Abstract
Organisms on islands provide a revealing window into the process of adaptation. Populations that colonize islands often evolve substantial differences in body size from their mainland relatives. Although the ecological drivers of this phenomenon have received considerable attention, its genetic basis remains poorly understood. We use house mice (subspecies: Mus musculus domesticus) from remote Gough Island to provide a genetic portrait of rapid and extreme size evolution. In just a few hundred generations, Gough Island mice evolved the largest body size among wild house mice from around the world. Through comparisons with a smaller-bodied wild-derived strain from the same subspecies (WSB/EiJ), we demonstrate that Gough Island mice achieve their exceptional body weight primarily by growing faster during the 6 weeks after birth. We use genetic mapping in large F2 intercrosses between Gough Island mice and WSB/EiJ to identify 19 quantitative trait loci (QTL) responsible for the evolution of 16-week weight trajectories: 8 QTL for body weight and 11 QTL for growth rate. QTL exhibit modest effects that are mostly additive. We conclude that body size evolution on islands can be genetically complex, even when substantial size changes occur rapidly. In comparisons to published studies of laboratory strains of mice that were artificially selected for divergent body sizes, we discover that the overall genetic profile of size evolution in nature and in the laboratory is similar, but many contributing loci are distinct. Our results underscore the power of genetically characterizing the entire growth trajectory in wild populations and lay the foundation necessary for identifying the mutations responsible for extreme body size evolution in nature.
Keywords: island syndrome, island evolution, phenotypic extreme, body size, complex trait
THE question of how organisms adapt to new environments continues to captivate biologists. Because adaptation requires genetic change, discovering the mutations responsible for adaptive phenotypes is a key step toward understanding the mechanisms of this process. There is a growing list of traits for which adaptive differences in nature have been directly traced to specific genes. Examples include adaptive coloration in pocket mice (Nachman et al. 2003), deer mice (Hoekstra et al. 2006), Drosophila melanogaster (Rebeiz et al. 2009), and peppered moths (van’t Hof et al. 2011); armor plate patterning and pelvic spine reduction in stickleback fish (Colosimo et al. 2005; Chan et al. 2010; Jones et al. 2012); and defense chemistry in Boechera stricta (Prasad et al. 2012). Despite these advances, the genetic architecture of adaptation in nature remains poorly understood. Most progress has focused on traits with simple genetic bases, where one or a few loci explain observed phenotypic variation (Rockman 2012). But the majority of trait differences between populations inhabiting contrasting environments are quantitative, suggesting that adaptation often involves more complex inheritance.
While it is notoriously difficult to demonstrate adaptation (Endler 1986), populations that rapidly evolve to phenotypic extremes following major environmental shifts provide a revealing window into the process. Evidence for adaptive change is often observed in island populations, where the limited geographic scope, sharp boundaries, and simplified biotas of islands facilitate the interpretation of evolutionary patterns (Losos and Ricklefs 2009). Insular mammals show elevated rates of morphological evolution (Pergams and Ashley 2001; Millien 2006) and include several examples of gigantism and dwarfism (Stock 1935; Freudenthal 1972; Roth 1992; Moncunill-Solé et al. 2014). Populations that colonize islands often experience substantial changes in predation risk, competition, and resource availability that together generate strong selection for shifts in body size (Sondaar 1977; Case 1978; Heaney 1978; Lawlor 1982; Lomolino 1985; Lomolino et al. 2012). Although the question of whether island mammals in general follow directional patterns in the evolution of body size (Foster 1964; Van Valen 1973; Lomolino 1985) has inspired debate (Lawlor 1982; Lomolino 1985, 2005; Meiri et al. 2004, 2008, 2011; Lomolino et al. 2005, 2012; Raia and Meiri 2006; Bromham and Cardillo 2007), murid rodents usually evolve larger sizes on islands (Adler and Levins 1994; Meiri et al. 2008). This pattern, combined with the remarkable success of house mice (Mus musculus) in colonizing islands from around the world, positions these rodents as an especially promising system for understanding adaptive size change. Considerable morphological diversity within and among island populations of house mice has been documented, especially in the research of R. J. Berry (Berry 1964, 1965, 1968, 1996; Berry and Jakobson 1975; Berry et al. 1987, 1978a,b, 1979, 1982; Berry and Scriven 2005).
The largest wild house mice in the world reside on Gough Island (GI), a remote volcanic island in the middle of the South Atlantic Ocean (Rowe-Rowe and Crafford 1992). Gough Island mice weigh approximately twice as much as their mainland relatives (Jones et al. 2003). Mice were introduced to Gough Island by teams of seal hunters (Wace 1961; Jones et al. 2003) between 130 and 200 years ago (Verrill 1895; Gray et al. 2014), suggesting that body size evolution has been very rapid.
Gough Island mice belong to the same subspecies as the laboratory mouse (Mus musculus domesticus) (Gray et al. 2014), providing access to an expansive genetic toolkit for investigating their phenotypic evolution. In this article, we focus on the rapid evolution of body size, for two reasons. First, body size is highly correlated with aspects of physiology, life history, morphology, behavior, and ecology (Peters 1983; Calder 1984; Schmidt-Nielsen 1984). As a result, size is perhaps the best singular phenotypic measurement of an organism’s overall adaptive profile (Damuth and MacFadden 1990). Second, body size is a canonical complex trait from a genetic perspective. Induced mutations in a large fraction of genes noticeably alter size (Reed et al. 2008), many quantitative trait loci (QTL) contribute to existing size differences among laboratory strains (Corva and Medrano 2001), and a variety of environmental factors influence growth. By genetically dissecting body size in Gough Island mice, we can learn how an island population in a new environment rapidly and recently achieved a marked evolutionary shift in a complex trait.
The major characteristics and determinants of body size growth in mice are understood. The growth trajectory is sigmoidal, with an increasing rate during the first few weeks after birth, peak growth between 3 and 5 weeks, and a decreasing rate thereafter (Cheverud 2005). Growth in utero and soon after birth is primarily promoted by insulin-like growth factor 2 (IGF2) (Fowden 2003), whereas the growth hormone (GH)-IGF1 axis drives growth after weaning (Lupu et al. 2001). Early growth mostly reflects increases in cell number, while later growth is mostly due to expansions in cell size (Riska and Atchley 1985; Atchley et al. 2000; Lui and Baron 2011). Genetic correlations between age-specific body weights decline with increasing time between ages (Cheverud et al. 1983a; Riska et al. 1984). Many QTL that contribute to body weight differences between laboratory mouse strains have been identified (Dragani et al. 1995; Cheverud et al. 1996; Keightley et al. 1996; Rance et al. 1997; Brockmann et al. 1998, 2004; Kirkpatrick et al. 1998; Morris et al. 1999; Vaughn et al. 1999; Corva and Medrano 2001; Rocha et al. 2004; Bennett et al. 2005; Kenney-Hunt et al. 2006; Shao et al. 2007; Casellas et al. 2009; Mollah and Ishikawa 2011; Ishikawa and Okuno 2014). A common pattern is that strain differences in weight and growth rate are due to a large number of loci with modest phenotypic effects. Consistent with the physiology of mouse growth, QTL effects vary among ages, an observation that motivates the genetic dissection of size evolution throughout the entire ontogenetic trajectory.
Whether similar genetic patterns characterize exceptional size evolution in nature remains an open empirical question. Two key contrasts prevent extrapolation of findings in laboratory mice to evolution in the wild. First, the dynamics of selection differ. Artificial selection for divergent body sizes in laboratory mice has generated impressive responses (Goodale 1938; MacArthur 1944; Falconer 1953; Eisen 1989; Bünger and Hill 1999), providing useful systems for mapping QTL. But wild mice encounter selective agents that are completely absent from the laboratory environment, and it is unclear whether artificial and natural selection are similar in intensity. Different selective pressures and intensities could lead to different genetic architectures. Second, laboratory mice have a complex history, with uncertain connections to wild mice (Silver 1995). Many genetic variants observed in natural populations are missing from the panel of commonly used inbred strains (Salcedo et al. 2007), suggesting that evolution in nature and in the laboratory could involve different sets of mutations. Here, we use mice from Gough Island to provide a genetic portrait of rapid and extreme size evolution in a wild, island population.
Materials and Methods
Gough Island mice
Gough Island belongs to the United Kingdom Overseas Territory of Tristan da Cunha, has an area of 65 km2, and is situated in the South Atlantic Ocean almost halfway between South Africa and South America (40° 19′S and 9° 55′W). Fifty mice were live trapped on Gough Island near the research station in September of 2009. Four mice died and two litters were born (five pups) during transport from Gough Island to the Charmany Instructional Facility at the University of Wisconsin-Madison. Twenty-five mature female mice and 21 mature male mice were used to start a breeding colony.
All mice were housed in microisolator cages and separated by sex. Males were grouped only if they were from the same litter and had not previously mated. The room was temperature controlled (68–72° F) and set on a 12 hr light/dark cycle. Corn cobs ground to 1/8th inch were used for bedding (Waldschmidt and Sons, Madison, WI). Water and Teklad 6% fat mouse/rat diet was provided ad libitum (Harlan Laboratories, Madison, WI). Irradiated sunflower seeds (Harlan Laboratories) and nesting material were provided as enrichment and were replaced weekly during cage changes.
Selected individuals were mated after 8 weeks of age. Breeders were supplemented with extra bedding and red huts and were fed breeder chow (Teklan Global 19% protein/9% fat; Harlan Laboratories) ad libitum. Mated pairs were only disturbed during weekly cage changes. Weekly weights to the nearest milligram were recorded, beginning 1 week after birth and ending at 16 weeks. To individually identify pups starting at 1 week, toe tattooing was performed using sterile lancets and tattoo paste. Ear punches were given at weaning (3–4 weeks) to individually identify adult mice.
Once the mice arrived in Madison, we first performed a random breeding common garden experiment with the wild founders. The purposes of this experiment were to establish a colony and to verify the large weight of Gough Island (GI) mice was inherited and not solely the result of environmental factors.
