Abstract
HLA-DRB1 is a sarcoidosis risk gene, and the *03:01 allele is strongly associated with disease resolution in European sarcoidosis cases. Whereas the HLA-DRB1 variation is associated with sarcoidosis susceptibility in African Americans, DRB1 risk alleles are not as well defined, and associations with disease resolution have not been studied. Associations between genotyped and imputed HLA-DRB1 alleles and disease susceptibility/resolution were evaluated in a sample of 1,277 African-American patients with sarcoidosis and 1,467 control subjects. In silico binding assays were performed to assess the functional significance of the associated alleles. Increased disease susceptibility was associated with the HLA-DRB1 alleles *12:01 (odds ratio [OR], 2.11; 95% confidence interval [CI], 1.65–2.69; P = 3.2 × 10−9) and *11:01 (OR, 1.69; 95% CI, 1.42–2.01; P = 3.0 × 10−9). The strongest protective association was found with *03:01 (OR, 0.56; 95% CI, 0.44–0.73; P = 1.0 × 10−5). The African-derived allele *03:02 was associated with decreased risk of persistent radiographic disease (OR, 0.52; 95% CI, 0.37–0.72; P = 1.3 × 10−4), a finding consistent across the three component studies comprising the analytic sample. The DRB1*03:01 association with disease persistence was dependent upon local ancestry, with carriers of at least one European allele at DRB1 at a decreased risk of persistent disease (OR, 0.36; 95% CI, 0.14–0.94; P = 0.037). Results of in silico binding analyses showed that DRB1*03:01 consistently demonstrated the highest binding affinities for six bacterial peptides previously found in sarcoidosis granulomas, whereas *12:01 displayed the lowest binding affinities. This study has identified DRB1*03:01 and *03:02 as novel alleles associated with disease susceptibility and course in African Americans. Further investigation of DRB1*03 alleles may uncover immunologic factors that favor sarcoidosis protection and resolution among African Americans.
Keywords: HLA-DRB1 chains, molecular docking simulation, pulmonary sarcoidosis, SNPs, association studies, genetic
Clinical Relevance
African Americans suffer disproportionately from sarcoidosis incidence and severity of disease. The current study reveals evidence of novel/distinct HLA-DRB1 alleles associated with susceptibility and disease course in African Americans that may provide insight into how distinct HLA backgrounds direct the pathogenesis of sarcoidosis differently according to ancestral genetic origin. If confirmed, our results indicating an association between HLA-DRB1*03:02 allele and a resolving course of disease may have direct clinical relevance as a potential marker for contraindications for treatment of sarcoidosis in African Americans, similar to the previous recommendations made for European HLA-DRB1*03:01 carriers with a greater risk of Löfgren's syndrome.
Sarcoidosis is caused by a dysregulated immune response to one or more yet-unidentified antigens (1). Human leukocyte antigen (HLA) class II molecules present peptide ligands from these antigens to T lymphocytes, initiating the immune response (2), which results in the formation of characteristic noncaseating granulomas. Pulmonary involvement ranges from spontaneous resolution with no impairment to fibrotic lung disease with respiratory failure. Patterns of familial clustering and differences in incidence by race and ethnicity suggest that genetic factors contribute to disease susceptibility, presentation, and resolution (1).
The strongest genetic associations with sarcoidosis are found in the HLA class II region of Chromosome 6p; among these genes, HLA-DRB1 is the most widely validated, with numerous associated risk variants (3–11). In the United States, African Americans suffer disproportionately from higher sarcoidosis prevalence and from more severe disease (12–14). Unfortunately, studies of DRB1 associations with sarcoidosis have been confined almost exclusively to populations of European origin. One multiethnic study did find that HLA-DRB1*11:01 is associated with increased sarcoidosis susceptibility across race-ethnicity groups, whereas risk associations with DRB1*12:01 and *15:01 were specific to African Americans and European Americans, respectively (7).
In addition to disease susceptibility, variation at DRB1 is associated with organ-specific involvement (7, 11) and disease course (3, 8, 9, 15–17). The allele DRB1*03 belongs to the European-derived “8.1 ancestral haplotype,” made up primarily of *03:01 carriers (18). Grunewald and colleagues demonstrated that 95% of patients with Löfgren’s syndrome (19) (an acute subtype of sarcoidosis) who were *03-positive experienced disease resolution within 2 years, whereas disease resolved for only half of *03-negative patients (20). However, such associations have not been evaluated in African Americans.
The class II molecules bind peptides on the surface of antigen-presenting cells; these peptides are subsequently recognized by CD4+ T cells. The docking of an antigenic peptide is facilitated by polymorphic residues in binding pockets (21, 22); a given protein prefers specific amino acid residues at distinct positions within a binding pocket while excluding others. For DRB1, anchor residue preferences for pockets 1, 4, 6/7, and 9 determine binding affinities that vary across 21 variable anchor residues. In sarcoidosis, T cell–mediated immune responses to mycobacterial antigens have been shown to be dependent upon DRB1 genotype (23). In silico predictions of peptide binding affinities (24) can be used to predict which DRB1 allele/antigen combinations, which are likely to initiate the most robust immune response in patients with sarcoidosis (25).
Capitalizing on our large sample of African-American patients with sarcoidosis and control subjects (n = 2,774) with available genome-wide genotyping data (26) and a subset (n = 325) also possessing DRB1 two-field allele (with the colon separating the first and second fields for each allele) typing (7), we used an imputation strategy (27) to estimate two-field DRB1 alleles in the full sample. The goals of this study were (1) to assess HLA–DRB1 associations with sarcoidosis susceptibility and clinical phenotypes in African-American patients and to determine whether these associations are dependent upon local West African/European ancestry at DRB1 and (2) to use bioinformatics approaches to assess their functional relevance to a T cell–mediated immune response.
Materials and Methods
Study Sample and Disease Phenotypes
Our complete analytic sample includes 2,744 self-identified African Americans (1,277 patients with sarcoidosis and 1,467 control subjects) with genome-wide genotyping from our previous genome-wide association study (26). The sample was assembled from the following three studies: (1) A Case-Control Etiologic Study of Sarcoidosis (ACCESS) (28), (2) a multisite affected-sibling sarcoidosis linkage study (29), and (3) a nuclear family–based sample ascertained through a single affected individual (30). The varied sampling schemes across studies resulted in a final analytic sample of related and unrelated cases and control subjects (see Table E1 in the online supplement for a more complete description of the study sample). Diagnostic criteria for patients with sarcoidosis and follow-up for radiographic resolution of pulmonary disease were described previously (31). Across all three studies, patients with sarcoidosis met diagnostic criteria for “definite” or “highly probable” sarcoidosis (29). “Definite” cases had histological confirmation of noncaseating granulomas and evidence of disease in either the thorax or two or more other organ systems; “highly probable” cases lacked histologic confirmation but had characteristic chest radiographs (bilateral symmetrical hilar adenopathy) and either a history of erythema nodosum or at least 2 years observation during which time no other disease was found to explain radiographic abnormalities. In ACCESS, cases were limited to those with definite sarcoidosis, whereas the other two studies included definite and highly probable cases.
Where possible, cases were phenotyped as to the resolution or persistence of radiographic evidence for lung disease after the initial date of diagnosis with a minimum of 2 years of follow-up to confirm persistent disease. Data collection was retrospective except for the ACCESS cases recruited during the first 2 years of enrollment, where the study protocol dictated a 2-year follow-up exam (13). Patients who presented with a Scadding stage IV chest radiograph (evidence of lung fibrosis or scarring) did not require follow-up X-rays because stage IV indicates permanent lung scarring. Organ involvement was assessed using ACCESS study criteria (32). For analytic purposes, any of the three defined levels of certainty (definite, probable, or possible) were considered positive for organ involvement.
All study protocols were approved by the Henry Ford Health System (Detroit, MI) Institutional Review Board. Subjects provided informed consent to all study procedures.
Genotyping and HLA-DRB1 Classical Allele Imputation
HLA typing was conducted for the class II HLA-DRB1 gene in 325 self-identified African-American subjects (156 patients with sarcoidosis and 169 control subjects) who were part of the ACCESS sample with genome-wide genotyping; details are reported in Rossman and colleagues (7). Intermediate (two-field) resolution typing was performed with sequence-specific oligonucleotide probes available through Orchid Diagnostics (Princeton, NJ) (7). Genome-wide genotyping was performed on the full sample using the Illumina HumanOmni1-Quad (San Diego, CA) (26).
