Abstract
Premature ovarian failure (POF) is a heterogeneous disease. Though dozens of candidate genes have been identified for the genetic etiology of POF, it is largely unexplained in majority of patients. Recently, Wt1+/R394W mice was found to present POF-like phenotype, which indicates that WT1 might be a plausible candidate gene for non-syndromic POF. The coding region of WT1 gene was screened in 384 patients with POF and 6 novel variations were identified, including two missense mutations (p. Pro126Ser in exon1 and p. Arg370His in exon7) and four intronic variants (c.647-27C > T, c.647-13G > C, c.647-13G > A in intron1 and c.950 + 14T > C in intron 4). In vitro experiments showed that both mutant p. Pro126Ser and p. Arg370His repressed the expression of Amh and Cdh1, and induced the expression of Fshr and Cyp19 in mRNA level (P < 0.05). The expression changes of AMH, FSHR, CYP19 and CDH1 were confirmed by western blot. These genes (AMH, FSHR, CYP19 and CDH1) are required for granular cells (GCs) proliferation, differentiation and oocyte-GCs interaction. The novel mutant p. P126S and p. R370H in the WT1 gene potentially impaired GCs differentiation and oocyte-GCs interaction, which might result in loss of follicles prematurely. Therefore, WT1 is a plausible causal gene for POF.
Premature ovarian failure (POF), also known as premature menopause or primary ovarian insufficiency (POI), is a heterogeneous disorder characterized by cessation of normal ovarian function before the age of 40 years. It affects approximately 1% of women under 40 yrs and only 0.1% or less under 301,2. Genetic factors are considered to be an important facet of the etiologies for POF. A positive family history has been reported in 12.7% of idiopathic POF cases3. In addition, a number of variants in different genes have been identified functionally causative, e.g., NOBOX, PGRMC1, BMP15 and FOXL24,5,6,7. More recently, the largest cohort of genome-wide association study (GWAS) in 791 patients with POF has discovered a susceptible locus at 8q22.3, however, it was a gene desert region and considered as an important yet undefined long distance regulatory region affecting oogenesis8. Therefore, the etiology in major cases remains unclear and the underlying mechanisms are largely unknown.
The Wilms’ tumors gene WT1 encodes an essential transcription factor, which functions in mammalian urogenital development9. It was initially identified as a tumor suppressor associated with Wilms’ tumors (a recessive childhood nephroblastoma)10, and the mutation or overexpression of WT1 may lead to glomerulosclerosis of the kidney, gonadal dysgenesis and leukemia11,12. In adult reproductive organs, WT1 is expressed in Sertoli cells of the tests and granulosa cells (GCs) of ovarian follicles13. In rat, the expression of WT1 in GCs decreased along with the follicular development. WT1 was speculated as a repressor of genes responsible for GCs proliferation and differentiation14. More recently, the Wt1+/R394W mice (a common WT1 missense mutation in Denys-Drash syndrome) has been shown to be subfertile due to defect in follicle development. Compared to controls, the Wt1+/R394Wmice have smaller ovaries and significantly fewer follicles, which is similar to the characteristics of patients with POF15. These findings indicate that the WT1 gene play a critical role in follicle development and might be a plausible candidate for POF. Therefore, we screened the coding region of WT1 gene in 384 Chinese women with POF to determine its contribution involved in the etiology of POF.
Results
Clinical characteristics of patients and two novel missense mutations identified within gene WT1
Clinical characteristics of the 384 subjects were showed in Table 1. Two novel heterozygous missense mutations c.376C > T (p. Pro126Ser, p. P126S) in exon 1 and c.1109G > A (p. Arg370His, p. R370H) in exon 7 were identified in two patients, respectively (Fig. 1A). The two variants were absent in either dbSNP database or 384 control females. The amino acids involved were highly conserved among WT1 orthologs (Fig. 1B).
