Background: MeCP2 is a key nuclear protein that binds methylated DNA to regulate the chromatin state.
Results: KPNA3 and KPNA4 bind to overlapping domains within MeCP2 to traffic it into the nucleus.
Conclusion: KPNA3 permits the nuclear localization of truncated forms of MeCP2, reconciling discrepancies between predictions and experimental observations.
Significance: Understanding the mechanism of MeCP2 nuclear import is critical for analyzing genotype-phenotype correlations in patients with Rett syndrome.
Keywords: autism, DNA methylation, epigenetics, nuclear transport, nucleus, MeCP2, Rett syndrome, genotype-phenotype
Abstract
Methyl-CpG binding protein 2 (MeCP2) is a nuclear protein with important roles in regulating chromatin structure and gene expression, and mutations in MECP2 cause Rett syndrome (RTT). Within the MeCP2 protein sequence, the nuclear localization signal (NLS) is reported to reside between amino acids 255–271, and certain RTT-causing mutations overlap with the MeCP2 NLS, suggesting that they may alter nuclear localization. One such mutation, R270X, is predicted to interfere with the localization of MeCP2, but recent in vivo studies have demonstrated that this mutant remains entirely nuclear. To clarify the mechanism of MeCP2 nuclear import, we isolated proteins that interact with the NLS and identified karyopherin α 3 (KPNA3 or Kap-α3) and karyopherin α 4 (KPNA4 or Kap-α4) as key binding partners of MeCP2. MeCP2-R270X did not interact with KPNA4, consistent with a requirement for an intact NLS in this interaction. However, this mutant retains binding to KPNA3, accounting for the normal localization of MeCP2-R270X to the nucleus. These data provide a mechanism for MeCP2 nuclear import and have implications for the design of therapeutics aimed at modulating the function of MeCP2 in RTT patients.
Introduction
RTT3 is caused by mutations in the X-linked gene MECP2 (1). Affected patients develop a neurological disorder starting between 6 and 18 months of age (2). Clinical features include loss of acquired speech and motor skills, microcephaly, stereotypic activity, seizures, and severe cognitive deficits. Since the initial identification of MECP2 mutations in RTT, a large number of causative alleles have been identified (3). These include truncating and missense mutations, many of which cluster within two major domains of the MeCP2 protein: the methyl-CpG binding domain and the transcriptional repression domain (3). Certain truncating mutations within the transcriptional repression domain are associated with a poorer clinical outcome. One of these mutations, R270X, is among the most commonly found in RTT patients, accounting for ∼6% of all cases (RettBASE). This mutation is located within the MeCP2 nuclear localization signal (NLS), which maps to amino acids 255–271 (4). When compared with more C-terminal mutations, R270X has been associated with poorer ambulation, earlier development of stereotypies, more severe social deficits, and earlier mortality (5–8). Because it disrupts the NLS, the R270X mutation has been predicted to prevent normal nuclear localization of MeCP2 (7). However, we and others have shown that this mutant form of the protein (MeCP2-R270X) localizes entirely to the nucleus both in cultured cells and in vivo (9, 10). How this mutant protein traffics into the nucleus without the fully intact NLS is currently unknown.
The MeCP2 NLS belongs to a family of targeting sequences termed classical nuclear localization signals (cNLSs) (11). cNLSs occur in two general patterns, monopartite and bipartite. Both are comprised of short sequences of basic amino acids. Monopartite cNLSs occur as a single stretch of amino acids conforming to the consensus K(K/R)X(K/R) (12). Bipartite cNLSs typically consist of an N-terminal sequence of at least two basic amino acids, followed by a stretch of 10–12 variable amino acids and then a second stretch where at least three of the following five amino acids are basic (12). Structural studies have revealed that both types of cNLSs are recognized by members of the importin-α/karyopherin-α (KPNA) family by direct interaction with these basic amino acids (13). In humans, the KPNA family consists of seven members that can be divided into three subfamilies sharing structural and functional overlap: the α1 family (KPNA1/5/6), the α2 family (KPNA2/7), and the α3 family (KPNA3/4) (14). KPNA proteins act as adaptors, forming a bridging interaction between substrate cargo and the karyopherin-β·Ran complex, which mediates import into the nucleus (15).
The initial characterization of the MeCP2 NLS was performed by deletion mapping (4). This identified a consensus bipartite cNLS that was needed for nuclear import (amino acids 255RKAEADPQAIPKKRGRK271). However, the protein factor(s) that bind(s) to this motif remain unknown. Because MeCP2-R270X localizes normally to the nucleus, we hypothesized that components of the nuclear import machinery mediate the nuclear import of MeCP2-R270X, similar to the wild-type protein. We set out to identify the transport proteins that ferry MeCP2 into the nucleus and their binding sites within the MeCP2 primary sequence. We found that both KPNA3 and KPNA4 bind to the canonical MeCP2 NLS. Interestingly, the binding of MeCP2 to KPNA3 is disrupted, but not abolished, by the R270X mutation. These data provide a mechanism for MeCP2 nuclear import and are important to consider when designing therapies for patients with mutations in MeCP2 that may alter nuclear localization.
