Abstract
Plant breeding populations exhibit varying levels of structure and admixture; these features are likely to induce heterogeneity of marker effects across subpopulations. Traditionally, structure has been dealt with as a potential confounder, and various methods exist to “correct” for population stratification. However, these methods induce a mean correction that does not account for heterogeneity of marker effects. The animal breeding literature offers a few recent studies that consider modeling genetic heterogeneity in multibreed data, using multivariate models. However, these methods have received little attention in plant breeding where population structure can have different forms. In this article we address the problem of analyzing data from heterogeneous plant breeding populations, using three approaches: (a) a model that ignores population structure [A-genome-based best linear unbiased prediction (A-GBLUP)], (b) a stratified (i.e., within-group) analysis (W-GBLUP), and (c) a multivariate approach that uses multigroup data and accounts for heterogeneity (MG-GBLUP). The performance of the three models was assessed on three different data sets: a diversity panel of rice (Oryza sativa), a maize (Zea mays L.) half-sib panel, and a wheat (Triticum aestivum L.) data set that originated from plant breeding programs. The estimated genomic correlations between subpopulations varied from null to moderate, depending on the genetic distance between subpopulations and traits. Our assessment of prediction accuracy features cases where ignoring population structure leads to a parsimonious more powerful model as well as others where the multivariate and stratified approaches have higher predictive power. In general, the multivariate approach appeared slightly more robust than either the A- or the W-GBLUP.
Keywords: genomic selection, multivariate models, GBLUP, plant breeding, GenPred, population structure, shared data resource
THE impact of population structure has been investigated for genome-wide association studies (GWAS) and genome-based prediction, where it was found to lead to false positive detected quantitative trait loci (QTL) and to have important influences on prediction accuracy (Windhausen et al. 2012; Albrecht et al. 2014; Guo et al. 2014). Population structure occurs naturally in animal, plant, and human populations due to geographic adaptation and natural selection. Especially in plant breeding programs substructure is generated between breeding groups or programs due to artificial selection and drift. Additionally in hybrid breeding, lines of different or same subpopulations may be crossed to different testers, which further complicates genome-based prediction tasks (Albrecht et al. 2014). Thus, structure and/or admixture are ubiquitous in plant breeding populations.
A common approach to account for population structure has been to include marker-derived principal components into GWAS (Price et al. 2006) or into genome-based best linear unbiased prediction (GBLUP) models (Yang et al. 2010; Janss et al. 2012). However, inclusion of principal components induces a mean correction that does not account for the fact that marker effects may be different across populations (de los Campos and Sorensen 2014). From a classical quantitative genetics theory perspective it is reasonable to expect that allele substitution effects may vary between populations due to, for example, differences in allele frequency (Falconer and Mackay 1996). Additionally, even when QTL allele substitution effects are constant across subpopulations, marker effects may vary between subpopulations due to differences in marker–QTL linkage disequilibrium (LD) patterns. Therefore, there are sound theoretical reasons to believe marker effects are different between subpopulations. This brings up the question of how data from structured populations should be dealt with. So far, less attention was paid to this issue in the plant breeding genomic selection literature.
Stratified analysis
To account for heterogeneity of marker effects across subpopulations, they can simply be estimated within each population separately. However, this reduces sample size and therefore the accuracy of estimated marker effects. If marker effects are correlated across subpopulations, data of each subpopulation provide information for the estimation of marker effects of the correlated populations. This borrowing of information between subpopulations can be achieved by using data from multiple subpopulations in a combined data analysis.
Combined analysis assuming constant effects across subpopulations
The simplest approach for a combined analysis consists of assuming that marker effects are constant across populations. This has been used, for instance, in dairy cattle where it has been suggested that combining data from different breeds in the training set for genome-based prediction may increase prediction accuracy, especially for small breeds (De Roos et al. 2009). However, results on experimental data have not shown a clear advantage of combining different breeds over prediction within breeds (Brøndum et al. 2011; Erbe et al. 2012), perhaps suggesting that the assumption of constant marker effects across subpopulations may be too strong
Multivariate approach
An intermediate approach between the two extremes discussed above can be obtained by using multivariate models where marker effects are allowed to be different but correlated across subpopulations. This approach has been considered in animal breeding applications involving multibreed analysis where it did not lead to a consistent improvement of prediction performance (Karoui et al. 2012; Olson et al. 2012; Makgahlela et al. 2013). However, less effort has been made to investigate the impact of population structure on estimation of marker effects in the context of genome-based prediction in plant breeding. While in animal breeding large clearly separated breeds exist, in plant breeding population structure can have very different forms and origins, including combined analysis of data from multiple, connected, breeding programs, diversity panels, and differently structured mating designs that lead to various forms of multiparental populations.
For genome-based prediction in multiple biparental maize families, Schulz-Streeck et al. (2012) proposed to extend the standard genomic prediction model that assumes homogeneous marker effects with interactions between markers and subpopulations. Using this approach the authors reported very similar prediction accuracies for the model including interaction effects and the model including only main effects. However, the approach adopted in the study of Schulz-Streeck et al. (2012) imposed important restrictions on the within- and between-subpopulation covariance parameters as no specific covariance for each pair of subpopulations is modeled. This imposes, for instance, a constant genomic variance within clusters and constant covariance (and hence correlation) between groups. A more general approach can be obtained by implementing multivariate models with unrestricted covariance matrices. Therefore, in this article we consider a multivariate Gaussian model with an unstructured covariance matrix for genomic effects. The model yields estimates of covariance parameters that can be used to quantify genetic correlations between subpopulations. Furthermore, it provides predictions of breeding values and of marker effects that can be used to predict the genetic merit of each individual when mated with randomly chosen individuals of any of the subpopulations. We implement the model in a Bayesian framework and apply this multivariate approach to three plant data sets that have different degrees of genetic differentiation between groups. The first data set is a rice (Oryza sativa) diversity panel that contains four subpopulations from different origins (Zhao et al. 2011); the materials in the different origins are fairly distant from a genetic standpoint. Therefore, results from this data set are relevant for cases where the researcher considers whether to analyze jointly or separately materials of highly differentiated origins. The second data set originates from the Centro Internacional de Mejoramiento de Maíz y Trigo (CIMMYT) wheat (Triticum aestivum L.) breeding program (Crossa et al. 2010) and contains 1279 wheat lines that can be clustered into two subpopulations. The two subpopulations have significant levels of genetic differentiation but are not as differentiated as in the case of the rice data set. Results from this data set are relevant for cases when one considers whether to analyze jointly or separately data from different breeding programs that maintain some level of exchange of genetic materials. This situation can be encountered in organizations such as CIMMYT or large corporations that have more than one breeding program. Finally, our third data set is a half-sib maize (Zea mays L.) panel including 10 biparental families with one common parent (Lehermeier et al. 2014). In all three data sets we compare the multivariate approach with the stratified analysis and with a combined analysis that assumes homogeneity of marker effects.
The multivariate approach allows estimating genomic correlations between subpopulations for each trait; these parameters can be used as a trait-specific measure of genetic similarity between subpopulations. We analyzed a diverse array of situations, including very distantly related and more closely related subpopulations, and, although we found an association between genomic correlations and genetic distance, we also found important differences between traits, suggesting that the degree of similarity in the underlying genetic architecture between subpopulations varies between traits. In terms of prediction accuracy, our results indicate that no single method is best across traits and subpopulations. Importantly, the multivariate method either was the best-performing method or, more often, tended to perform close to the best method.
Materials and Methods
We begin this section with a description of three methods that can be used for the analysis of data from structured populations. This is followed by a description of the data used and the data analysis conducted.
Statistical methods
Assume we have phenotypes and genotypes for p markers collected at n individuals, and suppose that these individuals can be clustered into K genetic groups each of size nk Our objective is to estimate marker effects and functions thereof, and to this end we consider three possible approaches: the first approach is whole-genome regression assuming that marker effects are constant across groups (A-GBLUP). This approach uses all available data; however, estimates of marker effects might be inconsistent because they are forced to be identical for each subpopulation. A second possible approach (W-GBLUP, a “stratified analysis”) consists of regressing phenotypes on genotypes within groups. This allows for population-specific marker effects. However, a stratified analysis reduces sample size, which reduces the precision of estimates. A third approach (MG-GBLUP) would be to regress phenotypes on markers, using multivariate methods that allow for population-specific marker effects that can be correlated between subpopulations. This approach combines desirable features of A-GBLUP and W-GBLUP: it allows for subpopulation-specific marker effects and, at the same time, exploits the full sample size by borrowing of information between subpopulations.