Table 1. Clinical characteristic of 384 Han Chinese women with POF.
| Characteristics | Mean ± S.D./N (%) |
|---|---|
| Age at diagnosis (yr) | 29.81 ± 5.27 |
| Age at menarche (yr)a | 14.43 ± 1.72 |
| Age of amenorrhea (yr)a | 25.72 ± 5.77 |
| FSH (IU/L) | 72.98 ± 32.66 |
| E2 (pmol/L) | 27.76 ± 23.59 |
| Family historyb | 21 (5.5%) |
| Parental consanguinity | 3 (0.8%) |
aRefer to patients with secondary amenorrhea.
bA positive family history was considered if another first or second degree female family member had POF or early menopause (menopause before 45 years old).
Figure 1. Mutations in WT1 gene associated with POF and the alternative splices were depicted in red.
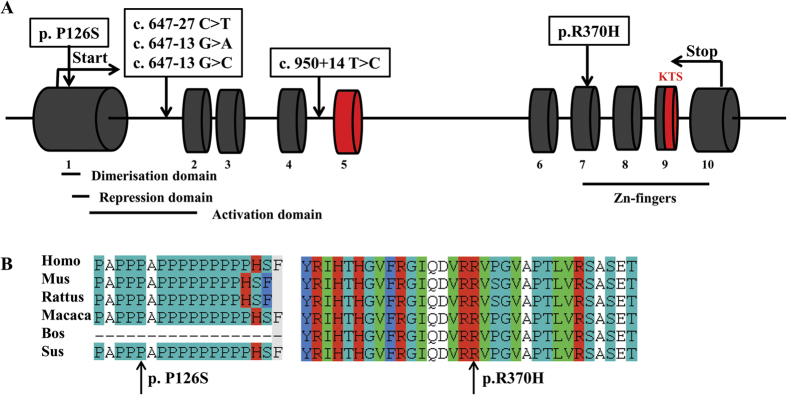
(A) Schematic presentation of WT1 mutations associated with POF. (B) Sequence alignment of WT1 among orthologs with arrow heads indicated.
The patient carrying p. P126S mutation was 28 years old. She suffered from primary amenorrhea with elevated serum FSH (77.23 IU/L). The other patient with p. R370H developed spontaneous menarche at the age of 16, and experienced irregular menses and subsequent amenorrhea at the age of 31. Trans-vaginal ultrasonography showed normal uterus and small ovaries with severe fibrosis. No other family member of the two carriers had POF or a form of 46, XY disorders of sex development (DSD).
In addition, 10 known single nucleotide polymorphisms (SNPs) and 4 novel intronic variants were found (Table 2, Fig. 1A). The 10 known SNPs included 8 intronic variants (rs2234585, rs1799933, rs5030170, rs5030171, rs2295081, rs5030277, rs192734605 and rs1799937) and 2 synonymous variants (c.330T > C and c.1107G > A). However, the genotype and allelic frequencies of the 10 SNPs between cases and the Asian population showed no significant differences. Of the 4 novel intronic variants, 3 variants were in intron 1 (c.647-27C >T , c.647-13G > A and c.647-13G > C) and 1 was in intron 4 (c.950 + 14T > C). They were not presented in controls.
Table 2. Known single nucleotide polymorphisms (SNPs) and novel intron variants identified in patients with POF.
