Background: Characteristics of ES cells are controlled by gene regulatory networks, and ETV4 and -5 participate in the network.
Results: Proliferation, induction of ectodermal marker genes, and expression of stem cell-related genes were impaired in Etv4/5 double KO ES cells.
Conclusion: ETV4 and -5 crucially define properties of ES cells.
Significance: Gene regulatory networks are dysregulated in the double KO ES cells.
Keywords: cell proliferation, embryonic stem cell, gene regulation, oncogene, transcription factor, differentiation
Abstract
The pluripotency and self-renewal capacity of embryonic stem (ES) cells is regulated by several transcription factors. Here, we show that the ETS-related transcription factors Etv4 and Etv5 (Etv4/5) are specifically expressed in undifferentiated ES cells, and suppression of Oct3/4 results in down-regulation of Etv4/5. Simultaneous deletion of Etv4 and Etv5 (Etv4/5 double knock-out (dKO)) in ES cells resulted in a flat, epithelial cell-like appearance, whereas the morphology changed into compact colonies in a 2i medium (containing two inhibitors for GSK3 and MEK/ERK). Expression levels of self-renewal marker genes, including Oct3/4 and Nanog, were similar between wild-type and dKO ES cells, whereas proliferation of Etv4/5 dKO ES cells was decreased with overexpression of cyclin-dependent kinase inhibitors (p16/p19, p15, and p57). A differentiation assay revealed that the embryoid bodies derived from Etv4/5 dKO ES cells were smaller than the control, and expression of ectoderm marker genes, including Fgf5, Sox1, and Pax3, was not induced in dKO-derived embryoid bodies. Microarray analysis demonstrated that stem cell-related genes, including Tcf15, Gbx2, Lrh1, Zic3, and Baf60c, were significantly repressed in Etv4/5 dKO ES cells. The artificial expression of Etv4 and/or Etv5 in Etv4/5 dKO ES cells induced re-expression of Tcf15 and Gbx2. These results indicate that Etv4 and Etv5, potentially through regulation of Gbx2 and Tcf15, are involved in the ES cell proliferation and induction of differentiation-associated genes in ES cells.
Introduction
The self-renewal capacity and pluripotency of murine embryonic stem (ES)4 cells are maintained through stimulations of both leukemia inhibitory factor (LIF) and BMP4 (bone morphogenic protein 4) in vitro (1–3). LIF stimulation activates JAK-STAT3 and PI3K-AKT pathways and maintains pluripotency of ES cells (4). In fact, artificial activation of either STAT3 or AKT maintains self-renewal of ES cells in the absence of LIF (5, 6). The transcription factors KLF4 and TBX3 are downstream regulators of STAT3 and AKT, respectively, and are involved in the maintenance of pluripotency (4). BMP induces expression of the Id (inhibitory of DNA binding) genes via the Smad pathway, and ID proteins suppress differentiation and sustain self-renewal of ES cells in collaboration with STAT3 (1).
In addition to the signal transduction pathways, including JAK-STAT, PI3K-AKT, and BMP-SMAD, several transcription factors, including OCT3/4, SOX2, and NANOG, are known to be major regulators of self-renewal. Oct3/4-deficient embryos develop to the blastocyst stage, but their inner cell mass from which ES cells are established loses pluripotency (7). A conditional Oct3/4-knock-out ES cell experiment showed that Oct3/4 deficiency promotes differentiation of ES cells into extraembryonic trophectodermal cells (7, 8). Sox2-deficient blastocysts form abnormal inner cell masses and fail to undergo outgrowth, and Sox2-null ES cells differentiate into trophoblast cells (9, 10). Although Nanog-deficient ES cells retain the capacity for self-renewal (11), deletion of the Nanog gene causes early embryonic lethality, whereas forced expression of Nanog in ES cells accelerates their self-renewal in a LIF-independent manner (12, 13). Furthermore, other transcriptional regulators, including ESRRB (14–16), DAX1 (17–19), SALL4 (20–22), ZIC3 (23), KLF4 (24), MYC (25, 26), and MAX (27), have been identified as key regulators of the self-renewal capacity and pluripotency of ES cells.
High-throughput analyses revealed that these transcription factors form a complex network of regulatory and/or feed-forward loops in ES cells. For example, chromatin immunoprecipitation experiments showed that OCT3/4, NANOG, SOX2, and other ES cell-specific transcription factors co-occupy target genes in ES cells and participate in regulatory loops that maintain self-renewal and pluripotency (24, 28–33). Protein-protein interaction networks centered on OCT3/4, NANOG, and MYC are thought to be involved in the maintenance of ES cell characteristics (34–37). Recent studies have shown that ES cells and tumor cells often possess similar characteristics, including rapid cell proliferation, self-renewal capacity in the undifferentiated state, and gene expression signatures (38, 39), indicating that genes involved in oncogenesis may also play role(s) in the constitution of ES cell characteristics.
The ETS transcription factors of the PEA3 group, including ETV1 (also called ER81), ETV4 (also called PEA3), and ETV5 (also called ERM), are involved in critical physiological processes, such as early development, organogenesis, and morphogenesis (40). ETV4 and ETV5 often have similar functions during morphogenesis, but ETV1 is thought to be different. A single knockout of either Etv4 or Etv5 is not sufficient to cause kidney defects, but Etv4/5 double knock-out mice do not develop kidneys, suggesting that ETV4 and ETV5 are functionally redundant (41). These transcription factors also function as oncoproteins in several tumor cells and promote cell proliferation (42). Interestingly, the BioGPS Database, as well as several studies, indicates that Etv4 and Etv5 are expressed in ES cells (32, 33, 43), indicating that ETV4 and ETV5 could be involved in the self-renewal capacity and/or pluripotency of ES cells. In the present study, we discovered that the expression of Etv4 and Etv5 is regulated by OCT3/4, and investigations of Etv4 and Etv5 double knock-out ES cells clarified that these two molecules are involved in the proliferation and differentiation of ES cells.
Experimental Procedures
Cell Culture
ES cell lines PE9 (control wild-type ES cells), PE15-2 (Etv4 and Etv5 double knock-out (Etv4/5 dKO) ES cells), and ZHBTc4 (conditional Oct3/4-knock-out ES cells) are derived from previous investigations (8, 44). These cells were cultured on gelatin-coated dishes with LIF-supplemented Dulbecco's modified Eagle's medium (DMEM) as described previously (5, 45). For the 2i culture, PE9 and PE15-2 ES cells were cultured in a chemically defined medium (DMEM/F-12 with N2B27 supplement plus 2i (2 μm mitogen-activated protein kinase/extracellular signal-regulated kinase (PD0325901) and 3 μm glycogen synthase kinase 3 (CHIR99021) inhibitors)) (Wako Pure Chemical, Osaka, Japan) as described previously (46). For the LIF depletion culture, PE9 and PE15-2 ES cells were cultured for 3 days in the absence of LIF in gelatin-coated dishes. To control Oct3/4 expression, ZHBTc4 ES cells were cultured with or without 1 μg/ml tetracycline (Tet) (Sigma-Aldrich) for 24 to 48 h. To restore Oct3/4 expression, the culture medium of Tet-treated cells was changed to a Tet-free medium, and the cells were cultured for another 24 h. For the embryoid body (EB) formation assay, ES cells were cultured by a hanging drop method (1 × 104 cells/20 μl). After 3 days, the EBs were transferred to ultra-low attachment tissue culture plates (Corning, Inc.) and then cultured for 6–9 days in the absence of LIF.
Plasmid Construction and Transfection
Construction of mammalian expression vectors pCAGIP-Myc and pCAGIP-FLAG has been described previously (18, 47). A hygromycin-resistant pCAG vector was constructed by inserting the puromycin resistance gene of pCAGIP-Myc into the hygromycin resistance gene. The plasmid was named pCAGIH-Myc. Coding regions of Etv4 and Etv5 were amplified by PCR using specific primers listed in supplemental Table S1. pCAGIP-Myc-Etv4, pCAGIP-Myc-Etv5, pCAGIP-FLAG-Etv5, and pCAGIH-Myc-Etv4 were constructed by inserting corresponding coding sequences into the expression vectors. For reporter plasmid construction, the promoter region (from −2019 to −2486; the transcription start site is considered as +1) of the Etv4 gene and the intron 6 region of the Etv5 gene were amplified by PCR using specific primers listed in supplemental Table S1. To introduce mutation of an OCT3/4-binding sequence of these regions, mutated OCT3/4 binding sequences were constructed by PCR using specific primers. PCR products were cloned into pGL4-promoter (48) and termed pGL4-promoter-Etv4 (−2486/−2019)-wild type, -Etv4 (−2486/−2019)-mutant, -Etv5 intron 6-wild type, and -Etv5 intron 6-mutant, respectively.
