Background: The functional importance of inter-keratin disulfide bonding is underappreciated in epithelial biology.
Results: Cysteines 4 and 40 in K14 participate in intermolecular disulfide bonding.
Conclusion: Residues Cys-4, Cys-40, and Cys-367 in K14 and their dependent disulfide bonds regulate the assembly, organization, and dynamics of keratin filaments.
Significance: Inter-keratin disulfide bonding plays an important role in regulating keratin intermediate filaments in skin keratinocytes.
Keywords: cytoskeleton, epidermis, keratin, keratinocyte, nucleus, disulfide bonding
Abstract
We recently showed that inter-keratin disulfide bonding plays an important role in the assembly, organization, and dynamics of keratin intermediate filaments in skin keratinocytes. In particular, cysteine 367 located in the central α-helical rod domain of keratin 14 is necessary for the formation of a stable perinuclear network of keratin filaments (with type II partner keratin 5) in skin keratinocytes analyzed by static and live cell imaging. Here, we show that two additional cysteine residues located in the non-helical head domain of K14, Cys-4 and Cys-40, also participate in inter-keratin disulfide bonding and tandemly play a key role complementary to that of Cys-367 in the assembly, organization, and dynamics of keratin filaments in skin keratinocytes in primary culture. Analysis of K14 variants with single or multiple substitutions of cysteine residues points to a spatial and temporal hierarchy in how Cys-4/Cys-40 and Cys-367 regulate keratin assembly in vitro and filament dynamics in live keratinocytes in culture. Our findings substantiate the importance and complexity of a novel determinant, namely inter-keratin disulfide bonding, for the regulation of several aspects of keratin filaments in surface epithelia.
Introduction
Ten-nanometer keratin intermediate filaments (IFs)3 figure prominently in the cytoplasm of keratinocytes in epidermis and related epithelia (1), where they impact cytoarchitecture and are essential to the maintenance of physical integrity (2, 3). Accordingly, disruption of the assembly or organization of keratin filaments (e.g. through mutation) results in cell and tissue fragility and underlies several inherited blistering diseases (2–4). Although individually rare, these diseases are often devastating, and there is unfortunately no effective treatment for disorders rooted in defective IFs (5, 6). In part, the inability to therapeutically intervene reflects a limited understanding of the assembly and detailed structure of all types of intermediate filaments and of the mechanisms defining their intracellular organization and dynamics.
Although highly variable in size (40–240 kDa) and primary structure, all IF proteins share a tripartite domain structure, consisting of a central α-helical (rod) domain of conserved length and substructure, flanked by non-helical N-terminal head and C-terminal tail domains of variable length and primary structure (7, 8). IF assembly is driven largely by the central rod domain, given long-range heptad repeats mediating coiled-coil dimerization and higher order interactions, with variable contributions from the head and tail domains (depending of the IF system), notably with regard to post-translational modifications and interactions with associated proteins (9). In vivo, IF proteins assemble into 10-nm filaments either as obligate (e.g. keratins, neurofilaments) or facultative (e.g. vimentin, glial fibrillary acidic protein) heteropolymers (8). IF assembly appears complex and even polymorphic but can be readily recapitulated from purified proteins in vitro (10–12). Further, live imaging studies over the last decade suggest that, in cultured epithelial cells, keratin IF assembly is initiated at the cell periphery and is completed as filament precursors become incorporated into a network that continuously flows toward the center of the cell, culminating in the formation of a robust perinuclear array of filaments (13–15). Despite this progress, much remains to be learned about the processes that preside over keratin assembly and network organization, in vitro as well as in vivo.
Disulfide bonding recently came to the fore as a novel and important mechanism that regulates the assembly, organization, and dynamics of keratin 14 (a type I keratin expressed in the progenitor basal layer of stratified epithelia)-containing intermediate filaments in skin keratinocytes (16, 17). Particularly, residue Cys-367 in the central α-helical rod domain of human K14 mediates a trans-dimer homotypic disulfide bond in a crystal of the interacting K5/K14 rod domains as well as in epidermal keratinocytes in vivo (16). This cysteine and its associated disulfide bonds play a key role in the formation and/or maintenance of a perinuclear-concentrated network of keratin IFs and in the centripetal keratin filament cycle observed in epidermal keratinocytes in primary culture (17). The data in hand also show that, albeit important, Cys-367 does not account for all of the disulfide bonding-dependent properties of K14 in vitro and in vivo. In this study, we uncover complementary roles for two additional cysteine residues located in the non-helical head domain of K14, Cys-4 and Cys-40. As is the case for Cys-367, K14 Cys-4 and Cys-40 play key roles toward both the assembly and organization of keratin IFs. Further, we present evidence that all three cysteine residues act in concert to foster the execution of a normal keratin cycle in live cultures of epidermal keratinocytes.
Experimental Procedures
Plasmids Used in This Study
The human K14 (NP_000517) and K5 (NP_000415) protein sequences can be accessed at the Human Intermediate Filament Database and the National Center for Biotechnology Information. Based on the simple nucleotide polymorphism database (dbSNP 142), human K14 (gene ID 3861) has an “A→G” SNP (dbSNP ID rs6503640), resulting in a missense variant (Tyr-63 → Cys) (UCSC Genome Browser Database). Because this SNP is relatively rare (0.739%; given 37 Cys and 4971 Tyr alleles among 5008 human genomes surveyed), it was not investigated in our study. Plasmids pT7HMT-K14WT, pT7HMT-K14CF, pT7HMT-K14CF-C367, and pT7HMT-K5WT (for in vitro studies); pBK-CMV His-GFP-K14WT, pBK-CMV His-GFP-K14CF, pBK-CMV His-GFP-K14C367A, pBK-CMV His-GFP-K14CF-C367, and pBK-CMV His-GFP-K14C389A (for expression in mammalian cells); and pShuttle-CMV-H2B-mCherry (live cell imaging study) have been described (16, 17). Targeted cysteine-to-alanine replacements (e.g. C4A, C18A, C40A, C4A/C40A, and the triple C4A/C40A/C367A variant) were introduced into the K14WT cDNA using the QuikChange lightning site-directed mutagenesis kit (Agilent Technologies). Selective cysteine restorations (e.g. Cys-4, Cys-40, Cys-4/Cys-40, and Cys-4/Cys-40/Cys-367) were created on the K14 cysteine-free (CF) backbone using the same mutagenesis kit (Agilent Technologies).
Antibodies and Other Reagents
The antibodies used include anti-K5 (rabbit, 1:1000 for immunofluorescence) from Covance, anti-GFP (mouse, 1:1000) from Abcam, IRDye 800CW goat anti-mouse IgG (1:20,000) from LI-COR Biosciences, and Alexa Fluor 594 goat anti-rabbit IgG (1:1000) from Life Technologies, Inc. DAPI was from Life Technologies, tris(2-carboxyethyl) phosphine (TCEP) was from Thermo Scientific, and N-ethylmaleimide was from Sigma-Aldrich. Dulbecco's modified Eagle's medium (DMEM)/calcium-free/low glucose and DMEM/calcium-free/low glucose/without phenyl red, used in the preparation of flavin adenine dinucleotide (FAD) medium and FAD imaging medium, were from United States Biological. Restriction endonucleases were from New England Biolabs. All other chemicals were obtained from Sigma-Aldrich unless noted otherwise.
Cell Culture, Transfection, and Calcium Switch Assays
308 mouse skin keratinocytes (18) were used to identify cysteine residues participating in disulfide bonding in the K14 protein. Cells were cultured in DMEM/low glucose supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Life Technologies). Transfection of GFP-tagged K14 cysteine variants was performed using FuGENE HD (Promega) according to the manufacturer's instructions. At 24 h after transfection, cells were switched to Keratinocyte Basal Medium-2 without Ca2+ (KBM-2, Lonza; so-called low calcium medium) for an additional 24 h and then switched to KBM-2 supplemented with 1.8 mm calcium and 10% FBS (high-calcium medium). Keratinocytes were harvested 4 h after high-calcium switch.
Preparation of Cell Lysates, Protein Gel Electrophoresis, and Immunoblotting Analysis
To prepare whole cell lysates, cells were lysed in cold urea lysis buffer (pH 7.0) (6.5 m urea; 50 mm Tris-HCl; 150 mm sodium chloride; 5 mm EDTA; 0.1% Triton X-100; 50 μm N-ethylmaleimide; 1 mm PMSF; 1 μg/ml each of chymostatin, leupeptin, and pepstatin; 10 μg/ml each of aprotinin and benzamidine; 2 μg/ml antipain; and 50 mm sodium fluoride), and then pelleted via centrifugation (14,000 rpm for 30 min at 4 °C) in order to remove cell debris.
