Background: Point mutation P222L in PKR activator PACT causes movement disorder dystonia.
Results: PACT mutant P222L causes delayed and prolonged PKR and eIF2α phosphorylation in response to the ER stress.
Conclusion: Altered kinetics of PKR activation in response to the ER stress enhances apoptosis in dystonia cells.
Significance: This is the first study of molecular mechanisms involved in dystonia 16.
Keywords: apoptosis, cell death, endoplasmic reticulum stress (ER stress), interferon, protein kinase RNA-activated (PKR)
Abstract
PACT is a stress-modulated activator of the interferon-induced double-stranded RNA-activated protein kinase (PKR). Stress-induced phosphorylation of PACT is essential for PACT's association with PKR leading to PKR activation. PKR activation leads to phosphorylation of translation initiation factor eIF2α inhibition of protein synthesis and apoptosis. A recessively inherited form of early-onset dystonia DYT16 has been recently identified to arise due to a homozygous missense mutation P222L in PACT. To examine if the mutant P222L protein alters the stress-response pathway, we examined the ability of mutant P222L to interact with and activate PKR. Our results indicate that the substitution mutant P222L activates PKR more robustly and for longer duration albeit with slower kinetics in response to the endoplasmic reticulum stress. In addition, the affinity of PACT-PACT and PACT-PKR interactions is enhanced in dystonia patient lymphoblasts, thereby leading to intensified PKR activation and enhanced cellular death. P222L mutation also changes the affinity of PACT-TRBP interaction after cellular stress, thereby offering a mechanism for the delayed PKR activation in response to stress. Our results demonstrate the impact of a dystonia-causing substitution mutation on stress-induced cellular apoptosis.
Introduction
PKR is an interferon-induced serine/threonine kinase expressed ubiquitously that mediates interferon's antiviral actions and regulates cellular survival and apoptosis in response to stress (1). PKR2 kinase activity requires binding to one of its activators leading to its autophosphorylation and enzymatic activation (2). Double-stranded RNA (dsRNA), a replication intermediate for several viruses, binds to PKR's two dsRNA binding motifs (dsRBMs) (3–5) and activates PKR by unmasking the ATP-binding site (6) leading to its autophosphorylation (7). The two dsRBMs also mediate dsRNA-independent protein-protein interactions with other proteins that carry similar domains (8, 9). Among these are proteins TRBP (human immunodeficiency virus (HIV)-1 transactivation-responsive (TAR) RNA-Binding Protein) that inhibits PKR activity (10) and PKR activator protein PACT (11).
PACT's association with PKR activates PKR in the absence of dsRNA (11, 12). PACT contains three copies of dsRBM (Fig. 1A), of which the two amino-terminal motifs (M1 and M2) bind to the dsRBMs of PKR. The third, carboxyl-terminal motif 3 (M3) is dispensable for interaction with PKR but is essential for PKR activation and interacts with a specific region in its kinase domain (13, 14). Although purified, recombinant PACT can activate PKR by direct interaction in vitro (11), PACT-dependent PKR activation in cells occurs in response to stress signals (12, 15–17) such as arsenite, peroxide, growth factor withdrawal, thapsigargin, and tunicamycin and leads to phosphorylation of the translation initiation factor eIF2α and cellular apoptosis (12, 15, 16). PACT (and its murine homolog RAX) is phosphorylated in response to stress, leading to its increased association with PKR (12, 15, 16).
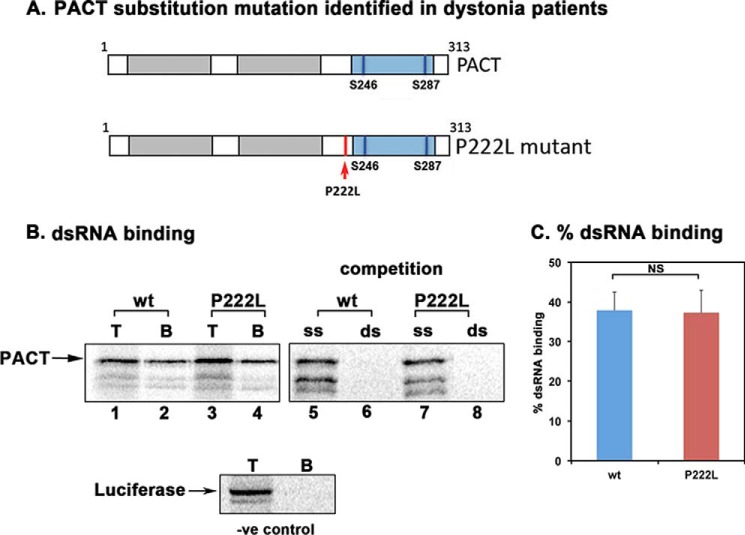
FIGURE 1.
Effect of P222L mutation on dsRNA binding. A, domain structure of PACT. Gray boxes, dsRNA binding M1 and M2 motifs; blue box, M3 motif that does not bind dsRNA but is in essential for PKR activation. Vertical blue lines, phosphoserines 246 and 287. Red arrow, P222L mutation. B, dsRNA binding assay. dsRNA binding activity of WT PACT and P222L was measured by a poly(I)·poly(C)-agarose binding assay with in vitro translated 35S-labeled proteins. T, total input; B, proteins bound to poly(I)·poly(C)-agarose. -ve, negative. Competition lanes: competition with 100-fold molar excess of single-stranded RNA (ss) or dsRNA (ds). The bands below the parent PACT bands represent products of in vitro translation from internal methionine codons and thus are not produced in similar quantities in all translation reactions and thus are of variable intensity in lanes 1–8. C, quantification of the dsRNA binding assay. Bands were quantified by phosphorimaging analysis, and % bound was calculated. Error bars: S.D. from three independent experiments. The p value (0.43) calculated using statistical analyses indicated no significant difference between % dsRNA-binding of WT (blue bar) and P222L mutant (red bar) as indicated by the bracket marked as NS.
Similar to PACT, TRBP is a dsRNA-binding protein, but unlike PACT it inhibits PKR. In uninfected cells and in the absence of cellular stress TRBP inhibits PKR by direct binding (18) and by forming heterodimers with PACT (19). Recently we showed that cellular stress signals cause PACT to dissociate from TRBP leading to PACT-mediated PKR activation. TRBP-PACT heterodimers present in unstressed cells dissociate, as PACT is phosphorylated on Ser-287 in M3 in response to oxidative stress, serum starvation, and endoplasmic reticulum (ER) stress (20, 21) by a protein kinase yet to be identified. Stress-induced phosphorylation at serine 287 has a dual role in PACT-mediated PKR activation as it causes dissociation of the PACT-TRBP complex and at the same time increases PACT affinity for PKR (21). Two PACT molecules can also interact via the conserved dsRBMs, and phosphorylation of serine 287 enhances PACT-PACT interactions (22). The PACT-PACT homodimers interact strongly with PKR, leading to catalytically active PKR. Thus stress-induced phosphorylation of serine 287 of PACT serves to enhance PACT-PACT and PACT-PKR interactions in addition to reducing PACT-TRBP interactions. Consequently, apoptosis in response to stress signals is regulated by various PACT-TRBP-PKR interactions, with each partner capable of forming homomeric interactions as well as interacting with the other two proteins.
Camargos et al. (23) described a recessively inherited form of early-onset generalized dystonia due to a homozygous missense mutation in PACT (PRKRA). The dystonias are a heterogeneous group of movement disorders in which affected individuals develop sustained, often painful involuntary muscle contractions and twisted postures that can have devastating consequences (24). For DYT16, the affected members from the two unrelated families have the same P222L mutation in PACT gene (25). This point mutation lies between the conserved motifs M2 and M3 within PACT (26). The other mutation reported in PACT that causes dystonia is a frameshift mutation that results in truncation of the protein after 88 amino acids (27). Recently, three more recessive mutations (C77S, C213F, and C213R) were found in DYT16 patients (28–30). The three most recent mutations reported in Polish and German families (T34S, N102S, and c.-14A→G) indicate a worldwide involvement of PACT (PRKRA) gene in dystonia (31).
Despite the identification of many genetic mutations that lead to dystonia, the molecular mechanisms involved in disease onset or progression have remained largely unknown (32). In this report we have analyzed the effect of P222L mutation on PACT's biochemical properties such as dsRNA binding, PKR interaction, and PKR activation. P222L mutation does not affect PACT's ability to bind dsRNA or its ability to interact with PKR in vitro. However, in mammalian cells the P222L mutant protein interacts with higher affinity with TRBP, forms homodimers more efficiently, and in response to stress causes a delayed but much prolonged activation of PKR. In accordance to the altered biochemical properties of P222L protein, dystonia patient cells exhibit enhanced apoptosis in response to ER stressor tunicamycin. These results indicate that dystonia-causing PACT mutation alters the kinetics and duration of eIF2α phosphorylation in response to ER stress and has deleterious implications on cell survival.
Experimental Procedures
Reagent, Cell Lines, and Antibodies
Patient B-lymphoblast cell lines were cultured in RPMI 1640 medium, and HeLa M cells were cultured in Dulbecco's modified Eagle's medium, both containing 10% fetal bovine serum and penicillin/streptomycin. The WT and DYT16 dystonia patient lymphoblast cell lines were Epstein-Barr Virus-transformed to create stable cell lines as previously described (33). The other reagents were tunicamycin (Santa Cruz) and phosphatase inhibitor mixture (Sigma). The antibodies used are as follows: anti-FLAG monoclonal M2 (Sigma A8592), anti-PKR (human) monoclonal (71/10, R&D Systems), anti-phospho-PKR polyclonal (Thr-451) (Cell Signaling, 3075), anti-eIF2α polyclonal (Invitrogen, AH01182), anti-phospho-eIF2α (Ser-51) polyclonal (Epitomics, 1090-1; Cell Signaling 9721), anti-PACT rabbit monoclonal (abcam 75749), and anti-myc monoclonal (Santa Cruz 9E10).
Generation of P222L Substitution Mutation
Point mutation P222L was generated using a mutagenic primer for PCR amplification to change the codon for proline 222 from CCT to CTT. The primer sequences were: upstream mutagenic primer, 5′-CCTTGAGGAATTCTCTTGGTGAAAAGATCAAC-3′; downstream primer, 5′-GGGGATCCTTACTTTCTTTCTGCTATTATC-3′.