Plasmids were introduced into cultured cells using Lipofectamine 2000 (Life Technologies, Inc.) according to the manufacturer's protocol. ETV4- or ETV5-expressing ES cells were established by introduction of pCAGIP-Myc-Etv4 or -Etv5 into Etv4/5 dKO (PE15-2) ES cells and cultured in the presence of 0.5 μg/ml puromycin (Nacalai Tesque, Kyoto, Japan). ETV4/5-expressing PE15-2 ES cells were established as follows. First, ETV4-expressing PE15-2 ES cells were established by introducing pCAGIH-Myc-Etv4 into PE15-2 ES cells. They were subsequently cultured in the presence of 300 μg/ml hygromycin (Nacalai Tesque). After that, pCAGIP-FLAG-Etv5 was introduced into the cells. Cells were selected by puromycin, and the cell line was named PE15-2 (ETV4/ETV5) 8. A control cell line was established by introduction of empty vectors (both pCAGIH-Myc and pCAGIP-FLAG) into PE15-2 ES cells in parallel. The cell line was named PE15-2 (ev/ev) 1.
Biotin-labeled DNA Pull-down Assay
Oligonucleotides containing an OCT3/4-binding site of the Etv4 gene and ETV4/5-binding site of the Gbx2 gene were synthesized. The oligonucleotide sequences are shown in supplemental Table S1. For a biotin-labeled DNA pull-down assay, 5 μm 3′-biotinylated double-stranded oligonucleotide was incubated overnight at 4 °C with 1 mg of either Myc-Oct3/4-, Myc-Etv4-, or Myc-Etv5-transfected HEK293 cell extracts in the presence of streptavidin-agarose (EMD Millipore, Darmstadt, Germany). For the competition assays, non-labeled oligonucleotide, either wild-type or mutant, was added in 25-fold molar excess. The beads were washed three times with a washing buffer (50 mm Tris-HCl (pH 7.5), 2 mm MgCl2, and 150 mm NaCl). Samples were examined by Western blot analysis.
Immunofluorescent Staining, Western Blot Analysis, and Determination of Alkaline Phosphatase Activity
For immunostaining analysis, ES cells were fixed with 4% paraformaldehyde at 4 °C for 30 min. After permeabilization with 0.5% Triton X-100 in phosphate-buffered saline (PBS), the cells were preincubated with 1% BSA in PBS. The cells were then incubated with antibodies against NANOG (SC1000, Merck Millipore, Darmstadt, Germany), OCT3/4 (sc-9081, Santa Cruz Biotechnology, Inc.), or ESRRB (PP-H6705-00, Perseus Proteomics, Tokyo, Japan) at 4 °C overnight, followed by incubation with 100-fold diluted goat anti-rabbit or anti-mouse IgG FITC (Santa Cruz Biotechnology). For Western blot analysis, samples were mixed with an SDS sample buffer (5× buffer: 50 mm Tris-HCl (pH 6.8), 30% glycerol, 10% SDS, 250 mm dithiothreitol, 10 mm EDTA, and 0.01% Coomassie Brilliant Blue R250), subjected to SDS-10% PAGE, and transferred to a nitrocellulose membrane. The membrane was incubated with antibodies for NANOG, OCT3/4, ESRRB, α-tubulin (M175-3, MBL, Nagoya, Japan), or c-Myc (sc-40; Santa Cruz Biotechnology), followed by horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit IgG (EMD Millipore). The blot was visualized using enhanced chemiluminescence reagents (PerkinElmer Life Sciences) with an LAS-1000 image analyzer (Fuji Film). Activity of alkaline phosphatase was determined using either a quantitative alkaline phosphatase ES characterization kit (Merck Millipore) or a vector blue substrate kit (Vector Laboratories, Inc., Burlingame, CA) according to the manufacturer's protocols.
For the reporter assay, ZHBTc4 ES cells were transfected with pGL4 reporter plasmids, and these cells were divided into two dishes 24 h after transfection and cultured with or without 1 μg/ml Tet for 24 h. Cells were lysed in the cell lysis buffer, and luciferase activity in the extracts was measured using a luciferase assay kit (Promega, Madison, WI) with an AB-2200 (ATTO, Tokyo, Japan).
Preparation of Nuclear Extracts and the Electromobility Shift Assay
Nuclear extracts were isolated as described before (16). Briefly, ZHBTc4 ES cells treated with or without 1 μg/ml Tet for 3 days were resuspended in a buffer (20 mm HEPES (pH 7.9), 20% glycerol, 10 mm NaCl, 0.2 mm EDTA (pH 8.0), 1.5 mm MgCl2, 1 mm dithiothreitol (DTT), 10 μg/ml leupeptin, 10 μg/ml pepstatin, and 10 μg/ml aprotinin). Samples were incubated on ice for 15 min and spun down at 1,000 rpm for 10 min. The pellet was resuspended in the same buffer at 2.5 × 107 nuclei/ml, and then 62.5 μl of 5 m NaCl was added to the 1 ml of sample. After incubation at 4 °C for 30 min, the sample was centrifuged at 10,000 rpm, and the supernatant was used as a nuclear extract.
Oligonucleotides containing the OCT3/4-binding site of the Etv5 gene were synthesized. The oligonucleotide sequences are shown in supplemental Table S1. Double-stranded probes were end-labeled with [γ-32P]ATP (MP Biomedicals, Santa Ana, CA) and purified by Mini Quick Spin Oligo columns (Roche Diagnostics, Basel, Switzerland). For a binding reaction, 5 μg of nuclear extracts were incubated with a radiolabeled probe, 2 μg of poly(dI-dC), and 300 μg/ml BSA either with or without a 200-fold excess of cold probe, either wild-type or mutated oligonucleotides, as a competitor. Also, either 2–4 μg of anti-OCT3/4 antibody (sc-5279; Santa Cruz Biotechnology) or an anti-FLAG antibody (F3165; Sigma-Aldrich) were added to the reaction mixture for a supershift assay. After incubation on ice for 40 min, the samples were subjected to electrophoresis in a nondenaturing 6% acrylamide gel at 100 V for 2 h. The gel was dried, and radiolabeled bands were visualized with an image analyzer (BAS-2000, Fuji Film, Tokyo, Japan).
Reverse Transcription-PCR (RT-PCR), Quantitative RT-PCR, and Oligonucleotide-based DNA Microarray Analysis
Total RNAs were isolated from cells with Sepasol-RNA I Super G (Nacalai Tesque) and converted to cDNAs by ReverTraAce (Toyobo, Osaka, Japan) with oligo(dT)12–18 primers (Nippongene, Tokyo, Japan). Gene expression was determined by either regular RT-PCR or quantitative RT-PCR using SYBR Green (Mx3005P; Agilent Technologies, Santa Clara, CA). For quantitative RT-PCR, all samples were tested in triplicate, and the results from each sample were normalized relative to glyceraldehyde-3-phosphate dehydrogenase with the ΔΔCt method. Primer sequences have been described previously (16, 49) or are listed in supplemental Table S1.
The 3D-Gene Mouse Oligo chip 24k (Toray Industries Inc., Tokyo, Japan) was used for the oligonucleotide-based DNA microarray analysis. Briefly, one pair of RNA samples was collected from control wild-type and Etv4/5 dKO ES cells. Total RNA was labeled with either Cy3 or Cy5, and Cy3 or Cy5 signals were obtained using the 3D-Gene Scanner (Toray Industries Inc.) and processed by 3D-Gene Extraction (Toray Industries Inc.). Detected signals for each gene were normalized by a global normalization method (Cy3/Cy5 ratio median = 1). Gene ontology term analysis and pathway analysis were done. The raw data has been deposited in the Gene Expression Omnibus (GEO) database, under number GSE47225.
Cell Proliferation Assay
The number of viable cells was measured by a direct cell count and a WST-1 assay. The WST-1 assay was performed according to the manufacturer's protocol (Roche Diagnostics). Briefly, cells were cultured in a 96-well plate for 3 days. Ten microliters of WST-1 reagent were added to each well, followed by incubation at 37 °C. The absorbance of the culture medium was measured at 450 and 630 nm using a microplate reader (Tecan, Männedorf, Switzerland).