A-GBLUP (regression assuming homogeneity):
A-GBLUP is defined assuming that data come from a homogeneous population; therefore this model assumes common marker effects for all subpopulations; that is,
where yk (k = 1,…, K) is the nk-dimensional vector of phenotypic values of subpopulation k, Xk (k = 1,…, K) is the -dimensional matrix of genotype scores of subpopulation k, µk (k = 1,…, K) are subpopulation-specific intercepts, is the p-dimensional vector of marker effects common for all K subpopulations, and is an nk-dimensional vector of residuals belonging to subpopulation k. Marker genotypes were centered and standardized by subtracting from the original genotype codes the sample mean and dividing by the sample standard deviation. In A-GBLUP, residuals are assumed to be uncorrelated within subpopulations as well as across subpopulations. Further, they are assumed to follow a normal distribution with mean 0 and subpopulation-specific variance In a Bayesian context different prior settings have been proposed for the estimation of the marker effects (de los Campos et al. 2013). We concentrate here on the Ridge regression model, where marker effects are assumed to follow a conditional normal distribution with common variance for all effects (j = 1,…, p): This model can be reformed in the analogous GBLUP model,
where gk (k = 1,…, K) is the nk-dimensional vector of genomic effects of subpopulation k. Here, the complete vector is assumed to follow where is a genomic relationship matrix with X as the complete marker matrix defined as before and is a genomic variance. For we assign a scaled inverse chi-square prior distribution with df1 degrees of freedom and scale parameter S1: The other variables are defined as above. For the residual variances also a scaled inverse chi-square prior distribution was used:
W-GBLUP (stratified analysis):
W-GBLUP estimates marker effects within each subpopulation k separately:
Here, and are defined as before, and is the p-dimensional vector of marker effects specific for subpopulation k. As in A-GBLUP, marker effects and model residuals are assumed to be normally distributed. Model residuals are assumed to be uncorrelated within and across subpopulations as in A-GBLUP. Similarly, for the fully stratified analysis also marker effects are assumed to be uncorrelated across subpopulations; therefore,
As with A-GBLUP, the stratified model can be parameterized as a GBLUP model, but here the vectors of genomic effects for each subpopulation are assumed to follow different independent normal distributions, where is a genomic relationship matrix describing realized relationships at markers among individuals of the kth subpopulation and is a genomic variance parameter. As in A-GBLUP, scaled inverse chi-square prior distributions are assigned to the variances and to the residual variances,
MG-GBLUP (multivariate analysis):
The multivariate model estimates population-specific marker effects like the stratified model; however, the multivariate analysis allows for correlations of effects between groups. The data equations are
As with the stratified analysis, residuals are assumed to be uncorrelated and to follow a normal distribution with subpopulation-specific residual variances Subpopulation-specific residual variance components are assigned the same scaled inverse chi-square prior distributions for all k=1,…, K: The joint prior distribution for the complete vector of residuals becomes
In the multivariate model, marker effects are assumed to follow a multivariate normal distribution with here, ⊗ denotes the Kronecker product, and B represents a within-marker covariance matrix of effects,
where (j = 1,…, K; k = 1,…, K; j ≠ k) is the covariance between marker effects of subpopulations j and k. This model can also be parameterized as a GBLUP model. Here we need to form an augmented -dimensional vector containing the genomic values of each individual in each group; that is, here where is a matrix containing all the genotypes in the sample. Therefore, the augmented vector of genomic values can be expressed as Because is a linear combination of a vector of marker effects that follows a MVN distribution, itself follows a MVN distribution, and the mean and covariance matrices of this distribution are and respectively. Therefore where is the genomic variance–covariance matrix among subpopulations:
Only some of the entries of are linked to phenotypes; therefore,
where are matrices linking phenotypes with the entries of each of these matrices has rows and n columns. To complete the Bayesian model an inverse-Wishart prior distribution is assigned to with scale matrix Ψ and degrees of freedom v.
It is worth noting that the W-GBLUP and A-GBLUP models can be seen as special cases of the MG-GBLUP. Indeed, the W-GBLUP and MG-GBLUP models are equivalent when is a diagonal matrix. And the MG-GBLUP and A-GBLUP become equivalent when all entries of are equal; that is, this corresponds to constant genomic variance and unit correlations between subpopulations. In the MG-GBLUP all those parameters are estimated from data; therefore, in principle, the MG-GBLUP has potential for accommodating all possible situations, from the W-GBLUP to the A-GBLUP and situations in between.
Data Sets
We investigated the three models described above using three differently structured data sets, which are described in the following subsections.
The rice data set:
This data set comes from a worldwide collected diversity panel of 413 inbred accessions of O. sativa (Zhao et al. 2011), which is publicly available under http://www.ricediversity.org/data/ and has been investigated for genome-based prediction before (Wimmer et al. 2013; Isidro et al. 2014). Zhao et al. (2011) classified the rice lines in five subpopulations [AUS, Indica (IND), Temperate Japonica (TEJ), Tropical Japonica (TRJ), and Aromatic] and one admixed group of lines that could not be assigned to a specific group. For our analyses we excluded the 14 lines of the smallest subgroup aromatic and the 62 admixed lines that could not be assigned to specific groups. The rice varieties were genotyped with an Affymetrix 44K single-nucleotide polymorphism (SNP) chip. After quality control and imputation of missing values by Zhao et al. (2011) and Wimmer et al. (2013), 36,858 polymorphic SNPs were available for the 312 lines used in this study. The diversity panel was phenotyped for 34 traits; in our study we concentrate on the traits plant height, flowering time, and panicle length, which were also analyzed in Wimmer et al. (2013).
The maize data set:
The maize data set was recently analyzed and published by Bauer et al. (2013) and Lehermeier et al. (2014). In this study we concentrate on the 841 phenotyped and genotyped lines from the dent heterotic group as described in Lehermeier et al. (2014). Here, doubled haploid (DH) progenies were generated from crosses with the common dent parental line F353 and 10 diverse dent founder lines, leading to 10 half-sib families with 53–104 full-sib progenies per family. DH lines were phenotyped as testcrosses with one common flint tester line (UH007). Field trial design and phenotypic data analysis are described in detail in Lehermeier et al. (2014). In this study, we concentrate on adjusted means of the traits dry matter yield (DMY) (decitonnes per hectare) and dry matter content (DMC) (percentage). The DH lines and parental lines were genotyped with the Illumina MaizeSNP50 BeadChip. Quality control and imputation of missing values are described in Lehermeier et al. (2014). A total of 32,801 high-quality polymorphic SNP markers were used for further analyses.
The wheat data set:
The wheat data set contains 599 wheat lines from the CIMMYT Global Wheat Breeding program and is publicly available within the BGLR R package (Pérez and de los Campos 2014). The lines were genotyped with 1447 binary Diversity Array Technology (DArT) markers (Wenzl et al. 2004). Markers with allele frequency <0.05 were removed and missing genotypes were imputed using Bernoulli samples as described by Crossa et al. (2010). A total of 1279 markers were used for further analyses. The trait average grain yield was collected for all lines in four environments (E1–E4). While for the rice and the maize data the assignment to subpopulations was predefined by Zhao et al. (2011) and by the crossing scheme, respectively, in the wheat data set subpopulations were inferred from the marker genotypes, using the PSMix software (Wu et al. 2006). PSMix infers clusters, using the EM algorithm applied to genotypes; for confirmatory purposes we computed the first two eigenvectors of the matrix G, using the eigen() function of R, and plotted with cluster-specific colors the loadings in the first two marker-derived principal components.
Data analysis
In all three data sets, phenotypes and markers were standardized across subpopulations to a zero mean and sample variance of one. A-GBLUP and W-GBLUP were implemented using the BGLR (v 1.04) R package (Pérez and de los Campos 2014). The scripts used for fitting these models are provided in Supporting Information, File S1. MG-GBLUP was implemented using a Gibbs sampler programmed in R (R Core Team 2014). The algorithm uses the eigenvalue decomposition of matrix G; this leads to an orthogonal representation of the model that simplifies computations greatly (de los Campos et al. 2010; Janss et al. 2012). The software is available in github (https://github.com/QuantGen/MTM). Further details about the algorithm and scripts that illustrate how models were fitted are provided in File S1.
The hyperparameters of the prior distributions of the variance components were chosen to obtain relatively flat priors with prior mode of the residual and genomic variances equal to 0.5 (50% of the sample variance of phenotypes). To achieve a relatively uninformative prior, in A-GBLUP and W-GBLUP the degree of freedom hyperparameters of the scaled inverse chi-square distributions assigned to variance parameters were all set to 4. In the parameterization used in BGLR the prior mode of the scaled inverse chi-square is therefore, in the univariate models, the scale parameters were set to In MG-GBLUP the degrees of freedom of the inverse-Wishart distribution were set to where K is the number of subpopulations, and the scale matrix Ψ was diagonal with entries equal to This choice of hyperparameters yields marginal prior distributions equal to the scaled inverse chi-square prior distributions used in A-GBLUP and W-GBLUP.
MCMC samples from the posterior distributions were obtained using Gibbs sampling. A total of 300,000 samples were collected; the first 100,000 samples were discarded as burn-in and the remaining 200,000 samples were thinned by keeping only every other sample of the post-burn-in samples. Thus, posterior means and posterior standard deviations were estimated using 100,000 samples.