For immunoblotting, the Bradford protein assay (Bio-Rad) was used to determine protein concentration in cell lysates. Samples for gel electrophoresis were prepared in LDS sample buffer (Life Technologies) in the presence or absence of 20 mm TCEP. TCEP treatment was performed at room temperature for 1 h in order to fully reduce disulfide bonds. Equal amounts of cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membrane (0.45 μm; Bio-Rad). Quantitative infrared immunoblotting analysis (LI-COR Biosciences) was performed using primary antibodies, followed by secondary antibodies conjugated to infrared fluorescent dye IRDye800 (1).
Krt14−/− Mouse Keratinocytes in Primary Culture and Transfection Assays
All studies involving mice were reviewed and approved by the Johns Hopkins Institutional Animal Care and Use Committee. Freshly isolated keratinocytes from 1- or 2-day-old Krt14−/− newborn mouse skin (19) were transfected by the pBK-CMV His-GFP-K14 cysteine variants using P1 primary cell 4D-Nucleofection X kit (Lonza). CnT57 (low calcium, 0.07 mm; CELLnTEC) and FAD media were used for keratin IF organization and live cell imaging studies, respectively (17). Transfected cells were fixed using prechilled methanol at 72 h post-transfection and processed for immunofluorescence staining to assess the organization of keratin IFs. Imaging of live keratinocytes was initiated at 48 h post-transfection. pShuttle-CMV-H2BmCherry was co-transfected in order to monitor the nucleus in transfected cells.
Immunofluorescence Microscopy
Keratinocytes were fixed by prechilled methanol; blocked in 10% normal goat serum, 0.1% Triton X-100, PBS for 1 h at room temperature; incubated in anti-K5 antibody solution for 1 h; washed in PBS; incubated in Alexa Fluor 594 goat anti-rabbit IgG solution; counterstained in DAPI; and mounted in FluorSave Reagent mounting medium (Calbiochem). Pictures were acquired as single focal planes using an Axio Observer.Z1 fluorescence microscope equipped with an ApoTome II attachment and AxioCam MRm camera (Carl Zeiss Microscopy). Focal planes were selected by focusing on the center of the nuclei. Pictures were taken using a 63× plan-apochromat oil immersion objective, with the system controlled by the AxioVision version 4.8.2 software (Carl Zeiss Microscopy). Distinct modes of keratin filament network organization were analyzed by blindly counting transfected mouse keratinocytes and re-examined based on quantitative criteria for each of the three categories of keratin filament network organization (see Fig. S3 in Ref. 17).
Live Cell Imaging and Image Processing
Imaging of live keratinocytes and associated data processing were as described (17). A single-point laser scanning confocal microscope (LSM780-FCS, Carl Zeiss Microscopy), fitted with a 40× PlanNeofluar oil immersion objective, was used to record pictures. Cells in a closed culture dish (35 mm; MatTek Corp.) were kept in an environment chamber at 37 °C with 5% CO2 and 25 °C humidity. Excitation was set at 488 nm for GFP and 561 nm for mCherry. Recording intervals were 5 min, color depth was 12 bit, and image resolution was 1024 × 1024 pixels. Seven focal planes (optical slice thickness: 0.43 μm/stack) were recorded for each time point, and the movie recording lasted 6 h. Maximum intensity projections were generated by compressing three-dimensional stacks of time series using the Zen software (Carl Zeiss Microscopy) and used for preparing figures and conversion into QuickTime movies (Apple). Reconstruction of 2.5-dimentional images from Z stacks was achieved using the Zen software. Measurements of filament length and of the fluorescence intensity of individual keratinocytes were performed using the ImageJ freeware (20). Tracking of nuclei was performed using the Imaris software (Bitplane).
Keratin Purification and Assembly of Keratin Filaments in Vitro
Plasmids pT7HMT-K5WT, pT7HMT-K14WT, pT7HMT-K14CF, pT7HMT-K14CF-C367, pT7HMT-K14CF-C4/C40, and pT7HMT-K14CF-C4/C40/C367 were transformed into the Escherichia coli strain BL21(DE3) to produce the corresponding recombinant human keratin proteins as inclusion bodies. Keratin proteins were purified using HiTrapQ and MonoQ columns (GE Healthcare) as described (21). Heterotypic complexes containing K5 and K14 in an equimolar ratio were obtained using MonoQ column chromatography (21). The concentration of purified keratin complexes was adjusted to 0.15 mg/ml, and keratin filaments were assembled under either reducing or oxidizing buffer conditions as described (17). For certain experiments, filaments assembled under oxidizing buffer conditions were subjected to a reducing environment at the postassembly stage.
Polymerization Efficiency and Morphological Analysis of Keratin Filament Assemblies
Physical states of the keratin polymers were analyzed using a high-speed sedimentation assay (150,000 × g for 30 min) as described previously (22). Polymerization efficiency of filament assembly was estimated on SDS-PAGE through densitometry with the ImageJ freeware (20). The ultrastructure of assembled keratin filaments was examined by negative staining (1% uranyl acetate) and transmission electron microscopy using HU-12A instruments (Hitachi). Measurements of filament length and diameter were performed on recorded micrographs (80,000× nominal magnification) using the AmtV602 capture engine software (Hitachi) and ImageJ freeware (20).
Statistical Analyses
Statistical analyses were performed using Microsoft Excel software 2011 (Microsoft Office) and Prism version 5 (GraphPad Software, Inc.). Student's t test analysis was used to compare two conditions or samples (see Figs. 1C, 5D, and 6 (A, C, and E)). One-way analysis of variance was used to compare three conditions or samples (see Figs. 2C and 3C). A statistical p value of <0.05 was considered significant.
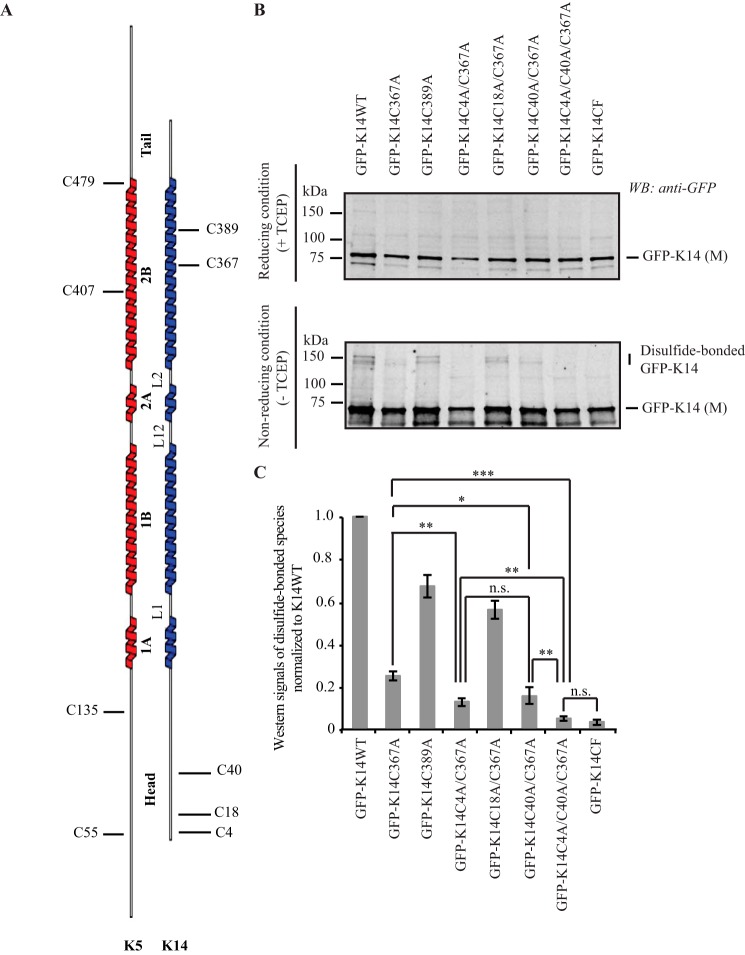
FIGURE 1.
