Abstract
DNA methylation is a complex, tissue-specific phenomenon that can reflect both endogenous factors and exogenous exposures. Buccal brushings represent an easily accessible source of DNA, which may be an appropriate surrogate tissue in the study of environmental exposures and chronic respiratory diseases. Buccal brushings were obtained from a subset of current and former smokers from the COPDGene study. Genome-wide DNA methylation data were obtained in the discovery cohort (n = 82) using the Illumina HumanMethylation450K array. Empirical Bayes methods were used to test for differential methylation by current smoking status at 468,219 autosomal CpG sites using linear models adjusted for age, sex, and race. Pyrosequencing was performed in a nonoverlapping replication cohort (n = 130). Current smokers were significantly younger than former smokers in both the discovery and replication cohorts. Seven CpG sites were associated with current smoking at a false discovery rate less than 0.05 in the discovery cohort. Six of the seven significant sites were pyrosequenced in the replication cohort; five CpG sites, including sites annotated to CYP1B1 and PARVA, were replicated. Correlations between cumulative smoke exposure and time since smoking cessation were observed in a subset of the significantly associated CpG sites. A significant correlation between reduced lung function and increased radiographic emphysema with methylation at cg02162897 (CYP1B1) was observed among female subjects. Site-specific methylation of DNA isolated from buccal mucosa is associated with exposure to cigarette smoke, and may provide insights into the mechanisms underlying differential susceptibility toward the development of smoking-related chronic respiratory diseases.
Keywords: DNA methylation, smoking, buccal mucosa
Clinical Relevance
Site-specific methylation of DNA isolated from buccal mucosa is easily accessible and can reflect environmental exposures, such as cigarette smoking. Differential methylation by cigarette smoke exposure may provide insights into the mechanisms underlying differential susceptibility toward the development of chronic respiratory diseases, such as chronic obstructive pulmonary disease.
Tobacco smoke is a well-established contributor to the development of respiratory diseases (1). However, our understanding of the mechanisms and loci that are associated with differential susceptibility toward disorders such as lung cancer and chronic obstructive pulmonary disease (COPD) remains incomplete. Gene expression studies have documented extensive changes in transcription in the aerodigestive tract in response to cigarette smoke (2–4). Because gene expression is tissue specific, the majority of these studies have been performed using epithelial cells isolated from the lower respiratory tract and lung tissue, both of which must be obtained invasively and are not suitable for studies involving large populations. Samples isolated from buccal mucosa swabs and biopsies have been investigated as a noninvasive lung surrogate tissue; however, concern regarding the degradation of samples from salivary RNAses has limited the widespread use of this sample type for gene expression studies (4–6).
In contrast to RNA, DNA isolated from buccal samples is relatively stable, and has been used for a wide variety of applications, including genotyping and DNA methylation analysis (7). Buccal DNA methylation patterns have been shown to correlate with prenatal tobacco smoke exposure (7) and particulate air pollution levels in children (8, 9); subgroup analyses in these studies have been suggestive of links between buccal methylation patterns and childhood wheeze/asthma and exhaled nitric oxide phenotypes. To date, the majority of studies using buccal methylation patterns to study environmental exposures have examined global methylation (10) (including Alu and LINE-1 repetitive elements [7]) or candidate gene methylation (7–9, 11). Studies examining the effects of tobacco smoking on genome-wide, site-specific methylation in buccal mucosa from adults have not been reported. Some of the results in the current article have been previously reported in the form of an abstract (12).
Materials and Methods
Cohort and Samples
A subset of current and former smokers participating the COPDGene Study (www.clinicaltrials.gov [NCT000608764]), recruited at Brigham and Women’s Hospital and Morehouse School of Medicine, were included in this study. Enrollment and exclusion criteria have been previously described (13). Briefly, all study participants were self-described non-Hispanic white or African American, between 45 and 80 years of age with 10 or more pack-years of smoking. All subjects completed questionnaire data, pre- and post-bronchodilator spirometry, as well as inspiratory and expiratory computed chest tomography. Current smoking was defined as an affirmative answer to the question “Do you currently smoke cigarettes (as of 1 month ago)?” Quantitative percent emphysema was calculated as the percentage of voxels with an attenuation less than −950 Hounsfield units (13). A buccal sample for DNA extraction was collected at the time of enrollment. This study was approved by the Institutional Review Board of each participating center, and informed consent was obtained from all subjects.
DNA Methylation—Sample Preparation, Data Processing, and Analysis
DNA was collected and extracted from buccal brushes using the Gentra Puregene Buccal Cell Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. DNA (1 μg) was bisulfite converted using the EZ DNA Methylation Gold Kit (Zymo Research, Irvine, CA). DNA methylation at 485,512 CpG sites throughout the genome was obtained in the discovery cohort (n = 82) using the Illumina (San Diego, CA) HumanMethylation450K array. Data preprocessing and quality control were performed using the Bioconductor packages minfi (v1.4.0) (14) and sva (v3.4.0) (15), as implemented in the R programming language (release 2.15.0); a summary of the data-cleaning process is provided in Table E1 in the online supplement.
Quantile normalization, adjustment for batch effects using the ComBat function (v3.4.0) (15), and subsequent analyses were performed separately by probe chemistry. Empirical Bayes mediated models were used to test for differential methylation by current smoking status at 468,219 autosomal CpG sites that passed quality control. All models were adjusted for age, sex, and race. A false discovery rate (FDR) of less than 0.05 was used to denote significance in the discovery cohort. Pyrosequencing was performed on bisulfite-converted DNA from a nonoverlapping replication cohort (n = 130) at EpigenDx, Inc. (Hopkinton, MA). A Student’s t test was used to test for differential methylation; a P value less than 0.05 with a consistent direction of effect was considered significant.
In Silico Analysis of Public Gene Expression Data
Using publically available data downloaded from the Gene Expression Omnibus repository (datasets GSE7895, GSE17913, GSE43079, GSE34517, GSE27002, GSE994, GSE37147, GSE19407, GSE2125, GSE32537), we explored the impact of current smoking on gene expression in various aerodigestive tissues at the differentially methylated loci identified from our analyses. A Student’s t test was used to assess for differences by current smoking status in the log-transformed transcript levels.