The PCR product was subcloned into pGEMT-easy vector (Promega). Once the sequence of the point mutant (P222L) was verified, we generated full-length P222L ORF in pcDNA3.1− by a three-piece ligation of XbaI-EcoRI restriction piece from FLAG-PACT/BSIIKS+, an EcoRI-BamHI piece from P222L point mutant/pGEMT-easy, and XbaI-BamH1 cut pcDNA3.1−. The full-length P222L mutant in pcDNA3.1− has an amino-terminal FLAG or myc tag. The full-length P222L mutant was subcloned into mammalian two-hybrid system vectors and pET15b (Novagen). TRBP constructs were as described before (21).
dsRNA Binding Assay
The in vitro translated, 35S-labeled PACT proteins were synthesized using the TNT-T7-coupled reticulocyte lysate system from Promega, and the dsRNA binding activity was measured by using the previously established poly(I)·poly(C)-agarose binding assay (3, 11). 4 μl of in vitro translation products were diluted with 25 μl of binding buffer (20 mm Tris, pH 7.5, 0.3 m NaCl, 5 mm MgCl2, 1 mm DTT, 0.1 mm PMSF, 0.5% Nonidet P-40, 10% glycerol) and incubated with 25 μl of poly(I)·poly(C)-agarose beads at 30 °C for 30 min. The beads were washed 4 times with 500 μl of binding buffer, and the bound proteins were analyzed by SDS-PAGE and fluorography. For a competition assay with soluble single- or double-stranded RNA, 1 μg of poly(C) or poly(I)·poly(C) was incubated with the proteins for 15 min at 30 °C before the addition of poly(I)·poly(C)-agarose beads. To ascertain specific interactions between PACT proteins and poly(I)·poly(C)-agarose beads, in vitro translated, 35S-labeled firefly luciferase protein was assayed for binding to the poly(I)·poly(C)-agarose beads using same conditions. The T lanes represent total radioactive proteins in the reticulocyte lysate, and the B lanes represent the proteins that remain bound to poly(I)·poly(C)-agarose beads after washing. The poly(I)·poly(C)-agarose binding was quantified on Typhoon FLA7000 by analyzing the band intensities in T and B lanes. The percentage of PACT proteins bound to poly(I)·poly(C)-agarose was calculated from these values (% binding = 100 × band intensity in B lane/band intensity in T lane) and was plotted as bar graphs.
Co-immunoprecipitation with in Vitro Translated Proteins
In vitro translated, 35S-labeled PKR, and FLAG epitope-tagged PACT proteins were synthesized using the TNT-T7 coupled reticulocyte system from Promega, and the co-immunoprecipitation assay was performed as described before (11).
Co-immunoprecipitation Assay in HeLa Cells
For PACT/P222L and PKR co-immunoprecipitations, HeLa cells were co-transfected in 6-well culture dishes with 500 ng of FLAG-PACT/pcDNA3.1− or FLAG-P222L/pcDNA3.1− using the Effectene (Qiagen). For PACT/P222L-TRBP, PACT-PACT, and P222L-P222L co-immunoprecipitations, HeLa cells were co-transfected in 6-well culture dishes with 250 ng each of (i) myc-TRBP/pcDNA3.1- and FLAG-WTPACT/pcDNA3.1-, (ii) myc-TRBP/pcDNA3.1- and FLAG-P222L pcDNA3.1-, (iii) FLAG-PACT/pcDNA3.1- and myc-PACT/pcDNA3.1-, and (iv) FLAG-P222L/pcDNA3.1- and myc-P222L/pcDNA3.1-. At 24 h post-transfection, co-immunoprecipitations were performed as described before.
PKR Kinase Activity Assays
PKR kinase activity assays were performed using HeLa M cell extracts as described before (11, 21). One μg/ml of poly(I)·poly(C) was used as the standard activator. Purified PACT or P222L proteins in amounts varying from 50 pg to 100 ng were tested for their effect on PKR activity.
DNA Fragmentation Analysis
DNA fragmentation analysis was performed as described before (17). 5 × 106 lymphoblast cells established from WT individuals and dystonia patients were treated with 0.5 μg/ml tunicamycin for 48 h followed by DNA fragmentation analysis. To quantify the DNA fragmentation, the fluorescence image was inverted, and the total band intensities in the entire lanes were computed with ImageQuant software on Typhoon FLA 7000 PhosphorImager (GE Healthcare and Life Sciences) and compared with untreated samples as well as between WT and patient cells. The band intensities in WT untreated samples were considered as 1.0, and -fold increases in band intensities with respect to WT untreated samples were calculated and subjected to statistical analysis. A statistical analysis from four different experiments was performed to calculate p values to determine significant differences between WT and patient untreated and treated samples.
Flow Cytometry Analysis
5 × 105 lymphoblast cells established from WT individuals and dystonia patients were treated with 0.5 μg/ml tunicamycin for 48 h. After treatment, cells were washed once in 1× PBS, resuspended in 70% ethanol, and kept at −20 °C overnight. Cells were rinsed with 1× PBS and resuspended in 0.5 ml 1× PBS, and 1 ml of PC buffer (50 mm Na2HPO4, 85 mm sodium citrate, 0.1% Triton X, pH 7.8) was added dropwise followed by incubation at room temperature for 35 min. Cells were rinsed with 1× PBS, labeled with propidium iodide (PI) (25 μg/ml PI in PBS, 10 μg/ml RNase A, 0.1% Triton X) for 30 min. Cell cycle analysis was performed by the Flow Cytometry core at Flow Cytometry core at University of South Carolina School of Pharmacy School of Pharmacy.
Caspase 3/7 Assay
Patient and WT B-lymphoblast cells were plated at 2 × 105 and either left untreated or treated with 0.5 μg/ml tunicamycin. Aliquots of untreated and treated cells were collected at the indicated time points. After collection, aliquots were mixed with equal parts of Promega Caspase-Glo 3/7 reagent and incubated for 45 min. Luciferase activity was measured with a negative control of cell culture medium alone used to normalize all readings.
Western Blot Analysis
Western blot analysis was performed using primary antibodies as described under reagents. Western blots were quantified using ImageQuant LAS 4000.
Expression and Purification of Recombinant WT PACT and P222L
The protein coding regions (WT or P222L mutant) were subcloned into pET15b (Novagen) to generate PACT/pET15b and P222L/pET15b, resulting in the in-frame fusion of PACT ORF to the histidine tag. The recombinant proteins were expressed and purified as described (11, 21).
Mammalian Two-hybrid Interaction Assay
The WT PACT and P222L ORFs were subcloned into pSG424 (Addgene) such that it produced an in-frame fusion to GAL4 DBD and in VP16 AD vector pVP16AASV19N (8, 34) such that it produced an in-frame fusion to VP16 AD. Fusion proteins were tested for interaction in various combinations. COS-1 cells were transfected with 250 ng of each of the three (two test plasmids encoding proteins to be tested for interaction and reporter plasmid pG5Luc) and 1 ng of pRLNull (Promega) to normalize the transfection efficiencies using Effectene (Qiagen). Cells were harvested 24 h after transfection and assayed for luciferase activity. Western blot analysis was done using the anti-GAL4 DBD and -VP16 AD antibodies.
Yeast Two-hybrid Interaction Assay
PACT and its point mutants were expressed as GAL4 DNA binding domain fusion proteins from pGBKT7 vector, whereas PACT and its point mutants, TRBP, and PKR was expressed as GAL4 activation domain fusion proteins from pGADT7 vector. Each pGBKT7 and pGADT7 construct was co-transformed into Saccharomyces cerevisiae strain AH109 (Clontech) and selected on double dropout SD minimal medium lacking tryptophan and leucine. Transformation of each of the PACT constructs in pGBKT7 and empty vector pGADT7 served as negative controls. To check for the transformants' ability to grow on quadruple dropout histidine-, leucine-, tryptophan-, and adenine-lacking medium, 10 μl of serial dilutions (of A600 = 10, 1.0, 0.1, 0.01) of an overnight liquid culture were spotted for each of the transformants on quadruple dropout SD medium plates lacking adenine, tryptophan, leucine, and histidine containing 25 mm 3-amino-1,2,4-triazole. Plates were incubated for 3 days at 30 °C.
Quantifications and Statistics
All Western blot images and radioactive gel scans (Typhoon FLA7000) were quantified using GE Life Sciences ImageQuant TL software. To determine the statistical significance of the results of the Western blots as well as DNA fragmentation, flow cytometry profiles, and caspase assays, a two-tailed Student's t test was performed, assuming equal variance. Each figure legend indicates p values as denoted by brackets and special characters. Note that our alpha level was p = 0.05.