Experimental Procedures
Generation of Vectors
The full-length human MECP2-e2 coding sequence was cloned into pENTR using the Gateway cloning system as described previously (16). The MECP2 cDNA was then tagged on the N terminus with GST by subcloning into pDEST27 or on the C terminus with GFP by subcloning into pDEST47. The human MECP2-e1 coding sequence was cloned into pHAGE-mRFP using standard restriction cloning. Subsequent mutants were generated by recombination using the QuikChange XL site-directed mutagenesis kit according to the instructions of the manufacturer (Stratagene). A list of primers can be found in supplemental Table 1. In brief, for C-terminal GFP-tagged constructs, N-terminal truncations were generated by substituting an initiator methionine and alanine in the context of a Kozak consensus for the deleted amino acids, including the indicated codon position (ΔN-248, deletes amino acids 1–248). For C-terminal truncations, conventional notation is followed, e.g. R270X indicates that amino acids 270–486 are deleted and that amino acid 269 is directly fused to a linker, followed by GFP. For N-terminally tagged GST constructs, GST was directly fused to a linker, followed by the indicated amino acids. For the ΔNLS construct, amino acids 255–271 (inclusive) were deleted. For the in vitro interaction assay, human KPNA3 and KPNA4 cDNAs were subcloned into pGEX-5×3 for bacterial expression of N-terminally tagged GST fusions. For generation of recombinant full-length human MeCP2, pTXB1-MECP2 was used as described previously (10).
Live Imaging
Neuro-2a (N2a) cells were plated in Lab-Tek chambered coverglass (Nunc) and cultured at 37 °C and 5% CO2 for 36–48 h before imaging. Cells were transfected with MeCP2 constructs tagged at the C terminus with GFP or tagged at the N terminus with RFP using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. For fluorescent protein only controls, cells were transfected with pEGFP-C1 or empty pHAGE-mRFP vector. Cells were treated with Hoechst 33342 at 5 μg/ml for 20 min at 37 °C and imaged using a Leica SP5 confocal microscope. Image data were processed using ImageJ software.
GST Affinity Purification
Deletion constructs tagged with N-terminal GST were transfected in N2a cells as indicated in Fig. 2 using Lipofectamine 2000. Cells were cultured at 37 °C and 5% CO2 for 36–48 h before harvesting. Cells were collected in ice-cold PBS and pelleted in a tabletop centrifuge. The cell pellet was then lysed by rotation in 500 μl of ice-cold cell lysis buffer (0.5% Nonidet P-40, 20 mm Tris Cl (pH 8.0), 180 mm NaCl, 1 mm EDTA, and Complete protease inhibitor (Roche)) for 15 min. The lysate was cleared by spinning at maximum speed in a tabletop centrifuge, and the cleared lysate (input) was added to prewashed glutathione-Sepharose 4B beads (GE Healthcare) for 1–3 h. Beads were then washed five times in cell lysis buffer, and bound complexes were eluted by boiling in 2× Laemmli buffer prior to loading on NuPAGE 4–12% BisTris gels followed by electrophoresis. Gels were then silver-stained to visualize coprecipitated components. For mass spectrometry, samples were prepared as above, except that the electrophoresed gel was stained with the Colloidal Blue staining kit (Invitrogen) according to the instructions of the manufacturer. The band indicated in Fig. 2 was then cut along with an adjacent control gel piece, and both pieces were submitted to the Baylor College of Medicine Mass Spectrometry Core for protein identification.
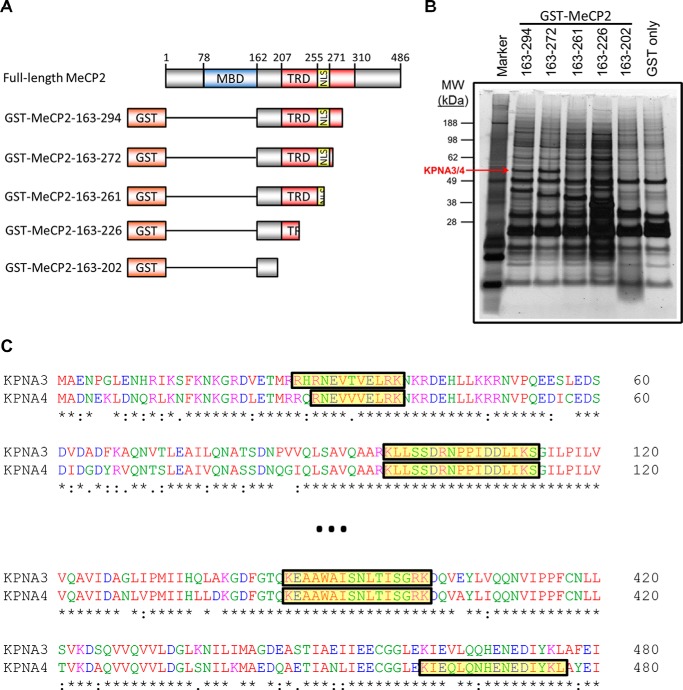
FIGURE 2.
Isolation of KPNA3 and KPNA4 by affinity purification with MeCP2. A, schematic of MeCP2 fusion constructs with GST used in affinity purification experiments. The amino acid numbers indicated on the left are inclusive. MBD, methyl-CpG binding domain (amino acids 78–162); TRD transcriptional repression domain (amino acids 207–310), NLS, the NLS (amino acids 255–271) as described in Ref. 4. The schematic is not to scale. B, N2a cells were transiently transfected with fragments of N-terminally GST-tagged MeCP2. Two days later, cells were lysed, and the MeCP2 fragments were isolated by affinity purification with glutathione beads. Bound proteins were eluted, separated by SDS-PAGE, and silver-stained. The band corresponding to KPNA3 and KPNA4 that was identified later by mass spectrometry is indicated (KPNA3/4). The amino acids included in each MeCP2 fragment are indicated above the corresponding lane. A protein marker is included in the first lane, and the corresponding molecular weights (MW) of the marker bands are indicated in kilodalton (n = 3). C, a partial Clustal Omega alignment of the mouse KPNA3 and KPNA4 protein sequences. Peptides identified by trypsin digestion and mass spectrometry of the band indicated in B are highlighted in yellow along the alignment.