Functional Annotation Clustering
Functional annotation clustering under the high-stringency classification option was performed using the Database for Annotation, Visualization, and Integrated Discovery (version 6.7) (16, 17); gene names annotated to CpG sites with an association P value less than 0.001 from our primary analyses were used as input.
Results
Technical replicates that passed bisulfite conversion (n = 4) demonstrated very high reproducibility at all probes assayed, with a minimum correlation coefficient of 0.996 between replicates (Figure E1). Characteristics of the subjects included in our analyses are summarized in Table 1. Current smokers in both cohorts were significantly younger than former smokers. There were significantly fewer African American subjects in the replication cohort relative to the discovery cohort. There were no differences in sex distribution, spirometric lung function, or prevalence of COPD by current smoking status in either cohort.
Table 1.
Cohort Characteristics by Current Smoking Status
| Discovery |
Replication |
|||
|---|---|---|---|---|
| Current Smokers | Former Smokers | Current Smokers | Former Smokers | |
| n | 30 | 52 | 43 | 87 |
| Age, yr | 60.4 (8.8)* | 66.8 (8.4) | 59.7 (8.1)* | 65.3 (8.1) |
| Sex, % male | 66.7 | 48.1 | 43.7 | 46.5 |
| African American, % | 26.7 | 26.9 | 14.0* | 2.3 |
| Pack-years | 52.6 (41.6) | 42.4 (29.0) | 51.4 (26.3)* | 41.2 (23.5) |
| Time since quit, yr | — | 17.3 (11.8) | — | 16.7 (12.3) |
| Body mass index | 26.4 (5.1)* | 29.7 (6.6) | 28.8 (5.3) | 29.0 (7.2) |
| FEV1 % predicted† | 79.8 (25.6) | 73.6 (26.6) | 78.7 (19.7) | 76.9 (27.1) |
| FVC % predicted† | 89.0 (20.0) | 84.8 (18.9) | 88.9 (14.6) | 87.4 (19.4) |
| FEV1/FVC† | 0.67 (0.14) | 0.65 (0.16) | 0.68 (0.13) | 0.65 (0.16) |
| COPD (≥GOLD 2), % | 33.3 | 42.3 | 37.2 | 35.6 |
| % emphysema (−950 HU) | 4.4 (0.5)* | 9.7 (0.5) | 5.0 (6.3) | 9.3 (12.1) |
Definition of abbreviations: COPD, chronic obstructive pulmonary disease; GOLD, Global Initiative for Chronic Obstructive Lung Disease; HU, Hounsfield units.
Data are presented as mean (SD) or percent.
P < 0.05 relative to former smokers.
Post-bronchodilator measurements.
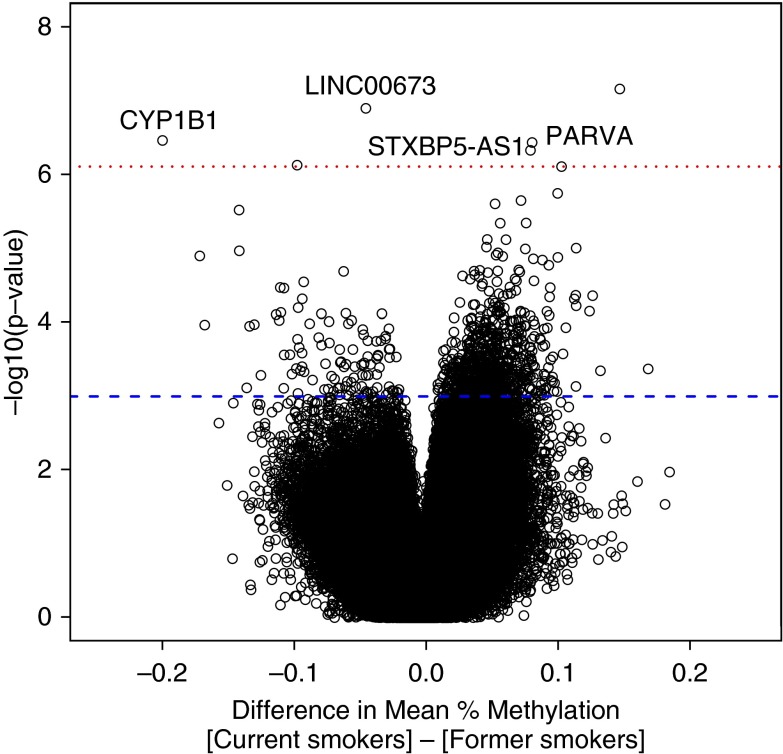
Seven CpG sites were significantly associated with current smoking in the discovery cohort. The sites most strongly associated with smoking in the Infinium I and Infinium II analyses are shown in Tables E2a and E2b, respectively. A qualitative representation of the analysis is presented in Figure 1 (volcano plot) and Figure E2 (Manhattan plot). Adjusting the models for post-bronchodilator FEV1 % predicted did not significantly change the top results (Table E3). When we examined both type I and II probes together, six probes, including cg02162897 (CYP1B1) and cg03126561 (PARVA), were significant at an FDR less than 0.05 (Table E4). Two probes that had previously been significant in the Infinium II–only analysis, cg16187635 and cg2131809 (FRMD4A), had an FDR of 0.05 in the combined analysis.
Figure 1.
Volcano plot of methylation by current smoking status in Infinium II analysis. The difference in mean methylation for each CpG site is plotted on the x axis, whereas the log-transformed P value is plotted on the y axis. Each point represents an individual CpG site. The dotted red line denotes the threshold for significance at a false discovery rate less than 0.05, whereas the blue dashed line corresponds to an unadjusted P value less than 0.001.
Although our original analyses included a covariate adjustment for race, we explored alternative approaches to account for the impact of race on our results. When we examined non-Hispanic white subjects only (n = 60), three of the seven significantly associated sites remained significant at an FDR < 0.05 (Table E5). Among African American subjects (n = 22), there were no significant differentially methylated sites (Table E6); however, one site in the African American–only analysis (cg04396288 annotated to the non-SMC element homolog 1 (NSMCE1) gene) had an FDR of 0.06. Whether this site represents a race-specific signal will need to be explored in future studies. We performed a sample-size weighted meta-analysis of the separate non-Hispanic white and African American analyses using METAL software (18). Many of the most highly associated sites in the original analysis remain strongly associated in the race-stratified meta-analysis (Table E7).