Inferences on variances and covariances:
Models were fitted to the entire data sets to obtain estimates of variance components. For every model we report posterior mean estimates of the genomic variance the residual variance and the genomic heritability for each subpopulation k (k = 1,…, K). Posterior means and standard deviations of genomic heritabilities (k = 1,…, K) were calculated based on the 100,000 thinned post-burn-in samples of and by For A-GBLUP is common for all subpopulations. For MG-GBLUP we calculated posterior means of genomic correlations (j = 1,…, K; k = 1,…, K) based on the 100,000 Gibbs samples as
Assessment of prediction accuracy:
To investigate the prediction performance of the three models we randomly sampled 50% of the lines of each subpopulation to form the training set and the rest of the lines formed the test set. Genotypic and phenotypic data of the training set were used to estimate model parameters. The prediction performance of each model was then evaluated within each subpopulation k (k = 1,..., K) as correlation of observed phenotypic values and predicted genomic values of the lines of subpopulation k that were in the test set. MG-GBLUP estimates K different genomic values for each line, where the estimated values are specific for subpopulation k and were used as estimated values for the lines belonging to subpopulation k. This cross-validation procedure was randomly repeated 100 times; thus, for every trait, subpopulation, and model we had 100 estimates of the within-subpopulation correlation between predictions and observed phenotypes. From this we report the average correlation and approximate P-values for pairwise comparisons of prediction accuracy between models based on paired t-tests applied after transformation of the correlations, using Fisher’s Z transforms.
Data Availability
All three data sets used in this study have already been published. The rice data can be downloaded from http://www.ricediversity.org/data/. The genotypic data of the maize lines can be downloaded from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50558 and the phenotypic data are available as Supporting Information, File S1 of Lehermeier et al. (2014). The wheat data set is available within the R package BGLR (http://cran.r-project.org/web/packages/BGLR/index.html). Details on the R code for fitting the models are given in the Supporting Information, File S1 of this article.
Results
We begin this section by presenting descriptive information about the genotypes and phenotypes in each of the data sets. Subsequently we introduce estimates of variance components and of prediction accuracy.
Assessment of population substructure
Rice data set:
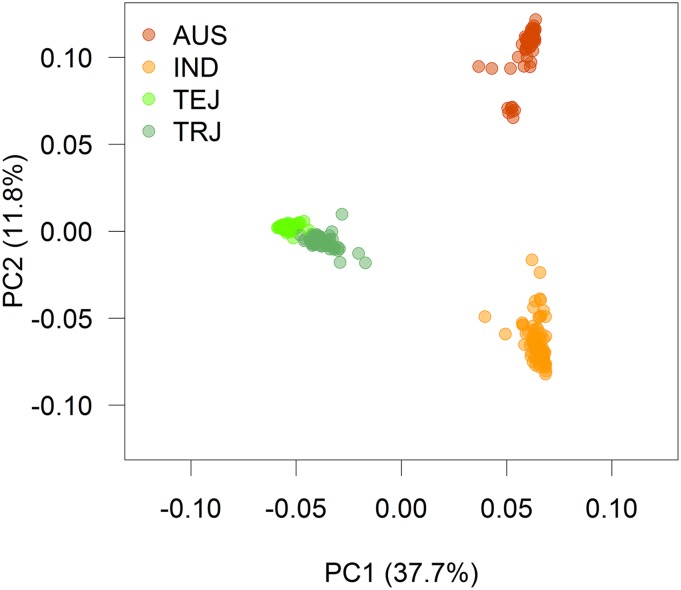
Figure 1 displays the loadings of each of the rice lines on the first two marker-derived principal components (PCs) of the G matrix and Figure S1 shows the eigenvalues of G, expressed as cumulative sum of variance explained. The four rice subpopulations AUS, IND, TEJ, and TRJ can be clearly identified based on the first two PCs. The first PC, which explains 37.7% of the genotypic variance (see Figure S1), clearly separates TEJ and TRJ from AUS and IND, which can also be identified as two separate subgroups based on PC1, but seem to represent a continuum rather than two separate clusters. The second PC separates AUS and IND. The heat map of the relationship matrix G (Figure S2) also shows a higher mean relatedness within subpopulations than between subpopulations, whereas relatedness between AUS and IND (mean 0.23) as well as between TEJ and TRJ (mean 0.15) is higher than across those two main groups (mean −0.32). In this data set, the first two PCs explained 50% of the sum of the eigenvalues, indicating the presence of a clear and strong substructure in the rice data.
Figure 1.
Loadings on the first (PC1) and second (PC2) marker-derived components of the rice data. Each point corresponds to a rice line, and colors indicate the different subpopulations [AUS, Indica (IND), Temperate Japonica (TEJ), Tropical Japonica (TRJ)].
Maize data set:
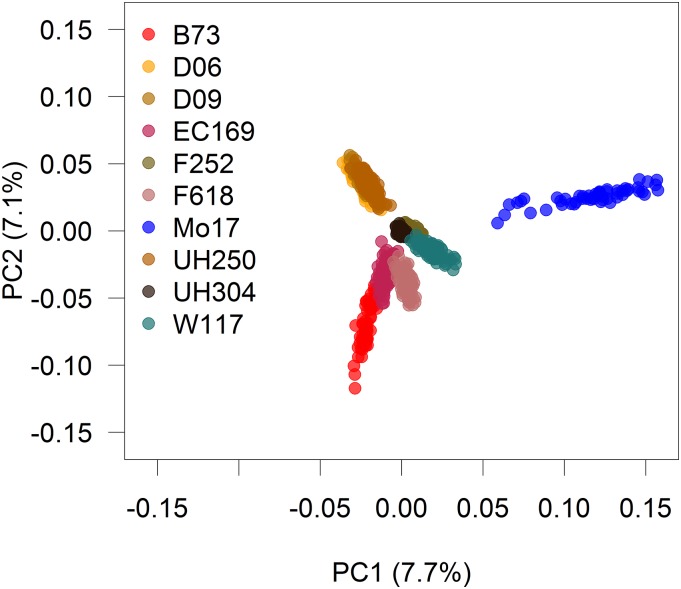
Genotypic relatedness of the maize dent lines is described in detail in Bauer et al. (2013) and Lehermeier et al. (2014). Figure 2 shows the loadings of each of the lines in the first two PCs of the G matrix. In brief, the DH lines clustered mainly according to the origin of their respective founder lines, where the first PC separated families descending from the Hohenheim breeding program (D06, D09, and UH250) from the remaining families. The heat map of the relationship matrix (Figure S3) shows, as one could predict, a higher relatedness among lines originating from the same cross than among lines originating from different crosses. Lines among families D06, D09, and UH250 show a relatively high relatedness with each other as these founder lines are related. The percentage of variance explained by the first two (five) PCs in this data set were 15% (30%); therefore, although this data set also shows a clear genetic substructure, the proportion of molecular variance explained by this substructure is much smaller than in the rice data set.
Figure 2.
Loadings on the first (PC1) and second (PC2) marker-derived components of the maize data. Each point corresponds to a maize line, and colors indicate the different families (families are specified by their specific parental line).
Wheat data set:
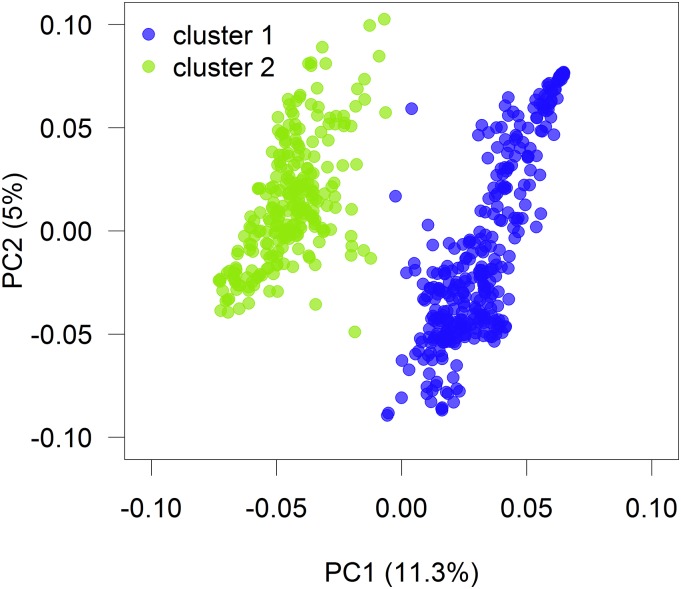
The first two PCs of the G matrix from the wheat lines are visualized in Figure 3. Both identified clusters were mainly separated by the first PC, explaining 11.3% of the molecular variance (see Figure S1). The mean genotypic relationship (heat map of G shown in Figure S4) is considerably higher within clusters than across the two clusters, whereas on average lines within cluster 2 (0.14) are higher related than within cluster 1 (0.07). Although the wheat data set also shows a clear substructure, among the three data sets included in this study, this is the one with the least proportion of variance explained by the top five PCs (26%, see Figure S1).
Figure 3.
Loadings on the first (PC1) and second (PC2) marker-derived components of the wheat data. Each point corresponds to a wheat line, and colors indicate two clusters.
Phenotypic analysis
Rice data set:
The distribution of the phenotypic traits flowering time, plant height, and panicle length for the four rice subpopulations is visualized with boxplots in Figure S5. Population AUS shows the earliest mean flowering time with the smallest range across all subpopulations. IND has the latest mean flowering time and together with TEJ the largest range with >190 days. IND shows also for the other traits a high variation, especially for plant height, whereas AUS shows considerably less variation for plant height than the other subpopulations. Lines of subpopulation TEJ show, on average, shorter plant height and panicle length compared to the other subpopulations. Phenotypic correlations between traits (Figure S6) were positive within all subpopulations, but quite low for population IND among all traits. Generally, correlations between flowering time and the other two traits were quite low and in a medium range between plant height and panicle length.
