Abstract
The classical view of neuronal protein synthesis is that proteins are made in the cell body and then transported to their functional sites in the dendrites and the dendritic spines. Indirect evidence, however, suggests that protein synthesis can directly occur in the distal dendrites, far from the cell body. We are developing protocols for dual labeling of RNA and proteins using 15N-uridine and 18O- or 13C-leucine pulse chase in cultured neurons to identify and localize both protein synthesis and fate of newly synthesized proteins. Pilot experiments show discrete localization of both RNA and newly synthesized proteins in dendrites, close to dendritic spines. We have for the first time directly imaged and measured the production of proteins at the subcellular level in the neuronal dendrites, close to the functional sites, the dendritic spines. This will open a powerful way to study neural growth and synapse plasticity in health and disease.
Keywords: MIMS, Multi-isotope Imaging Mass Spectrometry, Neurons, Dendritic spines, RNA, Protein synthesis
Introduction
Because the molecular mechanisms of neuropsychiatric conditions are still largely unclear, current therapy is typically based on symptoms. Medications have important side effects and often require long treatment before showing results; some patients do not respond to existing drugs. Studies during the past decade have identified psychiatric disorders as the single largest health-cost in the modern society1. Patients often get sick at a young age and then suffer from a life-long condition. The direct health care costs are high and the total cost for society is even higher, when one takes into account the lost income and production capacity of not only the sick person but also, in many cases, of a family member2.
A basic view of psychiatric conditions is that they arise when signaling in the brain goes wrong. For example, in schizophrenia it is established that not only a loss of spine density and size3 but also an imbalance of different dopamine receptors4, as well as an imbalance between the dopaminergic and glutamatergic systems, lead to symptoms we associate with the disease. Present pharmacology is based on compensating the system at the neurotransmitter level. To find better strategies, we need to understand how synthesis and transport of new proteins participate in the regulation.
The strength of a synaptic signal is based on both the amount of transmitter molecules and the number of available receptors; a host of other proteins regulate the function of the receptor or of its downstream signals. The post-synaptic signal processing is initiated in the dendritic spine. A typical spine has a thin neck branching from a dendrite (50-150 nm thick, 200-2000 nm long) ending in a spherical head (200-500 nm in diameter). The thin neck partially isolates the local spine environment from the dendrite. Within the spine head, many different proteins act in concert to decode the chemical signals. The small dimensions of the spine combined with the fact that there are only a few copies of each protein, even for proteins that are believed to be abundant such as the Na/K-ATPase, as we have recently shown5, suggest that production of proteins in dendrites can play a major role in tuning synaptic strength.
Recent studies6 based on deep sequencing of the synaptic neuropil in hippocampus have identified a relatively large pool of 2550 mRNAs resident in the dendrites that code for known synaptic proteins. Those results reveal the potential for a production of proteins in dendrites but do not provide direct evidence for the actual synthesis of proteins. Deep sequencing cannot reveal the localization or the regulation of protein synthesis.
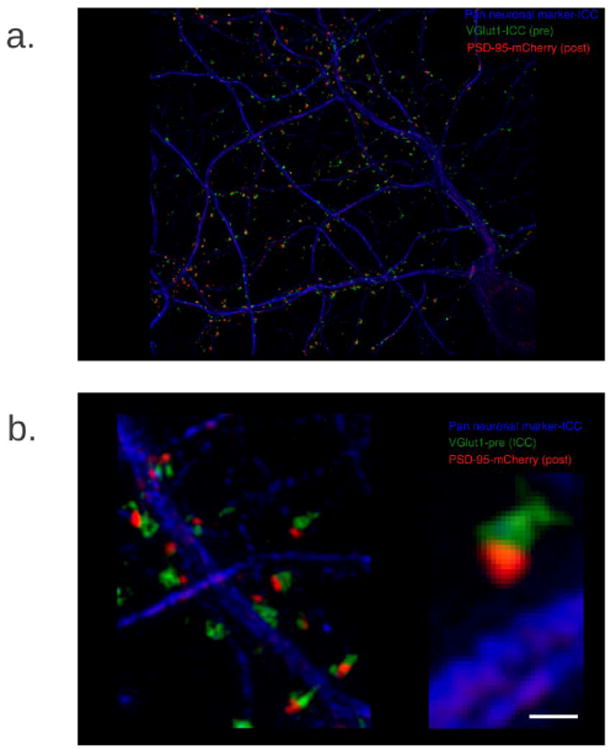
We have established super-resolution microscopy techniques and developed methodology to analyze colocalization of individual proteins with unprecedented detail in dendritic spines (Figure 1)7. Super-resolution microscopy, however, cannot reveal where or when proteins are synthesized. MIMS methodology has opened up entirely new possibilities for analysis of protein synthesis and protein turnover8, which is the key component for success in this project. With NRIMS, we are developing protocols for dual labeling of RNA and proteins using 15N-uridine and 18O- or 13C-leucine pulse chase in cultured neurons to identify and localize both protein synthesis and fate of newly synthesized proteins. We will demonstrate the presence of RNA, then visualize and measure local protein synthesis in localized segments of dendrites and/or spines. This will open a powerful way to study neural growth and synapse plasticity in health and disease.