Results
Expression of Etv4 and Etv5 mRNA Correlates with Oct3/4 Expression
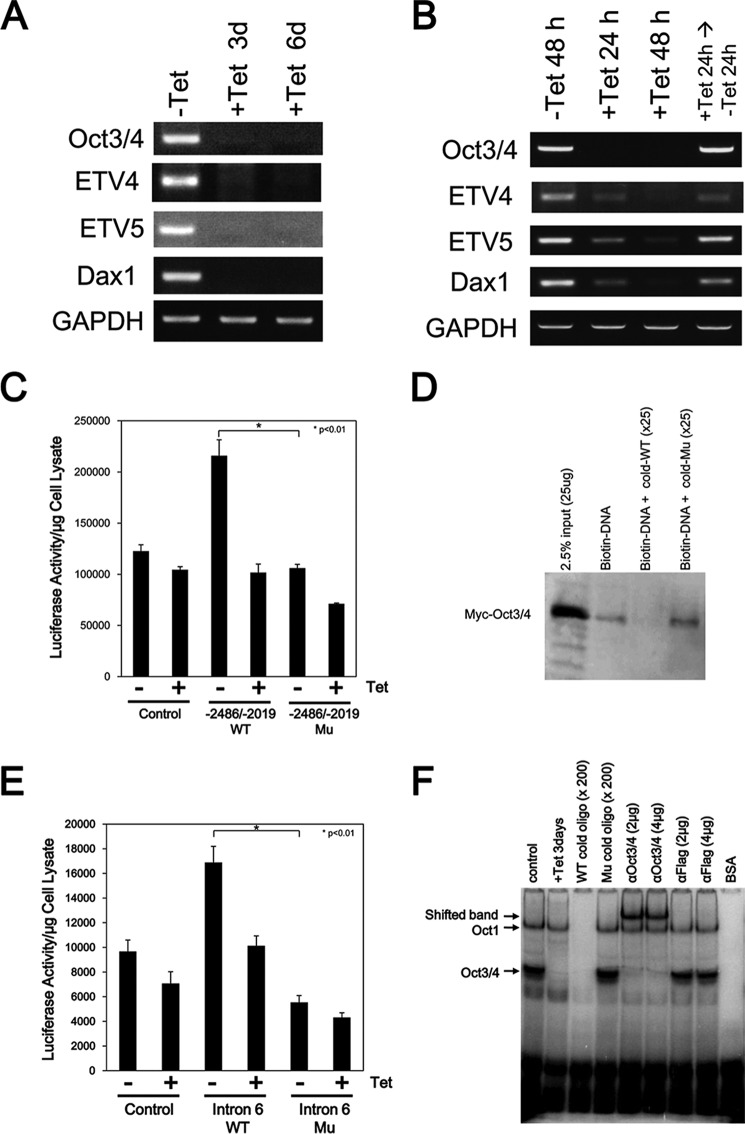
The transcription factor OCT3/4 is one of the major regulators of the self-renewal of ES cells. ZHBTc4 ES cells are Tet-inducible Oct3/4-conditional knock-out ES cells, which differentiate into trophectoderm after Oct3/4 depletion (8). Here we examined the expression of Etv4 and Etv5 mRNA in ZHBTc4 ES cells. Oct3/4 was expressed in ZHBTc4 ES cells in the absence of Tet, and the expression was repressed by Tet treatment for 3–6 days (Fig. 1A). Etv4 and Etv5 as well as Dax1, which is a direct downstream target of OCT3/4 (50), were also expressed in control ZHBTc4 ES cells, and their expression was also repressed after the treatment with Tet (Fig. 1A), indicating that Etv4 and Etv5 are expressed in undifferentiated ES cells and that their expression is repressed in trophectodermal cells.
FIGURE 1.
Expression of Etv4 and Etv5 mRNA in ZHBTc4 ES cells. A, reduction of Etv4 and Etv5 mRNA expression after repression of Oct3/4 expression. ZHBTc4 ES cells were cultured with or without 1 μg/ml Tet for 3 or 6 days. Expression of the indicated genes was examined by RT-PCR. B, induction of Etv4 and Etv5 mRNA expression by Oct3/4 expression. ZHBTc4 ES cells were cultured with or without 1 μg/ml Tet for 24–48 h. To restore Oct3/4 expression, the culture medium of Tet-treated cells was changed to a Tet-free medium, and the cells were cultured for another 24 h. Dax1 was used for a positive control as an OCT3/4 target gene. Gapdh was used as an internal control. All results are representative of three independent experiments. C, determination of an OCT3/4-responsive element in the Etv4 gene by a luciferase assay. The promoter region of the Etv4 gene contains a consensus sequence for an OCT3/4 binding site (ATTTGCAT). ZHBTc4 ES cells were transfected with either pGL4-promoter (control), pGL4-promoter-Etv4 (−2486/−2019)-wild type (−2486/−2019 WT), or pGL4-promoter-Etv4 (−2486/−2019)-mutant (−2486/−2019 Mu). ZHBTc4 ES cells were divided into two dishes 24 h after transfection and cultured with (+) or without (−) 1 μg/ml Tet for 24 h, and luciferase activity was measured. The sequence of the OCT3/4-responsive element was changed from ATTTGCAT to GACGTGGG. Shown are means (bars) and S.D. (error bars) of triplicate assays. D, OCT3/4 directly binds to the putative binding sequences. The association of DNA and OCT3/4 was examined by a biotin-labeled DNA pull-down assay. Biotin-labeled oligonucleotide containing the putative binding site was incubated with cell extracts from Myc-Oct3/4-transfected HEK293 cells either with or without 25-fold non-labeled WT or mutated (Mu) oligonucleotide. The result shown is representative of three separate experiments. E, determination of an OCT3/4-responsive element in the Etv5 gene by a luciferase assay. The intron 6 region of the Etv5 gene contains the consensus sequence of an OCT3/4 binding site (ATGCAAAT). ZHBTc4 ES cells were transfected with either pGL4-promoter (control), pGL4-promoter-Etv5 intron 6-wild type (Intron 6 WT), or pGL4-promoter-ETV5 intron 6-mutant (Intron 6 Mu). ZHBTc4 ES cells were divided into two dishes 24 h after transfection and cultured with (+) or without (−) 1 μg/ml Tet for 24 h, and luciferase activity was measured. The sequence of the OCT3/4-responsive element was changed from ATGCAAAT to GTGCCGCT. Shown are the means (bars) and S.D. (error bars) of triplicate assays. F, OCT3/4 directly binds to the putative binding sequences. An association of DNA and OCT3/4 was examined by an EMSA. Radiolabeled oligonucleotide containing the putative binding site was incubated with nuclear extracts from ZHBTc4 ES cells either with or without 200-fold cold WT or cold mutated (Mu) oligonucleotides, an anti-OCT3/4 antibody, or an anti-FLAG antibody. The result shown is representative of three separate experiments. Note that a band of OCT1-DNA complex can be detected in this assay as described before (66).
We also examined the effects of short term exposure to Tet on the expression of Etv4 and Etv5 in ZHBTc4 ES cells. Oct3/4 expression was repressed by Tet treatment within 24 h, and its expression was recovered by the removal of Tet (Fig. 1B). Expression of Etv4 and Etv5, as well as Dax1, was reduced after treatment with Tet for 24–48 h and recovered by the removal of Tet (Fig. 1B), showing that expression of Etv4 and Etv5 mRNA correlates with Oct3/4 expression in ES cells.
Next, we investigated whether expression of Etv4 and Etv5 is regulated by OCT3/4 in ES cells. The consensus sequence of an OCT3/4-binding site is known to be ATGCAAAT. The Etv4 gene contains several consensus OCT3/4-binding sequences, and the promoter region (from −2486 to −2019, transcription start site considered as +1) of the gene contains the consensus sequences and had enhancer activity in ES cells (Fig. 1C). The enhanced activity was repressed after Tet stimulation (Oct3/4-depleted condition) in ZHBTc4 ES cells, and no enhancer activity was detected when the consensus sequences were mutated (from ATTTGCAT to GACGTGGG). An association of the consensus sequences and OCT3/4 was observed by a biotin-labeled DNA pull-down assay. Myc-tagged OCT3/4 was precipitated with a biotin-labeled oligonucleotide contacting the OCT3/4-binding site of the Etv4 gene, and the association was competed by an excess of unlabeled oligonucleotide, but not by unlabeled mutated oligonucleotide (Fig. 1D).
Because intron 5 and intron 6 of the Etv5 gene contain the consensus sequences for OCT3/4, we also performed a luciferase reporter assay and found that the intron 6 region, but not the intron 5 region, had an enhancer activity in ES cells (Fig. 1E) (data not shown). Similar to the result with the Etv4 gene, the enhanced activity was reduced after Tet stimulation in ZHBTc4 ES cells. No enhancer activity was detected when the consensus sequences were mutated from ATGCAAAT to GTGCCGCT. An electrophoretic mobility shift assay (EMSA) revealed that the 25-bp oligonucleotide containing the OCT3/4 binding site of intron 6 formed a complex with OCT3/4 in vitro (Fig. 1F). The complex was not detected when a nuclear extract from Tet-treated ZHBTc4 ES cells was examined. The unlabeled competitor (200-fold excess) inhibited the formation of the complex, but it was not competed by an excess of unlabeled mutated oligonucleotide. Also, the band was supershifted by the addition of an anti-OCT3/4 antibody but not an anti-FLAG antibody as a negative control. These results indicate that OCT3/4 binds to the Etv4 and Etv5 genes and regulates their expression.
Etv4 and Etv5 dKO ES Cells Maintain an Undifferentiated State
ETV4 and ETV5 have similar molecular structures and exhibit overlapping expression, and redundant activities of ETV4 and ETV5 have been noted in kidney development (41). Also, RNAi depletion of Etv5 in ES cells did not result in obvious phenotypes (51). Thus, we analyzed Etv4/5 dKO ES cells to explore the functions of ETV4 and ETV5 in ES cells.
