechnol. 129:1–17. [DOI] [PubMed] [Google Scholar]
- Harichandan A, Sivasubramaniyan K, Hennenlotter J, Schwentner C, Stenzl A, Buhring HJ. 2013. Isolation of adult human spermatogonial progenitors using novel markers. J Mol Cell Biol. 5:351–353. [DOI] [PubMed] [Google Scholar]
- Hirohashi S. 1998. Inactivation of the E-cadherin-mediated cell adhesion system in human cancers. Am J Pathol. 153:333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung TC, Lin CW, Hsu TL, Wu CY, Wong CH. 2013. Investigation of SSEA-4 binding protein in breast cancer cells. J Am Chem Soc. 135:5934–5937. [DOI] [PubMed] [Google Scholar]
- Iwabuchi K, Yamamura S, Prinetti A, Handa K, Hakomori S. 1998. GM3-enriched microdomain involved in cell adhesion and signal transduction through carbohydrate-carbohydrate interaction in mouse melanoma B16 cells. J Biol Chem. 273:9130–9138. [DOI] [PubMed] [Google Scholar]
- Kallakury BV, Sheehan CE, Ross JS. 2001. Co-downregulation of cell adhesion proteins alpha- and beta-catenins, p120CTN, E-cadherin, and CD44 in prostatic adenocarcinomas. Hum Pathol. 32:849–855. [DOI] [PubMed] [Google Scholar]
- Kasahara K, Sanai Y. 2000. Functional roles of glycosphingolipids in signal transduction via lipid rafts. Glycoconj J. 17:153–162. [DOI] [PubMed] [Google Scholar]
- Katagiri YU, Kiyokawa N, Nakamura K, Takenouchi H, Taguchi T, Okita H, Umezawa A, Fujimoto J. 2005. Laminin binding protein, 34/67 laminin receptor, carries stage-specific embryonic antigen-4 epitope defined by monoclonal antibody Raft.2. Biochem Biophys Res Commun. 332:1004–1011. [DOI] [PubMed] [Google Scholar]
- Katagiri YU, Ohmi K, Katagiri C, Sekino T, Nakajima H, Ebata T, Kiyokawa N, Fujimoto J. 2001. Prominent immunogenicity of monosialosyl galactosylgloboside, carrying a stage-specific embryonic antigen-4 (SSEA-4) epitope in the ACHN human renal tubular cell line-a simple method for producing monoclonal antibodies against detergent-insoluble microdomains/raft. Glycoconj J. 18:347–353. [DOI] [PubMed] [Google Scholar]
- Kaucic K, Liu Y, Ladisch S. 2006. Modulation of growth factor signaling by gangliosides: positive or negative? Methods Enzymol. 417:168–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. 2003. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 169:1122–1133. [DOI] [PubMed] [Google Scholar]
- Konantz M, Andre MC, Ebinger M, Grauer M, Wang H, Grzywna S, Rothfuss OC, Lehle S, Kustikova OS, Salih HR, et al. 2013. EVI-1 modulates leukemogenic potential and apoptosis sensitivity in human acute lymphoblastic leukemia. Leukemia. 27:56–65. [DOI] [PubMed] [Google Scholar]
- Le PU, Nabi IR. 2003. Distinct caveolae-mediated endocytic pathways target the Golgi apparatus and the endoplasmic reticulum. J Cell Sci. 116:1059–1071. [DOI] [PubMed] [Google Scholar]
- Liu JC, Egan SE, Zacksenhaus E. 2013. A Tumor initiating cell-enriched prognostic signature for HER2+:ERalpha- breast cancer; rationale, new features, controversies and future directions. Oncotarget. 4:1317–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber CP, Runyan RB. 1990. A comparison of fibronectin, laminin, and galactosyltransferase adhesion mechanisms during embryonic cardiac mesenchymal cell migration in vitro. Dev Biol. 140:401–412. [DOI] [PubMed] [Google Scholar]
- Malecki M, Tombokan X, Anderson M, Malecki R, Beauchaine M. 2013. TRA-1–60, SSEA-4, POU5F1, SOX2, NANOG Clones of Pluripotent Stem Cells in the Embryonal Carcinomas of the Testes. J Stem Cell Res Ther. 31 pii: 1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuzuka K, Handa K, Satoh M, Arai Y, Hakomori S. 2005. A specific microdomain (“glycosynapse 3”) controls phenotypic conversion and reversion of bladder cancer cells through GM3-mediated interaction of alpha3beta1 integrin with CD9. J Biol Chem. 280:35545–35553. [DOI] [PubMed] [Google Scholar]
- Mutoh T, Tokuda A, Miyadai T, Hamaguchi M, Fujiki N. 1995. Ganglioside GM1 binds to the Trk protein and regulates receptor function. Proc Natl Acad Sci USA. 92:5087–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto Z, Yoshida T, Okabe M, Koike C, Fathy M, Tsuno H, Tomihara K, Arai N, Noguchi M, Nikaido T. 2013. CD44 and SSEA-4 positive cells in an oral cancer cell line HSC-4 possess cancer stem-like cell characteristics. Oral Oncol. 49:787–795. [DOI] [PubMed] [Google Scholar]
- Ostermeyer AG, Beckrich BT, Ivarson KA, Grove KE, Brown DA. 1999. Glycosphingolipids are not essential for formation of detergent-resistant membrane rafts in melanoma cells. methyl-beta-cyclodextrin does not affect cell surface transport of a GPI-anchored protein. J Biol Chem. 274:34459–34466. [DOI] [PubMed] [Google Scholar]