| Variation | Location | Amino acid variation | dpSNP ID | Genotype frequency (%) | Allele frequency (%) | P-valuec | Referenced | ||
|---|---|---|---|---|---|---|---|---|---|
| POF | Asian Populationa/controlsb | POF | Asian Populationa/controlsb | ||||||
| c.330T > C | Exon 1 | Synonymous | rs1799925 | TT(46.0) | NA | T(68.5) | T(67.5) | >0.05 | f, g |
| TC(44.9) | NA | C(31.5) | C(32.5) | ||||||
| CC(9.1) | NA | ||||||||
| c.647-27C > T | Intron 1 | Novel | CC(99.7) | CC(100) | C(99.9) | C(100) | >0.05 | f, g | |
| TC(0.3) | TC(0) | T(0.1) | T(0) | ||||||
| TT(0) | TT(0) | ||||||||
| c.647-13G > A | Intron1 | Novel | GG(99.7) | GG(100) | G(99.9) | G(100) | >0.05 | f, g | |
| GA(0.3) | GA(0) | A(0.1) | A(0) | ||||||
| AA(0) | AA(0) | ||||||||
| c.647-13G > C | Intron1 | Novel | GG(99.7) | GG(100) | G(99.9) | G(100) | >0.05 | f, g | |
| GC(0.3) | GC(0) | C(0.1) | C(0) | ||||||
| CC(0) | CC(0) | ||||||||
| c.770-57T > C | Intron 2 | rs2234585 | TT(45.3) | NA | T(69.8) | T(65.8) | >0.05 | f, g | |
| TC(49.0) | NA | C(30.2) | C(34.2) | ||||||
| CC(5.7) | NA | ||||||||
| c.872 + 16G > A | Intron 3 | rs1799933 | GG(97.4) | GG(95.6) | G(98.5) | G(97.8) | >0.05 | f, g | |
| GA(2.2) | GA(4.4) | A(1.5) | A(2.2) | ||||||
| AA(0.4) | AA(0) | ||||||||
| c.872 + 82T > G | Intron 3 | rs5030170 | TT(50.9) | NA | T(71.5) | T(65.8) | >0.05 | f, g | |
| TG(41.2) | NA | G(28.5) | G(34.2) | ||||||
| GG(7.9) | NA | ||||||||
| c.872 + 85C > G | Intron3 | rs5030171 | CC(45.6) | NA | C(67.1) | C(65.8) | >0.05 | f, g | |
| CG(43.0) | NA | G(32.9) | G(34.2) | ||||||
| GG(11.4) | NA | ||||||||
| c.950 + 14T > C | Intron 4 | Novel | TT(99.7) | TT(100) | T(99.9) | T(100) | >0.05 | f, g | |
| TC(0.3) | TC(0) | C(0.1) | C(0) | ||||||
| CC(0) | CC (0) | ||||||||
| c.950 + 85G > A | Intron 4 | rs2295081 | GG(50) | GG(52.2) | G(71.7) | G(71.7) | >0.05 | f, g | |
| AG(43.4) | AG(39.1) | A(28.3) | A(28.3) | ||||||
| AA(6.6) | AA(8.7) | ||||||||
| c.1107G > A | Exon 7 | Synonymous | rs16754 | GG(49.6) | GG(63.4) | G(71.4) | G(76.8) | >0.05 | f, g |
| AG(43.6) | AG(26.8) | A(28.6) | A(23.2) | ||||||
| AA(6.8) | AA(9.8) | ||||||||
| c.1249 + 134A > T | Intron 7 | rs5030277 | AA(51.3) | AA(56.1) | A(71.1) | A(73.2) | >0.05 | f, g | |
| AT(39.5) | AT(34.1) | T(28.9) | T(26.8) | ||||||
| TT(9.2) | TT(9.8) | ||||||||
| c.1432 + 42T > A | Intron 9 | rs192734605 | TT(99.2) | NA | T(99.6) | T(99.0) | >0.05 | f, g | |
| TA(0.8) | NA | A(0.4) | A(1.0) | ||||||
| AA(0) | NA | ||||||||
| c.1433-49 C > T | Intron 9 | rs1799937 | CC(56.0) | CC(63.4) | C(74.5) | C(76.8) | >0.05 | f, g | |
| CT(37.0) | CT(26.8) | T(25.5) | T(23.2) | ||||||
| TT(7.0) | TT(9.8) | ||||||||
NA: Not available.
aData from http://www.ncbi.nlm.nih.gov/SNP/.
bThe SNPs were compared with Asian population and the novel variants were compared with the controls.
cComparison of the allele frequency between patients with POF and Asian population/controls.
df = Ensembl accession number ENST00000332351. g = Ensembl accession number ENSP00000331327.
