Abstract
The conserved protein UNC-16 (JIP3) inhibits the active transport of some cell soma organelles, such as lysosomes, early endosomes, and Golgi, to the synaptic region of axons. However, little is known about UNC-16’s organelle transport regulatory function, which is distinct from its Kinesin-1 adaptor function. We used an unc-16 suppressor screen in Caenorhabditis elegans to discover that UNC-16 acts through CDK-5 (Cdk5) and two conserved synapse assembly proteins: SAD-1 (SAD-A Kinase), and SYD-2 (Liprin-α). Genetic analysis of all combinations of double and triple mutants in unc-16(+) and unc-16(−) backgrounds showed that the three proteins (CDK-5, SAD-1, and SYD-2) are all part of the same organelle transport regulatory system, which we named the CSS system based on its founder proteins. Further genetic analysis revealed roles for SYD-1 (another synapse assembly protein) and STRADα (a SAD-1-interacting protein) in the CSS system. In an unc-16(−) background, loss of the CSS system improved the sluggish locomotion of unc-16 mutants, inhibited axonal lysosome accumulation, and led to the dynein-dependent accumulation of lysosomes in dendrites. Time-lapse imaging of lysosomes in CSS system mutants in unc-16(+) and unc-16(−) backgrounds revealed active transport defects consistent with the steady-state distributions of lysosomes. UNC-16 also uses the CSS system to regulate the distribution of early endosomes in neurons and, to a lesser extent, Golgi. The data reveal a new and unprecedented role for synapse assembly proteins, acting as part of the newly defined CSS system, in mediating UNC-16’s organelle transport regulatory function.
Keywords: Caenorhabditis elegans, axonal transport, JIP3, Cdk5, Liprin, SAD-A, dynein
NEURONS have a unique architecture consisting of a cell soma, one or more dendrites that receive information, and a single axon, which can be a single process or intricately branched. Part of the axon is specialized to form synapses, which integrate and transmit information to other neurons or muscle cells via synaptic vesicles and dense core vesicles. This unique architecture and cell biology places extraordinary demands on the membrane-trafficking and transport machinery. In addition to transporting synaptic vesicles and dense core vesicles long distances into axons, motor neurons must also restrict, or even prevent, the flow of some organelles, including Golgi, lysosomes, and endosomes, into the synaptic region of their axons, which, under normal conditions, are relatively devoid of these organelles compared to cell somas. However, there may be special conditions, such as the need for axon repair or growth, where neurons require cell soma organelles in their axons, so the regulatory system for organelle transport must include components that inhibit, as well as promote, axonal transport. The association of mutations in the axonal transport machinery with neurodegenerative disorders in mice and humans underscores the importance of a properly functioning transport system for the long-term viability of neurons (Holzbaur 2004; El-Kadi et al. 2007; De Vos et al. 2008; Millecamps and Julien 2013; Maday et al. 2014).
The neuron’s axonal transport system consists of a system of microtubule tracks and motor proteins that carry cargos along the tracks, as well as proteins that regulate the system. Microtubules have an intrinsic plus- and minus-end polarity, and axonal microtubules are oriented with their plus-ends pointing outward toward the synaptic region (Burton and Paige 1981; Heidemann et al. 1981; Baas and Lin 2011). Plus-end-directed motors from the large family of kinesins carry synaptic vesicles and organelles outward, while the minus-end-directed motor dynein moves them in the opposite direction (Vale 2003; Holzbaur 2004; Hirokawa et al. 2009, 2010). The transport of synaptic vesicles, dense core vesicles, and at least some organelles is bidirectional (Kumar et al. 2010; Ou et al. 2010; Wong et al. 2012; Edwards et al. 2013; Hoover et al. 2014), suggesting that regulated directional biases determine their steady-state distributions.
Based on its strong interactions with Kinesin-1 in multiple invertebrate and vertebrate species, and its biochemical ability to enhance Kinesin-1 motility, several studies have concluded that UNC-16 (JIP3) functions mainly as an adaptor that connects Kinesin-1 to specific cargos to promote plus-end-directed transport into axons (Bowman et al. 2000; Cavalli et al. 2005; Sun et al. 2011). Indeed, there is genetic evidence in mice that JIP3 can function as an adaptor that promotes the Kinesin-1-mediated transport of TrkB receptors as well as one or more unidentified cargos relevant to axon elongation and neuronal degeneration (Huang et al. 2011; Sun et al. 2013; Sato et al. 2015; Watt et al. 2015).
However, in multiple species, both invertebrates and vertebrates, a major neuronal phenotype of mutants lacking UNC-16 (JIP3) is the massive accumulation of cell soma organelles in axons (Bowman et al. 2000; Brown et al. 2009; Drerup and Nechiporuk 2013; Edwards et al. 2013), which is also accompanied by the mislocalization of synaptic vesicle proteins (Byrd et al. 2001). There is no evidence that this anti-accumulation function of UNC-16 involves a Kinesin-1 adaptor function or the stimulation of Kinesin-1 activity. Indeed, the massive accumulation of organelles at microtubule plus ends in mutants lacking UNC-16 (JIP3) shows that this function of UNC-16 normally inhibits, rather than promotes, the plus-end-directed transport of those organelles, which include lysosomes, early endosomes, and Golgi (Edwards et al. 2013).
An attempt to explain the massive organelle accumulation in UNC-16 (JIP3) mutant axons in terms of a defective UNC-16–Kinesin-1 adaptor function led to the stalled transport model (Bowman et al. 2000). This model was based on the fact that fly Sunday Driver (JIP3) mutants die as larvae and thus must be derived from heterozygous mothers. The model posited that a small amount of wild-type JIP3 from the maternal contribution of messenger RNA allows organelles to enter axons, but that transport stalls as the maternal JIP3 supply is depleted (Bowman et al. 2000). However, this hypothesis was never tested until Caenorhabditis elegans studies proved it likely to be wrong by demonstrating that the axonal organelle accumulation also occurs in unc-16 homozygous mutants derived from homozygous parents (Brown et al. 2009; Edwards et al. 2013). Furthermore, the organelle accumulation in unc-16 mutants occurs in the absence of Kinesin-1 (Brown et al. 2009; Edwards et al. 2013). A zebrafish study showed that this organelle transport regulatory function of UNC-16 is conserved in both vertebrates and invertebrates (Drerup and Nechiporuk 2013).
Little is known about this regulatory function of UNC-16. In this study, we used an unbiased forward genetic screen in C. elegans to discover a previously unknown system through which UNC-16 acts to inhibit the active transport of organelles to axons. The proteins in this system had not previously been implicated in the active transport of cell soma organelles in neurons. Instead, previous studies have suggested that they have highly specialized roles in synapse assembly/ and stability. In an analysis of this new system, we combined the genetic strengths of C. elegans with quantitative imaging techniques and time-lapse imaging of lysosomes in motor neurons of living animals. The data revealed that UNC-16’s organelle transport regulatory function is unexpectedly mediated by synapse assembly proteins acting as part of a newly defined organelle transport regulatory system: the CSS system. In an accompanying article in this issue, we show that several CSS system proteins also regulate synaptic vesicle transport, further underscoring the counterintuitive dual roles of some synapse assembly proteins in regulating active transport and synapse assembly/stability (Edwards et al. 2015).
Materials and Methods
Worm culture and strains
Worm culture and manipulation essentially followed previously described methods (Brenner 1974; Sulston and Hodgkin 1988; Stiernagle 2006). Briefly, culture media was Nematode Growth Medium Light Only Back-peptone (NGM-LOB; Hoover et al. 2014). Prior studies defined the culture plate types “spread plates,” “streak plates,” and “locomotion plates” (Miller et al. 1999; Edwards et al. 2008). We produced “24-well plates” by dispensing 2.3 ml NGM-LOB media into wells of 24-well untreated tissue culture plates (Corning 3524), seeding each well 2 days later with 6 μl of OP-50 culture, immediately drying the plates with their lids off for 1 hr 15 min in a 37° room, and keeping the plates at room temperature lid side up for an additional 5 days before wrapping them in plastic wrap in sets of six and storing at 4°. To produce 96-well solid media culture plates, we prepared NGM-LOB media supplemented with three times the normal amount of peptone and 1 g/liter of yeast extract. We used a WellMate Microplate Dispenser (Matrix) to dispense 375 μl of media to each well using standard bore tubing. Water for prewarming and priming the input lines and the molten media were kept in a 73° water bath while dispensing to prevent solidification. After solidification of the wells, we used the WellMate to dispense 10 μl of OP-50 bacteria culture to each well with small-bore tubing. We then dried the plates with their lids off for 1 hr 15 min in a 37° room before putting their lids back on, repacking and sealing them in their original packaging, and incubating them at 37° for 64 hr. At the end of this time, we unpackaged the plates, removed the condensation from each lid using a Kimwipe, repackaged them, and stored them at 4° for up to 1 month before use. Supporting Information, File S1 lists the genotypes and molecular lesions of all strains used in this study.
unc-16 forward genetic suppressor screen
We mutagenized 9000 L4’s of the strain KG3035 ceIs134 (unc-17::CTNS-1-RFP, unc-17::GFP) with 27.6 mM EMS in M9 supplemented with OP-50 bacteria for 4 hr at 20°. After growing this P0 generation for 72 hr at 14° on four spread plates, we bleach-treated the cultures to release the F1 generation eggs, dispensed the eggs to a single locomotion plate, and allowed them to hatch overnight at 23°. F1 hatchlings were collected and counted, and 3000 were plated on each of 16 spread plates and grown for 24 hr at 14° + 54 hr at 23° to produce gravid F1 adults. We then bleach-treated the F1 adults to release the F2 generation eggs, dispensed the eggs onto three unseeded NGM-LOB plates, and then allowed them to hatch overnight at 20°. F2 hatchlings were collected in M9 buffer and counted, and 5000 were plated on each of 18 spread plates. Half of these plates were grown for 48 hr at 20° and half were grown for 48 hr at 14° + 24 hr at 20° so that screening could be done on 2 successive days. These growth times produced mid-L4 stage synchronous F2 grandprogeny. For each half day of screening, we harvested animals from three of these plates by adding 5 ml of sterile PBS per plate and transferring the suspension to 8 ml of sterile PBS, stirring slowly in a 50-ml beaker. We counted 12, 50-μl aliquots of this suspension, obtained an average concentration, and adjusted the concentration to 24 worms per 50 μl. At 20-min intervals, we pipetted 50 μl (∼24 worms) from the stirring suspension into each of the 12 center wells of a 96-well Mat-Tek glass-bottom plate (MatTek, Ashland, MA; P96G-1.5-F-F) that had been preloaded with 50 μl of 300 μM Levamisole per well. We began screening 10 min after loading the first well and screened the animals in the 12 wells over the next 20 min using a 0.75-numerical aperture, ×20 dry objective on a Nikon TE-2000E inverted microscope with a Universal stage containing an inset for 96-well plates. In each half day of screening, over 2 successive days per week, we screened nine such rows of 12 wells. We screened animals in each well for decreased CTNS-1-RFP fluorescent puncta in the dorsal cord. At the end of each 20 min screening session, after noting the wells containing mutants, we pipetted the contents of each mutant-bearing well onto a predried streak plate using a Pasteur pipette; rinsed the well with 100 μl of M9; and then, immediately after the liquid dried in, clonally distributed up to 24 animals to a 96-well solid-media culture plate containing a thick layer of OP-50 bacteria (see Worm culture and strains). Depending on the relative size of the mutant animal, we often did not plate animals that were outside the relative size range observed for the mutant. After 4 d at 20°, we used a sterile toothpick to pick approximately six L4 larvae from each well into Mat-Tek wells containing 150 μM Levamisole. After rescreening on the inverted microscope, we noted wells with 100% mutant phenotype and used the corresponding well on the 96-well plate to score behavioral and other phenotypes and to set up stocks. We screened 24,192 F2 animals for a calculated 3.5-fold genomic coverage, taking into account an experimentally determined 23% loss of animals during recovery of the mutant.
Complementation tests and mapping of new mutations
For complementation tests we crossed unc-16(ce483); ceIs134 males to mut-1; unc-16(ce483); ceIs134 hermaphrodites (where mut-1 is the unknown mutation). After 3 d at 23° we tested for X-linkage by scoring L4 males from the cross for the CTNS-1-RFP phenotype in Mat-Tek wells on the inverted microscope. After allowing cross-progeny to grow an additional day at 20°, we crossed mut-1/+; ce483; ceIs134 males to mut-2; unc-16(ce483); dpy-11(e224) ceIs134 (using dpy-11 as a marker for self-progeny), where mut-2 was ce749. After 3 days at 23°, we determined whether the two mutations complemented or noncomplemented by scoring L4 hermaphrodite progeny from this cross in Mat-Tek wells on the inverted scope for the CTNS-1-RFP suppressor phenotype. By this method we determined that ce748, ce749, ce755, and ce757 are allelic, that ce759 is an allele of a different X-linked gene, and that ce752 is an allele of a different autosomal gene. To map selected mutations, we crossed the ceIs134 transgene into the CB4856 mapping strain 12 times and then used ceIs134 [CB4856] males to cross the CB4856 polymorphisms through a mut-1; ce483; ceIs134 strain (where mut-1 is the unknown mutation). We cloned 20 non-Unc-16 L4 hermaphrodites from this cross and, after 4 days at 23°, cloned 96 Unc-16 or Mut-1; Unc-16 animals from the next generation to 24-well plates. After 3 days at 23°, we then scored the L4-stage progeny of these animals in Mat-Tek wells on the inverted microscope for the CTNS-1-RFP phenotype to identify the 24-well plate addresses containing homozygous mut-1 animals that had been crossed through the CB4856 mapping strain. We then used such lines to map the mutation relative to single nucleotide polymorphisms (SNPs) as previously described (Schade et al. 2005). By this method we mapped ce749 and ce753 to the same subregion of X and ce759 to a different subregion of X, and we also determined that ce752 is loosely linked to unc-16 on chromosome III.
Whole-genome sequencing and identification of new mutations
To produce genomic DNA for whole-genome sequencing, we plated ∼25 larvae onto each of two spread plates and grew them to starvation (8 days at 23°). We harvested larvae by adding 4 ml TEEN (24 g Tris base, 37 g EDTA, 23 g NaCl/liter; pH 7.5 with 1 N HCl) per plate and collecting them by centrifugation for 2 min at 1500 × g into a 15-ml conical tube. We then removed the supernatant by vacuum suction, washed twice with 12 ml of TEEN, spinning as above after each wash, and resuspended the final worm pellet in ∼200 μl of TEEN in a 1.5-ml snap-cap tube. After centrifuging for 30 sec at 14,000 × g, we removed the supernatant, froze the tubes in liquid nitrogen, and thawed them at room temp for 1.5 min followed by incubation in a 65° temperature block for 1 min. We then followed the manufacturer’s instructions for DNEasy preps (Qiagen) to purify genomic DNA from the samples. To produce libraries for whole-genome sequencing, we quantified the DNA concentrations using the Qubit fluorimeter (Invitrogen) and the Qubit dsDNA BR Assay Kit (Invitrogen), sheared the DNAs to ∼350 bp by transferring 1080 ng of each DNA in 60 μl of water to Covaris Microtubes using two 25-sec cycles in a Covaris S2 at 4°, and then followed the manufacturer’s instructions for the NEBNext Ultra DNA Library Prep Kit for Illumina (NEB E7370S) and NEBNext Multiplex Oligos for Illumina (Index Primers set 1 and set 2; E7335S and E7500S). We analyzed the average size and concentration of the fragments in each library using the Agilent 2200 Tape Station and adjusted the concentration of each library to equal that of the lowest concentration library. We then combined 5 μl of each library in a 250-μl screw-cap tube and submitted the combined samples for sequencing using the Rapid Run PE100 protocol on a single flow cell on a HiSequation 2500 (Illumina). We analyzed the sequences using “Whole Genomes,” which is a set of web-based applications produced by Bob Barstead for analyzing whole-genome sequences. For sequence alignment to C. elegans sequences, we used the analysis pipeline “BWA/ GATK C. elegans WS220 SNP/Indels single RG dups allowed.”.= We then used the resulting alignment file (in VCF format) to produce annotated lists of mutations by using the “Annotate Points” feature of Whole Genomes. We used the “Comparison” feature to subtract-out mutations that were common to another mutant isolated in the same genetic screen and “Filters” to specify homozygosity of mutations, high-quality “PASS” sequences, and nonsilent GC-to-AT mutations (as are produced by the EMS mutagen) and to restrict the analysis to the specific intervals of the genome where we mapped the mutations.