The 325 individuals with both genome-wide and two-field HLA-DRB1 allele genotyping were used as reference to impute the HLA-DRB1 classical alleles for the remainder of the sample. Imputation was conducted using the HLA Genotype Imputation with Attribute Bagging (HIBAG) method (27). Briefly, HIBAG takes advantage of the broader haplotype structure present in the HLA region to construct multiple classifiers that predict classical HLA alleles based on genotyped single nucleotide polymorphisms (SNPs) available from genome-wide genotyping platforms. The results from multiple classifiers are combined to form the HIBAG consensus best genotype prediction and posterior probability estimates for all possible HLA genotypes. In addition to the genotyped HLA-DRB1 alleles, the prediction model was constructed using 3,144 SNPs in a region spanning 500 kb on either side of the largest HLA-DRB1 mRNA transcript present in the NCBI RefSeq database, which mapped to human genome build 19 chromosome 6 base-pair position 32,546,547 to 32,557,613. Twenty-five classifiers were used to construct the final model for imputation in the remaining 2,417 subjects with genome-wide genotyping only. We have previously reported on this African-American DRB1 prediction model (33); the accuracy of the model was evaluated via cross-validation with an overall allele prediction accuracy of 91%. The allele-specific sensitivities (i.e., the percentage of a particular allele correctly predicted) for the top DRB1 alleles associated with risk and/or persistent disease in the current study were 100% (*03:01), 96% (*03:02), 100% (*11:01), and 90% (*12:01).
Statistical Analyses
Associations were assessed using HLA-DRB1 two-field allele dosage estimates, calculated based on the HIBAG posterior genotype probabilities for each subject. For the 325 subjects with direct HLA-DRB1 genotyping, genotypes were assumed to be known, with probability equal to 1. For each individual and allele, a dosage estimate was calculated as a weighted average of 0, 1, or 2 copies of each classical allele, where the weights (p0, p1, and p2, respectively) were calculated as the sum of the HIBAG genotype posterior probability estimates corresponding to 0, 1, and 2 copies of a particular allele. For example, the weight “p2” for two copies of the two-field *11:01 allele is the posterior probability estimate for the homozygous genotype (*11:01/*11:01), and the weight “p1” for one copy of the *11:01 allele is the summation of all heterozygous genotype (i.e., *11:01−, where “−” represents any allele other than *11:01) posterior probabilities. The weight “p0” is the summation of the probabilities of all remaining genotypes (i.e., −/−). The *11:01 allele dosage for a subject is then calculated as 2*p2 + 1*p1 + 0*p0.
HLA-DRB1 allele associations were estimated for risk of disease (case-control) and for disease course based on radiographic evidence of disease (resolved, persistent, and the subset of those with persistent Scadding stage IV disease) after a minimum of 2 years of follow-up in sarcoidosis cases only. To account for relatedness within the sample, HLA-DRB1 allele odds ratio (OR) estimates adjusted for sex and genome-wide percent West African ancestry were calculated using generalized estimating equations (GEE) fit with a logistic model treating each family as a cluster and using an independence working correlation structure, which we have shown appropriately controls the type 1 error rate for this sample (34). Because the majority of relationships in the two family-based studies were first-degree relationships, an exchangeable correlation structure was used for study-specific models. As a further check for cryptic relatedness, we calculated statistical significance using the linear mixed model implemented in the Efficient Mixed-Model Association eXpedited (EMMAX) method (35), which assumes a linear relationship between the proportion of risk and HLA-DRB1 alleles. The model accounts for the known and unknown relationships through an empirical kinship matrix calculated based on identity-by-state sharing of the genome-wide SNPs between all pairs of members of the study. The statistical significance between GEE and EMMAX results were remarkably consistent, and we limit the inclusion of the EMMAX P values to Table E2, which also contains the GEE-based results for all alleles. Both the additive (allele dosage) and dominant (dichotomized dosage, with estimates ≥0.5 defined as carriers) genetic models were used to estimate the OR for each HLA-DRB1 allele; a one-degree-of-freedom Wald test was used to evaluate statistical significance. Because allele frequency estimates were low (generally <0.05) and the results from both models were consistent, the results from only the dominant model are reported. To test for heterogeneity of each allelic association by study, multiplicative interaction terms comprising the specific allele and study terms were included in each model, with statistical significance evaluated using a two-degree-of-freedom Wald test. Heterogeneity by local West African ancestry at DRB1 (0, 1, or 2 African segments) was evaluated in the same manner, where local West African ancestry was estimated using the Local Ancestry in Admixed Populations method (36) as part of a prior publication (37). Except where otherwise noted, all analyses were performed using the R programming language (version 2.15.1; R Foundation for Statistical Computing, Vienna, Austria).
In Silico Binding Assays
We used Immune Epitope Database version 2.4 (38), which contains 18,133 HLA class II peptide-binding measurements across 31 full-typed HLA loci. To estimate peptide binding affinities for a given DRB1 molecule, the prediction tool uses a consensus approach to identify the best performing algorithm for a specific allele or combines multiple algorithms for superior consensus predictions (39). We primarily used NN_align (40) and NetMHCIIpan (41) for binding affinity predictions; NN_align uses an artificial neural network–based algorithm trained individually for each allele, whereas NetMHCIIpan is a pan-specific method that takes into account the antigenic peptide and HLA sequence to predict binding affinities where no experimental data exist. For each peptide, a percentile rank is generated by comparing the peptide's binding affinity score against the scores of five million random 15-mer peptides selected from the SWISSPROT database (http://www.ebi.ac.uk/uniprot). A low percentile rank indicates high binding affinity. In addition to the percentile rank, we report the half-maximal inhibitory concentration (IC50) of each possible 15-mer across the peptide sequence. As a rough guideline, IC50 values of less than 50 indicate high affinity peptides, values greater than or equal to 50 and less than 500 nM indicate intermediate affinity, and values greater than or equal to 500 and less than 5,000 nM indicate low affinity.
Results
Association of DRB1 Alleles with Sarcoidosis Susceptibility and Disease Course
Among the full sample of 2,744 self-identified African Americans (1,277 patients with sarcoidosis and 1,467 control subjects; see Table E1 for descriptive statistics of the sample), the HLA-DRB1 alleles most significantly associated with sarcoidosis susceptibility were *11:01, *12:01, and *03:01 (Table 1). Increased sarcoidosis susceptibility was observed among carriers of either *11:01 (OR, 1.69; 95% confidence interval [CI], 1.42–2.01; P = 3.0 × 10−9) or *12:01 (OR, 2.11; 95% CI, 1.65–2.69; P = 3.2 × 10−9). Conversely, decreased risk of sarcoidosis development was observed among *03:01 allele carriers (OR, 0.56; 95% CI, 0.44–0.73; P = 1.0 × 10−5). Nominally significant (P < 0.05) associations with increased (*03:02) and decreased (*01:01, *07:01, *09:01, *13:04, and *15:03) disease susceptibility were also observed (Table 1). None of these associations showed significant heterogeneity in results across the three study samples from which they were originally derived.
Table 1.
Associations* between HLA–DRB1 Two-Field Alleles and Sarcoidosis Risk
| Allele Frequency |
Full Sample |
Study-Specific Odds Ratios (95% CI) |
||||||
|---|---|---|---|---|---|---|---|---|
| Allele | Controls (n = 1,467) | Cases (n = 1,277) | OR* (95% CI)* | P Value* | ACCESS† | SAGA† | HFH† | Phet |
| 01:01 | 0.025 | 0.014 | 0.56 (0.37–0.85) | 0.006 | 0.60 (0.28–1.29) | 0.57 (0.24–1.36) | 0.74 (0.35–1.56) | 0.894 |
| 03:01 | 0.069 | 0.042 | 0.56 (0.44–0.73) | 1.0 × 10−5 | 0.74 (0.47–1.18) | 0.69 (0.38–1.26) | 0.57 (0.37–0.89) | 0.730 |
| 03:02 | 0.074 | 0.091 | 1.26 (1.03–1.56) | 0.027 | 1.28 (0.86–1.93) | 2.07 (1.20–3.59) | 1.07 (0.77–1.50) | 0.136 |
| 07:01 | 0.082 | 0.063 | 0.74 (0.59–0.93) | 0.009 | 0.93 (0.62–1.41) | 0.67 (0.40–1.11) | 0.81 (0.56–1.17) | 0.620 |
| 09:01 | 0.026 | 0.016 | 0.64 (0.43–0.95) | 0.027 | 0.61 (0.27–1.37) | 1.69 (0.62–4.59) | 0.43 (0.21–0.90) | 0.118 |
| 11:01 | 0.113 | 0.167 | 1.69 (1.42–2.01) | 3.0 × 10−9 | 1.80 (1.28–2.53) | 1.09 (0.75–1.57) | 1.55 (1.16–2.07) | 0.088 |
| 12:01 | 0.043 | 0.077 | 2.11 (1.65–2.69) | 3.2 × 10−9 | 2.06 (1.29–3.28) | 2.75 (1.49–5.06) | 1.35 (0.91–2.01) | 0.139 |
| 13:04 | 0.020 | 0.012 | 0.55 (0.33–0.92) | 0.023 | 0.50 (0.18–1.44) | 0.82 (0.27–2.45) | 0.45 (0.16–1.25) | 0.632 |
| 15:03 | 0.123 | 0.101 | 0.79 (0.66–0.95) | 0.011 | 0.68 (0.46–1.00) | 0.69 (0.46–1.02) | 0.89 (0.66–1.20) | 0.505 |
Definition of abbreviations: ACCESS, A Case-Control Etiologic Study of Sarcoidosis; CI, confidence interval; HFH, Henry Ford Hospital; OR, odds ratio; Phet, P value for heterogeneity; SAGA, Sarcoidosis Genetic Analysis.