Intercross experiments
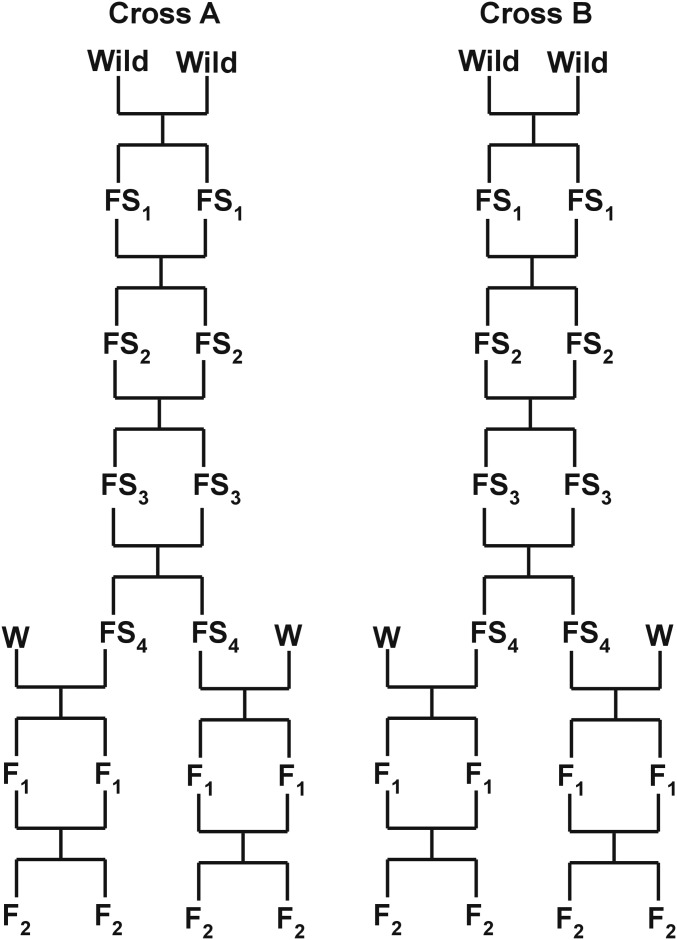
Beginning with the laboratory-born offspring of the wild mice, we created several partially inbred lines by brother–sister mating for four generations. While inbreeding was far from complete, four generations of brother–sister mating is expected to increase homozygosity to 60% within lines. We chose one pair of male and female littermates (full siblings) from each of two partially inbred lines (denoted line/cross A and B). These four mice were each crossed with WSB/EiJ (The Jackson Laboratory, Bar Harbor, ME; subsequently abbreviated as WSB) to generate four independent F2 intercrosses (Figure 1). Although we focus on QTL that are fixed between the panel of four GI mice and WSB in this study, our design will allow us to detect loci that are polymorphic within the GI population in the future. WSB was chosen because it is a wild-derived strain, has a body size typical of wild house mice, belongs to the same subspecies as GI mice, is fully inbred, has a sequenced genome (Keane et al. 2011), and is featured in large-scale efforts to genetically dissect complex traits such as the Collaborative Cross (Churchill et al. 2004). We generated a total of 1374 F2 mice: 497 from cross A (WSB × GI = 279 and GI × WSB = 218; maternal strain denoted first) and 877 from cross B (WSB × GI = 494 and GI × WSB = 383).
Figure 1.
Design of crosses for genetic mapping. Wild, first generation of Gough Island mice born in the laboratory; FS, filial self-generation; W, WSB.
Phenotyping
Weekly weights were collected for all mice from 1 week through 16 weeks of age, unless they were paired for breeding. Once mice reached 16 weeks (±1 day), they were killed by CO2 asphyxiation or by decapitation. Liver samples were collected and stored at −80° C until genetic analysis. This study was approved by the Institutional Animal Care and Use Committee at the University of Wisconsin (protocol no. V01447).
Genotyping
All mice were genotyped on the Mega Mouse Universal Genotyping Array (MegaMUGA; http://www.neogen.com/Agrigenomics/pdf/Slicks/MegaMUGAFlyer.pdf). The MegaMUGA is an Illumina Infinium array that contains ∼77,800 markers. Markers are spaced at ∼33 kb across the genome and are mostly single nucleotide polymorphisms (SNPs) with a few structural variants and transgenic markers. The markers cover all autosomes, sex chromosomes, and the mitochondria. Marker ascertainment was carried out to be maximally informative for studies involving the Collaborative Cross (Aylor et al. 2011; Threadgill and Churchill 2012), Diversity Outbred Cross (Svenson et al. 2012), wild populations of house mice, and other Mus species. Liver tissue was sent to GeneSeek (NeoGene, Lincoln, NE) for DNA extraction and genotyping with the MegaMUGA arrays. A total of 1536 samples were sent, including controls and mice that died before 16 weeks of age (which are still informative for genetic map estimation).
Multiple controls were used during extraction and genotyping to increase our ability to identify technical and biological errors. Liver tissue was organized into 16 (96 deep well) plates in such a way as to minimize array batch effects on related sets of samples. Tissue from WSB was placed in identical wells on every plate to account for plate extraction effects. The four GI parental samples were replicated four times each across the 16 plates. Lastly, replicate samples of the first well of each plate were placed in a more distant well and run on different arrays.
Genotype diagnostics
We examined the genotypes to identify biological, technical, and data entry errors. We omitted markers with high rates of missing data and those that were not informative in our crosses. We removed a small number of individuals that appeared to be unresolved sample mix-ups based on large numbers of Mendelian inconsistencies or mismatched sex (inferred from array intensities of markers on the X and Y chromosomes). We also removed a few mice with high rates of missing genotype data. Following these initial screens, the cleaned data included the four GI parents of the crosses, 70 F1 individuals, 1346 F2 individuals, and 33,191 markers.
In all subsequent analyses, we focused on a subset of 11,833 markers that were fixed in the four GI parents and therefore segregated as in a standard F2 intercross between inbred lines. We estimated the intermarker genetic distances assuming a genotyping error rate of 0.2% and converted estimated recombination fractions to map distances with the Carter–Falconer map function (Carter and Falconer 1951).
Smoothed phenotypes
We omitted F2 mice missing body weights at ≥4 of the 16 time points, resulting in a final set of 1212 F2 mice with nearly complete data. To reduce measurement error in the body weights and to interpolate some remaining missing values, we used the R package pspline to fit a 7 degrees-of-freedom cubic spline to the data for each mouse. The degrees of freedom were chosen by visual inspection, considering the trade-off between smoothness and the fidelity of the underlying curves. We estimated growth rate as the first derivative of the fitted cubic splines.
QTL analysis
QTL analysis was performed by Haley–Knott regression (Haley and Knott 1992) on a 0.5-cM grid across the genome. We used conditional genotype probabilities (given the available marker data) calculated assuming a genotyping error rate of 0.2% and the Carter–Falconer map function. While we considered data on 11,833 markers, QTL calculations were ultimately based on a fixed grid of 2665 pseudomarkers. Smoothed body weights and estimated growth rates were analyzed separately.
Single-QTL analysis was performed at each of the 16 time points, individually, including indicators for each F1 mother (to account for sibship differences) and sex as fixed additive covariates. The results across time points were combined by averaging the LOD scores across time points at each genomic position to give SLOD scores (Kwak et al. 2014). Thresholds for statistical significance were calculated using a permutation test (Churchill and Doerge 1994), permuting the rows in the phenotype data relative to the rows in the genotype data (that is, maintaining the correlation structure within the phenotype data and within the genotype data, but breaking the association between the genotypes and phenotypes). Autosome- and X-chromosome-specific significance thresholds were derived using the approach of Broman et al. (2006), with 1500 permutation replicates for the autosomes and 28,200 replicates for the X chromosome.
Multiple-QTL analysis was performed using strictly additive QTL models, using the penalized-SLOD criterion of Kwak et al. (2014), an extension of the penalized-LOD score criterion of Broman and Speed (2002). Consider a multiple-QTL model γ (a vector specifying the locations of some set of QTL). We considered each time point individually and calculated a LOD score measuring the quality of fit of the model at time point i, LODi(γ). We then obtained an overall measure of the fit of the model by taking the average, SLOD(γ) = ∑iLODi(γ)/16, where the sum is over the 16 time points. We subsequently considered a penalized version of this statistic to balance quality of fit with model size. The penalized statistic was pSLOD(γ) = SLOD(γ) − T|γ|, where |γ| is the number of QTL in the model γ, and T is a penalty, taken to be a quantile from the null distribution of SLOD in a single-QTL genome scan, estimated by a permutation test (here using a common threshold for the autosomes and the X chromosome). Model searching was performed by a stepwise algorithm: forward selection up to a model with 15 QTL, followed by backward elimination to the null model. The chosen model was that which maximized pSLOD, among all models visited. As with the single-QTL analysis, the multiple-QTL analysis included indicators for each F1 mother and sex as fixed additive covariates.
To indicate the strength of evidence for the QTL in the context of the selected multiple-QTL model, we calculated profile SLOD curves (Kwak et al. 2014) based on an idea from Zeng et al. (2000). We varied the position of each QTL, keeping all other QTL fixed at their estimated locations, and compared the full model with that QTL in varying position to the reduced model with that QTL omitted. To define approximate confidence intervals for the QTL, we fit the inferred multiple-QTL model at each time point and calculated 1.5-LOD support intervals (using the profile LOD scores) for the location of each QTL at each time point. For each QTL, we cite the interval corresponding to the time point for which the evidence for that QTL is maximum (that is, the time point at which the profile LOD score is highest).
To assess the relationship between sample size and the number of inferred QTL, we took random subsets of the F2 mice and applied the stepwise QTL analysis, using SLOD with the 5% significance threshold calculated for the full data. (The significance threshold is not much influenced by sample size.) We performed 37–38 replicates at sample sizes of 300, 600, 800, and 1000 F2 mice.
Data availability
Phenotypes and genotypes from this study are available from the QTL Archive at the Jackson Laboratory, at http://phenome.jax.org/db/q?rtn=projects/projdet&reqprojid=539. Supplemental information includes: p-values from strain comparisons (Figure S1), histograms of F2 weights (Figure S2), average F2 weights and growth rates (Figure S3), p-values from comparisons among crosses and mothers (Figure S4), genetic and physical maps of markers used in QTL analyses (Figure S5), F2 frequency of WSB allele (Figure S6), and candidate genes in QTL regions (Table S1).
Results
Patterns of phenotypic variation
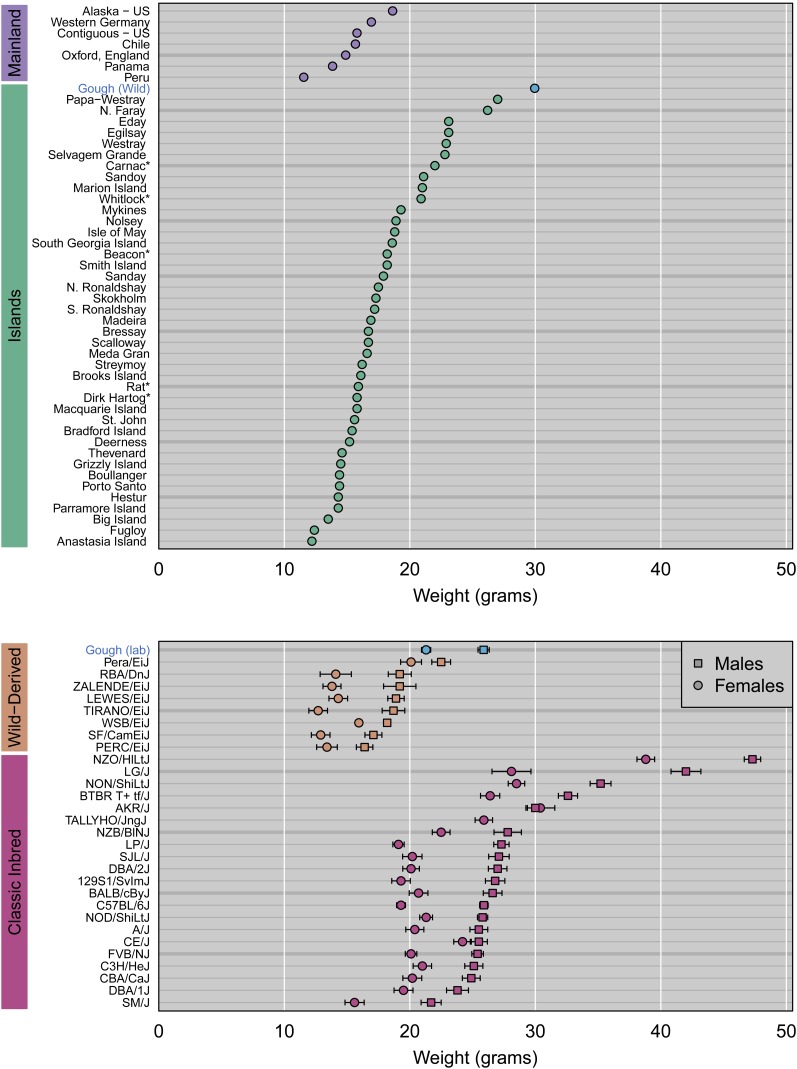
Wild-caught GI mice are exceptionally heavy compared to wild mice of the same subspecies (M. m. domesticus) from the mainland and other islands (Figure 2). GI mice raised in the laboratory are heavier than wild-derived inbred strains of M. m. domesticus (Figure 2). Lab-reared GI mice are similar in weight to classical inbred strains (Figure 2), which are bigger than most wild mice (presumably because of artificial selection during domestication). The weight difference between GI mice sampled from nature and those raised in the lab likely reflects the contrasting environments these mice experienced and the older ages of the wild-caught individuals.