Plasmids
File S1 lists all of the plasmids used in this study along with sources and/or construction details. In all constructs involving the cloning of PCR fragments, we sequenced the inserts and used clones containing no mutations in the fragment of interest to make the final stock.
Production of transgenes and genomic insertions
We prepared plasmids for microinjection using the Qiagen Tip-20 system according to the manufacturer’s instructions, except that we added a 0.1 M potassium acetate/2 vol ethanol precipitation step after resuspending the isopropanol-precipitated pellet. We produced transgenic strains bearing extrachromosomal arrays by the method of Mello et al. (1991). For the cdk-5 rescue experiment, the host was KG2430 ceIs56, and we crossed the ceEx450 transgene into unc-16(ce483); ceIs56. For the sad-1 and syd-2 rescue experiments, the hosts were KG4498 unc-16(ce483); sad-1(ce749) ceIs56 and KG4563 unc-16(ce483); syd-2(ok217) ceIs56, respectively. For all other injection experiments, N2 was the host. We used pBluescript carrier DNA to bring the final concentration of DNA in each injection mixture to 175 ng/μl and integrated the transgenes into the genome using 9100 Rads of gamma rays as described (Reynolds et al. 2005). File S1 lists all the transgenes in this study, their DNA contents, and the injection concentration of each DNA. We mapped the insertion sites of ceIs259 and ceIs267 by crossing the integrant through CB4856, re-isolating and cloning homozygous animals in the F2 generation, and using the resulting mapping lines to map the integration sites relative to SNPs as described (Schade et al. 2005).
Strain constructions
We outcrossed the strains bearing the ce749, ce753, ce759, and ok217 mutations two, two, two, and three times, respectively, before using them for the experiments in this article. File S1 includes a table of all the mutations used in this study and the methods we used to genotype/confirm homozygosity of each mutation during strain constructions. To cross an integrated transgene into a mutant background, we typically crossed a male integrant strain made by the heat-shock method (Sulston and Hodgkin 1988) to the mutant, although in some cases we used integrant/+ heterozygous males. After incubating 3 days at 20°, we cloned five L4-stage cross-progeny carrying the fluorescently marked transgene. After 4 days at 23°, we cloned 12 bright fluorescent (putatively homozygous for the transgene) mutant adult hermaphrodites and grew them for 4 days at 23° before choosing one homozygous mutant line (genotyped/confirmed as indicated in File S1) that was also homozygous for the transgene to make the stock. To construct strains carrying two mutations plus a transgene, we typically first crossed the insertion into each single mutant and then used homo- or hemizygous integrant males for the first cross to mutant A, allowing the entire construction to be done in a background that is homozygous for the insertion. We constructed these strains, as well as the double mutants used for the locomotion assays (the latter lacking transgenes), using the standard method of crossing heterozygous males of mutant A with homozygous hermaphrodites of mutant B and cloning virgin F1 cross-progeny. From plates segregating mutant A in their F2 progeny, we cloned mutant A and/or mutant B animals and looked for segregation of the double mutant in the next generation using behavioral phenotypes (for doubles involving unc-16, syd-1, and/or syd-2) or genotyping as indicated in File S1. After making a strain composed of two or more mutations, we confirmed the homozygosity of each mutation using the methods described in the mutations table in File S1.
Quantitative fluorescence imaging and image analysis of organelle marker distributions
File S1 provides the detailed procedures that we used for quantitative fluorescence imaging and image analysis, including growth and mounting of strains, image acquisition, processing images, quantifying images, and producing representative images.
Time-lapse video microscopy of organelle movements in live animals
File S1 provides the detailed procedures that we used for time-lapse imaging, including growth and mounting of strains, image acquisition, processing time-lapse images, quantifying movements from kymographs, pause analysis, and producing time-lapse movies.
Locomotion rate assays
Previous studies described the basal locomotion rate assays (Miller et al. 1999; Reynolds et al. 2005). We kept the temperature at the assay location at 22°–23.5°.
Statistical analysis
We performed all statistical comparisons using the unpaired t-test, Welch-corrected (for comparisons between two selected groups), or ANOVA followed by the Tukey–Kramer post-test (for comparisons involving three or more groups) or Fisher’s exact test (as indicated) using Graphpad Instat 3 (Graphpad Software). The P-value cutoff for statistical significance was ≤0.05.
Data availability
Strains and reagents are available upon request. File S1 contains Supplemental Materials and Methods and a detailed description of all of the strains, plasmids, and transgenes produced in this study.
Results
A forward genetic screen reveals a role for CDK-5, SAD-1, and SYD-2 in promoting the axonal accumulation of lysosomes in unc-16 mutants
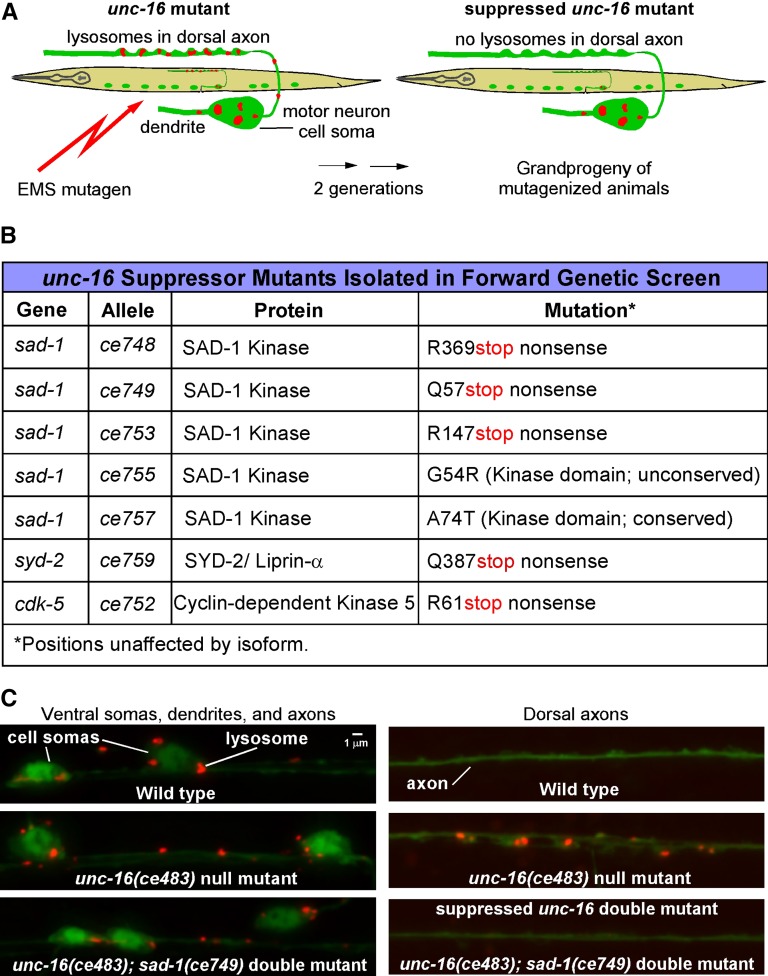
To identify proteins that contribute to the axonal accumulation of organelles in unc-16 mutants, we screened the grandprogeny of EMS-mutagenized unc-16 mutants for suppressor mutants in which lysosomes in the dorsal axons were reduced or absent. To mark lysosomes, we used a genomically integrated transgene that expresses CTNS-1-RFP in C. elegans cholinergic motor neurons. Prior studies confirmed that this marker, which encodes a lysosome cysteine transporter (Kalatzis et al. 2001), specifically localizes to lysosomes in C. elegans (Mangahas et al. 2008; Edwards et al. 2013). The transgene also expresses soluble GFP in the same neurons to monitor expression effects (Figure 1A). A screen of ∼24,200 F2 animals (∼3.5-fold genomic coverage) netted seven mutants with few or no detectable lysosomes in their dorsal axons and normal GFP levels. Complementation tests and mapping revealed that the seven mutations represent five alleles of one gene and single alleles of two other genes. Sequencing the genomes of these mutants and analyzing the mutations in the mapped intervals revealed that the group with five alleles had single mutations in sad-1 (three with premature stop codon mutations and two with missense mutations), while the other two mutants had premature stop codons in syd-2 and cdk-5, suggesting that the suppressor phenotypes of these mutants are all associated with loss of function. The three genes encode proteins known as SAD-1 (SAD-A Kinase), SYD-2 (Liprin-α), and CDK-5 (Cyclin-dependent Kinase 5) (Figure 1, B and C).
Figure 1.
Forward genetic screen for mutations that suppress axonal lysosome accumulation in unc-16 mutants. (A) Drawings illustrate the forward genetic screen for unc-16 suppressor mutants. The screen used EMS to mutagenize an unc-16 null mutant carrying the integrated transgene ceIs134, which uses the unc-17β promoter to co-express CTNS-1-RFP (to mark lysosomes) and GFP (as an expression control) in the ventral cord cholinergic motor neurons. We screened the F2 grandprogeny of mutagenized animals on 96-well glass-bottom Mat-Tek plates using an inverted microscope and selected animals with few or no lysosomes in their dorsal axons and normal GFP expression. (B) Summary of mutations identified in the unc-16 suppressor screen. “Conserved” means that the amino acid is conserved in the human ortholog of SAD-1, known as SAD-A/B or BRSK2. ce753 and ce755 are weaker suppressors of unc-16 than the sad-1 nonsense mutants, and they contain some lysosomes in their dorsal axons. (C) Representative images of wild-type, the unc-16 mutant, and a double mutant carrying the unc-16 null mutation in combination with one of the suppressor mutations. Lysosomes (red puncta) are inside GFP-labeled cholinergic motor neurons (green).
CDK-5, SAD-1, and SYD-2 act together to promote axonal lysosome accumulation in an unc-16 null mutant
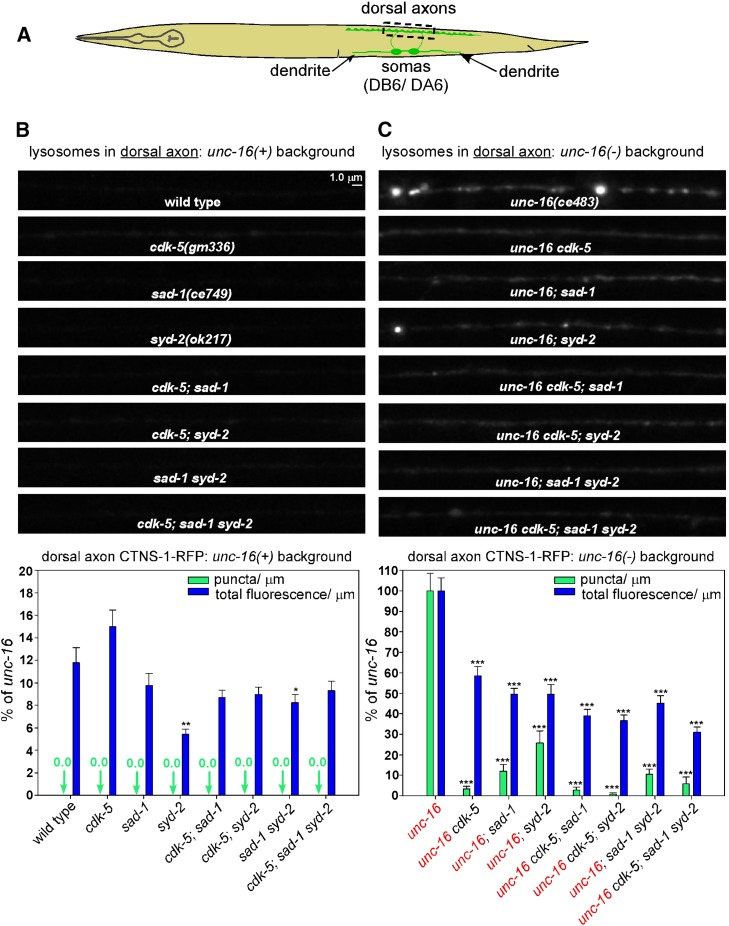
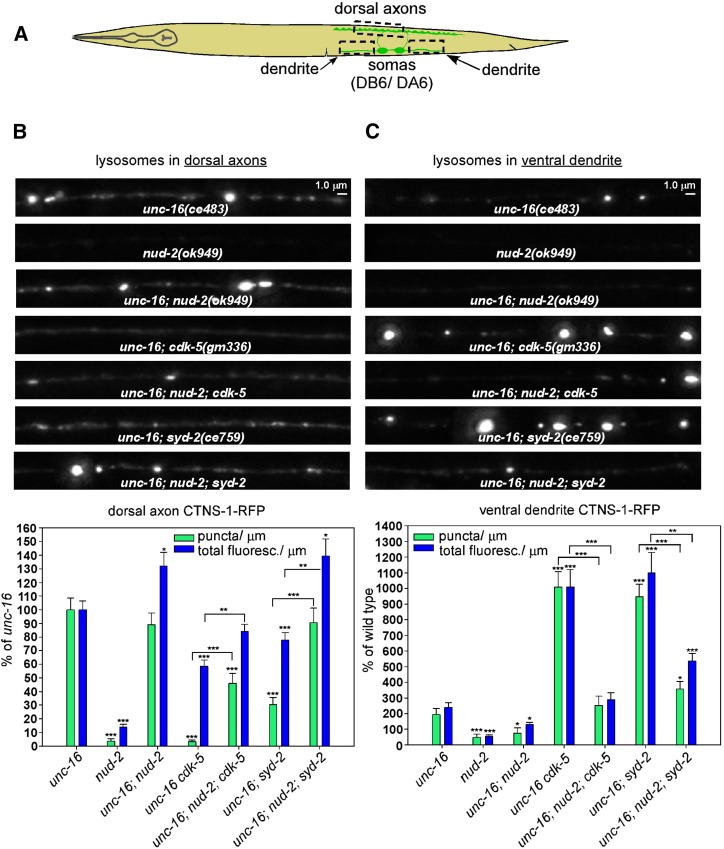
To quantify the extent to which eliminating the function of each of these proteins affects the distribution of CTNS-1-RFP lysosomes in neurons in unc-16(+) and unc-16(−) backgrounds, we crossed putative null mutations of each gene into an integrated transgene background that expresses CTNS-1-RFP in a subset of nine cholinergic motor neurons in which the axons, cell somas, and dendrites of each motor neuron are segregated and thus easily identifiable (Figure 2A). For these experiments, we used the independently isolated deletion alleles cdk-5(gm336) and syd-2(ok217) along with the sad-1(ce749) premature stop codon allele, and we quantified both the number of puncta per micrometer exceeding a predefined threshold (lysosome punctal density) and the total fluorescence per micrometer of axon length (CTNS-1-RFP fluorescence/μm). The total fluorescence/μm parameter includes both lysosome puncta and nonpunctal CTNS-1-RFP fluorescence.
Figure 2.
CDK-5, SAD-1, and SYD-2 act together to promote axonal lysosome accumulation in an unc-16 null mutant. (A) Drawing illustrates the location and anatomy of the cholinergic motor neurons imaged in this figure. Dashed box outlines the region imaged. (B and C) Representative, identically scaled images and quantification of CTNS-1-RFP lysosomal puncta and total fluorescence per micrometer of the indicated genotypes in an unc-16(+) background (B) or an unc-16(−) background (C). CTNS-1-RFP is expressed from the integrated transgene ceIs56. Data are means and SEMs from 13–14 animals. Asterisks indicate values significantly different from wild type (B) or unc-16 (C): *P < 0.05; **P < 0.01; ***P < 0.0001. For lysosome punctal density in an unc-16(−) background, none of the CCS mutant combinations differed significantly from the component single mutants. However, for the fluorescence/μm parameter, the unc-16 cdk-5; sad-1 triple mutant and the unc-16 cdk-5; sad-1 syd-2 quadruple mutant showed small but significant reductions when compared to the unc-16; sad-1 double mutant (P = 0.02 and <0.0001), and the unc-16 cdk-5; syd-2 triple mutant showed a small but significant reduction when compared to unc-16; syd-2 (P = 0.03).