Associations with P < 0.05. Models were estimated using generalized estimating equations, and the allele odds ratios are estimated assuming a dominant model adjusted for sex and genome-wide percent African ancestry. P values were calculated using a Wald test.
The study-specific sample sizes are as follows: ACCESS (n = 1,034; 224 patients with sarcoidosis and 810 control subjects), SAGA (n = 741; 566 patients with sarcoidosis and 175 control subjects), and HFH (n = 969; 487 patients with sarcoidosis and 482 control subjects).
We tested the association of DRB1 alleles with disease course (resolved versus persistent) in our subsample of 932 African-American patients with sarcoidosis (308 with resolved and 624 with persistent disease) with greater than or equal to 2 years of follow-up (Table 2). The *03:02 allele was significantly more common in cases with resolved versus persistent sarcoidosis, with carriers of the *03:02 allele approximately half as likely (OR, 0.52; 95% CI, 0.37–0.72; P = 1.3 × 10−4) to have persistent radiographic evidence for disease. There was no heterogeneity in the *03:02 association across the component studies of our analytic sample, with the direction and statistical significance of this association remarkably consistent across all three (Table 2). Carriers of *11:01 not only had a higher risk of disease (Table 1) but also were more likely to have persistent disease after 2 years (OR, 1.41; 95% CI, 1.04–1.91; P = 0.028). Nominally significant (P < 0.05) associations were also observed for HLA-DRB1*01:02 and *15:01, with decreased and increased risk for persistent disease, respectively (Table 2). As with the DRB1*03:02 result, none of the additional DRB1 associations with persistent radiographic evidence for disease showed significant heterogeneity in results across the three component study samples. Table E2 provides a complete summary of all the HLA-DRB1 two-field allelic associations with sarcoidosis susceptibility and radiographic resolution for the complete analytic sample and the three component studies.
Table 2.
| Allele Frequency |
Full Sample |
Study-Specific ORs (95% CI) |
||||||
|---|---|---|---|---|---|---|---|---|
| Allele | Resolved (n = 308) | Persistent (n = 624) | OR* (95% CI)* | P Value* | ACCESS‡ | SAGA‡ | HFH‡ | Phet |
| 01:02 | 0.064 | 0.046 | 0.63 (0.41–0.96) | 0.032 | –§ | 0.55 (0.30–1.02) | 0.75 (0.41–1.39) | 0.761 |
| 03:02 | 0.130 | 0.078 | 0.52 (0.37–0.72) | 1.3 × 10−4 | 0.27 (0.08–0.90) | 0.62 (0.38–0.99) | 0.46 (0.27–0.79) | 0.418 |
| 11:01 | 0.139 | 0.175 | 1.41 (1.04–1.91) | 0.028 | 4.16 (1.12–15.50) | 1.19 (0.77–1.83) | 1.53 (0.96–2.43) | 0.271 |
| 15:01 | 0.016 | 0.034 | 2.25 (1.07–4.74) | 0.033 | –§ | 3.16 (0.89–11.28) | 1.70 (0.65–4.45) | 0.507 |
For definition of abbreviations, see Table 1.
Associations with P < 0.05. Models were estimated using generalized estimating equations, and the allele odds ratios are estimated assuming a dominant model adjusted for sex and genome-wide percent African ancestry. P values were calculated using a Wald test.
Based on chest radiographs assessed after a minimum of 2 years of follow–up.
The study-specific sample sizes are as follows: ACCESS (n = 78; 24 resolved and 54 persistent), SAGA (n = 469; 142 resolved and 327 persistent), and HFH (n = 385; 142 resolved and 243 persistent).
Not estimable.
The Impact of Local African/European Ancestry on Allelic Associations with Susceptibility and Disease Course
To determine the role local ancestry at the HLA-DRB1 locus played in the most significant associations observed for disease susceptibility and course, we tested for heterogeneity of allelic association by the number of African alleles (0, 1, or 2) at HLA-DRB1 and calculated risk estimates (Table 3). For disease susceptibility, we detected significant evidence for heterogeneity by local African ancestry for DRB1*03:01 (P = 0.050) and *03:02 (P = 0.009). DRB1*03:01 was only associated with disease protection in individuals with one or two African alleles, whereas individuals with zero African alleles (conversely, two European alleles) demonstrated a nonstatistically significant increased risk of disease (Table 3). DRB1*03:02 was strongly associated with increased risk (OR, 2.39; 95% CI, 1.90–2.99; P = 0.002) only when local ancestry at DRB1 was mixed (one African and one European allele). For disease course, suggestive heterogeneity by local ancestry was observed for DRB1*03:01 (P = 0.146). DRB1*03:01 was inversely associated with persistent disease in carriers of with one or zero African alleles at DRB1 (OR, 0.36; 95% CI, 0.14–0.94; P = 0.037).
Table 3.
HLA-DRB1 Two-Field Sarcoidosis Risk Allele Associations Stratified by Local African Ancestry* at the DRB1 Locus
| Allele | Local Ancestry* | Case | Control | OR (95% CI) | P Value | Persistent | Resolved | OR (95% CI) | P Value |
|---|---|---|---|---|---|---|---|---|---|
| 03:01 | 2 | 0.034 | 0.058 | 0.54 (0.39–0.75) | 2.1 × 10−4 | 0.034 | 0.031 | 1.10 (0.59–2.07) | 0.759 |
| 1 | 0.061 | 0.106 | 0.54 (0.34–0.860) | 0.009 | 0.028 | 0.078 | 0.29 (0.09–0.90) | 0.032 | |
| 0 | 0.173 | 0.095 | 2.18 (0.72–6.60) | 0.166 | 0.167 | 0.200 | –† | –† | |
| Pint = 0.050 |
Pint = 0.146 |
||||||||
| 03:02 | 2 | 0.097 | 0.089 | 1.10 (0.88–1.38) | 0.415 | 0.085 | 0.137 | 0.56 (0.39–0.80) | 0.002 |
| 1 | 0.069 | 0.031 | 2.39 (1.36–4.18) | 0.002 | 0.056 | 0.108 | 0.44 (0.18–1.10) | 0.080 | |
| 0 | 0 | 0 | –† | –† | 0 | 0 | –† | –† | |
| Pint = 0.009 |
Pint = 0.656 |
||||||||
| 11:01 | 2 | 0.191 | 0.130 | 1.73 (1.43–2.09) | 2.1 × 10−8 | 0.197 | 0.159 | 1.32 (0.95–1.83) | 0.100 |
| 1 | 0.094 | 0.060 | 1.68 (1.07–2.64) | 0.024 | 0.120 | 0.059 | 2.80 (1.01–7.74) | 0.048 | |
| 0 | 0.058 | 0.071 | 0.72 (0.16–3.31) | 0.678 | 0.083 | 0 | –† | –† | |
| Pint = 0.583 |
Pint = 0.172 |
||||||||
| 12:01 | 2 | 0.083 | 0.043 | 2.08 (1.58–2.73) | 1.4 × 10−7 | 0.087 | 0.081 | 1.02 (0.67–1.56) | 0.915 |
| 1 | 0.057 | 0.027 | 2.31 (1.25–4.26) | 0.007 | 0.060 | 0.059 | 0.93 (0.33–2.63) | 0.889 | |
| 0 | 0.019 | 0.012 | 1.25 (0.10–15.69) | 0.861 | 0 | 0 | –† | –† | |
| Pint = 0.939 | Pint = 0.970 | ||||||||
Definition of abbreviations: CI, confidence interval; OR, odds ratio; Pint, Wald test P value for the allele-by–local ancestry interaction term(s).