Figure 2.
Comparison of body weight in Gough Island mice to other mice from the wild, from wild-derived inbred strains, and from classic inbred strains. Weights were compiled from sources listed in Table S1.
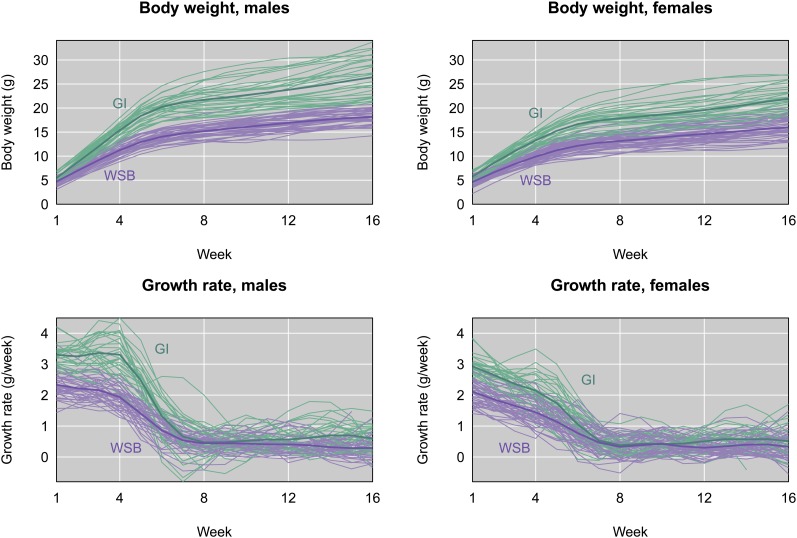
GI and WSB mice raised in the same environment exhibit contrasting growth trajectories (Figure 3), demonstrating that trait divergence has a heritable component. Weights are significantly different at all ages in both sexes (Supporting Information, Figure S1; t-tests, maximum P-values across time points; total: P < 10−8; females: P < 10−5; males: P < 10−3). The rate of growth differs over a narrower time window, pinpointing early growth as the primary driver of phenotypic divergence. Growth rates estimated from the first derivatives of fitted growth curves in individual mice are significantly faster in GI mice from 1–6 weeks (Figure S1; total: P < 10−4; females: P < 0.05; males: P < 0.01). At ∼4–6 weeks, the growth rate difference between GI and WSB mice begins to decline. GI and WSB mice also show divergent growth rates at 11–16 weeks (total: P < 0.05; males: P < 0.05), though females are not significantly different at 14–16 weeks. Male GI mice are heavier than female GI mice beginning at 3 weeks (Figure S1; P < 10−3), and grow significantly faster at 1–5 weeks (P < 10−3). Phenotypic patterns are similar in the partially inbred GI mice (and their offspring) we used for genetic mapping.
Figure 3.
Body weight (top panels) and growth rate (bottom panels) for males (left) and females (right), as a function of age in weeks, for a sample of Gough Island mice (GI, green) and WSB mice (purple) raised in the lab. Individual body weight curves were lightly smoothed using cubic splines; the growth rate curves were estimated as the first derivative of the fitted splines. Thicker curves follow the group averages.
F1 mean weights from crosses between GI and WSB mice are closer to GI means for the first 3 weeks (Figure S2). At subsequent ages, F1 weights are near the mid-parent value, with a bias toward WSB values (especially in males).
F2’s from crosses between GI and WSB mice vary widely in weight at all ages (Figure S2). F2 weight distributions are continuous and weight ranges exceed parental averages. Weight variances increase with age, whereas coefficients of variation are relatively stable across ages. Growth rate also shows substantial F2 variances at all ages, with coefficients of variation tending to increase with age. Collectively, these observations suggest that multiple mutations contributed to the evolution of weight and growth rate differences between GI and WSB mice.
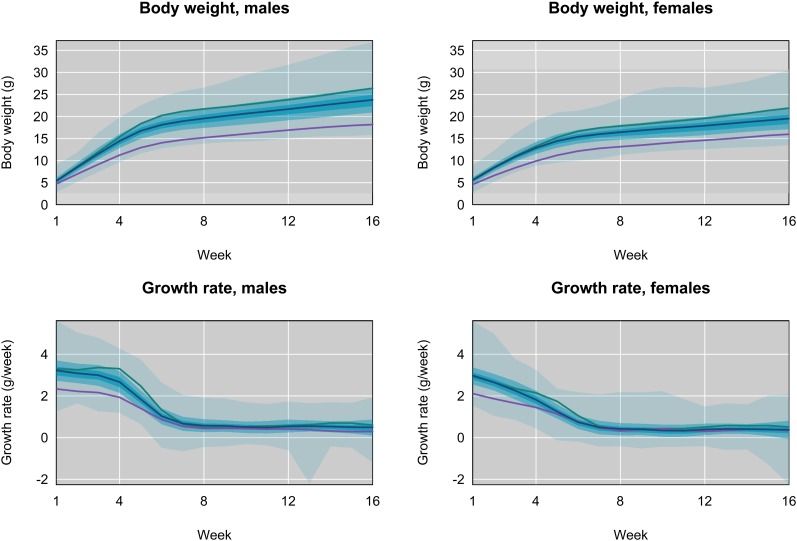
The collection of 16-week growth trajectories for individual F2’s is shown in Figure 4. Mirroring patterns in the parental lines, F2 males and females show large differences in body weight (Figure S3; P < 0.015; < 10−20 from week 3 onward) and growth rate at all ages (P < 10−4). Faster growth of males during weeks 1–5 (Figure 3) is primarily responsible for weight differences among the sexes. F2’s from different F1 mothers vary significantly in weight (Figure S4; P < 10−18) and growth rate (P < 0.05, with the exception of females at 14–15 weeks) at all ages, providing evidence for maternal effects (which could be genetic or environmental). F2 weights at all ages depend on GI parent (Figure S4; P < 10−8), as do growth rates at most ages (P < 10−5 in weeks 1–4; P < 0.037, aside from weeks 6–7 and 13–14). These results motivated us to account for effects of sex, mother, and line in all QTL analyses.
Figure 4.
Body weight (top panels) and growth rate (bottom panels) for males (left) and females (right), as a function of age in weeks, for the F2 mice. In each panel there are three shaded regions; the darkest region covers the middle third of the individuals; the next-darkest two-thirds, and the lightest region all mice. The blue curve is for the average of the F2 mice. The green and purple curves are for the averages of Gough Island and WSB mice, respectively, as in Figure 3.
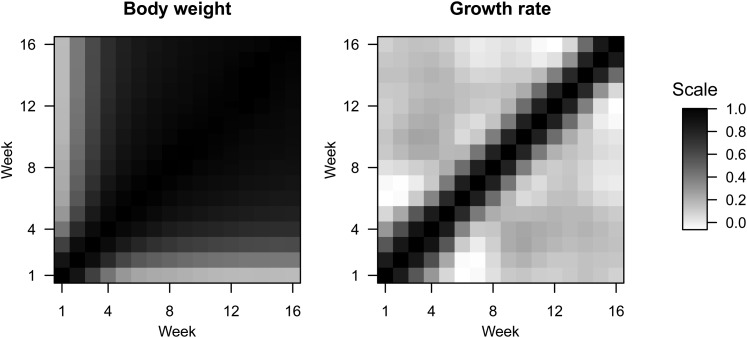
F2 weights show strong pairwise correlations across ages, with a steady decline in autocorrelation as the difference in ages increases (Figure 5). In contrast, correlations between growth rates are only high for neighboring ages (Figure 5). For example, the growth rate at one time point is nearly uncorrelated with the growth rate at an age 4 weeks later, whereas weights are highly correlated at these ages.
Figure 5.
Pairwise correlations across ages for body weight (left) and growth rate (right). Black pixels indicate a correlation near 1; white pixels indicate a correlation near 0.
Genetic map characteristics
The genetic map constructed from F2 genotypes (Figure S5) is 1327 cM in total length. Characteristics of the map resemble known recombination patterns in house mice (Cox et al. 2009), including an average of 26 crossovers per meiosis, elevated rates in subtelomeric regions, and decreased rates in subcentromeric regions. A region on chromosome 2 shows remarkable transmission ratio distortion in favor of the WSB allele (allele frequency = 0.65; peak marker located at 79.6 Mb) in F2’s descended from one GI line but not the other (Figure S6). Biased transmission of the WSB allele in this interval is also observed in the Collaborative Cross (Aylor et al. 2011) and seems to be generated by maternal meiotic drive (Didion et al. 2015).
QTL number and location
By combining information across ages, our multiple QTL mapping approach allowed us to identify sets of QTL that affect divergence in overall growth trajectories. We focus on results from this method. Major patterns are similar to those obtained by single QTL mapping at individual ages (results not shown).
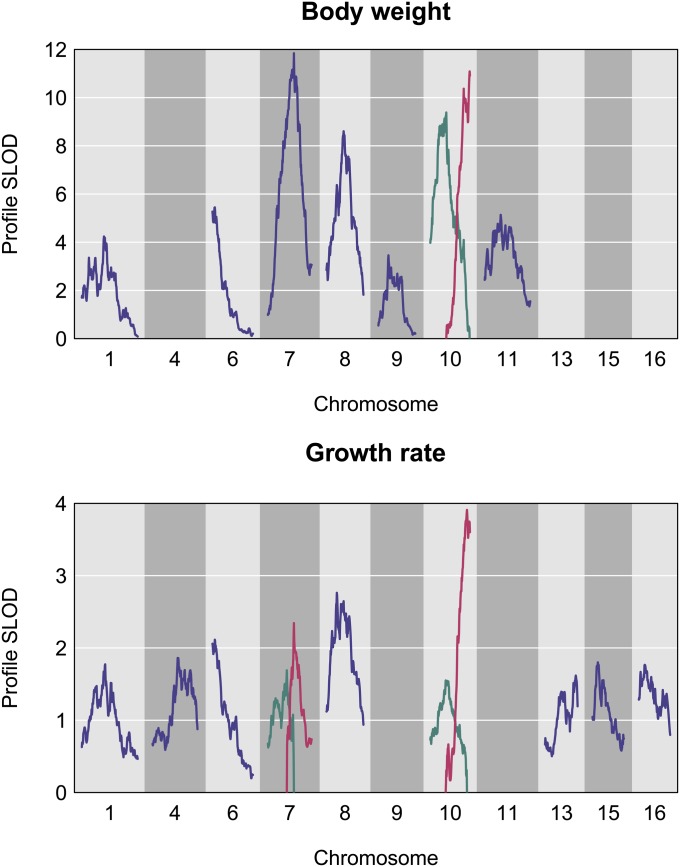
Eight QTL contribute to weight differences between GI and WSB mice when a 5% genome-wide significance threshold is used (Figure 6; Table 1). For growth rate, 11 QTL are detected (Figure 6; Table 1). Five weight QTL overlap with growth rate QTL on chromosomes 1, 6, 7, and 10 (two QTL) (Table 1).