Coimmunoprecipitation
Coimmunoprecipitation from cultured N2a cells was performed similar to GST affinity purification (above), except that MeCP2 was tagged at the C terminus with GFP and complexes were precipitated using protein G Dynabeads saturated with rabbit anti-GFP serum (Abcam, catalog no. ab290). Following electrophoresis, gels were transferred onto PVDF membranes and probed with goat anti-KPNA3 (1:2000, Abcam, catalog no. ab6038), rabbit anti-KPNA4 (1:1000, GeneTex, catalog no. GTX106325), affinity-purified rabbit anti-GFP (1:5000, Abcam, catalog no. ab6556), and mouse anti-GAPDH 6C5 (1:20,000, Advanced Immunochemicals, catalog no. 2-RGM2). Secondary antibodies were mouse anti-rabbit HRP (1:3000, Jackson ImmunoResearch Laboratories, catalog no. 211-032-171), donkey anti-mouse HRP (1:10,0000, Jackson ImmunoResearch Laboratories, 715-035-150), and donkey anti-goat HRP (1:100,000, Jackson ImmunoResearch Laboratories, 705-035-003).
For coimmunoprecipitation from mouse brain tissue, adult mice of the indicated genotype were sacrificed under anesthesia, and the brains were removed quickly. One hemibrain was lysed in ice-cold cell lysis buffer by Dounce homogenization and allowed to rotate for 15 min. Lysates were cleared by centrifugation at maximum speed in a tabletop centrifuge, and cleared lysates (input) were added to prewashed protein G Dynabeads saturated with rabbit anti-GFP serum (Abcam, catalog no. ab290). Complexes were allowed to bind at 4 °C for 45 min to 1 h prior to four washes in cell lysis buffer and elution by boiling in 2× Laemmli buffer. Bound complexes were analyzed by Western blot as described above.
In Vitro Interaction
Recombinant human MeCP2 was generated by Rosetta 2(DE3) Escherichia coli cells containing pTXB1-MECP2. Following 1 mm isopropyl 1-thio-β-d-galactopyranoside induction, the protein was purified using the IMPACT kit (New England Biolabs) according to the recommendations of the manufacturer. The protein was dialyzed into 0.33× PBS, and purity was assessed by Coomassie staining. Recombinant human KPNA3 and KPNA4 were isolated as above but using pGEX-5×3 vectors and purified on glutathione-Sepharose 4B beads (GE Healthcare). For the interaction assay, equal amounts of MeCP2 were added to glutathione-Sepharose 4B beads containing equimolar amounts of GST, GST-KPNA3, and GST-KPNA4. Samples were rotated at 4 °C for 1 h prior to five PBS washes and elution by boiling in 2× Laemmli buffer. Interaction was analyzed by Western blot as above but using rabbit antiserum raised against the MeCP2 N terminus (1:3000, Zoghbi laboratory, no. 0535) and rabbit anti-GST (1:2000, Sigma, catalog no. G7781).
Results
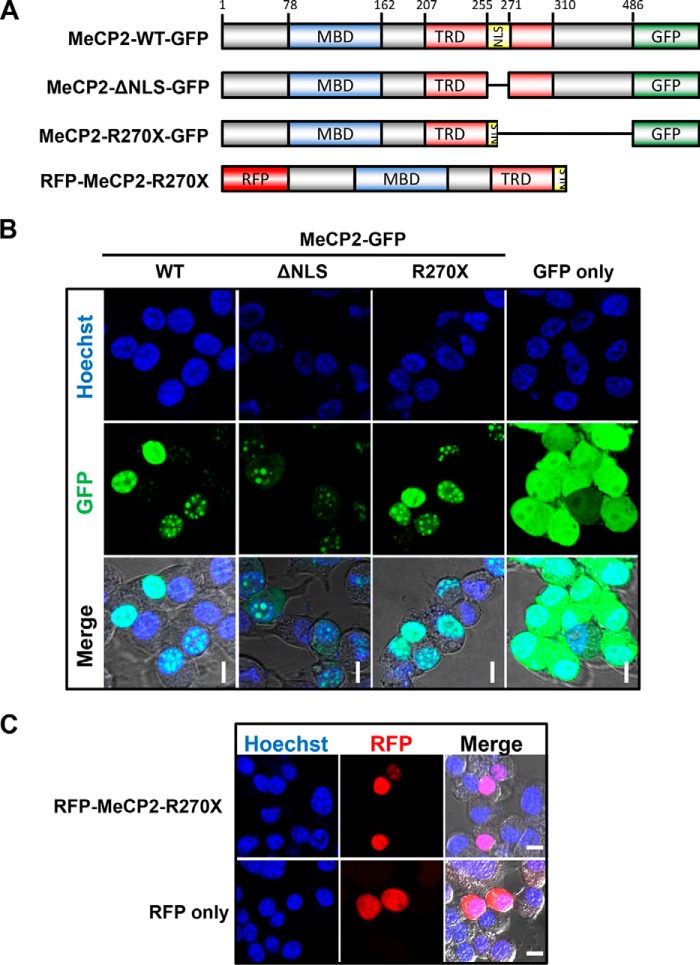
We began our study by live-imaging MeCP2 tagged with GFP in a mouse neuroblastoma cell line (N2a). Consistent with previous reports, the wild-type MeCP2 (MeCP2-WT) protein localized to the nucleus (Fig. 1, A and B). When we expressed MeCP2 with a deletion of the NLS sequence (amino acids 255–271, MeCP2-ΔNLS) we found that the protein was distributed between both the cytoplasm and the nucleus (Fig. 1B). We then expressed MeCP2 truncated at Arg270 (MeCP2-R270X) in N2a cells and found that it localized entirely to the nucleus, similar to MeCP2-WT (Fig. 1B). Importantly, GFP alone appeared diffuse throughout both the cytoplasm and nucleus (Fig. 1B). The MeCP2-R270X fusion construct used in these experiments featured a C-terminal GFP tag, whereas, in vivo, the R270X mutation would lead to an abrupt termination of the protein. To assess whether C-terminal fusion with GFP alters localization, we live-imaged MeCP2 with an N-terminal RFP tag instead (Fig. 1C). Similar to C-terminally tagged MeCP2, RFP-MeCP2-R270X localized exclusively to the nucleus, in contrast to RFP alone (Fig. 1C).