Figure 1.
Method
Primary cultures of hippocampal cells from embryonic mice were established on Si chips. Culture media was supplemented with 15N-uridine and 18O-leucine (or 13C-leucine) individually and in combination starting at a series of different days in culture. Through MIMS analysis, RNA is identified by the 15N signal; synthesized proteins are identified by the 18O signal. Quantitative and spatial analysis were used to assess the time course and localization of protein synthesis as well as the fate of synthesized proteins. The greatest technical challenges are to preserve synapses, to find them in the preparations, and to analyze them with good resolution and precision.
Results
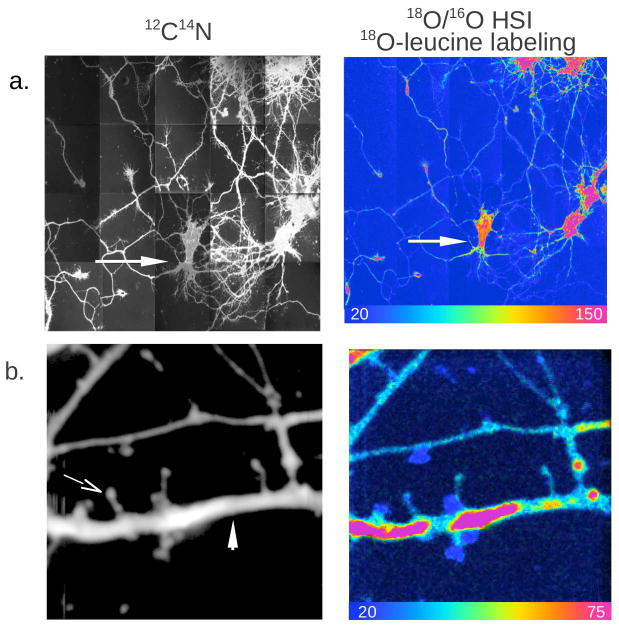
MIMS analysis of rat embryo cultured hippocampal cells is shown in Figure 2. In panel 2a cell bodies, axons, and dendrites can clearly be seen in the 12C14N image. The Hue Saturation Intensity (HSI) image of the 18O/16O ratio reveals regions of 18O-leucine incorporation indicating newly formed protein. The scale spans 1x the natural ratio of 18O/16O (blue) to over 7x the natural ratio (red). In panel 2b dendrites and typical spines, a thin neck branching from the dendrite and ending in a spherical head, are clearly visible in the 12C14N image. Along the marked dendrite (arrow) regions of high newly formed protein (high18O-leucine incorporation, over 4x the natural ratio) are separated by zones of lower incorporation.
Figure 2.
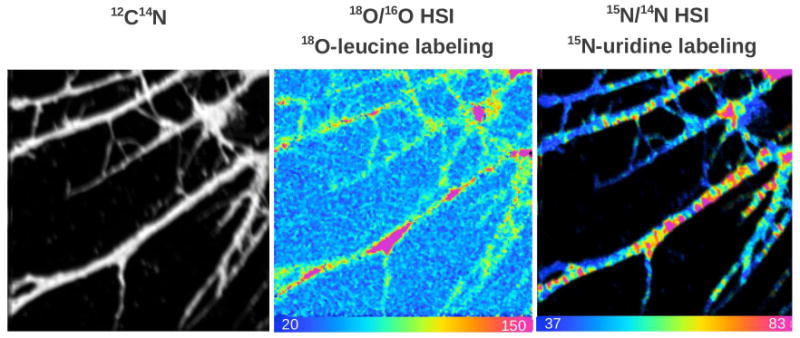
In Figure 3 dendrites are visible in the 12C14N image along with regions of newly formed protein seen in the 18O/16O HSI image. The 15N/14N HSI image shows regions of accumulation of 15N-uridine, a specific RNA precursor, revealing RNA localization. These results show for the first time that dendrites contain the machinery to synthesize protein and the presence of newly synthesized protein as was indirectly suspected, for example see recent study (6).