Figure 6.
Profile SLOD curves for the selected multiple-QTL models for body weight and growth rate. The location of each QTL was varied, one at a time, with all other QTL fixed at their estimated locations, and the multiple-QTL model was compared to the model with the given QTL omitted.
Table 1. Genomic positions of QTL.
| Phenotype | Chromosome | cM | cM lower CI | cM upper CI | Mb | Mb lower CI | Mb upper CI |
|---|---|---|---|---|---|---|---|
| Weight | 1 | 37.5 | 22.0 | 53.5 | 78.9 | 42.4 | 136.9 |
| Weight | 6 | 4.0 | 0.0 | 6.0 | 17.9 | 3.4 | 23.5 |
| Weight | 7 | 44.0 | 35.5 | 45.0 | 118.9 | 86.1 | 121.9 |
| Weight | 8 | 29.0 | 25.5 | 39.0 | 81.5 | 69.1 | 106.1 |
| Weight | 9 | 17.0 | 15.0 | 40.5 | 47.3 | 42.7 | 97.5 |
| Weight | 10 | 27.0 | 23.0 | 31.0 | 68.1 | 58.6 | 77.4 |
| Weight | 10 | 66.5 | 55.0 | 67.0 | 128.1 | 119.0 | 129.6 |
| Weight | 11 | 27.0 | 13.5 | 42.5 | 51.5 | 31.5 | 81.5 |
| Growth rate | 1 | 39.5 | 33.5 | 41.5 | 84.3 | 73.0 | 92.9 |
| Growth rate | 4 | 42.5 | 40.5 | 65.5 | 104.6 | 103.1 | 141.6 |
| Growth rate | 6 | 4.0 | 3.5 | 8.5 | 17.9 | 17.0 | 27.3 |
| Growth rate | 7 | 32.0 | 13.5 | 36.0 | 80.1 | 38.6 | 86.9 |
| Growth rate | 7 | 44.0 | 47.5 | 55.5 | 118.9 | 125.4 | 137.0 |
| Growth rate | 8 | 17.5 | 11.0 | 34.0 | 45.9 | 32.4 | 92.7 |
| Growth rate | 10 | 26.0 | 4.0 | 34.5 | 66.3 | 16.9 | 88.7 |
| Growth rate | 10 | 62.0 | 59.5 | 67.0 | 125.3 | 123.3 | 129.6 |
| Growth rate | 13 | 52.0 | 47.0 | 54.5 | 116.5 | 111.2 | 119.3 |
| Growth rate | 15 | 9.0 | 3.5 | 12.0 | 36.4 | 19.6 | 42.2 |
| Growth rate | 16 | 10.0 | 0.0 | 13.0 | 26.4 | 3.5 | 29.8 |
Confidence interval is the 1.5-LOD interval for the age with the highest LOD score.
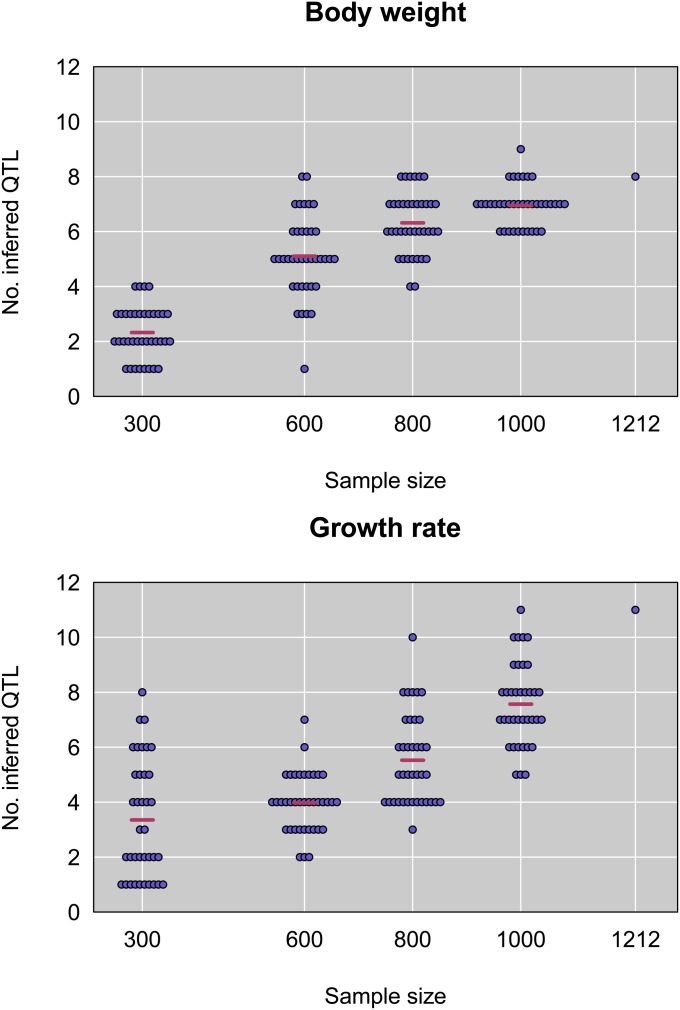
To gauge our ability to accurately estimate the number of QTL, we randomly subsampled F2’s, reran QTL analyses, and counted the number of QTL in the best fitting model (at the 5% significance threshold). Two key patterns emerge (Figure 7). First, the number of detected QTL steadily increases with sample size for both weight and growth rate, suggesting that a large number of loci with small effects contribute to trait divergence between GI and WSB. Second, there is considerable variance in the number of QTL detected with different F2 samples of the same size. Collectively, these results underscore the difficulty of estimating QTL number, even with large sample sizes such as ours.
Figure 7.
Number of inferred QTL as a function of sample size, for random subsets of the data, and for the full dataset, for body weight (top) and growth rate (bottom).
QTL effects
We measured QTL effects using two parameters: the additive effect of substituting one allele (calculated as half the difference in genotypic means between alternative homozygotes), and the dominance effect (calculated as the difference between the genotypic mean of the heterozygote and the average genotypic mean of alternative homozygotes).
Additive and dominance effects for all QTL are presented in Table 2 and Table 3. Individual QTL effects are small-to-moderate in size relative to the difference in mean phenotypes between GI and WSB mice. The largest additive effect across all weight QTL and ages is 0.66 g (chromosome 8 QTL at 16 weeks); homozygotes carrying the GI vs. WSB allele at this QTL differ by an average of 1.32 g. Additive effects of weight QTL tend to increase with age, whereas age-related trends in additive effects of growth rate QTL are less apparent. Looking across development, chromosomes 10, 7, and 8 harbor QTL with the largest effects for both weight and growth rate.
Table 2. Additive and dominance effects (in grams) of weight QTL.
| Week | 1@37.5 | 6@4.0 | 7@44.0 | 8@29.0 | 9@17.0 | 10@27.0 | 10@66.5 | 11@27.0 | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | a | −0.008 | −0.045 | −0.057 | 0.024 | −0.033 | 0.177 | 0.043 | 0.127 |
| d | 0.054 | 0.054 | 0.009 | 0.009 | 0.106 | 0.043 | 0.029 | 0.028 | |
| 2 | a | 0.045 | −0.004 | 0.047 | 0.090 | −0.057 | 0.200 | 0.171 | 0.159 |
| d | 0.054 | 0.076 | 0.061 | −0.035 | 0.157 | 0.050 | 0.043 | 0.007 | |
| 3 | a | 0.117 | 0.050 | 0.197 | 0.148 | −0.044 | 0.226 | 0.337 | 0.182 |
| d | 0.074 | 0.078 | 0.118 | −0.109 | 0.210 | 0.061 | 0.030 | −0.030 | |
| 4 | a | 0.214 | 0.146 | 0.381 | 0.206 | −0.016 | 0.281 | 0.453 | 0.221 |
| d | 0.074 | 0.040 | 0.148 | −0.137 | 0.239 | 0.056 | 0.014 | −0.037 | |
| 5 | a | 0.291 | 0.237 | 0.519 | 0.241 | 0.052 | 0.351 | 0.503 | 0.266 |
| d | 0.063 | 0.001 | 0.143 | −0.149 | 0.234 | 0.046 | 0.001 | −0.046 | |
| 6 | a | 0.321 | 0.298 | 0.561 | 0.284 | 0.120 | 0.406 | 0.507 | 0.307 |
| d | 0.047 | −0.013 | 0.095 | −0.173 | 0.226 | 0.049 | −0.001 | −0.068 | |
| 7 | a | 0.334 | 0.334 | 0.554 | 0.337 | 0.169 | 0.445 | 0.510 | 0.342 |
| d | 0.035 | −0.008 | 0.027 | −0.206 | 0.239 | 0.057 | 0.001 | −0.083 | |
| 8 | a | 0.344 | 0.359 | 0.538 | 0.390 | 0.206 | 0.471 | 0.521 | 0.365 |
| d | 0.039 | −0.004 | −0.022 | −0.235 | 0.269 | 0.048 | 0.001 | −0.081 | |
| 9 | a | 0.347 | 0.389 | 0.533 | 0.443 | 0.235 | 0.488 | 0.535 | 0.377 |
| d | 0.055 | −0.023 | −0.033 | −0.250 | 0.309 | 0.035 | −0.005 | −0.072 | |
| 10 | a | 0.333 | 0.431 | 0.538 | 0.501 | 0.250 | 0.509 | 0.535 | 0.380 |
| d | 0.071 | −0.066 | −0.020 | −0.251 | 0.343 | 0.035 | −0.010 | −0.069 | |
| 11 | a | 0.316 | 0.473 | 0.551 | 0.549 | 0.245 | 0.534 | 0.519 | 0.382 |
| d | 0.082 | −0.106 | −0.007 | −0.249 | 0.365 | 0.039 | −0.011 | −0.083 | |
| 12 | a | 0.315 | 0.509 | 0.564 | 0.586 | 0.233 | 0.570 | 0.505 | 0.385 |
| d | 0.102 | −0.128 | 0.003 | −0.244 | 0.365 | 0.037 | −0.012 | −0.102 | |
| 13 | a | 0.327 | 0.536 | 0.575 | 0.615 | 0.235 | 0.603 | 0.509 | 0.389 |
| d | 0.134 | −0.133 | 0.014 | −0.229 | 0.348 | 0.044 | −0.015 | −0.114 | |
| 14 | a | 0.332 | 0.537 | 0.601 | 0.639 | 0.251 | 0.624 | 0.526 | 0.383 |
| d | 0.154 | −0.145 | 0.005 | −0.206 | 0.345 | 0.058 | −0.008 | −0.135 | |
| 15 | a | 0.341 | 0.546 | 0.626 | 0.660 | 0.267 | 0.617 | 0.556 | 0.396 |
| d | 0.187 | −0.162 | −0.004 | −0.182 | 0.342 | 0.076 | −0.008 | −0.147 | |
| 16 | a | 0.346 | 0.546 | 0.644 | 0.660 | 0.257 | 0.592 | 0.619 | 0.419 |
| d | 0.224 | −0.168 | −0.006 | −0.159 | 0.347 | 0.109 | −0.045 | −0.157 |
Effects are adjusted for variation due to sex, line, and mother. QTL are designated by chromosome and cM position (chromosome@cM).