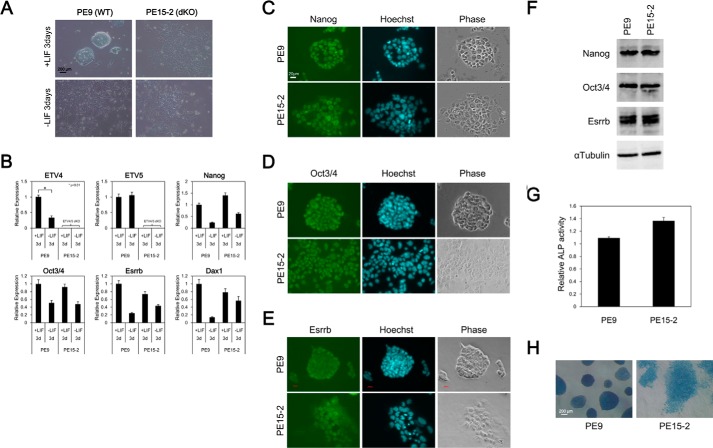
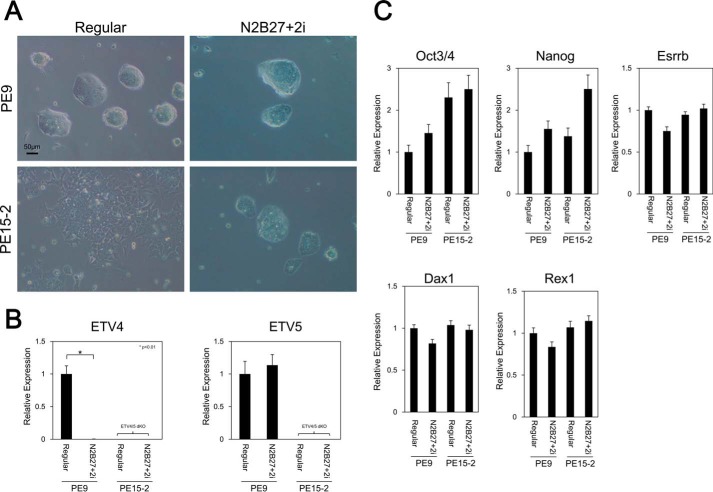
One major characteristic of ES cells is the capacity for self-renewal; they have the potential to proliferate in an undifferentiated state. First, we examined whether Etv4/5 dKO ES cells maintain an undifferentiated state or not. Control wild-type ES cells (PE9 ES cells) formed a compact colony, which is typical of undifferentiated ES cells, in the presence of LIF. In contrast, Etv4/5 dKO ES cells (PE15-2 ES cells) failed to form a compact colony but showed an epithelial cell-like morphology even in the presence of LIF (Fig. 2A). After removing LIF from the culture medium, both control and the dKO ES cells showed epithelial cell-like morphology. Expression analysis showed that Nanog, Oct3/4, Esrrb, and Dax1 were expressed in Etv4/5 dKO ES cells as well as control cells and down-regulated after depletion of LIF from culture medium (Fig. 2B). We also examined protein expression of self-renewal markers, including NANOG, OCT3/4, and ESRRB, in Etv4/5 dKO ES cells. Immunofluorescent staining and Western blot analysis revealed that NANOG, OCT3/4, and ESRRB were expressed in Etv4/5 dKO ES cells (Fig. 2, C–F). In addition, relative alkaline phosphatase activity, which is a marker of undifferentiated pluripotent stem cells, was comparable between wild-type and Etv4/5 dKO ES cells (Fig. 2, G and H). Self-renewal capacity is maintained by LIF stimulation followed by STAT3 activation. A recent study showed that inhibition of GSK3 and MEK/ERK pathways by small molecule inhibitors (2i) achieves LIF/STAT3-independent self-renewal of ES cells without serum (46), and naive ES cells can be cultured in the 2i medium, whereas primed epiblast stem cells cannot (52). Here, we examined whether Etv4/5 dKO ES cells can be cultured in the 2i medium. As shown in Fig. 3A, PE9 wild-type and PE15-2 Etv4/5 dKO ES cells grew in the 2i medium, and the epithelial-like morphology of PE15-2 ES cells was changed into a compact colony in the 2i medium. In PE9 wild-type ES cells, expression of Etv5 mRNA was maintained in the 2i medium; however, Etv4 mRNA was drastically repressed, suggesting that ETV4 is dispensable for the native status of ES cells. Quantitative RT-PCR analysis demonstrated that undifferentiated marker genes (Oct3/4, Nanog, and Esrrb), as well as naive marker genes (Dax1 and Rex1) were expressed in PE9 and PE15-2 ES cells in the 2i medium (Fig. 3C), suggesting that Etv4/5 dKO ES cells are able to maintain their naive status. Taken together, these results suggest that ETV4 and ETV5 are dispensable for the maintenance of the undifferentiated state of ES cells.
FIGURE 2.
Etv4/5 dKO ES cells express self-renewal marker genes. A, morphological signatures of PE9 (WT) and PE15-2 (Etv4/5 dKO) ES cells. PE9 and PE15-2 ES cells were cultured in the presence or absence of LIF for 3 days. Morphological signatures were observed. B, expression of self-renewal marker genes in PE9 and PE15-2 ES cells. Expression of the indicated self-renewal marker genes in the conditions of A in PE9 and PE15-2 ES cells was examined by quantitative RT-PCR. Expression of Etv4 and Etv5 mRNA was only shown in PE9 ES cells. All samples were analyzed in triplicate, and the data were normalized to Gapdh expression. C–E, immunofluorescent stain in PE9 and PE15-2 ES cells. Protein expression of the self-renewal marker genes, including NANOG (C), OCT3/4 (D), and ESRRB (E), in PE9 and PE15-2 ES cells was examined by immunofluorescent stain. Nuclei were stained with the Hoechst stain. F, Western blot analysis in PE9 and PE15-2 ES cells. Protein expression of the self-renewal marker genes (NANOG, OCT3/4, and ESRRB) was examined by Western blot analysis. α-Tubulin was used as a loading control. G, determination of alkaline phosphatase activity. Relative activity of alkaline phosphatase was determined in PE9 and PE15-2 ES cells. Shown are the means (bars) and S.D. (error bars) of triplicate plates. H, alkaline phosphatase stain in PE9 and PE15-2 ES cells. Alkaline phosphatase of ES cells was detected by the VECTOR Blue alkaline phosphatase substrate kit. Morphological signatures were observed. All results are representative of several independent experiments.
FIGURE 3.
Etv4/5 dKO ES cells maintain the undifferentiated state in N2B27 serum-free medium plus two inhibitors. A, morphological signatures of PE9 (WT) and PE15-2 (Etv4/5 dKO) ES cells. PE9 and PE15-2 ES cells were cultured in either a regular serum- and LIF-containing medium or a chemically defined medium (DMEM/F-12 with N2B27 supplement plus 2i (mitogen-activated protein kinase/extracellular signal-regulated kinase (PD0325901) and glycogen synthase kinase 3 (CHIR99021) inhibitors) (2i condition)). Morphological signatures were observed. B, expression of Etv4 and Etv5 in the 2i condition. Expression of Etv4 and Etv5 in the conditions of A in PE9 and PE15-2 ES cells was examined by quantitative RT-PCR. C, expression of self-renewal marker genes in PE9 and PE15-2 ES cells. Expression of the indicated self-renewal marker genes in the conditions of A in PE9 and PE15-2 ES cells was examined by quantitative RT-PCR. All samples were analyzed in triplicate, and the data were normalized to Gapdh expression. Error bars, S.D.
ETV4 and ETV5 Regulate Proliferation of ES Cells
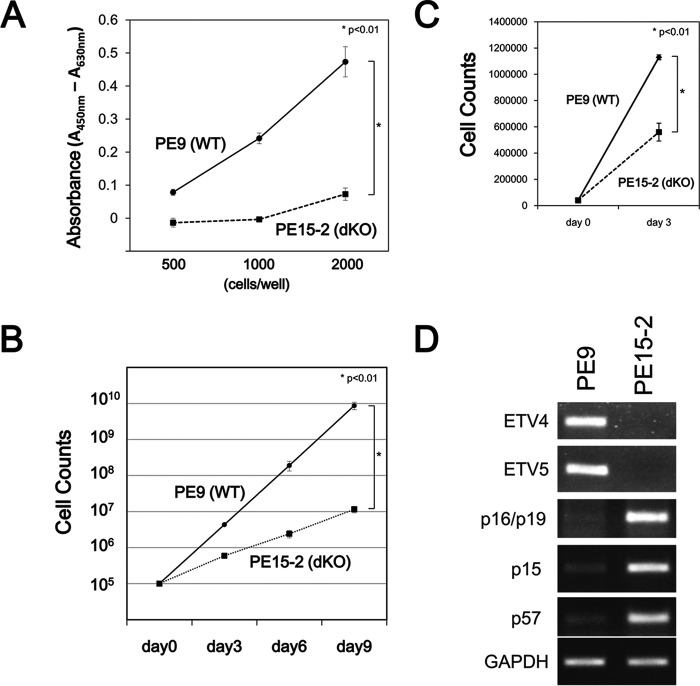
After finding that Etv4/5 dKO ES cells expressed self-renewal marker genes, next we compared the proliferation of control wild-type and Etv4/5 dKO ES cells. A WST-1 assay demonstrated that cell proliferation was reduced in Etv4/5 dKO ES cells (Fig. 4A). Direct cell counting also showed that proliferation of Etv4/5 dKO ES cells was significantly decreased compared with wild-type ES cells (Fig. 4B). We also compared cell counts between wild-type and Etv4/5 dKO ES cells cultured in the 2i medium. As shown in Fig. 4C, proliferation of Etv4/5 dKO ES cells is slower than wild-type ES cells, indicating that proliferation of the dKO ES cells is not recovered even in the 2i condition. Of note, expression of cyclin-dependent kinase inhibitors, including p16-INK4/p19-ARF, p15, and p57, was completely repressed in wild-type ES cells. By contrast, their expression was enhanced in Etv4/5 dKO ES cells (Fig. 4D). These results demonstrate that ETV4 and ETV5 regulate the proliferation of ES cells by controlling cyclin-dependent kinase inhibitors.