Maize data set:
The distribution of adjusted means of DMY and DMC of the maize lines is visualized by boxplots in Figure S7. A similar variation of phenotypic values was observed for all families. Mean yield ranged from 176.16 (W117) to 194.70 (F618) and mean DMC from 33.08 (B73) to 37.99 (F252). Generally, DMY and DMC are negatively correlated (Figure S8), but within some families the correlation was close to zero.
Wheat data set:
Figure S9 shows the distribution of standardized grain yield values of both wheat clusters in the four environments by boxplots. On average, cluster 1 shows higher grain yield in all environments. Both clusters show a quite similar variation of phenotypic values; only in environment 3 does cluster 2 show a higher variation in phenotypic values than cluster 1. Figure S10 shows the correlations of phenotypic values between grain yields across environments. Environment 1 performs quite differently from the other three environments, where correlations of phenotypic values in environment 1 with the other environments are very low or even negative. Phenotypic values of the other three environments are positively correlated (), especially environments 2 and 3 showing a high correlation ().
Variance components
Rice data set:
Genomic and residual variance components estimates as well as genomic heritability estimates obtained with each of the models are presented for the rice data in Table 1. A-GBLUP estimated one common genomic variance component for all subpopulations, so estimates here are identical for all subpopulations. However, results from W-GBLUP and MG-GBLUP show that the assumption of common genomic variance components for all subpopulations does not hold; indeed, results from these two models suggest important differences in genomic variances for a given trait across subpopulations. This is particularly clear for flowering time. Here, subpopulation TEJ shows a noticeably higher genomic variance compared to the other groups. The estimated residual variances were very similar for the W-GBLUP and MG-GBLUP; however, as one would expect, given the relatively low estimated genomic correlations the A-GBLUP model tended to give consistently higher estimates of the residual variance, reflecting worse fit than the W- and MG-GBLUPs. Overall, estimates of variance components and heritabilities did not show large differences between the models W-GBLUP and MG-GBLUP, but MG-GBLUP yielded for all subpopulations and traits consistently higher heritability estimates than W-GBLUP.
Table 1. Posterior mean estimates (± posterior standard deviation estimates) of genomic variance (), residual variance (), and heritability () for each rice subpopulation (Pop) [AUS, Indica (IND), Temperate Japonica (TEJ), Tropical Japonica (TRJ)] and from the three different models [across-group GBLUP (A-GBLUP), within-group GBLUP (W-GBLUP), multigroup GBLUP (MG-GBLUP)].
| Traita | Pop | A-GBLUPb | W-GBLUP | MG-GBLUP | A-GBLUP | W-GBLUP | MG-GBLUP | A-GBLUP | W-GBLUP | MG-GBLUP |
|---|---|---|---|---|---|---|---|---|---|---|
| Flow | AUS | 0.41 ± 0.14 | 0.17 ± 0.05 | 0.27 ± 0.07 | 0.13 ± 0.04 | 0.12 ± 0.03 | 0.12 ± 0.03 | 0.74 ± 0.07 | 0.59 ± 0.08 | 0.69 ± 0.07 |
| IND | 1.26 ± 0.31 | 1.33 ± 0.31 | 0.64 ± 0.20 | 0.27 ± 0.10 | 0.26 ± 0.09 | 0.40 ± 0.13 | 0.82 ± 0.08 | 0.83 ± 0.07 | ||
| TEJ | 5.45 ± 1.42 | 5.21 ± 1.37 | 1.33 ± 0.25 | 0.36 ± 0.14 | 0.36 ± 0.14 | 0.24 ± 0.08 | 0.93 ± 0.04 | 0.93 ± 0.04 | ||
| TRJ | 0.46 ± 0.18 | 0.65 ± 0.21 | 0.31 ± 0.06 | 0.30 ± 0.06 | 0.29 ± 0.06 | 0.56 ± 0.10 | 0.59 ± 0.11 | 0.68 ± 0.09 | ||
| PH | AUS | 0.77 ± 0.14 | 0.29 ± 0.09 | 0.40 ± 0.10 | 0.17 ± 0.05 | 0.17 ± 0.04 | 0.16 ± 0.04 | 0.82 ± 0.05 | 0.62 ± 0.09 | 0.70 ± 0.07 |
| IND | 1.16 ± 0.32 | 1.23 ± 0.31 | 0.53 ± 0.15 | 0.44 ± 0.14 | 0.42 ± 0.12 | 0.60 ± 0.09 | 0.71 ± 0.10 | 0.74 ± 0.09 | ||
| TEJ | 1.24 ± 0.48 | 1.36 ± 0.45 | 0.33 ± 0.07 | 0.28 ± 0.07 | 0.26 ± 0.07 | 0.70 ± 0.07 | 0.80 ± 0.10 | 0.83 ± 0.07 | ||
| TRJ | 1.05 ± 0.27 | 1.12 ± 0.27 | 0.25 ± 0.05 | 0.24 ± 0.05 | 0.24 ± 0.05 | 0.75 ± 0.06 | 0.81 ± 0.06 | 0.82 ± 0.05 | ||
| PaLe | AUS | 0.78 ± 0.18 | 0.61 ± 0.22 | 0.76 ± 0.23 | 0.31 ± 0.10 | 0.35 ± 0.11 | 0.31 ± 0.10 | 0.71 ± 0.09 | 0.63 ± 0.12 | 0.70 ± 0.10 |
| IND | 0.59 ± 0.22 | 0.76 ± 0.23 | 0.33 ± 0.10 | 0.38 ± 0.11 | 0.32 ± 0.10 | 0.70 ± 0.09 | 0.60 ± 0.14 | 0.69 ± 0.11 | ||
| TEJ | 1.43 ± 0.65 | 1.63 ± 0.60 | 0.44 ± 0.09 | 0.37 ± 0.10 | 0.34 ± 0.09 | 0.63 ± 0.08 | 0.76 ± 0.12 | 0.81 ± 0.09 | ||
| TRJ | 1.28 ± 0.48 | 1.40 ± 0.46 | 0.52 ± 0.11 | 0.44 ± 0.11 | 0.42 ± 0.10 | 0.60 ± 0.09 | 0.73 ± 0.11 | 0.75 ± 0.09 | ||
Traits are flowering time (Flow), plant height (PH), and panicle length (PaLe).
For A-GBLUP, estimates are common for all subpopulations.
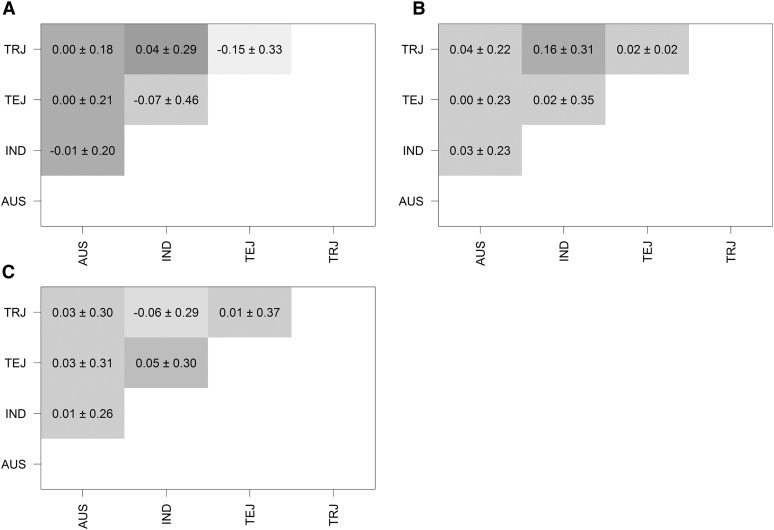
Posterior mean estimates of genomic correlations of the four rice subpopulations from MG-GBLUP are shown for the three traits in Figure 4. Estimated genomic correlations are close to zero and have high posterior standard deviations. Only for the trait plant height, IND and TRJ show an estimated genomic correlation higher than 0.1 (0.16 ± 0.31). For flowering time TEJ and TRJ had a negative genomic correlation estimate (−0.15) but the posterior standard deviation is high (0.33); therefore this estimate cannot be regarded as smaller than zero.
Figure 4.
Posterior mean estimates (± posterior standard deviation estimates) of genomic correlations derived from of rice subpopulations [AUS, Indica (IND), Temperate Japonica (TEJ), Tropical Japonica (TRJ)] from the multivariate model (MG-GBLUP). (A–C) Estimates for traits flowering time (A), plant height (B), and panicle length (C).
Maize data set:
The estimated variance components and genomic heritabilities obtained with this data set are given in Table 2. In A-GBLUP the common genomic variance for all maize families was estimated to be 0.75 (±0.11) for DMY and 0.54 (±0.07) for DMC. Although there were differences in the estimated error variance of the different models, the magnitudes of the differences were considerably smaller than those observed in the rice data set. This is likely to happen because in this population the estimated genomic correlations were relatively high (see next paragraph); this makes the A-GBLUP not very different from the MG-GBLUP. MG-GBLUP yielded higher estimates of genomic heritability than the other two models. For DMY, genomic heritability estimates varied from 0.49 (0.64) for family W117 to 0.78 (0.83) for family F252 estimated by W-GBLUP (MG-GBLUP), respectively. For DMC, EC169 showed the lowest heritability with 0.60 (0.76) estimated using W-GBLUP (MG-GBLUP). Finally, F618 showed the highest heritability estimate with 0.79 (0.85) estimated by W-GBLUP (MG-GBLUP).
