Abstract
DNA methylation, a major epigenetic mechanism, may regulate coordinated expression of multiple genes at specific time points during alveolar septation in lung development. The objective of this study was to identify genes regulated by methylation during normal septation in mice and during disordered septation in bronchopulmonary dysplasia. In mice, newborn lungs (preseptation) and adult lungs (postseptation) were evaluated by microarray analysis of gene expression and immunoprecipitation of methylated DNA followed by sequencing (MeDIP-Seq). In humans, microarray gene expression data were integrated with genome-wide DNA methylation data from bronchopulmonary dysplasia versus preterm and term lung. Genes with reciprocal changes in expression and methylation, suggesting regulation by DNA methylation, were identified. In mice, 95 genes with inverse correlation between expression and methylation during normal septation were identified. In addition to genes known to be important in lung development (Wnt signaling, Angpt2, Sox9, etc.) and its extracellular matrix (Tnc, Eln, etc.), genes involved with immune and antioxidant defense (Stat4, Sod3, Prdx6, etc.) were also observed. In humans, 23 genes were differentially methylated with reciprocal changes in expression in bronchopulmonary dysplasia compared with preterm or term lung. Genes of interest included those involved with detoxifying enzymes (Gstm3) and transforming growth factor-β signaling (bone morphogenetic protein 7 [Bmp7]). In terms of overlap, 20 genes and three pathways methylated during mouse lung development also demonstrated changes in methylation between preterm and term human lung. Changes in methylation correspond to altered expression of a number of genes associated with lung development, suggesting that DNA methylation of these genes may regulate normal and abnormal alveolar septation.
Keywords: lung development, premature infant, epigenetics, bronchopulmonary dysplasia
Clinical Relevance
DNA methylation is an important regulator of gene expression. Coordinated expression of multiple genes occurs at specific time points during alveolar septation in lung development. In this study, we identified multiple genes that are potentially regulated by DNA methylation during normal alveolar septation in the mouse and human lung, and in bronchopulmonary dysplasia compared to preterm or term lung.
Lung development is a critically important biological process that is highly complex and is divided into five stages—embryonic, pseudoglandular, canalicular, saccular, and alveolar (1). Precise regulation of gene expression is important for normal lung development to occur, and interruptions during key periods in its development can have serious consequences. Preterm infants are especially at risk, as they are born with immature lungs in the saccular stage, and are exposed to a multitude of environmental stressors that can disrupt the transition to the alveolar stage, leading to a form of chronic lung disease, known as bronchopulmonary dysplasia (BPD), characterized by inhibition of alveolar septation and varying degrees of fibrosis and vascular remodeling (2).
The exact molecular mechanisms that govern normal lung development and its arrest in BPD remain unknown (3). Previous studies have used microarray analysis to report gene expression profiles in normal lung development from murine (4) as well as human tissue samples (5). A recent study by Bhattacharya and colleagues (6) has also provided data on gene expression patterns observed in human BPD versus control lung tissue samples. However, despite the wealth of gene expression data from these studies, there are few studies that look at the underlying molecular mechanisms that regulate and maintain the changes in gene expression observed in normal and altered lung development.
DNA methylation is one of the most extensively studied epigenetic mechanisms that regulate gene expression. It is well established that changes in DNA methylation and differences in gene regulation are causally related, with hypomethylation generally leading to gene expression, and hypermethylation resulting in gene silencing (7). DNA methylation is known to be important in a number of biological processes, including normal differentiation and development of tissues and organs (8). Recent studies have demonstrated the significance of DNA methylation in regulating development of various organ systems, including the brain (9), bone (10), prostate (11), and hematopoietic system (12). It is likely that DNA methylation plays an important role in both normal and abnormal lung development.
The purpose of this study was to identify genes likely regulated by DNA methylation during normal alveolar septation in mice, and during disordered alveolar septation in human BPD. We performed an integrative analysis of both genome-wide DNA methylation and gene expression data, and focused on genes with inverse changes in expression and methylation to highlight genes, the expression of which is likely regulated by DNA methylation. We limited our study to alveolar septation, as BPD is a disease characterized by an arrest in alveolar development.
Materials and Methods
See the online supplement for details.
Lung Tissue Samples
Mouse
C57BL/6 mouse lungs were isolated at Postnatal Day 3 (P3) and at 6 weeks of age (P42) to allow comparison between newborn lungs before initiation and adult lungs after full completion of alveolar septation (13). The protocol was approved by the Institutional Animal Care and Use Committee at the University of Alabama at Birmingham (UAB).
Human
Formalin-fixed, paraffin-embedded (FFPE), deidentified autopsy lung tissue samples were obtained after Institutional Review Board approval. The study group consisted of preterm infants with BPD (postmenstrual age, 28–42 wk; n = 6). For comparison, preterm stillbirths (24- to 26-wk gestation; n = 5) and term stillbirths (36–40 wk; n = 6) were used as control groups.
DNA Isolation and DNA Methylation Profiling
Mouse
Mouse genomic DNA was extracted, and methylated DNA fragments were immunoprecipitated using a monoclonal antibody specific to 5-methylcytosine (anti–5-methylcytosine, MABE146; Millipore). High-throughput sequencing was then performed using the enriched methylated fragments (MeDIP-Seq) (14).
Human
Genomic DNA was isolated using the QIAmp FFPE Tissue Kit (Qiagen, Valencia, CA) and methylation profile assayed using the Illumina HumanMethylation450 DNA Analysis BeadChip (Illumina).
RNA Isolation and Gene Expression Profiling
Mouse
Total RNA was isolated using the QiagenRNeasy Mini kit (Qiagen) and then profiled using the Agilent Whole-Mouse Genome Microarrays Kit (G4122F; Agilent).
Human
RNA isolated using Qiagen RNeasy FFPE kit (Qiagen) was used for validation of selected genes by real-time RT-PCR (n = 4/group).
Data Analyses
DNA methylation data
Mouse
MeDIP-Seq data were mapped to the mouse reference genome using Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/). The Model-based Analysis of MeDIP-Seq software program (http://liulab.dfci.harvard.edu/MACS/) identified peak regions with P less than 0.05.
Human
HumanMethylation450 data were analyzed using the “minfi” package in Bioconductor (http://www.bioconductor.org/packages/release/bioc/html/minfi.html). Data were normalized using the subset-quantile within array normalization (15). F-test compared the three groups, and t tests were used for pair-wise contrasts to identify differential methylation at each cytosine-phosphate-guanine dinucleotide (CpG) site based on M values (logit transformation of β values). False discovery rate (FDR) of less than 0.05 was used to select significant CpG sites in BPD compared with preterm and term groups (16).
Gene expression data
Mouse
Data were analyzed using GeneSpring GX version 11 software (Agilent). A stringent FDR with a cutoff of 0.01, and Bonferroni correction for multiple comparisons was used, and genes with a fold change two or greater were identified as differentially expressed. Data reported in this paper are in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo; accession no. GSE41412 [time series expression data]) (17).
Human
We used a previously published dataset describing genome-wide expression using the Affymetrix HG-U133 plus array in lung tissue from BPD (n = 11) or controls (n = 9) (6). Ingenuity pathway analysis (IPA; Ingenuity Systems, CA) identified canonical pathways. Gene set enrichment analysis (GSEA) was performed using robust multiarray average-normalized expression intensities of all probe sets.
Integrating gene expression and DNA methylation
Data obtained from gene expression analysis was compared with DNA methylation analysis to identify genes that were significantly altered in both DNA methylation and gene expression. Pathway analysis of these genes was performed with IPA.
Results
Mouse Lung
Gene expression analysis reveals 1,040 differentially expressed genes between newborn versus adult mouse lungs
Gene expression profiles were obtained from total RNA isolated from newborn and adult mouse lungs. Of the more than 40,000 genes on the microarray chip, there were 1,040 genes with a P value less than 0.01 and fold change of two or greater between newborn and adult mouse lungs. Of these, 424 genes were up-regulated and 616 genes were down-regulated in adult compared with newborn lungs.
Integrating gene expression with DNA methylation analysis demonstrates 209 differentially methylated and expressed genes, with 95 genes having an inverse correlation
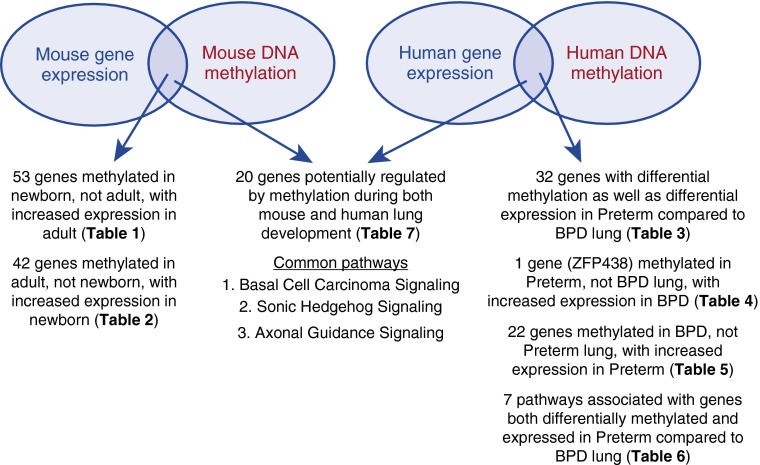
Genome-wide methylation profiles were obtained from DNA isolated from newborn and adults lungs. Differentially methylated genes between the two groups (P < 0.05) were identified and combined with data from gene expression profiles. Of the 1,040 genes with differential gene expression between newborn and adult mouse lungs, 209 genes also had differential DNA methylation. Of these, 95 had an inverse relationship between DNA methylation status and gene expression. A total of 53 genes had increased DNA methylation in newborn compared with adult lungs, with corresponding decreased gene expression in newborn lungs, representing genes possibly silenced by DNA methylation in the newborn lung (Table 1; Figure 1). On the other hand, 42 genes had increased DNA methylation with corresponding decreased gene expression in adult compared with newborn lungs, representing genes possibly silenced by DNA methylation in adult lungs (Table 2; Figure 1).