FIGURE 4.
ETV4 and ETV5 are involved in the proliferation of ES cells. A, proliferation of PE9 (WT) and PE15-2 (Etv4/5 dKO) ES cells. Either PE9 or PE15-2 ES cells were seeded at 500, 1000, or 2000 cells/well in a 96-well dish and subjected to a WST-1 assay. The results represent the means and S.D. (error bars) of triplicate assays. B, growth curves of PE9 and PE15-2 ES cells. PE9 and PE15-2 ES cells were cultured in the presence of LIF, and the cells were counted at the indicated days. The results represent the means and S.D. of four independent assays. C, direct cell counts of PE9 and PE15-2 ES cells in the 2i condition. PE9 and PE15-2 ES cells were cultured in the 2i condition, and the cells were counted at the indicated days. The results represent the means and S.D. of triplicate experiments. D, enhanced expression of cyclin-dependent kinase inhibitors in Etv4/5 dKO ES cells. Expression of cyclin-dependent kinase inhibitors, p16/p19, p15, and p57 mRNA in PE9 and PE15-2 ES cells was examined by RT-PCR. Gapdh was used as an internal control. The results are representative of three independent experiments.
Etv4/5 dKO ES Cells Are Impaired in Ectoderm Marker Gene Induction
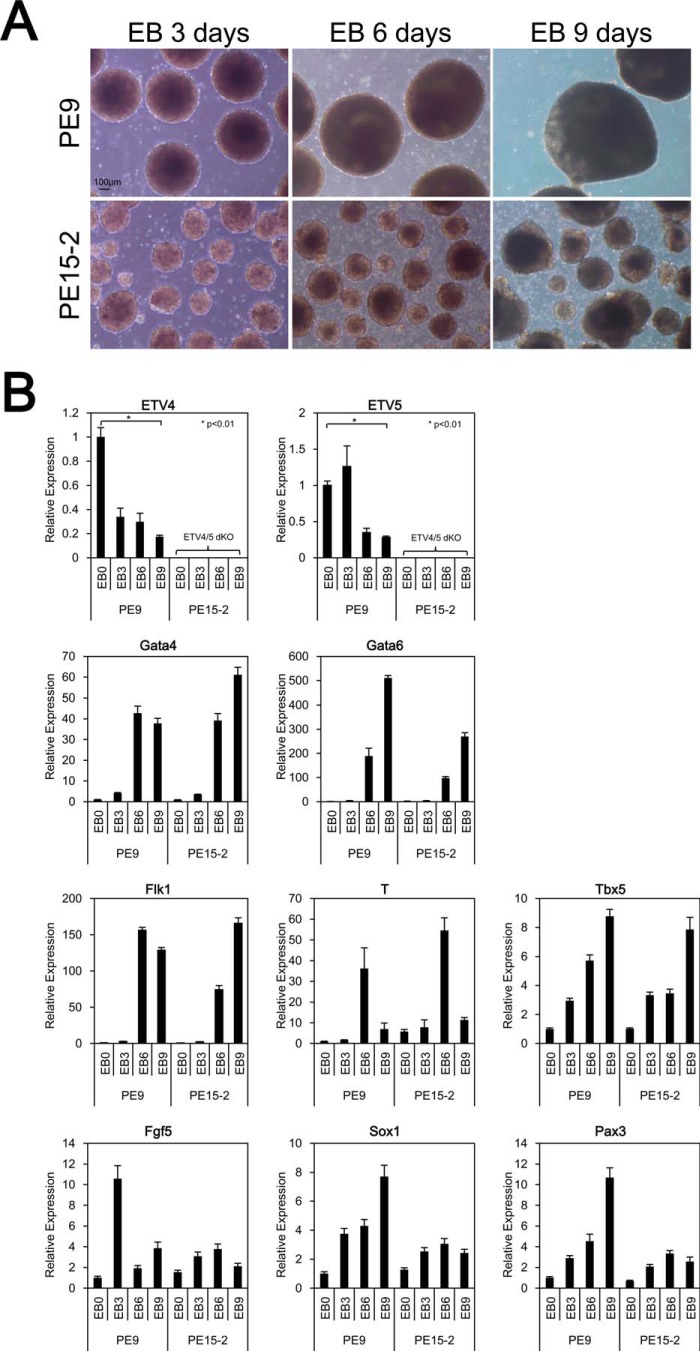
Next, we assessed differentiation ability, the second characteristic of ES cells, in Etv4/5 dKO ES cells within a system of EBs in suspension culture, which mimics early embryogenesis in vitro. Control wild-type ES cells formed EBs normally, whereas the EBs derived from Etv4/5 dKO ES cells were much smaller than control cells (Fig. 5A). Quantitative RT-PCR analysis revealed that expression of endoderm (Gata4 and Gata6) and mesoderm (Flk1, T, and Tbx5) markers was very similar between PE9 and PE15-2 cells (Fig. 5B). In contrast, expression patterns of ectoderm markers (Fgf5, Sox1, and Pax3) differed. Fgf5 expression was induced in an EB sample that had been incubated for 3 days, and it was repressed immediately in PE9 ES cells, whereas its expression was not induced in PE15-2 ES cells. Strong expression of Sox1 and Pax3 was found in an EB sample of PE9 ES cells that had been incubated for 9 days, but they were not induced in PE15-2 cells. These results suggest that Etv4/5 dKO ES cells have an impaired ability to induce ectoderm marker gene expression in vitro.
FIGURE 5.
Etv4/5 dKO ES cells have impaired abilities to induce ectoderm marker gene expression. A, morphological signatures of EBs derived from PE9 (WT) and PE15-2 (Etv4/5 dKO) ES cells. PE9 and PE15-2 ES cells were cultured in suspension without LIF for 3–9 days. Morphological signatures were observed. Scale bar, 100 μm. B, expression of differentiation marker genes in PE9 and PE15-2 EBs. Expression of the indicated differentiation marker genes in the conditions of A in PE9 and PE15-2 EBs was examined by quantitative RT-PCR. Expression of Etv4 and Etv5 mRNA was only shown in PE9 ES cells. All samples were analyzed in triplicate, and the data were normalized to Gapdh expression. Error bars, S.D.
Gene Expression Signatures of Etv4/5 dKO ES Cells
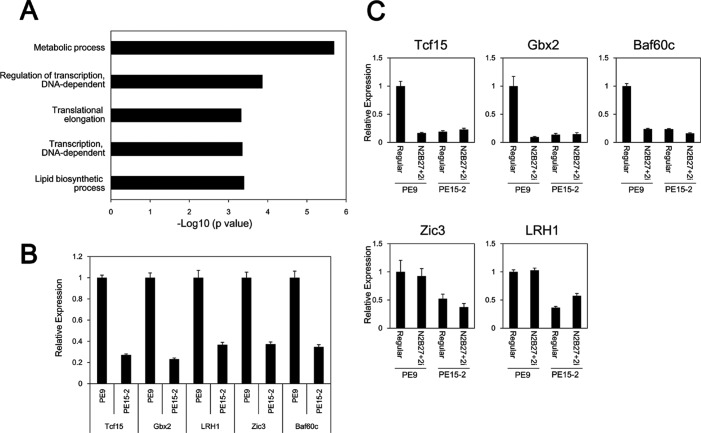
Recent studies indicate that gene regulatory and/or transcription factor networks are involved in determining the characteristics of ES cells. Because Etv4/5 dKO ES cells showed reduced proliferation and an impaired ability to induce an ectoderm marker, it is possible that the networks are changed in part. We compared the transcriptional profiles of wild-type and Etv4/5 dKO ES cells using oligonucleotide-based DNA microarray analysis. Pathway analysis showed that several pathways were significantly changed (supplemental Table S2). Furthermore, gene ontology (GO) term analysis revealed that genes involved in metabolic processes; regulation of transcription, DNA-dependent; translation elongation; transcription, DNA-dependent; and lipid biosynthetic were down-regulated in Etv4/5 dKO ES cells (Fig. 6A). Of note, gene expression-related GO terms (regulation of transcription, DNA-dependent; translation elongation; and transcription, DNA-dependent) were enriched. Genes with the GO term, “regulation of transcription, DNA-dependent,” are listed in Table 1. Several stem cell-related transcription factors, including Tcf15, Lrh1/Nr5a2, Gbx2, Baf60c/Smarcd3, and Zic3, were repressed in Etv4/5 dKO ES cells. Reduced expression of these genes in Etv4/5 dKO ES cells was validated by quantitative RT-PCR (Fig. 6B), indicating that the transcriptional network is impaired in Etv4/5 dKO ES cells. We also examined the expression of these genes in the 2i medium. In wild-type ES cells, expression of Tcf15, Gbx2, and Baf60c was repressed, whereas Zic3 and Lrh1 were unchanged, indicating that Tcf15, Gbx2, and Baf60c are dispensable for the native status of ES cells (Fig. 6C). In Etv4/5 dKO ES cells, the expression of these genes was still repressed compared with wild-type ES cells, suggesting that repressed genes in the dKO ES cells are not recovered by 2i medium.