Local African ancestry indicates the inferred number of African ancestral alleles present at the DRB1 locus for each subject.
Not estimable.
Differences in Associations for DRB1*03:01 and *03:02 with Organ Presentation and Amino Acids that Differ between the Two Alleles
We next tested for associations with disease presentation by organ involvement (Table 4). Carriers of the *03:02 allele were more likely to present with skin involvement (OR, 1.50; 95% CI, 1.12–2.02; P = 0.007). In contrast, *03:01 carriers were less likely to present with multiple different organs involved, including the most common organ manifestations of sarcoidosis: lung (OR, 0.48; 95% CI, 0.34–0.67; P = 1.6 × 10−5), eye (OR, 0.34; 95% CI, 0.19–0.61; P = 2.6 × 10−4), or skin (OR, 0.60; 95% CI, 0.41–0.88; P = 0.009).
Table 4.
Associations between HLA-DRB1*03:01 and *03:02 Alleles and Organ Involvement at Disease Presentation
| Cases |
||||||
|---|---|---|---|---|---|---|
| Allele | Organ Involvement | n | Allele Frequency | OR* | 95% CI | P Value |
| 03:01 | Lung | 749 | 0.037 | 0.48 | 0.34–0.67 | 1.6 × 10−5 |
| Nonthoracic | 729 | 0.041 | 0.55 | 0.40–0.75 | 1.7 × 10−4 | |
| Nonthoracic lymph node | 192 | 0.020 | 0.21 | 0.09–0.48 | 2.2 × 10−4 | |
| Ocular | 258 | 0.028 | 0.34 | 0.19–0.61 | 2.6 × 10−4 | |
| Skin | 384 | 0.043 | 0.60 | 0.41–0.88 | 0.009 | |
| Liver | 176 | 0.037 | 0.54 | 0.30–0.96 | 0.037 | |
| 03:02 | Skin | 384 | 0.103 | 1.50 | 1.12–2.02 | 0.007 |
Definition of abbreviations: CI, confidence interval; OR, odds ratio.
Statistical comparisons based on 1,467 unaffected controls with allele frequencies of 0.069 and 0.072 for DRB1*03:01 and *03:02, respectively.
To more thoroughly investigate the differential associations between risk of disease and persistent disease course exhibited by the HLA DRB1*03:01 and *03:02 alleles, respectively, we performed association analyses between the four amino acid residues that differ between these two alleles and sarcoidosis clinical phenotypes. The most significant association was observed for glutamic acid at the 28 position (Glu28) and disease resolution (Table 5). The Glu28 allele was associated with significant protection against persistent disease (OR, 0.68; 95% CI, 0.52–0.90; P = 0.007) and consistently with an increased risk for the resolved disease phenotype when compared with unaffected control subjects (OR, 1.61; 95% CI, 1.26–2.07; P = 1.8 × 10−4). The only other amino acid residue that reached nominal statistical significance was tyrosine at the 47 position (Tyr47), which was associated with decreased disease susceptibility (OR, 0.84; 95% CI, 0.72–0.99; P = 0.038).
Table 5.
Associations between the Four HLA-DRB1 Amino Acids that Differ between Alleles *03:02 and *03:01 and Disease Course and Susceptibility*
| Amino Acids |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phenylalanine, Position 26 |
Glutamate, Position 28 |
Tyrosine, Position 47 |
Glycine, Position 86 |
||||||
| Cases/Controls | n | OR† (95% CI) | P Value‡ | OR† (95% CI) | P Value‡ | OR† (95% CI) | P Value‡ | OR† (95% CI) | P Value‡ |
| Resolved cases | 315 | Referent | Referent | Referent | Referent | ||||
| Persistent | 635 | 1.05 (0.57–1.95) | 0.877 | 0.68 (0.52–0.90) | 0.007 | 0.78 (0.58–1.05) | 0.095 | 0.93 (0.68–1.28) | 0.678 |
| Stage IV | 193 | 1.19 (0.52–2.73) | 0.673 | 0.69 (0.47–0.99) | 0.046 | 0.76 (0.51–1.14) | 0.184 | 0.96 (0.63–1.46) | 0.851 |
| Unaffecteds | 1,467 | Referent | Referent | Referent | Referent | ||||
| All | 1,277 | 1.10 (0.79–1.53) | 0.566 | 1.15 (0.99–1.34) | 0.064 | 0.84 (0.72–0.99) | 0.038 | 1.08 (0.91–1.29) | 0.377 |
| Resolved | 315 | 1.08 (0.63–1.84) | 0.784 | 1.61 (1.26–2.07) | 1.76 × 10−4 | 1.09 (0.83–1.42) | 0.531 | 1.14 (0.86–1.51) | 0.377 |
| Persistent | 635 | 1.12 (0.73–1.73) | 0.591 | 1.09 (0.90–1.31) | 0.383 | 0.83 (0.68–1.02) | 0.077 | 1.06 (0.85–1.31) | 0.624 |
| Stage IV | 193 | 1.27 (0.63–2.56) | 0.508 | 1.10 (0.81–1.49) | 0.542 | 0.83 (0.59–1.15) | 0.261 | 1.09 (0.76–1.54) | 0.645 |
For definition of abbreviations, see Table 4.
Results are presented for patients with sarcoidosis with at least 2 years of follow-up. Individuals with disease resolution (“Resolved”) within this time served as controls; individuals with persistent disease (“Persistent”) served as cases. Persistent cases were further broken down to the subset with Scadding stage IV disease (“Stage IV”). Allele association results are also presented for categories of sarcoidosis cases (All, Resolved, Persistent, and Stage IV) compared with unaffected controls.
Models were estimated using generalized estimating equations, and the allele ORs are estimated assuming a dominant model, adjusted for sex and genome-wide percent African ancestry.
P values were calculated using a Wald test.
In Silico DRB1 Binding Studies
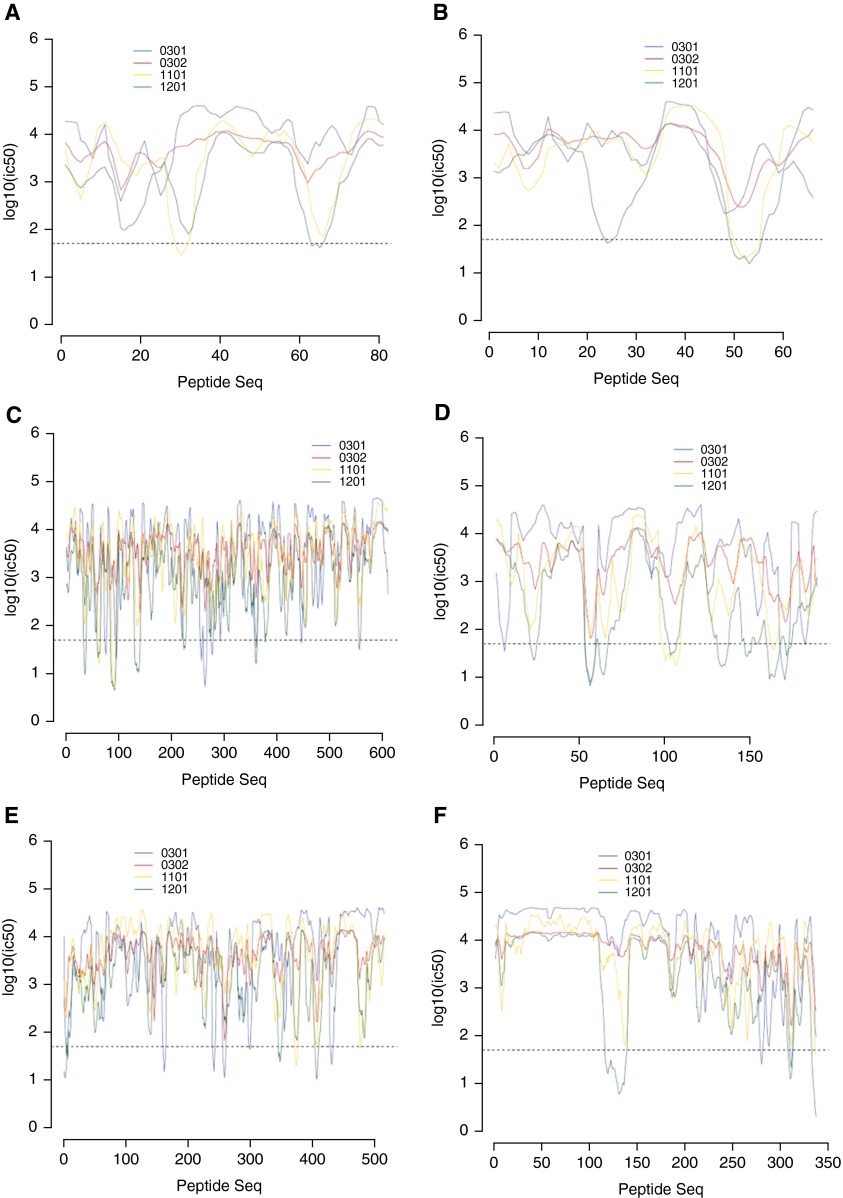
We analyzed the binding affinities of the HLA-DRB1*03:01, *03:02, *11:01, and *12:01 molecules for selected peptides previously associated with sarcoidosis. Table 6 lists the 15-mer peptides with the predicted best binding affinity and the corresponding percentile rank for the four DRB1 alleles of interest. The *03:01 allele consistently demonstrated the highest binding affinities for all six peptides, whereas the *12:01 allele displayed the lowest binding affinities. *03:02 had relatively strong binding affinities for the proteins SodA (Mycobacterium tuberculosis) and trigger factor (Propionibacterium acnes). *11:01 had strong binding affinities for the HSP70 (M. tuberculosis), trigger factor, and MAP2694 (Mycobacterium avium) proteins. Figure 1 shows the plotted IC50 binding affinity values for the full peptide sequences with the DRB1 molecules. HSP70, SodA, and the trigger factor proteins show the most promiscuous binding of the four DRB1 alleles; *12:01 binds the highest number of peptide sequences (n = 8) with high affinity across HSP70 and sodA, whereas *03:01 is most promiscuous across the trigger factor protein. Despite having similar IC50 binding affinity values across the 95-amino-acid peptide sequences of ESAT6, *11:01 and *12:01 bind two different distinct regions of this peptide.