FIGURE 1.
The MeCP2 NLS is critical for nuclear localization, and MeCP2-R270X localizes exclusively to the nucleus. A, schematic of MeCP2 fusion constructs with GFP and RFP used in localization experiments. MBD, methyl-CpG binding domain (amino acids 78–162); TRD transcriptional repression domain (amino acids 207–310), NLS, the NLS (amino acids 255–271) as described in Ref. 4. The schematic is not to scale. B, N2a cells were transiently transfected with MeCP2 tagged on the C terminus with GFP and live-imaged using endogenous GFP excitation with Hoechst 33342 as a nuclear counterstain. Individual channels are indicated, and a merged image overlaid on the differential interference contrast (DIC) channel is shown to indicate nuclear and cytoplasmic boundaries. The MeCP2 variants used are indicated. ΔNLS, full-length MeCP2 with amino acids 255–271 deleted. Scale bars = 10 μm. n = 3. C, a similar experiment as described in B except that N2a cells were transfected with MeCP2 tagged at its N terminus with RFP. Scale bars = 10 μm. n = 3.
To identify the binding partners that interact with the region overlapping the NLS, we tagged MeCP2 with GST and affinity-purified fragments from N2a cells (Fig. 2A). Progressive truncation identified a single band that was present in longer fragments but was lost when the MeCP2 NLS was removed (Fig. 2B, compare lanes 3 and 4). This band was isolated from a gel and subjected to MS, which identified peptides corresponding to KPNA3 and KPNA4 (Fig. 2C).
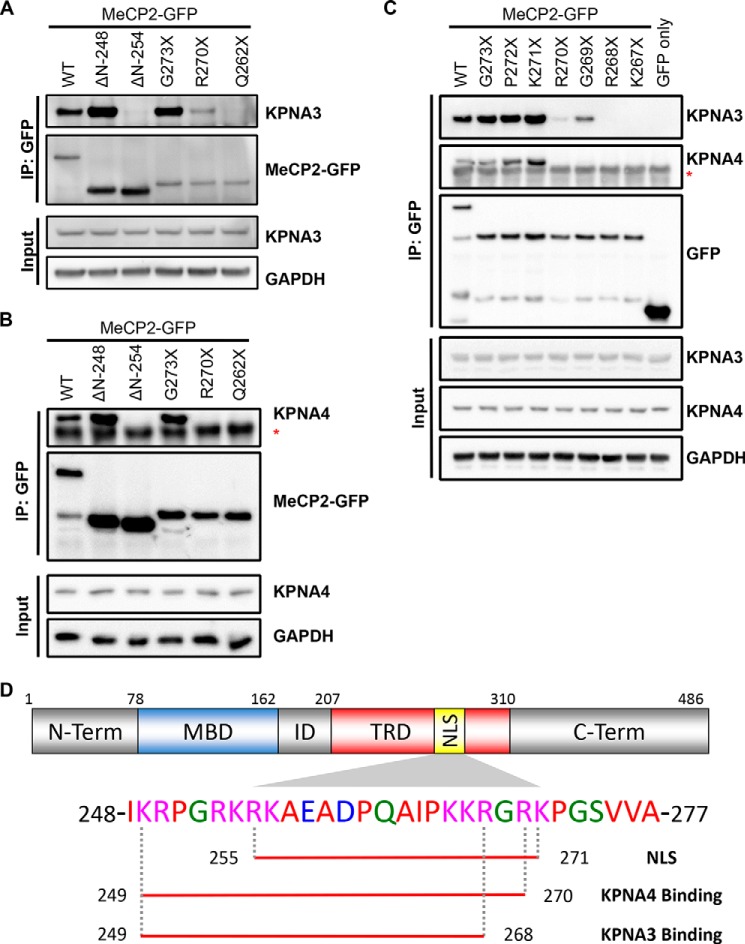
We then sought to map the portion of MeCP2 that interacts with endogenous KPNA3 and KPNA4 by coimmunoprecipitation from N2a cells. Initial deletion mapping indicated that only fragments of MeCP2 containing amino acids 249–272 demonstrated full binding to KPNA3 and KPNA4, consistent with the bipartite NLS described previously (Fig. 3, A and B). However, we noticed that the R270X mutant still retained partial binding to KPNA3 (Fig. 3A). This interaction, however, was not observed when MeCP2 was truncated at Gln262 (MeCP2-Q262X), suggesting that amino acids 262–269 are important for KPNA3 binding. Interestingly, KPNA4 was not bound by MeCP2-R270X, suggesting that KPNA4 requires Arg270 or, potentially, additional C-terminal amino acids for binding (Fig. 3B). To more precisely define the C-terminal extent of the KPNA3 and KPNA4 binding domains, we performed fine mapping by generating sequentially shorter truncations of the MeCP2 NLS (i.e. P272X, K271X, R270X, etc.). Coimmunoprecipitation with these MeCP2 mutants revealed that KPNA4 binding was lost when the NLS was truncated at or before Arg270 (Fig. 3C). In contrast, KPNA3 binding remained detectable until Arg268 was removed, although binding was reduced by deleting Arg270 and Gly269 (Fig. 3C). These data indicate that an NLS sequence consisting of amino acids 249–268, which is retained by MeCP2-R270X, is sufficient for reduced but functional binding to KPNA3, permitting exclusive localization of MeCP2 to the nucleus (Fig. 3D).