Mutant p. P126S and p. R370H impaired the regulations of AMH, FSHR, CYP19 and CDH1 expression
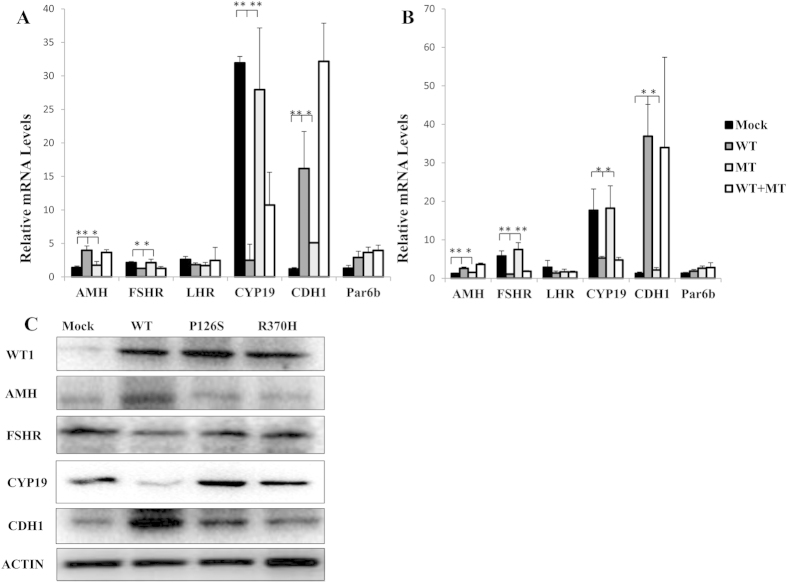
To explore the effect of mutant p. P126S and p. R370H on transcription, adenovirus vectors containing Wt1 gene were generated and transfected to differentiated GCs from three-weeks-mouse. Transcription level of down-stream genes including Amh, Fshr, Lhr, Cyp19, Cdh1 and Par6b were detected by qRT-PCR. As shown in Fig. 2A,B, in the wild type (WT) Wt1-overexpressing differentiated GCs, the expression of Amh, Cdh1, and Par6b was up-regulated while Fshr, Lhr, and Cyp19 down-regulated by Wt1, which was consistent with previous studies (15). In both p. P126S and p. R370H transfected GCs, the expression of Amh and Cdh1 was decreased and expression of Fshr and Cyp19 was increased (P < 0.05). Given the heterozygous status of two patients carrying p. P126S and p. R370H, dominant negative effect of the mutant over WT protein was further investigated. Co-transfection of WT with mutant p. P127S and p. R387H did not interfere with the transcription of WT protein (Fig. 2A,B), which did not support the dominant negative effect.
Figure 2. Transfection of empty (Mock), wild-type (WT), or mutant (MT) Wt1-expressing adenovirus was performed in differentiated granulosa cells.
The potential dominant negative effect of the mutants was assessed by co-transfecting WT expressing adenovirus with MT adenovirus (1:1). The results of p. P126S mutant were showed in (A) and p. R370H in (B). *P < 0.05, **P < 0.01. (C). The GCs were transfected with empty (Mock), wild-type (WT), mutant p. P126S and p. R370H Wt1-expressing adenovirus separately. Western blot analyzed nuclear extracts from GCs using antibodys against ACTIN, WT1, AMH, FSHR, CYP19 and CDH1, respectively. Blot images were cropped for comparison.