Table 1.
Genes with Increased DNA Methylation and Decreased Messenger RNA Expression in Newborn Mouse Lung Compared with Adult Mouse Lung, Arranged in Order of Increasing P Value
| Gene Symbol | Probe Name | Gene Title | Entrez Gene | Adjusted P Value | Fold Change in Gene Expression | Fold Enrichment in Methylation |
|---|---|---|---|---|---|---|
| Chi3l3 | A_51_P167292 | Chitinase 3–like 3 | 12655 | 7.4E−08 | −58.9 | 19.3 |
| Nrn1 | A_51_P308844 | Neuritin 1 | 68404 | 1.7E−07 | −11.4 | 22.6 |
| Lrp2 | A_52_P285470 | Low-density lipoprotein receptor–related protein 2 | 14725 | 2.4E−07 | −5.1 | 20.7 |
| Inmt | A_51_P162162 | Indolethylamine N-methyltransferase | 21743 | 4.7E−07 | −176.5 | 41.4 |
| Scube2 | A_55_P2232988 | Signal peptide, CUB domain, EGF-like 2 | 56788 | 5.5E−07 | −3.3 | 41.4 |
| Mmp3 | A_51_P255699 | Matrix metallopeptidase 3 | 17392 | 7.2E−07 | −144.4 | 11.09 |
| Ggct | A_52_P641758 | γ-Glutamylcyclotransferase | 110175 | 1.2E−06 | −2.2 | 8.28 |
| Cd209a* | A_55_P2018061 | CD209a antigen | 170786 | 1.8E−06 | −19.7 | 8.775 |
| Aox3 | A_52_P16752 | Aldehyde oxidase 3 | 71724 | 2.8E−06 | −8.3 | 33.11 |
| Sod3 | A_55_P2077558 | Superoxide dismutase 3, extracellular | 20657 | 2.9E−06 | −11.3 | 37.94 |
| Bcl6 | A_52_P161495 | B-cell leukemia/lymphoma 6 | 12053 | 4.3E−06 | −4.2 | 45.07 |
| Crebl2 | A_52_P541270 | cAMP responsive element binding protein–like 2 | 232430 | 4.8E−06 | −4.2 | 45.07 |
| Zbtb4 | A_55_P2165655 | Zinc finger and BTB domain containing 4 | 75580 | 1.2E−05 | −2.2 | 45.07 |
| Ccr6 | A_55_P2108943 | Chemokine (C-C motif) receptor 6 | 12458 | 2.2E−05 | −9.2 | 6.34 |
| H2-Ob* | A_55_P2082929 | Histocompatibility 2, O region β locus | 15002 | 2.2E−05 | −14.4 | 11.04 |
| Sp110 | A_55_P2152566 | Sp110 nuclear body protein | 109032 | 2.8E−05 | −3.2 | 45.07 |
| Lilrb3 | A_55_P2097279 | Leukocyte Ig–like receptor, subfamily B (with TM and ITIM domains), member 3 | 18733 | 1.1E−04 | −2.5 | 14.28 |
| St3gal1 | A_65_P03728 | ST3 β-galactoside α-2,3-sialyltransferase 1 | 20442 | 1.2E−04 | −2.1 | 15.57 |
| Efemp1 | A_55_P2048767 | Epidermal growth factor–containing fibulin-like extracellular matrix protein 1 | 216616 | 1.3E−04 | −4.9 | 9.03 |
| Adra1a | A_52_P424778 | Adrenergic receptor, α 1a | 11549 | 1.6E−04 | −20.5 | 7.95 |
| B2 m | A_51_P129012 | β-2 microglobulin | 12010 | 1.7E−04 | −4.2 | 19.66 |
| BC006779 | A_55_P1959953 | cDNA sequence BC006779 | 229003 | 2.1E−04 | −2.3 | 12.17 |
| Prdx6 | A_55_P2103561 | Peroxiredoxin 6 | 11758 | 2.1E−04 | −2.8 | 41.38 |
| Angpt2 | A_51_P201982 | Angiopoietin 2 | 11601 | 2.2E−04 | −2.6 | 9.44 |
| Lims2 | A_55_P2174203 | LIM and senescent cell antigen–like domains 2 | 225341 | 2.6E−04 | −3.5 | 41.38 |
| Prkcq | A_52_P72587 | Protein kinase C, θ | 18761 | 2.6E−04 | −3.0 | 9.36 |
| Scml4 | A_55_P2020338 | Sex comb on midleg–like 4 (Drosophila) | 268297 | 3.3E−04 | −4.3 | 41.38 |
| Aldh1a1 | A_51_P334942 | Aldehyde dehydrogenase family 1, subfamily A1 | 11668 | 3.6E−04 | −4.9 | 9.22 |
| Gbp5 | A_52_P327664 | Guanylate binding protein 5 | 229898 | 3.9E−04 | −4.0 | 54.09 |
| Slc9a4 | A_55_P1973588 | Solute carrier family 9 (sodium/hydrogen exchanger), member 4 | 110895 | 4.0E−04 | −15.5 | 33.11 |
| Sh3bp5 | A_55_P2049479 | SH3-domain binding protein 5 (BTK-associated) | 24056 | 4.0E−04 | −2.4 | 54.09 |
| Tmcc3* | A_52_P16877 | Transmembrane and coiled coil domains 3 | 319880 | 4.8E−04 | −2.8 | 9.7 |
| Cd209a* | A_51_P133884 | CD209a antigen | 170786 | 5.7E−04 | −17.9 | 8.76 |
| Gria1 | A_55_P2113758 | Glutamate receptor, ionotropic, AMPA1 (α 1) | 14799 | 7.8E−04 | −7.8 | 33.84 |
| Rasgrp1 | A_55_P1969650 | RAS guanyl releasing protein 1 | 19419 | 8.4E−04 | −5.2 | 22.15 |
| Kcnj15 | A_55_P2164090 | Potassium inwardly rectifying channel, subfamily J, member 15 | 16516 | 8.4E−04 | −2.3 | 45.07 |
| Cobl | A_55_P1995243 | Cordon-bleu | 12808 | 1.0E−03 | −2.4 | 11.38625 |
| Heatr1 | A_66_P108019 | HEAT repeat containing 1 | 217995 | 1.1E−03 | −2.0 | 7.57 |
| Blnk | A_55_P2002757 | B cell linker | 17060 | 1.1E−03 | −5.4 | 16.63 |
| Igfbp3 | A_52_P253179 | Insulin-like growth factor binding protein 3 | 16009 | 1.3E−03 | −6.1 | 12.41 |
| Adam3 | A_55_P2025721 | A disintegrin and metallopeptidase domain 3 (cyritestin) | 11497 | 2.1E−03 | −14.4 | 45.07 |
| Gm281 | A_55_P1970110 | Predicted gene 281 | 238939 | 2.3E−03 | −11.1 | 6.03 |
| Klrb1b | A_55_P2106681 | Killer cell lectin-like receptor subfamily B member 1B | 80782 | 2.5E−03 | −5.1 | 24.83 |
| Spnb5 | A_55_P1994504 | Spectrin β 5 | 640524 | 3.7E−03 | −4.0 | 13.79 |
| Sh2d1a | A_55_P1958146 | SH2 domain protein 1A | 20400 | 4.2E−03 | −6.5 | 15.76 |
| Wisp2 | A_51_P390804 | WNT1 inducible signaling pathway protein 2 | 22403 | 4.4E−03 | −15.0 | 14.77 |
| Tmcc3* | A_51_P189272 | Transmembrane and coiled coil domains 3 | 319880 | 4.8E−03 | −3.7 | 9.7 |
| Phf15 | A_55_P2139753 | PHD finger protein 15 | 76901 | 4.9E−03 | −3.3 | 20.69 |
| Cdhr3 | A_66_P122115 | Cadherin-related family member 3 | 68764 | 6.1E−03 | −4.1 | 49.58 |
| Stat4 | A_51_P177092 | Signal transducer and activator of transcription 4 | 20849 | 6.2E−03 | −5.7 | 8.28 |
| Cd19 | A_55_P2079079 | CD19 antigen | 12478 | 7.1E−03 | −5.2 | 33.11 |
| H2-Ob* | A_66_P130916 | Histocompatibility 2, O region β locus | 15002 | 7.6E−03 | −8.7 | 11.04 |
| Mtmr7 | A_66_P117578 | Myotubularin-related protein 7 | 54384 | 7.8E−03 | −2.6 | 18.02 |
| Ccl5 | A_52_P638459 | Chemokine (C-C motif) ligand 5 | 20304 | 9.1E−03 | −11.2 | 33.86 |
| Ampd3 | A_52_P502754 | Adenosine monophosphate deaminase 3 | 11717 | 9.5E−03 | −2.7 | 82.76 |
Definition of abbreviations: AMPA1, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; BTB, BR-C, ttk, and bab domain; BTK, Bruton's tyrosine kinase; CUB, complement C1r/C1s, Uegf, Bmp1 domain; EGF, epidermal growth factor; HEAT, Huntingtin, elongation factor 3, the PR65/A subunit of protein phosphatase 2A, and the lipid kinase Tor; ITIM, immunoreceptor tyrosine-based inhibition motif; LIM, Lin11, Isl-1, and Mec-3 proteins; PHD, plant homeodomain protein; TM, transmembrane; RAS, rat sarcoma; SH, Src Homology 2; ST3, sialyltransferase 3; WNT, wingless-type MMTV integration site family.