Figure 3.
Conclusion
We have for the first time directly imaged and measured the production of proteins at the subcellular level in the neuronal dendrites, close to the functional sites, the dendritic spines. The central hypothesis of this work is that local protein synthesis in the dendritic spines of neurons is a determinant of synaptic strength/signaling. This adds a new conceptual layer to the fine regulation of the nervous system. Based on the latest developments of MIMS, we will be able to image production and fate of proteins on the subcellular level in neurons and extend this work to the in vivo study of animal models. This work can also be expanded with the use of gold nanoparticle immuno-reporter tags9 so that one can localize and quantitative the turnover of a specific protein in spines, e.g. post-synaptic protein PSD-95 (Figure 1).
An understanding of the dendritic protein synthesis machinery will open pathways for development of new pharmacology and new therapeutic strategies for psychiatric disorders. The methods developed will be applicable to the study of both long-term potentiation and long-term depression of synaptic activity. These results, in turn, will be applied to the study of single gene disorders that intersect with Autism Spectrum Disorders (ASD), genetic defects that impact the brain such as in fragile X syndrome. Of particular interest are mutations in FMR1, the major cause of inherited ASD, which results in the absence of Fragile X mental retardation protein (FMRP), a major regulator of activity dependent synaptic protein synthesis.
Acknowledgments
C. Lechene is funded by the NIH (5P41EB001974-13, AG034641, R01 AG040019, R21AG034641-01, R01 AG040209), Human Frontier Science Program (RGP0048) and the Ellison Medical Foundation (AG-SS-2215-08). H. Brismar is supported by Faculty resources grant (KTH), Swedish government, VR-NT, Swedish research council, VR-RFI, Swedish research council, EU-FP7 and The Erling Persson foundation. A. Aperia is funded by Family Erling-Persson Foundation, Swedish Research Council, Heart Lung Foundation, Söderberg Foundation and Faculty Support.
References
- 1.Brundtland GH. JAMA. 2001;286(19):2391. doi: 10.1001/jama.286.19.2391. [DOI] [PubMed] [Google Scholar]
- 2.Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jonsson B, Olesen J, Allgulander C, Alonso J, Faravelli C, Fratigloni L, Jennum P, Lieb R, Maercker A, van Os J, Preisig M, Salvador-Carulla L, Simon R, Steinhausen HC. Eur Neuropsychopharmacol. 2011;21(9):655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Penzes P, Cahill ME, Hones KA, VanLeeuwen JE, Woolfrey KM. Nat Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beaulieu JM, Gainetdinov RR. Pharmacol Rev. 2011;63(1):182–217. doi: 10.1124/pr.110.002642. [DOI] [PubMed] [Google Scholar]
- 5.Blom H, Ronnlund D, Scott L, Spicarova Z, Widengren J, Bondar A, Aperia A, Brismar H. BMC Neurosci. 2011;12:16. doi: 10.1186/1471-2202-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. Neuron. 2012;74(3):453–466. doi: 10.1016/j.neuron.2012.02.036. E.M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blom H, Ronnlund D, Scott L, Spicarova Z, Rantanen V, Widengren J, Aperia A, Brismar H. Microsc Res Tech. 2011;75(2):220–228. doi: 10.1002/jemt.21046. [DOI] [PubMed] [Google Scholar]
- 8.Zhang DS, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, Prosser HM, Ervasti JM, Corey DP, Lechene CP. Nature. 2012;481(7382):520–524. doi: 10.1038/nature10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiery-Lavenant G, Guillermier C, Wang M, Lechene CP. Surface and Interface Analysis. 2014 doi: 10.1002/sia.5596. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]