In an unc-16(+) background, wild-type and all three single mutants had lysosome punctal densities of zero. Wild type’s CTNS-1-RFP fluorescence/μm was ∼12% of the unc-16 null mutant, and neither the cdk-5 nor the sad-1 single mutant was significantly different from the wild type. The syd-2 mutant was slightly, but significantly, lower at ∼6% of the unc-16 mutant (Figure 2B).
In an unc-16(−) background, the lysosomal punctal density was strongly reduced in the unc-16cdk-5, unc-16; sad-1, and unc-16; syd-2 doubles relative to unc-16 single mutants, thus confirming the suppression phenotype that we detected in the genetic screen. The strongest reduction occurred in the unc-16cdk-5 double (∼3% of unc-16), whereas sad-1 and syd-2 doubles showed reductions to 12 and 26% of unc-16, respectively. All three doubles showed similar but smaller reductions in fluorescence/μm (to ∼50% of wild type; Figure 2C). Independently isolated alleles of sad-1 and syd-2 showed quantitatively similar reductions in lysosome punctal density and fluorescence (data not shown). In addition, expressing cdk-5, sad-1, or syd-2 transgenically in the same neurons significantly rescued the suppression phenotype in unc-16cdk-5, unc-16; sad-1, and unc-16; syd-2 double mutant axons, demonstrating that all three proteins function in the motor neurons that express the unc-16 suppression phenotype (Figure S1B).
To determine whether the cdk-5, sad-1, and syd-2 mutations disrupt the same or different systems regulating axonal lysosome content, we analyzed all possible combinations of the mutants in unc-16(+) and unc-16(−) backgrounds and compared their axonal lysosome density and fluorescence/μm parameters to the component single mutants. In an unc-16(+) background, none of the mutant combinations differed significantly from the component mutants from which they were constructed for either parameter (i.e., they all showed zero puncta and had low fluorescence levels similar to wild type; Figure 2B). In an unc-16(−) background, none of the mutant combinations, including the unc-16cdk-5; sad-1syd-2 quadruple mutant, showed a significant difference in lysosomal punctal density relative to the lowest level of any individual component mutant. However, for the fluorescence/μm parameter, the unc-16cdk-5; sad-1 triple mutant and the unc-16cdk-5; sad-1syd-2 quadruple mutant showed significant reductions when compared to the unc-16; sad-1 double mutant, and the unc-16cdk-5; syd-2 triple mutant showed a significant reduction when compared to unc-16; syd-2. However, these reductions were relatively minor, decreasing from 50% of the unc-16 single mutant to 39, 31, and 31%, respectively, and none of the other mutant combinations showed a significant difference when compared to the lowest level of the component mutants (Figure 2C).
In summary, although these results show that the three proteins do not have equally important roles (CDK-5 apparently having the strongest role, at least for lysosomal punctal density), they do suggest that CDK-5, SAD-1, and SYD-2 largely act together and cell-autonomously as part of a system that promotes axonal lysosome accumulation in the absence of UNC-16. In addition, because elimination of CDK-5, SAD-1, or SYD-2 strongly suppresses axonal lysosome accumulation in unc-16 mutants, the data are consistent with UNC-16 directly or indirectly inhibiting the CDK-5/SAD-1/SYD-2 system from promoting axonal accumulation of lysosomes.
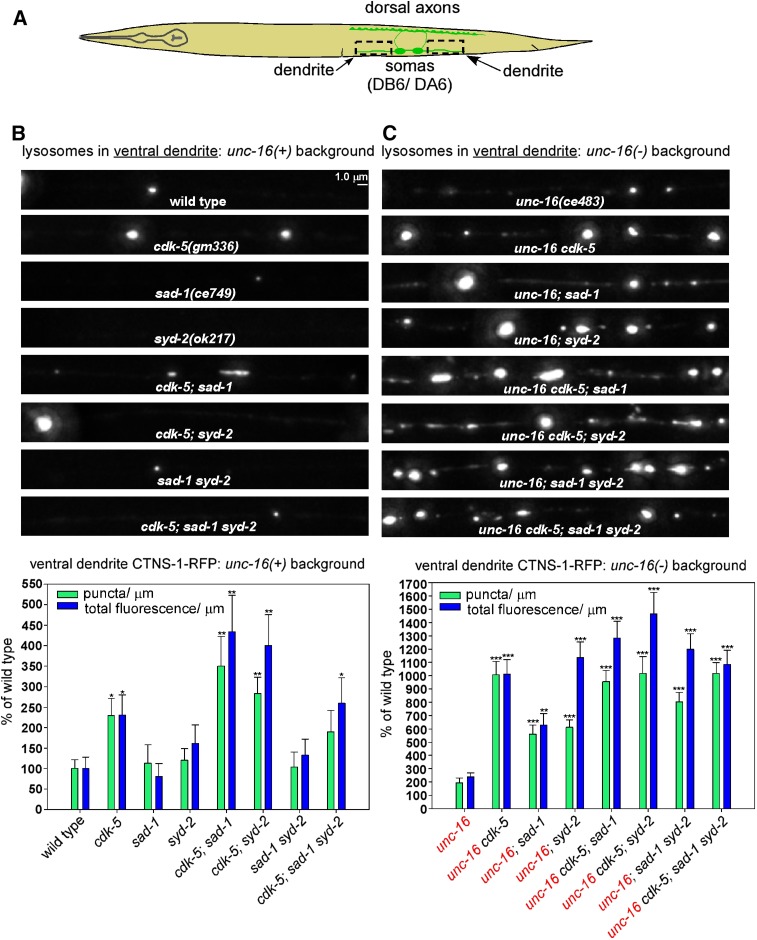
CDK-5, SAD-1, and SYD-2 act together to inhibit dendritic lysosome accumulation in an unc-16 null mutant
We next investigated how the same mutations affect the lysosome content of dendrites. In an unc-16(+) background, wild type had a relatively low lysosomal punctal density in its dendrites, averaging ∼1 puncta every 52 μm in the region bounding the DA6/DB6 motor neurons. Dendritic lysosomal puncta density and fluorescence/μm for sad-1 and syd-2 single mutants were not significantly different from wild type; however, the levels of puncta and fluorescence in cdk-5 single mutants were significantly higher (approximately two-fold), demonstrating that CDK-5 has a role in regulating dendritic lysosome content even in the presence of wild-type UNC-16 (Figure 3, A and B).
Figure 3.
CDK-5, SAD-1, and SYD-2 act together to inhibit dendritic lysosome accumulation in an unc-16 null mutant. (A) Drawing illustrates the location and anatomy of the cholinergic motor neurons imaged in this figure. Dashed boxes outline the regions imaged (both dendrites were imaged together and then combined for quantification). (B and C) Representative, identically scaled images and quantification of CTNS-1-RFP lysosomal puncta and total fluorescence per micrometer of the indicated genotypes in an unc-16(+) background (B) or an unc-16(−) background (C). CTNS-1-RFP is expressed from the integrated transgene ceIs56. Data are means and SEMs from 13–14 animals. Asterisks indicate values significantly higher than wild type (B) or unc-16 (C): *P < 0.05; **P < 0.01; ***P < 0.0001. None of the mutant combinations in unc-16(+) or unc-16(−) backgrounds, including the unc-16 cdk-5; sad-1 syd-2 quadruple mutant, showed a significant difference in dendritic lysosomal punctal density compared to the highest level of any individual component mutant. However, for the fluorescence/μm parameter, the unc-16 cdk-5; syd-2 triple mutant was significantly higher than the unc-16; cdk-5 double mutant (P = 0.03).
Similar to cdk-5 single mutants, unc-16 mutants had dendritic levels of lysosomal puncta and fluorescence that were ∼2-fold higher than wild type. However, the unc-16cdk-5 double showed a nonadditive synthetic effect in which dendritic puncta and fluorescence levels increased to ∼10-fold higher than wild type. Furthermore, even though sad-1 and syd-2 single mutants had wild-type levels of lysosomes in their dendrites, their doubles with unc-16 had levels that were 6–10-fold higher than wild type (Figure 3C). Independently isolated alleles of sad-1 and syd-2 in doubles with unc-16 showed quantitatively similar dendritic accumulations of lysosomes (data not shown). In addition, expressing cdk-5, sad-1, or syd-2 transgenically in the same neurons significantly rescued the suppression phenotype in unc-16cdk-5, unc-16; sad-1, and unc-16; syd-2 double mutant dendrites, demonstrating that all three proteins function in the motor neurons that express the unc-16 suppression phenotype (Figure S1C).
To determine whether the cdk-5, sad-1, and syd-2 mutations disrupt the same or different systems for regulating dendritic lysosome content, we analyzed all possible combinations of the mutants in unc-16(+) and unc-16(−) backgrounds and compared their dendritic puncta and fluorescence levels to the component single mutants. None of the mutant combinations in unc-16(+) or unc-16(−) backgrounds, including the unc-16cdk-5; sad-1syd-2 quadruple mutant, showed a significant difference in dendritic lysosomal punctal density compared to the highest level of any individual component mutant. However, for the fluorescence/μm parameter, the unc-16cdk-5; syd-2 triple mutant was 1400% of wild type, which was a significant increase compared to the unc-16; cdk-5 double mutant (1000% of wild type). However, none of the other mutant combinations in unc-16(+) or unc-16(−) backgrounds, including the quadruple mutant, showed a significant difference in fluorescence/μm compared to the highest level in the component mutants (Figure 3, B and C).
We also analyzed cell soma levels of CTNS-1-RFP-labeled lysosomes in the same set of mutants and found that cdk-5, sad-1, and syd-2 loss-of-function mutations are associated with significant, albeit mild-to-moderate, decreases in lysosomes (∼60–90% of the unc-16 single mutant; Figure S2). This is consistent with a relatively slow loss of lysosomes to the dendrite in these mutants (i.e., at a rate that allows cell soma levels to remain relatively stable) and also shows that effects on expression of the transgene are not the cause of the increased dendritic lysosome levels.
In summary, although the results again show that the three proteins do not have equally important roles (CDK-5 and SYD-2 apparently having the strongest roles), they do suggest that CDK-5, SAD-1, and SYD-2 largely act together as part of the same general system that inhibits dendritic lysosome accumulation in the absence of UNC-16.
SYD-1 and STRD-1 (STRADα) act with the CDK-5/SAD-1/SYD-2 system to regulate axonal and dendritic lysosome trafficking in an unc-16 null mutant
A previous study found that CDK-5 acts in parallel with the cyclin-dependent Pctaire kinase PCT-1 to regulate the trafficking of synaptic vesicles in some, but not all, classes of motor neurons in C. elegans (Ou et al. 2010). To determine if CDK-5 and PCT-1 also act in parallel to regulate the distribution of lysosomes in cholinergic motor neurons, we compared unc-16 single mutants to unc-16cdk-5 and unc-16; pct-1 doubles and the unc-16cdk-5; pct-1 triple. For this analysis we used the null deletion allele pct-1(tm2175), which removes the kinase domain in all three isoforms (Ou et al. 2010). The unc-16; pct-1 double did have a significantly lower lysosomal punctal density in its axons compared to the unc-16 single mutant (∼50%); however, the percentage decrease was much less than the unc-16; cdk-5 double, and its CTNS-1 fluorescence/μm was not significantly different from the unc-16 single mutant (Figure S3, A and B). The unc-16; pct-1 double also had a small but significant increase in fluorescence/μm in its dendrites compared to the unc-16 single mutant (approximately two-fold), but its punctal density was not significantly different from the unc-16 single mutant. Also, in the unc-16cdk-5; pct-1 triple mutant, the severity of the axonal and dendritic phenotypes was not significantly increased relative to the unc-16cdk-5 double mutant. These results thus show that CDK-5 is the most important/relevant cyclin-dependent kinase in these neurons for lysosomal trafficking and also suggest that CDK-5 can mostly substitute for lack of PCT-1, but the reverse is not true.
Previous studies in C. elegans and Drosophila found that SYD-1 acts in the same pathway as SYD-2 to regulate synapse assembly and positioning of active zone components in motor neurons (Dai et al. 2006; Patel et al. 2006; Li et al. 2014). To determine whether SYD-1 and SYD-2 also act in the same system to regulate the trafficking of lysosomes in motor neurons, we compared unc-16 single mutants to unc-16; syd-1 and unc-16; syd-2 doubles and the unc-16; syd-1; syd-2 triple. For this analysis we used the null deletion allele syd-1(tm6234). In the unc-16; syd-1 double, the lysosomal punctal density in axons was significantly lower than the unc-16 single mutant (∼60% of unc-16). This suppression phenotype was significantly weaker than the unc-16; syd-2 double, which had a lysosome punctal density that was ∼25% of unc-16. In dendrites, the lysosomal punctal density and fluorescence/μm for the unc-16; syd-1 double were significantly higher than the unc-16 single mutant, and, in this region, the lysosomal punctal density of the unc-16; syd-1 double was not significantly different from the unc-16; syd-2 double. The unc-16; syd-1; syd-2 triple was not significantly different from the unc-16; syd-2 double for either parameter in axons or dendrites (Figure S3, A–C). We conclude that SYD-1 functions in the same system as SYD-2 to regulate the trafficking of lysosomes in axons and dendrites and that SYD-2 remains partially functional in the absence of SYD-1 in its axonal lysosome trafficking role, but the reverse is not true. Neither protein is functional without the other for dendritic lysosome trafficking.
We applied a similar analysis to test for a role for NAB-1 (Neurabin) in lysosome trafficking. Previous C. elegans studies found that NAB-1 is an F-actin-binding protein that also directly binds SAD-1 (Hung et al. 2007) and SYD-1 (Chia et al. 2012), and that SYD-2 may form a complex with SYD-1 and NAB-1 (Chia et al. 2012). For this analysis, we used the nab-1(ok943) null deletion allele (Hung et al. 2007). In axons, the unc-16; nab-1 double showed no significant difference when compared to the unc-16 single for lysosomal punctal density or fluorescence/μm. In dendrites, the unc-16; nab-1 double had a lysosomal punctal density that was approximately threefold higher than wild type (significantly higher than the approximately two-fold increase of the unc-16 single) with a similar significant increase in fluorescence/μm. The unc-16; sad-1; nab-1 and unc-16; syd-2; nab-1 triples were not significantly different from the corresponding unc-16; sad-1 and unc-16syd-2 doubles for either parameter in axons or dendrites (Figure S4, A–C). We conclude that NAB-1 does not have a nonredundant role in preventing the axonal accumulation of lysosomes in unc-16 mutants. In dendrites, it appears to have a minor role in inhibiting lysosome accumulation in unc-16 mutants, and this minor function overlaps with the functions of SAD-1 and SYD-2.
Continuing our search for other proteins that may act with the CDK-5/SAD-1/SYD-2 system to regulate lysosome trafficking in neurons, we applied a similar analysis to test for a role for STRD-1 (STRADα), a STE-20-related pseudokinase. A previous C. elegans study found that STRD-1 forms a complex with SAD-1 and that loss-of-function mutations in strd-1 and sad-1 show similar neuronal phenotypes (Kim et al. 2010). For this analysis, we used the strd-1(ok2283) null deletion allele (Kim et al. 2010). Like the sad-1 single mutant, strd-1 single mutants in an unc-16(+) background showed no significant increase in dendritic lysosomes for either parameter (S. L. Edwards and K. G. Miller, unpublished results). In axons, similar to the unc-16; sad-1 double mutant, the unc-16; strd-1 double showed significantly lower levels than the unc-16 single mutant for lysosomal punctal density and fluorescence/μm. However, its suppressor phenotype, which was ∼38 and 65% of the unc-16 single mutant for lysosomal punctal density and fluorescence/μm, respectively, was significantly weaker than that conferred by the sad-1 mutant (which was ∼12 and 50% of the unc-16 single mutant, respectively). In dendrites, the unc-16; sad-1 and unc-16; strd-1 doubles were not significantly different for either parameter. The unc-16; sad-1; strd-1 triple mutant showed no significant differences for either parameter in axons or dendrites when compared to unc-16; sad-1 (Figure S5, B and C). We conclude that STRD-1 (STRADα) and SAD-1 are part of the same system to regulate the trafficking of lysosomes in axons and dendrites. The data further show that neither protein can function without the other in regulating dendritic lysosome trafficking, but SAD-1 remains partially functional in the absence of STRD-1 for axonal lysosome trafficking.