Results
P222L Mutation Did Not Affect PACT's dsRNA Binding but Increased Interaction with PKR
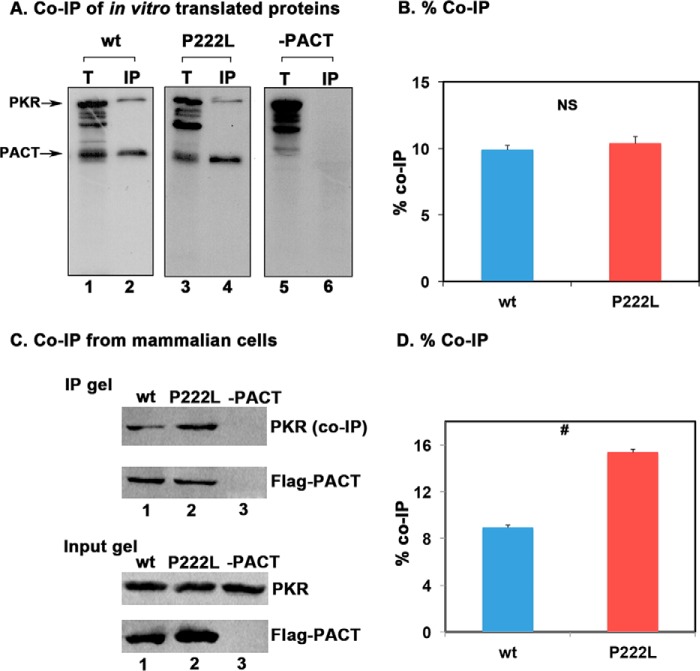
The P222L point mutation is between the conserved M2 and M3 motifs (Fig. 1 A). Currently there is no structural information on PACT to predict possible changes due to P222L mutation. To determine if the mutation affects PACT's dsRNA binding activity, an in vitro dsRNA binding assay previously well established for PKR and PACT (11) was performed (Fig. 1, B and C). As seen in Fig. 1B, both the WT PACT (lane 2) and P222L mutant (lane 4) bound to dsRNA. The binding to dsRNA immobilized on the beads could be competed out by exogenously added dsRNA (lanes 6 and 8) but not single-stranded RNA (lanes 5 and 7). Firefly luciferase, a protein that does not bind dsRNA used as a negative control showed no binding to the beads. The quantification of percentage binding indicates that both WT PACT and P222L appear to have a similar affinity for dsRNA under the condition that the assay was carried out (Fig. 1C). We next examined if P222L mutation affects PACT's interaction with PKR using co-immunoprecipitation assays (Fig. 2, A and B). [35S]Methionine-labeled FLAG-PACT, FLAG-P222L mutant, and untagged PKR were in vitro translated using the rabbit reticulocyte system. The FLAG-tagged PACT or P222L mutant was immunoprecipitated using anti-FLAG mAb-agarose, and the co-immunoprecipitation of untagged WT PKR was measured. As seen in Fig. 2, A and B, both FLAG-PACT and FLAG-P222L mutants appear to have a similar affinity for PKR under the conditions in which the assay was carried out (lanes 2 and 4), and PKR was not immunoprecipitated with FLAG-mAb-agarose in the absence of FLAG-PACT (lane 6). In this assay, using in vitro translated proteins, no significant difference in the strength of interaction with PKR was observed between WT and P222L mutant PACT proteins under the condition that the assay was carried out (Fig. 2B). To assay for PACT-PKR interaction in a mammalian cells, FLAG-PACT and FLAG-P222L mutant were expressed in HeLa cells, and co-immunoprecipitation of endogenous WT PKR with the FLAG-tagged proteins was assayed. Both FLAG-PACT and FLAG-P222L were able to pull down WT PKR, and FLAG-P222L consistently showed a higher efficiency for co-immunoprecipitating PKR as compared with FLAG-PACT (Fig. 2C, lanes 1 and 2). In the absence of transfected FLAG-PACT proteins, no immunoprecipitation of endogenous PKR was observed with anti-FLAG mAb-agarose (lane 3). The % co-immunoprecipitation was quantified from three separate experiments by analyzing band intensities (Fig. 2D), and the results consistently showed a significantly stronger interaction of FLAG-P222L with PKR as compared with FLAG-PACT interaction with PKR. These results establish that P222L mutant retains the ability to bind to PKR and, compared with WT PACT, exhibits an enhanced PKR interaction in mammalian cells.
FIGURE 2.
Effect of P222L mutation on PACT-PKR interaction. A, co-IP of in vitro translated proteins. 5 μl of in vitro translated, 35S-labeled FLAG-tagged WT PACT and P222L proteins were mixed with 5 μl of in vitro translated, 35S-labeled WT PKR. FLAG-PACT proteins were immunoprecipitated using anti-FLAG mAb-agarose, and WT PKR co-immunoprecipitation was analyzed by SDS-PAGE. T, total input (20% of the IP samples). B, quantification of data in panel A. The radioactivity present in the bands was measured by phosphorimaging analysis, and the % co-IP was calculated as (radioactivity present in the co-immunoprecipitated PKR band/the radioactivity present in the PKR band in the total lane) × 100. This value was normalized to the amount of radioactivity present in the PACT bands in IP lanes to correct for differences in translation/immunoprecipitation. Error bars: S.D. from three independent experiments. The p value (0.391) calculated using statistical analyses indicated no significant difference between % co-IP of WT (blue bar) and P222L mutant (red bar) with PKR as indicated by the bracket marked as NS. C, co-IP of WT PACT and P222L proteins from HeLa cells. HeLa cells were transfected with FLAG-PACT/pCDNA3.1− (lanes 1 and 2) or pCDNA3.1− (lane 3). 24 h after transfection, FLAG-tagged PACT protein was immunoprecipitated for 1 h using anti-FLAG mAb-agarose. The immunoprecipitates were analyzed by Western blot analysis with anti-PKR monoclonal antibody (PKR (co-IP panel)). The blot was stripped and re-probed with monoclonal anti-FLAG M2 antibody (FLAG-PACT panel). The input gel (20% of IP lanes) shows Western blot analysis of total proteins in the extract without immunoprecipitation to ascertain equal amounts of FLAG-PACT expression after transfection in and that an equal amount of PKR was present in both samples. D, quantification of Co-IP in panel C. The relevant bands were quantified by phosphorimaging analysis, and % co-IP was calculated from the band intensities in input and IP lanes. These values were normalized to band intensities of FLAG-PACT in IP gel. Error bars: S.D. was calculated from three independent experiments. The p value (0.0027) calculated using statistical analyses indicated a significant difference between % co-IP of WT (blue bar) and P222L mutant (red bar) with PKR as indicated by the bracket marked by #.
P222L Mutation Makes PACT a More Efficient PKR Activator
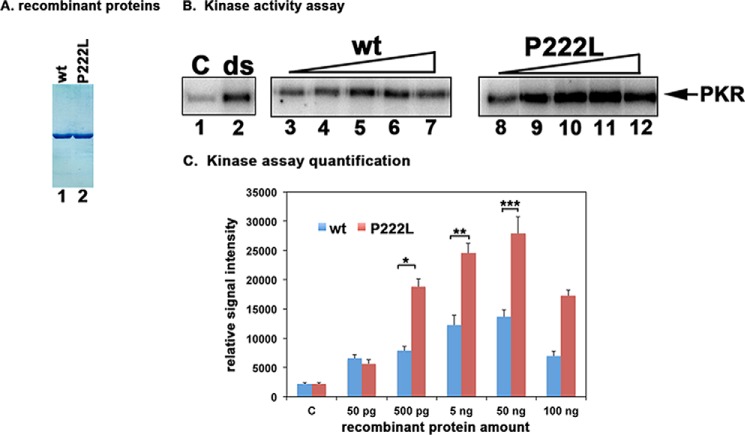
The effect of P222L mutation on PACT's ability to activate PKR was assayed by using the in vitro PKR activity assay. Hexahistidine-tagged WT PACT and P222L proteins expressed in bacteria were purified on nickel affinity beads (Fig. 3A). These purified recombinant proteins were used as activators for an in vitro PKR kinase activity assay using PKR immunoprecipitated from HeLa cells (11). The assay measures autophosphorylation and activation of PKR. As seen in Fig. 3B, there was a basal level of PKR activity seen in the absence of exogenous PKR activator (lane 1) and significantly elevated PKR activity in the dsRNA (ds) positive control (lane 2). There was a dose-dependent increase in PKR autophosphorylation in response to increasing amounts of WT PACT (lanes 3–7). In response to increasing amounts of P222L there was significantly higher autophosphorylation of PKR (lanes 8–12) as compared with that seen with equivalent amounts of WT PACT (lanes 3–7). The quantification of radioactive signal in PKR band using phosphorimaging analysis is shown in Fig. 3C. Statistical analyses were performed on results from five independent experiments, and all concentrations of WT and P222L protein showed significant PKR activation. Three of these p values are as indicated in Fig. 3C with brackets. These results indicate that the P222L mutant activates PKR more efficiently in vitro as compared with WT PACT.
FIGURE 3.
Effect of P222L mutation on PKR activation. A, purification of recombinant PACT proteins. 500 ng of purified, recombinant hexahistidine-tagged WT PACT and P222L proteins were analyzed by SDS-PAGE and Coomassie Blue staining. B, kinase activity assay. PKR immunoprecipitated from HeLa cell extracts using PKR monoclonal antibody, which immunoprecipitates total PKR (R&D Systems), was used to measure PKR kinase activity without any activator (C) or activators added as indicated above the lanes. Lane 1, PKR activity without any activator, lane 2, 100 ng/ml poly(I)·poly(C) as activator. Purified recombinant WT PACT or P222L in amounts of 50 pg (lanes 3 and 8), 500 pg (lanes 4 and 9), 5 ng (lanes 5 and 10), 50 ng (lanes 6 and 11), and 100 ng (lanes 7 and 12). C, quantification of kinase assay. The radioactivity present in the bands was measured by phosphorimaging analysis, and the relative signal intensities are plotted. Error bars: S.D. from five independent experiments performed with two independent preparations of recombinant WT PACT and P222L proteins. Student's t tests performed indicated that the relative signal intensity increases in radioactive PKR bands as compared with control lanes at all WT and P222L protein concentrations were very significant with all p values lower than 0.05. Three of these values were also further analyzed to investigate if the differences observed between WT and P222L activation of PKR were significant and are as indicated: 500 pg (bracket *) = 0.0015; 5 ng (bracket **) = 0.0017; 50 ng (bracket ***) = 0.003, n = 5.