Genes with methylation in more than one locus.
Figure 1.
Overall approach in this project with summary and location of results. BPD, bronchopulmonary dysplasia.
Table 2.
Genes with Increased DNA Methylation and Decreased Messenger RNA Expression in Adult Mouse Lung Compared with Newborn Mouse Lung, Arranged in Order of Increasing P Value
| Gene Symbol | Probe Name | Gene Title | Entrez Gene | Adjusted P Value | Fold Change in Gene Expression | Fold Enrichment In Methylation |
|---|---|---|---|---|---|---|
| Col27a1 | A_52_P480088 | Collagen, type XXVII, α 1 | 373864 | 4.5E−09 | −4.0 | 26.39 |
| Slc27a6 | A_55_P2039379 | Solute carrier family 27 (fatty acid transporter), member 6 | 225579 | 1.3E−06 | −18.4 | 26.39 |
| Skp2 | A_66_P134481 | S-phase kinase–associated protein 2 (p45) | 27401 | 5.6E−06 | −4.0 | 43.98 |
| Camk1 g | A_55_P2020331 | Calcium/calmodulin–dependent protein kinase I γ | 215303 | 8.7E−06 | −42.8 | 43.98 |
| Rrm1 | A_51_P502082 | Ribonucleotide reductase M1 | 20133 | 1.0E−05 | −3.9 | 20.16 |
| Col6a3 | A_52_P479262 | Collagen, type VI, α 3 | 12835 | 1.3E−05 | −2.4 | 48.39 |
| Raet1e | A_55_P2115955 | Retinoic acid early transcript 1E | 379043 | 1.4E−05 | −2.6 | 10.16 |
| Shisa2 | A_51_P408649 | Shisa homolog 2 (Xenopus laevis) | 219134 | 2.0E−05 | −5.1 | 35.19 |
| E2f7 | A_55_P2062598 | E2F transcription factor 7 | 52679 | 3.2E−05 | −10.8 | 32.26 |
| E2f8 | A_55_P2000833 | E2F transcription factor 8 | 108961 | 3.3E−05 | −10.3 | 52.78 |
| Wif1 | A_51_P484526 | Wnt inhibitory factor 1 | 24117 | 3.9E−05 | −7.6 | 40.32 |
| Auts2 | A_51_P398191 | Autism susceptibility candidate 2 | 319974 | 5.7E−05 | −2.8 | 52.23 |
| Cdh11 | A_55_P2181356 | Cadherin 11 | 12552 | 6.0E−05 | −3.0 | 43.98 |
| Rrm2 | A_55_P2173982 | Ribonucleotide reductase M2 | 20135 | 7.3E−05 | −9.2 | 9.87 |
| Eln | A_52_P609972 | Elastin | 13717 | 8.3E−05 | −3.8 | 52.78 |
| Pdlim3 | A_52_P579531 | PDZ and LIM domain 3 | 53318 | 8.3E−05 | −6.9 | 48.39 |
| Spag5 | A_51_P513530 | Sperm-associated antigen 5 | 54141 | 1.0E−04 | −11.1 | 16.13 |
| Ptx3 | A_51_P374726 | Pentraxin-related gene | 19288 | 1.2E−04 | −5.1 | 43.98 |
| 2810417H13Rik | A_66_P133404 | RIKEN cDNA 2810417H13 gene | 68026 | 1.3E−04 | −13.6 | 9.6 |
| Unc5c | A_66_P129566 | UNC-5 homolog C (Caenorhabditis elegans) | 22253 | 1.6E−04 | −7.1 | 61.57 |
| Dgki | A_51_P136411 | Diacylglycerol kinase, iota | 320127 | 2.3E−04 | −5.5 | 61.57 |
| Dctd* | A_52_P183826 | dCMP deaminase | 320685 | 2.7E−04 | −6.2 | 32.26 |
| Kcnma1 | A_55_P2378486 | Potassium large conductance calcium-activated channel, subfamily M, α member 1 | 16531 | 3.7E−04 | −2.7 | 48.38 |
| Tnc* | A_55_P2421597 | Tenascin C | 21923 | 3.9E−04 | −26.7 | 52.78 |
| Dctd* | A_52_P301085 | dCMP deaminase | 320685 | 5.2E−04 | −13.0 | 32.26 |
| Gm6531 | A_55_P2057142 | Predicted gene 6531 | 624855 | 5.2E−04 | −6.3 | 48.39 |
| Ext1 | A_51_P416689 | Exostoses (multiple) 1 | 14042 | 5.8E−04 | −2.3 | 32.26 |
| Rfc5 | A_51_P246339 | Replication factor C (activator 1) 5 | 72151 | 6.6E−04 | −3.8 | 43.98 |
| Ttf2 | A_55_P2062836 | Transcription termination factor, RNA polymerase II | 74044 | 1.1E−03 | −2.1 | 52.78 |
| Prdm8 | A_55_P2024525 | PR domain–containing 8 | 77630 | 1.2E−03 | −2.4 | 61.57 |
| Plac1 | A_55_P1962279 | Placental specific protein 1 | 56096 | 1.3E−03 | −27.3 | 32.26 |
| Shisa3 | A_55_P2181104 | Shisa homolog 3 (Xenopus laevis) | 330096 | 1.4E−03 | −14.8 | 40.32 |
| Lrrc4b* | A_55_P1953503 | Leucine rich repeat containing 4B | 272381 | 1.5E−03 | −5.5 | 56.45 |
| Lrrc4b* | A_55_P2125982 | Leucine rich repeat containing 4B | 272381 | 1.7E−03 | −5.5 | 56.45 |
| Gm7092 | A_55_P2016079 | Predicted gene 7092 | 632778 | 2.0E−03 | −2.1 | 43.98 |
| Shisa6 | A_55_P2154749 | Shisa homolog 6 (Xenopus laevis) | 380702 | 2.2E−03 | −5.2 | 48.38 |
| Ctnna2* | A_55_P2075726 | Catenin (cadherin-associated protein), α 2 | 12386 | 2.6E−03 | −3.6 | 52.78 |
| Eme1 | A_51_P401263 | Essential meiotic endonuclease 1 homolog 1 (Schizosaccharomyces pombe) | 268465 | 2.6E−03 | −7.6 | 24.19 |
| Ermap | A_51_P391716 | Erythroblast membrane-associated protein | 27028 | 2.7E−03 | −7.7 | 40.32 |
| Robo1 | A_55_P1985070 | Roundabout homolog 1 (Drosophila) | 19876 | 3.5E−03 | −3.0 | 24.19 |
| Sox9 | A_52_P214630 | SRY-box containing gene 9 | 20682 | 4.4E−03 | −6.8 | 61.57 |
| Tnc* | A_52_P355169 | Tenascin C | 21923 | 4.6E−03 | −54.3 | 52.78 |
| Dido1 | A_55_P2430472 | Death inducer–obliterator 1 | 23856 | 4.6E−03 | −3.4 | 40.32 |
| Ctnna2* | A_51_P438841 | Catenin (cadherin-associated protein), α 2 | 12386 | 4.9E−03 | −17.0 | 52.78 |
| 4833423E24Rik | A_66_P106783 | RIKEN cDNA 4833423E24 gene | 228151 | 5.2E−03 | −8.3 | 6.82 |
| Tmsb10* | A_55_P2168990 | Thymosin, β 10 | 19240 | 7.3E−03 | −2.4 | 9.86 |
| Tmsb10* | A_55_P2092085 | Thymosin, β 10 | 19240 | 8.6E−03 | −2.3 | 9.86 |
Defintion of abbreviations: dCMP, deoxycytidine monophosphate; PDZ, post-synaptic density protein, Drosophila disc large tumor suppressor, and zonula occludens-1 protein; SRY, sex determining region Y.
Genes with methylation in more than one locus.
The remaining 114 genes had the same direction of DNA methylation and gene expression. Of these, 83 genes had both increased DNA methylation and increased gene expression in newborn compared with adult lungs (see Table E1 in the online supplement), and 31 genes had both increased DNA methylation and increased gene expression in adult compared with newborn lungs (Table E2).
Validation of gene expression of matrix metallopeptidase (MMP) 3 (increased DNA methylation, decreased mRNA in newborn) and tenascin C (TNC; increased DNA methylation and decreased mRNA expression in adult) confirmed the direction of gene expression seen on microarray (newborn versus adult mRNA/glyceraldehyde 3-phosphate dehydrogenase: MMP3, 0.009 ± 0.003 versus 1.33 ± 0.19; TNC, 35.4 ± 6.6 versus 2.40 ± 0.5; all, P < 0.05). DNA methylation of MMP3 and TNC was additionally confirmed by bisulfite conversion of newborn and adult mouse lung DNA, followed by PCR and sequencing (see Figure E1 in the online supplement). DNA methylation of transmembrane and coiled-coil domain family 3 and CMP deaminase was confirmed by pyrosequencing (Figure E2).
Pathway analysis
IPA of genes possibly silenced by DNA methylation in newborn mouse lungs revealed six gene networks (Table E3 and Figures E3–E8), whereas IPA of the genes possibly silenced by DNA methylation in adult lungs revealed four gene networks (Table E4 and Figures E9–E12). The top networks were associated with several relevant functional categories, including cellular development, cellular growth and proliferation, cellular assembly and organization, cell-to-cell signaling and interaction, organ morphology, and inflammatory response.