K14 cysteine residues involved in disulfide bonding. A, schematic representation of the location of cysteine residues in human K5 and K14. B, immunoblotting analysis shows that replacing Cys in K14 with Ala at positions 4, 40, and 367 results in a complete loss of disulfide-bonded dimers of the GFP-K14 upon transfection in 308 mouse keratinocytes (top gel, reducing conditions (+TCEP); bottom gel, non-reducing conditions (−TCEP)). Reducing and non-reducing samples were loaded on SDS-PAGE and analyzed simultaneously using the same exposure time when scanned on the infrared imaging system (LI-COR Biosciences). C, quantification of disulfide-bonded species of GFP-K14WT and cysteine variants by densitometry-based analysis of Western signals of GFP-K14 as reported in B. n = 3 experiments were performed. Western signals of the TECP-treated GFP-K14 (monomer) as the loading control. Error bars, S.D. *, p < 0.5; **, p < 0.01; ***, p < 0.001; n.s., not significant (p > 0.5).
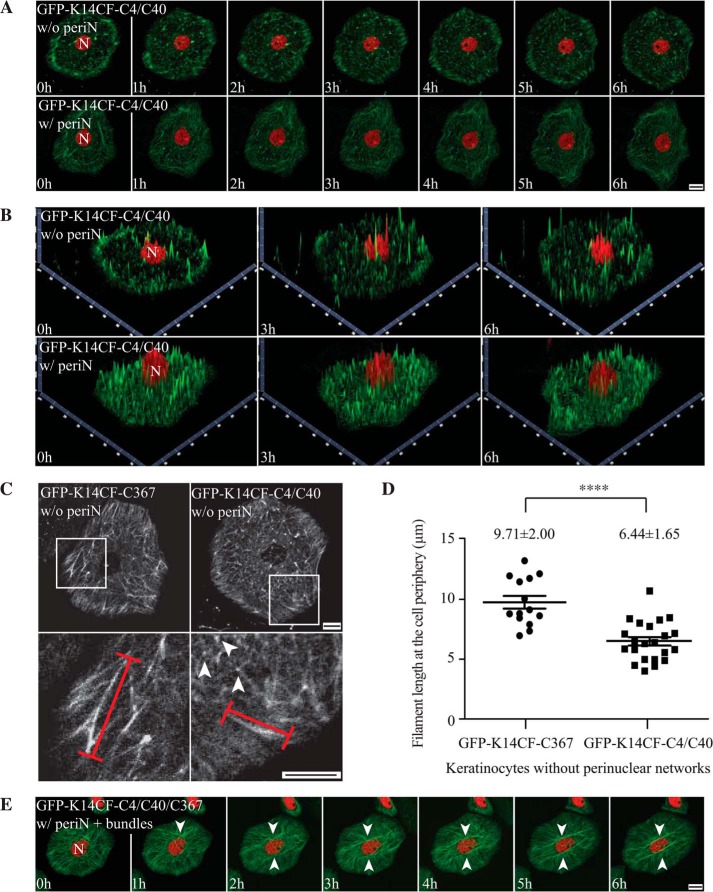
FIGURE 5.
Residues Cys-4 and Cys-40 in the K14 head domain foster a normal keratin filament cycle in live mouse keratinocytes. A, still fluorescence images from movie recordings of live mouse keratinocytes in primary culture expressing GFP-K14CF-C4/C40 (green channel). H2B-mCherry (red channel) was co-transfected to localize nuclei. The images shown correspond to seven time points from 0 to 6 h in time lapse movies recorded using laser scanning confocal microscopy (see supplemental Movies 1 and 2). Maximum intensity projections identify two different populations of mouse keratinocytes expressing the GFP-K14CF-C4/C40 with (w/) or without (w/o) perinuclear (periN) concentrated networks of keratin filaments (20% versus 80%, respectively). Bar, 10 μm. B, 2.5-dimensional views of Z stacked images corresponding to the two-dimensional images at time 0, 3 h, and 6 h in A. C, comparison of GFP-K14CF-C367- and GFP-K14CF-C4/C40-containing filaments in live mouse keratinocytes without a perinuclear network. Top row panels, still fluorescence images corresponding to the 6 h time point in recorded movies. Bottom panels, magnified views of the boxed areas in the top row panels. Representative individual filaments in the cell periphery analyzed were marked using red lines. Arrowheads denote very short filaments showing spotted fluorescence intensity. D, length measurements of GFP-K14CF-C367- and GFP-K14CF-C4/C40-containing filaments at the cell periphery in live mouse keratinocytes without a perinuclear network. Each symbol represents the average filament length in the cell periphery in individual keratinocytes, in which 20 filaments were chosen randomly, and their lengths were measured. n = 14 cells for GFP-CF-C367 without a perinuclear network; n = 22 cells for GFP-CF-C4/C40 without a perinuclear concentrated network. ****, p < 0.0001. Of note, the diameters of the cytoplasmic space of GFP-K14CF-C367- and GFP-K14CF-C4/C40-expressing cells are statistically the same (∼20 μm). Error bars, S.D. E, still fluorescence images from movie recordings of live mouse Krt14−/− keratinocytes in primary culture expressing GFP-K14CF-C4/C40/C367 (green channel) and H2B-mCherry (red channel) and displaying a normal filament cycle and formation of a perinuclear network along with accumulation of keratin filament bundles in the perinuclear region (see supplemental Movie 4). Arrowheads denote keratin bundles. Bar, 10 μm.
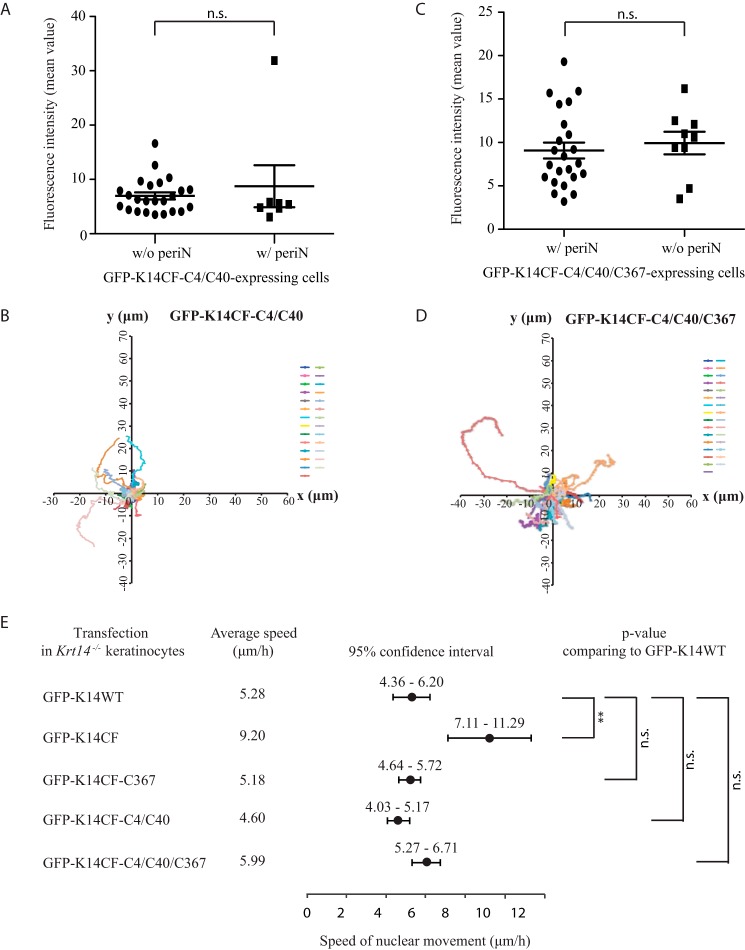
FIGURE 6.