Patient Lymphoblasts Homozygous for P222L Mutation Exhibit Enhanced Sensitivity to the ER Stressor Tunicamycin
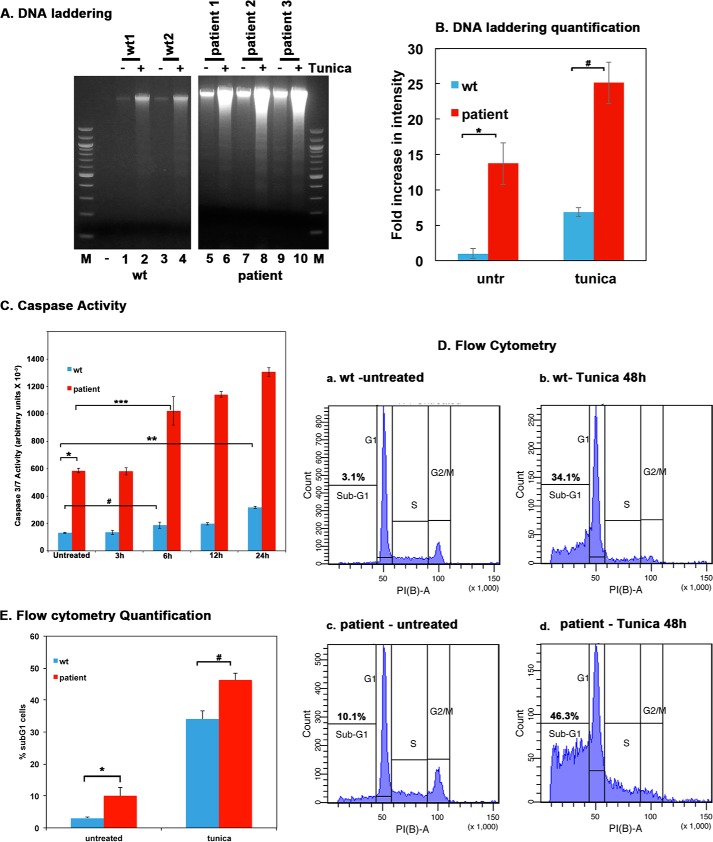
To analyze the effect of P222L mutant's increased ability to activate PKR in a cellular context, apoptosis in response to the ER stressor tunicamycin was evaluated in patient and WT lymphoblasts. Tunicamycin inhibits protein glycosylation in the ER, causing misfolded proteins to accumulate, thereby inducing the ER stress response pathway (35). Apoptosis was measured using DNA fragmentation, flow cytometry, and caspase 3 and 7 activity. As seen in Fig. 4A, the WT lymphoblast cells showed some increase in fragmented DNA with tunicamycin treatment (lanes 2 and 4). In contrast to this, the patient lymphoblasts showed markedly increased DNA fragmentation in response to tunicamycin (lanes 6, 8, and 10), indicating enhanced sensitivity to the ER stress. Quantification of DNA fragmentation was performed to calculate -fold increases in band intensities after tunicamycin treatment compared with untreated samples (Fig. 4B). Statistical analysis revealed that the patient lymphoblasts show significantly more DNA fragmentation as compared with WT lymphoblast in response to tunicamycin (bracket marked by #). In addition, the patient lymphoblasts also show significantly more DNA fragmentation as compared with WT lymphoblasts even in untreated samples (bracket marked by *). These results indicate that patient lymphoblasts are more sensitive to apoptosis in response to the stress and show significantly higher levels of apoptosis even in the absence of any deliberately applied external ER stress. To further compare the apoptotic response of WT and patient lymphoblasts, we measured the caspase activity in response to tunicamycin. As seen in Fig. 4C, there is a dramatic increase in caspase 3/7 activity in patient lymphoblasts as compared with the WT lymphoblasts, even in the absence of the ER stress (bracket *). In addition, a significant increase in caspase activity was seen 6 h after treatment in the patient lymphoblasts (bracket ***), whereas the WT lymphoblasts showed a very modest increase in caspase activity at 6 h (bracket #). Although there was a more significant increase in caspase activity in WT lymphoblasts at 24 h (bracket **), the overall caspase activity in the WT lymphoblasts was fairly low even at 24 h after treatment as compared with the patient lymphoblasts. These results further establish that patient lymphoblasts exhibit enhanced sensitivity to the ER stressor tunicamycin, and in agreement with our DNA fragmentation, results (Fig. 4A) also show higher caspase activity without any external stressor. To further quantify the difference in apoptosis between WT and patient lymphoblasts, flow cytometry analysis was performed. As seen in Fig. 4D, only 3.1% of the WT cells had a subG1 DNA content in the absence of the ER stress (panel a), whereas 10.1% of the patient cells had a subG1 DNA content in the absence of the ER stress (panel c). When the lymphoblast lines were treated with tunicamycin for 48 h before analysis, the subG1 population increased to 34.1% for WT lymphoblast (panel b) and to 48.3% in patient lymphoblast (panel d). Fig. 4E shows a bar graph representing these data with the error bars and statistical analyses performed for four different experiments. The difference in % apoptosis between WT and dystonia patient cells both in untreated as well as in tunicamycin-treated samples is statistically significant as indicated by the brackets * and #. These results demonstrate that the patient cells exhibit significantly higher levels of apoptosis in the absence of an extrinsic stress signal and are markedly more sensitive to the ER stressor tunicamycin.
FIGURE 4.
Tunicamycin-induced apoptosis is enhanced in dystonia patient lymphoblasts. A, DNA fragmentation analysis. Lymphoblast cell lines established from 2 normal (WT) individuals and 3 dystonia patients were treated with 0.5 μg/ml tunicamycin for 48 h. The DNA fragmentation was analyzed as described under “Experimental Procedures.” Lanes 1–4: WT (normal) lymphoblasts; lanes 5–10, dystonia patient (affected) lymphoblasts. − lanes (1, 3, 5, 7, and 9), untreated cells; + lanes, (lanes 2, 4, 6, 8, and 10), tunicamycin-treated cells. Lane M, 100-bp ladder. B, quantification of DNA fragmentation. The inverted images from all four experiments as in panel A were analyzed by GE Healthcare ImageQuant TL software to calculate total band intensities in each lane from top to bottom. Blue bars, normal (WT) lymphoblasts; red bars, dystonia patient lymphoblasts. Band intensities in WT untreated samples were considered as 1.0, and all other samples were expressed as -fold increases compared with that value. Student's t tests were performed, and p values are as follows. * = 0.00012 (significant), # = 0.00015 (significant), n = 4. C, Caspase-Glo 3/7 assay. Lymphoblast cell lines established from 2 normal (WT) individuals and 3 dystonia patients were treated with 0.5 μg/ml tunicamycin for the indicated time points. Caspase 3 and 7 activities were measured. Blue bars, normal (WT) lymphoblasts; red bars, dystonia patient lymphoblasts. Student's t tests were performed, and p values are as follows. * = 0.00016 (significant), ** = 0.00041 (significant), # = 0.002 (significant), and *** = 0.0037 (significant), n = 4. D, flow cytometry analysis. Lymphoblast from 2 normal (WT) or 3 affected (dystonia patient) individuals were treated with 0.5 μg/ml tunicamycin. Cells were harvested at 48 h after the treatment and subjected to flow cytometry analysis. The sub-G0/G1 cell population represents the dying cells. The sub-G0/G1 percentages are displayed in each panel. These experiments were repeated four times. The most representative profiles from a single experiment with one WT and one patient line are shown. E, flow cytometry quantification and statistical analysis. The percentage of sub-G1 cells from the experiment in panel D is represented as a bar graph, and the percentages of sub-G1 cells from four different experiments were subjected to Student's t tests. p values are as follows. * = 0.00022 (significant), # = 0.00025 (significant).
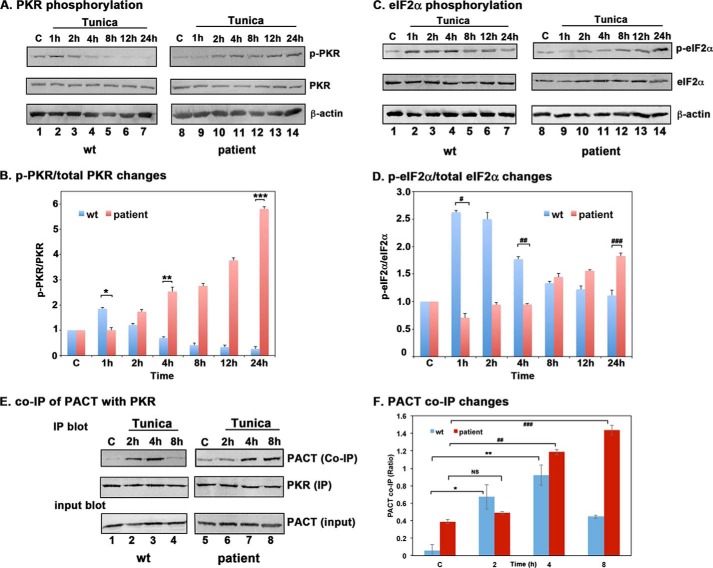
Patient Lymphoblasts Homozygous for P222L Mutation Show a Delayed but Enhanced and Prolonged PKR Activation in Response to Tunicamycin
To understand the mechanism of enhanced sensitivity of patient lymphoblasts to the ER stress, we compared PKR activation kinetics by Western blot analysis in WT and patient lymphoblasts at the indicated time points after tunicamycin treatment. As shown in Fig. 5A, PKR phosphorylation was observed at 1 h (lane 2) and 2 h (lane 3) after treatment, began to decrease at 4 h (lane 4), and was barely detectable after 8 h (lanes 5–7) in WT cells. In contrast to this, the patient cells did not show an increase in PKR phosphorylation until 2 h after treatment (lane 9), but this phosphorylation was maintained and continually increased until 24 h after treatment (lanes 10–13). These results demonstrated that PKR activation follows a slow, albeit prolonged kinetics in patient cells as opposed to rapid and short duration in WT cells. The ratio of phosphorylated PKR to total PKR was calculated based on phosphorimaging quantification of band intensities and is represented in Fig. 5B, and the p values calculated from Student's t tests indicate a significant difference in PKR phosphorylation between WT and patient lymphoblasts as represented by the brackets marked *, **, and ***. We next examined if eIF2α phosphorylation in response to tunicamycin followed kinetics similar to PKR activation. Similar to the kinetics of PKR activation, eIF2α phosphorylation is seen as early as 1 h (Fig. 5A, lane 2) after treatment and is maintained at high until 4 h (lanes 2–4), after which the levels gradually decreased (lanes 5–7) in WT cells. In patient lymphoblasts, a comparable increase was not seen until 8 h after treatment (lane 12), and these levels slowly rose until 24 h after treatment (lanes 11–14). These results indicate that eIF2α phosphorylation is delayed in response to tunicamycin in patient lymphoblasts but persists for a prolonged duration compared with WT cells. The ratio of phosphorylated eIF2α to total eIF2α was calculated based on phosphorimaging quantification of band intensities and is represented in Fig. 5D, and the p values calculated from Student's t tests indicate a significant difference in eIF2α phosphorylation between WT and patient lymphoblasts as represented by the brackets marked #, ##, and ###. We next evaluated the association of PACT with PKR in response to tunicamycin using a co-immunoprecipitation analysis. PKR was immunoprecipitated from WT and patient lymphoblast cell lysates at the indicated time points after tunicamycin treatment using a PKR monoclonal antibody. The immunoprecipitates were analyzed by Western blot analysis using anti-PACT antibody. As seen in Fig. 5E, in WT cells, compared with control (lane 1), PACT association with PKR was increased at 2 h and 4 h (lanes 2 and 3) after tunicamycin treatment and returned to basal levels by 8 h (lane 4). However, in patient cells PACT association with PKR stayed at basal levels at 2 h after treatment (lane 6) similar to control (lane 5) and only showed increases at 4 h (lane 7) and 8 h (lane 8). These results suggest that PACT association with PKR shows delayed kinetics in patient lymphoblasts in response to tunicamycin. It is interesting to note that in the absence of tunicamycin an increase in P222L association with PKR was seen in patient cells as compared with PACT association with PKR in WT cells, further confirming increased association of P222L with PKR in absence of stress (Fig. 2C). PACT's co-immunoprecipitation with PKR was quantified from three separate experiments and is shown as a bar graph in Fig. 5F. The ratio of PACT co-IP signals to PACT input signals was normalized to PKR IP signals for each lane. The delayed kinetics of PACT association with PKR in response to tunicamycin in dystonia patient cells is further confirmed by this analysis. The p values calculated from Student's t tests are as indicated in the figure legend and demonstrate that the difference observed in kinetics of PACT-PKR association between WT and patient cells is statistically significant. The data shown are representative of results from three different patient samples, all of which showed a similar trend.