Human Lung
As we used two separate cohorts of infants with BPD (the gene expression dataset from the study of Bhattacharya and colleagues [6], Rochester cohort, and the samples used for DNA methylation analyses from UAB), we first compared gene expression in these two cohorts. Expression of the four genes with the maximum fold change increase in the Rochester cohort was highly increased in the UAB cohort as well (carboxypeptidase A3—Rochester, 7.14-fold increase; UAB, 12.2-fold increase; chemokine [C-C motif] ligand [CCL] 18—Rochester, 6.74-fold increase; UAB, 17.7-fold increase; Ig heavy constant α 1—Rochester, 6.9-fold increase; UAB, 12.2-fold increase; tryptase α/β 1—Rochester, 6.16-fold increase; UAB, 4.5-fold increase).
DNA methylation studies identify 149 genes differentially methylated in BPD
DNA methylation profiles were obtained from total DNA isolated and restored from BPD and control (term and preterm) FFPE lung tissue samples. By comparing β-subset–quantile within array normalization values, 149 genes located in 286 loci were identified to have differential methylation in BPD lungs compared with preterm lungs with FDR of less than 0.05.
Integrating DNA methylation with gene expression data reveals 32 genes differentially methylated and expressed, with 23 genes having an inverse correlation
Gene expression for the 149 identified genes residing within 286 loci of abnormal methylation was assessed using a total of 432 probe sets; 32 of 149 differentially methylated genes (22%) were also significantly dysregulated by gene expression in BPD lung tissue, representing a significant fourfold enrichment (Fisher exact P < 0.001; Table 3; Figure 1). The magnitude of expression changes for these genes was modest (≤2-fold). Three genes were independently identified as differentially expressed by multiple probe sets: KIAA1217 (×3), EH-domain containing 4 (×2), and BAI1-associated protein 2 (×2).
Table 3.
Genes with Differential DNA Methylation as well as Differential Messenger RNA Expression in Bronchopulmonary Dysplasia Lung Compared with Preterm Lung, Arranged in Order of Decreasing Fold Change
| Gene Symbol | Probeset ID | Gene Title | Entrez Gene | Adjusted P Value | Fold Change in Gene Expression | Location | % Me Preterm | % Me Term | % Me BPD |
|---|---|---|---|---|---|---|---|---|---|
| GFI1 | 206589_at | Growth factor independent 1 | 2672 | 4.34E−02 | 1.87 | cg01766941; chr1: 92945907–92952609 | 0.57 | 0.69 | 0.69 |
| ANKRD55 | 220112_at | Ankyrin repeat domain 55 | 79722 | 4.90E−02 | 1.42 | cg22709202 | 0.58 | 0.62 | 0.66 |
| TP53RK | 225402_at | TP53 regulating kinase | 112858 | 3.63E−02 | 1.39 | cg20598650 | 0.24 | 0.38 | 0.37 |
| NRP2* | 223510_at | Neuropilin 2 | 8828 | 3.85E−02 | 1.32 | cg10307632 | 0.87 | 0.91 | 0.91 |
| NRP2* | 223510_at | Neuropilin 2 | 8828 | 3.85E−02 | 1.32 | cg19795793; chr2: 206549599–206551316 | 0.54 | 0.73 | 0.73 |
| CDH23 | 232845_at | Cadherin-like 23 | 64072 | 1.98E−02 | 1.30 | cg15077792 | 0.07 | 0.13 | 0.15 |
| ZNF438 | 1563333_at | Zinc finger protein 438 | 220929 | 3.49E−02 | 1.27 | cg01656216 | 0.71 | 0.57 | 0.58 |
| TRPS1* | 224218_s_at | Trichorhinophalangeal syndrome I | 7227 | 4.03E−02 | 1.21 | cg06368590; chr8: 116679698–116679936 | 0.06 | 0.15 | 0.16 |
| TRPS1* | 224218_s_at | Trichorhinophalangeal syndrome I | 7227 | 4.03E−02 | 1.21 | cg16821992; chr8: 116679698–116679936 | 0.12 | 0.27 | 0.27 |
| TRPS1* | 224218_s_at | Trichorhinophalangeal syndrome I | 7227 | 4.03E−02 | 1.21 | cg12396523; chr8: 116681380–116681623 | 0.07 | 0.13 | 0.11 |
| MYBPC1 | 214087_s_at | Myosin binding protein C, slow type | 4604 | 6.98E−03 | 1.14 | cg21726618; chr12:102036253-102036461 | 0.32 | 0.41 | 0.42 |
| PRDM16 | 237965_at | PR domain containing 16 | 63976 | 2.10E−02 | 1.11 | cg17033287; chr1: 3056541–3056772 | 0.22 | 0.33 | 0.33 |
| PHC1 | 225958_at | Polyhomeotic homolog 1 (Drosophila) | 1911 | 2.78E−02 | 0.91 | cg09912793; chr12: 9066946–9067480 | 0.16 | 0.25 | 0.23 |
| RYBP | 201845_s_at | RING1 and YY1 binding protein | 23429 | 2.56E−02 | 0.89 | cg16990174; chr3: 72495853–72496852 | 0.20 | 0.28 | 0.28 |
| BAIAP2† | 207832_at | BAI1-associated protein 2 | 10458 | 3.96E−02 | 0.87 | cg00026327; chr17: 79051892–79052534 | 0.72 | 0.58 | 0.59 |
| KIAA1217† | 1560115_a_at | KIAA1217 | 56243 | 2.47E−02 | 0.85 | cg26229043 | 0.12 | 0.21 | 0.20 |
| CASZ1 | 233863_at | Castor homolog 1, zinc finger (Drosophila) | 54897 | 3.57E−02 | 0.81 | cg10234511 | 0.40 | 0.56 | 0.61 |
| ACHE | 205377_s_at | Acetylcholinesterase (Yt blood group) | 43 | 2.72E−02 | 0.79 | cg10611760; chr7: 100492217–100494941 | 0.79 | 0.89 | 0.89 |
| BAIAP2† | 1556145_a_at | BAI1-associated protein 2 | 10458 | 2.53E−03 | 0.79 | cg00026327; chr17: 79051892–79052534 | 0.72 | 0.58 | 0.59 |
| KCNH2 | 205262_at | Potassium voltage-gated channel, subfamily H (eag-related), member 2 | 3757 | 3.50E−02 | 0.79 | cg24830730; chr7: 150655108–150655643 | 0.15 | 0.33 | 0.31 |
| STXBP6 | 230560_at | Syntaxin binding protein 6 (amisyn) | 29091 | 4.57E−02 | 0.77 | cg11775837; chr14:25518424–25519612 | 0.07 | 0.11 | 0.11 |
| BTC | 207326_at | Betacellulin | 685 | 1.68E−02 | 0.74 | cg09346617; chr4: 75719101–75719740 | 0.08 | 0.11 | 0.12 |
| FLJ22536 | 229280_s_at | Hypothetical locus LOC401237 | 401237 | 4.77E−02 | 0.72 | cg03854238 | 0.14 | 0.25 | 0.25 |
| EHD4† | 1556607_at | EH-domain containing 4 | 30844 | 4.23E−02 | 0.72 | cg21824733; chr15: 42192913–42193255 | 0.22 | 0.29 | 0.28 |
| EHD4† | 1556608_a_at | EH-domain containing 4 | 30844 | 3.63E−02 | 0.71 | cg21824733; chr15: 42192913–42193255 | 0.22 | 0.29 | 0.28 |
| MYOM2 | 205826_at | Myomesin (M-protein) 2, 165 kD/myomesin (M-protein) 2, 165kDa | 9172 | 2.36E−03 | 0.70 | cg05241134 | 0.18 | 0.28 | 0.25 |
| PLEKHB1* | 209504_s_at | Pleckstrin homology domain containing, family B (evectins) member 1 | 58473 | 4.32E−02 | 0.70 | cg26776957 | 0.21 | 0.36 | 0.35 |
| PLEKHB1* | 209504_s_at | Pleckstrin homology domain containing, family B (evectins) member 1 | 58473 | 4.32E−02 | 0.70 | cg25288155 | 0.27 | 0.36 | 0.36 |
| KCNC3 | 230531_at | Potassium voltage-gated channel, Shaw-related subfamily, member 3 | 3748 | 2.96E−02 | 0.70 | cg00740020; chr19: 50833813–50834128 | 0.10 | 0.16 | 0.13 |
| NRG2 | 208062_s_at | Neuregulin 2 | 9542 | 2.05E−02 | 0.70 | cg19583819; chr5: 139283350-139284282 | 0.27 | 0.41 | 0.41 |
| C13orf26* | 243884_at | Chromosome 13 open reading frame 26 | 122046 | 2.91E−02 | 0.70 | cg13614409 | 0.07 | 0.14 | 0.14 |
| C13orf26* | 243884_at | Chromosome 13 open reading frame 26 | 122046 | 2.91E−02 | 0.70 | cg00424169 | 0.22 | 0.36 | 0.36 |
| EYA4 | 207327_at | Eyes absent homolog 4 (Drosophila) | 2070 | 4.06E−02 | 0.69 | cg08917489; chr6: 133562086–133563586 | 0.21 | 0.27 | 0.27 |
| KIAA1217† | 244147_at | KIAA1217 | 56243 | 4.02E−02 | 0.69 | cg26229043 | 0.12 | 0.21 | 0.20 |
| NBEA | 226439_s_at | Neurobeachin | 26960 | 1.19E−02 | 0.68 | cg21392700; chr13: 36049570–36050159 | 0.24 | 0.32 | 0.31 |
| KIAA1217† | 232762_at | KIAA1217 | 56243 | 1.34E−02 | 0.67 | cg26229043 | 0.12 | 0.21 | 0.20 |
| NCALD | 1556952_at | Neurocalcin delta | 83988 | 5.39E−04 | 0.67 | cg00680551; chr8: 103135914–103136775 | 0.08 | 0.17 | 0.17 |
| GSTM3 | 235867_at | Glutathione S-transferase M3 (brain) | 2947 | 1.70E−02 | 0.66 | cg10807101; chr1: 110282351–110283306 | 0.22 | 0.36 | 0.40 |
| ADCY1 | 213245_at | Adenylate cyclase 1 (brain) | 107 | 4.40E−02 | 0.62 | cg08619378; chr7: 45613386–45615504 | 0.67 | 0.75 | 0.75 |
| DSCAML1* | 232059_at | Down syndrome cell adhesion molecule like 1 | 57453 | 4.87E−02 | 0.62 | cg05223210 | 0.50 | 0.62 | 0.62 |
| DSCAML1* | 232059_at | Down syndrome cell adhesion molecule like 1 | 57453 | 4.87E−02 | 0.62 | cg11776727 | 0.62 | 0.76 | 0.77 |
| DSCAML1* | 232059_at | Down syndrome cell adhesion molecule like 1 | 57453 | 4.87E−02 | 0.62 | cg07533617 | 0.84 | 0.92 | 0.93 |
| BMP7 | 209590_at | Bone morphogenetic protein 7 (osteogenic protein 1) | 655 | 3.48E−02 | 0.53 | cg11598935; chr20: 55839287–55839766 | 0.48 | 0.63 | 0.68 |
Definition of abbreviations: BAI1, brain-specific angiogenesis inhibitor 1; BPD, bronchopulmonary dysplasia; EH, Eps15 homology; Me, methylation; PR, positive regulatory; RING, Really Interesting New Gene; TP53, tumor protein 53; Yt, Cartwright; YY1, Yin yang 1.