KIF1A (UNC-104) is not the motor that carries lysosomes to the axonal synaptic region in unc-16 mutants
In the accompanying article in this issue, we found that the synapse assembly proteins SYD-1, SYD-2, and SAD-1 promote the plus-end-directed trafficking of synaptic vesicles by the kinesin KIF1A (UNC-104), preventing their accumulation in dendrites and cell somas in C. elegans cholinergic motor neurons (Edwards et al. 2015). However, UNC-104 does not contribute to the accumulation of lysosomes in unc-16 mutant axons because, when we compared unc-16 single mutants to unc-16; unc-104 doubles, the cell soma and axonal levels of lysosomes in motor neurons in the unc-16; unc-104 double were not significantly lower than the unc-16 single mutant (Figure S6). For this analysis we used the unc-104(e1265) allele, which contains a missense mutation in the cargo-binding domain that causes a strong loss of function (Kumar et al. 2010), but we also obtained the same result using the motor domain temperature-sensitive allele unc-104(ce782) (Edwards et al. 2015), which we analyzed at the restrictive temperature of 20° (data not shown).
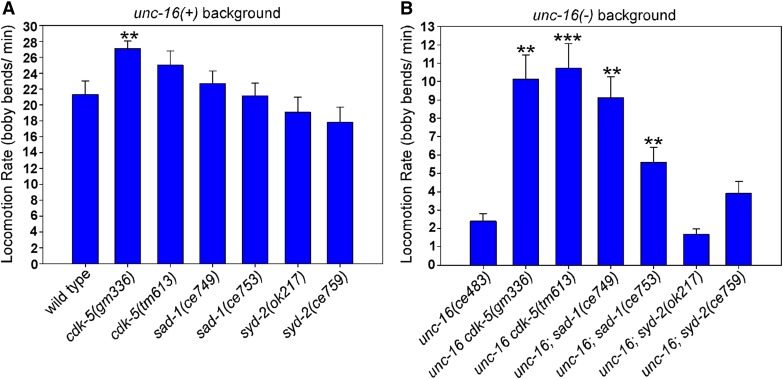
cdk-5 and sad-1 null mutations also suppress the sluggish locomotion of an unc-16 null mutant
unc-16 null mutants have sluggish spontaneous locomotion rates that are ∼10% of wild type (Edwards et al. 2013). We considered the possibility that the abnormal presence of high levels of cell soma organelles in motor neuron axons might directly or indirectly impair neuronal function and, in so doing, might contribute to the sluggish locomotion phenotype. If this is the case, then mutations that suppress axonal organelle accumulation in unc-16 mutants should also improve their locomotion. To test this, we quantified the locomotion rates of wild type and cdk-5, sad-1, and syd-2 mutants in unc-16(+) and unc-16(−) backgrounds. For this analysis, we tested two independently isolated null or loss-of-function alleles of each gene. In an unc-16(+) background, none of the single mutants had locomotion rates that were significantly slower than wild type, although one of the cdk-5 mutants had a slightly, but significantly, higher locomotion rate (Figure 4A). However, depending on the allele used, unc-16cdk-5 and unc-16; sad-1 double mutants showed significant ∼2.3–4.5-fold improvements in their locomotion rates compared to unc-16 single mutants (Figure 4B). unc-16; syd-2 double mutants did not show improved locomotion compared to the unc-16 single. However, we also noted that unc-16; syd-2 doubles had synthetic phenotypes such as slower growth and short body length, and the extent to which these and other possible synthetic phenotypes that affect neuronal function in unknown ways might contribute to their overall locomotion rate is unclear.
Figure 4.
Eliminating CDK-5 or SAD-1 improves the locomotion rate of an unc-16 null mutant. (A and B) Locomotion rates of the indicated genotypes in unc-16(+) (A) and unc-16(−) (B) backgrounds. Data are means and SEMs from 10 animals. **P < 0.01 and ***P < 0.0001, respectively, when compared to wild type (A) or unc-16(ce483) (B). Unmarked bars are not significantly different from the wild-type (A) or unc-16(ce483) (B) controls.
In summary, although such whole-animal behavioral data cannot allow us to make a causal conclusion about the extent to which the accumulation of cell soma organelles in axons impairs neuronal function, they demonstrate a significant correlation.
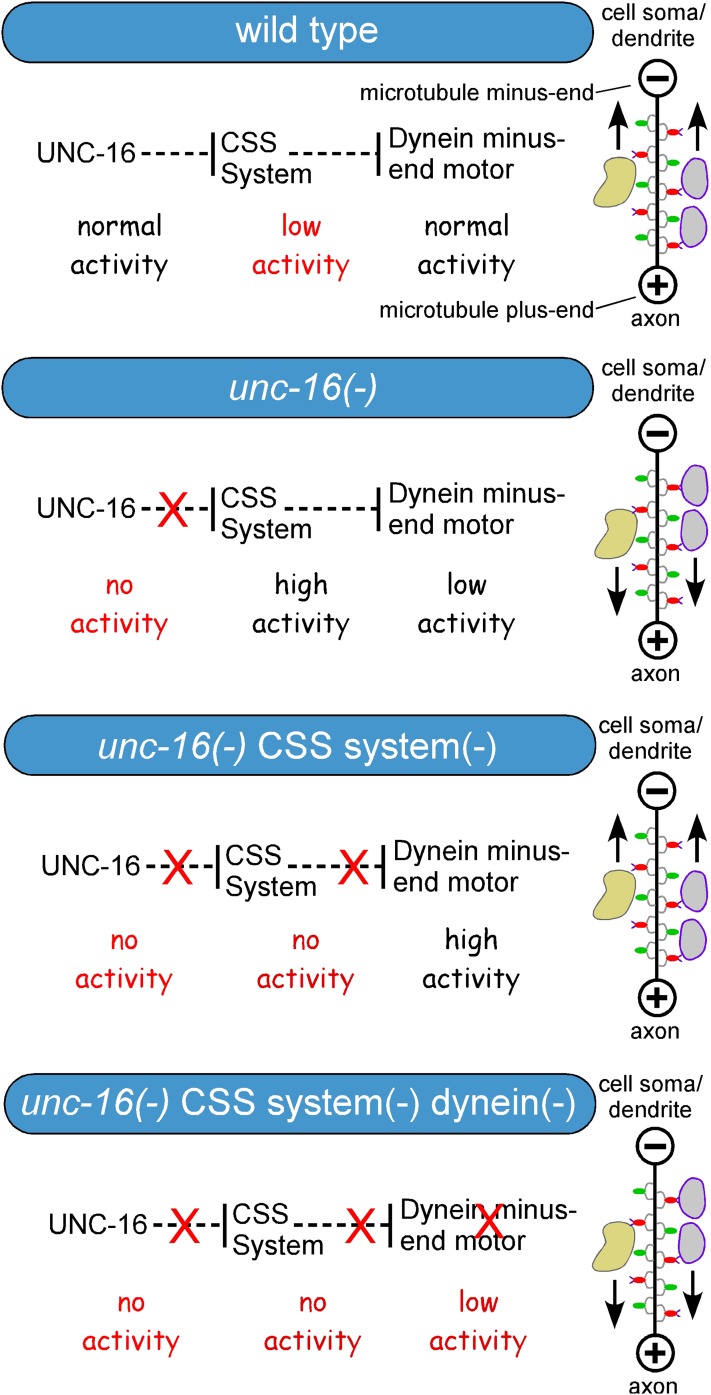
Accumulation of lysosomes in the dendrites of unc-16 cdk-5 and unc-16; syd-2 double mutants is dynein-dependent
In C. elegans motor neurons, including the DA6/DB6 motor neuron pair that we imaged in this study, axonal microtubules are primarily plus-end-out and dendritic microtubules are largely minus-end-out (Goodwin et al. 2012; Yan et al. 2013). Because the unc-16 suppressor mutations lead to the depletion of lysosomes from axons and their accumulation in dendrites, we tested the hypothesis that this redistribution is dependent on the minus-end motor dynein by comparing unc-16; cdk-5 and unc-16; syd-2 mutants in nud-2(+) and nud-2(−) backgrounds. nud-2 encodes the C. elegans ortholog of the Aspergillus nidulans NUDE protein (known as NUDEL and Nde1 in vertebrates), which forms a complex with the dynein regulator LIS1 and dynein heavy chain (Niethammer et al. 2000; Sasaki et al. 2000) that may stimulate the transport of acidic organelles upon phosphorylation by Cdk5 (Pandey and Smith 2011). For this analysis we used the nud-2(ok949) null deletion allele (Fridolfsson et al. 2010), which is homozygous viable.
When we analyzed CTNS-1-RFP in axons, the unc-16; nud-2 double did not differ from the unc-16 single mutant for lysosomal punctal density. However, in the absence of both unc-16 and cdk-5, the additional genetic ablation of nud-2 (i.e., the unc-16; nud-2; cdk-5 triple) resulted in a 14-fold increase in axonal lysosomal punctal density, to ∼60% of the unc-16 single, and increased axonal fluorescence/μm to ∼82% of the unc-16 single. Similarly, ablating nud-2 restored the axonal lysosomal punctal density of the unc-16; syd-2 double mutant from ∼30% of the unc-16 single to a level not significantly different from the unc-16 single and increased its fluorescence/μm to a level significantly higher than the unc-16 single (Figure 5B).
Figure 5.
CDK-5 and SYD-2 prevent the dynein-dependent accumulation of lysosomes in dendrites in the absence of UNC-16. (A) Drawing illustrates the location and anatomy of the cholinergic motor neurons imaged in this figure. Dashed boxes outline the regions imaged. (B and C) Representative, identically scaled images and quantification of CTNS-1-RFP lysosomal puncta and total fluorescence per micrometer in the dorsal axons (B) and ventral dendrites (C) of the indicated genotypes in an unc-16(−) background. CTNS-1-RFP is expressed from the integrated transgene ceIs56. Data are means and SEMs from 13–14 animals per region. Asterisks over bars indicate values significantly different from unc-16. Asterisks over brackets denote P-values for the selected comparisons: *P < 0.05; **P < 0.01; ***P < 0.0001.
In dendrites, we found that ablating nud-2 restored the approximately two-fold increase seen in unc-16 single mutants to levels not significantly different from wild type for both punctal density and fluorescence/μm. However, we observed a much larger effect of the nud-2 ablation on the dendritic lysosome content of the unc-16cdk-5 double, where CTNS-1-RFP levels decreased from >10-fold higher than wild type in the unc-16cdk-5 double to levels not significantly different from the unc-16 single in the unc-16; nud-2; cdk-5 triple. The nud-2 mutation had similar effects on the unc-16; syd-2 double, although dendritic lysosome levels in the unc-16; nud-2; syd-2 triple were still significantly higher than the unc-16 single (Figure 5D). Lysosome levels in the cell soma were not significantly affected in the strains used for this experiment (Figure S7, A and B).
In summary, these data suggest that the loss of lysosomes from the axons of unc-16cdk-5 and unc-16; syd-2 double mutants, and the accumulation of lysosomes in the dendrites of the same mutants, is dependent on the minus-end motor dynein.
CDK-5, SAD-1, and SYD-2 regulate dendritic lysosome transport even in an UNC-16(+) background
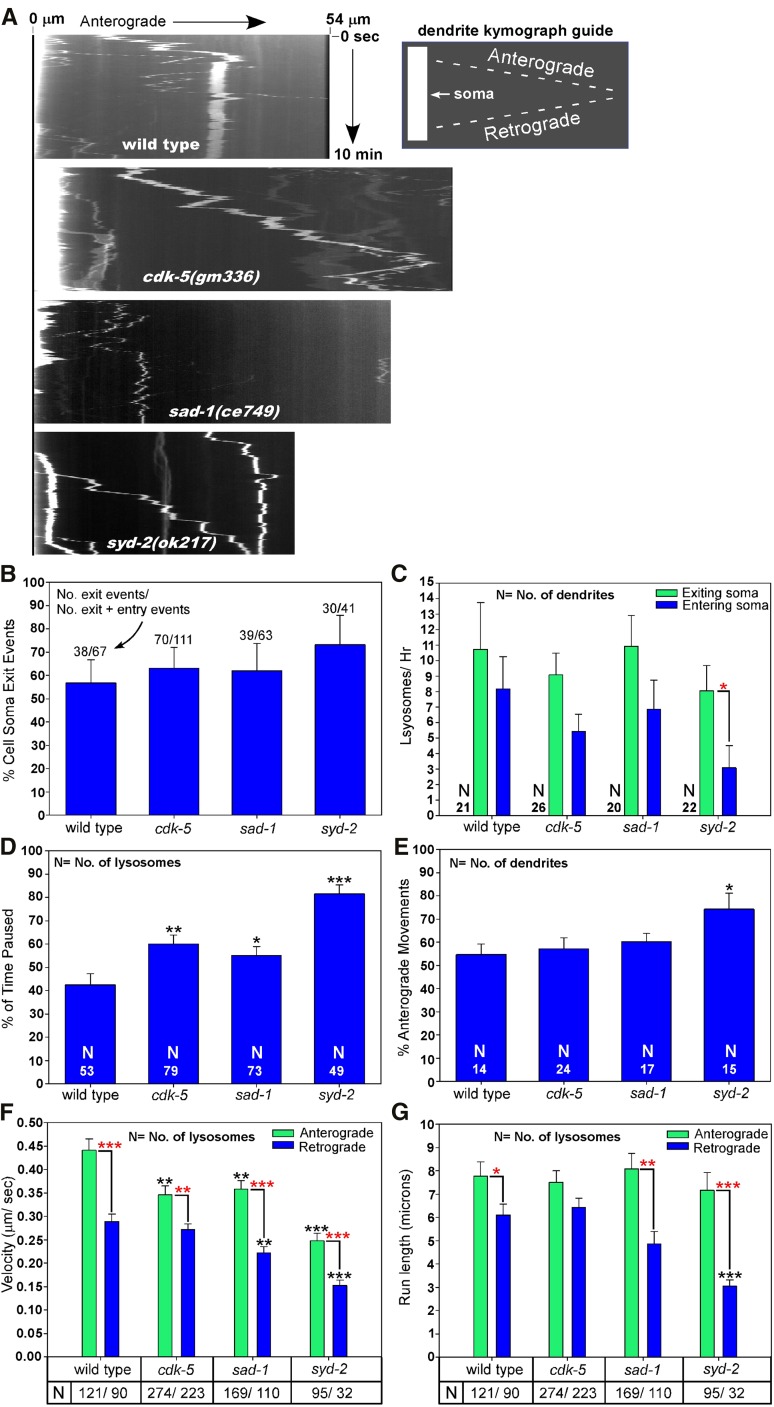
In a previous study we found that, in wild type, lysosomes are not restricted to the cell soma, but instead move back and forth between the cell soma and the axon initial segment, only rarely moving beyond the initial segment, and even more rarely attaining the distal synaptic region in wild-type motor neurons (Edwards et al. 2013). However, that study did not examine lysosome movements between the cell soma and dendrite. We therefore used time-lapse imaging in living animals to view the movements of CTNS-1-RFP-tagged lysosomes both between the DB7 cholinergic motor neuron cell soma and dendrite and within the DB7 dendrite in wild-type, cdk-5, sad-1, and syd-2 null mutants. Similar to the axon/soma boundary, we found that lysosomes in wild-type animals frequently exited the cell soma and entered the dendrite (and vice versa), either in the form of spherical (symmetrical) puncta or large multi-micrometer-long tubular organelles (Figure 6A; movie File S2). The number of exit and entry events was approximately equal, and each occurred at approximately the same rate of 8–10 events per hour at 22° (Figure 6, B and C). None of the three single mutants differed significantly from wild type in their percentage of cell-soma-to-dendrite exit events (i.e., the proportion of all exit/entry events that were exit events) nor in the rates at which lysosomes entered/exited the soma to/from the dendrite (Figure 6, B and C). However, syd-2 mutants showed a significantly lower rate of lysosomes entering the cell soma from the dendrite compared to the rate at which lysosomes exited the cell soma to the dendrite in the same strain (Figure 6C).