Residues Cys-4, Cys-40, and Cys-367 in K14 contribute to regulating keratinocyte motility. Fluorescence intensity comparison of GFP-K14CF-C4/C40-expressing (A) and GFP-K14CF-C4/C40/C367-expressing (C) mouse keratinocytes with (w/) or without (w/o) perinuclear (periN)-concentrated networks. Each symbol represents the average fluorescence intensity (mean value) of individual keratinocytes at seven time points (from 0 to 6 h) during movie recordings from a single experiment. n = 24 cells for GFP-CF-C4/C40 without a perinuclear concentrated network; n = 7 cells for GFP-CF-C4/C40 with a perinuclear concentrated network. n = 22 cells for GFP-K14CF-C4/C40/C367 with a perinuclear concentrated network; n = 9 cells for GFP-K14CF-C4/C40/C367 without a perinuclear concentrated network. Error bars, S.D. Nuclear movement was obtained by tracking the position of the nucleus in GFP-K14CF-C4/C40-expressing (B) and GFP-K14CF-C4/C40/C367-expressing (D) cells. Each color trace represents an individual cell. n = 27 GFP-K14CF-C4/C40-transfected cells, and n = 31 GFP-K14CF-C4/C40/C367-transfected cells were analyzed. E, comparison of nuclear and whole cell movement of Krt14−/− keratinocytes expressing GFP-K14CF (n = 41), GFP-K14CF-C367 (n = 21), GFP-K14CF-C4/C40 (n = 27), or GFP-K14CF-C4/C40/C367 (n = 24) with their wild-type counterpart GFP-K14WT (n = 28) by tracking the speed of the nuclear movement. Mean speed (●) and 95% confidence interval (horizontal bars) of the nuclear movement are presented here. **, p < 0.01; n.s., not significant, p > 0.5.
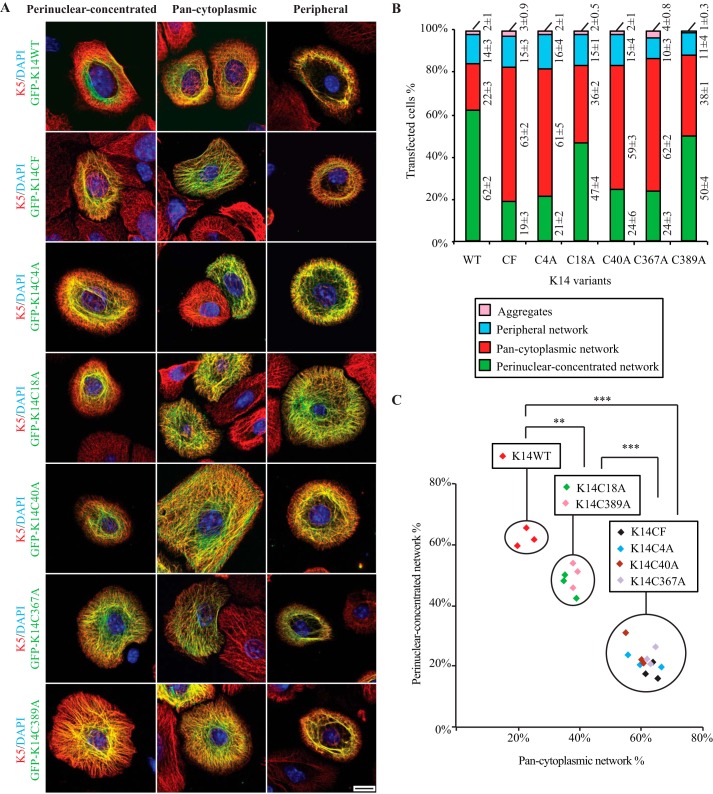
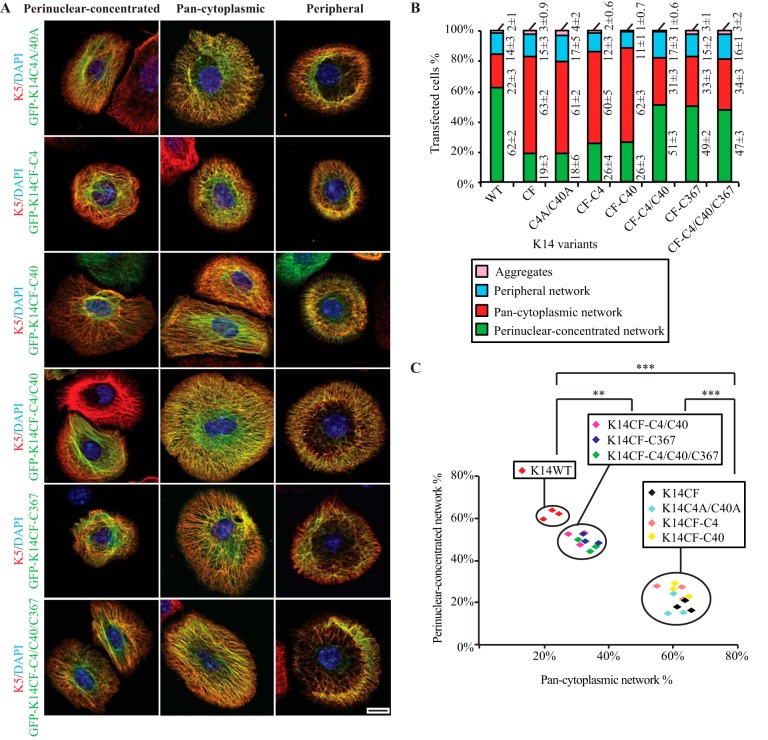
FIGURE 2.
Formation and/or maintenance of a perinuclear concentrated network is compromised by loss of cysteine at position 4, 40, or 367 in K14. A, immunofluorescence imaging shows three modes of keratin network organization resulting from the expression of GFP-tagged K14WT, K14CF, C14C4A, K14C18A, K14C40A, K14C367A, and K14C389A in Krt14−/− mouse keratinocytes in primary culture. Bar, 10 μm. B, population frequencies for each type of keratin IF network illustrated in A. Data are presented as average ± S.D. from three independent experiments. C, analysis of K14 WT and K14 cysteine variants by relating the percentage of cells with perinuclear concentrated networks (y axis) to those with pan-cytoplasmic networks (x axis). Each symbol represents one independent experiment; n = 3 experiments were performed per construct. Within here, the types of keratin filament organization promoted by the K14 variants being expressed are statistically the same. **, p < 0.01; ***, p < 0.001.
FIGURE 3.
Residues Cys-4 and Cys-40 in the K14 head domain and Cys-367 in the rod domain function separately in regulating the formation and/or maintenance of a perinuclear concentrated network. A, immunofluorescence imaging shows three modes of keratin network organization resulting from overexpression of GFP-tagged K14C4A/C40A, K14CF-C4, K14CF-C40, K14CF-C4/C40, K14CF-C367, and K14CF-C4/C40/C367 in Krt14−/− mouse keratinocytes in primary culture. Bar, 10 μm. B, population frequency of each type of IF network as illustrated in A. Data are presented as the average ± S.D. from three independent experiments. C, analysis of K14 WT and various K14 cysteine variants by relating the percentage of cells with perinuclear concentrated networks (y axis) to those with pan-cytoplasmic networks (x axis). Each symbol represents one independent experiment; n = 3 experiments were performed per construct. Within here, the types of keratin filament organization promoted by the K14 variants being expressed are statistically the same. **, p < 0.01; ***, p < 0.001.
Results
Residues Cys-4 and Cys-40 in the K14 Head Domain Participate in Intermolecular Disulfide Bonding in Keratinocytes
Human K14 contains five cysteine residues, with Cys-4, Cys-18, and Cys-40 located in its N-terminal head domain and Cys-367 and Cys-389 in its coiled-coil-forming rod domain (Fig. 1A). Previously (16), we showed that replacing Cys with Ala at position 367 (K14C367A variant) decreased but did not eliminate disulfide-bonded K14 species in transfected 308 mouse keratinocytes, suggesting that another cysteine(s) in the K14 head is also involved in intermolecular disulfide bonding (also see Ref. 17). We generated a series of variants by replacing cysteine residues with alanines at specific positions within the K14 protein (e.g. K14C4A/C367A, K14C18A/C367A, and K14C40A/C367A (Table 1)) in order to examine which cysteine(s) in the K14 head domain participate(s) in disulfide bonding and may play functional roles in keratin filaments. Upon expression in 308 mouse keratinocytes followed by immunoblotting analysis of total protein extracts, the GFP-K14C4A/C367A and GFP-K14C40A/C367A variants result in a significant decrease in the yield of disulfide-bonded K14 species, as is the case for GFP-K14C367A, relative to control GFP-K14WT (Fig. 1, B and C). In addition, the triple cysteine variant GFP-K14C4A/C40A/C367A yields no disulfide-bonded species of molecular masses greater than monomer size (same as the GFP-K14CF variant; Fig. 1C), suggesting that Cys-4, Cys-40, and Cys-367 in K14 are involved in intermolecular disulfide bonding in mouse skin keratinocytes.