Table 3. Additive and dominance effects (in grams per week) of growth rate QTL.
| Week | 1@39.5 | 4@42.5 | 6@4.0 | 7@32.0 | 7@44.0 | 8@17.5 | 10@26.0 | 10@62.0 | 13@52.0 | 15@9.0 | 16@10.0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | a | 0.047 | 0.031 | 0.021 | 0.072 | 0.025 | 0.047 | 0.001 | 0.124 | −0.034 | 0.065 | 0.069 |
| d | 0.019 | 0.033 | 0.039 | −0.027 | 0.052 | 0.002 | 0.007 | 0.040 | 0.006 | 0.036 | 0.047 | |
| 2 | a | 0.071 | 0.018 | 0.045 | 0.093 | 0.058 | 0.049 | −0.001 | 0.165 | −0.050 | 0.075 | 0.077 |
| d | 0.036 | 0.046 | 0.028 | −0.058 | 0.093 | 0.000 | 0.004 | 0.006 | −0.022 | 0.077 | 0.040 | |
| 3 | a | 0.099 | −0.003 | 0.084 | 0.108 | 0.097 | 0.052 | 0.025 | 0.152 | −0.091 | 0.053 | 0.070 |
| d | 0.024 | 0.057 | −0.015 | −0.061 | 0.081 | −0.001 | −0.012 | −0.006 | −0.043 | 0.071 | 0.025 | |
| 4 | a | 0.097 | −0.018 | 0.102 | 0.061 | 0.132 | 0.050 | 0.063 | 0.091 | −0.108 | 0.022 | 0.030 |
| d | 0.001 | 0.043 | −0.041 | −0.038 | 0.042 | −0.012 | −0.021 | −0.013 | −0.035 | 0.043 | 0.008 | |
| 5 | a | 0.052 | −0.016 | 0.077 | −0.033 | 0.118 | 0.045 | 0.071 | 0.014 | −0.073 | 0.005 | −0.013 |
| d | −0.016 | −0.003 | −0.028 | 0.007 | −0.029 | −0.025 | −0.009 | −0.012 | −0.006 | 0.014 | −0.015 | |
| 6 | a | 0.013 | 0.001 | 0.044 | −0.078 | 0.066 | 0.047 | 0.054 | −0.018 | −0.023 | −0.009 | −0.032 |
| d | −0.014 | −0.054 | −0.007 | 0.024 | −0.078 | −0.025 | 0.003 | −0.014 | 0.013 | 0.004 | −0.026 | |
| 7 | a | 0.004 | 0.019 | 0.024 | −0.056 | 0.021 | 0.043 | 0.040 | −0.014 | 0.000 | −0.031 | −0.038 |
| d | −0.001 | −0.073 | 0.002 | 0.012 | −0.072 | −0.014 | −0.007 | −0.018 | −0.002 | −0.005 | −0.023 | |
| 8 | a | 0.000 | 0.029 | 0.022 | −0.008 | −0.011 | 0.041 | 0.026 | −0.005 | 0.001 | −0.042 | −0.030 |
| d | 0.011 | −0.061 | −0.010 | 0.010 | −0.035 | −0.005 | −0.022 | −0.020 | −0.017 | −0.006 | −0.024 | |
| 9 | a | −0.011 | 0.037 | 0.033 | 0.025 | −0.023 | 0.047 | 0.022 | −0.004 | −0.001 | −0.031 | −0.021 |
| d | 0.013 | −0.036 | −0.037 | 0.008 | 0.002 | 0.014 | −0.013 | −0.017 | 0.001 | −0.002 | −0.022 | |
| 10 | a | −0.022 | 0.042 | 0.040 | 0.023 | −0.013 | 0.052 | 0.025 | −0.013 | 0.001 | −0.023 | −0.017 |
| d | 0.008 | −0.013 | −0.046 | −0.016 | 0.027 | 0.034 | 0.001 | −0.011 | 0.025 | 0.005 | −0.016 | |
| 11 | a | −0.013 | 0.042 | 0.035 | −0.003 | 0.013 | 0.053 | 0.032 | −0.016 | −0.004 | −0.034 | −0.020 |
| d | 0.010 | 0.005 | −0.028 | −0.039 | 0.038 | 0.030 | 0.000 | −0.004 | 0.024 | 0.018 | −0.006 | |
| 12 | a | 0.004 | 0.043 | 0.027 | −0.042 | 0.042 | 0.052 | 0.038 | −0.004 | −0.012 | −0.037 | −0.033 |
| d | 0.021 | 0.014 | −0.007 | −0.039 | 0.039 | 0.019 | 0.006 | 0.000 | 0.014 | 0.013 | 0.006 | |
| 13 | a | 0.011 | 0.044 | 0.017 | −0.067 | 0.060 | 0.042 | 0.028 | 0.014 | −0.008 | −0.031 | −0.034 |
| d | 0.039 | 0.004 | 0.001 | −0.022 | 0.030 | 0.024 | 0.019 | −0.003 | 0.004 | −0.010 | 0.015 | |
| 14 | a | 0.009 | 0.026 | 0.007 | −0.056 | 0.064 | 0.027 | 0.008 | 0.025 | 0.004 | −0.022 | −0.024 |
| d | 0.042 | −0.016 | −0.013 | −0.024 | 0.014 | 0.021 | 0.021 | −0.001 | −0.018 | −0.016 | 0.019 | |
| 15 | a | 0.006 | 0.019 | 0.006 | −0.033 | 0.051 | 0.016 | −0.020 | 0.038 | 0.010 | −0.007 | −0.002 |
| d | 0.042 | −0.021 | −0.016 | −0.038 | 0.013 | 0.033 | 0.026 | −0.007 | −0.029 | −0.023 | 0.029 | |
| 16 | a | 0.000 | 0.025 | 0.000 | −0.026 | 0.033 | 0.010 | −0.043 | 0.059 | 0.018 | 0.003 | 0.011 |
| d | 0.042 | −0.031 | −0.005 | −0.047 | 0.025 | 0.072 | 0.034 | −0.041 | −0.012 | −0.031 | 0.026 |
Effects are adjusted for variation due to sex, line, and mother. QTL are designated by chromosome and cM position (chromosome@cM).
QTL vary in the timing of their largest effects. For example, QTL on chromosomes 10 and 11 affect weight beginning at week 1, whereas the chromosome 9 weight QTL has little effect until week 6. Once QTL begin to affect weight, they continue to do so at subsequent ages. In contrast, the biggest effects of growth rate QTL tend to be concentrated early in ontogeny. The growth rate QTL found distally on chromosome 10 exhibits its strongest effects during the first 4 weeks, and the QTL on chromosome 7 principally affects growth at 3–5 weeks of age. These patterns underscore the power of dissecting the genetics of size throughout the entire growth trajectory.
Most dominance effects of detected QTL are small. The two exceptions are weight QTL on chromosomes 8 and 9. The weight QTL on chromosome 9 shows the largest dominance deviations, which exceed its additive effects at all ages. Heterozygotes at this locus are shifted toward the GI mean. QTL with the biggest phenotypic effects primarily act in an additive fashion.
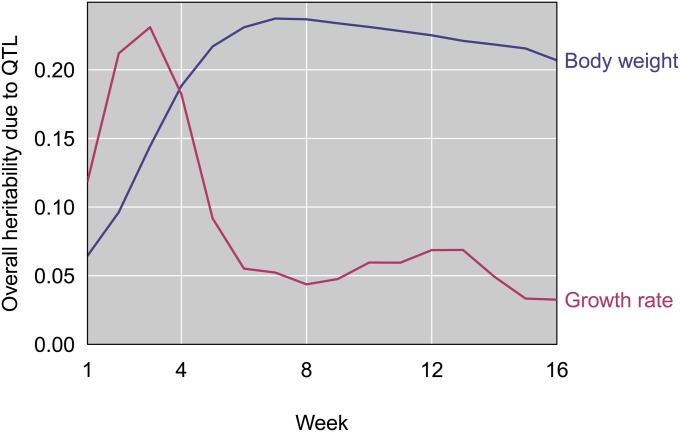
Collectively, the eight weight QTL explain between 6% and 24% of the F2 variance in weight across ages (Figure 8). Overall heritability due to weight QTL starts low (at 1 week), increases to its maximum at 7 weeks, and decreases slightly through 16 weeks. The 11 growth rate QTL jointly explain between 3% and 20% of the F2 variance (Figure 8). In contrast to weight QTL, the overall heritability due to growth rate QTL peaks at 3 weeks, quickly drops during weeks 4 and 5, and remains low through 16 weeks. This pattern mirrors the temporal divergence in growth rate between GI and WSB mice (Figure 3).
Figure 8.
Estimated heritability due to the set of QTL in the multiple-QTL models for body weight (blue) and growth rate (pink).
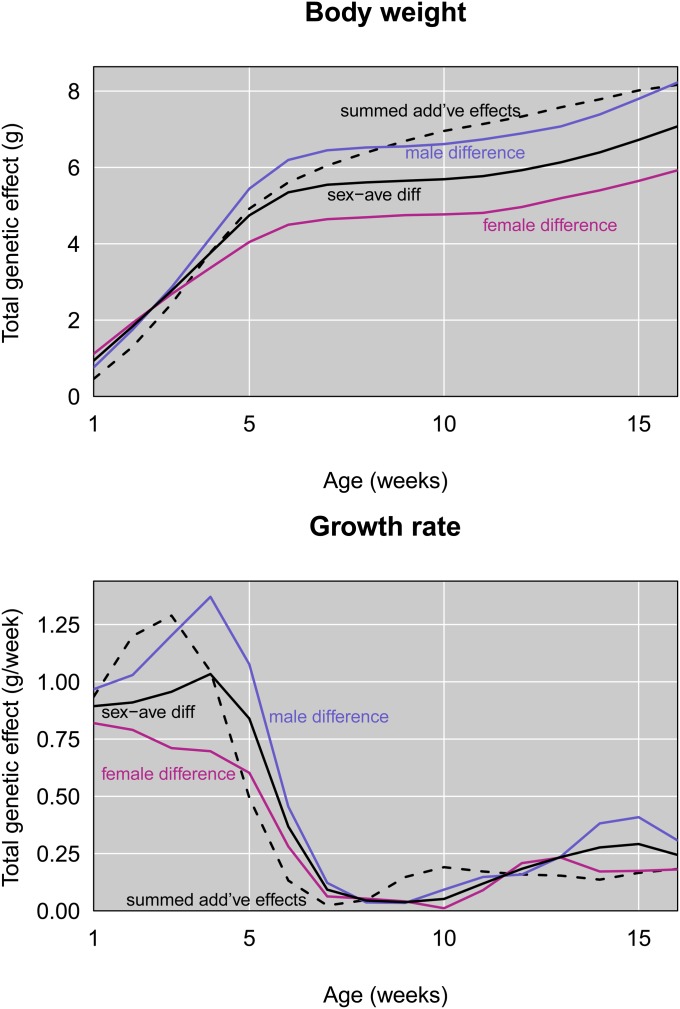
We used an additional approach to measure the ability of detected QTL to explain body size evolution. Phenotypic means of GI and WSB mice represent their genetic values. In a purely additive model, the difference between phenotypic means of these parents at a particular age should equal twice the sum of the additive effects of all mutations that contribute to weight or growth rate differences. Figure 9 compares the summed additive effects of detected QTL to phenotypic differences between GI and WSB. These results raise the possibility that detected QTL collectively explain a substantial proportion of the genetic difference in size between GI and WSB mice.
Figure 9.
Comparison of differences in average GI and WSB values with summed additive effects of QTL.