FIGURE 5.
PACT-PKR interaction, PKR activation, and eIF2α phosphorylation in response to tunicamycin in normal and dystonia patient lymphoblasts. A, PKR phosphorylation kinetics. Lymphoblasts from normal (WT) or affected (dystonia patient) individuals were treated with 0.5 μg/ml tunicamycin. The cell extracts were prepared at the indicated times and analyzed by Western blot analysis. The same blot was stripped and re-probed with all antibodies. The analysis was repeated four times with each line, and the best representative blots are shown. B, quantification of p-PKR and total PKR signals. The ratio of phosphorylated PKR to total PKR, calculated using the ImageQuant software is represented. The value for ratio obtained for control samples was considered 1.0 for each experiment, and all other ratios obtained with treated samples were expressed relative to this value for comparisons between experiments. The -fold increases in ratios after tunicamycin treatment compared with the control samples were calculated at each time point. Error bars represent S.D. from three experiments. C, eIF2α phosphorylation kinetics in response to tunicamycin. Same as in A, except eIF2α phosphorylation and total eIF2α was analyzed. D, quantification of p- eIF2α and total eIF2α signals. The ratio of phosphorylated eIF2α to total eIF2α, calculated using the ImageQuant software and phosphorimaging, is represented. The analysis was done as in panel B. E, co-immunoprecipitation of endogenous PKR and PACT proteins. Lymphoblasts from normal (WT) or dystonia patients (P222L) were treated with 0.5 μg/ml tunicamycin. The cell extracts were prepared at the indicated times, and endogenous PKR protein was immunoprecipitated using anti-PKR mAb, which immunoprecipitates total PKR and protein A-Sepharose. The immunoprecipitates were analyzed by Western blot analysis with anti-PACT monoclonal antibody (PACT Co-IP panel). The blot was stripped and re-probed with anti-PKR mAb to ascertain an equal amount of PKR in each lane (PKR panel). Input blot: Western blot analysis of total proteins in the extract with anti-PACT mAb showing equal amount of PACT in all samples. F, PACT-PKR co-IP quantification and statistical analysis. The ratio of PACT co-IP signals to total input PACT signals were calculated using GE Healthcare ImageQuant TL software and was further normalized to PKR IP signals. The averages from three experiments are plotted as bar graphs. The error bars represent S.E., and the data from different experiments were subjected to Student's t tests. p values are as follows. * = 0.0025 (significant), NS = 0.0531 (not significant), ** = 0.0003 (significant), ## = 0.0002 (significant), and ### = 0.0006 (significant).
P222L Mutation Enhances PACT-TRBP as Well as PACT-PACT Interactions
We next examined if the P222L mutation affects PACT's interaction with TRBP, consequently resulting in altered kinetics of PKR association and activation. In the absence of stress, PKR-TRBP and PACT-TRBP heterodimers prevail and prevent PACT from activating PKR. In response to stress, PACT is phosphorylated on serine 287, which decreases its affinity for TRBP, thus reducing TRBP-PACT heterodimers (21). This phosphorylation also increases PACT's binding affinity for PKR while simultaneously stabilizing PACT-PACT interactions. Thus we reasoned that the altered kinetics of PKR activation in patient cells may result from P222L's effect on PACT-TRBP interaction. We examined the interactions between PACT-TRBP and PACT-PACT using co-immunoprecipitation assays with PACT and TRBP marked by two different epitope tags. Fig. 6A represents analysis of PACT-TRBP interaction. As seen in lanes 1 and 3, myc-TRBP interacted with both FLAG-WT as well as FLAG-P222L. The interaction between FLAG-WT PACT and myc-TRBP was significantly reduced to almost undetectable levels 2 h after tunicamycin treatment (lane 2). In contrast to this, FLAG-P222L stayed associated with myc-TRBP strongly at 2 h after treatment (lane 4). In the absence of FLAG-PACT, no myc-TRBP was immunoprecipitated (lanes 5 and 6), thereby indicating that myc-TRBP immunoprecipitation is solely due to interaction with FLAG-PACT. These results indicate that the delayed activation of PKR in patient cells could result from an enhanced interaction of P222L with TRBP that sequesters P222L mutant away from PKR. Because we observed altered PKR phosphorylation as well as eIF2α phosphorylation kinetics in patient lymphoblasts as compared with WT lymphoblasts, we examined if this change in kinetics could result from a lack of TRBP re-association with PACT at later time points after ER stress. Fig. 6B represents analysis of PACT-TRBP interaction at 8 and 24 h after tunicamycin treatment. As seen in lanes 1 and 4, myc-TRBP interacts with both FLAG-WT as well as FLAG-P222L in untreated cells. The FLAG-WT PACT and myc-TRBP begin to re-associate 8 h after tunicamycin treatment (lane 2). In contrast to this, myc-TRBP stayed dissociated from FLAG-P222L at 8 h after treatment (lane 5). At 24 h after tunicamycin treatment myc-TRBP showed a strong re-association with FLAG-WT (lane 3), and in contrast to this, myc-TRBP showed no re-association with FLAG-P222L even at 24 h. These results indicate that the delayed but prolonged activation of PKR in patient cells could result from an enhanced interaction of P222L with TRBP that initially sequesters P222L mutant away from PKR, but once dissociated from TRBP, the P222L mutant does not re-associate rapidly with TRBP and thus continues to activate PKR even at 24 h after the ER stress. Because PKR activation in response to stress is efficiently achieved by PACT-PACT homodimers after serine 287 phosphorylation, we examined if P222L mutation affects formation of PACT-PACT homodimers in response to tunicamycin. As seen in Fig. 6C, myc-WT PACT and FLAG-WT PACT did not associate with high affinity in the absence of stress (lane 1), but their interactions was detected 2 h after tunicamycin treatment (lane 2). In contrast, FLAG-P222L bound strongly to myc-P222L molecules even in absence of stress (lane 3), and this interaction was somewhat intensified at 2 h after treatment (lane 4) and was significantly stronger than the interaction between WT PACT molecules. In the absence of FLAG-PACT, no myc-PACT was immunoprecipitated, thereby validating that myc-PACT was immunoprecipitated solely by its interaction with FLAG-PACT (lanes 5 and 6). These results suggest that the prolonged activation of PKR seen in patient cells results from more efficient formation of PACT homodimers that may be stable for a longer duration after initial onset of stress. Because we detected an enhanced interaction between P222L molecules in the absence of stress, we further examined this by using a mammalian two-hybrid assay to determine the relative increase in homomeric interactions due to P222L mutation in comparison with the WT PACT homomeric interactions. As shown in Fig. 6D, P222L-P222L homomeric interaction (P222L/P222L) activated the reporter gene 1.5-fold higher than WT PACT-WT PACT homomeric interactions (WT/WT), thus validating that P222L mutation enhances interaction between PACT molecules. Student's t tests indicated the observed differences to be statistically significant as indicated in the figure legend. It is worth noting that the WT PACT-WT PACT as well as P222L-P222L interactions are statistically significant as compared with all negative controls (brackets *, **, #, and ##), and the difference between WT PACT-WT PACT and P222L-P222L interactions is also statistically significant (bracket ###). Fig. 6E shows the results of a Western blot analysis to ensure that WT and P222L proteins were expressed at similar levels in cells, and the observed enhanced P222L-P222L interaction was not due to differences in expression levels of WT PACT and P222L.
FIGURE 6.
P222L mutation affects TRBP-PACT and PACT-PACT interactions. A, co-immunoprecipitation of FLAG-PACT with myc-TRBP at early time point after ER stress. HeLa cells were transfected with either FLAG- WT PACT or FLAG-P222L mutant and myc-TRBP. 24 h after transfection cells were either left untreated or treated with 50 μg/ml tunicamycin. 2 h after treatment cells were harvested, FLAG-PACT was immunoprecipitated using anti-FLAG mAb-Sepharose, and co-immunoprecipitation of myc-TRBP was analyzed by Western blot analysis with anti-myc mAb (myc-TRBP panel). The blot was stripped and re-probed with anti-FLAG mAb to ascertain an equal amount of FLAG-PACT protein in each lane (FLAG-PACT panel). Input blot: Western blot analysis of total proteins in the extract with anti-myc mAb. B, co-immunoprecipitation of FLAG-PACT with myc-TRBP at late time points after ER stress. HeLa cells were transfected with either FLAG-WT PACT or FLAG-P222L mutant and myc-TRBP. 24 h after transfection, cells were either left untreated or treated with 50 μg/ml tunicamycin. AT 8 and 24 h after tunicamycin treatment, cells were harvested, FLAG-PACT was immunoprecipitated using anti-FLAG mAb-Sepharose, and co-immunoprecipitation of myc-TRBP was analyzed by Western blot analysis with anti-myc mAb (myc-TRBP panel). The blot was stripped and re-probed with anti-FLAG mAb to ascertain an equal amount of FLAG-PACT protein in each lane (FLAG-PACT panel). Input blot: Western blot analysis of total proteins in the extract with anti-myc mAb. C, co-immunoprecipitation of FLAG-PACT and myc-PACT. HeLa cells were transfected with either FLAG-WT PACT and myc-WT PACT or FLAG-P222L mutant and myc-P222L mutant. 24 h after transfection, cells were either left untreated or treated with 50 μg/ml tunicamycin. 2 h after treatment, cells were harvested, FLAG-PACT was immunoprecipitated using anti-FLAG mAb-Sepharose, and co-immunoprecipitation of myc-PACT was analyzed by Western blot analysis with anti-myc mAb (myc-PACT panel). The blot was stripped and re-probed with anti-FLAG mAb to ascertain an equal amount of FLAG-PACT protein in each lane (FLAG-PACT panel). Input blot: Western blot analysis of total proteins in the extract with anti-myc mAb. D, mammalian two-hybrid assay. COS-1 cells were transfected with 250 ng of each of the two test plasmids encoding proteins to be tested for interaction, 50 ng of the reporter plasmid pG5Luc, and 1 ng of plasmid pRL-Null to normalize transfection efficiency. Cells were harvested 24 h after transfection, and cell extracts were assayed for luciferase activity. The plasmid combinations are as indicated. C1, WT PACT/GAL4DBD and VP16AD EV (negative control); C2, GAL4DBD EV and WT PACT/VP16AD (negative control); WT/WT, WT PACT/GAL4DBD and WT PACT/VP16AD; C3, P222L/GAL4DBD and VP16AD EV (negative control); C4, GAL4DBD EV and P222L/VP16AD (negative control); mut/mut, P222L/GAL4DBD and P222L/VP16AD. The experiment was repeated twice with each sample in triplicate, and the averages with S.E. bars are presented. Student's t tests performed to calculate p values indicated that the differences observed between the negative controls and test values (C1, C2 and WT/WT, C3, C4 and mut/mut) were highly significant: bracket * (0.0071), bracket ** (0.0001), bracket # (0.0037), bracket ## (0.0002). The difference between WT/WT and mut/mut interaction was also highly significant as indicated by the p value represented by bracket ### (0.0001). RLU, relative light units. E, Western blot analysis. COS-1 cell extracts were examined by Western blot analysis using an anti-GAL4-DBD mAb (Santa Cruz), anti-VP16AD Ab (Santa Cruz), and anti-β-actin mAb. The samples are indicated on top of the lanes.