TABLE 1.
List of K14 cysteine variants analyzed in this study
Ten cysteine-replacing variants (K14C4A, K14C18A, K14C40A, K14C367A, K14C389A, K14C4A/C367A, K14C18A/C367A, K14C40A/C367A, K14C4A/C40A, and K14C4A/C40A/C367A) were generated by site-directed mutagenesis in the K14WT backbone. Five cysteine-restoring variants (K14CF-C4, K14CF-C40, K14CF-C367, K14CF-C4/C40, and K14CF-C4/C40/C367) were engineered by putting cysteine(s) back in a cysteine-free K14 (K14CF) backbone (see Ref. 17).
| Parental backbone | Associated variants |
|---|---|
| K14WT | K14C4A |
| K14C18A | |
| K14C40A | |
| K14C367A | |
| K14C389A | |
| K14C4A/C367A | |
| K14C18A/C367A | |
| K14C40A/C367A | |
| K14C4A/C40A | |
| K14C4A/C40A/C367A | |
| K14CF | K14CF-C4 |
| K14CF-C40 | |
| K14CF-C4/C40 | |
| K14CF-C367 | |
| K14CF-C4/C40/C367 |
Residues Cys-4 and Cys-40 in K14 Together Contribute to the Formation and/or Maintenance of a Perinuclear Concentrated Network of Keratin Filaments
We recently showed that replacing Cys with Ala at position 367 (K14C367A) interferes with the formation and/or maintenance of perinuclear IF networks in transfected Krt14−/− mouse keratinocytes in primary culture (17). Conversely, the presence of a single cysteine at position 367 in a completely cysteine-free K14 backbone (K14CF-C367) partially rescues this phenotype (17). To test whether K14 Cys-4 and Cys-40 also contribute to generating stable perinuclear IFs, we analyzed primary cultures of Krt14−/− mouse keratinocytes transfected with either the GFP-K14C4A or GFP-K14C40A variant. Such transfected cells show a significant decrease in the frequency of perinuclear IF networks (seen in only 21 and 24% of cells expressing GFP-K14C4A and GFP-K14C40A, respectively; Fig. 2), similar to GFP-K14CF-expressing cells (Fig. 2C). Thus, the loss of a cysteine and/or disulfide bonding at either position 4 or 40 in K14 interferes with formation and/or maintenance of a perinuclear network, even in the presence of Cys-367. Conversely, the selective loss of Cys-367 in K14 also impairs the perinuclear organization of keratin IFs (even with Cys residues present at positions 4 and 40; see Ref. 17). Altogether, this suggests that Cys-4, Cys-40, and Cys-367 play important but potentially distinct roles in this regard.
Compared with GFP-K14WT-expressing cells, Krt14−/− keratinocytes transfected with GFP-K14C18A or GFP-K14C389A show a very modest decrease in the frequency of perinuclear IF networks (∼15 and ∼12% reduction, respectively, p < 0.01; Fig. 2, B and C), suggesting that K14 Cys-18 and Cys-389 may play a modest role in the genesis of the perinuclear network of keratin IFs although they are not apparently involved in intermolecular disulfide bonding (Fig. 1B). The properties of these two Cys variants have not been further analyzed in this study.
Additional variants were produced by site-directed mutagenesis at positions Cys-4 and Cys-40, either singly or in combination, in either the wild type (K14) or cysteine-free K14 backbone (Table 1), and their properties were assessed after transfection in Krt14−/− mouse keratinocytes in primary culture. The main mode of keratin organization in either GFP-K14CF-C4- or GFP-K14CF-C40-expressing cells is pan-cytoplasmic (showing 60 and 62% frequencies, respectively; Fig. 3), whereas, as expected (17), the majority (62%) of GFP-K14WT-expressing cells show perinuclear concentrated networks (see Fig. 3, B and C). This outcome is similar to that observed in cells expressing the single replacement variants GFP-K14C4A or GFP-K14C40A (given 61 and 59%, respectively, of cells showing pan-cytoplasmic networks; Fig. 2B) and in cells expressing the double-replacement variant GFP-K14C4A/C40A (61% of cells; Fig. 3, B and C). By comparison, restoring both Cys-4 and Cys-40 in the K14CF backbone leads to 51% of cells exhibiting a perinuclear concentrated array of keratin filaments (Fig. 3), similar to what is seen for GFP-K14CF-C367-expressing cells (49% of cells; Fig. 3, B and C). Together, these findings suggest that the presence of both Cys-4 and Cys-40 in the K14 head domain is necessary and sufficient to foster the formation and/or maintenance of a perinuclear network of keratin filaments in the absence of Cys-367.
Because both sets of cysteines, Cys-4 and Cys-40, in the K14 head and Cys-367 in the K14 rod can each foster the formation of a perinuclear network of keratin filaments, we next tested whether they function synergistically or separately to this end. A new variant was generated by restoring these three cysteines in the K14CF backbone (K14CF-C4/C40/C367) and then analyzed by transfection in Krt14−/− mouse keratinocytes. Relative to GFP-K14CF-C4/C40 and GFP-K14CF-C367 (51 and 49% frequencies, respectively), expression of the triple-cysteine-restoring variant GFP-K14CF-C4/C40/C367 showed no further improvement in terms of yielding perinuclear concentrated keratin networks (47% of cells; Fig. 3). This outcome suggests that Cys-4 and Cys-40 in the K14 head, on the one hand, and Cys-367 in the K14 rod, on the other, may function at different stages during the genesis of a perinuclear concentrated IF network. We also find that expression of cysteine-restoring variants, including the triple variant GFP-K14CF-C4/C40/C367, does not completely replicate the ability of GFP-K14WT to rescue perinuclear IF organization in Krt14−/− mouse keratinocytes (47% versus 62%; p < 0.01; Fig. 3, B and C), providing additional evidence that the other cysteines in K14 (Cys-18 and Cys-389) make a contribution to this phenomenon.
Formation of K14CF-C4/C40-mediated Inter-disulfide Bonds Plays an Important Role in the Assembly of Keratin Filaments in Vitro
We recently reported that the K14CF-C367 variant (having a single Cys residue at position 367) yielded very aberrant IFs having abnormally large and variable diameters (8–27 nm) when co-assembled with K5 in vitro (17). Given this, we next tested whether the formation of Cys-4 and Cys-40-dependent disulfide bonds plays a role in 10-nm filament assembly in vitro. Under standard polymerization buffer conditions (in the presence of a reducing agent) and with K5 as the type II pairing partner, the K14CF-C4/C40 variant shows a high assembly efficiency (∼98%) (Fig. 4, A and B) and forms normal 10-nm filaments, as seen by electron microscopy (data not shown). Under an oxidation-prone environment, however, the K5/K14CF-C4/C40 pairing yields a subnormal assembly efficiency (∼30% reduction) and filaments that are only short to intermediate in length (0.2–1-μm range) but display a normal and fairly uniform diameter (9–11 nm) (Fig. 4). Unlike the case for Cys-367, therefore, formation of disulfide bonds involving Cys-4 and/or Cys-40 does not interfere with either the control of subunit number per filament or their compaction or maturation during assembly in vitro.
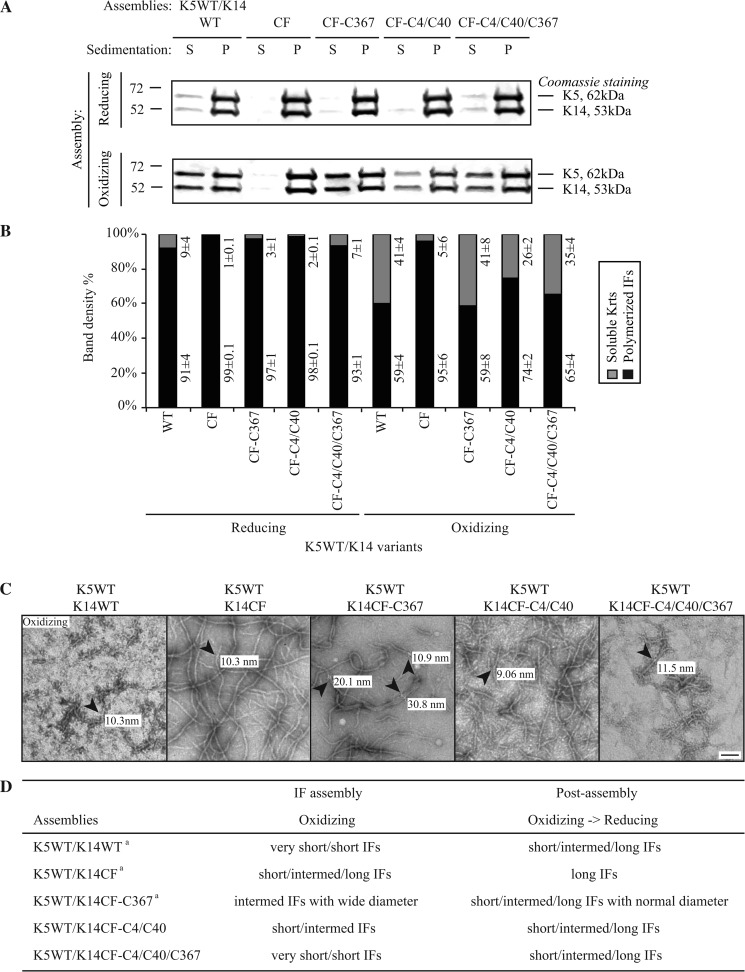
FIGURE 4.