Table 6.
Listing of Specific Peptides in Putative Sarcoidosis-Causing Organisms with the Strongest In Silico Computed Binding Affinities to Selected HLA-DRB1 Alleles
| Organism | Protein | Position | Peptide | Rank* | Position | Peptide | Rank* |
|---|---|---|---|---|---|---|---|
| *03:01 |
*03:02 |
||||||
| Mycobacterium tuberculosis | ESAT6 | 63–77 | TELNNALQNLARTIS | 4.45 | 15–29 | ASAIQGNVTSIHSLL | 11.06 |
| M. tuberculosis | KatG | 9–23 | ATWLGDERYSGKRDL | 1.97 | 51–65 | AAVDIRETFRRMAMN | 3.34 |
| M. tuberculosis | HSP70 | 358–372 | GEVKDVLLLDVTPLS | 0.01 | 141–155 | VLRIVNEPTAAALAY | 2.28 |
| M. tuberculosis | SodA | 56–70 | HSAILLNEKNLAFNL | 0.19 | 56–70 | HSAILLNEKNLAFNL | 0.52 |
| Propionibacterium acnes | Trigger factor | 256–270 | FDTVDEMRADLRTAL | 0.13 | 259–273 | VDEMRADLRTALENM | 0.54 |
| Mycobacterium avium ssp. paratuberculosis | MAP2694 | 311–325 | VTIIAVDTKPVSVFI | 0.22 | 311–325 | VTIIAVDTKPVSVFI | 1.99 |
| *11:01 |
*12:01 |
||||||
| M. tuberculosis | ESAT6 | 63–77 | TELNNALQNLARTIS | 1.64 | 63–77 | TELNNALQNLARTIS | 21.65 |
| M. tuberculosis | KatG | 53–67 | VDIRETFRRMAMNDV | 3.42 | 23–37 | LENPLAAVQMGLIYV | 19.55 |
| M. tuberculosis | HSP70 | 90–104 | EISARILMKLKRDAE | 0.32 | 93–107 | ARILMKLKRDAEAYL | 7.27 |
| M. tuberculosis | SodA | 100–114 | ADAFGSFDKFRAQFH | 1.82 | 56–70 | HSAILLNEKNLAFNL | 5.58 |
| P. acnes | Trigger factor | 264–278 | ADLRTALENMARLDQ | 0.65 | 2–16 | PSSLEKLSTNRVKLT | 10.72 |
| M. avium ssp. paratuberculosis | MAP2694 | 128–142 | LVVLAVIALVATLVV | 0.32 | 337–351 | GLIGKIIAALKVAKS | 1.34 |
Ranking of peptide–allele binding affinity as a percentile among binding affinities of all DRB1 alleles and all possible protein peptides.
Figure 1.
Half-maximal inhibitory concentration (IC50) values for class II HLA-DRB1*03:01, *03:02, *11:01, and *12:01 allelic epitopes for selected proteins previously shown to be associated with sarcoidosis. IC50 values are plotted for all possible 15-mer peptides across the amino acid sequences of ESAT6 (Mycobacterium tuberculosis) (A), T KatG (M. tuberculosis) (B), HSP70 (M. tuberculosis) (C), soda (M. tuberculosis) (D), trigger factor (Propionibacterium acnes) (E), and MAP2694 (Mycobacterium avium ssp. paratuberculosis K-10) (F). The dashed line on each graph represents an IC50 binding threshold of 50 nM, which is considered strong binding.
Discussion
Our results indicate that the HLA-DRB1 alleles *11:01 and *12:01 confer significantly increased risk for sarcoidosis susceptibility in African Americans, with *12:01 associated with a doubling of disease risk. We found that DRB1*15:01 was associated with increased risk for persistent disease, similar to what has been previously reported (6, 16, 17, 42). In Europeans, the *03:01 allele is associated with increased sarcoidosis susceptibility but also with a resolving course of disease (20). This study identified *03:01 as a novel susceptibility allele in African Americans that confers a decreased risk for disease overall and found that this association is dependent upon local ancestry, with individuals who are genetically European at DRB1 (i.e., an African-American individual with two European alleles at this gene) having an increased risk of disease, consistent with the findings in Europeans. The association between *03:01 and persistent disease was also dependent upon local ancestry, with African-American carriers of one or two European alleles at DRB1 more likely to have a resolving disease course. This result is consistent with studies in European populations finding *03:01 associated with resolving disease (3, 9, 20, 43). In addition, we report that the novel finding that the African-derived *03:02 allele was associated with increased sarcoidosis susceptibility and with disease resolution.