FIGURE 3.
Fine mapping of the KPNA3- and KPNA4-interacting domain within MeCP2. A and B, N2a cells were transiently transfected with the indicated GFP-tagged MeCP2 construct. Two days later, cells were lysed, and the MeCP2 fragments were precipitated with anti-GFP antibodies. Bound proteins were eluted and identified by Western blotting with the indicated antibodies, along with an aliquot of the input protein. Blots are shown for KPNA3 (A) and KPNA4 (B). ΔN, N-terminal deletion fragments where the coding sequence up to and including the amino acid listed is replaced with an initiator methionine; asterisk, location of the immunoglobulin heavy chain band below the KPNA4 band. n = 3 (KPNA3) and 2 (KPNA4). IP, immunoprecipitation. C, similar to the above experiments, the C-terminal extent of the KPNA3- and KPNA4-interacting domain was mapped along MeCP2 by coimmunoprecipitation from N2a cells. Asterisk, location of the immunoglobulin heavy chain band below the KPNA4 band. n = 3. D, schematic of MeCP2 indicating the location of the NLS (amino acids 255–271) as identified in Ref. 4. The amino acids required for interaction with KPNA3 and KPNA4 as determined in A–C are underlined below the corresponding sequence. ID, the interdomain between the methyl-CpG binding domain (MBD) and the transcriptional repression domain (TRD). N-term, N terminus; C-term, C terminus.
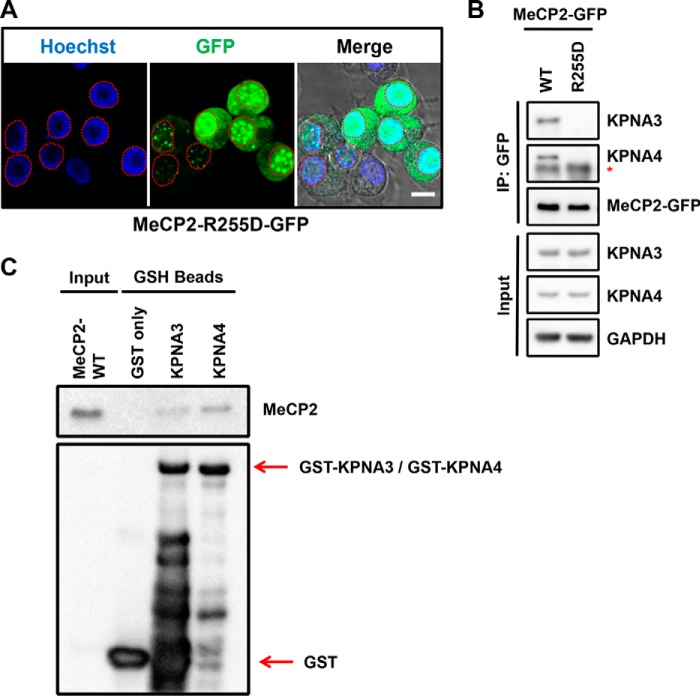
To assess whether KPNA3 and KPNA4 bind specifically to this region in the context of full-length MeCP2, we also tested a novel point mutation, R255D, which leads to the appearance of MeCP2 within the cytoplasm (Fig. 4A). We found that MeCP2-R255D also failed to interact with KPNA3 or KPNA4 (Fig. 4B). The fact that Arg255 is located within the MeCP2 NLS suggests that these proteins bind MeCP2 directly at this site. However, we considered that this interaction could be indirect. To test this possibility, we purified recombinant MeCP2-WT along with GST-tagged KPNA3 and KPNA4 from a bacterial source. We used these recombinant proteins to perform an in vitro interaction assay, which revealed that indeed both KPNA3 and KPNA4 bind directly to MeCP2 (Fig. 4C). Taken together with our deletion mapping, these data suggest that KPNA3 and KPNA4 interact exclusively with a small region of MeCP2 overlapping Arg255 (amino acids 249–268 and 249–270, respectively) and that this region is critical in the context of the full-length protein for exclusive nuclear localization.
FIGURE 4.