Comparison of K14WT-, K14CF-, K14CF-C367-, K14CF-C4/C40-, and K14CF-C4/C40/C367-containing filaments assembled in vitro under either reducing or oxidizing buffer conditions. A, high-speed sedimentation assay (150,000 × g, 30 min) followed by SDS-PAGE analysis of supernatant (S) and pellet (P) fractions. B, quantitation of polymerization efficiency using densitometry-based analysis of fractions shown in A. Data are presented as average ± S.D. from three independent experiments. C, negative staining and transmission electron microscopy were performed to compare the ultrastructural features of the keratin pairing shown on the top of each micrograph. Diameters of representative individual filaments (denoted by arrowheads) assembled in vitro under oxidizing buffer conditions are labeled in each micrograph. Bar, 100 nm. D, summary of in vitro assembly data for various keratin combinations under different buffer conditions. For the K5WT/K14WT, K5WT/K14CF, and K5WT/K14CF-C367 pairings, similar findings were originally reported in Ref. 17. We repeated these analyses in the current study in order to carry out a comparison with the K5WT/K14CF-C4/C40 and K5WT/K14CF-C4/C40/C367. Very short IFs, <150 nm; short IFs, 200–500 nm; Intermed. IFs, 500–1000 nm; long IFs, >1 μm.
Restoring Cys-4, Cys-40, and Cys-367 in the K14CF backbone (K14CF-C4/C40/C367 variant) also yields subnormal assembly efficiency along with filaments that are quite short after co-polymerization with K5 in vitro under oxidizing buffer conditions (Fig. 4). This is largely similar to the properties of K5/K14WT assemblies (Fig. 4, A and B; also see Ref. 17). This outcome suggests that the inhibition of filament elongation under oxidizing conditions is due to formation of disulfide bonds predominantly mediated by Cys-4, Cys-40, and Cys-367 in K14. Remarkably, these K5/K14CF-C4/C40/C367 filaments display a normal diameter (9–12 nm; see Fig. 4C). Thus, restoring Cys-4 and Cys-40 in addition to Cys-367 in the K14CF backbone results in the nearly complete normalization of the diameter in the resulting filaments, suggesting that the intermolecular disulfide bonds mediated by Cys-4 and Cys-40 act in a dominant fashion over those mediated by Cys-367 to regulate the number of subunits per filament cross-section or to foster filament compaction and/or maturation in vitro.
K14 Cys-4 and Cys-40 Foster a Normal Keratin Filament Cycle in Live Mouse Keratinocytes
We reported that, upon its transfection in Krt14−/− mouse keratinocytes, GFP-K14WT behaves in a manner consistent with the keratin cycle originally described by Leube and colleagues (13–15). In brief, short filaments first form at the cell periphery, elongate and integrate into the keratin IF network as they start moving toward the centrally located nucleus, and finally become part of the perinuclear concentrated network (17). Loss of all K14-bound cysteines and/or disulfide bonds or even the single loss of Cys-367 impairs this normal keratin filament cycle and disrupts perinuclear IF organization in live keratinocytes. This said, restoration of a single cysteine residue at position 367 in the K14CF backbone incompletely rescues the keratin filament cycle and perinuclear IF organization (45% rescue efficiency, as opposed to 59% when testing K14WT; see Ref. 17). Such findings provide further support for the notion that cysteine(s) other than Cys-367 in K14 participate in these cellular processes.
To examine this possibility, we transfected Krt14−/− mouse keratinocytes with GFP-K14CF-C4/C40 and subjected them to live cell recordings. Two types of outcome were observed, irrespective of transgene expression levels (Fig. 6A). Most transfected cells (∼80%) exhibit an impaired keratin filament cycle and fail to form a perinuclear concentrated IF network (supplemental Movie 1). However, a small population of cells (∼20%) displays normal keratin filament cycling and formation of a perinuclear filament network (supplemental Movie 2 and Fig. 5, A and B), similar to GFP-K14WT-transfected cells (supplemental Movie 3). Comparing non-rescued keratinocytes expressing either GFP-K14CF-C367 or GFP-K14CF-C4/C40, two differences stood out: (i) the occurrence of long (Cys-367) versus short (Cys-4/Cys-40) filaments at the cell periphery (a location where keratins are more likely to be seen as single filaments) (13, 15) and (ii) filaments showing uniform (Cys-367) versus spotted (Cys-4/Cys-40) fluorescence intensity in the cytoplasm (Fig. 5, C and D). Accordingly, although both sets of cysteines foster the establishment of a normal keratin filament cycle, it appears that they function at different stages, with Cys-4 and Cys-40 involved early on (e.g. during nucleation and/or elongation) and Cys-367 playing a more defining role at later stages (e.g. when the perinuclear network of filaments is being formed; see “Discussion”). Besides, we observed that 85% of GFP-K14CF-C4/C40-expressing keratinocytes fail to exhibit the aberrant cell motility behavior characteristic of Krt14−/− keratinocytes (Fig. 6, B and E; note that 90% rescue in this behavior was reported for GFP-K14CF-C367 in Ref. 17), suggesting that residues Cys-4, Cys-40, and Cys-367 in K14 contribute to regulating keratinocyte motile properties.
Next, we expressed the GFP-K14CF-C4/C40/C367 variant in Krt14−/− mouse keratinocytes and performed live cell imaging to assess the combined contributions of Cys-4, Cys-40, and Cys-367 to the keratin filament cycle. Greater than 70% of cells expressing GFP-K14CF-C4/C40/C367 display a normal keratin cycle, culminating in the formation of a perinuclear concentrated network, whereas only ∼30% of cells exhibit an impaired keratin cycle and failure to form a perinuclear keratin network (data not shown). Analysis of fluorescence intensity suggests that this difference in outcome is not rooted in transgene expression levels in individual cells (Fig. 6C). Whereas this rescue efficiency is superior to that seen for the GFP-K14CF-C367 (45%) (17) and GFP-K14CF-C4/C40 variants (20%) (this study), these findings indicate that residues Cys-4, Cys-40, and Cys-367 in K14 are necessary but not sufficient to promote a completely normal keratin cycle and formation of a stable perinuclear network. Also of interest, the majority of Krt14−/− keratinocytes expressing GFP-K14CF-C4/C40/C367 exhibit parallel arrays of prominent keratin filament bundles apposed against the nucleus (supplementalMovie4 and Fig. 5E). Many factors could account for enhanced filament bundling in that region of the cell, including but not limited to altered disulfide bonding between keratin filaments and/or keratins and interacting proteins and/or reduced turnover of keratin filaments in the perinuclear region. Besides, 92% of GFP-K14CF-C4/C40/C367-expressing Krt14−/− keratinocytes show a marked normalization of the abnormal cell motility exhibited by Krt14−/− keratinocytes (Fig. 6, D and E), consistent with observations made when analyzing GFP-K14CF-C367- and GFP-K14CF-C4/C40-expressing cells (17) (this study) and further supporting the notion that cysteines 4, 40, and 367 in K14 contribute to regulating aspects of keratinocyte motility.