% Me indicates methylation level of CpG locus.
Genes with methylation in more than one locus.
Genes with differential expression identified by multiple probe sets.
Of the 32 genes identified, 23 genes had opposite directions of methylation and expression changes. One gene, zinc finger protein 438 (ZNF438), had decreased methylation associated with increased expression in BPD (Table 4; Figure 1), and 22 genes had increased methylation corresponding with decreased expression in BPD (Table 5; Figure 1).
Table 4.
Genes with Decreased DNA Methylation and Increased Messenger RNA Expression in Bronchopulmonary Dysplasia Lung Compared with Preterm Lung
| Gene Symbol | Probe Name | Gene Title | Entrez Gene | Adjusted P Value | Fold Change | Location | % Me Preterm | % Me Term | % Me BPD | Location of CpG Site |
|---|---|---|---|---|---|---|---|---|---|---|
| ZNF438 | 1563333_at | Zinc finger protein 438 | 220929 | 3.49E−02 | 1.27 | cg01656216 | 0.71 | 0.57 | 0.58 | Enhancer |
Definition of abbreviations: BPD, bronchopulmonary dysplasia; CpG, cytosine-phosphateguanine dinucleotide; Me, methylation.
% Me indicates methylation level of CpG locus.
Table 5.
Genes with Increased DNA Methylation and Decreased Messenger RNA Expression in Bronchopulmonary Dysplasia Lung Compared with Preterm Lung, Arranged in Order of Decreasing Fold Change
| Gene Symbol | Probe Name | Gene Title | Entrez Gene | Adjusted P value | Fold change | Location | % Me Preterm | % Me Term | % Me BPD | Location of CpG Site |
|---|---|---|---|---|---|---|---|---|---|---|
| PHC1 | 225958_at | Polyhomeotic homolog 1 (Drosophila) | 1911 | 2.78E−02 | 0.91 | cg09912793; chr12: 9066946–9067480 | 0.16 | 0.25 | 0.23 | N_Shore |
| RYBP | 201845_s_at | RING1 and YY1 binding protein | 23429 | 2.56E−02 | 0.89 | cg16990174; chr3: 72495853–72496852 | 0.20 | 0.28 | 0.28 | S_Shore |
| KIAA1217* | 1560115_a_at | KIAA1217 | 56243 | 2.47E−02 | 0.85 | cg26229043 | 0.12 | 0.21 | 0.20 | Enhancer |
| CASZ1 | 233863_at | Castor homolog 1, zinc finger (Drosophila) | 54897 | 3.57E−02 | 0.81 | cg10234511 | 0.40 | 0.56 | 0.61 | Enhancer |
| ACHE | 205377_s_at | Acetylcholinesterase (Yt blood group) | 43 | 2.72E−02 | 0.79 | cg10611760; chr7: 100492217–100494941 | 0.79 | 0.89 | 0.89 | N_Shore |
| KCNH2 | 205262_at | Potassium voltage-gated channel, subfamily H (eag-related), member 2 | 3757 | 3.50E−02 | 0.79 | cg24830730; chr7: 150655108–150655643 | 0.15 | 0.33 | 0.31 | N_Shore |
| STXBP6 | 230560_at | Syntaxin binding protein 6 (amisyn) | 29091 | 4.57E−02 | 0.77 | cg11775837; chr14: 25518424–25519612 | 0.07 | 0.11 | 0.11 | Island |
| BTC | 207326_at | Betacellulin | 685 | 1.68E−02 | 0.74 | cg09346617; chr4: 75719101–75719740 | 0.08 | 0.11 | 0.12 | Island |
| FLJ22536 | 229280_s_at | Hypothetical locus LOC401237 | 401237 | 4.77E−02 | 0.72 | cg03854238 | 0.14 | 0.25 | 0.25 | Enhancer |
| EHD4* | 1556607_at | EH-domain containing 4 | 30844 | 4.23E−02 | 0.72 | cg21824733; chr15: 42192913–42193255 | 0.22 | 0.29 | 0.28 | S_Shelf |
| EHD4* | 1556608_a_at | EH-domain containing 4 | 30844 | 3.63E−02 | 0.71 | cg21824733; chr15: 42192913–42193255 | 0.22 | 0.29 | 0.28 | S_Shelf |
| MYOM2 | 205826_at | Myomesin (M-protein) 2, 165 kD/myomesin (M-protein) 2, 165 kD | 9172 | 2.36E−03 | 0.70 | cg05241134 | 0.18 | 0.28 | 0.25 | Enhancer |
| PLEKHB1 | 209504_s_at | Pleckstrin homology domain containing, family B (evectins) member 1 | 58473 | 4.32E−02 | 0.70 | cg25288155 | 0.27 | 0.36 | 0.36 | Body |
| KCNC3 | 230531_at | Potassium voltage-gated channel, Shaw-related subfamily, member 3 | 3748 | 2.96E−02 | 0.70 | cg00740020; chr19: 50833813–50834128 | 0.10 | 0.16 | 0.13 | N_Shore |
| NRG2 | 208062_s_at | Neuregulin 2 | 9542 | 2.05E−02 | 0.70 | cg19583819; chr5: 139283350–139284282 | 0.27 | 0.41 | 0.41 | N_Shore |
| C13orf26 | 243884_at | Chromosome 13 open reading frame 26 | 122046 | 2.91E−02 | 0.70 | cg00424169 | 0.22 | 0.36 | 0.36 | Body |
| EYA4 | 207327_at | Eyes absent homolog 4 (Drosophila) | 2070 | 4.06E−02 | 0.69 | cg08917489; chr6: 133562086–133563586 | 0.21 | 0.27 | 0.27 | S_Shore |
| KIAA1217* | 244147_at | KIAA1217 | 56243 | 4.02E−02 | 0.69 | cg26229043 | 0.12 | 0.21 | 0.20 | Enhancer |
| NBEA | 226439_s_at | Neurobeachin | 26960 | 1.19E−02 | 0.68 | cg21392700; chr13: 36049570–36050159 | 0.24 | 0.32 | 0.31 | N_Shore |
| KIAA1217* | 232762_at | KIAA1217 | 56243 | 1.34E−02 | 0.67 | cg26229043 | 0.12 | 0.21 | 0.20 | Enhancer |
| NCALD | 1556952_at | Neurocalcin delta | 83988 | 5.39E−04 | 0.67 | cg00680551; chr8: 103135914–103136775 | 0.08 | 0.17 | 0.17 | N_Shore |
| GSTM3 | 235867_at | Glutathione S-transferase M3 (brain) | 2947 | 1.70E−02 | 0.66 | cg10807101; chr1: 110282351–110283306 | 0.22 | 0.36 | 0.40 | N_Shore |
| ADCY1 | 213245_at | Adenylate cyclase 1 (brain) | 107 | 4.40E−02 | 0.62 | cg08619378; chr7: 45613386–45615504 | 0.67 | 0.75 | 0.75 | S_Shore |
| DSCAML1† | 232059_at | Down syndrome cell adhesion molecule like 1 | 57453 | 4.87E−02 | 0.62 | cg05223210 | 0.50 | 0.62 | 0.62 | Enchancer |
| DSCAML1† | 232059_at | Down syndrome cell adhesion molecule like 1 | 57453 | 4.87E−02 | 0.62 | cg11776727 | 0.62 | 0.76 | 0.77 | Enchancer |
| DSCAML1† | 232059_at | Down syndrome cell adhesion molecule like 1 | 57453 | 4.87E−02 | 0.62 | cg07533617 | 0.84 | 0.92 | 0.93 | Enchancer |
| BMP7 | 209590_at | Bone morphogenetic protein 7 (osteogenic protein 1) | 655 | 3.48E−02 | 0.53 | cg11598935; chr20: 55839287–55839766 | 0.48 | 0.63 | 0.68 | N_Shore |
Definition of abbreviations: BPD, bronchopulmonary dysplasia; CpG, cytosine-guanine dinucleotide; Eag, ether-a-go-go; EH, Eps15 homology; Me, methylation; RING, Really Interesting New Gene; Yt, Cartwright; YY1, Yin yang 1.
% Me indicates methylation level of CpG locus.
Genes with differential expression identified by multiple probe sets.
Genes with methylation in more than one locus.