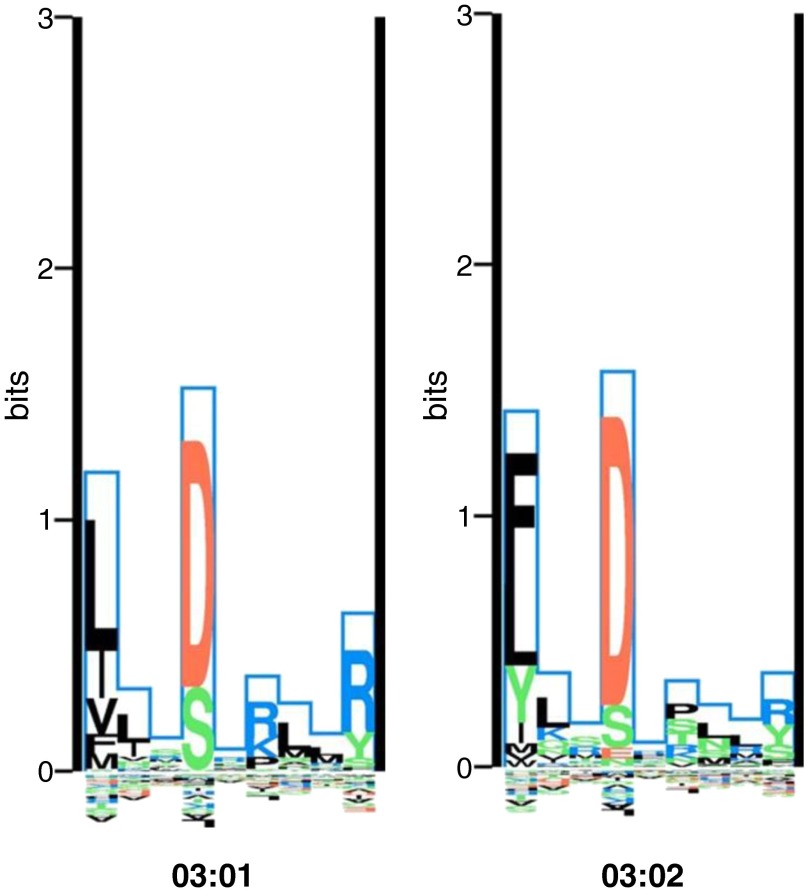
The differences in *03:01 and *03:02 binding pocket motifs are subtle (Figure 2). Differences at four exon 2–encoded amino acid positions (26, 28, 47, and 86) reside in three different binding pockets (1, 4, and 6/7). At position 47 (pocket 6/7), *03:02 has a neutral tyrosine and *03:01 has a hydrophobic phenylalanine. Tyrosine at position 47 has been associated with decreased sarcoidosis susceptibility in African Americans (7) and a resolving disease phenotype in individuals of primarily European ancestry (3); phenylalanine at position 26 (pocket 4) has been associated with decreased risk of sarcoidosis with stage I radiographic presentation in Europeans (44). In our study sample, we found that glutamic acid at position 28 (pocket 6) had the strongest association with disease resolution. Position 28 is in contact with five of the nine amino acids of nonamer peptides that bind to this molecule, whereas the other three positions that differ between *03:01 and *03:02 are each in contact with only one. Although the two different amino acid residues that *03:01 and *03:02 code for at position 28—aspartic acid and glutamic acid—are similar in size and charge, this amino acid change does affect some differences in binding preferences in anchor pocket 6 (Figure 2), which could account for this residue having the strongest association with disease resolution in our study. However, although the tyrosine at position 47 and glutamic acid at position 28 had the strongest associations with disease risk and resolution, respectively, among the single amino acids tested, the *03:01 and *03:02 alleles had stronger overall associations, suggesting that the combined effects of the four amino acids that differ between the *03:01 and *03:02 alleles collectively play a greater role in disease outcomes than any of the component single amino acids.
Figure 2.
The binding motifs of the HLA-DRB1*03:01 and 03:02 alleles visualized using a logo-plot. The Kullback–Leibler (KL) sequence logo representation of the binding motifs of the HLA-DRB1*03:01 and *03:02 alleles is taken from the MHC Motif Viewer website (http://www.cbs.dtu.dk/biotools/MHCMotifViewer/Home.html). The KL information content is plotted along the nine-amino-acid binding core. Amino acids with greater binding preference are plotted on the positive y axis, whereas amino acids with a negative influence on binding are plotted on the negative y axis. The height of each amino acid represents its relative binding specificity. Color-coding of amino acids represent the physicochemical properties of acidic (red), basic (blue), hydrophobic (black), and neutral (green). Of the primary anchor positions (P1, P4, P6, and P9), positions 1 and 4 show the most dramatic differences in amino acid preferences between the two HLA molecules. At P1, *03:01 prefers hydrophobic amino acids, whereas *03:02 prefers hydrophobic but will also bind neutral tyrosine. The allele *03:01 has a preference for basic amino acids at P6, whereas *03:02 is more promiscuous, preferentially binding with hydrophobic or neutral amino acids at the P6 anchor.
A previous in silico analysis found that patients with Löfgren’s syndrome express HLA-DR alleles capable of binding a significantly higher number of bacterial epitopes than other HLA-DR alleles (25). DRB1*03:01 shows the highest predicted binding to M. tuberculosis epitopes (45). Given the affinity for *03:01-encoded epitopes to bind to M. tuberculosis–derived peptides, a potential model for exposure to this antigen involving effective clearance and subsequent self-limiting Löfgren’s disease emerges. We observed that the African-derived *03:02 allele is associated with resolving disease in African Americans; our in silico binding studies showed that *03:01 has a higher binding affinity for sarcoidosis-associated M. tuberculosis–derived peptides than *03:02, suggesting that a different array of antigens that preferentially bind to DRB1*03:02 may uniquely trigger the onset of sarcoidosis that resolves within 2 years in African-American carriers of *03:02.
Studies of the functional effect of HLA-DRB1 microvariation on peptide binding (46, 47) shed some light on the binding differences between *03:01 and *03:02. Using the observation that HLA-DRB1*03:01-encoded DR3 molecules bind M. tuberculosis heat shock protein (HSP) peptides, whereas *03:02-encoded DR3 molecules do not, the authors found that amino acid residues 28 and 86 were critical in determining the specificity of HSP 3 to 13 and HSP 4 to 15 binding to *03:01 and *03:02. The differences in peptide binding affinities of the DR3 proteins encoded by *03:01 and *03:02 alleles may explain the observed differences in disease associations for these alleles. In African Americans, *03:02 confers protection against type I diabetes, whereas *03:01 increases risk (48). DRB1*03:01 is a strong risk factor for systemic erythematous lupus in whites (49), but neither *03:01 nor *03:02 confers risk for systemic erythematous lupus in African Americans (50). In the most extensive examination of the HLA-DRB1 variation and sarcoidosis risk in African Americans to date, Rossman and colleagues found *03:02 over-represented in African-American cases, but this association did not reach statistical significance (7). DRB1 variation was not examined in relation to disease course in this report. In sarcoidosis and in other immune-related diseases, the greater variation of HLA-DRB1 in populations of African descent suggests that differences in immune responses to the same exposures may exist, resulting in heterogeneity of allelic associations and phenotypic variation across groups.
HLA genes do not act in isolation, and, in terms of sarcoidosis risk allelic variants at the HLA class II locus, HLA-DRB1 alleles in particular have been shown to interact with other HLA risk variants (17). Because HLA genes are known to be inherited in haplotype blocks, information about the ancestral background at a HLA locus may include more than the risk effect of an allele at that locus. In analyses stratified by local ancestry, we found that the associations of HLA-DRB1*03:01 and *03:02 with susceptibility and DRB1*03:01 with persistent disease were dependent on local ancestry at DRB1. Dependency on ancestral background for an allelic association to be manifest in an admixed population is not without precedent, although we know of no previous study that has shown such an effect for an HLA association. For example, the importance of an admixed heterozygous ancestral background was manifest for the chromosome 6q14.1 SNP rs1361549, which was found to show an association with asthma only in Puerto Rican subjects who were heterozygous for African and non-African ancestry at the risk locus (51). Our finding that HLA-DRB1 allele associations with disease susceptibility and course are dependent upon local ancestral background raises the question of possible trans effects of HLA loci on sarcoidosis risk.
Our study has several limitations regarding the genotyping and phenotyping of study cases. For most of the sample, DRB1 alleles were not directly typed but rather were imputed from high-density SNP data. The use of imputation of classical HLA alleles to study associations with disease is becoming more common (52), and the analytic technique we used has been shown to be accurate in populations of African descent (27, 33). Further, although our study results focus on particular DRB1 alleles, genotypic combinations of alleles with similar antigen-binding capabilities may also contribute to disease risk and outcome. However, the frequency of most genotypes was less than 0.5%, making the estimation of genotypic risk inaccurate given the sample size of the study.
Although the phenotype data used across the three original studies were collected using a standardized instrument, differences in study design resulted in retrospective and prospective data collection across studies. Nevertheless, we found no evidence for heterogeneity of results across the three studies in terms of HLA-DRB1 associations with either disease risk or radiographic resolution. For all three studies, if a case had a minimum of 2 years of follow-up after initial diagnosis, chest radiographic status was reassessed at the later follow-up date. Although the recent report of the World Association of Sarcoidosis and Other Granulomatous Diseases Task Force on clinical outcome status in sarcoidosis recommended 5 years of follow-up, this recommendation was based on observations at only one clinical site and involved a more global view of disease status related to the need for therapy (53). Studies that have systematically followed patients for resolution of pulmonary involvement over the course of 5 years have found that, although the majority of patients who radiographically resolve do so within 2 years, as many as 10–20% of patients resolve between 2 and 5 years after diagnosis (54–56). However, data on long-term disease course in African-American patients with sarcoidosis is lacking—one observational study that followed a cohort of African-American patient cohort for up to 12 years found that the majority of disease organ involvement was identified within 2 years of follow-up (57).