Discussion
Human K14 contains five cysteine residues that are perfectly conserved in its mammalian orthologs and in the highly homologous human K16 (17). Additionally, select cysteines (e.g. Cys-367) are conserved in other type I keratin proteins expressed in skin epithelia (e.g. K10 and K17). Building on two previous reports (16, 17), we show here that three of the five cysteine residues occurring in K14 are extensively involved in interkeratin disulfide bonding, in a manner that is readily detectable when using a combination of transfection of reporter constructs in cultured keratinocytes and immunoblotting analyses of protein extracts. These three residues are Cys-4 and Cys-40 (this study), located in the head domain of K14, and Cys-367 (17), located within a stutter segment in the 2B portion of the central α-helical rod domain (16). All three cysteines play significant roles (i) during 10-nm filament assembly in vitro; (ii) in fostering the formation of a perinuclear network of keratin filaments in cultured skin keratinocytes; iii) during the centripetal keratin filament cycle that typifies epithelial cells in culture. Further, our findings highlight clear differences in the contributions of Cys-4 and Cys-40, on the one hand, and Cys-367, on the other, to these processes and hint at the existence of a spatial and temporal hierarchy with regard to the involvement of these residues and their disulfide-bonding capabilities.
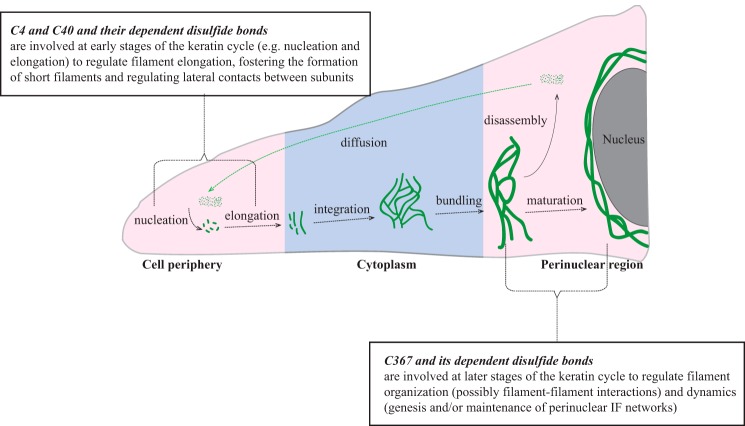
A popular model in the field conveys that, as IF proteins polymerize, short 100-nm-long (unit-length) filaments (ULFs) first form and then anneal to form long but wide (16-nm) filaments, which then undergo collapse or compaction in their cross-sectional axis to give rise to mature 10-nm filaments (8). If this model indeed applies to keratin assemblies in their native context, then inter-keratin (inter-subunit?) disulfide bonding may function as a significant regulatory step during keratin assembly. The combination of in vitro and transfection-based findings amassed to date lead us to propose a model, schematized in Fig. 7, that also takes into account differences in redox potential across the cell (23, 24). We propose that K14 Cys-4 and Cys-40 make a significant contribution to the regulation of filament elongation, specifically the annealing of ULFs, and this selectively takes place at sites of new filament formation. In epithelial cells in culture, such sites preferentially occur at the cell periphery (Fig. 7) (see Ref. 25), where the redox environment is reportedly oxidizing (23). Formation of Cys-4- and Cys-40-dependent disulfide bonds would inhibit the annealing of ULFs but also protect against their lateral growth beyond ∼10 nm, thus contributing to define the number and/or sites of filaments to be further elongated (26). Movement of ULFs toward the center of the cell (e.g. in the context of the keratin cycle) would entail exposure to a more reducing environment (27), thereby reducing Cys-4- and Cys-40-dependent disulfide bonds and fostering the annealing of ULFs and the formation of long 10-nm filaments. As cycling filaments reach the more oxidizing environment prevailing in the perinuclear region (23, 28), Cys-367-dependent disulfide bonding, in particular, would play a significant role in the regulation of filament-filament interactions and dynamics (Fig. 7) (16, 17). The model should also account for the fact that Cys-367 is also influential during the annealing of ULFs, although less so than Cys-4/Cys-40 (17) (this study). This model is obviously largely conceptual at this stage. Major differences in the contribution of disulfide bonding-forming cysteines are to be expected, depending, for instance, on the type of cell, the density and distribution of cysteine residues in the IF proteins involved, and the prevailing biological context. Much experimentation along with better insight into the high-resolution structure of keratin filaments is needed to test these ideas.
FIGURE 7.
Model suggesting roles for K14-dependent disulfide bonding toward the organization and dynamics of keratin IFs in skin keratinocytes. We propose that Cys-4 and Cys-40 and their dependent disulfide bonding make a significant contribution to regulating the early stages of filament assembly (e.g. nucleation and elongation), whereas Cys-367 and its dependent disulfide bonding play a significant role in the genesis and/or maintenance of perinuclear IF networks as part of the keratin filament cycle (15). Pink and blue shades represent regions of relatively oxidizing and reducing environment within the cell, respectively, based on evidence from the literature (23). This schematic is inspired and adapted from Ref. 15.
Formation of intramolecular disulfide bonds in K14, either in vitro or in vivo, cannot be ruled out, given the limits of the methods and assays used in our study. Formation of intramolecular disulfide bonding facilitates the correct folding and contributes to the stability of proteins (29, 30). Intramolecular disulfide bonding is unlikely for Cys-389, given that its side chain faces inward in the coiled-coiled interface between K5 and K14 (16). The finding that expression of K14C389A has a modest but consistent impact on perinuclear keratin networks thus serves as a reminder that some of the cysteine-to-alanine substitutions introduced may have an impact on the local structure of the K14 protein. Likewise, the rationale for the similarly modest impact of the K14C18A variant on the genesis of a perinuclear network awaits investigation. Also of significance, we cannot account for contributions by other type I keratins, K16 and K17 in particular, to the behaviors observed in keratinocytes transfected with various K14 cysteine variants.
Two glaring questions arise when the findings reported here are combined with previous studies (16, 17). What are the mechanisms responsible for disulfide bond formation in keratinocytes? And what is their functional importance in vivo? Mechanistically, disulfide bonds could form enzymatically or non-enzymatically, and there is evidence that both can occur in the cytoplasm of keratinocytes. Sulfhydryl oxidases that are involved in catalyzing the formation of disulfide bonds in the late differentiating layers of epidermis (31, 32) also occur in the basal layer of mouse epidermis (both mRNA and protein products were detected in mouse skin keratinocytes in primary culture).4 In addition, changes in redox potential and/or intracellular oxidative stress were recently implicated in terminal differentiation of epidermal keratinocytes, providing another likely mechanism for the formation of keratin disulfide bonding (33–35). Regarding the issue of physiological relevance, our findings show that the disulfide bonding-dependent formation of a K14-containing perinuclear network of keratin filaments is important for stably positioning the nucleus in keratinocytes in culture (this study) (17) and also impacts the size and shape of the nucleus (16). Whether any of this plays out in the epidermis in situ and the associated significance are unknown. Whereas there is no report of disease-causing mutation affecting any of the K14 cysteine residues (4, 6), a G61C variant in the head domain of K8 predisposes to liver injury (36, 37), also the R125C mutation in K14 (38) is a recurring allele in K14 (4, 6, 39) and other type I keratins and is associated with severe disease presentations. To best address this issue, select K14 cysteine variants (e.g. C4A, C40A, and C367A alone or in combination) should be knocked in at the Krt14 locus in the mouse genome in order to ascertain the functional relevance of the findings reported so far.
Author Contributions
X. F. and P. A. C. designed the research. X. F. performed the experiments. X. F. and P. A. C. analyzed data and wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to members of the Coulombe laboratory for support; to Jill Hakim and Janet Folmer for technical assistance; to the School of Medicine Microscope Facility for live cell imaging experiments (National Institutes of Health Grant S10 OD016374); and to Valeria Culotta, Michael Matunis, Andrew Ewald, and Ryan Hobbs for advice.
This work was supported, in whole or in part, by National Institutes of Health, NIAMS, Grant R01 AR042047 (to P. A. C.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movies 1–4.
X. Feng and P. A. Coulombe, unpublished data.
- IF
- intermediate filament
- CF
- cysteine-free
- TCEP
- tris(2-carboxyethyl) phosphine
- ULF
- unit-length filament.