Figure 6.
Regulation of dendritic lysosome transport by CDK-5, SAD-1, and SYD-2 in an unc-16(+) background. (A) Representative kymographs of CTNS-1-RFP-tagged lysosome movements in the DB7 dendrite of animals with the indicated genotypes. The bright region at the left of the top three images is the cell soma. CTNS-1-RFP is expressed from the integrated transgene ceIs56. See also movies in File S2, File S3, File S4, and File S5. (B–G) Graphs plotting various indicated parameters extracted from the kymographs and time-course analyses. Error bars in B are 95% confidence intervals derived from Fisher’s exact test. All other error bars are standard errors of the means. *P < 0.05, **P < 0.01, and ***P < 0.0001. Black asterisks compare the marked bar to the wild-type value. Red asterisks compare the indicated two bars in a group. Unmarked bars are not significantly different from wild type or the other group member. Total recorded minutes used for exit/entry analyses (in order of strains as shown): 213, 356, 229, and 220. Total recorded minutes used for movement analyses (in order of strains as shown): 199, 311, 195, 207.
Upon entering the dendrite, wild-type lysosomes spent ∼42% of their time in a paused state. However, lysosomes in cdk-5, sad-1, and syd-2 mutants spent a significantly higher percentage of their time in a paused state (60, 57, and 80%, respectively), suggesting that lysosomes in all three mutants have decreased processivity in the dendrite.
We also analyzed the directionality of lysosome movements within the dendrite. Because the dendrites of these motor neurons are oriented with their minus-ends out (Goodwin et al. 2012; Yan et al. 2013), anterograde movements (movements outward from the soma) are minus-end-directed and retrograde movements (movements toward the soma) are plus-end-directed. This analysis revealed that 55–60% of the movements in wild-type, cdk-5, and sad-1 were in an anterograde (minus-end) direction, whereas lysosomes in the syd-2 mutant moved anterogradely (toward minus ends) 75% of the time (a difference that was significant compared to wild type; Figure 6E).
When not in a paused state, lysosomes in wild type moved anterogradely at a mean velocity of 0.45 μm/sec and retrogradely at a significantly slower mean velocity of 0.28 μm/sec. Although all three mutants shared the wild-type pattern of having retrograde movements that were significantly slower than anterograde movements, their mean anterograde and retrograde velocities were significantly slower than wild type, with the exception of the retrograde velocity in cdk-5 mutants, which was not significantly different from wild type. Correlating with a higher percentage of time in the paused state, lysosomes in syd-2 mutants had the slowest velocities: both anterograde and retrograde velocities were ∼55% of wild type (Figure 6F). Despite these slower velocities, the mean anterograde run lengths of wild type and the three mutants were not significantly different. The mean retrograde run length of the cdk-5 mutant was also not significantly different from wild type; however, the sad-1 mutant had a shorter retrograde run length that approached statistical significance (P = 0.08), and the syd-2 mutant had a mean retrograde run length that was ∼50% of wild type (a highly significant difference; Figure 6G).
In summary, all three mutants showed significant differences from wild type in two or more parameters for the dendritic transport of lysosomes. Dendritic lysosomes in all three mutants spend a higher percentage of their time in a paused state and have slower anterograde (minus-end-directed) velocities when they are not in a paused state. sad-1 and syd-2 mutants also have slower retrograde (plus-end-directed) velocities and shorter retrograde run lengths. These phenotypes are strongest in the syd-2 mutant, which also was the only one of the three mutants in which lysosomes entered the soma from the dendrite at a significantly lower rate than the rate in the reverse direction.
In an unc-16(−) background, decreased processivity and a bias toward anterograde (minus-end) movements may cause lysosomes to accumulate in dendrites in cdk-5, sad-1, and syd-2 null mutants
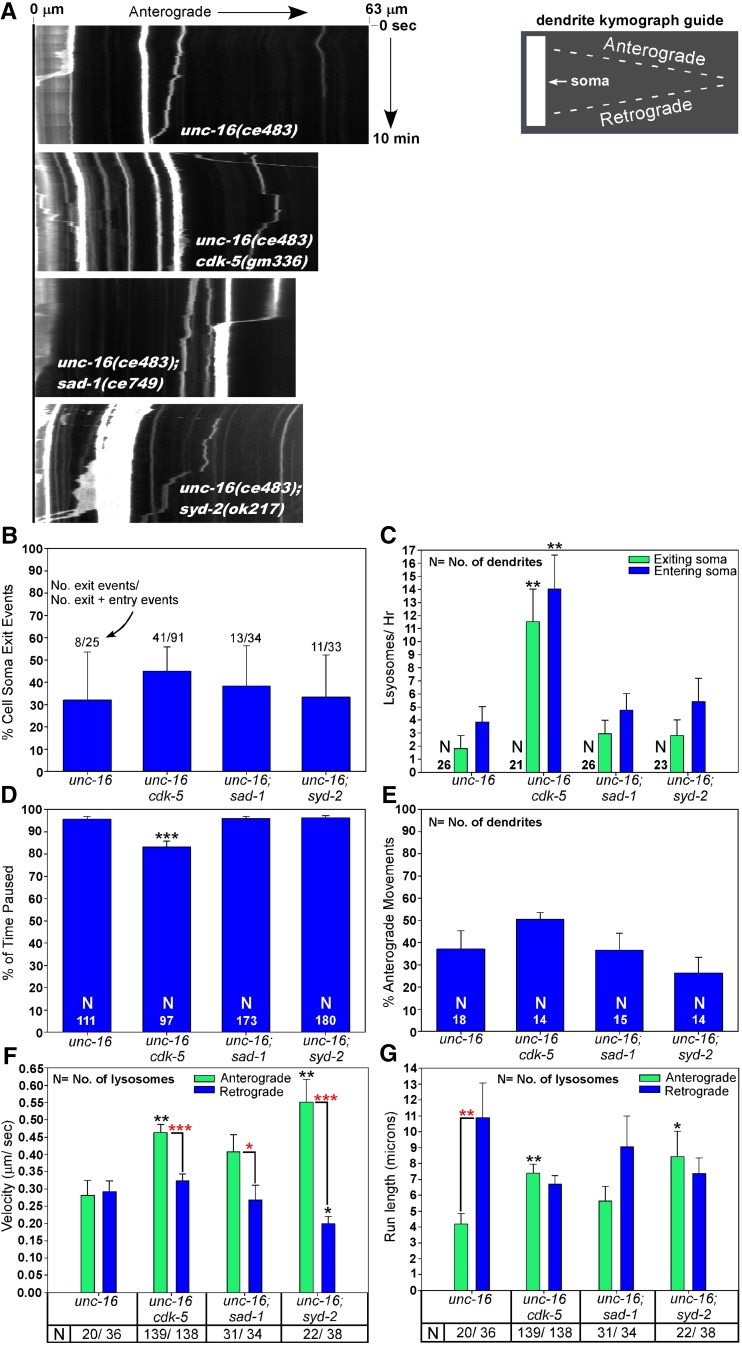
To investigate why lysosomes accumulate at high levels in the dendrites of unc-16 mutants that also lack cdk-5, sad-1, or syd-2, we performed the same time-lapse imaging experiments on unc-16 and all three double mutants. Similar to the results in the unc-16(+) background, none of the three suppressor mutants significantly affected the percentage of cell-soma-to-dendrite exit events compared to unc-16 single mutants (Figure 7B), although all three double mutants had a lower percentage of cell soma exit events than the corresponding strains in an unc-16(+) background (comparing Figure 6B and Figure 7B; P < 0.03 for all three mutant comparisons; P = 0.06 when comparing wild-type and the unc-16 single mutant). This latter result could be explained by the increased concentrations of dendritic lysosomes in the double mutants, which would tend to bias toward dendrite-to-soma entry events.
Figure 7.
Regulation of dendritic lysosome transport by CDK-5, SAD-1, and SYD-2 in an unc-16(−) background. (A) Representative kymographs of CTNS-1-RFP-tagged lysosome movements in the DB7 dendrite of animals with the indicated genotypes. CTNS-1-RFP is expressed from the integrated transgene ceIs56. See also movies in File S6, File S7, File S8, and File S9. (B–G) Graphs plotting various indicated parameters extracted from the kymographs and time-course analyses. Error bars in B are 95% confidence intervals derived from Fisher’s exact test. All other error bars are standard errors of the means. Numbers at the base of the bars in D are the number of lysosomes analyzed. *P < 0.05, **P < 0.01, and ***P < 0.0001. Black asterisks compare the marked bar to the wild-type value. Red asterisks compare the indicated two bars in a group. Unmarked bars are not significantly different from unc-16(ce483) or the other group member. Some statistical comparisons to the corresponding unc-16(+) strains (Figure 6) are stated in the text. Total recorded minutes used for exit/entry analyses (in order of strains as shown): 264, 214, 264, and 234. Total recorded minutes used for movement analyses (in order of strains as shown): 244, 128, 263, and 223.
In unc-16 single mutants the rate at which lysosomes exited the cell soma to the dendrite was ∼20% of the wild-type rate (P = 0.01), although the rate at which lysosomes entered the cell soma from the dendrite was not significantly different from wild type. This is consistent with previous findings that lysosomes tend to move toward microtubule plus ends in unc-16 single mutants and thus accumulate in axons (Edwards et al. 2013). In unc-16cdk-5 double mutants, both the entry and exit rates were strongly and significantly increased compared to unc-16 single mutants such that they were not significantly different from the wild-type entry and exit rates (Figure 7B; compare to wild type in Figure 6B). However, the entry and exit rates of unc-16; sad-1 and unc-16; syd-2 double mutants were not significantly different from unc-16 single mutants (Figure 7B).
Upon entering the dendrite, lysosomes in unc-16 single mutants spent a significantly higher percentage of their time in a paused state compared to wild type (95 vs. 42% for wild type). In unc-16cdk-5 doubles, lysosomes spent significantly less of their time in a paused state (80%) compared to unc-16 single mutants. However, lysosomes in both unc-16; sad-1 and unc-16; syd-2 double mutants were not significantly different from unc-16 single mutants in percentage of time spent paused.
Analyzing the directionality of lysosome movements within the dendrite revealed what appeared to be a lower percentage of movements in the anterograde (minus-end) direction in unc-16 single mutants compared to wild type (38 vs. 53%), but this was not quite statistically significant (comparing Figure 6E and Figure 7E). None of the double mutants significantly differed from the unc-16 single mutant in this parameter.
The high fraction of immobile dendritic puncta in the unc-16(−) background hindered our ability to acquire high numbers of moving puncta for velocity and run-length analyses; however, we were able to collect enough data to see significant and potentially insightful differences. When not in a paused state, lysosomes in unc-16 single mutants had mean velocities of 0.28 μm/sec (in either direction), which is nearly identical to the mean retrograde velocity of wild type (compare Figure 6F and Figure 7F). However, unlike wild type, in which anterogradely moving lysosomes had mean velocities ∼60% faster than retrogradely moving lysosomes (Figure 6F), there was no significant difference in the mean retrograde and anterograde velocities of dendritic lysosomes in unc-16 single mutants (Figure 7F). Consistent with having less resistance for anterograde movements and suppressing the unc-16 null phenotype, all three double mutants had mean anterograde (minus-end-directed) velocities that were 43–96% higher than unc-16 single mutants. Although this pattern was not quite statistically significant for the unc-16; sad-1 double (P = 0.066), it was highly significant for the unc-16cdk-5 and unc-16; syd-2 doubles, and lysosomes in all three doubles showed the wild-type pattern of moving significantly faster in the anterograde direction than in the retrograde direction. This phenotype was quite strong in the unc-16; syd-2 double. This was the only one of the three doubles that had a mean retrograde velocity that was significantly slower than the unc-16 single mutant, and its mean anterograde velocity was 275% higher than its mean retrograde velocity (Figure 7F).
Despite the fact that anterograde and retrograde velocities were not significantly different in unc-16 single mutants, retrograde (plus-end-directed) run lengths in these dendrites were almost three times longer than anterograde run lengths in unc-16 single mutants (Figure 7G). This is consistent with previous findings that lysosomes tend to move toward microtubule plus ends in unc-16 single mutants and thus accumulate in axons (Edwards et al. 2013), where microtubules are oriented with their plus ends out (Goodwin et al. 2012; Yan et al. 2013). Consistent with having less resistance for anterograde movements and suppressing the unc-16 null phenotype, two of the three double mutants (unc-16cdk-5 and unc-16; syd-2) had anterograde run lengths that were significantly longer than unc-16 single mutants, and unlike unc-16 single mutants, all three double mutants showed no significant difference between anterograde and retrograde run lengths when compared in the same strain (Figure 7G).
In summary, several of the lysosome active transport phenotypes in the double mutants would be expected to promote dendritic lysosome accumulation. In the unc-16cdk-5 double, the strongly increased rate at which lysosomes flux in and out of the soma (compared to unc-16 single mutants), combined with spending 80% of their time in a paused state while in the dendrite, would promote dendritic trapping and accumulation of lysosomes, as would the increased anterograde (minus-end-directed) velocity and run lengths in the same strain. Similarly, although lysosomes in the unc-16; sad-1 and unc-16; syd-2 doubles do not differ significantly in their soma-dendrite flux rates compared to the unc-16 single, they spend >95% of their time in a paused state upon entering the dendrite. In addition, like the unc-16cdk-5 double, their anterograde (minus-end-directed) movements within the dendrite are faster and longer than the unc-16 single mutant (strongest in the unc-16; syd-2 double).
Synapse assembly proteins also regulate the transport of early endosomes in neurons
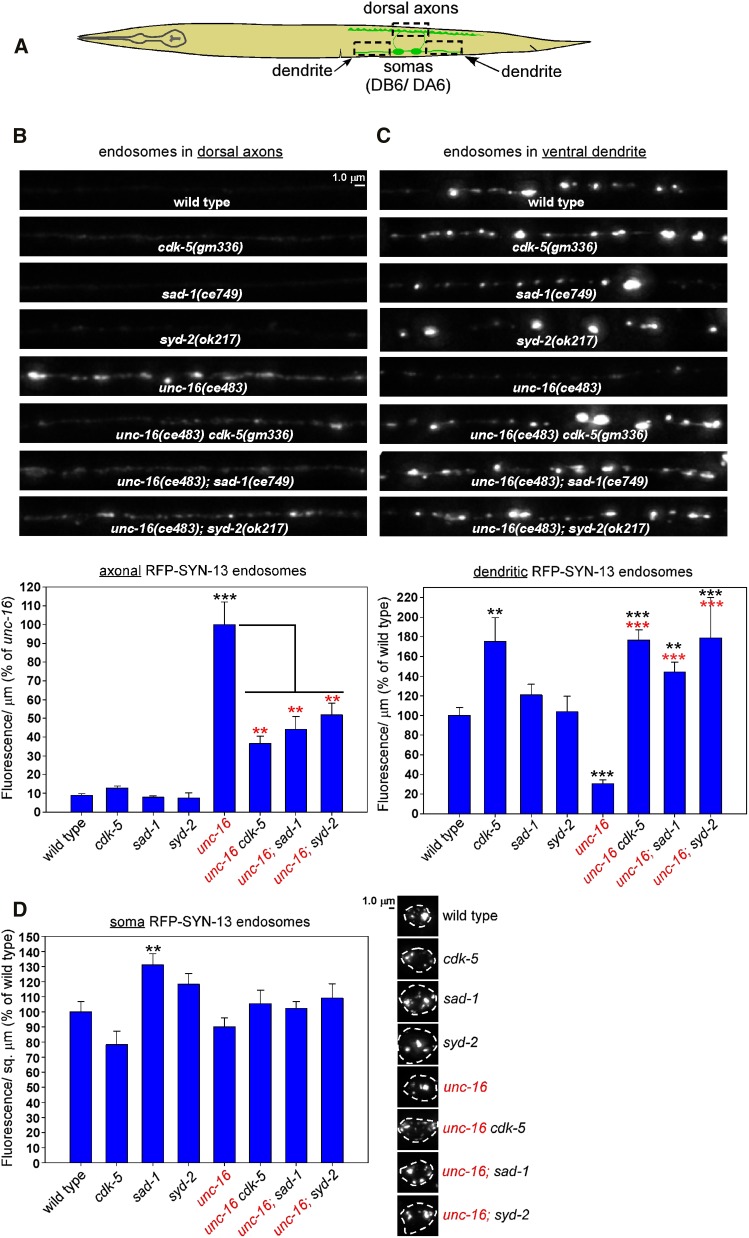
In a prior study we found that, in addition to lysosomes, unc-16 null mutants also accumulate similarly high levels of early endosomes in their axons (Edwards et al. 2013). To determine the extent to which CDK-5, SAD-1, and SYD-2 regulate early endosome transport in neurons, we crossed the ceIs259 genomically integrated transgene into cdk-5, sad-1, and syd-2 null mutants in unc-16(+) and unc-16(−) backgrounds. ceIs259 expresses RFP-SYN-13 at a low level in the same motor neurons as the CTNS-1-RFP marker that we used for the lysosome studies. SYN-13 localizes to early/recycling endosomes (Prekeris et al. 1998; Chun et al. 2008) and was previously shown to precisely colocalize with YFP-RAB-5, another early endosome marker (Edwards et al. 2013). Because, unlike lysosomes, the endosomal puncta were often not well separated from each other, we quantified total integrated fluorescence/μm of axon or dendrite length in DA6/DB6 motor neurons and did not additionally quantify punctal density (Figure 8A shows the regions imaged).