Table 2. Posterior mean estimates (± posterior standard deviation estimates) of genomic variance (), residual variance (), and heritability () for each maize family (families are specified by their specific parental line) and from the three different models [across-group GBLUP (A-GBLUP), within-group GBLUP (W-GBLUP), multigroup GBLUP (MG-GBLUP)].
| Traita | Family | A-GBLUPb | W-GBLUP | MG-GBLUP | A-GBLUP | W-GBLUP | MG-GBLUP | A-GBLUP | W-GBLUP | MG-GBLUP |
|---|---|---|---|---|---|---|---|---|---|---|
| DMY | B73 | 0.75 ± 0.11 | 0.67 ± 0.26 | 1.09 ± 0.31 | 0.45 ± 0.11 | 0.50 ± 0.13 | 0.42 ± 0.11 | 0.63 ± 0.07 | 0.56 ± 0.12 | 0.71 ± 0.08 |
| D06 | 0.78 ± 0.31 | 1.30 ± 0.37 | 0.42 ± 0.07 | 0.46 ± 0.09 | 0.38 ± 0.08 | 0.64 ± 0.05 | 0.61 ± 0.11 | 0.76 ± 0.07 | ||
| D09 | 0.81 ± 0.30 | 1.26 ± 0.34 | 0.31 ± 0.06 | 0.32 ± 0.07 | 0.27 ± 0.06 | 0.71 ± 0.05 | 0.69 ± 0.10 | 0.82 ± 0.06 | ||
| EC169 | 0.66 ± 0.29 | 1.19 ± 0.36 | 0.41 ± 0.09 | 0.46 ± 0.11 | 0.37 ± 0.10 | 0.64 ± 0.06 | 0.57 ± 0.13 | 0.75 ± 0.08 | ||
| F252 | 1.31 ± 0.41 | 1.55 ± 0.39 | 0.40 ± 0.08 | 0.34 ± 0.08 | 0.31 ± 0.07 | 0.65 ± 0.06 | 0.78 ± 0.08 | 0.83 ± 0.06 | ||
| F618 | 0.46 ± 0.18 | 0.85 ± 0.23 | 0.29 ± 0.05 | 0.31 ± 0.06 | 0.27 ± 0.05 | 0.72 ± 0.05 | 0.58 ± 0.11 | 0.75 ± 0.07 | ||
| Mo17 | 0.70 ± 0.29 | 1.13 ± 0.34 | 0.54 ± 0.15 | 0.59 ± 0.18 | 0.47 ± 0.15 | 0.58 ± 0.08 | 0.53 ± 0.14 | 0.70 ± 0.10 | ||
| UH250 | 0.80 ± 0.34 | 1.38 ± 0.40 | 0.50 ± 0.09 | 0.54 ± 0.11 | 0.44 ± 0.09 | 0.60 ± 0.06 | 0.58 ± 0.12 | 0.75 ± 0.07 | ||
| UH304 | 0.68 ± 0.29 | 1.11 ± 0.34 | 0.41 ± 0.08 | 0.40 ± 0.08 | 0.36 ± 0.08 | 0.64 ± 0.06 | 0.61 ± 0.12 | 0.74 ± 0.07 | ||
| W117 | 0.64 ± 0.26 | 1.06 ± 0.31 | 0.58 ± 0.11 | 0.63 ± 0.13 | 0.56 ± 0.11 | 0.56 ± 0.06 | 0.49 ± 0.11 | 0.64 ± 0.08 | ||
| DMC | B73 | 0.54 ± 0.07 | 0.37 ± 0.12 | 0.66 ± 0.16 | 0.22 ± 0.05 | 0.22 ± 0.05 | 0.20 ± 0.05 | 0.72 ± 0.05 | 0.62 ± 0.10 | 0.77 ± 0.06 |
| D06 | 0.64 ± 0.20 | 0.90 ± 0.21 | 0.18 ± 0.03 | 0.17 ± 0.04 | 0.15 ± 0.03 | 0.75 ± 0.04 | 0.77 ± 0.07 | 0.85 ± 0.04 | ||
| D09 | 0.44 ± 0.13 | 0.73 ± 0.17 | 0.17 ± 0.03 | 0.17 ± 0.03 | 0.15 ± 0.03 | 0.76 ± 0.04 | 0.71 ± 0.08 | 0.82 ± 0.05 | ||
| EC169 | 0.51 ± 0.19 | 0.90 ± 0.25 | 0.27 ± 0.06 | 0.31 ± 0.07 | 0.26 ± 0.07 | 0.66 ± 0.06 | 0.60 ± 0.11 | 0.76 ± 0.07 | ||
| F252 | 0.70 ± 0.23 | 0.99 ± 0.25 | 0.24 ± 0.05 | 0.24 ± 0.05 | 0.21 ± 0.05 | 0.69 ± 0.05 | 0.73 ± 0.09 | 0.82 ± 0.05 | ||
| F618 | 0.73 ± 0.21 | 0.95 ± 0.22 | 0.19 ± 0.04 | 0.18 ± 0.04 | 0.17 ± 0.03 | 0.73 ± 0.05 | 0.79 ± 0.07 | 0.85 ± 0.05 | ||
| Mo17 | 0.38 ± 0.12 | 0.66 ± 0.16 | 0.21 ± 0.06 | 0.22 ± 0.06 | 0.20 ± 0.06 | 0.72 ± 0.06 | 0.62 ± 0.10 | 0.76 ± 0.07 | ||
| UH250 | 0.72 ± 0.24 | 1.04 ± 0.26 | 0.25 ± 0.05 | 0.24 ± 0.05 | 0.21 ± 0.04 | 0.68 ± 0.05 | 0.73 ± 0.09 | 0.83 ± 0.05 | ||
| UH304 | 0.82 ± 0.28 | 1.10 ± 0.29 | 0.26 ± 0.05 | 0.25 ± 0.06 | 0.23 ± 0.05 | 0.68 ± 0.05 | 0.75 ± 0.08 | 0.82 ± 0.05 | ||
| W117 | 0.66 ± 0.22 | 0.95 ± 0.24 | 0.30 ± 0.06 | 0.32 ± 0.07 | 0.29 ± 0.06 | 0.64 ± 0.06 | 0.66 ± 0.10 | 0.76 ± 0.06 | ||
Results are given for the traits dry matter yield (DMY) and dry matter content (DMC).
For A-GBLUP, estimates are common for all families.
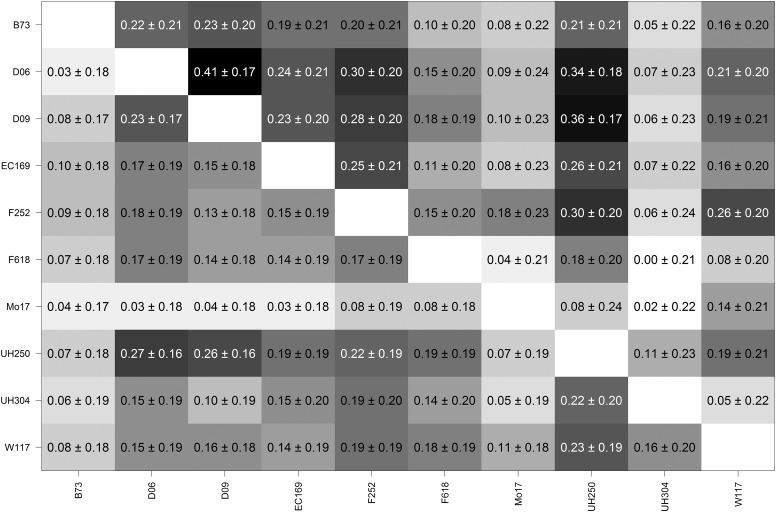
Estimated genomic correlations between the maize families are given in Figure 5 for both traits DMY and DMC. The estimated genomic correlations between families were much higher than those obtained with the rice data set; however, in all cases the estimated genomic correlations were considerably <0.5, suggesting a great extent of marker-by-family interaction. The estimated genomic correlations between families were of the order of 0.2 for most comparisons for the trait DMY. Correlation estimates >0.3 were observed for D06, D09, and UH250. Family UH304 showed genomic correlations close to zero with all other families and Mo17 also showed quite low correlations with the other families. Generally, families showed lower genomic correlations for DMC than for DMY with the exception of UH304. Due to small within-family sample size, and relatively weak genomic relationships between families, all the estimated genomic correlations had large posterior standard deviations.
Figure 5.
Posterior mean estimates (± posterior standard deviation estimates) of genomic correlations derived from of maize dent families (families are specified by their specific parental line) from the multivariate model (MG-GBLUP) for traits dry matter yield (upper diagonal) and dry matter content (lower diagonal).
Wheat data set:
For the wheat data, estimated variance components, genomic heritabilities, and genomic correlations between both wheat clusters are given in Table 3 for all four environments. The estimated error variances were slightly higher in A-GBLUP, relative to the other two models. Genomic variances estimated with A-GBLUP varied from 0.49 (E3) to 0.57 (E1) across environments. Results from W-GBLUP and MG-GBLUP show that variance components of both clusters are different. In environment 3, cluster 1 shows a higher genomic variance and heritability estimate than cluster 2. Whereas in the other three environments the estimated heritability is higher in cluster 2 than in cluster 1. Similar to what we observed in the rice and the maize data, MG-GBLUP yielded always higher estimates of genomic variances () than W-GBLUP and A-GBLUP and lower or similar estimates of residual variance (); therefore, MG-GBLUP always gave higher heritability estimates than W-GBLUP and A-GBLUP. The estimated genomic correlations between clusters were higher in the wheat data set than in the other two data sets and ranged from 0.15 (E4) to 0.49 (E2).