FIGURE 6.
Several transcription factors are down-regulated in Etv4/5 dKO ES cells. A, GO term analysis of down-regulated candidates in Etv4/5 dKO ES cells. Genes involved in metabolic processes; regulation of transcription, DNA-dependent; translation elongation; transcription, DNA-dependent; and lipid biosynthetic are enriched significantly. GO categories are ranked according to their p value. B, expression of several transcription-related genes in PE9 and PE15-2 ES cells. Expression of transcription-related genes, including Tcf15, Gbx2, Lrh1, Zic3, and Baf60c, is down-regulated in PE15-2 ES cells, as shown by microarray analysis. Expression of the indicated genes in PE9 and PE15-2 ES cells was examined by quantitative RT-PCR. C, expression of several transcription-related genes in PE9 and PE15-2 ES cells in the 2i condition. PE9 and PE15-2 ES cells were cultured in the 2i condition, and expression of the indicated genes was examined by quantitative RT-PCR. All samples were analyzed in triplicate, and the data were normalized to Gapdh expression. Error bars, S.D.
TABLE 1.
Down-regulated genes (GO:0006355; regulation of transcription, DNA-dependent) in Etv4/5 dKO ES cells
Genes whose expression difference is log2 (ratio (PE15-2/PE9)) < −2.0 are listed.
| Symbol | Description | Relative -fold changes, log2 |
|---|---|---|
| Zfp386 | Zinc finger protein 386 (Kruppel-like) | −4.02 |
| Tcf15 | Transcription factor 15 | −3.64 |
| Trps1 | Trichorhinophalangeal syndrome I (human) | −3.34 |
| Lrh1/Nr5a2 | Nuclear receptor subfamily 5, group A, member 2 | −3.31 |
| Gbx2 | Gastrulation brain homeobox 2 | −3.29 |
| Bcl11a | B-cell CLL/lymphoma 11A (zinc finger protein) | −2.98 |
| Nkx6-3 | NK6 transcription factor related, locus 3 (Drosophila) | −2.94 |
| Baf60c/Smarcd3 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 3 | −2.58 |
| Dbp | D site albumin promoter binding protein | −2.51 |
| Rhox10 | −2.43 | |
| Tcea3 | Transcription elongation factor A (SII), 3 | −2.42 |
| Suhw3 | Suppressor of hairy wing homolog 3 (Drosophila) | −2.22 |
| Nkx6-2 | NK6 transcription factor related, locus 2 (Drosophila) | −2.21 |
| Zic3 | Zinc finger protein of the cerebellum 3 | −2.19 |
| Zfp459 | Zinc finger protein 459 | −2.11 |
| A930001N09Rik | RIKEN cDNA A930001N09 gene | −2.04 |
| Spic | Spi-C transcription factor (Spi-1/PU.1-related) | −2.03 |
| Supt16h | Suppressor of Ty 16 homolog (Saccharomyces cerevisiae) | −2.01 |
Both ETV4 and ETV5 Are Able to Rescue the Epithelial-like Morphology and Expression of Tcf15 and Gbx2 in dKO ES Cells
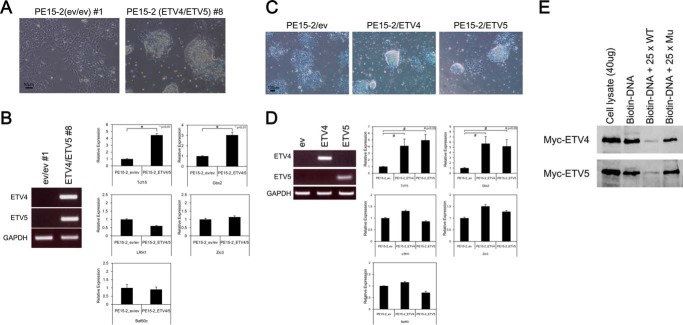
Next, we established ETV4/5-expressing PE15-2 ES cells, where exogenous ETV4 and ETV5 were stably expressed in Etv4/5 dKO ES cells. As shown in Fig. 7A, artificial expression of ETV4 and ETV5 in the dKO ES cells resulted in compact colony formation, in contrast to the control cells, which remained flat in shape. When we examined gene expression in these cells, expression of Tcf15 and Gbx2, which are known to regulate ES cell characteristics (see “Discussion”), was recovered in the ETV4/5-expressing PE15-2 ES cells (Fig. 7B).
FIGURE 7.
Both ETV4 and ETV5 are able to rescue the epithelial like morphology and expression of Tcf15 and Gbx2 in dKO ES cells. A, overexpression of both ETV4 and ETV5 in PE15-2 (Etv4/5 dKO) ES cells. ETV4/5-expressing PE15-2 ES cells and control cells were established as described under “Experimental Procedures.” Morphological signatures were observed. Scale bar, 50 μm. B, induction of Tcf15 and Gbx2 expression in ETV4/5-expressing PE15-2 ES cells. Expression of Tcf15 and Gbx2 mRNA, as well as Lrh1, Zic3, and Baf60c mRNA, in the PE15-2 ES cell lines was examined by quantitative RT-PCR. All samples were analyzed in triplicate, and the data were normalized to Gapdh expression. Expression of Etv4 and Etv5 mRNA was examined by regular RT-PCR. C, overexpression of either ETV4 or ETV5 in PE15-2 (Etv4/5 dKO) ES cells. ETV4- or ETV5-expressing PE15-2 ES cells and control cells were established as described under “Experimental Procedures.” Morphological signatures were observed. Scale bar, 100 μm. D, induction of Tcf15 and Gbx2 expression in ETV4- and ETV5-expressing PE15-2 ES cells. Expression of Tcf15 and Gbx2 mRNA as well as Lrh1, Zic3, and Baf60c mRNA in the PE15-2 ES cell lines was examined by quantitative RT-PCR. All samples were analyzed in triplicate, and the data were normalized to Gapdh expression. Expression of Etv4 and Etv5 mRNA was examined by regular RT-PCR. E, ETV4 and ETV5 directly bind to the putative binding sequences of the Gbx2 gene. Biotin-labeled oligonucleotide containing the putative binding site was incubated with cell extracts from Myc-ETV4- or Myc-ETV5-transfected HEK293 cells either with or without 25-fold non-labeled WT or mutated (Mu) oligonucleotide. The result shown is representative of three separate experiments. Error bars, S.D.
Because redundant activities of ETV4 and ETV5 have been reported, we examined whether single overexpression of either ETV4 or ETV5 in PE15-2 ES cells is enough to recover the morphology and gene expression of Tcf15 and Gbx2. Etv4/5 dKO ES cells were transfected with either an ETV4 or ETV5 expression vector, and the expression of the dysregulated genes in Etv4/5 dKO ES cells was examined. As shown in Fig. 7C, single overexpression of either ETV4 or ETV5 in dKO ES cells resulted in the cells forming a compact colony. Also, expression of Tcf15 and Gbx2 was recovered in these cells (Fig. 7D). The Gbx2 promoter contains the ETV4/5 binding site (from −1283 to −1266; transcription start site considered as +1). An association of the consensus sequences and either ETV4 or ETV5 was observed by a biotin-labeled DNA pull-down assay. Either Myc-tagged ETV4 or Myc-tagged ETV5 was precipitated with a biotin-labeled oligonucleotide contacting the ETV4/5-binding site of the Gbx2 gene, and the association was competed by an excess of unlabeled oligonucleotide but not by unlabeled mutated oligonucleotide (Fig. 7E). Taken together, these results suggest that the morphological phenotype of Etv4/5 dKO ES cells is rescued by either ETV4 or ETV5, and Gbx2 could be a downstream target of ETV4 and ETV5 in ES cells.
Discussion
ETV4 and ETV5 play crucial roles during organogenesis and morphogenesis. ETV4 is important for neuronal development, evidenced by the fact that Etv4-deficient mice have defects in neuromuscular innervation (53). Also, Etv4-deficient male mice fail to reproduce, probably because of dysfunctions in erection or ejaculation (54). Phenotypes of Etv5-deficient mice vary depending on the gene targeting constructions. Mice homozygous for a deletion of exons 2–5 of the Etv5 gene are viable but display disruptions of spermatogenesis in males and functional disorders of the ovary in females (55, 56). A much more severe phenotype occurs when exons 11 and 12 of the Etv5 gene are deleted (the same as our Etv4/5 dKO ES cell construct). In that case, the homozygotes are embryonic lethal at embryonic day 8.5 (41). In addition, ETV4 and ETV5 have essential functions for kidney development. A single knockout of either the Etv4 or Etv5 gene had little effect on mouse kidney development, whereas kidney development failed completely in Etv4/5 dKO mice (41, 44). Because our results show that expression patterns of ectoderm markers differ between wild-type and dKO ES cells in an EB assay, ETV4 and ETV5 are probably involved in early embryogenesis as well as organogenesis.