Figure 8.
CDK-5, SAD-1, and SYD-2 regulate the distribution of endosomes in unc-16 mutant neurons. (A) Drawing illustrates the location and anatomy of the cholinergic motor neurons imaged in this figure. Dashed box outlines regions imaged (both dendrite regions were collected and quantified together). (B–D) Representative, identically scaled images and quantification of RFP-SYN-13 endosomes expressed from the integrated transgene array ceIs259 in the indicated genotypes in dorsal axons (B), ventral dendrites (C), or cell somas (D). Data are means and SEMs from 13–14 animals per region. **P < 0.01 and ***P < 0.0001. Black asterisks compare the marked bar with the wild-type value. Red asterisks compare the marked bar with the unc-16 single-mutant value. Unmarked bars do not differ significantly from wild type.
In axons in an unc-16(+) background, wild type had very low, barely detectable levels of the early endosomes, but this level increased ∼10-fold in an unc-16(−) background (Figure 8B). Thus, UNC-16 normally inhibits the accumulation of early endosomes in axons. None of the three single mutants differed significantly from wild type in their axonal early endosome levels; however, in an unc-16(−) background, all three double mutants had significantly reduced levels of axonal early endosomes relative to unc-16 single mutants (38–50% of unc-16; Figure 8B), which was similar to, or slightly greater than, the total fluorescence μm reductions that we observed for lysosomes in the same genetic backgrounds (Figure 2C). Thus, UNC-16’s activity in inhibiting the axonal accumulation of early endosomes is 50–62% dependent on the individual activities of CDK-5, SAD-1, and SYD-2.
In dendrites in an unc-16(+) background, wild type had a relatively high endosomal density, in contrast to the relatively low levels of lysosomes in wild-type motor neuron dendrites (compare representative images in Figure 3B and Figure 8C). Unlike lysosomes, which showed a two-fold increase in unc-16 mutant dendrites, the early endosome level in unc-16 single-mutant dendrites significantly decreased to ∼30% of wild type, consistent with the heavy endosome accumulation in unc-16 mutant axons. Thus, UNC-16 normally promotes the accumulation of early endosomes in dendrites. In cdk-5 single mutants, dendritic early endosome levels were significantly increased by ∼1.75-fold over wild type (similar to the fold-increase of lysosomes in the same mutant as shown in Figure 3B), but this level was not further increased in an unc-16(−) background (Figure 8C). In sad-1 and syd-2 single mutants, dendritic early endosome levels were not significantly different from wild type. However, even though unc-16 single mutants had low early endosomes levels that were just 30% of wild type, the unc-16; sad-1 and unc-16; syd-2 doubles were either not significantly different (unc-16; sad-1) or ∼1.8-fold higher (unc-16; syd-2; P = 0.001) than the corresponding single mutants (Figure 8C). Thus, in dendrites, the cdk-5, sad-1, and syd-2 null mutant early endosome distribution phenotype (high, similar to wild type) was completely epistatic to unc-16’s early endosome distribution phenotype (low/∼30% of wild type). This suggests that UNC-16’s activity in promoting the dendritic accumulation of early endosomes is completely dependent on the individual activities of CDK-5, SAD-1, and SYD-2.
In cell somas, none of the single- or double-mutant combinations differed significantly from wild type in their early endosome levels, with the exception of the sad-1 single mutant, which had slightly (but significantly) higher levels of the marker (Figure 8D). Thus, any differences in the distribution of this marker in axons and dendrites are unlikely to be explained by changes in transgene expression.
In summary, these results suggest that UNC-16 has a strong role in inhibiting the trafficking of early endosomes in the direction of microtubule plus ends (the anterograde direction in axons) and/or in promoting movement toward microtubule minus ends (the anterograde direction in dendrites). In axonal trafficking, for the fluorescence/μm parameter of the early endosome marker, UNC-16’s activity is ∼50–60% dependent on the individual activities of CDK-5, SAD-1, and SYD-2, whereas in dendritic trafficking, UNC-16’s activity in promoting the dendritic localization of early endosomes appears completely dependent on the CDK-5/SAD-1/SYD-2 system.
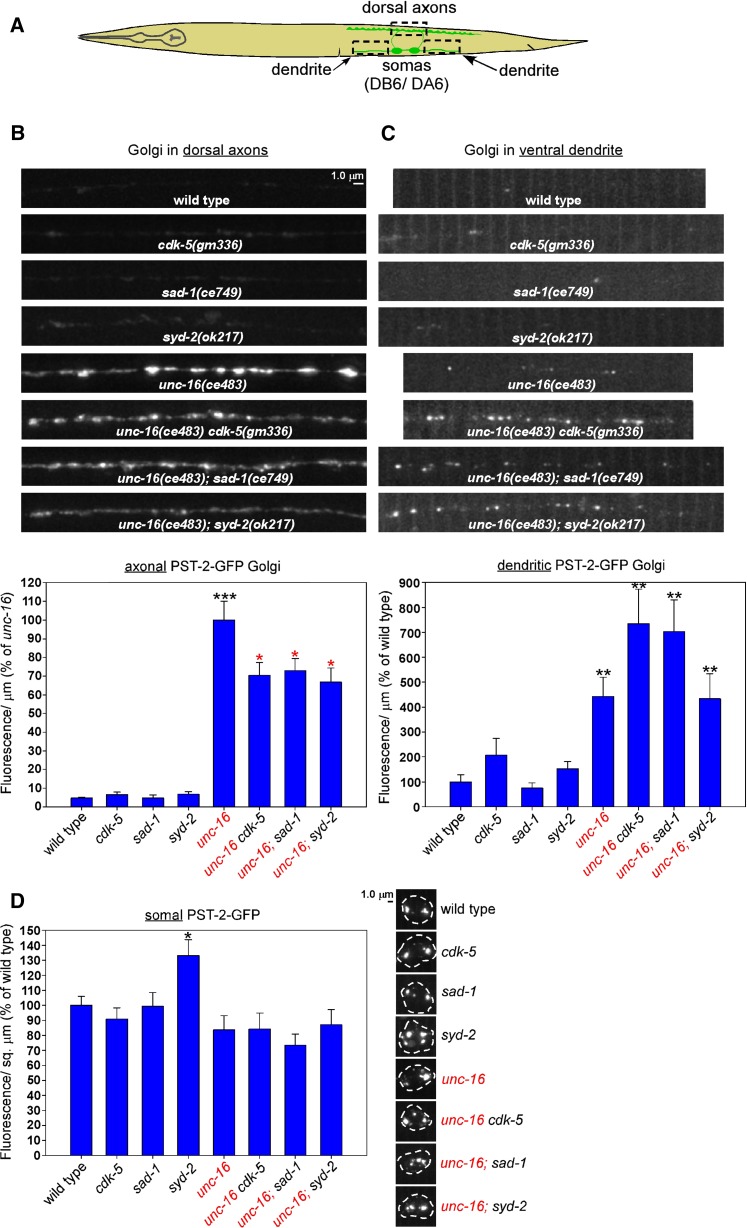
Synapse assembly proteins are less important for Golgi transport in unc-16 mutant neurons
In a prior study we found that, in addition to lysosomes and early endosomes, unc-16 null mutants also accumulate similarly high levels of Golgi in their axons (Edwards et al. 2013). To determine the extent to which CDK-5, SAD-1, and SYD-2 regulate Golgi transport in neurons, we crossed the ceIs267 genomically integrated transgene into cdk-5, sad-1, and syd-2 null mutants in unc-16(+) and unc-16(−) backgrounds. ceIs267 expresses PST-2-GFP at a low level in the same motor neurons as the other markers that we have used in this study (Figure 9A). PST-2 is a C. elegans ortholog of a Golgi-resident 3′-phosphoadenosine 5′-phosphosulfate transporter (Dejima et al. 2010). A previous study (Edwards et al. 2013) showed that PST-2-GFP colocalizes with AMAN-2 (α-Mannosidase II) to Golgi mini-stacks (the form that Golgi takes in invertebrates).
Figure 9.
CDK-5, SAD-1, and SYD-2 regulate the distribution of Golgi in unc-16 mutant neurons. (A) Drawing illustrates location and anatomy of the cholinergic motor neurons imaged in this figure. Dashed box outlines regions imaged (both dendrite regions were collected and quantified together). (B–D) Representative, identically scaled images and quantification of RFP-SYN-13 endosomes expressed from the integrated transgene array ceIs259 in the indicated genotypes in dorsal axons (B), ventral dendrites (C), or cell somas (D). Data are means and SEMs from 13–14 animals per region. *P < 0.05, **P < 0.01, and ***P < 0.0001. Black asterisks compare the marked bar with the wild-type value. Red asterisks compare the marked bar with the unc-16 single-mutant value. Unmarked bars do not differ significantly from their control bar (which is either wild type or the unc-16 single mutant).
The axons of wild-type worms had very low (barely detectable) levels of the PST-2-GFP marker, but this level increased ∼20-fold in an unc-16(−) background (Figure 9B). None of the three single mutants differed significantly from wild type in their axonal PST-2-GFP levels. In an unc-16(−) background, however, all three double mutants had significantly reduced levels of axonal PST-2-GFP relative to unc-16 single mutants, but the reductions amounted to 67–73% of the unc-16 single mutant (Figure 9B), which are less significant reductions than the lysosome and early endosome reductions.
The dendrites of wild-type worms had low levels of Golgi, but this level increased significantly by ∼4.5-fold in the unc-16 single mutant (Figure 9C). None of the three single mutants (cdk-5, sad-1, or syd-2) differed significantly from wild type in their dendritic Golgi levels; and, similarly, none of the three double mutants with unc-16 differed significantly from unc-16 single mutants in their dendritic Golgi levels, although the unc-16cdk-5 and unc-16; sad-1 doubles approached statistical significance for having levels that were ∼two-fold higher than unc-16 single mutants (Figure 9C; P = 0.08 and 0.054, respectively).
In cell somas, none of the single- or double-mutant combinations differed significantly from the control strains in their PST-2-GFP fluorescence/μm2, with the exception of the syd-2 single mutant, which had slightly (but significantly) higher levels of the marker (Figure 9D). Thus, the decreased levels of this marker in the double-mutant axons are unlikely to be explained by changes in transgene expression.
In summary, these results confirm previous findings that UNC-16 has a strong role in inhibiting the trafficking of Golgi to axons, but these new data show that this activity is only 27–33% dependent on the individual activities of CDK-5, SAD-1, or SYD-2, in contrast to the heavier dependence of lysosome and early endosome trafficking on these three proteins. To a lesser but significant extent, UNC-16 also inhibits the trafficking of Golgi to dendrites by a mechanism that does not require the individual activities of CDK-5, SAD-1, or SYD-2, nor does elimination of any one of these proteins in combination with UNC-16 significantly enhance the trafficking of Golgi to dendrites, in strong contrast with dendritic lysosome trafficking.
Discussion
In this study we used an unbiased forward genetic screen in C. elegans to discover a previously unknown system through which UNC-16 acts to regulate the active transport of lysosomes, early endosomes, and Golgi in neurons. The screen identified three conserved proteins as key regulators in this system: CDK-5, SAD-1, and SYD-2. Because the genetic results showed that the three proteins are all part of the same organelle transport regulatory system, we named it the CSS system based on the first letter of each founder protein. Further genetic analysis revealed important roles for the conserved proteins SYD-1 and STRD-1 in the CSS system. None of these proteins had previously been implicated in inhibiting the plus-end-directed transport of cell soma organelles to axons. Instead, previous studies have suggested that they that have highly specialized roles in regulating synapse assembly (SYD-1, SYD-2, and SAD-1) and/or synaptic vesicle transport (CDK-5 and SYD-2). In an accompanying article in this issue, we provide evidence that SAD-1 and SYD-1 also regulate synaptic vesicle transport (Edwards et al. 2015). In the current study, we have combined the genetic strengths of C. elegans with quantitative imaging techniques and time-lapse imaging of lysosomes in motor neurons to gain insights into the roles of CDK-5, SAD-1, and SYD-2 in organelle transport and the extent to which their functions overlap or intersect.
The CSS system promotes axonal accumulation and inhibits dendritic accumulation of lysosomes in unc-16 null mutants
The results showed that the heavy accumulation of lysosomal puncta in unc-16 mutant axons is 74–97% dependent on the individual activities of the CSS proteins, with CDK-5 having a significantly more important role than SAD-1 or SYD-2. Null mutations in all three of the genes reduced the total fluorescence/μm of the lysosomal marker by similar amounts, to ∼50% of the unc-16 single mutant. The total fluorescence/μm includes both the puncta and a diffuse nonpunctal fluorescence from the lysosomal marker, the latter of which is also present in the unc-16 single mutant. The source of the membrane(s) from which the diffuse fluorescence originates is unclear, but apparently its transport is regulated differently from the puncta in axons. In dendrites there was no such difference.
Because none of the individual CSS mutations completely suppressed the axonal accumulation of lysosomal puncta or total fluorescence of the lysosomal marker, we first hypothesized that the three proteins may be part of two or three different systems for regulating lysosomal transport in neurons. However, in our analysis of all possible combinations of double and triple mutants in unc-16(+) and unc-16(−) backgrounds we did not see significant additive effects of the mutations in either axons or dendrites, suggesting that all three proteins function within the same system. This does not mean that the proteins interact to mediate the same function nor that they operate in a single linear pathway. The genetic results are also consistent with the CSS proteins mediating two or more parallel but interdependent functions that ultimately converge to promote axonal accumulation and inhibit dendritic accumulation of lysosomes in unc-16 null mutants. Consistent with this latter possibility, our time-lapse studies identified active transport phenotypes in both unc-16(+) and unc-16(−) backgrounds where cdk-5 mutants differed from sad-1 and syd-2 mutants, although most active transport phenotypes were shared between the three mutants.
Other players
Our genetic screen was clearly not saturating: even though we found five sad-1 alleles, we only isolated one allele each of cdk-5 and syd-2. This suggests that further rounds of screening will identify other important players within this system. However, for this study, we began by testing null mutants that eliminate the functions of proteins known to interact with one or more of the CSS proteins. This complementary reverse genetics approach was fruitful, resulting in the identification of SYD-1 and STRD-1 (STRADα) as important determinants of lysosome distribution in unc-16 mutant neurons. In addition, our results suggest that PCT-1 has a minor role in this system in both axons and dendrites, although CDK-5 appears mostly able to substitute for lack of PCT-1 in this context. NAB-1 (Neurabin) also appears to have a minor role, although only in unc-16 mutant dendrites.