Six of the seven CpG sites with an FDR less than 0.05 in the discovery cohort (Table 2[b]) were pyrosequenced in a nonoverlapping replication cohort (n = 130). Five of six sites assayed demonstrated significant differential methylation by current smoking status with a consistent direction of effect (Table 2). Of the six sites assayed, only cg21371809 (FRMD4A) did not demonstrate a significant association with current smoking status; however, relative hypermethylation in current smokers (consistent with the direction observed in the discovery cohort) was observed (data not shown). Use of nonparametric tests (Wilcoxon rank-sum) did not significantly change the results.
Table 2.
Replicated CpG Sites with Differential Methylation by Current Smoking Status in Buccal Mucosa
| Discovery Cohort |
Replication Cohort |
|||||
|---|---|---|---|---|---|---|
| CpG Site | Gene Symbol | Chromosome | Difference in Mean Methylation* | P Value† | Difference in Mean Methylation* | P Value‡ |
| cg09853702 | 12 | +0.15 | 6.84 × 10−08 | +0.08 | 7.04 × 10−3 | |
| cg16323911 | LINC00673 | 17 | −0.04 | 1.26 × 10−07 | −0.07 | 4.52 × 10−3 |
| cg02162897 | CYP1B1 | 2 | −0.20 | 3.41 × 10−07 | −0.22 | 5.80 × 10−8 |
| cg03126561 | PARVA | 11 | +0.08 | 3.64 × 10−07 | +0.06 | 1.46 × 10−2 |
| cg16199747 | STXBP5-AS1 | 6 | +0.08 | 4.64 × 10−07 | +0.04 | 5.38 × 10−3 |
Difference in mean methylation defined as: (mean methylation in current smokers) − (mean methylation in former smokers) (i.e., plus symbol denotes relative hypermethylation in current smokers).
P value from Bayes mediated linear models, adjusted for age, sex, and race.
P value from Student’s t test.
We explored the correlation between additional smoking metrics and the seven sites associated with current smoking in the discovery cohort. Four CpG sites demonstrated a significant (P < 0.05) correlation with cumulative pack-years (Table 3), with a fifth site, cg16199747, demonstrating a trend toward significance (P = 0.06). The strength of association between cumulative smoke exposure and CpG methylation appeared to vary by current smoking status (Figure E3); at cg03126561 and cg16199747, the associations appeared stronger in current smokers, whereas, at cg16187635 and cg2131809, the associations were primarily driven by former smokers.
Table 3.
Correlations between Site-Specific Methylation and Cumulative Smoke Exposure (Pack-Years) in the Discovery Cohort (n = 82)
| All Subjects |
Current Smokers |
Former Smokers |
||||
|---|---|---|---|---|---|---|
| CpG Site | r* | P Value | r* | P Value | r* | P value |
| cg09853702 | 0.12 | 0.27 | −0.03 | 0.88 | 0.12 | 0.40 |
| cg16323911 (LINC00673) | −0.17 | 0.14 | −0.26 | 0.16 | 0.14 | 0.31 |
| cg02162897 (CYP1B1) | −0.23 | 0.04 | −0.29 | 0.13 | −0.08 | 0.60 |
| cg03126561 (PARVA) | 0.35 | 1.3 × 10−3 | 0.52 | 3.6 × 10−3 | 0.21 | 0.14 |
| cg16199747 (STXBP5-AS1) | 0.21 | 0.06 | 0.42 | 0.02 | −0.09 | 0.52 |
| cg16187635 | −0.37 | 5.3 × 10−4 | −0.28 | 0.14 | −0.43 | 1.5 × 10−3 |
| cg21371809 (FRMD4A) | 0.32 | 3.0 × 10−3 | 0.20 | 0.28 | 0.38 | 5.9 × 10−3 |
Pearson’s product-moment correlation coefficient.
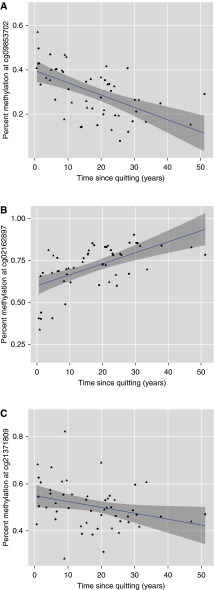
Although our cohort was cross-sectional in nature, we assessed the association between methylation levels with the time since smoking cessation among former smokers in the discovery cohort to explore the possibility of dynamic changes in methylation. Three CpG sites demonstrated significant correlations with time since quitting (Figure 2). In each instance, the direction of effect was the opposite of that observed in the current smoking analysis—this is suggestive of dynamic reversal of the effects of current smoking at these loci (19).
Figure 2.
Correlation between buccal DNA methylation and time since smoking cessation in former smokers (n = 52) in the discovery cohort. Time since smoking cessation (in years) is plotted on the x axis, whereas percent methylation is plotted on the y axis for (A) cg09853702 (Pearson’s r = −0.57; P = 9.4 × 10−6), (B) cg02162897 (Pearson’s r = 0.58; P = 6.2 × 10−6), and (C) cg21371809 (Pearson’s r = −0.30; P = 0.03). Males are represented by triangles, whereas females are represented by circles.
Because cigarette smoking may impact males and females differently, we examined each of the significant loci in the discovery cohort for differences in methylation by sex. At cg16323911 and cg02162897, significant correlations with cumulative smoke exposure were noted in females only (Figure E4). At cg16187635, a significant correlation between time since smoking cessation and percent methylation was observed in females only (Figure E5). Interestingly, methylation at cg02162897, which is located in the CpG shore of the cytochrome p450, family 1, subfamily B, polypeptide 1 (CYP1B1) gene, was correlated with FEV1 % predicted, FEV1/FVC ratio, and percent emphysema in females only (Table 4).
Table 4.