Western blot was performed to compare the expression of AMH, FSHR, CYP19 and CDH1 between WT Wt1 and mutant Wt1-overexpressed GCs. As shown in Fig. 2C, the expression of FSHR and CYP19 were repressed in WT Wt1-transfected GCs; whereas higher expression was observed in GCs transfected either with p. P126S or with p. R370H. The level of AMH and CDH1 was significantly lower in GCs overexpressed either with p. P126S or with p. R370H compared with WT Wt1. These findings confirmed the impaired regulations of mutant p. P126S and p. R370H on AMH, FSHR, CYP19 and CDH1.
Discussion
The p. P126S mutant located in the proline/glutamine rich DNA-binding domain and p. R370H was adjacent to the four zinc-finger domains. The zinc-finger domains are crucial for WT1 to acquire the ability to interact with transcription and splice factors16. In mice, Wt1 plays a key role in follicular development regulating these genes associated with GCs proliferation and differentiation, e.g. Amh, Fshr, Lhr and Cyp19, and GCs polarity, e.g. Cdh1 and Par6b15. Dysregulation of these genes could result in immature differentiation of GCs and impaired establishment of polarity, and then aberrant follicular development.
FSHR is detected in GCs from primary follicle and remains present throughout folliculogenesis17. CYP19, encoding aromatase cytochrome P450 (P450arom), is expressed in GCs of large follicles and corpus luteum18. FSHR and CYP19 trigger the proliferation and differentiation of GCs, which is important for normal follicular growth19,20,21. As a transcriptional repressor of FSHR and CYP19, WT1 inhibits the premature proliferation and differentiation of GCs. In this study, mutant p. P126S and p. R370H induced FSHR and CYP19 expression, resulting in possible impaired estrogen synthesis, immature proliferation and differentiation of GCs.
AMH is expressed in primary follicles and preantral/small antral follicles22,23. AMH inhibits the recruitment of primordial follicles and decreases the responsiveness of growing follicles to FSH24. Our results revealed that mutant p. P126S and p. R370H repressed the expression of AMH, which would result in increased sensitivity to FSH. Therefore, it was speculated that p. P126S and p. R370H increased the FSH-responsiveness of follicles and promote the activation of primordial follicles.
In the early growing follicle, adherent junctions (AJs) are key adhesion complex25. Cadherins and nectins are two important molecules associated with AJs. The most well- characterized cadherins are E-cadherin (official symbol CDH1) and N-cadherin (official symbol CDH2)26. CDH1 is a calcium-dependent cell adhesion protein localized at the adherent junctions and serves as a marker for epithelial differentiation27. It is essential for maintaining the cuboidal shape of GCs, providing a means of rotating the orientation of mitotic axes and enabling multi-layering of GCs28. In this study, mutant p. P126S and p. R370H repressed the expression of CDH1 protein. It is speculated that declined CDH1 levels could not maintain the oocyte-GC interaction and the loss of granulosa cell adhesion would result in follicular atresia25,29.
To our knowledge, this is the first study to explore the role of the WT1 gene in human POF. Two novel heterozygous mutations p. P126S and p. R370H were identified, and resulted in impaired transcription on downstream genes, including AMH, FSHR, CYP19 and CDH1. They could interfere with the proliferation and differentiation of GCs and disrupt the oocyte-GC interaction. Further studies focusing on the signaling pathways associated with WT1 in ovary are warranted to reveal the mechanism involved in POF.
Methods
Ethics statement
The study procedures were approved by the Institutional Review Board of Reproductive Medicine of Shandong University. All the methods described here were carried out in accordance with the guidelines and regulations approved by the Institutional Review Board of Reproductive Medicine of Shandong University.
Patients
A total of 384 unrelated Han Chinese women with POF were recruited. Inclusion criteria consisted of primary amenorrhea (PA) or secondary amenorrhea (SA) for at least 6 months before 40 years of age and at least two serum FSH measurements exceeding 40 IU/L. Women with known karyotype abnormalities, previous chemotherapy or radiotherapy, ovarian surgery, or autoimmune diseases were excluded. Patients accompanying with somatic anomalies were excluded, particularly any reported as associated with pleiotropic Mendelian disorders (e.g., blepharophimosis-ptosis-epicanthus inversus syndrome, neurosensory deafness and cerebellar ataxia). Three hundred and eighty-four Han Chinese women with regular menses and normal hormone level were included as controls. Written informed consent was obtained from each subject.