Table 3. Posterior mean estimates (± posterior standard deviation estimates) of genomic variance (), residual variance (), and heritability () for each wheat cluster (Clust) and from the three different models [across-group GBLUP (A-GBLUP), within-group GBLUP (W-GBLUP), multigroup GBLUP (MG-GBLUP)]. Additionally estimates of genomic correlations (Genomic corr.) between both clusters are given.
| Enva | Clust | A-GBLUPb | W-GBLUP | MG-GBLUP | A-GBLUP | W-GBLUP | MG-GBLUP | A-GBLUP | W-GBLUP | MG-GBLUP | Genomic corr. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E1 | 1 | 0.57 ± 0.10 | 0.55 ± 0.13 | 0.59 ± 0.14 | 0.60 ± 0.06 | 0.57 ± 0.07 | 0.56 ± 0.06 | 0.48 ± 0.05 | 0.48 ± 0.08 | 0.51 ± 0.07 | 0.30 ± 0.16 |
| 2 | 0.65 ± 0.13 | 0.66 ± 0.13 | 0.43 ± 0.06 | 0.35 ± 0.06 | 0.35 ± 0.06 | 0.57 ± 0.06 | 0.64 ± 0.07 | 0.65 ± 0.07 | |||
| E2 | 1 | 0.50 ± 0.09 | 0.59 ± 0.14 | 0.64 ± 0.14 | 0.65 ± 0.06 | 0.63 ± 0.07 | 0.61 ± 0.07 | 0.43 ± 0.06 | 0.48 ± 0.08 | 0.51 ± 0.07 | 0.49 ± 0.13 |
| 2 | 0.47 ± 0.11 | 0.54 ± 0.12 | 0.46 ± 0.06 | 0.46 ± 0.07 | 0.43 ± 0.06 | 0.52 ± 0.06 | 0.50 ± 0.08 | 0.55 ± 0.08 | |||
| E3 | 1 | 0.49 ± 0.10 | 0.59 ± 0.15 | 0.62 ± 0.15 | 0.54 ± 0.06 | 0.50 ± 0.06 | 0.49 ± 0.06 | 0.47 ± 0.07 | 0.54 ± 0.08 | 0.55 ± 0.08 | 0.33 ± 0.17 |
| 2 | 0.55 ± 0.15 | 0.58 ± 0.15 | 0.74 ± 0.09 | 0.66 ± 0.09 | 0.65 ± 0.09 | 0.40 ± 0.06 | 0.45 ± 0.09 | 0.47 ± 0.09 | |||
| E4 | 1 | 0.50 ± 0.10 | 0.57 ± 0.14 | 0.59 ± 0.13 | 0.63 ± 0.06 | 0.57 ± 0.06 | 0.56 ± 0.06 | 0.44 ± 0.06 | 0.50 ± 0.08 | 0.51 ± 0.07 | 0.15 ± 0.19 |
| 2 | 0.59 ± 0.14 | 0.62 ± 0.14 | 0.54 ± 0.07 | 0.44 ± 0.07 | 0.43 ± 0.07 | 0.48 ± 0.07 | 0.57 ± 0.08 | 0.58 ± 0.08 | |||
Results are given for the four environments (Env) (E1, E2, E3, and E4).
For A-GBLUP, estimates are common for both clusters.
Comparison of predicted genomic values
Rice data set:
The comparison of the predicted genomic values from a full model including all rice lines is shown for flowering time, plant height, and panicle length in Figure S11, Figure S12, and Figure S13, respectively. W-GBLUP and MG-GBLUP yielded highly similar predicted genomic values for all traits with a correlation close to 1. Predicted genomic values from A-GBLUP were slightly different from the ones obtained by W-GBLUP and MG-GBLUP, but showed also a correlation >0.9 with the predicted values from W-GBLUP and MG-GBLUP for the traits plant height and panicle length. An exception was subpopulation TEJ for the trait flowering time. Here, A-GBLUP estimated a higher residual variance than W-GBLUP and MG-GBLUP. Thus, predicted genomic values obtained with A-GBLUP were shrunk more strongly toward zero than those obtained with W-GBLUP and MG-GBLUP.
Maize data set:
For the maize lines the comparison of the predicted genomic values obtained by the three models is shown in Figure S14 and Figure S15 for traits DMY and DMC, respectively. Models A-GBLUP and W-GBLUP make very different assumptions about the genomic correlations between subpopulations: A-GBLUP assumes a correlation of 1, and W-GBLUP assumes a null correlation; therefore, it is not surprising to see that predictions from these two models are the least correlated. However, the correlation of predicted genomic values derived from these two models was always >0.9. For both traits, predicted genomic values from MG-GBLUP were more similar to those from A-GBLUP than to those from W-GBLUP.
Wheat data set:
The comparison of the predicted genomic values of the wheat lines from the different models is shown for the four environments in Figure S16, Figure S17, Figure S18, and Figure S19, respectively. As with the rice data the predicted genomic values obtained with the multivariate model (MG-GBLUP) are highly correlated with those obtained with the W-GBLUP (correlations were all close to one, across environments). However, predictions from these two models were also highly correlated with A-GBLUP, always >0.93, across environments.
Prediction accuracy
Pearson’s product–moment correlation between observed phenotypes and predictions was assessed for each data set, trait, model, subpopulation, and training–testing partition.
Rice data set:
Figure 6 shows the average testing correlations (across training–testing partitions) for the rice data by subpopulation and trait. Prediction accuracy varied greatly between traits and subpopulations within traits. For instance, for flowering time, prediction correlations ranged from between 0.1 and 0.25 in AUS and TRJ to values >0.59 in IND and TEJ. In this trait, the levels of prediction accuracy were higher for the subpopulations having highest heritability estimates (IND and TEJ). For plant height there were also important differences in prediction accuracy across subpopulations, and although the group showing highest prediction accuracy (TRJ) was also the one with the highest heritability estimate, the relationship between prediction accuracy and heritability estimate was not as clear for this trait. Finally, plant height also gave important differences in prediction accuracy across subpopulations.
Figure 6.
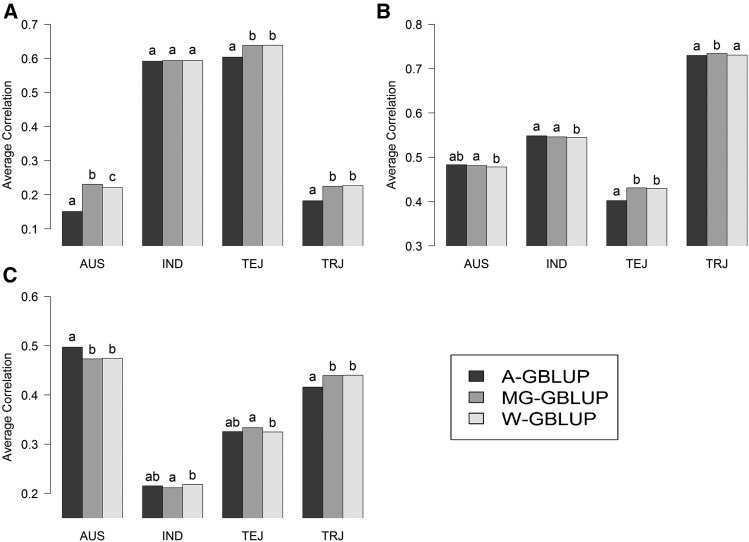
Average prediction correlations in testing sets from 100 random replications of across-group GBLUP (A-GBLUP), multigroup GBLUP (MG-GBLUP), and within-group GBLUP (W-GBLUP) evaluated within each of the four rice subpopulations [AUS, Indica (IND), Temperate Japonica (TEJ), Tropical Japonica (TRJ)]. Letters above bars indicate significant differences between models’ prediction correlations from paired t-tests evaluated within each subpopulation and trait, where estimates with the same letter are not significantly different (P < 0.05). (A) Flowering time. (B) Plant height. (C) Panicle length.
On average, all three models yielded very similar predictive abilities, especially W-GBLUP and MG-GBLUP. The largest differences between models were observed for flowering time in AUS and TRJ; here MG-GBLUP and W-GBLUP clearly outperformed A-GBLUP.