Etv4 and Etv5 are downstream targets of GDNF/Ret signaling in kidney development (41, 44) and FGF signaling in limb development (57). Here, we show that Etv4 and Etv5 are expressed in undifferentiated ES cells and that their expression is down-regulated after repression of Oct3/4. Because OCT3/4 is the major transcription factor in ES cells, it seems reasonable to suggest that ETV4/5 is involved in the stemness of ES cells. In fact, although Etv4/5 dKO ES cells expressed self-renewal marker genes, including Oct3/4 and Nanog, the cells failed to form a compact colony, which is typical of undifferentiated ES cells. Furthermore, reduction of Nanog, Esrrb, and Dax1 in Etv4/5 dKO was mild at −LIF 3 days (Fig. 2B), and expression of Oct3/4 and Nanog increased in Etv4/5 dKO ES cells cultured with the 2i condition (Fig. 3C). Molecular mechanisms of the phenotype remain to be clarified. Of note, the expression of several stem cell-related genes (Tcf15, Lrh1/Nr5a2, Gbx2, Baf60c/Smarcd3, and Zic3) was repressed, whereas the expression of differentiation-associated genes (Olig1, Olig2, Gata2, and Hand1) was enhanced in the cells from microarray analysis (data not shown). The self-renewal capacity and pluripotency of ES cells is thought to be controlled by gene regulatory networks, of which OCT3/4 and NANOG serve as master regulators. ETV5 is one of a large number of additional transcription factors in the network (33). Our investigation revealed that ETV4/5 are dispensable for self-renewal ability. However, induction of ectodermal gene expression was impaired in Etv4/5 dKO ES cells. Therefore, we conclude that Etv4/5 dKO ES cells are not completely differentiated and instead remain between an undifferentiated and a differentiated state or prodifferentiated state. Alternatively, because ES cells are thought to be a heterogeneous population, the Etv4/5 dKO ES cell line may represent a mixed population of undifferentiated and differentiated cells.
Overexpression of both ETV4 and ETV5, as well as single overexpression of either ETV4 or ETV5, in dKO ES cells led to morphological changes (formation into a compact colony) and reinduction of Tcf15 and Gbx2 expression, suggesting a redundant function of ETV4 and ETV5 in ES cells. The proliferation ratio was almost comparable between ETV4/5-expressing dKO ES cells and empty vector-transfected dKO ES cells (data not shown). Because the expression of exogenous Etv4 and Etv5 was driven by the CAG promoter, a strict regulation by their native promoters may be required for complete rescue of the phenotype of dKO ES cells.
Our analysis indicates that Tcf15 and Gbx2 are target genes of ETV4 and ETV5. TCF15 is a basic helix-loop-helix transcription factor and thought to be a key regulator of mesoderm differentiation (58). Furthermore, somite formation is disrupted in Tcf15-deficient mice (59). TCF15 primes pluripotent cells for entry to the somatic lineage, and forced expression of TCF15 in ES cells suppresses endodermal differentiation (60). Gbx2 is a homeobox gene and downstream target of LIF/STAT3 in ES cells (61). GBX2-overexpressing ES cells are able to maintain the undifferentiated state in the absence of LIF. Stat3-deficient ES cells differentiate even in the presence of LIF, although the cells are able to maintain the undifferentiated state by treatment with 2i (46). Importantly, GBX2 suffices to maintain the undifferentiated state in Stat3-deficient ES cells without 2i, suggesting that GBX2 is a regulator of ES cell self-renewal. Although Tcf15 and Gbx2 are downstream genes of ETV4 and ETV5 and could be involved in determining ES cell properties, these two molecules were insufficient to rescue the phenotypes of Etv4/5 dKO ES cells. We also overexpressed GBX2 in Etv4/5 dKO ES cells, but it appeared to have cytotoxic activity, and thus ES cell colonies were not obtained (data not shown). Overexpression of TCF15 in Etv4/5 dKO ES cells led to compact colony formation, but cell proliferation as well as expression of other repressed genes, including Zic3 and Gbx2, were not recovered (data not shown). Therefore, the compensatory factors for ETV4 and ETV5 remain to be clarified.
ETV4 and ETV5 are overexpressed in various tumors, including those affecting the breast, ovary, endometrium, prostate, and esophagus, and are involved in prognosis (40, 42). Ectopic forced expression of ETV4 in breast cancer cells promotes proliferation of MDA 231 breast cancer cells with induction of cyclin D3 expression (62), and functional inhibition of ETV4 or ETV5 by RNAi in mouse mammary tumor cells reduces cell proliferation with decreased expression of cyclin D1 and D2 (63). Proliferation of esophageal adenocarcinoma cells is also regulated by ETV4 (64). Our present data show reduced proliferation of Etv4/5 dKO ES cells. Expression levels of D-type and E-type cyclins were comparable between wild-type and Etv4/5 dKO ES cells from the microarray analysis; however, expression of p16/p19, p15, and p57 was significantly enhanced in Etv4/5 dKO ES cells. Indeed, ectopic expression of either P15 or P16 in human ES cells resulted in an accumulation of cells in the G1 phase (65). Although the regulation mechanisms of p16/p19, p15, and p57 by ETV4 and ETV5 remain to be elucidated, ETV4 and ETV5 can be involved in the proliferation of various types of cells by controlling cell cycle regulators.
In summary, Etv4/5 dKO ES cells express OCT3/4 and NANOG, which are core factors of the gene regulatory network, allowing their undifferentiated status to be maintained. In contrast, expression of some stem cell-related genes, including Tcf15, Gbx2, Zic3, and Lrh1, was dysregulated in Etv4/5 dKO ES cells, indicating that the network of the cells is partially impaired, resulting in a reduction of proliferation and a lack of induction of ectodermal gene expression. Further studies, including clarification of protein interaction partners and characterization of downstream targets of ETV4/5, will be helpful for understanding the functions of ETV4/5 in mouse ES cells.
Author Contributions
T. A. designed, performed, and analyzed the study and wrote the paper. S. K. and F. C. provided materials and wrote the paper. K. U. and H. K. analyzed data. T. Y. designed and analyzed the study and wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Hitoshi Niwa (RIKEN Center for Developmental Biology, Japan) for ZHBTc4 ES cells and the Center for Biomedical Research and Education at Kanazawa University for use of the DNA sequencer. We are also grateful to Milla Mikkola (Biomedicum Stem Cell Center-Core Facility Services, University of Helsinki, Finland) and our laboratories for helpful discussions. We thank Enago for the English language review.
This work was supported by Grant-in-Aid for Scientific Research (B) 22370050 (to T. Y.) and for Young Scientists (B) 21790268 (to T. A.) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and the Takeda Science Foundation (to T. A.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1 and S2.
- ES
- embryonic stem
- GO
- gene ontology
- LIF
- leukemia inhibitory factor
- dKO
- double knock-out
- Tet
- tetracycline
- EB
- embryoid body
- BMP
- bone morphogenic protein.