Mutation Screening of WT1 Gene
Genomic DNA was extracted from peripheral blood samples with QIAamp DNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. All 10 exons and exon-intron boundaries of the WT1 gene (NM_000378.4) were amplified using polymerase chain reaction (PCR). All WT1 primers and PCR conditions are presented in Supplementary Table S1. The PCR product was first analyzed by agarose gel electrophoresis and then sequenced on an automated sequencer (ABI 3730 × 1 DNA Analyzer; Applied Biosystems, Foster City, CA). The novel sequence variants were confirmed by three independent PCR runs, followed by sequencing in both forward and reverse directions. Nomenclature of variants identified was established according to Human Genome Variant Society (HGVS, www.hgvs.org/mutnomen).
Adenovirus vectors construction
The mutant c.376C > T (p. P126S) and c.1109G > A (p. R370H), which were found in patients with POF are homologous to c. 187C > T (p. P127S) and c. 905G > A (p. R387H) in mouse, respectively. Wt1 expression vectors containing c. 187C > T and c. 905G > A were generated by site-directed mutagenesis (QuikChange Lightning Site-Directed Mutagenesis Kit; Stratagene, LaJolla, CA) with WT mouse Wt1 expression vector as a template. A recombinant adenovirus plasmid was generated by GV adenovirus vector system, pHelper 1.0 vector and pHelper 2.0 vector (Genechem, Shanghai, CH) and then packaged by AdMax system (Microbix, CA). A high titer (1 × 1011 PFU/ml) of adenovirus was obtained by two rounds of amplification and purified.
Granulosa cells isolation and quantitative RT-PCR analysis
Female mouse at the age of three weeks were injected with 5 IU pregnant mare serum gonadotropin (PMSG), and the ovaries were harvested at 46–48 h after PMSG injection. GCs from pre-ovulatory follicles were collected and cultured overnight for adhesion. Twenty-four hours after adenovirus vectors containing Wt1 gene infected, GCs were harvested and RNA was extracted. After reverse transcription (Takara, Japan), quantitative real-time PCR (qRT-PCR) was performed on the Roche Lightcycle 480 using specific primers (listed in Supplementary Table S2) with PrimeScript RT Master Mix (Takara). The mRNA levels were normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and analyzed using the comparative cycle threshold method.
Western blot
The nuclear extracts from infected GCs of empty (Mock), wild-type (WT), mutant p. P126S and p. R370H Wt1-expressiong adenovirus separately were added to 5× sodium dodecyl sulfate (SDS) sample buffer and heated at 95 °C for 5 minutes to denature the proteins. The nuclear extracts were then separated on 10% or 6% SDS-PAGE gels and transferred to polyvinylidene difluoride membranes. The membranes were blocked, incubated with primary antibodies against ACTIN (catalog # 60008-1-lg, Proteintech, CH), WT1 (catalog # sc-192, Santa Cruz, USA), AMH (catalo # sc-166752, Santa Cruz), FSHR (catalog # 22665-1-AP, Proteintech), CYP19 (catalog # 16554-1-AP, Proteintech ) and CDH1 (catalog # 610182, BD Biosciences, USA ) overnight at 4 °C, washed, and then incubated with horseradish peroxidase-conjugated secondary antibodies. Chemiluminescent detection was performed using immobilon western chemiluminescent HRP substrate (Millipore Corporation, Billerica, USA).
Statistical analysis
Results of qRT-PCR were expressed by bar graphs plotted in MS Excel and represented the Mean ± S.D. of 3 independent experiments, each performed in triplicate. Experiments were repeated at least tree times. The data were performed by chi-squared test and one-way ANOVA. Statistical differences were considered significant when P < 0.05.