Further characterization of QTL
To better understand the biological context of QTL, we conducted targeted interaction tests involving sex, mother, line, and other loci. Though sex and mother are important contributors to F2 weight and growth rate, there is no evidence that QTL action depends on these factors. These results indicate that the QTL we detected are likely shared by males and females, and across F1 mothers. Most QTL also show no sign of being affected by line. One QTL—on chromosome 7—displays an interaction with line for body weight (but not growth rate). Because the two lines of GI mice that contributed to our intercross were founded by different wild parents, this result suggests that the chromosome 7 QTL may be polymorphic in the island population. Pairwise interaction tests revealed no evidence for epistasis between detected QTL. Comprehensive searches for epistasis involving other loci are difficult to conduct and interpret because we are considering the entire growth trajectory. Consequently, we focus on the additive and dominance effects of individual QTL in this article. To identify candidate genes, we looked for genes in the centers of QTL intervals (including 2 Mb on either side of the peak) that are known to affect body size or related processes. We used Co-Citer software (Qiao et al. 2013) to count references that mention each gene and “body weight,” “body mass,” or “growth.” Candidate genes are listed in Table S1.
QTL evolution
Additive effects indicate that GI alleles increase weight at most QTL and at most ages. To evaluate whether this pattern is consistent with neutral evolution of body size, we asked whether the number of QTL at which the GI allele increases size is consistent with chance expectations, conditioning on the observed weight difference between GI and WSB mice (Orr 1998). Considering the maximum effect for each weight QTL across ages reveals suggestive evidence for a contribution of natural selection (Orr’s test; P = 0.07), with GI alleles increasing weight at all eight QTL. Again using the maximum effect as a summary, GI alleles increase growth rate at 10 of 11 QTL (P = 0.15; 11 of 11 QTL would have yielded an estimated P = 0.02).
Discussion
The ability of house mice to invade islands from around the world and their status as a genetic model organism combine to make them an unusually powerful system for addressing the question of how natural populations evolve extreme phenotypes. Our results provide a rare genetic portrait of this process in a wild population that has achieved a dramatic increase in body size during just a few hundred generations.
The suite of QTL we identified and the large magnitude of our cross allow us to confidently conclude that GI mice did not evolve their unusual body size via a few genetic changes of major effect, perhaps contrary to intuition. Instead, multiple mutations with a range of modest, mostly additive effects were responsible. The contrast between this result and the simple genetic architecture underlying fitness-related phenotypic changes in other rodents and in stickleback fish (Nachman et al. 2003; Colosimo et al. 2005; Hoekstra et al. 2006) may primarily reflect differences in the focal traits rather than differences between species. One potential explanation for the genetic complexity of body size evolution is that weight and growth rate are both composite phenotypes affected by a diverse array of biological processes. Decomposing these traits into a series of intermediate phenotypes could reveal simpler genetic mechanisms.
From an evolutionary perspective, the distribution of QTL effects deserves further attention. The modest additive effects of individual QTL and the low percentage of F2 phenotypic variance jointly explained by all QTL suggest limited ability to predict body size from detected loci. Our subsampling experiment indicates that QTL with smaller effects were missed, and undetected epistasis also may have contributed to F2 variance. At the same time, there are reasons to be optimistic that detected QTL capture much of the genetic basis of body size evolution in this case. In addition to being affected by genetic variation, F2 phenotypes reflect uncontrolled environmental and developmental variance. For example, mice that ingested different amounts of food would be expected to grow to different sizes. The heritability of body size is the maximum fraction of phenotypic variance that QTL could theoretically explain. It is therefore interesting to note that the combined heritability for the group of body weight QTL we detected roughly matches overall heritability estimates for weight in laboratory mouse populations (range of point estimates = 0.22–0.43; Lynch 1992), although heritability is a population-specific quantity. In contrast to individual F2 phentoypes, average values for the partially inbred GI mice and the fully inbred WSB reflect genetic values, with random environmental deviations contributing to the variance about these averages. In this context, it is revealing that the summed additive effects of detected QTL account for most of the phenotypic differences between GI mice and WSB. An important caveat to this point is that QTL mapping generates upward biases in additive effect estimates (Beavis 1994, 1998). Nevertheless, the suite of QTL we discovered may explain much of the genetic component of body size evolution, despite its complexity.
When viewed in combination with previous findings, our results provide an unusual opportunity to compare the genetics of size evolution in nature to that in the laboratory. Here, we focus on a study that is especially comparable in terms of experimental design. Cheverud et al. (1996) identified QTL for age-specific (weekly) weights and growth rates up to 10 weeks in an F2 intercross between the large (LG) and small (SM) strains. These strains were developed by (i) crossing a variety of inbred lines, (ii) artificially selecting for increased (Goodale 1938) or decreased (MacArthur 1944) body weight at 60 days of age, and (iii) inbreeding for >125 generations (Chai 1956a,b; Festing 1996). Selection was very effective, producing a 20- to 24-g weight difference between LG and SM at 10 weeks of age (Chai 1956b; Kramer et al. 1998).
The divergence in growth trajectories between GI mice and WSB resembles that between LG and SM. In GI mice and LG, accelerated growth during the first 6 weeks is the primary driver of increased weight gain. The magnitude of the acceleration is greater in males. In F2’s, correlations between weekly weights decline as the time between ages increases. Early and late growth rates are weakly correlated.
The overall genetic architectures of LG–SM and GI–WSB size differences are also similar. The number of QTL affecting LG–SM weight differences ranges from 7 (1 week) to 17 (10 weeks), whereas 8 QTL control GI–WSB weight differences at some time during ontogeny. Between 11 (early) and 12 (late) QTL contribute to growth rate differences between LG and SM; 11 QTL are responsible for growth rate variation among GI and WSB. In both cases, summed genotypic effects of QTL reveal mostly additive action, with some dominance toward the larger strain at early ages. The average additive effect of a weight QTL ranges from 0.05 g (1 week) to 0.60 g (10 weeks) for LG–SM and from 0.03 g (1 week) to 0.51 g (16 weeks) for GI–WSB. QTL effects vary throughout ontogeny, with different QTL shaping early and later growth rates. The allele from the larger strain increases body weight at most QTL in both crosses. The overall genetic response to selection for increased body size—whether selection is natural or artificial—has a common profile.
Why might rapid size evolution show similar properties in nature and in the laboratory? The agents of selection are fundamentally distinct. It also seems likely that artificial selection targeting body size is usually much stronger than natural selection. Perhaps the common profile instead reflects shared genetic, developmental, and evolutionary constraints on growth. Laboratory studies of random-bred mice revealed higher heritabilities for growth rate between 2 and 5 weeks of age than during other intervals, as well as reduced genetic correlations between early and late growth (Cheverud et al. 1983a,b; Riska et al. 1984). These observations predict that responses to selection for increased size will often accelerate early growth. Furthermore, the pervasive correlations between body size and other traits suggest that mutations affecting size will routinely have deleterious pleiotropic consequences. This logic could explain why mutations with small effects contribute most of the response to selection in nature and in the laboratory.
Although these comparisons reiterate the value of artificial selection experiments, understanding the genetics of size evolution in nature will continue to require the examination of wild populations. An important demonstration of this fact comes from the comparison of QTL location. Only 7 of the 19 QTL intervals we discovered contain estimated QTL positions from Cheverud et al. (1996). Although our judgment of QTL overlap is rough, this result suggests that the evolution of body size involved different genetic changes in GI mice and laboratory strains. This conclusion is bolstered by comparisons to other QTL known to affect body weight in mice (Dragani et al. 1995; Cheverud et al. 1996; Keightley et al. 1996; Rance et al. 1997; Brockmann et al. 1998, 2004; Kirkpatrick et al. 1998; Morris et al. 1999; Vaughn et al. 1999; Corva and Medrano 2001; Rocha et al. 2004; Bennett et al. 2005; Kenney-Hunt et al. 2006; Shao et al. 2007; Casellas et al. 2009; Mollah and Ishikawa 2011; Ishikawa and Okuno 2014). Although a few of these QTL overlap with those we identified, most do not. For example, a major X-linked QTL for body weight discovered from another artificial selection experiment (Liu et al. 2001; Oliver et al. 2005) is not found in our study.
Nevertheless, those QTL that are shared across strains and populations may be repeatedly targeted by selection and represent promising loci for identifying the causative mutations (Chan et al. 2012). A series of genetic mapping studies identified a large number of loci that contribute to weight differences between M. m. castaneus from the Philippines (a smaller form) and the C57BL/6 laboratory strain (primarily M. m. domesticus in origin) (Ishikawa et al. 2000; Ishikawa and Namikawa 2004; Mollah and Ishikawa 2010), including one in the region of the QTL we identified in the distal part of chromosome 10. Although the considerable divergence time between the two subspecies M. m. castaneus and M. m. domesticus makes it difficult to associate QTL with island evolution in this case, the resemblance between these results and ours is encouraging. Furthermore, a population genetic study that compared the genomes of laboratory mice artificially selected for divergent body sizes discovered genomic regions showing consistent signatures of adaptive evolution and unusual patterns of variation in large mice from the Faroe Islands (Chan et al. 2012). One of these genomic regions is located within the distal QTL on chromosome 10. Genetic characterization of mice from other islands will reveal whether shared patterns of size evolution across islands reflect a common genetic basis.
QTL mapped in crosses between a single pair of strains could represent genetic changes along either lineage. As a result, some QTL we identified might have contributed to size evolution in WSB or its ancestors rather than conferring phenotypic changes in GI mice. If the time to the most recent common ancestor of GI mice and WSB was earlier than the colonization time of Gough Island, using WSB as a reference strain could have exacerbated this problem. Although the mice used to found the WSB strain (which were caught in Maryland) were likely descended from Western European mice, determining the precise geographic origin of the GI mice will help to elucidate the dynamics of size evolution on the island.
Our results usefully focus the search for the genetic, developmental, and molecular mechanisms that ultimately underlie the evolution of body size in nature. GI mice evolved extreme size by accelerating growth during the first 6 weeks of life. The observation that GI mice are heavier at 1 week of age suggests that accelerated growth begins in utero or soon after birth. In addition to eliminating alternative evolutionary scenarios—including the notion that size was increased by extending the growth trajectory—our results suggest that natural selection on size or size-correlated traits targeted early development. This research is a first, necessary, and foundational step toward pinpointing the genetic variants responsible for increased size in GI mice. Identifying the causative genes and mutations will allow several intriguing evolutionary questions to be answered. Did selection on Gough Island target standing variants or new mutations? Do the causative loci show signatures of adaptive evolution predicted by strong selection on individual loci or by selection spread across many genes?
How did the genetic changes that increased body size in GI mice evolve? Rapid phenotypic evolution in a novel environment suggests natural selection as the primary evolutionary force. The observed bias toward GI mouse alleles increasing body size provides modest support for this conclusion. Differences in the environments experienced by mice on Gough Island and on the mainland are potential selective agents. Mice on the island may experience significantly colder temperatures because they live outside year round (in contrast to mice on the mainland that often live indoors). It is possible that the availability of an energetically rich food resource in the winter period (albatross and petrel chicks) and the predatory behavior of mice to exploit this resource (Jones et al. 2003; Cuthbert and Hilton 2004; Wanless et al. 2007) were important selective factors in the rapid evolution of body size. The size increase of GI mice also could have been catalyzed by the removal of selective constraints. GI mice have few natural predators or interspecific competitors (Verrill 1895; Heaney and Holdgate 1957; Jones et al. 2003; Wanless et al. 2009). Both reduced predation and reduced competition have been invoked to explain the tendency of small mammals to grow larger on islands (Sondaar 1977; Heaney 1978; Lawlor 1982), and recent comparative work supports this conclusion (Lomolino et al. 2012). In principle, GI mice could have achieved their unusual body size without natural selection. Perhaps the evolution of body size was enabled by stronger genetic drift resulting from the reduction in population size that accompanied island colonization (Gray et al. 2014). Understanding the relative contributions of different evolutionary processes to extreme body size evolution in mice on Gough Island will ultimately require additional field studies.