- Park Y, Choi IY, Lee SJ, Lee SR, Sung HJ, Kim JH, Yoo YD, Geum DH, Kim SH, Kim BS. 2010. Undifferentiated propagation of the human embryonic stem cell lines, H1 and HSF6, on human placenta-derived feeder cells without basic fibroblast growth factor supplementation. Stem Cells Dev. 19:1713–1722. [DOI] [PubMed] [Google Scholar]
- Pulukuri SM, Gondi CS, Lakka SS, Jutla A, Estes N, Gujrati M, Rao JS. 2005. RNA interference-directed knockdown of urokinase plasminogen activator and urokinase plasminogen activator receptor inhibits prostate cancer cell invasion, survival, and tumorigenicity in vivo. J Biol Chem. 280:36529–36540. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. 2010. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 18:510–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. 2011. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nat Commun. 2:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Aoki H, Ito A, Ueno S, Wada T, Mitsuzuka K, Satoh M, Arai Y, Miyagi T. 2003. Human alpha2,3-sialyltransferase (ST3Gal II) is a stage-specific embryonic antigen-4 synthase. J Biol Chem. 278:26474–26479. [DOI] [PubMed] [Google Scholar]
- Sharma DK, Brown JC, Cheng Z, Holicky EL, Marks DL, Pagano RE. 2005. The glycosphingolipid, lactosylceramide, regulates beta1-integrin clustering and endocytosis. Cancer Res. 65:8233–8241. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al. 2005. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 174:6477–6489. [DOI] [PubMed] [Google Scholar]
- Sivasubramaniyan K, Harichandan A, Schumann S, Sobiesiak M, Lengerke C, Maurer A, Kalbacher H, Buhring HJ. 2013. Prospective isolation of mesenchymal stem cells from human bone marrow using novel antibodies directed against Sushi domain containing 2. Stem Cells Dev. 22:1944–1954. [DOI] [PubMed] [Google Scholar]
- Sivasubramaniyan K, Lehnen D, Ghazanfari R, Sobiesiak M, Harichandan A, Mortha E, Petkova N, Grimm S, Cerabona F, de Zwart P, et al. 2012. Phenotypic and functional heterogeneity of human bone marrow- and amnion-derived MSC subsets. Ann NY Acad Sci. 1266:94–106. [DOI] [PubMed] [Google Scholar]
- Stelling MP, Lages YM, Tovar AM, Mourao PA, Rehen SK. 2013. Matrix-bound heparan sulfate is essential for the growth and pluripotency of human embryonic stem cells. Glycobiology. 23:337–345. [DOI] [PubMed] [Google Scholar]
- Tondeur S, Assou S, Nadal L, Hamamah S, De Vos J. 2008. Biology and potential of human embryonic stem cells. Ann Biol Clin. 66:241–247. [DOI] [PubMed] [Google Scholar]
- Valeriote F, van Putten L. 1975. Proliferation-dependent cytotoxicity of anticancer agents: a review. Cancer Res. 35:2619–2630. [PubMed] [Google Scholar]
- Varma R, Mayor S. 1998. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 394:798–801. [DOI] [PubMed] [Google Scholar]
- Verga Falzacappa MV, Ronchini C, Reavie LB, Pelicci PG. 2012. Regulation of self-renewal in normal and cancer stem cells. FEBS J. 279:3559–3572. [DOI] [PubMed] [Google Scholar]
- Virant-Klun I, Skutella T, Hren M, Gruden K, Cvjeticanin B, Vogler A, Sinkovec J. 2013. Isolation of small SSEA-4-positive putative stem cells from the ovarian surface epithelium of adult human ovaries by two different methods. Biomed Res Int. 2013:690415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P. 2014. Surface markers of cancer stem cells in solid tumors. Curr Stem Cell Res Ther. 92:102–11. [DOI] [PubMed] [Google Scholar]
- Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, Zhang L, Yan J, Miller D, Zhang HG. 2012. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PLoS One. 7:e50781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, et al. 2005. Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol. 168:441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Takeo Y, Yoshida S, Kouchi Z, Nakamura Y, Fukami K. 2009. Lipid rafts and caveolin-1 are required for invadopodia formation and extracellular matrix degradation by human breast cancer cells. Cancer Res. 69:8594–8602. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Yoshida S, Muroi E, Yoshida N, Kawamura M, Kouchi Z, Nakamura Y, Sakai R, Fukami K. 2011. Phosphoinositide 3-kinase signaling pathway mediated by p110alpha regulates invadopodia formation. J Cell Biol. 193:1275–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudoh K, Matsui H, Kanamori M, Ohmori K, Tsuji H. 1995. Tumor cell attachment to laminin promotes degradation of the extracellular matrix and cell migration in high-metastatic clone cells of RCT sarcoma in vitro. Jpn J Cancer Res. 86:685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Fang H, Tsuruoka T, Tsuji T, Sasaki T, Hakomori S. 1993. Regulatory role of GM3 ganglioside in alpha 5 beta 1 integrin receptor for fibronectin-mediated adhesion of FUA169 cells. J Biol Chem. 268:2217–2222. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.