The remaining nine genes had the same direction of methylation and expression. One gene, BAI1-associated protein 2, had decreased methylation associated with decreased expression in BPD (Table E5), and eight genes had increased methylation and increased expression in BPD (Table E6). The location of methylated CpG sites in relation to the gene was summarized and showed that, in general, methylation of CpG sites located in enhancers and north shores tend to have an opposite direction of gene expression (Table E7).
Validation of gene expression of KIAA1217 and ZNF438 by real-time RT-PCR confirmed the direction of gene expression seen on microarray (preterm versus BPD messenger RNA/glyceraldehyde 3-phosphate dehydrogenase—KIAA1217: 3.00 ± 1.16 versus 0.67 ± 0.33 [4-fold decrease in BPD]; ZNF438: 0.00056 + 0.0003 versus 0.0058 + 0.007 [10-fold increase in BPD]; all P < 0.05).
Pathway analysis
Pathway analysis of gene expression of all 149 genes (represented by 432 probe sets) affected by methylation identified nine canonical pathways (Table E8). These pathways, some of which deal with progenitor cell differentiation, were not previously understood to be important in lung development and/or BPD. One pathway of potential significant interest was Wnt/β-catenin signaling. When the list was limited to the 32 significantly dysregulated genes (represented by 36 probe sets), seven pathways were identified (Table 6; Figure 1). A single pathway (cardiomyocyte differentiation) was common to both analyses. The remaining six pathways included ErbB receptor tyrosine kinase/neuregulin signaling (18), as well as a nitric oxide–related pathway, both of which have been previously implicated in lung development and BPD pathogenesis.
Table 6.
Pathways Associated with Genes Both Differentially Methylated and Expressed in Bronchopulmonary Dysplasia Lung Compared with Preterm Lung
| Canonical Pathways | −log(P Value) | Ratio | Molecules |
|---|---|---|---|
| ErbB signaling | 2.05E−00 | 2.22E−02 | NRG2, BTC |
| Neuregulin signaling | 2.03E−00 | 1.92E−02 | NRG2, BTC |
| RhoA signaling | 1.79E−00 | 1.64E−02 | NRP2, BAIAP2 |
| Cellular effects of sildenafil (Viagra) | 1.71E−00 | 1.29E−02 | ADCY1, KCNH2 |
| Cardiomyocyte differentiation via BMP receptors | 1.5E−00 | 4.55E−02 | BMP7 |
| Axonal guidance signaling | 1.47E−00 | 6.21E−03 | NRP2, BAIAP2, BMP7 |
| Glutathione-mediated detoxification | 1.34E−00 | 2.27E−02 | GSTM3 |
Definition of abbreviations: ADCY1, adenylate cyclase 1; BAIAP2, brain-specific angiogenesis inhibitor 1-associated protein 2; BMP, bone morphogenetic protein; BTC, betacellulin; ErbB, erbB receptor tyrosine kinase; GSTM3, glutathione S-transferase μ 3; KCNH2, potassium voltage-gated channel, subfamily H, member 2; NRG2, neuregulin 2; NRP2, neuropilin 2; RhoA, ras homolog gene family, member A.
GSEA
GSEA comparing gene expression of the 149 genes with altered methylation status from lung tissues from BPD (n = 11) to controls (n = 9) identified 74 gene sets as up-regulated in BPD out of 190 gene sets that passed the size threshold (minimum = 5; maximum = 500), five of which were significant (P < 0.05; Table E9). GSEA also identified 116 gene sets as up-regulated in controls compared with BPD out of 190 gene sets that passed the size threshold (minimum = 5; maximum = 500), seven of which were significant (P < 0.05; Table E10).
Comparing DNA methylation levels between preterm and term lung identified 437 genes with differential methylation potentially involved in normal alveolar septation
DNA methylation data obtained from preterm lung were compared with those from term lung. A total of 437 genes located in 483 loci were identified that were differentially methylated between preterm and term lung. Of these, 33 genes had increased methylation in preterm compared with term lung (Table E11), and 402 genes had increased methylation in term compared with preterm lung (Table E12 showing top 50 out of 402 genes). Two genes, myomesin 2 and copine family member IX, were methylated in two loci, with one locus having increased methylation in preterm lung and the other locus having increased methylation in term lung. A total of 34 other genes had methylation in more than one locus, all of which had increased methylation in term lung (Table E13).
Overlap of Mouse and Human Methylation Data
Methylation data obtained from murine lung tissue (adult versus newborn) were overlapped with methylation data obtained from control human lung (term versus preterm) to identify genes with conserved methylation across the two species during alveolar septation. Twenty genes were identified that were differentially methylated in adult versus newborn mouse lung and also differentially methylated in term versus preterm human lung (Table 7; Figure 1; P < 0.001). Evaluation of overlap at the pathway level using IPA identified (at P < 0.05) 16 pathways differentially methylated in mouse lung, and seven differentially methylated in human lung (Table E14). Three pathways were common to both mouse and human lung DNA methylation analyses (basal cell carcinoma signaling, axonal guidance signaling, sonic hedgehog signaling; Table E14).
Table 7.
Common Genes Differentially Methylated in Mouse Newborn versus Adult Lung and Human Preterm versus Term Lung
| Gene Symbol | Gene Title | % Methylation Preterm | % Methylation Term | P Value | Methylation in Mouse Newborn versus Adult Lung | P Value |
|---|---|---|---|---|---|---|
| DNMT3A | DNA methyltransferase 3A | 0.16 | 0.27 | 1.99E−05 | Decreased | 2.51E−03 |
| SHISA3 | Shisa family member 3 | 0.08 | 0.14 | 2.20E−05 | Decreased | 1.44E−03 |
| KCNMA1 | Potassium large conductance calcium-activated channel, subfamily M, α member 1 | 0.32 | 0.43 | 4.36E−05 | Decreased | 3.67E−04 |
| NRN1 | Neuritin 1 | 0.10 | 0.13 | 6.18E−05 | Decreased | 1.69E−07 |
| WDFY4 | WDFY family member 4 | 0.16 | 0.28 | 6.71E−05 | Decreased | 2.43E−03 |
| CASZ1 | Castor zinc finger 1 | 0.40 | 0.56 | 1.41E−04 | Decreased | 4.32E−03 |
| PITPNC1 | Phosphatidylinositol transfer protein, cytoplasmic 1 | 0.14 | 0.21 | 1.42E−04 | Decreased | 7.76E−03 |
| EXT1 | Exostosinglycosyltransferase 1 | 0.08 | 0.22 | 1.57E−04 | Decreased | 5.83E−04 |
| UBE2E2 | Ubiquitin-conjugating enzyme E2E 2 | 0.27 | 0.37 | 2.85E−04 | Decreased | 8.76E−04 |
| GRIA1 | Glutamate receptor, ionotropic, AMPA 1 | 0.10 | 0.17 | 2.93E−04 | Decreased | 7.84E−04 |
| TBX4 | T-box 4 | 0.16 | 0.21 | 3.42E−04 | Decreased | 2.65E−03 |
| ADAMTSL2 | ADAMTS-like 2 | 0.09 | 0.22 | 3.53E−04 | Decreased | 1.65E−03 |
| E2F8 | E2F transcription factor 8 | 0.11 | 0.15 | 3.63E−04 | Decreased | 3.30E−05 |
| PRDM8 | PR domain containing 8 | 0.23 | 0.35 | 4.21E−04 | Decreased | 1.23E−03 |
| ROBO1 | Roundabout homolog 1 | 0.08 | 0.10 | 4.35E−04 | Decreased | 3.55E−03 |
| IFNAR2 | interferon (α, β and ω) receptor 2 | 0.07 | 0.11 | 4.89E−04 | Decreased | 2.88E−04 |
| PRICKLE1 | Prickle homolog 1 | 0.11 | 0.18 | 6.37E−04 | Decreased | 1.23E−05 |
| BMPER | BMP-binding endothelial regulator | 0.07 | 0.11 | 8.51E−04 | Decreased | 5.96E−04 |
| CTNNA2 | Catenin (cadherin associated protein) | 0.18 | 0.30 | 9.08E−04 | Decreased | 2.61E−03 |
| AUTS2 | Autism susceptibility candidate 2 | 0.23 | 0.36 | 9.38E−04 | Decreased | 5.66E−05 |
Definition of abbreviations: ADAMTS, a disintegrin-like and metallopeptidase with thrombospondin; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; BMP, bone morphogenetic protein; PR, positive regulatory.
All genes had reduced methylation in human preterm lung and mouse newborn lung as compared with human term or mouse adult lung, respectively.
Discussion
In this study we integrated genome-wide DNA methylation and gene expression data from murine and human lung tissue samples to identify genes that are possibly regulated by DNA methylation during normal and abnormal alveolar septation. The concept that DNA methylation plays an important role in development is not new. Initial studies done in mice have demonstrated elegantly that, when genes encoding for DNA methyltransferases were silenced or overexpressed, affected mice developed abnormally and died in utero (19, 20). Other studies have shown that development follows a specific pattern of demethylation and remethylation (21), and that deviation from this normal methylation pattern results in abnormal development and increased fetal loss (22). Another study looked at differences in tissue-specific DNA methylation regions between fetal and adult lung tissues, and demonstrated that only 17% of tissue-specific DNA methylation regions present in the fetus were actually conserved into adulthood (23). These and numerous other studies provide evidence that extensive reprogramming of the epigenome by DNA methylation is important for normal development to occur. Our study extends these previous studies by demonstrating lung-specific alterations in DNA methylation in both murine and human samples, and identifying downstream genes that are potentially involved in lung development.