Additional Information
How to cite this article: Wang, H. et al. Novel WT1 Missense Mutations in Han Chinese Women with Premature Ovarian Failure. Sci. Rep. 5, 13983; doi: 10.1038/srep13983 (2015).
Supplementary Material
Acknowledgments
This work is supported by grants from the National Basic Research Program of China (973 program-2012CB944700), the National Natural Science Foundation of China (81471509, 81270662, 81370692). Ethical approval: This study was approved by the Institutional Review Board of Reproductive Medicine of Shandong University.
Footnotes
The authors declare no competing financial interests.
Author Contributions All authors were involved in the study. H.W., G.L., Y.Q., W.L., F.G. and Z.-.J.C. conceived and designed the experiments; H.W., G.L., J.Z. performed the experiments; H.W., G.L. and Y.Q. analyzed the data; H.W., Y.Q., F.G. and Z.-J.C. wrote the paper, H.W. undertook patient management and collection of samples.
References
- Coulam C. B., Adamson S. C. & Annegers J. F. Incidence of premature ovarian failure. Obstetrics and gynecology 67, 604–606 (1986). [PubMed] [Google Scholar]
- Goswami D. & Conway G. S. Premature ovarian failure. Hum Reprod Update 11, 391–410, 10.1093/humupd/dmi012 (2005). [DOI] [PubMed] [Google Scholar]
- van Kasteren Y. M. et al. Familial idiopathic premature ovarian failure: an overrated and underestimated genetic disease? Hum Reprod 14, 2455–2459 (1999). [DOI] [PubMed] [Google Scholar]
- Kuo F. T., Bentsi-Barnes I. K., Barlow G. M. & Pisarska M. D. Mutant Forkhead L2 (FOXL2) proteins associated with premature ovarian failure (POF) dimerize with wild-type FOXL2, leading to altered regulation of genes associated with granulosa cell differentiation. Endocrinology 152, 3917–3929, 10.1210/en.2010-0989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri M. R. et al. Alterations in the expression, structure and function of progesterone receptor membrane component-1 (PGRMC1) in premature ovarian failure. Human molecular genetics 17, 3776–3783, 10.1093/hmg/ddn274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. et al. NOBOX homeobox mutation causes premature ovarian failure. American journal of human genetics 81, 576–581, 10.1086/519496 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetti R. et al. BMP15 mutations associated with primary ovarian insufficiency cause a defective production of bioactive protein. Human mutation 30, 804–810, 10.1002/humu.20961 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y. et al. Association of 8q22.3 locus in Chinese Han with idiopathic premature ovarian failure (POF). Human molecular genetics 21, 430–436, 10.1093/hmg/ddr462 (2012). [DOI] [PubMed] [Google Scholar]
- Kreidberg J. A. et al. WT-1 is required for early kidney development. Cell 74, 679–691 (1993). [DOI] [PubMed] [Google Scholar]
- Gessler M. et al. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature 343, 774–778, 10.1038/343774a0 (1990). [DOI] [PubMed] [Google Scholar]
- Hohenstein P. & Hastie N. D. The many facets of the Wilms’ tumour gene, WT1. Human molecular genetics 15 Spec No 2, R196–201, 10.1093/hmg/ddl196 (2006). [DOI] [PubMed] [Google Scholar]
- Rosenfeld C., Cheever M. A. & Gaiger A. WT1 in acute leukemia, chronic myelogenous leukemia and myelodysplastic syndrome: therapeutic potential of WT1 targeted therapies. Leukemia 17, 1301–1312, 10.1038/sj.leu.2402988 (2003). [DOI] [PubMed] [Google Scholar]