Correlations between Percent Methylation at cg02162897 with Lung Function and Percent Emphysema by Sex in the Discovery Cohort (n = 82)
| Correlation of Methylation at cg02162897 (CYP1B1) with: | Pearson Correlation (ρ) | P Value |
|---|---|---|
| FEV1 % predicted* | ||
| Males | 0.06 | 0.72 |
| Females | 0.37 | 0.03 |
| FEV1/FVC* | ||
| Males | 0.09 | 0.54 |
| Females | 0.42 | 9.1 × 10−3 |
| Percent emphysema† | ||
| Males | 0.08 | 0.59 |
| Females | −0.51 | 2.7 × 10−3 |
Post-bronchodilator spirometry values.
Percent emphysema assessed as the percentage of voxels with an attenuation of less than −950 Hounsfield units on axial computed tomography of the chest.
To explore expression patterns at differentially methylated loci in the discovery cohort, we examined public data generated in airway and lung-related tissues available through the Gene Expression Omnibus (2, 3, 20–26). Transcripts demonstrating nominally significant differences in expression (Student’s t test P < 0.05) by current smoking status from selected studies are summarized in Table E8; at the majority of the loci examined, the direction of the expression changes were anticorrelated with the methylation changes observed in our analysis. A significant positive correlation between cumulative smoke exposure (pack-years) and expression levels of several CYP1B1 transcripts was noted in a study performed in small airway epithelial cells (Figure E6) (25). Interestingly, no correlation between pack-years and expression at the CYP1B1 locus was noted in a study by Steiling and colleagues (24) conducted in large airway epithelium. In this study, significant positive correlations between the expression of transcripts annotated to FRMD4A and PARVA, but not CYP1B1, were correlated with FEV1 % predicted (24) (Figure E7).
To explore the biological context of differential methylation by current smoking status observed in our analysis, we performed functional annotation clustering through the online Database for Annotation, Visualization, and Integrated Discovery resource (version 6.7) (16, 17). A total of 1,031 CpG sites had an unadjusted P value of less than 0.001 from our primary analyses in the discovery cohort in both Infinium I and Infinium II probes. From these, 283 CpG sites were not annotated to any known genes. After removing redundant entries, 713 unique gene names were used as input. Functional annotation clustering under high stringency showed significant enrichment (FDR, <0.05) of several functional categories and structural domains, including PXXP repeats, and cadherin and pleckstrin homology domains (Table E9).
Discussion
DNA methylation is a complex phenomenon, which can serve as a unique reflection of both the genetic and environmental factors that contribute to the changing phenotype of an organism. Analogous to gene expression, DNA methylation patterns are partially tissue specific; thus, appropriate sampling for the disease or exposure of interest is paramount. In this study, we examine site-specific changes in DNA methylation throughout the genome relative to current cigarette smoking and smoking-related phenotypes in buccal mucosa, a tissue that, in addition to being a major site of direct exposure, may have utility as a surrogate tissue in the study of smoking-related lung diseases, such as COPD (4, 27).
A number of the differentially methylated loci reported in our article are biologically plausible, and have previously been described as being associated with smoking or smoking-related phenotypes. The CYP1B1 gene product is involved in xenobiotic metabolism, including the activation of procarcinogens (28). Induction of expression of CYP1B1 in oral and respiratory tissues by cigarette smoke has been well established (3, 6). Relative hypomethylation with concurrent increases in gene expression at this locus has been previously described in small airway epithelium (21) and suggests a potential role for buccal methylation at CYP1B1 as a biomarker for lung tissue. In addition to xenobiotic metabolism, CYP1B1 is also known to play a role in estrogen metabolism, and may contribute to differential susceptibility by sex toward smoking-related diseases. Genetic polymorphisms in CYP1B1 have been associated with an increased risk of lung cancer and early menopause in women; notably, interactions between single-nucleotide polymorphisms in CYP1B1 and smoking history appear to increase the risk of both of these outcomes (29–31). The differential correlations by sex between methylation at CYP1B1 and lung function and radiographic emphysema reported in our study warrant further investigation.
Additional loci identified in this study, which have been associated with smoking or related phenotypes, include the FERM domain containing 4A (FRMD4A) and parvin, α (PARVA) genes. The gene product of FRMD4A is an epidermal stem cell marker (32); genetic polymorphisms at this locus have been associated with nicotine dependence in East Asian populations (33), and a smoking cessation genotype success score (34) in European populations. The product of the PARVA gene is a focal adhesion protein (35); relative hypermethylation at this locus has been reported in the small airway epithelium of smokers (21).
Several of the CpG sites exhibiting differential methylation by smoking status are not annotated to known protein-coding sequences. cg16323911 is located within long intervening non–protein coding RNA 673 (LINC00673), whereas cg16199747 is annotated to STXBP5 antisense RNA 1 (STXBP-AS1); both transcripts belong to a novel class of molecules known as long noncoding RNAs, which may play regulatory and other roles in the cell (36). cg09853702 maps to 12q13.11; although no transcripts have yet been identified for this region, the region is a DNase hypersensitivity region in 72 of 125 cell lines assayed through the Encyclopedia of DNA Elements (ENCODE) project, and has been identified as a potential binding region for multiple transcription factors (37, 38).The mechanisms by which differential methylation at these loci relate to cigarette smoke exposure remains unknown.
Notably, the most highly associated loci identified in our analyses appear distinct from the sites identified in epigenome-wide studies performed in peripheral blood or blood-derived tissues. Strong associations between methylation at the coagulation factor II receptor–like 3 (F2RL3) and aryl-hydrocarbon receptor repressor (AHRR) have been reported in independent populations (19, 39–42), some of which employed the same array-based platform used in our study (40, 42). A total of 27 sites annotated to AHRR and four sites annotated to F2RL3 were nominally associated (unadjusted P < 0.05) with current smoking in our analysis (data not shown); however, none of these sites were significant after adjustment for multiple comparisons. The results at these two loci were unchanged when we implemented a sliding window approach to identify differentially methylated regions (43). The different loci identified in studies performed in blood relative to tissues derived from the aerodigestive tract, as well as the magnitude of differential methylation observed at significant loci, may highlight tissue-specific effects of cigarette smoking on DNA methylation.