By comparing methylation and expression data from newborn and adult mouse lung tissue samples, we were able to identify 95 genes involved in normal alveolar septation that were also likely regulated by DNA methylation. These included genes involved with the immune system, antioxidant defense, extracellular matrix (ECM) formation, and lung cancer. Genes previously reported in other studies to be important in lung development (WNT1 inducible signaling pathway protein 2 [24],WNT inhibitory factor 1 [25], angiopoietin 2 [26], and sex-determining region Y-box 9 [27]) were also observed in our study, further validating their importance, as well as offering insight into the role DNA methylation may have in regulating their expression. A number of genes with less obvious significance to alveolar septation were also noted, and further evaluation of these targets is required to determine their role in septation.
We identified 13 genes known to function in inflammation and immune system response (chitinase-like 3 [Chi3l3], B-cell CLL/lymphoma 6 [Bcl6], chemokine [C-C motif] receptor 6 [Ccr6], signal transducer and activator of transcription 4 [Stat4], Ccl5, CD209a antigen [Cd209a], histocompatibility 2, O region β locus [H2-Ob], CD19 antigen [Cd19], SH2 domain containing 1A [Sh2 d1a], killer cell lectin-like receptor subfamily B member 1B [Klrb1b], B cell linker [Blnk], β-2 microglobulin [B2 m], and protein kinase C, θ [Prkcq]), the expression of which were decreased in newborn and increased in adult lung. Similar changes in inflammatory/immune system genes were also observed by Cortese and colleagues (28) in lung development and cancer. A similar pattern of regulation was also seen in superoxide dismutase 3, extracellular (Sod3) and peroxiredoxin 6 (Prdx6) expression, genes known to have important antioxidant activity in the lungs (29). Increased expression of genes involved in immune and antioxidant defense correlates with the expected exposure of the newborn lungs to numerous environmental stressors soon after birth. Possible regulation of these genes by methylation offers an interesting target for augmenting immune and antioxidant defense in the lungs, especially for premature infants who are at higher risk for developing oxidant-induced lung injury.
Genes involved in ECM formation and organization, such as elastin (Eln), tenascin C (Tnc), collagen (Col), type VI, α 3 (Col6a3), and Col, type XXVII, α 1 (Col27a1), were observed to have increased expression and decreased methylation in the newborn, with subsequent decreased expression and increased methylation in the adult. Such an expression profile is consistent with the rapid and marked increase in ECM seen during alveolar septation (13, 28); similar increases in ECM-related genes were also observed by Cortese and colleagues (28). Three genes (S-phase kinase–associated protein 2 [Skp2], ribonucleotide reductase M [Rrm] 1, and Rrm2) were identified, the increased expression of which is typically associated with various forms of lung cancer in adults (30–32). In our study, these genes were shown to have increased expression only in newborns, with subsequent decreased expression in adults. It has been hypothesized that dysregulation of genes involved in normal development may play an important role in carcinogenesis (33). Our study suggests that Skp2, Rrm1, and Rrm2 are normally expressed during alveolar septation in the newborn, but repressed upon alveolar maturation in the adult. It is speculated that abnormal re-expression of these genes in adult lung (a recapitulation of development) may contribute to the pathogenesis of lung cancer.
We compared methylation and expression data from human lung tissue samples to identify genes involved in abnormal alveolar septation seen in BPD. A total of 23 genes were identified that exhibited inverse correlation between methylation and expression, suggesting regulation by DNA methylation. A select number of these 23 genes are of particular interest. Glutathione S-transferase M3 (Gtsm3) is a detoxifying enzyme that metabolizes oxidative compounds from exposure to environmental toxins. Studies have shown that genetic variations of this gene are associated with poor lung growth and function in relation to environmental toxin exposure (34), and correlated with severity of lung diseases, such as cystic fibrosis (35). Betacellulin is an epidermal growth factor, the overexpression of which in mice resulted in severe pulmonary pathology, including thickening of alveolar septa, intra-alveolar accumulation of hemosiderin-containing macrophages, and nodular pulmonary remodeling—features commonly seen in BPD (36). Finally, bone morphogenetic protein (Bmp) 7 is a member of the transforming growth factor (TGF)-β superfamily that has been reported to oppose the activity of TGF-β in lungs (37). As increased TGF-β signaling in the lung results in pulmonary fibrosis and arrested lung development (38), Bmp7, as an endogenous antagonist for TGF-β, may be an interesting target for therapy in BPD.
We also compared methylation patterns of preterm versus term human lung with newborn versus adult mouse lung, and identified 20 genes with shared differential methylation in both human and mouse lung. Of these, six genes (Shisa3, Nrn1, Ext1, Gria1, E2f8, and Robo1) were identified in our study as potentially regulating gene expression. The relevance of the remaining 14 genes with shared methylation patterns between mouse and human lungs is unknown at present, and needs further study. It is possible that these genes with methylation differences during development, but without corresponding changes in gene expression, have other regulators of gene transcription (e.g., microRNA, transcription factors) that overrule or modify the effects of changes in DNA methylation. We also identified three pathways (basal cell carcinoma signaling, axonal guidance signaling, and sonic hedgehog signaling) that were common to both mouse and human lung DNA methylation analyses. Of these pathways, basal cell carcinoma signaling pathway represents constitutive activation of sonic hedgehog signaling. Molecules involved in sonic hedgehog signaling (e.g., sonic hedgehog homolog, patched, gliotactin) (39, 40) and axonal guidance (e.g., Netrins, Slits, and Robo receptors, Semaphorins, Ephrins) (41–45) are well known to be involved in lung development, and our results suggest that DNA methylation may help regulate them.
Our study focused on genes that showed an inverse relationship between methylation and expression, as this reflects the current mainstream understanding that DNA methylation of CpG sites located near promoter regions is associated with inhibition of gene expression. There is, however, increasing evidence that the function of DNA methylation in regulating gene expression may vary, depending upon the precise location and context of the CpG sites relative to the gene in question (i.e., gene body versus the shores, shelves, and enhancers surrounding the promoter CpG islands) (46), and that methylation and expression changes in the same direction may also be important. In light of this, we have also characterized and reported on genes with a direct correlation between methylation and expression, and have specified the location of the differentially methylated CpG sites in relation to the gene. These data may be helpful as we gain a better understanding of how methylation of specific CpG sites regulates gene expression.
Our study provides valuable insights into many aspects of normal and damaged lung development, but several inherent limiting factors warrant discussion. Although we have shown an inverse correlation between DNA methylation and gene expression for a number of genes involved in normal and abnormal alveolar septation, we cannot conclude that the changes in expression of these genes occurred solely because of changes in DNA methylation. To prove that the DNA methylation changes we identified drive the changes in gene expression, mechanistic studies that target these genes and their regulation by DNA methylation using in vivo models would be required. Our study was also conducted on whole lung tissue, containing DNA and RNA from multiple cell types and compartments. This could have resulted in contamination of results due to inclusion of cells that do not directly participate in alveolar septation. The magnitude of observed changes in DNA methylation may be lower due to the mixture of cells in the whole lung, as actual changes may be cell-type specific. Further studies, possibly using laser capture microdissection or cell sorting to isolate developing secondary septa, are needed for validation. Such studies would avoid contamination by gene expression and DNA methylation signals from other regions of the lung. Our study also did not investigate other known epigenetic regulators of gene expression, such as histone acetylation and microRNA. An increasing number of studies have shown that, in addition to DNA methylation, alterations in histone acetylation (47) and microRNA (17) are also important in lung development and disease. The present study had a limited number of human samples, which limits the power of the study. In addition, a limitation is that human gene expression was from a different set of samples from that used for assessment of methylation, although our validation studies show similar patterns of gene expression of selected genes in both samples.
Nevertheless, our study has several important strengths. The genome-wide approach used to identify candidate genes, as compared with the traditional single-pathway or gene approach, allowed unbiased validation of genes previously identified from other studies as important, and enabled identification of novel candidate genes for future research. This integration of gene expression data with DNA methylation data provides additional insight into epigenetic mechanisms regulating observed changes in gene expression. Our evaluation of genes with an inverse relationship between methylation and expression highlights key genes, the expression of which is likely regulated by DNA methylation; this process potentially could enable future studies that manipulate methylation to influence gene expression. The use of human BPD and preterm and term control lung tissue samples allowed the development of a unique and valuable data set not possible from murine or other animal models of BPD.
Further studies are required to validate candidate genes using mechanistic in vitro and subsequent in vivo animal studies. Overall, understanding how DNA methylation regulates gene expression during normal alveolar formation may not only yield new insights into normal lung development, but may also provide insights into how dysregulation of DNA methylation contributes to lung disorders.
Footnotes
This work was supported by National Institutes of Health grants R01 HL092906 (N.A.), U01 HL122626 (N.A. and N.K.), and R01 HL095397 (N.K.).