The MeCP2-R255D mutant disrupts nuclear localization and the interaction with KPNA3 and KPNA4. A, N2a cells were transiently transfected with a human MeCP2-R255D construct tagged on the C terminus with GFP and live-imaged using endogenous GFP excitation with Hoechst 33342 as a nuclear counterstain. Individual channels are indicated, and a merged image overlaid on the DIC channel is shown to indicate nuclear and cytoplasmic boundaries. A dashed red line indicates the nuclear outline of each transfected cell. Scale bars = 10 μm; n = 3. B, N2a cells were transiently transfected with either MeCP2 WT or MeCP2-R255D. Two days later, cells were lysed, and MeCP2 was isolated by precipitation with anti-GFP antibodies. Bound proteins were eluted and identified by Western blotting with the indicated antibodies, along with an aliquot of the input protein. Asterisk, the location of the immunoglobulin heavy chain band below the KPNA4 band. n = 3. C, GSH beads loaded with bacterially expressed GST-KPNA3, GST-KPNA4, or GST only were incubated with bacterially expressed MeCP2-WT in vitro. Bound proteins were eluted and identified by Western blotting with the indicated antibodies, along with an aliquot of input MeCP2 protein. n = 3.
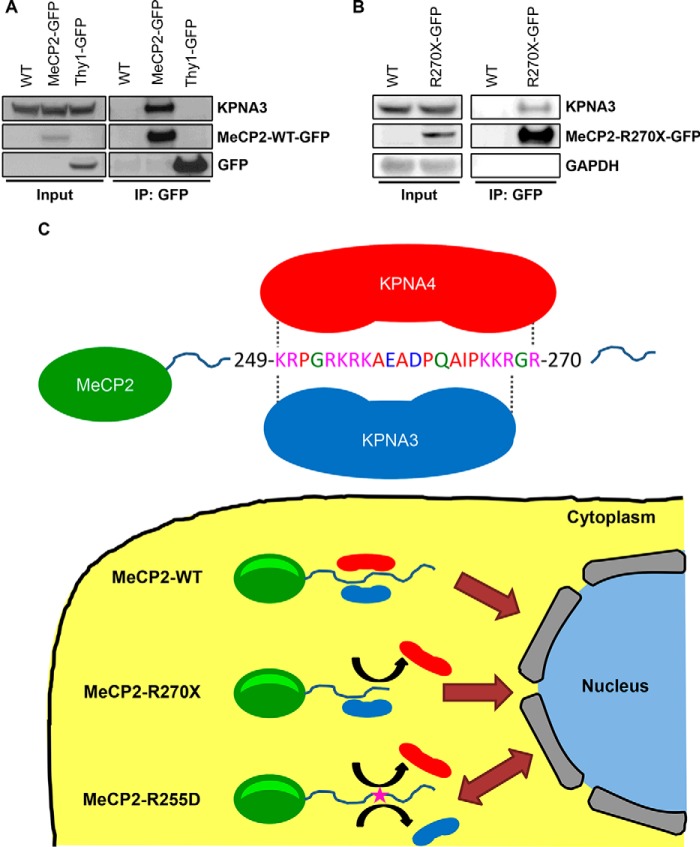
We next wanted to confirm that KPNA3 interacts with MeCP2 in vivo. We took advantage of a transgenic mouse line expressing a GFP-tagged MeCP2 within the brain (17). Immunoprecipitation of MeCP2 from mouse brain lysates revealed binding to KPNA3 (Fig. 5A). Importantly, transgenic mice expressing GFP alone (Thy1-GFP mice) (18) revealed no interaction of the tag with KPNA3 (Fig. 5A). Therefore, KPNA3 is a bona fide interacting partner of MeCP2 within the mouse brain. Given that MeCP2-R270X binding to KPNA3 was sufficient to confer exclusive nuclear localization in vitro, we sought to verify that this interaction occurs in vivo using previously derived R270X transgenic mice (10). When MeCP2-R270X was immunoprecipitated in whole brain lysates from transgenic animals, KPNA3 was found to interact with this truncated protein (Fig. 5B). Taken together, these data demonstrate that the MeCP2-KPNA3 interaction confers exclusive nuclear localization on MeCP2-R270X in vivo, providing a mechanism for the localization of this mutant protein, in contrast to predictions made on the basis of the previously annotated MeCP2 NLS.
FIGURE 5.
MeCP2 interacts with KPNA3 in the mouse brain, and a model of MeCP2 nuclear import. A, whole-brain extracts were made from male Mecp2+/y mice (WT), Tg(MECP2-GFP) transgenic mice (MeCP2-GFP), or Tg(Thy1-GFP) transgenic mice (Thy1-GFP) and immunoprecipitated (IP) with anti-GFP antibodies. Bound proteins were eluted and analyzed by Western blot with the indicated antibodies, along with an aliquot of the input protein. n = 2. B, whole-brain extracts were made from WT and Tg(MECP2-R270X-GFP) transgenic mice (R270X-GFP) and immunoprecipitated with anti-GFP antibodies. Bound proteins were eluted and analyzed by Western blot with the indicated antibodies, along with an aliquot of the input protein. WT mice serve as a negative control because they do not express a GFP-tagged protein. n = 3. C, the sequence surrounding the MeCP2 NLS within the C terminus is shown, indicating the location of amino acids required for KPNA3 and KPNA4 binding. MeCP2-WT includes the full KPNA3- and KPNA4-interacting domains and is efficiently imported into the nucleus, providing exclusive nuclear localization. The MeCP2-R270X mutation disrupts KPNA4 binding, but KPNA3 is still able to bind to MeCP2 and mediate nuclear import. The MeCP2-R255D mutation disrupts both the KPNA3 and KPNA4 interactions, causing a loss of active nuclear transport and leading to diffusion mediated distribution of MeCP2 between the nucleus and the cytoplasm.