Maize data set:
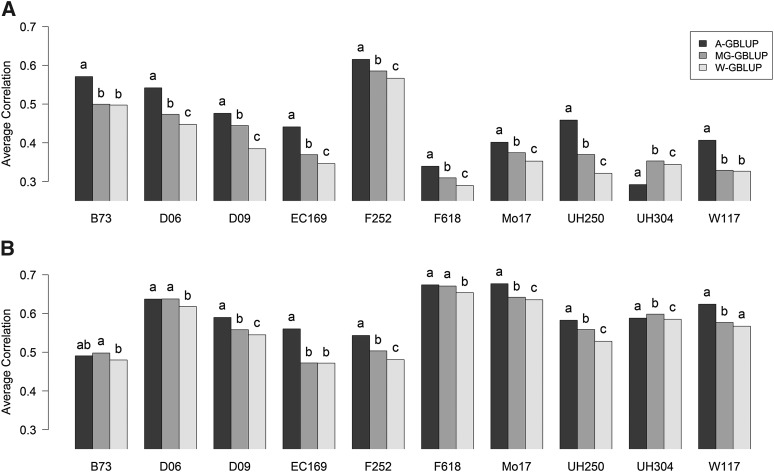
For maize, mean prediction correlations from 100 training–testing replicates evaluated within families and traits by model are shown in Figure 7. Prediction accuracy varied between traits and subpopulations and in this case more clearly between models. Overall prediction accuracy was higher for DMC than for DMY; this could be expected given the higher heritability estimate of DMC. The variability of prediction accuracy across families was also greater for DMY than for DMC and this also could be expected because for DMY there were important differences in estimates of genomic heritability across families. For instance, F252, which was a family with a very large estimated genomic heritability for DMY, also shows higher prediction accuracy than other families for that trait.
Figure 7.
Average prediction correlations in testing sets obtained from 100 random training–testing partitions, using models across-group GBLUP (A-GBLUP), multigroup GBLUP (MG-GBLUP), and within-group GBLUP (W-GBLUP) evaluated within each of the 10 maize dent families (families are indicated by their specific parental line). Letters above bars indicate significant differences between models’ prediction correlations from paired t-tests evaluated within each subpopulation and trait, where estimates with the same letter are not significantly different (P < 0.05). (A) Dry matter yield. (B) Dry matter content.
In this data set the A-GBLUP model consistently performed better than either of the other two models for the great majority of traits and families. The MG-GBLUP tended to perform in between the A-GBLUP and the W-GBLUP, which was the one with the lowest level of prediction accuracy. There were only a few exceptions to this pattern (e.g., UH304 for both traits or B73 for DMC) but in those cases the differences in prediction correlation were typically small.
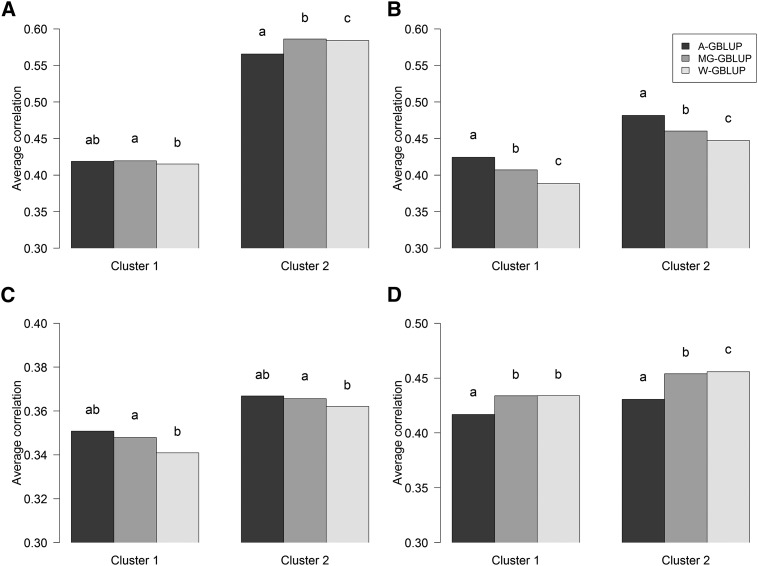
Wheat data set:
Figure 8 shows the mean prediction correlations of the three models for the wheat data evaluated within cluster and environment. As before, prediction accuracy varied between outcomes (environment in this case) and clusters. For instance, in E1, the average correlation was higher in cluster 2 than in cluster 1. There were also differences, albeit small in magnitude, between models; however, these were not consistent across clusters or environments. There were three types of cases: (i) for E1 (cluster 2) and E4 (both clusters), the MG-GBLUP and W-GBLUP performed similarly and clearly better than the A-GBLUP; on the other hand, (ii) for E2 (both clusters) and E3 (cluster 1), the A-GBLUP model performed much better than the W-GBLUP and the MG-GBLUP performed in between the two other models; and finally, (iii) in E1 (cluster 1) and E3 (cluster 2) there were no sizable differences between models.
Figure 8.
Average prediction correlations in testing sets from 100 random replications of across-group GBLUP (A-GBLUP), multigroup GBLUP (MG-GBLUP), and within-group GBLUP (W-GBLUP) evaluated within each wheat cluster. Letters above bars indicate significant differences between models’ prediction correlations from paired t-tests evaluated within each subpopulation and trait, where estimates with the same letter are not significantly different (P < 0.05). (A) Environment 1. (B) Environment 2. (C) Environment 3. (D) Environment 4.
Discussion
Structure and admixture are pervasive in plant breeding populations. Differences in allele frequencies and in LD patterns can make marker effects vary between groups in a population; therefore it is not clear how data from heterogeneous populations should be analyzed. Even though the use of structured populations is common practice in plant breeding, the genomic selection literature has largely overlooked this problem. On the other hand, the animal breeding literature has addressed this topic in multibreed settings.
In this article we address the problem of analyzing data from heterogeneous plant breeding populations, using three different plant data sets: a rice diversity panel comprising data from four distant rice populations, a wheat data set comprising lines from two interconnected subpopulations, and, finally, a maize data set comprising data from sib families. When analyzing these data, we considered three models: a model that assumes that marker effects are constant across subpopulations (A-GBLUP), a stratified (i.e., within subpopulations) analysis that allows for marker effects to change across subpopulations but does not permit borrowing of information between subpopulations (W-GBLUP), and, finally, a multivariate approach (MG-GBLUP) where data from all subpopulations are analyzed jointly using a model where marker effects are assumed to be different, but correlated, between subpopulations. These three models can be implemented either in a Bayesian framework, as we did in this article, or using a standard mixed-model approach with (co)variance parameters estimated with either maximum-likelihood or restricted maximum-likelihood methods followed by best linear unbiased estimation (BLUE)-BLUP equations for estimation/prediction of effects.
Model MG-GBLUP should be considered in between the other two approaches (A-GBLUP and W-GBLUP); indeed, if we set all genomic correlations equal to one (zero), we obtain A-GBLUP (W-GBLUP) as special cases of MG-GBLUP. In MG-GBLUP the estimates of covariance components determine whether the model behaves similarly to A-GBLUP or W-GBLUP or in between. These estimates not only control the extent of borrowing of information across groups, but also provide measures (e.g., genomic correlations between subpopulations) that can shed light on trait-specific degrees of genetic similarities/differences across groups. Our results show clearly that no single approach is best across data sets and traits and give explanations on the factors that affect the relative performance of the three methods considered.
Rice diversity panel
These data, which represent the most heterogeneous set of material analyzed here, have four genetically distant groups. Not surprisingly, the estimates of genomic correlations between subpopulations were close to zero, suggesting pronounced genetic heterogeneity between subpopulations. Consequently, MG-GBLUP gave predictions that were highly correlated with those of the W-GBLUP and both models achieved very similar levels of prediction accuracy. Because the extent of genetic heterogeneity in this data set is large, in general, the W- and MG-GBLUPs performed either similarly to or better than the A-GBLUP model; the only exception to these patterns was observed in population AUS for prediction of panicle length.
However, the differences in prediction accuracy between the A-GBLUP and the W-GBLUP or MG-GBLUP were relatively small in many traits and subpopulations. This may appear a bit surprising considering that the estimates of genomic correlations in this data set were in general small; thus, at first look one may expect a poor performance from a model like the A-GBLUP because this model assumes a perfect correlation between groups. We believe that this finding can be explained by the fact that in A-GBLUP the extent of borrowing of information between any two lines () from different subpopulations (say j and k) depends on the product of the genomic relationship between the two lines () times the genomic covariance, that is, The off-diagonal terms of G, of pairs of lines belonging to two different subpopulations are typically close to zero; therefore, there will not be borrowing of information even in the A-GBLUP model.
Maize
The maize data set represents a very different situation from that of the rice diversity panel; the DH lines of this data set share one common parent and have, as a second parent, lines that are either closely (e.g., D06, D09, and UH250) or distantly (Mo17 and B73) related. Because all lines share one of the parents, within families, allele frequencies are expected to be 0.5 for segregating loci. Thus, for segregating loci, allele substitution effects are not expected to vary strongly among families due to differences in allele frequencies. Furthermore, results presented in Lehermeier et al. (2014) suggest that the patterns of LD are quite consistent across families. Thus, it is not surprising that genomic correlations among maize families were higher than those obtained with the rice data set.
It was interesting to observe that the estimated genomic correlations were large between families derived from relatively close parents (D06, D09, and UH250) and close to zero for families derived from relatively distant parents (e.g., Mo17 and B73).
In terms of prediction accuracy, in the maize data set the A-GBLUP model tended to outperform the W-GBLUP model and MG-GBLUP had a predictive performance in between A-GBLUP and W-GBLUP; this is consistent with the fact that the estimated genomic correlations ranged from low to moderate.
Wheat
In this data set the estimated genomic correlations were all positive and ranged from low (E4) to moderate (E1–E3); in general, because the estimated correlations were not high, predictions from the MG-GBLUP were more correlated with those from the W-GBLUP than with those from the A-GBLUP. As one would expect, results showed that the lower the estimated genomic correlation among clusters, the higher the correlation among predicted genomic values obtained with W-GBLUP and MG-GBLUP.