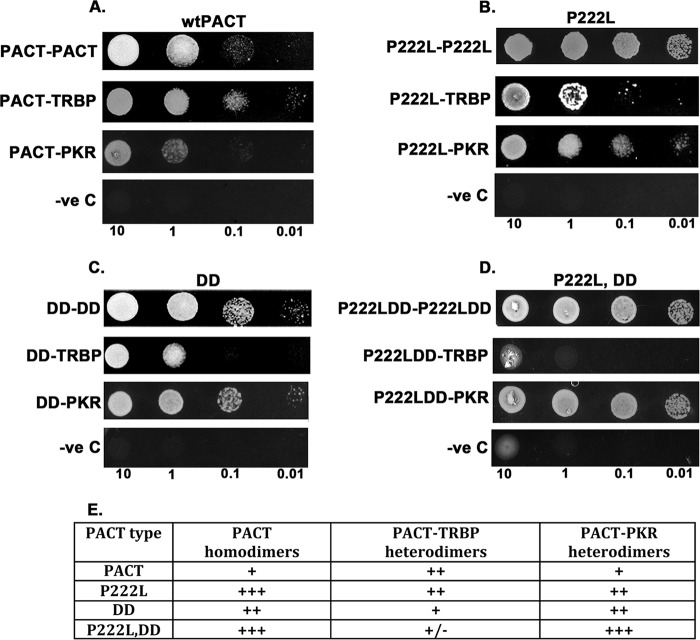
To ensure that the P222L mutation affects the affinity of PACT-PACT, PACT-TRBP, and PACT-PKR directly without the involvement of a third partner, we measured the relative affinities of these interactions using a yeast two-hybrid system. We have used this system extensively to demonstrate that stress-induced phosphorylation of PACT results in changes in affinity of its interaction with TRBP and PKR. Thus, this system is sensitive enough to detect changes in affinity between these proteins and measures direct interaction between two proteins. Such changes in relative affinities of the binding partners have been demonstrated to change the cellular outcome in response to a stressor. As seen in Fig. 7, A and B, in comparison to WTPACT, the P222L mutant showed a significant increase in interaction between two PACT molecules (P222L-P222L in panel B versus PACT-PACT in panel A). The P222L mutation also increased P222L-PKR (panel B) interaction as compared with WTPACT-PKR (panel A). In contrast, the interaction with TRBP (P222L-TRBP in panel B versus PACT-TRBP in panel A) was not affected to a significant extent by the P222L mutation. These results are in agreement with our co-immunoprecipitation data in Fig. 6. PACT is constitutively phosphorylated on serine 246 and gets phosphorylated at serine 287 in response to the ER stress. We previously established that a phosphomimetic double mutation S246D,S287D (termed DD) increased interactions with PKR (DD-PKR in panel C versus PACT-PKR in panel A) and another molecule of PACT (DD-DD in panel C versus PACT-PACT in panel A) while decreasing the interaction with TRBP (DD-TRBP in panel C versus PACT-TRBP in panel A). To test the effect of stress-induced phosphorylation on the molecular interactions under study, we combined the two mutants to generate the P222L,DD mutant. Thus, the P222L,DD mutant represents the P222L dystonia mutant under conditions of the ER stress caused by tunicamycin. As seen in panel D, P222L,DD mutant showed the strongest interaction with itself (P222L,DD-P222L,DD) and with PKR (P222L,DD-PKR) and the least interaction with TRBP (P222L,DD-TRBP) when compared with results in panels A–C. Thus, these results clearly indicate that the P222L mutation found in dystonia patients leads to increased homodimer formation and PKR interaction under conditions of the ER stress. The altered kinetics of PKR activation and eIF2a phosphorylation that we observe in dystonia patient cells can be attributed to these changes in relative affinities of the binding partners.
FIGURE 7.
Effect of P222L mutation on molecular interactions with PACT's binding partners in yeast two-hybrid assay. A, interaction of WTPACT with WTPACT, TRBP, and PKR. B, interaction of P222L mutant with P222L, TRBP, and PKR. C, interaction of PACT mutant S246D,S287D (DD) with DD, TRBP, and PKR. D, interaction of PACT mutant P222LDD with P222LDD, TRBP, and PKR. Various plasmid constructs as indicated under “Experimental Procedures” were transformed into yeast strain AH109 and selected on double dropout medium lacking tryptophan and leucine. 10 μl of serial dilutions (of A600 = 10, 1.0, 0.1, 0.01) were spotted for each transformant on quadruple dropout S.D. medium plate that lacked adenine, tryptophan, leucine, and histidine and had 25 mm 3-amino-1,2,4-triazole. Plates were incubated for 3 days at 30 °C. Transformation of PACT constructs in pGBKT7 and empty vector pGADT7 served as negative controls. -ve, negative. E, relative affinities of PACT-PACT, PACT-TRBP, and PACT-PKR interactions. The growth obtained in the yeast two-hybrid interaction assay was scored and is represented in the table. +++ indicates strong growth, ++ indicates moderate growth, + indicates good growth, and +/− indicates poor growth.
Discussion
Dystonia is a genetically heterogeneous neurological movement disorder that is characterized by intermittent or sustained muscle contractions causing abnormal repetitive movements and postures. In recent years, genetic diagnostic tools have led to identification of several monogenic forms of dystonia and dystonia-related disorders designated as DYT1–25 in OMIM (Online Mendelian Inheritance in Man), and many causative mutations in respective genes have been described (36). Of these, DYT16 is associated with mutations in PACT and was initially described in two unrelated Brazilian families (23) and a single German patient (27). The affected individuals displayed symptoms of generalized dystonia in childhood with the mean age being 9 years. All seven Brazilian patients shared the same substitution mutation (c.655C→T;P222L) in exon 7 of the PACT gene. The German patient had a heterozygous frameshift mutation (c.266–267delAT;p.H89fsX20) in exon 3. Recently another genetically confirmed case of DYT16 with an early presentation was described, and whole exome sequencing revealed two mutations in PACT (29). The first c.665C→T;P222L was inherited from the mother and was the same as the Brazilian cohort. The second mutation c.637T→C;C213R was a de novo event and was absent in both parents. Two other recessive mutations have been identified that manifest as substitution mutations in PACT C77S and C213F (30). Recently three additional point mutations were reported in Polish and German patients (31).
We examined the effect of the P222L mutation on cellular stress response as regulation of apoptosis in response to stress is PACT's well characterized function (12, 19, 21, 22). The lymphoblasts derived from three dystonia patients carrying the P222L mutation exhibited significantly enhanced apoptosis in response to ER stress. The mechanisms leading to the enhanced sensitivity to tunicamycin were explored, and our results indicated that the P222L mutation leads to PACT's increased interaction with TRBP and consequently causes altered kinetics of PKR and eIF2α phosphorylation in response to the ER stress. In patient cells, the dissociation of PACT from TRBP in response to the ER stress is delayed, and thus formation of PACT-PKR homodimers is delayed. However, PACT-PACT homodimers appear to be more stable, and thus PKR activation is enhanced and persists for a significantly longer duration after an ER stress event. Consequently eIF2α phosphorylation also is delayed but persists for a longer duration in patient cells. The reason for relative eIF2α phosphorylation levels in patient cells not reaching as high as that in WT cells is unclear but may be due to differences in the level of eIF2α phosphatases at early and late time points after the initial stress. The phosphorylation of eIF2α on serine 51, which is a single regulatory stress-induced modification, is of significant functional relevance (37). Our work presented here highlights the importance of regulating the speed and duration of eIF2α phosphorylation in determining cellular fate.
As represented in a schematic model in Fig. 8, in normal cells PACT-TRBP interaction has an inhibitory effect on PKR activation, and phosphorylation of PACT in response to stress disrupts TRBP-PACT interaction (19, 21). Free PACT then forms homodimers that associate with PKR to activate its kinase activity resulting in eIF2α phosphorylation (Fig. 8A) (22). The P222L mutant remains associated with TRBP longer under the ER stress conditions, thereby causing a delayed PKR activation (Fig. 8B). However, P222L associates with PKR with higher affinity and thus remains associated with PKR for a longer duration thereby causing stronger and persistent PKR activation and eIF2α phosphorylation. In accordance with this, P222L-P222L homodimers are more stable even in the absence of the ER stress (Fig. 6, B and C). It is interesting to note that although P222L-P222L homodimers form efficiently in the absence of a stress signal, the P222L homodimers are unable to cause PKR activation in the absence of a stress signal (Figs. 6B, 5A, and 7). Thus, stress-induced phosphorylation at serine 287 possibly serves an additional function than simply promoting formation of PACT-PACT homodimers. It is also interesting to note that P222L mutation enhances PACT's interaction with PKR as measured by cell-based co-immunoprecipitation assays from mammalian cell extracts (Fig. 2, C and D). However, co-immunoprecipitation assays performed with the in vitro translated proteins shows no enhancement of P222L interaction with PKR (Fig. 2, A and B). This indicates a possible role of post-translational modifications on P222L in enhancing interactions with PKR that possibly take place in mammalian cells but not in reticulocyte lysates. Further molecular analysis of other substitution mutations (C77S, C213F, C213R, T34S, N102S) recently described in DYT16 patients (30, 31) would be important to determine if an altered ER stress response is a common theme in DYT16 dystonia. Our preliminary results indicate that the frameshift mutant protein reported in dominantly inherited dystonia case causes potent PKR activation and also leads to cellular death in cell culture.3 For efficient recovery and cellular survival after the ER stress, it is essential that the temporary inhibition of protein synthesis caused by phosphorylated eIF2α is reversed and synthesis of survival-related proteins takes place at late time points (38). As depicted in Fig. 8C, the perturbation of this survival pathway due to the P222L mutation leading to delayed but prolonged PKR activation and eIF2α phosphorylation can result in increased cell death as recovery and survival mechanisms may not be induced optimally in dystonia patients.