Synapse assembly proteins as general regulators of active transport in neurons
Several of the CSS proteins and “other players” noted above have been found to be important for synapse assembly and stability. For example SYD-2, SYD-1, SAD-1, and NAB-1 regulate synapse assembly and positioning of active zone components in C. elegans HSN neurons (Dai et al. 2006; Patel et al. 2006; Chia et al. 2012; Li et al. 2014). High-pressure freezing electron microscopy followed by tomographic reconstructions of active zones show that SYD-2 has roles in tethering/clustering synaptic vesicles near the active zone (Stigloher et al. 2011; Kittelmann et al. 2013) and in determining the size of the dense projection to which the synaptic vesicles are tethered (Kittelmann et al. 2013). In syd-2 mutants, the rate at which synaptic vesicles dissociate from mature synapses is increased (Wu et al. 2013), and synaptic vesicles or clusters of vesicles accumulate in distal regions of the axon that are normally devoid of synapses and synaptic vesicles (Li et al. 2014; Edwards et al. 2015).
The phenotype of accumulation of synaptic vesicles in the distal asynaptic region of motor neurons, which could be considered diagnostic of reduced synaptic vesicle clustering activity at synapses, is also shared by syd-1 and sad-1 mutants (Li et al. 2014; Edwards et al. 2015), and prior studies also demonstrated defects in synaptic vesicle clustering at other synapses in C. elegans sad-1 mutants (Crump et al. 2001; Patel et al. 2006). Other studies have proposed that C. elegans sad-1 and syd-1 mutants also have defects in axon identity/fate, referred to as a “polarity defect,” based largely on the accumulation of synaptic vesicles in dendrites (Hallam et al. 2002; Hung et al. 2007; Kim et al. 2008, 2010). Indeed, SAD-1’s orthologs do have roles in specifying axon identity in mammalian forebrain neurons (Kishi et al. 2005), where SAD-A/B act downstream of the LKB1 kinase (Barnes et al. 2007). However, neurons outside of the cortex, including peripheral motor neurons and many other types of central neurons, do not require the SAD kinases or LKB1 for axon formation/identity (Lilley et al. 2013). In the current study, we show that sad-1 mutants have wild-type locomotion rates, a result that seems inconsistent with a motor neuron axon/dendrite identity defect.
In the accompanying article in this issue, we provide evidence that the accumulation of synaptic vesicles in sad-1, syd-1, and syd-2 motor neuron dendrites results from a defect in the regulation of synaptic vesicle transport, as opposed to an axon/dendrite identity defect (Edwards et al. 2015). Indeed, the regulation of synaptic vesicle and/or dense core vesicle transport appears to be another common theme that ties together several of CSS system proteins. Prior studies have also demonstrated a role for SYD-2 in synaptic vesicle and dense core vesicle transport in Drosophila (Miller et al. 2005) and C. elegans (Wagner et al. 2009; Goodwin and Juo 2013; Zheng et al. 2014), as well as for CDK-5 and PCT-1 in synaptic vesicle and dense core vesicle transport in C. elegans motor neurons (Ou et al. 2010; Goodwin et al. 2012).
Although Cdk5 has been shown to have a role in the active transport of acidic organelles in cultured cells (Pandey and Smith 2011), the current study is the first to show that several synapse assembly proteins (SYD-2, SYD-1, and SAD-1) also have general roles in regulating cell soma organelle transport in neurons. It is unclear whether there is a common mechanism that ties together the synapse assemble and active transport functions of SYD-1, SYD-2, and SAD-1 or whether they are simply hyper-adaptable to different functions. For example, SYD-2 belongs to a class of “intrinsically unstructured proteins” that can bind several partners in a structurally adaptive process (Wagner et al. 2009).
It is worth noting that SAD-1 and its mammalian orthologs also regulate axon termination (Crump et al. 2001) and terminal branching (Lilley et al. 2013) in certain classes of sensory neurons, and it is possible that this latter function might be related to the cell soma organelle transport function of SAD-1, since it would be reasonable to expect that organelle delivery, or lack thereof, could affect the length and branching of the axon. UNC-16 (JIP3) is also known to regulate axon elongation and branching as well as axonal transport and regeneration after injury (Cavalli et al. 2005; Bilimoria et al. 2010; Suzuki et al. 2010; Sun et al. 2013; Nix et al. 2014; Watt et al. 2015). However, it is unknown whether these axon growth functions of SAD-1 and UNC-16 are related and whether other CSS system proteins also participate.
Dynein redistributes lysosomes in unc-16 mutants when the function of the CSS system is blocked
We have not yet identified the plus-end-directed motor that carries organelles to axons in unc-16 null mutants, although we have ruled out UNC-116 (Kinesin-1) (Edwards et al. 2013) and UNC-104 (KIF1A; this study). However, our data show that the loss of lysosomes from the axons of unc-16cdk-5 and unc-16; syd-2 double mutants, as well as the accumulation of lysosomes in the dendrites of the same mutants, is dependent on NUD-2 (NUDEL/Nde1), a regulator of the minus-end motor dynein (Niethammer et al. 2000; Sasaki et al. 2000; Pandey and Smith 2011). In axons, crossing in the nud-2 mutation completely suppressed the unc-16; syd-2 phenotype back to the state of an unc-16 single mutant. In dendrites, crossing in the nud-2 mutation completely suppressed the unc-16; cdk-5 phenotype (heavy accumulation of lysosomes) back to the state of the unc-16 single mutant. In the other two cases (unc-16; cdk-5 axons and unc-16; syd-2 dendrites), the suppression was nearly complete (60–78% of the unc-16 single mutant). Our data are consistent with previous results showing that the accumulation of synaptic vesicles and dense core vesicles in cdk-5 mutant dendrites is completely dependent on dynein and/or NUD-2 (Ou et al. 2010; Goodwin et al. 2012), and the accumulation of dense core vesicles in syd-2 mutant dendrites is also completely dependent on dynein (Goodwin and Juo 2013). These epistatic or near-epistatic results seem consistent with a model in which the CSS system normally functions upstream of dynein and inhibits dynein (Figure 10). Thus, in an unc-16 null single mutant, dynein would be inhibited by the CSS system, and the unidentified plus-end motor would carry lysosomes to axons. Genetically blocking the activity of the CSS system in an unc-16 null mutant would then cause dynein to become overactive and bias movements toward the minus ends of microtubules, which would deplete organelles from axons and cause them to accumulate in dendrites (Figure 10). However, we are cautious to not overinterpret the epistatic suppression by the nud-2 mutation since this result is equally consistent with a model in which the normal function of the CSS system is to positively regulate a plus-end motor or to regulate both motors. For example, a mutation in the plus-end motor UNC-104 (KIF1A) causes a 50% depletion of synaptic vesicles in axons and a corresponding 15-fold increase in dendritic synaptic vesicles, and yet crossing in the nud-2 mutation epistatically redistributes synaptic vesicles to the wild-type state as well as restores wild-type locomotion (Edwards et al. 2015). By analogy, blocking the CSS system could decrease activity of the plus-end motor, thus allowing dynein-mediated minus-end movements to dominate and giving the appearance of hyperactive dynein, when in fact its activity could be unchanged. Indeed, a prior study used a GFP-tagged dynein subunit and time-lapse microscopy to look for hyperactive dynein in cdk-5 mutant motor neuron dendrites in C. elegans and found no such evidence (Goodwin et al. 2012).
Figure 10.
Graphic summary of genetic results. Shown is one possible model consistent with the genetic results. In wild-type animals, UNC-16 directly or indirectly inhibits the CSS system (vertical bars indicate inhibition). When CSS system activity is low, minus- and plus-end motor activity is properly balanced and cell soma organelles (lysosomes and endosomes; tan and purple shapes) remain in the soma and/or enter the dendrite at optimal rates. When CSS system activity is high, plus-end motor activity dominates due to inhibition of dynein, and organelles accumulate in axons. In other equally plausible models, the CSS system could positively regulate a plus-end motor or regulate both motors, and the effect on organelle transport would be the same.
A previous study found that UNC-16’s N terminus can interact with the dynein light intermediate chain DLI-1 in the yeast two-hybrid system. Full-length UNC-16 did not interact with DLI-1, however, and UNC-16 and DLI-1 could not be co-immunoprecipitated when co-expressed in COS-7 cells (Arimoto et al. 2011). Still, a study in vertebrates found that the dynactin subunits p50 and p150 could be co-immunoprecipitated with UNC-16 (JIP3) from the sciatic nerve (Cavalli et al. 2005). A later study in zebrafish observed cotransport of lysosomes and UNC-16 and, based on a decrease in the proportion of dynein-light intermediate chain-positive lysosomes moving in the minus-end direction in Jip3 mutants, concluded that UNC-16 functions as an important adaptor that links lysosomes to the dynein motor through the dynein light intermediate chain (Drerup and Nechiporuk 2013). However, in that study, 63–85% of lysosomes (depending on the age of the animal) moving in the minus-end direction showed no cotransport with UNC-16. Thus, if UNC-16 also functions as an adaptor, it appears to not be required for linking most lysosomes to dynein. Our results do not exclude direct interactions between UNC-16 and dynein or dynactin. Such interactions could serve to localize UNC-16 to its sites of action. However, rather than viewing UNC-16 solely as an adaptor protein, our data seem most consistent with a model in which one of UNC-16’s functions is direct or indirect regulation of the CSS system proteins, which in turn regulate the minus- and/or plus-end-directed motors (Figure 10).
Active transport defects in CSS system mutants promote the dendritic accumulation of lysosomes
Given that our static imaging data showed that the lysosome levels in the dendrites of sad-1 and syd-2 single mutants do not differ significantly from wild type, and that the levels in cdk-5 single mutants were only ∼2.3-fold higher than wild type, we expected that time-lapse imaging of lysosomes in the dendrites of these mutants might not reveal any defects in active transport in an unc-16(+) background; however, this was not the case. All three mutants showed significant differences from wild type in at least two parameters for the active transport of lysosomes in dendrites. Our finding that dendritic lysosomes in all three mutants spend a higher percentage of their time in a paused state compared to wild type suggests that the CSS proteins are important for maintaining the processivity of lysosome movements. Dendritic lysosomes in all three mutants also had significantly slower minus-end-directed velocities when not in a paused state, although sad-1 and syd-2 mutants also had slower plus-end-directed velocities that resulted in shorter plus-end run lengths. Consistent with these data, prior studies in C. elegans reported that both cdk-5 and syd-2 null mutants have increased numbers of stationary dense core vesicles in their dendrites, and, additionally, dendritic dense core vesicles in syd-2 null mutants have slower minus- and plus-end velocities (Goodwin et al. 2012; Goodwin and Juo 2013). The fact that we see significant effects of CSS mutations on dendritic lysosome transport in an unc-16(+) background shows that CSS proteins regulate the active transport of dendritic lysosomes even in wild type, and not just in an unc-16(−) background.
Our time-lapse analysis of dendritic lysosome movements in unc-16 single mutants revealed several transport phenotypes that explain the bias toward plus-end transport that ultimately results in the accumulation of lysosomes in unc-16 mutant axons. These include a strongly decreased rate of lysosomes exiting the soma to the dendrite (∼20% of wild type), a possibly lower percentage of movements in the minus-end direction (not quite significant), a slower minus-end velocity than wild type (with no difference in plus-end velocity), and plus-end run lengths that were almost three times longer than minus-end run lengths. We also saw a twofold increase in dendritic lysosomes in the unc-16 single mutant. This minus-end accumulation seems counterintuitive, given the heavy plus-end accumulation in axons; however, it could result from the trapping of the few lysosomes that enter the dendrite. Trapping could result from the high percentage of time that dendritic lysosomes spend in a paused state in the unc-16 single mutant (∼95%).
Given the heavy accumulation of lysosomes in the dendrites of CSS mutants in an unc-16(−) background, we expected to see an increased rate of soma-to-dendrite exit events in all of the double mutants relative to the unc-16 single mutant. That prediction, however, proved to be true only for the unc-16cdk-5 double, which had a soma-to-dendrite exit rate not significantly different from wild type (i.e., about five times higher than the unc-16 single mutant). Since soma-to-dendrite exiting is a minus-end-directed activity, this result is consistent with the strong nud-2 suppression of dendritic accumulation of lysosomes in the unc-16cdk-5; nud-2 triple. However, this strongly increased exit rate is balanced by a corresponding increase in the soma-to-dendrite entry rate to a level not significantly different from either its exit rate or that of wild type. Thus, looking at entry and exit rates alone, there is no reason to predict that lysosomes should accumulate in unc-16cdk-5 mutant dendrites since they should be no different from wild type. Instead, the accumulation does occur, and the data suggest that it results from trapping of lysosomes in the dendrite due to the high percentage of time they spend in a paused state once entering the dendrite (∼80 vs. ∼40% for wild type), combined with increased minus-end velocities and longer minus-end run lengths compared to unc-16 single mutants.
In contrast to the unc-16cdk-5 double, the unc-16; sad-1 and unc-16; syd-2 doubles were not significantly different from the unc-16 single in their soma-to-dendrite flux rates in either direction. Although they spent a higher percentage of their time in a paused state than the unc-16cdk-5 double (95 vs. 80%), this was not different from unc-16 single mutants. Thus, based on the soma-dendrite flux and pause parameters, there is no reason to predict that lysosomes should accumulate in the dendrites of these double mutants at levels any higher than the unc-16 single mutant (approximately two-fold). However, consistent with the strong nud-2 suppression of dendritic accumulation of lysosomes in the unc-16; syd-2; nud-2 triple, both double mutants (unc-16; sad-1 and unc-16; syd-2) had higher minus-end velocities and longer minus-end run lengths, similar to unc-16cdk-5 double mutants. We thus hypothesize that these minus-end-directed biases, combined with the high percentage of time spent in a paused state, leads to trapping and accumulation of lysosomes in the dendrites of unc-16; sad-1 and unc-16; syd-2 double mutants.
CSS system also regulates early endosome distribution in unc-16 mutant neurons but is less important for Golgi
A previous study in zebrafish proposed that UNC-16’s organelle transport function is specifically limited to lysosomes, because late endosomes and autophagosomes did not appear to accumulate in zebrafish axons lacking UNC-16 (JIP3) (Drerup and Nechiporuk 2013). However, that study did not test early endosomes or Golgi membranes. In addition to lysosomes, C. elegans unc-16 mutants also accumulate high levels of early endosomes and Golgi in their axons (Edwards et al. 2013). In this study, we found that the CSS system mediates the axonal accumulation of early endosomes in unc-16 null mutants to about the same extent that it mediates the axonal accumulation of lysosomes in unc-16 null mutants (∼50–60%, when using fluorescence/μm as the parameter). However, the 20-fold accumulation of Golgi in unc-16 mutant axons was significantly less dependent on the CSS system (∼30%). This suggests that the active transport of Golgi ministacks shares some, but not all, of the regulatory features of lysosomes and early endosomes.
Our analysis of early endosome and Golgi distributions in dendrites further emphasized this point and revealed additional regulatory differences between lysosomes and early endosomes. In contrast to lysosomes, early endosomes were relatively abundant in wild-type motor neuron dendrites, but were decreased four-fold in unc-16 mutant dendrites, which is consistent with increased plus-end movement of early endosomes into axons. This suggests that UNC-16 normally promotes the minus-end transport of early endosomes to dendrites, just as we hypothesize it does for lysosomes (Figure 10). However, lysosomes are increased approximately two-fold over wild type in unc-16 mutant dendrites. Based on our time-lapse data, we hypothesize that this increase results from rare lysosomes that enter the dendrite and become trapped in a paused state. Although we have not yet performed time-lapse imaging of early endosomes in unc-16 null mutants, we hypothesize that they may be less susceptible than lysosomes to pausing and thus not as vulnerable to becoming trapped in the dendrite.