The present study is the largest association study of HLA-DRB1 and sarcoidosis in African Americans and is the only study to examine DRB1 in the context of disease phenotype and outcomes. In Europeans, *03:01-positivity is considered a contraindication for treatment because of the high likelihood of a self-limiting disease course (20). We found that the DRB1*03:02 allele confers a similar likelihood of resolving disease in African-American patients with sarcoidosis. Although the DRB1*03:01 allele is found less frequently in African American than in white patients with sarcoidosis (7) and may therefore have limited impact on overall disease burden, the *03:02 allele in African Americans has a similar prevalence and level of association with disease resolution as the *03:01 allele in sarcoidosis patients of European ancestry, suggesting it could have similar clinical implications (58). Our results were consistent in direction of effect and significance across the three original study samples, but additional validation in an independent sample is needed before clinical practice recommendations are made concerning DRB1*03:02 in African-American patients with sarcoidosis. Likewise, immunologic studies that further characterize the mechanistic relationship between antigenic response and DRB1 risk alleles, including those unique to ancestral African populations, will improve our understanding of sarcoidosis pathogenesis as well as its variable presentation and course across race-ethnicity groups.
Acknowledgments
Acknowledgments
The authors acknowledge the NHLBI-supported ACCESS and SAGA study investigators and especially Drs. Milton Rossman and Dimitri Monos, who generated the original ACCESS HLA-DRB1 genotype data.
Footnotes
This work was supported by National Institutes of Health grants R56-AI072727 and R01-HL092576 (B.A.R.), R01-HL54306 and U01-HL060263 (M.C.I.), 1RC2HL101499 and R01HL113326 (C.G.M.), and P20GM103456 (I.A.).
Author Contributions: Conception and design: A.M.L., M.C.I., C.G.M., and B.A.R. Analysis and interpretation: A.M.L., I.A., I.D., W.P.D., J.L., C.G.M., and B.A.R. Drafting the manuscript for important intellectual content: A.M.L., M.C.I., S.T., W.P.D., C.G.M., and B.A.R.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0227OC on December 15, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Iannuzzi MC. Genetics of sarcoidosis. Semin Respir Crit Care Med. 2007;28:15–21. doi: 10.1055/s-2007-970330. [DOI] [PubMed] [Google Scholar]
- 2.Mangalam AK, Taneja V, David CS. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol. 2013;190:513–518. doi: 10.4049/jimmunol.1201891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wennerström A, Pietinalho A, Vauhkonen H, Lahtela L, Palikhe A, Hedman J, Purokivi M, Varkki E, Seppänen M, Lokki ML, et al. Finnish Sarcoidosis Study Group. HLA-DRB1 allele frequencies and C4 copy number variation in Finnish sarcoidosis patients and associations with disease prognosis. Hum Immunol. 2012;73:93–100. doi: 10.1016/j.humimm.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Shen L, Zhang Y, Jiang D, Li H. Human leukocyte antigen-A, -B, and -DRB1 alleles and sarcoidosis in Chinese Han subjects. Hum Immunol. 2011;72:571–575. doi: 10.1016/j.humimm.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Darlington P, Tallstedt L, Padyukov L, Kockum I, Cederlund K, Eklund A, Grunewald J. HLA-DRB1* alleles and symptoms associated with Heerfordt’s syndrome in sarcoidosis. Eur Respir J. 2011;38:1151–1157. doi: 10.1183/09031936.00025011. [DOI] [PubMed] [Google Scholar]
- 6.Grunewald J, Brynedal B, Darlington P, Nisell M, Cederlund K, Hillert J, Eklund A. Different hla-drb1 allele distributions in distinct clinical subgroups of sarcoidosis patients. Respir Res. 2010;11:25. doi: 10.1186/1465-9921-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossman MD, Thompson B, Frederick M, Maliarik M, Iannuzzi MC, Rybicki BA, Pandey JP, Newman LS, Magira E, Beznik-Cizman B, et al. ACCESS Group. HLA-DRB1*1101: a significant risk factor for sarcoidosis in blacks and whites. Am J Hum Genet. 2003;73:720–735. doi: 10.1086/378097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma SK, Balamurugan A, Pandey RM, Saha PK, Mehra NK. Human leukocyte antigen-DR alleles influence the clinical course of pulmonary sarcoidosis in Asian Indians. Am J Respir Cell Mol Biol. 2003;29:225–231. doi: 10.1165/rcmb.2003-0007OC. [DOI] [PubMed] [Google Scholar]
- 9.Bogunia-Kubik K, Tomeczko J, Suchnicki K, Lange A. HLA-DRB1*03, DRB1*11 or DRB1*12 and their respective DRB3 specificities in clinical variants of sarcoidosis. Tissue Antigens. 2001;57:87–90. doi: 10.1034/j.1399-0039.2001.057001087.x. [DOI] [PubMed] [Google Scholar]
- 10.Foley PJ, McGrath DS, Puscinska E, Petrek M, Kolek V, Drabek J, Lympany PA, Pantelidis P, Welsh KI, Zielinski J, et al. Human leukocyte antigen-DRB1 position 11 residues are a common protective marker for sarcoidosis. Am J Respir Cell Mol Biol. 2001;25:272–277. doi: 10.1165/ajrcmb.25.3.4261. [DOI] [PubMed] [Google Scholar]
- 11.Sato H, Woodhead FA, Ahmad T, Grutters JC, Spagnolo P, van den Bosch JM, Maier LA, Newman LS, Nagai S, Izumi T, et al. Sarcoidosis HLA class II genotyping distinguishes differences of clinical phenotype across ethnic groups. Hum Mol Genet. 2010;19:4100–4111. doi: 10.1093/hmg/ddq325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybicki BA, Major M, Popovich J, Jr, Maliarik MJ, Iannuzzi MC. Racial differences in sarcoidosis incidence: a 5-year study in a health maintenance organization. Am J Epidemiol. 1997;145:234–241. doi: 10.1093/oxfordjournals.aje.a009096. [DOI] [PubMed] [Google Scholar]
- 13.Judson MA, Baughman RP, Thompson BW, Teirstein AS, Terrin ML, Rossman MD, Yeager H, Jr, McLennan G, Bresnitz EA, DePalo L, et al. ACCESS Research Group. Two year prognosis of sarcoidosis: the ACCESS experience. Sarcoidosis Vasc Diffuse Lung Dis. 2003;20:204–211. [PubMed] [Google Scholar]
- 14.Swigris JJ, Olson AL, Huie TJ, Fernandez-Perez ER, Solomon J, Sprunger D, Brown KK. Sarcoidosis-related mortality in the United States from 1988 to 2007. Am J Respir Crit Care Med. 2011;183:1524–1530. doi: 10.1164/rccm.201010-1679OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mrazek F, Holla LI, Hutyrova B, Znojil V, Vasku A, Kolek V, Welsh KI, Vacha J, du Bois RM, Petrek M. Association of tumour necrosis factor-alpha, lymphotoxin-alpha and HLA-DRB1 gene polymorphisms with Löfgren’s syndrome in Czech patients with sarcoidosis. Tissue Antigens. 2005;65:163–171. doi: 10.1111/j.1399-0039.2005.00370.x. [DOI] [PubMed] [Google Scholar]
- 16.Voorter CE, Drent M, van den Berg-Loonen EM. Severe pulmonary sarcoidosis is strongly associated with the haplotype HLA-DQB1*0602-DRB1*150101. Hum Immunol. 2005;66:826–835. doi: 10.1016/j.humimm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Grunewald J, Eklund A, Olerup O. Human leukocyte antigen class I alleles and the disease course in sarcoidosis patients. Am J Respir Crit Care Med. 2004;169:696–702. doi: 10.1164/rccm.200303-459OC. [DOI] [PubMed] [Google Scholar]
- 18.Candore G, Lio D, Colonna Romano G, Caruso C. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev. 2002;1:29–35. doi: 10.1016/s1568-9972(01)00004-0. [DOI] [PubMed] [Google Scholar]
- 19.Löfgren S. Primary pulmonary sarcoidosis: I. Early signs and symptoms. Acta Med Scand. 1953;145:424–431. [PubMed] [Google Scholar]
- 20.Grunewald J, Eklund A. Löfgren’s syndrome: human leukocyte antigen strongly influences the disease course. Am J Respir Crit Care Med. 2009;179:307–312. doi: 10.1164/rccm.200807-1082OC. [DOI] [PubMed] [Google Scholar]
- 21.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 22.Murthy VL, Stern LJ. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: implications for the structural basis of the specificity of peptide binding. Structure. 1997;5:1385–1396. doi: 10.1016/s0969-2126(97)00288-8. [DOI] [PubMed] [Google Scholar]