Discussion
The proper localization and function of MeCP2 are essential for the central nervous system. MECP2 mutations found in RTT patients are located throughout the coding sequence, many of which are reported or predicted to interrupt its nuclear localization. In this study, we identified KPNA3 and KPNA4 as important mediators of MeCP2 nuclear import. In support of our findings, KPNA3 and KPNA4 also appear on a list of proteins, identified by mass spectrometry, that copurify with MeCP2 from mouse brain lysates, in addition to members of the NCoR complex (19). Here we demonstrate that KPNA3 and KPNA4 are, in fact, direct interactors of MeCP2 and, furthermore, we define the domain of interaction of each protein. Initially, truncating MeCP2 at Arg270 was predicted to lead to improper nuclear localization (7). However, this report and previous studies have found MeCP2-R270X to localize normally (9, 10). This finding has important implications for patients with this common mutation and the interpretation of genotype-phenotype studies. Girls with the R270X mutation exhibit a more severe disease course than girls with later-truncating mutations (e.g. R294X) (5). Because MeCP2-R270X can localize to the nucleus, this suggests that domains important for the function of MeCP2 lie between amino acids Arg270 and Arg294. Indeed, we characterized the MeCP2 AT-Hook 2 domain (amino acids 265–272), which overlaps this region and is rendered non-functional by truncation at Arg270 (10). This domain retains function when MeCP2 is truncated at Gly273, and, therefore, the loss of this domain provides a plausible explanation for the more severe phenotype of RTT patients with the R270X mutation when compared with R294X girls. Importantly, because these premature stop codons are introduced in the final exon of MECP2, it is unlikely the resulting RNA transcripts are subject to nonsense-mediated decay. Loss of the AT-Hook 2 domain could destabilize the protein (20), but we and others detected approximately wild-type levels of MeCP2-R270X in transgenic animals expressing MeCP2 from its endogenous human locus (9, 10).
Interestingly, although deletion of the MeCP2 NLS or mutation of Arg255 to aspartate leads to the appearance of these mutant proteins within the cytoplasm, both proteins still partially localize to the nucleus. The molecular mass of MeCP2 is ∼52 kDa, and, fused to GFP, the combined proteins have a mass of ∼79 kDa. Proteins of more than 60 kDa are often considered to have negligible diffusion through the nuclear pore (21). However, the early studies from which these predications are made, often included a limited number of proteins and frequently utilized bovine serum albumin (molecular mass, ∼68 kDa) to define the diffusion limit (22, 23). More recent analysis indicates that proteins as large as 110 kDa can localize to the nucleus without relying on active transport (24). Therefore, the partial localization of MeCP2-ΔNLS and MeCP2-R255D within the nucleus likely reflects passive diffusion, although we cannot exclude the presence of a second, weakly functional NLS.
We propose a model (Fig. 5C) in which MeCP2 is initially translated within the cytoplasm and is recognized by KPNA3 and KPNA4 through a sequence in the C terminus containing amino acids 249–270. Binding to these proteins leads to active transport of MeCP2 through the nuclear pore and exclusive localization of MeCP2 to the nucleus. When MeCP2 is truncated at amino acid Arg270, this leads to loss of KPNA4 binding. However, KPNA3 retains a partial interaction with amino acids 249–268 and facilitates MeCP2 nuclear import. In contrast, the MeCP2-R255D mutation abolishes the interaction of MeCP2 with both KPNA3 and KPNA4, ultimately leading to the distribution of MeCP2 between the nucleus and the cytoplasm. That the presence of a single amino acid change (namely, R255D) within the MeCP2 NLS (refined here to amino acids 249–270) leads to the improper localization of MeCP2 to the cytoplasm suggests that this region encompasses a minimal NLS that is retained by the MeCP2-R270X mutant protein.
Fine mapping of the KPNA3 and KPNA4 NLS interaction surfaces will hopefully facilitate the clinical interpretation of patient missense alleles in this region because failure of MeCP2 nuclear translocation should result in RTT-like neurological symptoms. Furthermore, understanding the mechanism of MeCP2 nuclear import will also be an important consideration for future therapeutic strategies. Many RTT-causing mutations are hypomorphic in nature and allow for production of the mutant product (2). Because many of these mutant alleles produce proteins with partial function, one potential therapeutic avenue may involve boosting the abundance of these mutants to restore a normal balance of MeCP2 function. When using therapies aimed at increasing the function of these hypomorphic alleles, one will need to consider whether the malfunctioning protein is targeted properly to the nucleus. Given that MeCP2 binds to KPNA3 using the N-terminal 268 amino acids, this interaction ensures that patients with later-truncating mutations will benefit from compounds designed to increase the concentration of MeCP2 within the neuronal nucleus.
Author Contributions
S. A. B. and H. Y. Z. conceived the study and wrote the manuscript. S. A. B. and L. M. L. performed the experiments. S. A. B., L. M. L., and H. Y. Z. analyzed the data, reviewed the results, and approved the final version of the manuscript.