- Buckler A. J., Pelletier J., Haber D. A., Glaser T. & Housman D. E. Isolation, characterization, and expression of the murine Wilms’ tumor gene (WT1) during kidney development. Mol Cell Biol 11, 1707–1712 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S. Y. et al. Wilms’ tumor protein WT1 as an ovarian transcription factor: decreases in expression during follicle development and repression of inhibin-alpha gene promoter. Mol Endocrinol 9, 1356–1366, 10.1210/mend.9.10.8544844 (1995). [DOI] [PubMed] [Google Scholar]
- Gao F. et al. Wt1 functions in ovarian follicle development by regulating granulosa cell differentiation. Human molecular genetics 23, 333–341, 10.1093/hmg/ddt423 (2014). [DOI] [PubMed] [Google Scholar]
- Ladomery M., Sommerville J., Woolner S., Slight J. & Hastie N. Expression in Xenopus oocytes shows that WT1 binds transcripts in vivo, with a central role for zinc finger one. Journal of cell science 116, 1539–1549 (2003). [DOI] [PubMed] [Google Scholar]
- Oktay K., Briggs D. & Gosden R. G. Ontogeny of follicle-stimulating hormone receptor gene expression in isolated human ovarian follicles. The Journal of clinical endocrinology and metabolism 82, 3748–3751, 10.1210/jcem.82.11.4346 (1997). [DOI] [PubMed] [Google Scholar]
- Suzuki T. et al. Temporal and spatial localization of steroidogenic enzymes in premenopausal human ovaries: in situ hybridization and immunohistochemical study. Molecular and cellular endocrinology 97, 135–143 (1993). [DOI] [PubMed] [Google Scholar]
- Kempisty B. et al. Association between the expression of LHR, FSHR and CYP19 genes, cellular distribution of encoded proteins and proliferation of porcine granulosa cells in real-time. Journal of biological regulators and homeostatic agents 28, 419–431 (2014). [PubMed] [Google Scholar]
- Nelson D. R. et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA and cell biology 12, 1–51 (1993). [DOI] [PubMed] [Google Scholar]
- Robker R. L. & Richards J. S. Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol Endocrinol 12, 924–940, 10.1210/mend.12.7.0138 (1998). [DOI] [PubMed] [Google Scholar]
- Munsterberg A. & Lovell-Badge R. Expression of the mouse anti-mullerian hormone gene suggests a role in both male and female sexual differentiation. Development 113, 613–624 (1991). [DOI] [PubMed] [Google Scholar]
- Taketo T., Saeed J., Manganaro T., Takahashi M. & Donahoe P. K. Mullerian inhibiting substance production associated with loss of oocytes and testicular differentiation in the transplanted mouse XX gonadal primordium. Biol Reprod 49, 13–23 (1993). [DOI] [PubMed] [Google Scholar]
- Durlinger A. L., Visser J. A. & Themmen A. P. Regulation of ovarian function: the role of anti-Mullerian hormone. Reproduction 124, 601–609 (2002). [DOI] [PubMed] [Google Scholar]
- Mora J. M. et al. Characterization and significance of adhesion and junction-related proteins in mouse ovarian follicles. Biol Reprod 86, 153, 151–114, 10.1095/biolreprod.111.096156 (2012). [DOI] [PubMed] [Google Scholar]
- Halbleib J. M. & Nelson W. J. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 20, 3199–3214, 10.1101/gad.1486806 (2006). [DOI] [PubMed] [Google Scholar]
- Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science 251, 1451–1455 (1991). [DOI] [PubMed] [Google Scholar]
- Da Silva-Buttkus P. et al. Effect of cell shape and packing density on granulosa cell proliferation and formation of multiple layers during early follicle development in the ovary. Journal of cell science 121, 3890–3900, 10.1242/jcs.036400 (2008). [DOI] [PubMed] [Google Scholar]
- Ryan P. L., Valentine A. F. & Bagnell C. A. Expression of epithelial cadherin in the developing and adult pig ovary. Biol Reprod 55, 1091–1097 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.