Whether changes in buccal mucosal methylation are representative of changes occurring in lung and lung-related tissues is a topic that warrants additional investigation. In silico changes in gene expression at protein coding loci identified in our analyses (CYP1B1, FRMD4A, and PARVA) are highly consistent across a variety of airway and lung-derived tissues (2, 3, 20–26). In addition, two of these loci (CYP1B1 and PARVA) were also reported to be differentially methylated in small airway epithelium (21). The association between methylation at these loci with smoking-related phenotypes, such as cumulative cigarette smoke exposure and time since smoking cessation, as well as spirometric lung function and radiographic emphysema, suggests that DNA methylation patterns in buccal source DNA may capture additional dimensions of cigarette smoke exposure relevant to the development of lung disease, including the differential susceptibility by sex reported in epidemiological studies (44–46). A separate study examining the DNA methylation patterns of small airways epithelium among former smokers with COPD demonstrated multiple loci associated with both quantitative lung function and cumulative smoker exposure (47).
Functional annotation clustering revealed a significant enrichment of several domains. The pleckstrin homology domain is comprised of approximately 100 amino acids organized into seven antiparallel β sheets with an α helix at the carboxy terminus surrounding a ligand binding pocket (48). Pleckstrin homology domains are contained in a wide range of proteins, and are believed to be instrumental in cell signaling pathways; their relationship to current smoking status has not previously been explored. In contrast, the cadherins represent a family of calcium-dependent transmembrane proteins involved in cell adhesion and morphogenesis (49)—as such, their role in establishing and maintaining barrier functions against environmental toxins, such as cigarette smoke, has been studied extensively. Exposure to cigarette smoke has been shown to alter methylation patterns (50) and to decrease the expression of epithelial cadherin (E-cadherin) in human lung-derived tissues (50–52); thus, our finding of an enrichment of cadherin-associated domains is biologically plausible.
We acknowledge the following limitations to our study. First, our sample size was limited, which may have reduced our power to detect moderate or small differences in methylation. We are, however, encouraged that many of the most highly associated sites demonstrated considerable differences in methylation; notably, the difference in methylation by current smoking status are greater than the differences reported in studies conducted in blood (19, 39). Second, although the impact of ancestral (racial) heterogeneity on site-specific DNA methylation patterns is incompletely characterized, the inclusion of African American subjects (which account for ∼27% of our cohort) in our analyses increases the generalizability of our findings. In addition, we contend that our results our robust, given: (1) the majority of the most highly associated sites remained robust when we performed a race-stratified analysis; and (2) the lack of known probe sequence polymorphisms (single-nucleotide polymorphisms, etc.) within 10 base pairs of the differentially methylated CpG sites reported. Third, the lack of simultaneous gene expression data limits our ability to directly assess associations between methylation changes and gene expression in our samples. We assert that the findings from publicly available datasets, including an integrated methylation and gene expression dataset from small airways (21), support our findings. Fourth, we rely upon self-reported smoking status and lack confirmatory serum/urine cotinine measurements. It should be noted, however, that, except in populations where current smoking may be perceived negatively (such as among teenagers or pregnant women), self-reported smoking status has been shown to be highly reliable (53–55). Fifth, we were unable to validate the array-based, site-specific methylation estimates using a second technology, such as pyrosequencing, due to lack of sufficient remaining biological material from the initial (discovery) cohort. We believe that this shortcoming is mitigated by the pyrosequencing performed in the independent replication cohort. Finally, the lack of longitudinal data and never-smokers in our cohort limits our ability to assess permanent changes in methylation due to ever smoking—these questions serve as the basis for future work in the COPDGene cohort. We conclude that buccal mucosal methylation can serve as a biomarker of environmental exposures relevant to chronic respiratory diseases. Future studies should strive to obtain contemporaneously ascertained buccal and lower airway tract samples to directly evaluate the correlation in methylation and expression patterns between these two sites.
Acknowledgments
The Members of the COPDGene Core Teams: Administrative Core: James Crapo, M.D. (Principal Investigator), Edwin Silverman, M.D., Ph.D. (Principal Investigator), Barry Make, M.D., Elizabeth Regan, M.D., Ph.D., Rochelle Lantz, Lori Stepp, Sandra Melanson. Genetic Analysis Core: Terri Beaty, Ph.D., Barbara Klanderman, Ph.D., Nan Laird, Ph.D., Christoph Lange, Ph.D., Michael Cho, M.D., Stephanie Santorico, Ph.D., John Hokanson, M.P.H., Ph.D., Dawn DeMeo, M.D., M.P.H., Nadia Hansel, M.D., M.P.H., Craig Hersh, M.D., M.P.H., Peter Castaldi, M.D., M.Sc., Merry-Lynn McDonald, Ph.D., Jing Zhou, M.D., Ph.D., Manuel Mattheissen, M.D., Ph.D., Emily Wan, M.D., Megan Hardin, M.D., Jacqueline Hetmanski, M.S., Margaret Parker, M.S., Tanda Murray, M.S. Imaging Core: David Lynch, M.B., Joyce Schroeder, M.D., John Newell, Jr., M.D., John Reilly, M.D., Harvey Coxson, Ph.D., Philip Judy, Ph.D., Eric Hoffman, Ph.D., George Washko, M.D., Raul San Jose Estepar, Ph.D., James Ross, M.Sc., Mustafa Al Qaisi, M.D., Jordan Zach, Alex Kluiber, Jered Sieren, Tanya Mann, Deanna Richert, Alexander McKenzie, Jaleh Akhavan, Douglas Stinson. “Pulmonary Function Test Quality Assurance” Core, “Intermountain Latter Day Saints (LDS)” Hospital, Salt Lake City, Utah: Robert Jensen, Ph.D. Biological Repository, Johns Hopkins University, Baltimore, Maryland: Homayoon Farzadegan, Ph.D., Stacey Meyerer, Shivam Chandan, Samantha Bragan. Data Coordinating Center and Biostatistics, National Jewish Health, Denver, Colorado: Douglas Everett, Ph.D., Andre Williams, Ph.D., Carla Wilson, M.S., Anna Forssen, M.S., Amber Powell, Joe Piccoli. Epidemiology Core, University of Colorado School of Public Health, Denver, Colorado: John Hokanson, M.P.H., Ph.D., Marci Sontag, Ph.D., Jennifer Black-Shinn, M.P.H., Gregory Kinney, M.P.H., Sharon Lutz, M.P.H., Ph.D.