FIGURE 8.
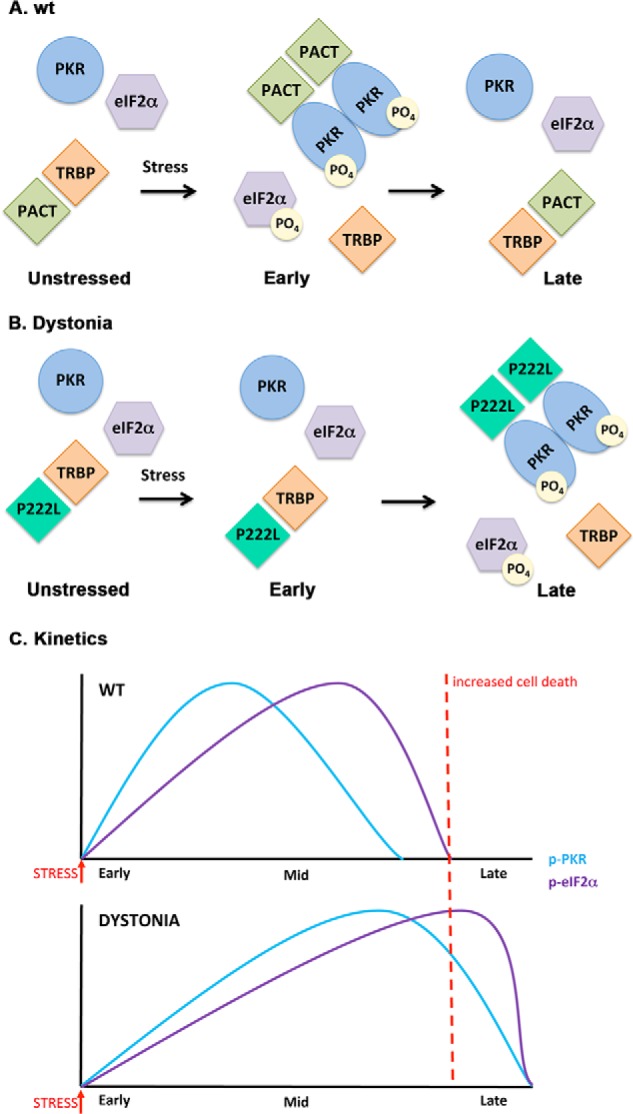
A schematic model of PKR activation in WT and dystonia cells. A, WT cells. As previously established (Refs. 19, 21, 22), in the absence of stress, PACT heterodimerizes with TRBP, PKR is catalytically inactive, and eIF2α is not phosphorylated. At early time points after ER stress, PACT dissociates from TRBP due to its phosphorylation, forms homodimers that bind to PKR with high affinity, and activates its kinase activity leading to eIF2α phosphorylation. At late time points after ER stress, cells recover by forming TRBP-PACT heterodimers and turning off PKR and eIF2α phosphorylation. B, dystonia cells. In the absence of stress, P222L mutant forms heterodimers with TRBP, PKR is catalytically inactive and eIF2α is not phosphorylated. At early time points after ER stress, P222L remains bound to TRBP, and PKR and eIF2α phosphorylation is inhibited. At late time points after ER stress, P222L dissociates from TRBP, forms homodimers that bind to PKR with high affinity and activate its kinase activity leading to eIF2α phosphorylation. Thus, at late time point cells are unable to recover efficiently from ER stress because PKR and eIF2α remain phosphorylated. C, schematic representation of the altered PKR activation and eIF2α phosphorylation kinetics in dystonia cells. Blue line, PKR phosphorylation; purple line, eIF2α phosphorylation. Red dotted line, a threshold time point. If PKR and eIF2α remain in their phosphorylated state beyond this time point, recovery from ER stress is prevented, resulting in increased apoptosis.
Our results presented here elucidate the cellular consequence of dystonia causing PACT mutations for the first time. Based on our results, it is conceivable to imagine that in P222L homozygotes, the neuronal cells may not cope well with cellular stress. Although apoptosis of neurons is a possible extreme outcome of such detrimental events, survival of neurons that may not function at an optimal level could also be equally detrimental. Analysis of whole exome sequencing in one particular patient (29) revealed two mutations within PACT gene. The first c.665C→T (p.P222L) was inherited from his mother and is the same mutation described in the Brazilian cohort and analyzed here. The second mutation c.637T→C (p.C213R) was not present in either parent and indicated a de novo event. Thus, this patient is heterozygous for each mutation but has both copies of PACT gene mutated and thus has no WT PACT protein present. Similar to other DYT16 patients, this patient developed dystonia symptoms in early childhood, and imaging revealed progressive MRI abnormalities with significant bilateral volume loss in the basal ganglia (28), which is interesting in view of the observed enhanced apoptosis in our experiments. This patient developed dystonia after a febrile illness, which could be a possible cellular stress event that may have triggered progressive cellular dysfunction or loss. Although our results are obtained using patient lymphoblasts, the PACT-PKR stress response pathway is present ubiquitously in all cell types including neurons (39–41). Although neurodegeneration would be the expected long term outcome of neuronal apoptosis, neither apoptosis not neurodegeneration has been systematically investigated in blood or brain of dystonia patients, but possible similarities and links between neurodegenerative Parkinson disease and dystonia have recently been noted (42). Thus, neurodegeneration has not been investigated in any form of dystonia patients, and this lack of information is usually interpreted as neurodegeneration generally being absent in dystonia patients. At the same time, PKR activation and ER stress due to misfolded proteins has also been observed in pathologies of many neurodegenerative diseases such as Alzheimer, Parkinson, Huntington, and amyotrophic lateral sclerosis (ALS) (43). Any contribution of PACT in PKR activation in these neurodegenerative diseases also remains unexplored at present. Nevertheless, possible neurodegeneration should be explored in DYT16 patients in the future especially in light of our results indicating increased apoptosis in P222L patient cells. It is also unknown at present if DYT16 patients exhibit any deficiencies in the innate immune system and respond differently to infections, and in the future this may be something worth investigating considering PACT's involvement in innate immunity. It is interesting to note that DYT16 patient lymphoblasts show higher levels of apoptosis in the absence of a stress signal as seen in the results of our flow cytometry analysis and caspase activity assays. It is unclear at present if this results from chronic low levels of PKR activation or is via a PKR-independent mechanism. Similarly, any effect of PACT on activation of PKR-like endoplasmic reticulum kinase (PERK) has not been explored and could have a yet unidentified role on higher levels of apoptosis observed in patient cells.
In addition to activating PKR in response to cellular stress, PACT is known to function in the RNAi (44) and innate immune pathways (45), and it is possible that the P222L mutation could affect these pathways. Although our research certainly does not rule out this possibility, it definitely establishes that at least one pathway regulated by the PACT-PKR interaction is significantly altered by the P222L mutation and results in major changes in the cell survival in response to the ER stress. The importance of such perturbation by the P222L mutation is further underscored by the fact that the change in response to ER stress is reflected in increased apoptosis in patient cells.
The involvement and importance of the ER stress response for dystonia has been noted before for two other genes. Torsin A mutations were the first mutations to be described as the genetic basis of DYT1 dystonias (46). Torsin A functions as a molecular chaperone within the ER/secretory pathways, and the mutant forms of torsin A have been shown to either misfold and trigger the ER stress response or cause a secretion defect causing a chronic, low level ER stress (47–49). THAP1 (thanatos-associated domain-containing apoptosis-associated protein-1) mutations were described as the genetic basis of DYT6 dystonia (50). THAP1 is a transcription factor and has a nuclear function; however, it is also localized to the cytoplasm and possesses an amino-terminal THAP domain that is homologous to the THAP0 domain found in p52rIPK or PRKRIR (protein kinase, interferon-inducible dsRNA-dependent inhibitor, repressor of p58 repressor) (32). Thus, THAP1 may be involved in the ER stress response pathway by regulating PKR activity by inhibiting the function of the PKR inhibitor protein p58. Functionally, THAP1 may work similarly to PACT if it enhanced PKR activation. This can be tested in the future in DYT6 patient cells. Our results strongly emphasize the importance of the proper regulation of the ER stress response pathway and underscore the possibility that any dysregulation may sensitize cells to apoptosis.
Author Contributions
L. S. V. performed all experiments, interpreted the results, and prepared the figures. D. C. B. and N. S. established the patient and WT lymphoblast cell lines. S. C. and F. C. were involved in collecting the blood samples from all patients, and R. C. P. planned, supervised, coordinated all work, and wrote the manuscript.
Acknowledgments
We thank Laurie Ozelius for sharing the genome sequencing data about newly discovered PACT mutations in dystonia patients. We also thank Madhurima Singh and Evelyn Chukwurah for help with yeast two hybrid experiments.
This work was supported in part by a Dystonia Medical Research Foundation grant (to R. P.). The work was also supported by National Institutes of Health Grant P50NS037409 (NINDS; to N. S.). The authors declare that they have no conflicts of interest with the contents of this article.
L. S. Vaughn and R. C. Patel, unpublished results.
- PKR
- protein kinase RNA-activated
- dsRNA
- double-stranded RNA
- ER
- endoplasmic reticulum
- dsRBM
- dsRNA binding motif
- TRBP
- human immunodeficiency virus (HIV)-1 transactivation-responsive RNA-binding protein
- PACT
- PKR activator protein
- THAP1
- thanatos-associated domain-containing apoptosis-associated protein-1
- M3
- motif 3
- DD
- S246D,S287D
- IP
- immunoprecipitate.