References
- 1. Feng X., Zhang H., Margolick J. B., Coulombe A. (2013) Keratin intracellular concentration revisited: implications of keratin function in surface epithelia. J. Invest. Dermatol. 133, 850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Omary M. B., Coulombe P. A., McLean W. H. (2004) Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 351, 2087–2100 [DOI] [PubMed] [Google Scholar]
- 3. Coulombe P. A., Kerns M. L., Fuchs E. (2009) Epidermolysis bullosa simplex: a paradigm for disorders of tissue fragility. J. Clin. Invest. 119, 1784–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szeverenyi I., Cassidy A. J., Chung C. W., Lee B. T., Common J. E., Ogg S. C., Chen H., Sim S. Y., Goh W. L., Ng K. W., Simpson J. A., Chee L. L., Eng G. H., Li B., Lunny D. P., Chuon D., Venkatesh A., Khoo K. H., McLean W. H., Lim Y. P., Lane E. B. (2008) The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 29, 351–360 [DOI] [PubMed] [Google Scholar]
- 5. McLean W. H., Moore C. B. (2011) Keratin disorders: from gene to therapy. Hum. Mol. Genet. 20, R189–R197 [DOI] [PubMed] [Google Scholar]
- 6. Coulombe P. A., Lee C. H. (2012) Defining keratin protein function in skin epithelia: epidermolysis bullosa simplex and its aftermath. J. Invest. Dermatol. 132, 763–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fuchs E., Weber K. (1994) Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 63, 345–382 [DOI] [PubMed] [Google Scholar]
- 8. Herrmann H., Aebi U. (2004) Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 73, 749–789 [DOI] [PubMed] [Google Scholar]
- 9. Snider N. T., Omary M. B. (2014) Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 15, 163–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steinert P. M., Idler W. W., Zimmerman S. B. (1976) Self-assembly of bovine epidermal keratin filaments in vitro. J. Mol. Biol. 108, 547–567 [DOI] [PubMed] [Google Scholar]
- 11. Aebi U., Fowler W. E., Rew P., Sun T. T. (1983) The fibrillar substructure of keratin filaments unraveled. J. Cell Biol. 97, 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aebi U., Haner M., Troncoso J., Eichner R., Engel A. (1988) Unifying principles in intermediate filament assembly. Protoplasma 145, 73–81 [Google Scholar]
- 13. Kölsch A., Windoffer R., Würflinger T., Aach T., Leube R. E. (2010) The keratin-filament cycle of assembly and disassembly. J. Cell Sci. 123, 2266–2272 [DOI] [PubMed] [Google Scholar]
- 14. Windoffer R., Leube R. E. (2004) Imaging of keratin dynamics during the cell cycle and in response to phosphatase inhibition. Methods Cell Biol. 78, 321–352 [DOI] [PubMed] [Google Scholar]
- 15. Windoffer R., Beil M., Magin T. M., Leube R. E. (2011) Cytoskeleton in motion: the dynamics of keratin intermediate filaments in epithelia. J. Cell Biol. 194, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee C. H., Kim M. S., Chung B. M., Leahy D. J., Coulombe P. A. (2012) Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat. Struct. Mol. Biol. 19, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng X., Coulombe P. A. (2015) A role for disulfide bonding in keratin intermediate filament organization and dynamics in skin keratinocytes. J. Cell Biol. 209, 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strickland J. E., Greenhalgh D. A., Koceva-Chyla A., Hennings H., Restrepo C., Balaschak M., Yuspa S. H. (1988) Development of murine epidermal cell lines which contain an activated rasHa oncogene and form papillomas in skin grafts on athymic nude mouse hosts. Cancer Res. 48, 165–169 [PubMed] [Google Scholar]
- 19. Lloyd C., Yu. Q. C., Cheng J., Turksen K., Degenstein L., Hutton E., Fuchs E. (1995) The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J. Cell Biol. 129, 1329–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee C. H., Coulombe P. A. (2009) Self-organization of keratin intermediate filaments into cross-linked networks. J. Cell Biol. 186, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma L., Yamada S., Wirtz D., Coulombe P. A. (2001) A “hot-spot” mutation alters the mechanical properties of keratin filament networks. Nat. Cell Biol. 3, 503–506 [DOI] [PubMed] [Google Scholar]
- 23. Clarke S. J., Hollmann C. A., Zhang Z., Suffern D., Bradforth S. E., Dimitrijevic N. M., Minarik W. G., Nadeau J. L. (2006) Photophysics of dopamine-modified quantum dots and effects on biological systems. Nat. Mater. 5, 409–417 [DOI] [PubMed] [Google Scholar]
- 24. Kojer K., Riemer J. (2014) Balancing oxidative protein folding: the influences of reducing pathways on disulfide bond formation. Biochim. Biophys. Acta 1844, 1383–1390 [DOI] [PubMed] [Google Scholar]
- 25. Kölsch A., Windoffer R., Leube R. E. (2009) Actin-dependent dynamics of keratin filament precursors. Cell Motil. Cytoskeleton 66, 976–985 [DOI] [PubMed] [Google Scholar]
- 26. Lichtenstern T., Mücke N., Aebi U., Mauermann M., Herrmann H. (2012) Complex formation and kinetics of filament assembly exhibited by the simply epithelial keratin K8 and K18. J. Struct. Biol. 177, 54–62 [DOI] [PubMed] [Google Scholar]
- 27. Saaranen M. J., Ruddock L. W. (2013) Disulfide bond formation in the cytoplasm. Antioxid. Redox Signal. 19, 46–53 [DOI] [PubMed] [Google Scholar]
- 28. Werner N. S., Windoffer R., Strnad P., Grund C., Leube R. E., Magin T. M. (2004) Epidermolysis bullosa simplex-type mutations alter the dynamics of the keratin cytoskeleton and reveal a contribution of actin to the transport of keratin subunits. Mol. Biol. Cell 15, 990–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Darby N., Creighton T. E. (1995) Disulfide bonds in protein folding and stability. Methods Mol. Biol. 40, 219–252 [DOI] [PubMed] [Google Scholar]
- 30. Wedemeyer W. J., Welker E., Narayan M., Scheraga H. A. (2000) Disulfide bonds and protein folding. Biochemistry 39, 4207–4216 [DOI] [PubMed] [Google Scholar]
- 31. Takamori K., Thorpe J. M., Goldsmith L. A. (1980) Skin sulfhydryl oxidase: purification and some properties. Biochim. Biophys. Acta 615, 309–323 [DOI] [PubMed] [Google Scholar]
- 32. Hashimoto Y., Suga Y., Matsuba S., Mizoguchi M., Takamori K., Seitz J., Ogawa H. (2000) Immunohistochemical localization of sulfhydryl oxidase correlates with disulfide crosslinking in the upper epidermis of rat skin. Arch. Dermatol. Res. 292, 570–572 [DOI] [PubMed] [Google Scholar]
- 33. Thiele J. J., Schroeter C., Hsieh S. N., Podda M., Packer L. (2001) The antioxidant network of the stratum corneum. Curr. Probl. Dermatol. 29, 26–42 [DOI] [PubMed] [Google Scholar]
- 34. Hamanaka R. B., Glasauer A., Hoover P., Yang S., Blatt H., Mullen A. R., Getsios S., Gottardi C. J., DeBerardinis R. J., Lavker R. M., Chandel N. S. (2013) Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci. Signal. 6, ra8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kennedy L. H., Sutter C. H., Leon Carrion S., Tran Q. T., Bodreddigari S., Kensicki E., Mohney R. P., Sutter T. R. (2013) 2,3,7,8-Tetrachlorodibenzo-p-dioxin-mediated production of reactive oxygen species is an essential step in the mechanism of action to accelerate human keratinocyte differentiation. Toxicol. Sci. 132, 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ku N. O., Gish R., Wright T. L., Omary M. B. (2001) Keratin 8 mutations in patients with cryptogenic liver disease. N. Engl. J. Med. 344, 1580–1587 [DOI] [PubMed] [Google Scholar]
- 37. Ku N. O., Omary M. B. (2006) A disease- and phosphorylation-related nonmechanical function for keratin 8. J. Cell Biol. 174, 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Coulombe P. A., Hutton M. E., Letai A., Hebert A., Paller A. S., Fuchs E. (1991) Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell 66, 1301–1311 [DOI] [PubMed] [Google Scholar]
- 39. Letai A., Coulombe P. A., McCormick M. B., Yu Q. C., Hutton E., Fuchs E. (1993) Disease severity correlates with position of keratin point mutations in patients with epidermolysis bullosa simplex. Proc. Natl. Acad. Sci. U.S.A. 90, 3197–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.