The COPDGene Investigators from the Participating Clinical Centers: Ann Arbor Department of Veterans Affairs, Ann Arbor, Michigan: Jeffrey Curtis, M.D., Ella Kazerooni, M.D. Baylor College of Medicine, Houston, Texas: Nicola Hanania, M.D., MS, Philip Alapat, M.D., Venkata Bandi, M.D., Kalpalatha Guntupalli, M.D., Elizabeth Guy, M.D., Antara Mallampalli, M.D., Charles Trinh, M.D., Mustafa Atik, M.D., Hasan Al-Azzawi, M.D., Marc Willis, D.O., Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., Anne Marie Marciel, M.D. Brigham and Women’s Hospital, Boston, Massachusetts: Dawn DeMeo, M.D., M.P.H., Craig Hersh, M.D., M.P.H., George Washko, M.D., Francine Jacobson, M.D., M.P.H., Hiroto Hatabu, M.D., Ph.D., Peter Clarke, M.D., Ritu Gill, M.D., Andetta Hunsaker, M.D., Beatrice Trotman-Dickenson, M.B.B.S., Rachna Madan, M.D. Columbia University, New York, New York: R. Graham Barr, M.D., Dr.PH., Byron Thomashow, M.D., John Austin, M.D., Belinda D’Souza, M.D. Duke University Medical Center, Durham, North Carolina: Neil MacIntyre, Jr., M.D., Lacey Washington, M.D., H. Page McAdams, M.D. Fallon Clinic, Worcester, Massachusetts: Richard Rosiello, M.D., Timothy Bresnahan, M.D., Joseph Bradley, M.D., Sharon Kuong, M.D., Steven Meller, M.D., Suzanne Roland, M.D. Health Partners Research Foundation, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H., Joseph Tashjian, M.D. Johns Hopkins University, Baltimore, Maryland: Robert Wise, M.D., Nadia Hansel, M.D., M.P.H., Robert Brown, M.D., Gregory Diette, M.D., Karen Horton, M.D. Los Angeles Biomedical Research Institute at Harbor University of California Los Angeles Medical Center, Los Angeles, California: Richard Casaburi, M.D., Janos Porszasz, M.D., Ph.D., Hans Fischer, M.D., Ph.D., Matt Budoff, M.D., Mehdi Rambod, M.D. Michael E. DeBakey Department of Veterans Affairs Medical Center, Houston, Texas: Amir Sharafkhaneh, M.D., Charles Trinh, M.D., Hirani Kamal, M.D., Roham Darvishi, M.D., Marc Willis, DO, Susan Pinero, M.D., Linda Fahr, M.D., Arun Nachiappan, M.D., Collin Bray, M.D., L. Alexander Frigini, M.D., Carlos Farinas, M.D., David Katz, M.D., Jose Freytes, M.D., Anne Marie Marciel, M.D. Minneapolis Department of Veterans Affairs, Minneapolis, Minnesota: Dennis Niewoehner, M.D., Quentin Anderson, M.D., Kathryn Rice, M.D., Audrey Caine, M.D. Morehouse School of Medicine, Atlanta, Georgia: Marilyn Foreman, M.D., MS, Gloria Westney, M.D., MS, Eugene Berkowitz, M.D., Ph.D. National Jewish Health, Denver, Colorado: Russell Bowler, M.D., Ph.D., David Lynch, MB, Joyce Schroeder, M.D., Valerie Hale, M.D., John Armstrong, II, M.D., Debra Dyer, M.D., Jonathan Chung, M.D., Christian Cox, M.D. Temple University, Philadelphia, Pennsylvania: Gerard Criner, M.D., Victor Kim, M.D., Nathaniel Marchetti, DO, Aditi Satti, M.D., A. James Mamary, M.D., Robert Steiner, M.D., Chandra Dass, M.D., Libby Cone, M.D. University of Alabama, Birmingham, Alabama: William Bailey, M.D., Mark Dransfield, M.D., Michael Wells, M.D., Surya Bhatt, M.D., Hrudaya Nath, M.D., Satinder Singh, M.D. University of California, San Diego, California: Joe Ramsdell, M.D., Paul Friedman, M.D. University of Iowa, Iowa City, Iowa: Alejandro Cornellas, M.D., John Newell, Jr., M.D., Edwin J. R. van Beek, M.D., Ph.D. University of Michigan, Ann Arbor, Michigan: Fernando Martinez, M.D., MeiLan Han, M.D., Ella Kazerooni, M.D. University of Minnesota, Minneapolis, Minnesota: Christine Wendt, M.D., Tadashi Allen, M.D. University of Pittsburgh, Pittsburgh, Pennsylvania: Frank Sciurba, M.D., Joel Weissfeld, M.D., M.P.H., Carl Fuhrman, M.D., Jessica Bon, M.D., Danielle Hooper, M.D. University of Texas Health Science Center at San Antonio, San Antonio, Texas: Antonio Anzueto, M.D., Sandra Adams, M.D., Carlos Orozco, M.D., Mario Ruiz, M.D., Amy Mumbower, M.D., Ariel Kruger, M.D., Carlos Restrepo, M.D., Michael Lane, M.D.
Acknowledgments
The authors acknowledge and thank the COPDGene Core teams and the COPDGene investigators from the participating Clinical Centers.
Footnotes
This work was supported by a Parker B. Francis Foundation Fellowship and a Career Development Award from the Brigham and Women’s Center for Faculty Development and Diversity (E.S.W.). The COPDGene Study is supported by National Institutes of Health (NIH) grants NIH R01 HL089856 and NIH R01 HL089897 (J.D.C.). The COPDGene project is also supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, and Sunovion. Additional support for this project was provided by NIH grants R01 HL089438 (D.L.D.) and NIH P01 HL105339.