Acknowledgments
We thank Trevor Glass and the administrator of Tristan da Cunha for permission to live capture and remove mice from Gough Island and Henk Louw and Paul Visser for capturing mice. We thank Lauren Brooks for sharing her unpublished compilation of body weights for mice from different islands. We thank Hannah Buchanan, Valeri Lapacek, Spencer Compton, Arielle Henderson, Elizabeth Linder, Lauren Hennelly, and Lauren Agnew for their devotion to mouse husbandry and phenotyping. We acknowledge staff at the Charmany Instructional Facility for assistance with all aspects of mouse care. We thank Mark Nolte for useful comments on the manuscript. Mark Nolte, Alan Attie, and Mark Keller provided helpful guidance about genetic mechanisms of mouse growth. We acknowledge Chris Vinyard for sharing ideas about the connection between fitness and body size. We thank Peicheng Jing for computational support. We appreciate enthusiasm and encouragement from Payseur lab members, including Mark Nolte, Beth Dumont, Michael White, Ryan Haasl, Jenny Andrie, Leslie Turner, John Hvala, Richard Wang, Lauren Brooks, April Peterson, and Peicheng Jing, throughout the course of this project. We thank Leonie Moyle and two anonymous reviewers for their constructive comments on the manuscript. The research was supported by National Institutes of Health (NIH) R01 GM100426A to B.A.P., NIH National Research Service Award 1F32GM090685 to M.M.G., and a National Science Foundation graduate research fellowship to M.D.P.
Footnotes
Communicating editor: L. C. Moyle
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177790/-/DC1.
Literature Cited
- Adler G. H., Levins R., 1994. The island syndrome in rodent populations. Q. Rev. Biol. 69: 473–490. [DOI] [PubMed] [Google Scholar]
- Atchley W. R., Wei R., Crenshaw P., 2000. Cellular consequences in the brain and liver of age-specific selection for rate of development in mice. Genetics 155: 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylor D. L., Valdar W., Foulds-Mathes W., Buus R. J., Verdugo R. A., et al. , 2011. Genetic analysis of complex traits in the emerging Collaborative Cross. Genome Res. 21: 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis, W. D., 1994 The power and deceit of QTL experiments: lessons from comparative QTL studies. In: Proceedings of the Forty-Ninth Annual Corn and Sorghum Industry Research Conference, Washington, D.C., pp. 250–266. [Google Scholar]
- Beavis, W. D., 1998 QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by A. H. Paterson. CRC Press, Boca Raton, FL. [Google Scholar]
- Bennett B., Carosone-Link P. J., Lu L., Chesler E. J., Johnson T. E., 2005. Genetics of body weight in the LXS recombinant inbred mouse strains. Mamm. Genome 16: 764–774. [DOI] [PubMed] [Google Scholar]
- Berry R. J., 1964. The evolution of an island population of the house mouse. Evolution 18: 468–483. [Google Scholar]
- Berry R. J., 1965. Genetical change in an island mouse population. Heredity 20: 651. [Google Scholar]
- Berry R. J., 1968. The ecology of an island population of house mouse. J. Anim. Ecol. 37: 445–470. [Google Scholar]
- Berry R. J., 1996. Small mammal differentiation on islands. Philos. Trans. R. Soc. B 351: 753–764. [DOI] [PubMed] [Google Scholar]
- Berry R. J., Jakobson M. E., 1975. Ecological genetics of an island population of house mouse (Mus-musculus). J. Zool. 175: 523–540. [Google Scholar]
- Berry R. J., Scriven P. N., 2005. The house mouse: a model and motor for evolutionary understanding. Biol. J. Linn. Soc. Lond. 84: 335–347. [Google Scholar]
- Berry R. J., Jakobson M. E., Peters J., 1978a The house mice of the Faroe Islands: a study in microdifferentiation. J. Zool. 185: 573–592. [Google Scholar]
- Berry R. J., Peters J., Van Aarde R. J., 1978b Sub-antarctic house mice: colonization, survival and selection. J. Zool. 184: 127–141. [Google Scholar]
- Berry R. J., Bonner W. N., Peters J., 1979. Natural-selection in house mice (Mus-musculus) from South Georgia (South-Atlantic Ocean). J. Zool. 189: 385–398. [Google Scholar]
- Berry R. J., Cuthbert A., Peters J., 1982. Colonization by house mice: an experiment. J. Zool. 198: 329–336. [Google Scholar]
- Berry R. J., Jakobson M. E., Peters J., 1987. Inherited differences within an island population of the house mouse (Mus-domesticus). J. Zool. (Lond.) 211: 605–618. [Google Scholar]
- Brockmann G. A., Haley C. S., Renne U., Knott S. A., Schwerin M., 1998. Quantitative trait loci affecting body weight and fatness from a mouse line selected for extreme high growth. Genetics 150: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann G. A., Karatayli E., Haley C. S., Renne U., Rottmann O. J., et al. , 2004. QTLs for pre- and postweaning body weight and body composition in selected mice. Mamm. Genome 15: 593–609. [DOI] [PubMed] [Google Scholar]
- Broman K. W., Speed T. P., 2002. A model selection approach for the identification of quantitative trait loci in experimental crosses. J. R. Stat. Soc., B 64: 641–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., Sen S., Owens S. E., Manichaikul A., Southard-Smith E. M., et al. , 2006. The X chromosome in quantitative trait locus mapping. Genetics 174: 2151–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L., Cardillo M., 2007. Primates follow the “island rule”: implications for interpreting Homo floresiensis. Biol. Lett. 3: 398–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bünger L., Hill W. G., 1999. Inbred lines of mice derived from long-term divergent selection on fat content and body weight. Mamm. Genome 10: 645–648. [DOI] [PubMed] [Google Scholar]
- Calder, W. A., 1984 Size, Function, and Life History. Harvard University Press, Cambridge, MA. [Google Scholar]
- Carter T. C., Falconer D. S., 1951. Stocks for detecting linkage in the mouse, and the theory of their design. J. Genet. 50: 307–323. [DOI] [PubMed] [Google Scholar]
- Case T. J., 1978. General explanation for insular body size trends in terrestrial vertebrates. Ecology 59: 1–18. [Google Scholar]
- Casellas J., Farber C. R., Gularte R. J., Haus K. A., Warden C. H., et al. , 2009. Evidence of maternal QTL affecting growth and obesity in adult mice. Mamm. Genome 20: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C., 1956a Analysis of quantitative inheritance of body size in mice. I. Hybridization and maternal influence. Genetics 41: 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C., 1956b Analysis of quantitative inheritance of body size in mice. II. Gene action and segregation. Genetics 41: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. F., Marks M. E., Jones F. C., Villarreal G., Shapiro M. D. et al, 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y. F., Jones F. C., McConnell E., Bryk J., Bünger L., et al. , 2012. Parallel selection mapping using artificially selected mice reveals body weight control loci. Curr. Biol. 22: 794–800. [DOI] [PubMed] [Google Scholar]
- Cheverud J. M., Rutledge J. J., Atchley W. R., 1983a Quantitative genetics of development: genetic correlations among age-specific trait values and the evolution of ontogeny. Evolution 37: 895–905. [DOI] [PubMed] [Google Scholar]
- Cheverud J. M., Leamy L., Rutledge J., Atchley W., 1983b Quantitative genetics and the evolution of ontogeny. I. Ontogenetic changes in quantitative genetic variance components in randombred mice. Genet. Res. 42: 65–75. [DOI] [PubMed] [Google Scholar]
- Cheverud J. M., Routman E. J., Duarte F. A. M., Van Swinderen B., Cothran K., et al. , 1996. Quantitative trait loci for murine growth. Genetics 142: 1305–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. M., 2005. Genetics of growth in the mouse, pp. 113–130 in The Mouse in Animal Genetics and Breeding Research, edited by Eisen E. J. Imperial College Press, Singapore. [Google Scholar]
- Churchill G. A., Doerge R. W., 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G. A., Airey D. C., Allayee H., Angel J. M., Attie A. D., et al. , 2004. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137. [DOI] [PubMed] [Google Scholar]
- Colosimo P. F., Hosemann K. E., Balabhadra S., Villarreal G., Dickson M., et al. , 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Corva P. M., Medrano J. F., 2001. Quantitative trait loci (QTLs) mapping for growth traits in the mouse: a review. Genet. Sel. Evol. 33: 105–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A., Ackert-Bicknell C., Dumont B. L., Ding Y., Tzenova Bell J., et al. , 2009. A new standard genetic map for the mouse. Genetics 182: 1335–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert R., Hilton G., 2004. Introduced house mice Mus musculus: A significant predator of threatened and endemic birds on Gough Island, South Atlantic Ocean? Biol. Conserv. 117: 483–489. [Google Scholar]
- Damuth J., MacFadden B. J., 1990. Body Size in Mammalian Paleobiology, Cambridge University Press, New York. [Google Scholar]
- Didion J. P., Morgan A. P., Clayshulte A. M.-F., Mcmullan R. C., Yadgary L., et al. , 2015. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet. 11: e1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragani T. A., Zeng Z. B., Canzian F., Gariboldi M., Ghilarducci M. T., et al. , 1995. Mapping of body weight loci on mouse Chromosome X. Mamm. Genome 6: 778–781. [DOI] [PubMed] [Google Scholar]
- Eisen E. J., 1989. Selection experiments for body composition in mice and rats: a review. Livest. Prod. Sci. 23: 17–32. [Google Scholar]
- Endler J. A., 1986. Natural Selection in the Wild, Princeton University Press, Princeton, NJ. [Google Scholar]
- Falconer D. S., 1953. Selection for large and small size in mice. Genetics 51: 470–501. [Google Scholar]
- Festing M. F. W., 1996. Origins and characteristics of inbred strains of mice, Genetic Variants and Strains of the Laboratory Mouse, edited by Lyon M. F., Raston S., Brown S. D. M. Oxford University Press, New York. [Google Scholar]
- Foster J. B., 1964. The evolution of mammals on islands. Nature 202: 234–235. [Google Scholar]
- Fowden A. L., 2003. The insulin-like growth factors and feto-placental growth. Placenta 24: 803–812. [DOI] [PubMed] [Google Scholar]
- Freudenthal M., 1972. Deinogalerix koenigswaldi nov. gen., nov. spec., a giant insectivore from the Neogene of Italy. Scr. Geol. 14: 1–19. [Google Scholar]
- Goodale H., 1938. A study of the inheritance of body weight in the albino mouse by selection. J. Hered. 29: 101–112. [Google Scholar]
- Gray M. M., Wegmann D., Haasl R. J., White M. A., Gabriel S. I., et al. , 2014. Demographic history of a recent invasion of house mice on the isolated Island of Gough. Mol. Ecol. 23: 1923–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley C. S., Knott S. A., 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69: 315–324. [DOI] [PubMed] [Google Scholar]
- Heaney J. T., Holdgate M. W., 1957. The Gough Island scientific survey. Geogr. J. 123: 20–31. [Google Scholar]
- Heaney L. R., 1978. Island area and body size of insular mammals: evidence from the tri-colored squirrel (Callosciurus prevosti) of Southeast Asia. Evolution 32: 29–44. [DOI] [PubMed] [Google Scholar]
- Hoekstra H. E., Hirschmann R. J., Bundey R. A., Insel P. A., Crossland J. P., 2006. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science 313: 101–104. [DOI] [PubMed] [Google Scholar]