References
- 1. Ying Q. L., Nichols J., Chambers I., Smith A. (2003) BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- 2. Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988) Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688–690 [DOI] [PubMed] [Google Scholar]
- 3. Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988) Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684–687 [DOI] [PubMed] [Google Scholar]
- 4. Niwa H., Ogawa K., Shimosato D., Adachi K. (2009) A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 460, 118–122 [DOI] [PubMed] [Google Scholar]
- 5. Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T., Yokota T. (1999) STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 18, 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe S., Umehara H., Murayama K., Okabe M., Kimura T., Nakano T. (2006) Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene 25, 2697–2707 [DOI] [PubMed] [Google Scholar]
- 7. Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 8. Niwa H., Miyazaki J., Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 9. Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S. H., Niwa H. (2007) Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- 11. Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- 12. Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 13. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 14. Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Göttgens B., Niwa H., Smith A. (2012) Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Festuccia N., Osorno R., Halbritter F., Karwacki-Neisius V., Navarro P., Colby D., Wong F., Yates A., Tomlinson S. R., Chambers I. (2012) Esrrb is a direct Nanog target gene that can substitute for Nanog function in pluripotent cells. Cell Stem Cell 11, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uranishi K., Akagi T., Sun C., Koide H., Yokota T. (2013) Dax1 associates with Esrrb and regulates its function in embryonic stem cells. Mol. Cell. Biol. 33, 2056–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khalfallah O., Rouleau M., Barbry P., Bardoni B., Lalli E. (2009) Dax-1 knockdown in mouse embryonic stem cells induces loss of pluripotency and multilineage differentiation. Stem Cells 27, 1529–1537 [DOI] [PubMed] [Google Scholar]
- 18. Sun C., Nakatake Y., Akagi T., Ura H., Matsuda T., Nishiyama A., Koide H., Ko M. S. H., Niwa H., Yokota T. (2009) Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol. Cell. Biol. 29, 4574–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fujii S., Nishikawa-Torikai S., Futatsugi Y., Toyooka Y., Yamane M., Ohtsuka S., Niwa H. (2015) Nr0b1 is a negative regulator of Zscan4c in mouse embryonic stem cells. Sci. Rep. 5, 9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elling U., Klasen C., Eisenberger T., Anlag K., Treier M. (2006) Murine inner cell mass-derived lineages depend on SaII4 function. Proc. Natl. Acad. Sci. U.S.A. 103, 16319–16324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakaki-Yumoto M., Kobayashi C., Sato A., Fujimura S., Matsumoto Y., Takasato M., Kodama T., Aburatani H., Asashima M., Yoshida N., Nishinakamura R. (2006) The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 133, 3005–3013 [DOI] [PubMed] [Google Scholar]
- 22. Zhang J., Tam W. L., Tong G. Q., Wu Q., Chan H. Y., Soh B. S., Lou Y., Yang J., Ma Y., Chai L., Ng H. H., Lufkin T., Robson P., Lim B. (2006) Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 8, 1114–1123 [DOI] [PubMed] [Google Scholar]
- 23. Lim L. S., Loh Y. H., Zhang W., Li Y., Chen X., Wang Y., Bakre M., Ng H. H., Stanton L. W. (2007) Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol. Biol. Cell 18, 1348–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang J., Chan Y. S., Loh Y. H., Cai J., Tong G. Q., Lim C. A., Robson P., Zhong S., Ng H. H. (2008) A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353–360 [DOI] [PubMed] [Google Scholar]
- 25. Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. (2005) LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132, 885–896 [DOI] [PubMed] [Google Scholar]
- 26. Singh A. M., Dalton S. (2009) The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 5, 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hishida T., Nozaki Y., Nakachi Y., Mizuno Y., Okazaki Y., Ema M., Takahashi S., Nishimoto M., Okuda A. (2011) Indefinite self-renewal of ESCs through Myc/Max transcriptional complex-independent mechanisms. Cell Stem Cell 9, 37–49 [DOI] [PubMed] [Google Scholar]
- 28. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 30. Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., Lipovich L., Kuznetsov V. A., Robson P., Stanton L. W., Wei C. L., Ruan Y., Lim B., Ng H. H. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- 31. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharov A. A., Masui S., Sharova L. V., Piao Y., Aiba K., Matoba R., Xin L., Niwa H., Ko M. S. H. (2008) Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics 9, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou Q., Chipperfield H., Melton D. A., Wong W. H. (2007) A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 104, 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010) An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van den Berg D. L. C., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 37. Kim J., Woo A. J., Chu J., Snow J. W., Fujiwara Y., Kim C. G., Cantor A. B., Orkin S. H. (2010) A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ben-Porath I., Thomson M. W., Carey V. J., Ge R., Bell G. W., Regev A., Weinberg R. A. (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clarke M. F., Fuller M. (2006) Stem cells and cancer: two faces of Eve. Cell 124, 1111–1115 [DOI] [PubMed] [Google Scholar]
- 40. de Launoit Y., Baert J. L., Chotteau-Lelievre A., Monte D., Coutte L., Mauen S., Firlej V., Degerny C., Verreman K. (2006) The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta 1766, 79–87 [DOI] [PubMed] [Google Scholar]
- 41. Lu B. C., Cebrian C., Chi X., Kuure S., Kuo R., Bates C. M., Arber S., Hassell J., MacNeil L., Hoshi M., Jain S., Asai N., Takahashi M., Schmidt-Ott K. M., Barasch J., D'Agati V., Costantini F. (2009) Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat. Genet. 41, 1295–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oh S., Shin S., Janknecht R. (2012) ETV1, 4 and 5: an oncogenic subfamily of ETS transcription factors. Biochim. Biophys. Acta 1826, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W. 3rd, Su A. I. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuure S., Chi X., Lu B., Costantini F. (2010) The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development. Development 137, 1975–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yoshida-Koide U., Matsuda T., Saikawa K., Nakanuma Y., Yokota T., Asashima M., Koide H. (2004) Involvement of Ras in extraembryonic endoderm differentiation of embryonic stem cells. Biochem. Biophys. Res. Commun. 313, 475–481 [DOI] [PubMed] [Google Scholar]
- 46. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Niwa H., Yamamura K., Miyazaki J. (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 [DOI] [PubMed] [Google Scholar]
- 48. Ura H., Usuda M., Kinoshita K., Sun C., Mori K., Akagi T., Matsuda T., Koide H., Yokota T. (2008) STAT3 and Oct-3/4 control histone modification through induction of Eed in embryonic stem cells. J. Biol. Chem. 283, 9713–9723 [DOI] [PubMed] [Google Scholar]
- 49. Akagi T., Usuda M., Matsuda T., Ko M. S. H., Niwa H., Asano M., Koide H., Yokota T. (2005) Identification of Zfp-57 as a downstream molecule of STAT3 and Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 331, 23–30 [DOI] [PubMed] [Google Scholar]
- 50. Sun C., Nakatake Y., Ura H., Akagi T., Niwa H., Koide H., Yokota T. (2008) Stem cell-specific expression of Dax1 is conferred by STAT3 and Oct3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 372, 91–96 [DOI] [PubMed] [Google Scholar]
- 51. Chen X., Fang F., Liou Y. C., Ng H. H. (2008) Zfp143 regulates Nanog through modulation of Oct4 binding. Stem Cells 26, 2759–2767 [DOI] [PubMed] [Google Scholar]
- 52. Nichols J., Smith A. (2009) Naive and primed pluripotent states. Cell Stem Cell 4, 487–492 [DOI] [PubMed] [Google Scholar]
- 53. Livet J., Sigrist M., Stroebel S., De Paola V., Price S. R., Henderson C. E., Jessell T. M., Arber S. (2002) ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron 35, 877–892 [DOI] [PubMed] [Google Scholar]
- 54. Laing M. A., Coonrod S., Hinton B. T., Downie J. W., Tozer R., Rudnicki M. A., Hassell J. A. (2000) Male sexual dysfunction in mice bearing targeted mutant alleles of the PEA3 ets gene. Mol. Cell. Biol. 20, 9337–9345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen C., Ouyang W., Grigura V., Zhou Q., Carnes K., Lim H., Zhao G. Q., Arber S., Kurpios N., Murphy T. L., Cheng A. M., Hassell J. A., Chandrashekar V., Hofmann M. C., Hess R. A., Murphy K. M. (2005) ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436, 1030–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Eo J., Shin H., Kwon S., Song H., Murphy K. M., Lim J. H. (2011) Complex ovarian defects lead to infertility in Etv5−/− female mice. Mol. Hum. Reprod. 17, 568–576 [DOI] [PubMed] [Google Scholar]
- 57. Mao J., McGlinn E., Huang P., Tabin C. J., McMahon A. P. (2009) Fgf-dependent Etv4/5 activity is required for posterior restriction of sonic hedgehog and promoting outgrowth of the vertebrate limb. Dev. Cell 16, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blanar M. A., Crossley P. H., Peters K. G., Steingrímsson E., Copeland N. G., Jenkins N. A., Martin G. R., Rutter W. J. (1995) MESO1, a basic-helix-loop-helix protein involved in mammalian presomitic mesoderm development. Proc. Natl. Acad. Sci. U.S.A. 92, 5870–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Burgess R., Rawls A., Brown D., Bradley A., Olson E. N. (1996) Requirement of the paraxis gene for somite formation and musculoskeletal patterning. Nature 384, 570–573 [DOI] [PubMed] [Google Scholar]
- 60. Davies O. R., Lin C.-Y., Radzisheuskaya A., Zhou X., Taube J., Blin G., Waterhouse A., Smith A. J. H., Lowell S. (2013) Tcf15 primes pluripotent cells for differentiation. Cell Rep. 3, 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tai C.-I., Ying Q.-L. (2013) Gbx2, a LIF/Stat3 target, promotes reprogramming to and retention of the pluripotent ground state. J. Cell Sci. 126, 1093–1098 [DOI] [PubMed] [Google Scholar]
- 62. Jiang J., Wei Y., Liu D., Zhou J., Shen J., Chen X., Zhang S., Kong X., Gu J. (2007) E1AF promotes breast cancer cell cycle progression via upregulation of Cyclin D3 transcription. Biochem. Biophys. Res. Commun. 358, 53–58 [DOI] [PubMed] [Google Scholar]
- 63. Firlej V., Ladam F., Brysbaert G., Dumont P., Fuks F., de Launoit Y., Benecke A., Chotteau-Lelievre A. (2008) Reduced tumorigenesis in mouse mammary cancer cells following inhibition of Pea3- or Erm-dependent transcription. J. Cell Sci. 121, 3393–3402 [DOI] [PubMed] [Google Scholar]
- 64. Keld R., Guo B., Downey P., Gulmann C., Ang Y. S., Sharrocks A. D. (2010) The ERK MAP kinase-PEA3/ETV4-MMP-1 axis is operative in oesophageal adenocarcinoma. Mol. Cancer 9, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ruiz S., Panopoulos A. D., Herrerías A., Bissig K. D., Lutz M., Berggren W. T., Verma I. M., Izpisua Belmonte J. C. (2011) A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr. Biol. 21, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yeom Y. I., Fuhrmann G., Ovitt C. E., Brehm A., Ohbo K., Gross M., Hübner K., Schöler H. R. (1996) Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122, 881–894 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.