- 23.Oswald-Richter K, Sato H, Hajizadeh R, Shepherd BE, Sidney J, Sette A, Newman LS, Drake WP. Mycobacterial ESAT-6 and katG are recognized by sarcoidosis CD4+ T cells when presented by the American sarcoidosis susceptibility allele, DRB1*1101. J Clin Immunol. 2010;30:157–166. doi: 10.1007/s10875-009-9311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saltini C, Pallante M, Puxeddu E, Contini S, Voorter CE, Drent M, Amicosante M. M. avium binding to HLA-DR expressed alleles in silico: a model of phenotypic susceptibility to sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2008;25:100–116. [PubMed] [Google Scholar]
- 26.Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, Parker R, Adler A, Kelly JA, Kaufman KM, Lessard CJ, et al. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PLoS One. 2012;7:e43907. doi: 10.1371/journal.pone.0043907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, Nelson MR, Weir BS. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J. 2014;14:192–200. doi: 10.1038/tpj.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Group AR ACCESS Research Group. Design of a case control etiologic study of sarcoidosis (ACCESS) J Clin Epidemiol. 1999;52:1173–1186. doi: 10.1016/s0895-4356(99)00142-0. [DOI] [PubMed] [Google Scholar]
- 29.Rybicki BA, Hirst K, Iyengar SK, Barnard JG, Judson MA, Rose CS, Donohue JF, Kavuru MS, Rabin DL, Rossman MD, et al. A sarcoidosis genetic linkage consortium: the sarcoidosis genetic analysis (SAGA) study. Sarcoidosis Vasc Diffuse Lung Dis. 2005;22:115–122. [PubMed] [Google Scholar]
- 30.Iannuzzi MC, Maliarik MJ, Poisson LM, Rybicki BA. Sarcoidosis susceptibility and resistance HLA-DQB1 alleles in African Americans. Am J Respir Crit Care Med. 2003;167:1225–1231. doi: 10.1164/rccm.200209-1097OC. [DOI] [PubMed] [Google Scholar]
- 31.Rybicki BA, Levin AM, McKeigue P, Datta I, Gray-McGuire C, Colombo M, Reich D, Burke RR, Iannuzzi MC. A genome-wide admixture scan for ancestry-linked genes predisposing to sarcoidosis in African-Americans. Genes Immun. 2011;12:67–77. doi: 10.1038/gene.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judson MA, Baughman RP, Teirstein AS, Terrin ML, Yeager H., Jr Defining organ involvement in sarcoidosis: the access proposed instrument. Access Research Group. A case control etiologic study of sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 1999;16:75–86. [PubMed] [Google Scholar]
- 33.Levin AM, Adrianto I, Datta I, Iannuzzi MC, Trudeau S, McKeigue P, Montgomery CG, Rybicki BA. Performance of HLA allele prediction methods in African Americans for class II genes HLA-DRB1, -DQB1, and -DPB1. BMC Genet. 2014;15:72. doi: 10.1186/1471-2156-15-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Yang J, Levin AM, Montgomery CG, Datta I, Trudeau S, Adrianto I, McKeigue P, Iannuzzi MC, Rybicki BA. Efficient generalized least squares method for mixed population and family-based samples in genome-wide association studies. Genet Epidemiol. 2014;38:430–438. doi: 10.1002/gepi.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am J Hum Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin AM, Iannuzzi MC, Montgomery CG, Trudeau S, Datta I, Adrianto I, Chitale DA, McKeigue P, Rybicki BA. Admixture fine-mapping in African Americans implicates XAF1 as a possible sarcoidosis risk gene. PLoS One. 2014;9:e92646. doi: 10.1371/journal.pone.0092646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang P, Sidney J, Dow C, Mothé B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen M, Lund O. NN-align: an artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10:296. doi: 10.1186/1471-2105-10-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4:e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grosser M, Luther T, Fuessel M, Bickhardt J, Magdolen V, Baretton G. Clinical course of sarcoidosis in dependence on HLA-DRB1 allele frequencies, inflammatory markers, and the presence of M. tuberculosis DNA fragments. Sarcoidosis Vasc Diffuse Lung Dis. 2005;22:66–74. [PubMed] [Google Scholar]
- 43.Grubić Z, Peros-Golubicić T, Stingl K, Zunec R. The investigation of HLA microsatellites influence in predisposition to sarcoidosis among Croatians. Sarcoidosis Vasc Diffuse Lung Dis. 2011;28:18–26. [PubMed] [Google Scholar]
- 44.Voorter CE, Amicosante M, Berretta F, Groeneveld L, Drent M, van den Berg-Loonen EM. HLA class II amino acid epitopes as susceptibility markers of sarcoidosis. Tissue Antigens. 2007;70:18–27. doi: 10.1111/j.1399-0039.2007.00842.x. [DOI] [PubMed] [Google Scholar]
- 45.Amicosante M, Puxeddu E, Saltini C. Reactivity to mycobacterial antigens by patients with Löfgren’s syndrome as a model of phenotypic susceptibility to disease and disease progression. Am J Respir Crit Care Med. 2009;180:685, author reply 685–686. doi: 10.1164/ajrccm.180.7.685. [DOI] [PubMed] [Google Scholar]
- 46.Posch PE, Hurley CK, Geluk A, Ottenhoff TH. The impact of DR3 microvariation on peptide binding: the combinations of specific DR beta residues critical to binding differ for different peptides. Hum Immunol. 1996;49:96–105. doi: 10.1016/0198-8859(96)00061-4. [DOI] [PubMed] [Google Scholar]
- 47.Posch PE, Araujo HA, Creswell K, Praud C, Johnson AH, Hurley CK. Microvariation creates significant functional differences in the DR3 molecules. Hum Immunol. 1995;42:61–71. doi: 10.1016/0198-8859(94)00074-z. [DOI] [PubMed] [Google Scholar]
- 48.Howson JM, Roy MS, Zeitels L, Stevens H, Todd JA. HLA class II gene associations in African American type 1 diabetes reveal a protective HLA-DRB1*03 haplotype. Diabet Meds. 2013;30:710–716. doi: 10.1111/dme.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reveille JD, Moulds JM, Ahn C, Friedman AW, Baethge B, Roseman J, Straaton KV, Alarcón GS. Systemic lupus erythematosus in three ethnic groups: I. The effects of HLA class II, C4, and CR1 alleles, socioeconomic factors, and ethnicity at disease onset. LUMINA Study Group. Lupus in minority populations, nature versus nurture. Arthritis Rheum. 1998;41:1161–1172. doi: 10.1002/1529-0131(199807)41:7<1161::AID-ART4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 51.Torgerson DG, Capurso D, Ampleford EJ, Li X, Moore WC, Gignoux CR, Hu D, Eng C, Mathias RA, Busse WW, et al. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J Allergy Clin Immunol. 2012;130:622–629. doi: 10.1016/j.jaci.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, Alfredsson L, Padyukov L, Klareskog L, Worthington J, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baughman RP, Nagai S, Balter M, Costabel U, Drent M, du Bois R, Grutters JC, Judson MA, Lambiri I, Lower EE, et al. Defining the clinical outcome status (COS) in sarcoidosis: results of WASOG Task Force. Sarcoidosis Vasc Diffuse Lung Dis. 2011;28:56–64. [PubMed] [Google Scholar]
- 54.Nagai S, Shigematsu M, Hamada K, Izumi T. Clinical courses and prognoses of pulmonary sarcoidosis. Curr Opin Pulm Med. 1999;5:293–298. doi: 10.1097/00063198-199909000-00005. [DOI] [PubMed] [Google Scholar]
- 55.Chappell AG, Cheung WY, Hutchings HA. Sarcoidosis: a long-term follow up study. Sarcoidosis Vasc Diffuse Lung Dis. 2000;17:167–173. [PubMed] [Google Scholar]
- 56.Hillerdal G, Nöu E, Osterman K, Schmekel B. Sarcoidosis: epidemiology and prognosis. A 15-year European study. Am Rev Respir Dis. 1984;130:29–32. doi: 10.1164/arrd.1984.130.1.29. [DOI] [PubMed] [Google Scholar]
- 57.Judson MA, Boan AD, Lackland DT. The clinical course of sarcoidosis: presentation, diagnosis, and treatment in a large white and black cohort in the United States. Sarcoidosis Vasc Diffuse Lung Dis. 2012;29:119–127. [PubMed] [Google Scholar]
- 58.Grunewald J. HLA associations and Löfgren’s syndrome. Expert Rev Clin Immunol. 2012;8:55–62. doi: 10.1586/eci.11.76. [DOI] [PubMed] [Google Scholar]