Disclaimer: The content of this manuscript is solely the responsibility of the authors; none of the above-named entities participated in the design or conduct of the study, the collection, management, analysis, or interpretation of the data, or the preparation, review, approval, or decision to submit the manuscript for publication. E.S.W. had full access to all of the data in this study, and takes complete responsibility for the integrity of the data and the accuracy of the data analysis.
Author Contributions: All authors participated in manuscript drafting and revisions; E.S.W. and D.L.D. contributed to the concept and design of the study; E.S.W. and J.M. performed the data analysis; W.Q and V.J.C. provided statistical support; D.L.D., H.B., M.G.F., J.E.H., and R.P.B. assisted with data collection; J.D.C. and D.L.D. provided funding support.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0103OC on December 17, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.U.S. Department of Health and Human Services The health consequences of smoking: a report of the surgeon general Washington, DC: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Office on Smoking and Health2004 [Google Scholar]
- 2.Boyle JO, Gümüs ZH, Kacker A, Choksi VL, Bocker JM, Zhou XK, Yantiss RK, Hughes DB, Du B, Judson BL, et al. Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev Res (Phila) 2010;3:266–278. doi: 10.1158/1940-6207.CAPR-09-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA. 2004;101:10143–10148. doi: 10.1073/pnas.0401422101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, Liu G, Dumas YM, Zhang X, Brody JS, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kupfer DM, White VL, Jenkins MC, Burian D. Examining smoking-induced differential gene expression changes in buccal mucosa. BMC Med Genomics. 2010;3:24. doi: 10.1186/1755-8794-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spivack SD, Hurteau GJ, Jain R, Kumar SV, Aldous KM, Gierthy JF, Kaminsky LS. Gene–environment interaction signatures by quantitative mRNA profiling in exfoliated buccal mucosal cells. Cancer Res. 2004;64:6805–6813. doi: 10.1158/0008-5472.CAN-04-1771. [DOI] [PubMed] [Google Scholar]
- 7.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breton CV, Salam MT, Wang X, Byun HM, Siegmund KD, Gilliland FD. Particulate matter, DNA methylation in nitric oxide synthase, and childhood respiratory disease. Environ Health Perspect. 2012;120:1320–1326. doi: 10.1289/ehp.1104439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, Gilliland FD. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2011;129:232–239.e1–7. doi: 10.1016/j.jaci.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabriel HE, Crott JW, Ghandour H, Dallal GE, Choi SW, Keyes MK, Jang H, Liu Z, Nadeau M, Johnston A, et al. Chronic cigarette smoking is associated with diminished folate status, altered folate form distribution, and increased genetic damage in the buccal mucosa of healthy adults. Am J Clin Nutr. 2006;83:835–841. doi: 10.1093/ajcn/83.4.835. [DOI] [PubMed] [Google Scholar]
- 11.Torrone Dz, Kuriakose J, Moors K, Jiang H, Niedzwiecki M, Perera F, Miller R. Reproducibility and intraindividual variation over days in buccal cell DNA methylation of two asthma genes, interferon γ (IFNγ) and inducible nitric oxide synthase (iNOS) Clin Epigenetics. 2012;4:3. doi: 10.1186/1868-7083-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan ES, Qiu W, Carey VJ, Bacherman H, Hokanson JE, Bowler RP, Crapo JD, DeMeo DL. The effects of current smoking on buccal DNA methylation in the COPDgene study [abstract] Am J Respir Crit Care Med. 2013;187:A5762. [Google Scholar]
- 13.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hansen K, Aryee M.minfi: analyze Illumina’s 450K methylation arrays. R package version 1.4.0; 2012
- 15.Leek J, Johnson E, Parker H, Jaffe A, Storey J.sva: surrogate variable analysis. R package version 3.4.0; 2012
- 16.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, Agusti A, Anderson W, Lomas DA, Demeo DL. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007;8:R201. doi: 10.1186/gb-2007-8-9-r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buro-Auriemma LJ, Salit J, Hackett NR, Walters MS, Strulovici-Barel Y, Staudt MR, Fuller J, Mahmoud M, Stevenson CS, Hilton H, et al. Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum Mol Genet. 2013;22:4726–4738. doi: 10.1093/hmg/ddt326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graff JW, Powers LS, Dickson AM, Kim J, Reisetter AC, Hassan IH, Kremens K, Gross TJ, Wilson ME, Monick MM. Cigarette smoking decreases global microRNA expression in human alveolar macrophages. PLoS One. 2012;7:e44066. doi: 10.1371/journal.pone.0044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philibert RA, Sears RA, Powers LS, Nash E, Bair T, Gerke AK, Hassan I, Thomas CP, Gross TJ, Monick MM. Coordinated DNA methylation and gene expression changes in smoker alveolar macrophages: specific effects on VEGF receptor 1 expression. J Leukoc Biol. 2012;92:621–631. doi: 10.1189/jlb.1211632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steiling K, van den Berge M, Hijazi K, Florido R, Campbell J, Liu G, Xiao J, Zhang X, Duclos G, Drizik E, et al. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am J Respir Crit Care Med. 2013;187:933–942. doi: 10.1164/rccm.201208-1449OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Ahmed J, Wang G, Hassan I, Strulovici-Barel Y, Hackett NR, Crystal RG. Down-regulation of the canonical Wnt β-catenin pathway in the airway epithelium of healthy smokers and smokers with COPD. PLoS One. 2011;6:e14793. doi: 10.1371/journal.pone.0014793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, Paquet AC, Erle DJ. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med. 2005;172:1383–1392. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe R, Gemma C, Beyan H, Hawa MI, Bazeos A, Leslie RD, Montpetit A, Rakyan VK, Ramagopalan SV. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8:445–454. doi: 10.4161/epi.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada T, Hayes CL, Yamazaki H, Amin S, Hecht SS, Guengerich FP, Sutter TR. Activation of chemically diverse procarcinogens by human cytochrome P-450 1B1. Cancer Res. 1996;56:2979–2984. [PubMed] [Google Scholar]