- Ishikawa A., Matsuda Y., Namikawa T., 2000. Detection of quantitative trait loci for body weight at 10 weeks from Philippine wild mice. Mamm. Genome 11: 824–830. [DOI] [PubMed] [Google Scholar]
- Ishikawa A., Namikawa T., 2004. Mapping major quantitative trait loci for postnatal growth in an intersubspecific backcross between C57BL/6J and Philippine wild mice by using principal component analysis. Genes Genet. Syst. 79: 27–39. [DOI] [PubMed] [Google Scholar]
- Ishikawa A., Okuno S., 2014. Fine mapping and candidate gene search of quantitative trait loci for growth and obesity using mouse intersubspecific subcongenic intercrosses and exome sequencing. PLoS One 9: e113233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. G., Chown S. L., Gaston K. J., 2003. Introduced house mice as a conservation concern on Gough Island. Biodivers. Conserv. 12: 2107–2119. [Google Scholar]
- Jones F. C., Grabherr M. G., Chan Y. F., Russell P., Mauceli E. et al, 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane T. M., Goodstadt L., Danecek P., White M. A., Wong K., et al. , 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Hardge T., May L., Bulfield G., 1996. A genetic map of quantitative trait loci for body weight in the mouse. Genetics 142: 227–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney-Hunt J. P., Vaughn T. T., Pletscher L. S., Peripato A., Routman E., et al. , 2006. Quantitative trait loci for body size components in mice. Mamm. Genome 17: 526–537. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick B. W., Mengelt A., Schulman N., Martin I. C., 1998. Identification of quantitative trait loci for prolificacy and growth in mice. Mamm. Genome 9: 97–102. [DOI] [PubMed] [Google Scholar]
- Kramer M. G., Vaughan T. T., Pletscher L. S., King-Ellison K., Adams E., et al. , 1998. Genetic variation in body weight gain and composition in the intercross of Large (LG/J) and Small (SM/J) inbred strains of mice. Genet. Mol. Biol. 21: 211–218. [Google Scholar]
- Kwak I.-Y., Moore C. R., Spalding E. P., Broman K. W., 2014. A simple regression-based method to map quantitative trait loci underlying function-valued phenotypes. Genetics 197: 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor T. E., 1982. The evolution of body size in mammals: evidence from insular populations in Mexico. Am. Nat. 119: 54–72. [Google Scholar]
- Liu X. J., Oliver F., Brown S. D. M., Denny P., Keightley P. D., 2001. High-resolution quantitative trait locus mapping for body weight in mice by recombinant progeny testing. Genet. Res. 77: 191–197. [DOI] [PubMed] [Google Scholar]
- Lomolino M. V., 1985. Body size of mammals on islands: the island rule reexamined. Am. Nat. 125: 310–316. [Google Scholar]
- Lomolino M. V., 2005. Body size evolution in insular vertebrates: generality of the island rule. J. Biogeogr. 32: 1683–1699. [Google Scholar]
- Lomolino M. V., Riddle B. R., Brown J. H., 2005. Biogeography, Sinauer Associates, Sunderland, MA. [Google Scholar]
- Lomolino M. V., Sax D. F., Palombo M. R., van der Geer A. A., 2012. Of mice and mammoths: evaluations of causal explanations for body size evolution in insular mammals. J. Biogeogr. 39: 842–854. [Google Scholar]
- Losos J. B., Ricklefs R. E., 2009. Adaptation and diversification on islands. Nature 457: 830–836. [DOI] [PubMed] [Google Scholar]
- Lui J. C., Baron J., 2011. Mechanisms limiting body growth in mammals. Endocr. Rev. 32: 422–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F., Terwilliger J. D., Lee K., Segre G. V., Efstatiadis A., 2001. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev. Biol. 229: 141–162. [DOI] [PubMed] [Google Scholar]
- Lynch C., 1992. Clinal variation in cold adaptation in Mus domesticus: verification of predictions from laboratory populations. Am. Nat. 139: 1219–1236. [Google Scholar]
- MacArthur J., 1944. Genetics of body size and related characters. I. Selection of small and large races of the laboratory mouse. Am. Nat. 78: 142–157. [Google Scholar]
- Meiri S., Dayan T., Simberloff D., 2004. Body size of insular carnivores: little support for the island rule. Am. Nat. 163: E469–E479. [DOI] [PubMed] [Google Scholar]
- Meiri S., Cooper N., Purvis A., 2008. The island rule: Made to be broken? Proc. Biol. Sci. 275: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiri S., Raia P., Phillimore A. B., 2011. Slaying dragons: limited evidence for unusual body size evolution on islands. J. Biogeogr. 38: 89–100. [Google Scholar]
- Millien V., 2006. Morphological evolution is accelerated among island mammals. PLoS Biol. 4: 1863–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollah B. R., Ishikawa A., 2010. A wild derived quantitative trait locus on mouse chromosome 2 prevents obesity. BMC Genet. 11: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollah M. B. R., Ishikawa A., 2011. Intersubspecific subcongenic mouse strain analysis reveals closely linked QTLs with opposite effects on body weight. Mamm. Genome 22: 282–289. [DOI] [PubMed] [Google Scholar]
- Moncunill-Solé B., Jordana X., Marín-Moratalla N., Moyà-Solà S., Köhler M., 2014. How large are the extinct giant insular rodents? New body mass estimations from teeth and bones. Integr. Zool. 9: 197–212. [DOI] [PubMed] [Google Scholar]
- Morris K. H., Ishikawa A., Keightley P. D., 1999. Quantitative trait loci for growth traits in C57BL/6J x DBA/2J mice. Mamm. Genome 10: 225–228. [DOI] [PubMed] [Google Scholar]
- Nachman M. W., Hoekstra H. E., D’Agostino S. L., 2003. The genetic basis of adaptive melanism in pocket mice. Proc. Natl. Acad. Sci. USA 100: 5268–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver F., Christians J. K., Liu X., Rhind S., Verma V., et al. , 2005. Regulatory variation at glypican-3 underlies a major growth QTL in mice. PLoS Biol. 3: e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pergams O. R. W., Ashley M. V., 2001. Microevolution in island rodents. Genetica 112: 245–256. [PubMed] [Google Scholar]
- Peters R. H., 1983. The Ecological Implications of Body Size, Cambridge University Press, Cambridge. [Google Scholar]
- Prasad K. V. S. K., Song B.-H., Olson-Manning C., Anderson J. T., Lee C.-R., et al. , 2012. A gain-of-function polymorphism controlling complex traits and fitness in nature. Science 337: 1081–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao N., Huang Y., Naveed H., Green C. D., Han J.-D. J., 2013. CoCiter: an efficient tool to infer gene function by assessing the significance of literature co-citation. PLoS One 8: e74074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raia P., Meiri S., 2006. The island rule in large mammals: paleontology meets ecology. Evolution 60: 1731–1742. [PubMed] [Google Scholar]
- Rance K. A., Hill W. G., Keightley P. D., 1997. Mapping quantitative trait loci for body weight on the X chromosome in mice. I. Analysis of a reciprocal F2 population. Genet. Res. 70: 117–124. [DOI] [PubMed] [Google Scholar]
- Rebeiz M., Pool J. E., Kassner V. A., Aquadro C. F., Carroll S. B., 2009. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326: 1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D. R., Lawler M. P., Tordoff M. G., 2008. Reduced body weight is a common effect of gene knockout in mice. BMC Genet. 9: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riska B., Atchley W. R., Rutledge J. J., 1984. A genetic analysis of targeted growth in mice. Genetics 107: 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riska B., Atchley W. R., 1985. Genetics of growth predict patterns of brain-size evolution. Science 229: 668–671. [DOI] [PubMed] [Google Scholar]
- Rocha J. L., Eisen E. J., Van Vleck L. D., Pomp D., 2004. A large-sample QTL study in mice: I. Growth. Mamm. Genome 15: 83–99. [DOI] [PubMed] [Google Scholar]
- Rockman M. V., 2012. The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth V. L., 1992. Inferences from allometry and fossils: dwarfing of elephants on islands. Oxford Surv. Evol. Biol. 8: 259–288. [Google Scholar]
- Rowe-Rowe D. T., Crafford J. E., 1992. Density, body size, and reproduction of feral house mice on Gough Island. S. Afr. J. Zool. 27: 1–5. [Google Scholar]
- Salcedo T., Geraldes A., Nachman M. W., 2007. Nucleotide variation in wild and inbred mice. Genetics 177: 2277–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Nielsen K., 1984. Scaling: Why Is Animal Size so Important? University of Cambridge Press, Cambridge. [Google Scholar]
- Shao H., Reed D. R., Tordoff M. G., 2007. Genetic loci affecting body weight and fatness in a C57BL/6J x PWK/PhJ mouse intercross. Mamm. Genome 18: 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver L. M., 1995. Mouse Genetics: Concepts and Applications. Oxford University Press, New York. [Google Scholar]
- Sondaar P. Y., 1977. Insularity and its effects on mammal evolution, pp. 671–707 in Major Patterns of Vertebrate Evolution, edited by Hecht M. K., Goody P. C., Hecht B. M. Plenum Press, New York. [Google Scholar]
- Stock C., 1935. Exiled elephants of the Channel Islands. Sci. Mon. 41: 205–214. [Google Scholar]
- Svenson K. L., Gatti D. M., Valdar W., Welsh C. E., Cheng R., et al. , 2012. High-resolution genetic mapping using the Mouse Diversity outbred population. Genetics 190: 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D. W., Churchill G. A., 2012. Ten years of the Collaborative Cross. Genetics 190: 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valen L., 1973. Pattern and the balance of nature. Evol. Theory 1: 31–49. [Google Scholar]
- van’t Hof A. E., Edmonds N., Dalíková M., Marec F., Saccheri I. J., 2011. Industrial melanism in British peppered moths has a singular and recent mutational origin. Science 332: 958–960. [DOI] [PubMed] [Google Scholar]
- Vaughn T. T., Pletscher L. S., Peripato A., King-Ellison K., Adams E., et al. , 1999. Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genet. Res. 74: 313–322. [DOI] [PubMed] [Google Scholar]
- Verrill G. E., 1895. Notes on birds and eggs from the islands of Gough, Kerguelen, and South Georgia. Trans. Connect. Acad. 11: 429–480. [Google Scholar]
- Wace N. M., 1961. Vegetation of Gough Island. Ecol. Monogr. 31: 337–367. [Google Scholar]
- Wanless R. M., Angel A., Cuthbert R. J., Hilton G. M., Ryan P. G., 2007. Can predation by invasive mice drive seabird extinctions? Biol. Lett. 3: 241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanless R. M., Ryan P. G., Altwegg R., Angel A., Cooper J., et al. , 2009. From both sides: dire demographic consequences of carnivorous mice and longlining for the critically endangered Tristan albatrosses on Gough Island. Biol. Conserv. 142: 1710–1718. [Google Scholar]
- Zeng Z. B., Liu J., Stam L. F., Kao C. H., Mercer J. M., et al. , 2000. Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154: 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Phenotypes and genotypes from this study are available from the QTL Archive at the Jackson Laboratory, at http://phenome.jax.org/db/q?rtn=projects/projdet&reqprojid=539. Supplemental information includes: p-values from strain comparisons (Figure S1), histograms of F2 weights (Figure S2), average F2 weights and growth rates (Figure S3), p-values from comparisons among crosses and mothers (Figure S4), genetic and physical maps of markers used in QTL analyses (Figure S5), F2 frequency of WSB allele (Figure S6), and candidate genes in QTL regions (Table S1).