Just as with lysosomes, cdk-5 single mutants were the only mutants that showed significantly elevated levels of early endosomes in dendrites in an unc-16(+) background. In dendrites, the cdk-5, sad-1, and syd-2 null mutant early endosome distribution phenotype (high, similar to wild type) was completely epistatic to unc-16’s early endosome distribution phenotype (low/∼30% of wild type). Thus, in dendritic trafficking, UNC-16’s activity in promoting the dendritic localization of early endosomes appears completely dependent on the CSS proteins, suggesting that UNC-16 directly or indirectly inhibits the CSS system to produce an appropriate ratio of plus- to minus-end motor activity that maintains early endosomes in the cell soma and/or the dendrite at optimal levels (Figure 10).
Unlike early endosomes, but similar to lysosomes, Golgi membranes are present only at very low levels in wild-type dendrites. Similar to lysosomes, the dendritic Golgi marker is increased in the unc-16 single mutant (∼4.5-fold over wild type). Although we have not yet performed time-lapse imaging of Golgi in unc-16 null mutants, we hypothesize that, similar to lysosomes, this accumulation results from rare Golgi ministacks that enter the dendrite and become trapped due to a high rate of pausing in dendrites in an unc-16(−) background. However, in an unc-16(−) background, eliminating individual CSS system proteins did not significantly increase the dendritic levels of Golgi, although two of the three doubles (unc-16cdk-5 and unc-16; syd-2) approached statistical significance for having ∼2-fold higher levels than the unc-16 single mutant. This again suggests that the active transport of Golgi ministacks is less regulated by the CSS system than lysosomes and early endosomes and thus shares some, but not all, of the regulatory features of lysosome and early endosome transport.
Conclusion
Using the strengths of the genetic model C. elegans, we have discovered a new, unprecedented, and counterintuitive role for synapse assembly proteins in regulating the transport of cell soma organelles in neurons, where they act downstream of the conserved protein UNC-16, as part of the newly defined CSS system. Key factors contributing to this discovery were an unbiased forward genetic screen, genetic tools and techniques that facilitated the construction of complex strains for analysis, and the ability to perform high-resolution, quantitative imaging of organelles in defined regions of identified motor neurons in live animals. Still, much remains to be learned about how UNC-16 interacts with the CSS system and how the CSS system in turn regulates minus- and/or plus-end motors. Notably, the relevant plus-end motor in this system remains unidentified. Future genetic studies, including new forward genetic screens, additional cycles of the current screen, and reverse genetic approaches, should identify the plus-end motor, as well as positive and negative regulators of this new system. Such studies should reveal general principles governing organelle transport in neurons and could provide important insights into motor neuron diseases that are associated with axonal transport defects.
Supplementary Material
Acknowledgments
We thank Graham Wiley and Patrick Gaffney of the Oklahoma Medical Research Foundation’s Next Generation Sequencing Core facility for providing important equipment, training, and advice for the whole-genome sequencing of the mutants in this study; Bob Barstead for his development of the user-friendly “Whole Genomes” suite of applications that we used for analyzing the whole-genome mutant sequences in this study; Peter Juo for providing cdk-5(gm336); the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440), for many critical strains in this work; and Shohei Mitani for providing important strains through the Japanese National Bioresource Project. This work was supported by grants from the National Institute of General Medical Sciences of the NIH (R01GM080765 to K.G.M) and from the Oklahoma Center for the Advancement of Science and Technology (HR14-003 to K.G.M.). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Note added in proof: See Edwards et al. 2015 (pp. 91–116) in this issue for a related work.
Footnotes
Communicating editor: M. V. Sundaram
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177345/-/DC1.
Literature Cited
- Arimoto M., Koushika S. P., Choudhary B. C., Li C., Matsumoto K., et al. , 2011. The Caenorhabditis elegans JIP3 protein UNC-16 functions as an adaptor to link kinesin-1 with cytoplasmic dynein. J. Neurosci. 31: 2216–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. W., Lin S., 2011. Hooks and comets: the story of microtubule polarity orientation in the neuron. Dev. Neurobiol. 71: 403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes A. P., Lilley B. N., Pan Y. A., Plummer L. J., Powell A. W., et al. , 2007. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell 129: 549–563. [DOI] [PubMed] [Google Scholar]
- Bilimoria P. M., de la Torre-Ubieta L., Ikeuchi Y., Becker E. B., Reiner O., et al. , 2010. A JIP3-regulated GSK3beta/DCX signaling pathway restricts axon branching. J. Neurosci. 30: 16766–16776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman A. B., Kamal A., Ritchings B. W., Philp A. V., McGrail M., et al. , 2000. Kinesin-dependent axonal transport is mediated by the sunday driver (SYD) protein. Cell 103: 583–594. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of C. elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown H. M., Van Epps H. A., Goncharov A., Grant B. D., Jin Y., 2009. The JIP3 scaffold protein UNC-16 regulates RAB-5 dependent membrane trafficking at C. elegans synapses. Dev. Neurobiol. 69: 174–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton P. R., Paige J. L., 1981. Polarity of axoplasmic microtubules in the olfactory nerve of the frog. Proc. Natl. Acad. Sci. USA 78: 3269–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd D. T., Kawasaki M., Walcoff M., Hisamoto N., Matsumoto K., et al. , 2001. UNC-16, a JNK-signaling scaffold protein, regulates vesicle transport in C. elegans. Neuron 32: 787–800. [DOI] [PubMed] [Google Scholar]
- Cavalli V., Kujala P., Klumperman J., Goldstein L. S., 2005. Sunday Driver links axonal transport to damage signaling. J. Cell Biol. 168: 775–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P. H., Patel M. R., Shen K., 2012. NAB-1 instructs synapse assembly by linking adhesion molecules and F-actin to active zone proteins. Nat. Neurosci. 15: 234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun D. K., McEwen J. M., Burbea M., Kaplan J. M., 2008. UNC-108/Rab2 regulates post-endocytic trafficking in C. elegans. Mol. Biol. Cell 19: 2682–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. G., Zhen M., Jin Y., Bargmann C. I., 2001. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 29: 115–129. [DOI] [PubMed] [Google Scholar]
- Dai Y., Taru H., Deken S. L., Grill B., Ackley B., et al. , 2006. SYD-2 Liprin-alpha organizes presynaptic active zone formation through ELKS. Nat. Neurosci. 9: 1479–1487. [DOI] [PubMed] [Google Scholar]
- Dejima K., Murata D., Mizuguchi S., Nomura K. H., Izumikawa T., et al. , 2010. Two Golgi-resident 3′-phosphoadenosine 5′-phosphosulfate transporters play distinct roles in heparan sulfate modifications and embryonic and larval development in Caenorhabditis elegans. J. Biol. Chem. 285: 24717–24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos K. J., Grierson A. J., Ackerley S., Miller C. C., 2008. Role of axonal transport in neurodegenerative diseases. Annu. Rev. Neurosci. 31: 151–173. [DOI] [PubMed] [Google Scholar]
- Drerup C. M., Nechiporuk A. V., 2013. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 9: e1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Charlie N. K., Milfort M. C., Brown B. S., Gravlin C. N., et al. , 2008. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 6: e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Yu S. C., Hoover C. M., Phillips B. C., Richmond J. E., et al. , 2013. An organelle gatekeeper function for Caenorhabditis elegans UNC-16 (JIP3) at the axon initial segment. Genetics 194: 143–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Yorks R. M., Morrison L. M., Hoover C. M., Miller K. G., 2015. Synapse-assembly proteins maintain synaptic vesicle cluster stability and regulate synaptic vesicle transport in Caenorhabditis elegans. Genetics 201: 91–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kadi A. M., Soura V., Hafezparast M., 2007. Defective axonal transport in motor neuron disease. J. Neurosci. Res. 85: 2557–2566. [DOI] [PubMed] [Google Scholar]
- Fridolfsson H. N., Ly N., Meyerzon M., Starr D. A., 2010. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev. Biol. 338: 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin P. R., Juo P., 2013. The scaffolding protein SYD-2/Liprin-alpha regulates the mobility and polarized distribution of dense-core vesicles in C. elegans motor neurons. PLoS One 8: e54763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin P. R., Sasaki J. M., Juo P., 2012. Cyclin-dependent kinase 5 regulates the polarized trafficking of neuropeptide-containing dense-core vesicles in Caenorhabditis elegans motor neurons. J. Neurosci. 32: 8158–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S. J., Goncharov A., McEwen J., Baran R., Jin Y., 2002. SYD-1, a presynaptic protein with PDZ, C2 and rhoGAP-like domains, specifies axon identity in C. elegans. Nat. Neurosci. 5: 1137–1146. [DOI] [PubMed] [Google Scholar]
- Heidemann S. R., Landers J. M., Hamborg M. A., 1981. Polarity orientation of axonal microtubules. J. Cell Biol. 91: 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y., Niwa S., 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10: 682–696. [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., Tanaka Y., 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68: 610–638. [DOI] [PubMed] [Google Scholar]
- Holzbaur E. L., 2004. Motor neurons rely on motor proteins. Trends Cell Biol. 14: 233–240. [DOI] [PubMed] [Google Scholar]
- Hoover C. M., Edwards S. L., Yu S. C., Kittelmann M., Richmond J. E., et al. , 2014. A novel CaM kinase II pathway controls the location of neuropeptide release from Caenorhabditis elegans motor neurons. Genetics 196: 745–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. H., Duan S., Sun T., Wang J., Zhao L., et al. , 2011. JIP3 mediates TrkB axonal anterograde transport and enhances BDNF signaling by directly bridging TrkB with kinesin-1. J. Neurosci. 31: 10602–10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W., Hwang C., Po M. D., Zhen M., 2007. Neuronal polarity is regulated by a direct interaction between a scaffolding protein, Neurabin, and a presynaptic SAD-1 kinase in Caenorhabditis elegans. Development 134: 237–249. [DOI] [PubMed] [Google Scholar]
- Kalatzis V., Cherqui S., Antignac C., Gasnier B., 2001. Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. EMBO J. 20: 5940–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Lilley B. N., Zhang C., Shokat K. M., Sanes J. R., et al. , 2008. A chemical-genetic strategy reveals distinct temporal requirements for SAD-1 kinase in neuronal polarization and synapse formation. Neural Dev. 3: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Hung W., Narbonne P., Roy R., Zhen M., 2010. C. elegans STRADalpha and SAD cooperatively regulate neuronal polarity and synaptic organization. Development 137: 93–102. [DOI] [PubMed] [Google Scholar]
- Kishi M., Pan Y. A., Crump J. G., Sanes J. R., 2005. Mammalian SAD kinases are required for neuronal polarization. Science 307: 929–932. [DOI] [PubMed] [Google Scholar]
- Kittelmann M., Hegermann J., Goncharov A., Taru H., Ellisman M. H., et al. , 2013. Liprin-alpha/SYD-2 determines the size of dense projections in presynaptic active zones in C. elegans. J. Cell Biol. 203: 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Choudhary B. C., Metpally R., Zheng Q., Nonet M. L., et al. , 2010. The Caenorhabditis elegans Kinesin-3 motor UNC-104/KIF1A is degraded upon loss of specific binding to cargo. PLoS Genet. 6: e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tian X., Zhu M., Bulgari D., Bohme M. A., et al. , 2014. Drosophila Syd-1, liprin-alpha, and protein phosphatase 2A B′ subunit Wrd function in a linear pathway to prevent ectopic accumulation of synaptic materials in distal axons. J. Neurosci. 34: 8474–8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley B. N., Pan Y. A., Sanes J. R., 2013. SAD kinases sculpt axonal arbors of sensory neurons through long- and short-term responses to neurotrophin signals. Neuron 79: 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A. E., Moughamian A. J., Holzbaur E. L., 2014. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 84: 292–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas P. M., Yu X., Miller K. G., Zhou Z., 2008. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J. Cell Biol. 180: 357–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10(12): 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S., Julien J. P., 2013. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 14: 161–176. [DOI] [PubMed] [Google Scholar]
- Miller K. E., DeProto J., Kaufmann N., Patel B. N., Duckworth A., et al. , 2005. Direct observation demonstrates that Liprin-alpha is required for trafficking of synaptic vesicles. Curr. Biol. 15: 684–689. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Emerson M. D., Rand J. B., 1999. Goα and diacylglycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron 24: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer M., Smith D. S., Ayala R., Peng J., Ko J., et al. , 2000. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28: 697–711. [DOI] [PubMed] [Google Scholar]
- Nix P., Hammarlund M., Hauth L., Lachnit M., Jorgensen E. M., et al. , 2014. Axon regeneration genes identified by RNAi screening in C. elegans. J. Neurosci. 34: 629–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C. Y., Poon V. Y., Maeder C. I., Watanabe S., Lehrman E. K., et al. , 2010. Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141: 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey J. P., Smith D. S., 2011. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J. Neurosci. 31: 17207–17219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. R., Lehrman E. K., Poon V. Y., Crump J. G., Zhen M., et al. , 2006. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat. Neurosci. 9: 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R., Klumperman J., Chen Y. A., Scheller R. H., 1998. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143: 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N. K., Schade M. A., Miller K. G., 2005. Convergent, RIC-8 dependent Gα signaling pathways in the C. elegans synaptic signaling network. Genetics 169: 650–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S., Shionoya A., Ishida M., Gambello M. J., Yingling J., et al. , 2000. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28: 681–696. [DOI] [PubMed] [Google Scholar]
- Sato T., Ishikawa M., Mochizuki M., Ohta M., Ohkura M., et al. , 2015. JSAP1/JIP3 and JLP regulate kinesin-1-dependent axonal transport to prevent neuronal degeneration. Cell Death Differ. 22: 1260–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade M. A., Reynolds N. K., Dollins C. M., Miller K. G., 2005. Mutations that rescue the paralysis of C. elegans ric-8 (Synembryn) mutants activate the Gαs pathway and define a third major branch of the synaptic signaling network. Genetics 169: 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans, WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Stigloher C., Zhan H., Zhen M., Richmond J., Bessereau J. L., 2011. The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J. Neurosci. 31: 4388–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J., 1988. Methods, pp. 596–597 in The Nematode Caenorhabditis elegans, edited by Wood W. B. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sun F., Zhu C., Dixit R., Cavalli V., 2011. Sunday Driver/JIP3 binds kinesin heavy chain directly and enhances its motility. EMBO J. 30: 3416–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Yu N., Zhai L. K., Li N., Zhang C., et al. , 2013. c-Jun NH2-terminal kinase (JNK)-interacting protein-3 (JIP3) regulates neuronal axon elongation in a kinesin- and JNK-dependent manner. J. Biol. Chem. 288: 14531–14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Arikawa C., Kuwahara Y., Itoh K., Watanabe M., et al. , 2010. The scaffold protein JIP3 functions as a downstream effector of the small GTPase ARF6 to regulate neurite morphogenesis of cortical neurons. FEBS Lett. 584: 2801–2806. [DOI] [PubMed] [Google Scholar]
- Vale R. D., 2003. The molecular motor toolbox for intracellular transport. Cell 112: 467–480. [DOI] [PubMed] [Google Scholar]
- Wagner O. I., Esposito A., Kohler B., Chen C. W., Shen C. P., et al. , 2009. Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc. Natl. Acad. Sci. USA 106: 19605–19610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt D., Dixit R., Cavalli V., 2015. JIP3 activates kinesin-1 motility to promote axon elongation. J. Biol. Chem. 290: 15512–15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M. Y., Zhou C., Shakiryanova D., Lloyd T. E., Deitcher D. L., et al. , 2012. Neuropeptide delivery to synapses by long-range vesicle circulation and sporadic capture. Cell 148: 1029–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. E., Huo L., Maeder C. I., Feng W., Shen K., 2013. The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron 78: 994–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Chao D. L., Toba S., Koyasako K., Yasunaga T., et al. , 2013. Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. eLife 2: e00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Ahlawat S., Schaefer A., Mahoney T., Koushika S. P., et al. , 2014. The vesicle protein SAM-4 regulates the processivity of synaptic vesicle transport. PLoS Genet. 10: e1004644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and reagents are available upon request. File S1 contains Supplemental Materials and Methods and a detailed description of all of the strains, plasmids, and transgenes produced in this study.