Author Contributions: A.C. and N.A.—conceptualized and designed the study, drafted the initial manuscript, revised the manuscript, and approved the final manuscript as submitted; B.H., D.K.C., X.C., K.P., S.B., A.A., and T.J.M.—assisted with the study design, acquisition of data, performed statistical analysis of the data, revised the manuscript, and approved the final manuscript as submitted; N.K., O.F.-P., and D.K.—assisted with the study design, critically reviewed the manuscript, and approved the final manuscript as submitted.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2014-0160OC on November 11, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coalson JJ. Pathology of bronchopulmonary dysplasia. Semin Perinatol. 2006;30:179–184. doi: 10.1053/j.semperi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Hagood JS, Ambalavanan N. Systems biology of lung development and regeneration: current knowledge and recommendations for future research. Wiley Interdiscip Rev Syst Biol Med. 2013;5:125–133. doi: 10.1002/wsbm.1205. [DOI] [PubMed] [Google Scholar]
- 4.Mariani TJ, Reed JJ, Shapiro SD. Expression profiling of the developing mouse lung: insights into the establishment of the extracellular matrix. Am J Respir Cell Mol Biol. 2002;26:541–548. doi: 10.1165/ajrcmb.26.5.2001-00080c. [DOI] [PubMed] [Google Scholar]
- 5.Kho AT, Bhattacharya S, Tantisira KG, Carey VJ, Gaedigk R, Leeder JS, Kohane IS, Weiss ST, Mariani TJ. Transcriptomic analysis of human lung development. Am J Respir Crit Care Med. 2010;181:54–63. doi: 10.1164/rccm.200907-1063OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharya S, Go D, Krenitsky DL, Huyck HL, Solleti SK, Lunger VA, Metlay L, Srisuma S, Wert SE, Mariani TJ, et al. Genome-wide transcriptional profiling reveals connective tissue mast cell accumulation in bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2012;186:349–358. doi: 10.1164/rccm.201203-0406OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 8.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 9.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delgado-Calle J, Sañudo C, Fernández AF, García-Renedo R, Fraga MF, Riancho JA. Role of DNA methylation in the regulation of the RANKL-OPG system in human bone. Epigenetics. 2012;7:83–91. doi: 10.4161/epi.7.1.18753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keil KP, Altmann HM, Mehta V, Abler LL, Elton EA, Vezina CM. Catalog of mRNA expression patterns for DNA methylating and demethylating genes in developing mouse lower urinary tract. Gene Expr Patterns. 2013;13:413–424. doi: 10.1016/j.gep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bröske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 13.Burri PH. Structural aspects of postnatal lung development—alveolar formation and growth. Biol Neonate. 2006;89:313–322. doi: 10.1159/000092868. [DOI] [PubMed] [Google Scholar]
- 14.Mohn F, Weber M, Schübeler D, Roloff TC. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 15.Maksimovic J, Gordon L, Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz MH, Pandit KV, Lino Cardenas CL, Ambalavanan N, Kaminski N, Bar-Joseph Z. Reconstructing dynamic microRNA-regulated interaction networks. Proc Natl Acad Sci USA. 2013;110:15686–15691. doi: 10.1073/pnas.1303236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purevdorj E, Zscheppang K, Hoymann HG, Braun A, von Mayersbach D, Brinkhaus MJ, Schmiedl A, Dammann CE. ErbB4 deletion leads to changes in lung function and structure similar to bronchopulmonary dysplasia. Am J Physiol Lung Cell Mol Physiol. 2008;294:L516–L522. doi: 10.1152/ajplung.00423.2007. [DOI] [PubMed] [Google Scholar]
- 19.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 20.Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 22.Kang YK, Park JS, Koo DB, Choi YH, Kim SU, Lee KK, Han YM. Limited demethylation leaves mosaic-type methylation states in cloned bovine pre-implantation embryos. EMBO J. 2002;21:1092–1100. doi: 10.1093/emboj/21.5.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuen RK, Neumann SM, Fok AK, Peñaherrera MS, McFadden DE, Robinson WP, Kobor MS. Extensive epigenetic reprogramming in human somatic tissues between fetus and adult. Epigenetics Chromatin. 2011;4:7. doi: 10.1186/1756-8935-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pongracz JE, Stockley RA. Wnt signalling in lung development and diseases. Respir Res. 2006;7:15. doi: 10.1186/1465-9921-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felcht M, Luck R, Schering A, Seidel P, Srivastava K, Hu J, Bartol A, Kienast Y, Vettel C, Loos EK, et al. Angiopoietin-2 differentially regulates angiogenesis through TIE2 and integrin signaling. J Clin Invest. 2012;122:1991–2005. doi: 10.1172/JCI58832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turcatel G, Rubin N, Menke DB, Martin G, Shi W, Warburton D. Lung mesenchymal expression of Sox9 plays a critical role in tracheal development. BMC Biol. 2013;11:117. doi: 10.1186/1741-7007-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cortese R, Hartmann O, Berlin K, Eckhardt F. Correlative gene expression and DNA methylation profiling in lung development nominate new biomarkers in lung cancer. Int J Biochem Cell Biol. 2008;40:1494–1508. doi: 10.1016/j.biocel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 30.Hung WC, Tseng WL, Shiea J, Chang HC. Skp2 overexpression increases the expression of MMP-2 and MMP-9 and invasion of lung cancer cells. Cancer Lett. 2010;288:156–161. doi: 10.1016/j.canlet.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 31.Pitterle DM, Kim YC, Jolicoeur EM, Cao Y, O’Briant KC, Bepler G. Lung cancer and the human gene for ribonucleotide reductase subunit M1 (RRM1) Mamm Genome. 1999;10:916–922. doi: 10.1007/s003359901114. [DOI] [PubMed] [Google Scholar]
- 32.Rahman MA, Amin AR, Wang D, Koenig L, Nannapaneni S, Chen Z, Wang Z, Sica G, Deng X, Chen ZG, et al. RRM2 regulates Bcl-2 in head and neck and lung cancers: a potential target for cancer therapy. Clin Cancer Res. 2013;19:3416–3428. doi: 10.1158/1078-0432.CCR-13-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonner AE, Lemon WJ, Devereux TR, Lubet RA, You M. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene. 2004;23:1166–1176. doi: 10.1038/sj.onc.1207234. [DOI] [PubMed] [Google Scholar]
- 34.Breton CV, Vora H, Salam MT, Islam T, Wenten M, Gauderman WJ, Van den Berg D, Berhane K, Peters JM, Gilliland FD. Variation in the GST mu locus and tobacco smoke exposure as determinants of childhood lung function. Am J Respir Crit Care Med. 2009;179:601–607. doi: 10.1164/rccm.200809-1384OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flamant C, Henrion-Caude A, Boëlle PY, Brémont F, Brouard J, Delaisi B, Duhamel JF, Marguet C, Roussey M, Miesch MC, et al. Glutathione-S-transferase M1, M3, P1 and T1 polymorphisms and severity of lung disease in children with cystic fibrosis. Pharmacogenetics. 2004;14:295–301. doi: 10.1097/00008571-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 36.Schneider MR, Dahlhoff M, Herbach N, Renner-Mueller I, Dalke C, Puk O, Graw J, Wanke R, Wolf E. Betacellulin overexpression in transgenic mice causes disproportionate growth, pulmonary hemorrhage syndrome, and complex eye pathology. Endocrinology. 2005;146:5237–5246. doi: 10.1210/en.2005-0418. [DOI] [PubMed] [Google Scholar]
- 37.Izumi N, Mizuguchi S, Inagaki Y, Saika S, Kawada N, Nakajima Y, Inoue K, Suehiro S, Friedman SL, Ikeda K. BMP-7 opposes TGF-beta1–mediated collagen induction in mouse pulmonary myofibroblasts through Id2. Am J Physiol Lung Cell Mol Physiol. 2006;290:L120–L126. doi: 10.1152/ajplung.00171.2005. [DOI] [PubMed] [Google Scholar]
- 38.Vicencio AG, Lee CG, Cho SJ, Eickelberg O, Chuu Y, Haddad GG, Elias JA. Conditional overexpression of bioactive transforming growth factor-β1 in neonatal mouse lung: a new model for bronchopulmonary dysplasia? Am J Respir Cell Mol Biol. 2004;31:650–656. doi: 10.1165/rcmb.2004-0092OC. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Kugler MC, Loomis CA, Samdani R, Zhao Z, Chen GJ, Brandt JP, Brownell I, Joyner AL, Rom WN, et al. Hedgehog signaling in neonatal and adult lung. Am J Respir Cell Mol Biol. 2013;48:703–710. doi: 10.1165/rcmb.2012-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, Wang H, Teng H, Shi J, Zhang Y. Expression of SHH signaling pathway components in the developing human lung. Histochem Cell Biol. 2010;134:327–335. doi: 10.1007/s00418-010-0738-2. [DOI] [PubMed] [Google Scholar]
- 41.Dalvin S, Anselmo MA, Prodhan P, Komatsuzaki K, Schnitzer JJ, Kinane TB. Expression of Netrin-1 and its two receptors DCC and UNC5H2 in the developing mouse lung. Gene Expr Patterns. 2003;3:279–283. doi: 10.1016/s1567-133x(03)00047-4. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Stein E, Oliver T, Li Y, Brunken WJ, Koch M, Tessier-Lavigne M, Hogan BL. Novel role for netrins in regulating epithelial behavior during lung branching morphogenesis. Curr Biol. 2004;14:897–905. doi: 10.1016/j.cub.2004.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greenberg JM, Thompson FY, Brooks SK, Shannon JM, Akeson AL. Slit and robo expression in the developing mouse lung. Dev Dyn. 2004;230:350–360. doi: 10.1002/dvdy.20045. [DOI] [PubMed] [Google Scholar]
- 44.Vadivel A, Alphonse RS, Collins JJ, van Haaften T, O’Reilly M, Eaton F, Thébaud B. The axonal guidance cue semaphorin 3C contributes to alveolar growth and repair. PLoS One. 2013;8:e67225. doi: 10.1371/journal.pone.0067225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vadivel A, van Haaften T, Alphonse RS, Rey-Parra GJ, Ionescu L, Haromy A, Eaton F, Michelakis E, Thébaud B. Critical role of the axonal guidance cue EphrinB2 in lung growth, angiogenesis, and repair. Am J Respir Crit Care Med. 2012;185:564–574. doi: 10.1164/rccm.201103-0545OC. [DOI] [PubMed] [Google Scholar]
- 46.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 47.Nagarajan P, Ge Z, Sirbu B, Doughty C, Agudelo Garcia PA, Schlederer M, Annunziato AT, Cortez D, Kenner L, Parthun MR. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013;9:e1003518. doi: 10.1371/journal.pgen.1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]