References
- 1. García M. A., Gil J., Ventoso I., Guerra S., Domingo E., Rivas C., Esteban M. (2006) Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70, 1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meurs E., Chong K., Galabru J., Thomas N. S., Kerr I. M., Williams B. R., Hovanessian A. G. (1990) Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62, 379–390 [DOI] [PubMed] [Google Scholar]
- 3. Patel R. C., Sen G. C. (1992) Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J. Biol. Chem. 267, 7671–7676 [PubMed] [Google Scholar]
- 4. Green S. R., Mathews M. B. (1992) Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 6, 2478–2490 [DOI] [PubMed] [Google Scholar]
- 5. McCormack S. J., Thomis D. C., Samuel C. E. (1992) Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2α protein kinase. Virology 188, 47–56 [DOI] [PubMed] [Google Scholar]
- 6. Nanduri S., Rahman F., Williams B. R., Qin J. (2000) A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 19, 5567–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cole J. L. (2007) Activation of PKR: an open and shut case? Trends Biochem. Sci. 32, 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel R. C., Stanton P., McMillan N. M., Williams B. R., Sen G. C. (1995) The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 8283–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang K. Y., Ramos A. (2005) The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 272, 2109–2117 [DOI] [PubMed] [Google Scholar]
- 10. Benkirane M., Neuveut C., Chun R. F., Smith S. M., Samuel C. E., Gatignol A., Jeang K. T. (1997) Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 16, 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel R. C., Sen G. C. (1998) PACT, a protein activator of the interferon-induced protein kinase, PKR. EMBO J. 17, 4379–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patel C. V., Handy I., Goldsmith T., Patel R. C. (2000) PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275, 37993–37998 [DOI] [PubMed] [Google Scholar]
- 13. Peters G. A., Hartmann R., Qin J., Sen G. C. (2001) Modular structure of PACT: distinct domains for binding and activating PKR. Mol. Cell Biol. 21, 1908–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang X., Hutchins B., Patel R. C. (2002) The C-terminal, third conserved motif of the protein activator PACT plays an essential role in the activation of double-stranded-RNA-dependent protein kinase (PKR). Biochem. J. 366, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito T., Yang M., May W. S. (1999) RAX, a cellular activator for double-stranded RNA-dependent protein kinase during stress signaling. J. Biol. Chem. 274, 15427–15432 [DOI] [PubMed] [Google Scholar]
- 16. Bennett R. L., Blalock W. L., Abtahi D. M., Pan Y., Moyer S. A., May W. S. (2006) RAX, the PKR activator, sensitizes cells to inflammatory cytokines, serum withdrawal, chemotherapy, and viral infection. Blood 108, 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singh M., Fowlkes V., Handy I., Patel C. V., Patel R. C. (2009) Essential role of PACT-mediated PKR activation in tunicamycin-induced apoptosis. J. Mol. Biol. 385, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cosentino G. P., Venkatesan S., Serluca F. C., Green S. R., Mathews M. B., Sonenberg N. (1995) Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9445–9449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daher A., Laraki G., Singh M., Melendez-Peña C. E., Bannwarth S., Peters A. H., Meurs E. F., Braun R. E., Patel R. C., Gatignol A. (2009) TRBP control of PACT-induced phosphorylation of protein kinase R is reversed by stress. Mol. Cell Biol. 29, 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peters G. A., Li S., Sen G. C. (2006) Phosphorylation of specific serine residues in the PKR activation domain of PACT is essential for its ability to mediate apoptosis. J. Biol. Chem. 281, 35129–35136 [DOI] [PubMed] [Google Scholar]
- 21. Singh M., Castillo D., Patel C. V., Patel R. C. (2011) Stress-induced phosphorylation of PACT reduces its interaction with TRBP and leads to PKR activation. Biochemistry 50, 4550–4560 [DOI] [PubMed] [Google Scholar]
- 22. Singh M., Patel R. C. (2012) Increased interaction between PACT molecules in response to stress signals is required for PKR activation. J. Cell. Biochem. 113, 2754–2764 [DOI] [PubMed] [Google Scholar]
- 23. Camargos S., Scholz S., Simón-Sánchez J., Paisán-Ruiz C., Lewis P., Hernandez D., Ding J., Gibbs J. R., Cookson M. R., Bras J., Guerreiro R., Oliveira C. R., Lees A., Hardy J., Cardoso F., Singleton A. B. (2008) DYT16, a novel young-onset dystonia-parkinsonism disorder: identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 7, 207–215 [DOI] [PubMed] [Google Scholar]
- 24. Geyer H. L., Bressman S. B. (2006) The diagnosis of dystonia. Lancet Neurol. 5, 780–790 [DOI] [PubMed] [Google Scholar]
- 25. Camargos S., Lees A. J., Singleton A., Cardoso F. (2012) DYT16: the original cases. J. Neurol. Neurosurg. Psychiatry 83, 1012–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Klein C. (2008) DYT16: a new twist to familial dystonia. Lancet Neurol. 7, 192–193 [DOI] [PubMed] [Google Scholar]
- 27. Seibler P., Djarmati A., Langpap B., Hagenah J., Schmidt A., Brüggemann N., Siebner H., Jabusch H. C., Altenmüller E., Münchau A., Lohmann K., Klein C. (2008) A heterozygous frameshift mutation in PRKRA (DYT16) associated with generalised dystonia in a German patient. Lancet Neurol. 7, 380–381 [DOI] [PubMed] [Google Scholar]
- 28. Brashear A. (2013) Commentary. Movement Disorders 28, 1939. [DOI] [PubMed] [Google Scholar]
- 29. Lemmon M. E., Lavenstein B., Applegate C. D., Hamosh A., Tekes A., Singer H. S. (2013) A novel presentation of DYT 16: acute onset in infancy and association with MRI abnormalities. Movement Disorders 28, 1937–1938 [DOI] [PubMed] [Google Scholar]
- 30. de Carvalho Aguiar P., Borges V., Ferraz H. B., Ozelius L. J. (2015) Novel compound heterozygous mutations in PRKRA cause pure dystonia. Movement Disorders 30, 877–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zech M., Castrop F., Schormair B., Jochim A., Wieland T., Gross N., Lichtner P., Peters A., Gieger C., Meitinger T., Strom T. M., Oexle K., Haslinger B., Winkelmann J. (2014) DYT16 revisited: exome sequencing identifies PRKRA mutations in a European dystonia family. Movement Disorders 29, 1504–1510 [DOI] [PubMed] [Google Scholar]
- 32. Bragg D. C., Armata I. A., Nery F. C., Breakefield X. O., Sharma N. (2011) Molecular pathways in dystonia. Neurobiol. Dis. 42, 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Anderson M. A., Gusella J. F. (1984) Use of cyclosporin A in establishing Epstein-Barr virus-transformed human lymphoblastoid cell lines. In vitro 20, 856–858 [DOI] [PubMed] [Google Scholar]
- 34. Takacs A. M., Das T., Banerjee A. K. (1993) Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. U.S.A. 90, 10375–10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schönthal A. H. (2012) Endoplasmic reticulum stress: its role in disease and novel prospects for therapy. Scientifica 2012, 857516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiao J., Vemula S. R., LeDoux M. S. (2014) Recent advances in the genetics of dystonia. Curr. Neurol. Neurosci. Rep. 14, 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Donnelly N., Gorman A. M., Gupta S., Samali A. (2013) The eIF2α kinases: their structures and functions. Cell. Mol. Life Sci. 70, 3493–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hetz C. (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 [DOI] [PubMed] [Google Scholar]
- 39. Chen G., Ma C., Bower K. A., Ke Z., Luo J. (2006) Interaction between RAX and PKR modulates the effect of ethanol on protein synthesis and survival of neurons. J. Biol. Chem. 281, 15909–15915 [DOI] [PubMed] [Google Scholar]
- 40. Paquet C., Mouton-Liger F., Meurs E. F., Mazot P., Bouras C., Pradier L., Gray F., Hugon J. (2012) The PKR activator PACT is induced by Aβ: involvement in Alzheimer's disease. Brain Pathol. 22, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vaughn L. S., Snee B., Patel R. C. (2014) Inhibition of PKR protects against tunicamycin-induced apoptosis in neuroblastoma cells. Gene 536, 90–96 [DOI] [PubMed] [Google Scholar]
- 42. Stoessl A. J., Lehericy S., Strafella A. P. (2014) Imaging insights into basal ganglia function, Parkinson's disease, and dystonia. Lancet 384, 532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marchal J. A., Lopez G. J., Peran M., Comino A., Delgado J. R., García-García J. A., Conde V., Aranda F. M., Rivas C., Esteban M., Garcia M. A. (2014) The impact of PKR activation: from neurodegeneration to cancer. FASEB J. 28, 1965–1974 [DOI] [PubMed] [Google Scholar]
- 44. Yong Y., Luo J., Ke Z. J. (2014) dsRNA binding protein PACT/RAX in gene silencing, development and diseases. Front. Biol. (Beijing) 9, 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heyam A., Lagos D., Plevin M. (2015) Dissecting the roles of TRBP and PACT in double-stranded RNA recognition and processing of noncoding RNAs. Wiley Interdiscip. Rev. RNA 6, 271–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bressman S. B. (2007) Genetics of dystonia: an overview. Parkinsonism Relat. Disord. 13, S347–S355 [DOI] [PubMed] [Google Scholar]
- 47. Gordon K. L., Glenn K. A., Gonzalez-Alegre P. (2011) Exploring the influence of torsinA expression on protein quality control. Neurochem. Res. 36, 452–459 [DOI] [PubMed] [Google Scholar]
- 48. Hewett J. W., Tannous B., Niland B. P., Nery F. C., Zeng J., Li Y., Breakefield X. O. (2007) Mutant torsinA interferes with protein processing through the secretory pathway in DYT1 dystonia cells. Proc. Natl. Acad. Sci. U.S.A. 104, 7271–7276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Thompson M. L., Chen P., Yan X., Kim H., Borom A. R., Roberts N. B., Caldwell K. A., Caldwell G. A. (2014) TorsinA rescues ER-associated stress and locomotive defects in C. elegans models of ALS. Dis. Model. Mech. 7, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuchs T., Gavarini S., Saunders-Pullman R., Raymond D., Ehrlich M. E., Bressman S. B., Ozelius L. J. (2009) Mutations in the THAP1 gene are responsible for DYT6 primary torsion dystonia. Nat. Genet. 41, 286–288 [DOI] [PubMed] [Google Scholar]