- 29.Butts SF, Sammel M.D., Greer C, Rebbeck TR, Boorman DW, Freeman EW. Cigarettes, genetic background, and menopausal timing: the presence of single nucleotide polymorphisms in cytochrome P450 genes is associated with increased risk of natural menopause in European-American smokers. Menopause. 2014;21:694–701. doi: 10.1097/GME.0000000000000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cote ML, Yoo W, Wenzlaff AS, Prysak GM, Santer SK, Claeys GB, Van Dyke AL, Land SJ, Schwartz AG. Tobacco and estrogen metabolic polymorphisms and risk of non–small cell lung cancer in women. Carcinogenesis. 2009;30:626–635. doi: 10.1093/carcin/bgp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timofeeva MN, Kropp S, Sauter W, Beckmann L, Rosenberger A, Illig T, Jäger B, Mittelstrass K, Dienemann H, Bartsch H, et al. LUCY-Consortium. CYP450 polymorphisms as risk factors for early-onset lung cancer: gender-specific differences. Carcinogenesis. 2009;30:1161–1169. doi: 10.1093/carcin/bgp102. [DOI] [PubMed] [Google Scholar]
- 32.Goldie SJ, Mulder KW, Tan DW, Lyons SK, Sims AH, Watt FM. FRMD4A upregulation in human squamous cell carcinoma promotes tumor growth and metastasis and is associated with poor prognosis. Cancer Res. 2012;72:3424–3436. doi: 10.1158/0008-5472.CAN-12-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon D, Kim YJ, Cui WY, Van der Vaart A, Cho YS, Lee JY, Ma JZ, Payne TJ, Li MD, Park T. Large-scale genome-wide association study of Asian population reveals genetic factors in FRMD4A and other loci influencing smoking initiation and nicotine dependence. Hum Genet. 2012;131:1009–1021. doi: 10.1007/s00439-011-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose JE, Behm FM, Drgon T, Johnson C, Uhl GR. Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. Mol Med. 2010;16:247–253. doi: 10.2119/molmed.2009.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korenbaum E, Olski TM, Noegel AA. Genomic organization and expression profile of the parvin family of focal adhesion proteins in mice and humans. Gene. 2001;279:69–79. doi: 10.1016/s0378-1119(01)00743-0. [DOI] [PubMed] [Google Scholar]
- 36.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking–related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P, Flanagan JM. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22:843–851. doi: 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- 41.Sun YV, Smith AK, Conneely KN, Chang Q, Li W, Lazarus A, Smith JA, Almli LM, Binder EB, Klengel T, et al. Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum Genet. 2013;132:1027–1037. doi: 10.1007/s00439-013-1311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du P, Bourgon R.methylanalysis: DNA methylation data analysis and visualization. R package 1.4.2, version 1.6.0; 2014
- 44.Gan WQ, Man SF, Postma DS, Camp P, Sin DD. Female smokers beyond the perimenopausal period are at increased risk of chronic obstructive pulmonary disease: a systematic review and meta-analysis. Respir Res. 2006;7:52. doi: 10.1186/1465-9921-7-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez FJ, Curtis JL, Sciurba F, Mumford J, Giardino ND, Weinmann G, Kazerooni E, Murray S, Criner GJ, Sin DD, et al. National Emphysema Treatment Trial Research Group. Sex differences in severe pulmonary emphysema. Am J Respir Crit Care Med. 2007;176:243–252. doi: 10.1164/rccm.200606-828OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sørheim IC, Johannessen A, Gulsvik A, Bakke PS, Silverman EK, DeMeo DL. Gender differences in COPD: are women more susceptible to smoking effects than men? Thorax. 2010;65:480–485. doi: 10.1136/thx.2009.122002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vucic EA, Chari R, Thu KL, Wilson IM, Cotton AM, Kennett JY, Zhang M, Lonergan KM, Steiling K, Brown CJ, et al. DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. Am J Respir Cell Mol Biol. 2014;50:912–922. doi: 10.1165/rcmb.2013-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ingley E, Hemmings BA. Pleckstrin homology (PH) domains in signal transduction. J Cell Biochem. 1994;56:436–443. doi: 10.1002/jcb.240560403. [DOI] [PubMed] [Google Scholar]
- 49.Maître JL, Heisenberg CP. Three functions of cadherins in cell adhesion. Curr Biol. 2013;23:R626–R633. doi: 10.1016/j.cub.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Word B, Lyn-Cook LE, Jr, Mwamba B, Wang H, Lyn-Cook B, Hammons G. Cigarette smoke condensate induces differential expression and promoter methylation profiles of critical genes involved in lung cancer in NL-20 lung cells in vitro: short-term and chronic exposure. Int J Toxicol. 2013;32:23–31. doi: 10.1177/1091581812465902. [DOI] [PubMed] [Google Scholar]
- 51.Forteza RM, Casalino-Matsuda SM, Falcon NS, Valencia Gattas M, Monzon ME. Hyaluronan and layilin mediate loss of airway epithelial barrier function induced by cigarette smoke by decreasing E-cadherin. J Biol Chem. 2012;287:42288–42298. doi: 10.1074/jbc.M112.387795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oldenburger A, Poppinga WJ, Kos F, de Bruin HG, Rijks WF, Heijink IH, Timens W, Meurs H, Maarsingh H, Schmidt M. A-kinase anchoring proteins contribute to loss of E-cadherin and bronchial epithelial barrier by cigarette smoke. Am J Physiol Cell Physiol. 2014;306:C585–C597. doi: 10.1152/ajpcell.00183.2013. [DOI] [PubMed] [Google Scholar]
- 53.Patrick DL, Cheadle A, Thompson DC, Diehr P, Koepsell T, Kinne S. The validity of self-reported smoking: a review and meta-analysis. Am J Public Health. 1994;84:1086–1093. doi: 10.2105/ajph.84.7.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong SL, Shields M, Leatherdale S, Malaison E, Hammond D. Assessment of validity of self-reported smoking status. Health Rep. 2012;23:47–53. [PubMed] [Google Scholar]
- 55.Yeager DS, Krosnick JA. The validity of self-reported nicotine product use in the 2001–2008 National Health and Nutrition Examination Survey. Med Care. 2010;48:1128–1132. doi: 10.1097/MLR.0b013e3181ef9948. [DOI] [PubMed] [Google Scholar]