Supplementary Material
This work was supported, in whole or in part, by NINDS/National Institutes of Health Grants NS057819 (to H. Y. Z.) and F30NS066527 (to S. A. B.). This work was also supported by the Howard Hughes Medical Institute and services from the cores of the Baylor College of Medicine IDDRC (HD024064). The authors declare they have no conflicts of interest with the contents of this article.

This article contains supplemental Table S1.
- RTT
- Rett syndrome
- NLS
- nuclear localization signal
- cNLS
- classical nuclear localization signal
- KPNA
- karyopherin α
- RFP
- red fluorescent protein.
References
- 1. Amir R. E., Van den Veyver I. B., Wan M., Tran C. Q., Francke U., Zoghbi H. Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 [DOI] [PubMed] [Google Scholar]
- 2. Chahrour M., Zoghbi H. Y. (2007) The story of Rett syndrome: from clinic to neurobiology. Neuron 56, 422–437 [DOI] [PubMed] [Google Scholar]
- 3. Miltenberger-Miltenyi G., Laccone F. (2003) Mutations and polymorphisms in the human methyl CpG-binding protein MECP2. Hum. Mutat. 22, 107–115 [DOI] [PubMed] [Google Scholar]
- 4. Nan X., Tate P., Li E., Bird A. (1996) DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell Biol. 16, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colvin L., Leonard H., de Klerk N., Davis M., Weaving L., Williamson S., Christodoulou J. (2004) Refining the phenotype of common mutations in Rett syndrome. J. Med. Genet. 41, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bebbington A., Anderson A., Ravine D., Fyfe S., Pineda M., de Klerk N., Ben-Zeev B., Yatawara N., Percy A., Kaufmann W. E., Leonard H. (2008) Investigating genotype-phenotype relationships in Rett syndrome using an international data set. Neurology 70, 868–875 [DOI] [PubMed] [Google Scholar]
- 7. Huppke P., Held M., Hanefeld F., Engel W., Laccone F. (2002) Influence of mutation type and location on phenotype in 123 patients with Rett syndrome. Neuropediatrics 33, 63–68 [DOI] [PubMed] [Google Scholar]
- 8. Jian L., Archer H. L., Ravine D., Kerr A., de Klerk N., Christodoulou J., Bailey M. E., Laurvick C., Leonard H. (2005) p.R270X MECP2 mutation and mortality in Rett syndrome. Eur. J. Hum. Genet. 13, 1235–1238 [DOI] [PubMed] [Google Scholar]
- 9. Kifayathullah L. A., Arunachalam J. P., Bodda C., Agbemenyah H. Y., Laccone F. A., Mannan A. U. (2010) MeCP2270 mutant protein is expressed in astrocytes as well as in neurons and localizes in the nucleus. Cytogenet. Genome Res. 129, 290–297 [DOI] [PubMed] [Google Scholar]
- 10. Baker S. A., Chen L., Wilkins A. D., Yu P., Lichtarge O., Zoghbi H. Y. (2013) A newly characterized AT-hook domain in MeCP2 determines clinical course of Rett syndrome and related disorders. Cell 152, 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Importin α: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 12. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 13. Conti E., Uy M., Leighton L., Blobel G., Kuriyan J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 [DOI] [PubMed] [Google Scholar]
- 14. Kelley J. B., Talley A. M., Spencer A., Gioeli D., Paschal B. M. (2010) Karyopherin α7 (KPNA7), a divergent member of the importin α family of nuclear import receptors. BMC Cell Biol. 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cingolani G., Petosa C., Weis K., Müller C. W. (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 [DOI] [PubMed] [Google Scholar]
- 16. Sakai Y., Shaw C. A., Dawson B. C., Dugas D. V., Al-Mohtaseb Z., Hill D. E., Zoghbi H. Y. (2011) Protein interactome reveals converging molecular pathways among autism disorders. Sci. Transl. Med. 3, 86ra49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heckman L. D., Chahrour M. H., Zoghbi H. Y. (2014) Rett-causing mutations reveal two domains critical for MeCP2 function and for toxicity in MECP2 duplication syndrome mice. eLife 3, e02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng G., Mellor R. H., Bernstein M., Keller-Peck C., Nguyen Q. T., Wallace M., Nerbonne J. M., Lichtman J. W., Sanes J. R. (2000) Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51 [DOI] [PubMed] [Google Scholar]
- 19. Lyst M. J., Ekiert R., Ebert D. H., Merusi C., Nowak J., Selfridge J., Guy J., Kastan N. R., Robinson N. D., de Lima Alves F., Rappsilber J., Greenberg M. E., Bird A. (2013) Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat. Neurosci. 16, 898–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pitcher M. R., Herrera J. A., Buffington S. A., Kochukov M. Y., Merritt J. K., Fisher A. R., Schanen N. C., Costa-Mattioli M., Neul J. L. (2015) Rett syndrome like phenotypes in the R255X Mecp2 mutant mouse are rescued by MECP2 transgene. Hum. Mol. Genet. 24, 2662–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nigg E. A. (1997) Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386, 779–787 [DOI] [PubMed] [Google Scholar]
- 22. Paine P. L., Feldherr C. M. (1972) Nucleocytoplasmic exchange of macromolecules. Exp. Cell Res. 74, 81–98 [DOI] [PubMed] [Google Scholar]
- 23. Bonner W. M. (1975) Protein migration into nuclei: I: frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J. Cell Biol. 64, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang R., Brattain M. G. (2007) The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 581, 3164–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.