In terms of prediction accuracy the wheat data set offered situations where all models performed similarly (e.g., E1/cluster 1), or the W- and MG-GBLUPs outperformed the A-GBLUP (E4), or the A-GBLUP outperformed the W-GBLUP (E2). In general, the MG-GBLUP was the best model or performed similarly to the best-performing model or in between the best- and worst-performing models; therefore, if we average across environments and clusters, there is a small advantage in favor of the MG-GBLUP.
Estimates of genomic heritability
In all data sets we tended to observe higher estimates of genomic heritability with the MG-GBLUP compared to W-GBLUP. This went along with a better fit of the MG-GBLUP model to the training data (e.g., higher correlation between predictions and phenotypes in the training data sets) than that obtained with the W-GBLUP model. The true genetic variances are not known; therefore one cannot establish whether this is reflecting better ability of the MG-GBLUP model to capture genetic signals or simply reflects overfitting.
Previous studies from the animal breeding literature that compare the W- and A-GBLUPs or similar Bayesian models have shown either null or small gain of the A-GBLUP relative to the W-GBLUP (Hayes et al. 2009). Other studies (e.g., Karoui et al. 2012; Olson et al. 2012; Zhou et al. 2014) that have compared MG-GBLUP with the W-GBLUP models have reported small gains in prediction accuracy with the MG-GBLUP methods. There are important differences between animal and plant breeding data sets that grant considerations. First, in many plant breeding programs data are from inbred lines, while in animal breeding data are from outbred individuals. When data are from inbred lines and when considering a fully biallelic system, marker effects represent contrasts between the two homozygous marker classes; these effects are expected to be more stable across subpopulations than effects of allele substitutions in outbred populations that can vary, due to unaccounted dominance, simply because of differences in allele frequency. However, even with inbred lines marker effects are allele frequency dependent due to unaccounted forms of epistasis.
A second important difference between the analyses presented in our study and previous studies from animal breeding is marker density; this was clearly seen in the case of the wheat data set where marker density was relatively low. With low marker density, differences in LD patterns between subpopulations can induce differences in marker effects, even if QTL effects are constant across subpopulations. Therefore, it was not surprising to observe estimates of genomic correlations that were rather low. Further research with higher marker density would be needed to disentangle whether the low correlations observed in our study are mostly due to genetic heterogeneity at the QTL level or simply due to differences in the marker–QTL patterns of LD.
A third important difference between the study presented here and previous studies from animal breeding is sample size. The A-GBLUP and W-GBLUP models have both advantages and disadvantages. On the one hand, A-GBLUP estimates marker effects making use of all the available data; however, this is achieved at the price of imposing homogeneity of marker effects. On the other hand the W-GBLUP model allows for heterogeneity of marker effects but does not allow for borrowing of information between subpopulations. The MG-GBLUP resides in between the other two. Borrowing of information is most important when the within-subpopulation sample size is small and this situation is very common in plant breeding where available phenotypic and genotypic data from individual plant breeding populations are typically small. With small sample sizes it may occur that a parsimonious model such as the A-GBLUP may outperform W-GBLUP or MG-GBLUP, even when heterogeneity is present. We believe that this is an important factor that explains why the A-GBLUP performed well even in cases where estimates of variance showed large differences between subpopulations.
However, even in situations where the multivariate model does not lead to better prediction performance, the MG-GBLUP provides estimates of genomic correlations between subpopulations that shed light on the extent of genetic heterogeneity between subpopulations. The estimated genomic correlations based on the variance–covariance matrix correspond to the marker effects correlations based on the variance–covariance matrix of the marker effects B. These measures of genetic similarity between subpopulations are trait specific. Because of the nature of the model, which is based on markers and not genotypes at causal loci, the genomic correlations are affected both by similarity/difference of QTL effects and by differences in marker–QTL LD between subpopulations. Those correlations between subpopulations are trait specific, as traits are assumed to be affected by different QTL with different contributions of epistasis and dominance (Karoui et al. 2012).
Beyond GBLUP
A rather strict assumption in MG-GBLUP is that marker effects among subpopulations are homogeneously correlated along the genome. One option would be to extend MG-GBLUP to Bayesian models with locus-specific correlations of effects. However, this largely increases the number of parameters that need to be estimated, leading to an even more underdetermined model. Another option, which would be more restrictive but where fewer parameters need to be estimated, would be to build clusters of markers and to assume different correlation structures for different marker clusters. This would allow that some markers are highly correlated among subpopulations, for example in regions where the marker–QTL LD is constant across subpopulations. Another group of markers might be very specific for each subpopulation, for example due to LD with differently segregating QTL.
Accommodating admixed individuals
The models discussed in this article assume that individuals cluster in homogeneous, clearly separable, groups; however, genetic variation sometimes is better described by a continuum in which some individuals belong to homogeneous subpopulations and others are admixed. Clearly, the models discussed in this article do not accommodate admixed lines adequately; further research is needed to develop methods that can deal with admixed individuals and structure simultaneously.
Conclusions
Multivariate models can be used to infer the extent to which marker effects vary among clusters in a heterogeneous population and to predict population-specific breeding values; this information allows characterizing genetic heterogeneity between subpopulations. Our results suggest that the extent to which effects change from cluster to cluster in a population depends not only on the genetic distance between clusters but also on the trait of interest.
From a prediction perspective, the choice of method seems to depend on the extent of genetic heterogeneity and on sample size. In highly differentiated populations (e.g., the rice data set) the W- and MG-GBLUPs tended to perform better. However, in more closely related subpopulations and with relatively small within-subpopulation sample size (e.g., the maize data set) the A-GBLUP model had a relatively good prediction performance. In general, one would expect benefits of using MG-GBLUP, relative to W- or A-GBLUP, in populations that exhibit a moderate degree of genetic differentiation between subpopulations and with small or intermediate sample size. As the within-sample size increases it would be reasonable to expect that the W-GBLUP model becomes increasingly competitive.
Supplementary Material
Acknowledgments
Researchers and institutions are acknowledged for developing the data sets and making them publicly available. The authors thank the editor Fred van Eeuwijk and two anonymous reviewers for the valuable comments provided during the review. This research was funded by the Federal Ministry of Education and Research (BMBF, Germany) within the AgroClustEr Synbreed–Synergistic plant and animal breeding (FKZ 0315528A). Gustavo de los Campos had financial support from two National Institutes of Health grants (R01GM099992 and R01GM101219) and a research grant sponsored by ARVALIS-Institut du Vegetal.
Footnotes
Communicating editor: F. van Eeuwijk
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.177394/-/DC1.
Literature Cited
- Albrecht T., Auinger H.-J., Wimmer V., Ogutu J. O., Knaak C., et al. , 2014. Genome-based prediction of maize hybrid performance across genetic groups, testers, locations, and years. Theor. Appl. Genet. 127: 1375–1386. [DOI] [PubMed] [Google Scholar]
- Bauer E., Falque M., Walter H., Bauland C., Camisan C., et al. , 2013. Intraspecific variation of recombination rate in maize. Genome Biol. 14: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brøndum R. F., Rius-Vilarrasa E., Strandén I., Su G., Guldbrandtsen B., et al. , 2011. Reliabilities of genomic prediction using combined reference data of the Nordic Red dairy cattle populations. J. Dairy Sci. 94: 4700–4707. [DOI] [PubMed] [Google Scholar]
- de los Campos G., Sorensen D., 2014. On the genomic analysis of data from structured populations. J. Anim. Breed. Genet. 131: 163–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G., Gianola D., Rosa G. J. M., Weigel K. A., Crossa J., 2010. Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet. Res. 92: 295–308. [DOI] [PubMed] [Google Scholar]
- de los Campos G., Hickey J. M., Pong-Wong R., Daetwyler H. D., Calus M. P. L., 2013. Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 193: 327–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., de los Campos G., Perez P., Gianola D., Burgueno J., et al. , 2010. Prediction of genetic values of quantitative traits in plant breeding using pedigree and molecular markers. Genetics 186: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roos A. P. W., Hayes B. J., Goddard M. E., 2009. Reliability of genomic predictions across multiple populations. Genetics 183: 1545–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbe M., Hayes B. J., Matukumalli L. K., Goswami S., Bowman P. J., et al. , 2012. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 95: 4114–4129. [DOI] [PubMed] [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics. Longman, Essex, England. [Google Scholar]
- Guo Z., Tucker D. M., Basten C. J., Gandhi H., Ersoz E., et al. , 2014. The impact of population structure on genomic prediction in stratified populations. Theor. Appl. Genet. 127: 749–762. [DOI] [PubMed] [Google Scholar]
- Hayes B. J., Bowman P. J., Chamberlain A. C., Verbyla K., Goddard M. E., 2009. Accuracy of genomic breeding values in multi-breed dairy cattle populations. Genet. Sel. Evol. 41: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidro J., Jannink J.-L., Akdemir D., Poland J., Heslot N., et al. , 2014. Training set optimization under population structure in genomic selection. Theor. Appl. Genet. 128: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janss L., de los Campos G., Sheehan N., Sorensen D., 2012. Inferences from genomic models in stratified populations. Genetics 192: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoui S., Carabaño M. J., Díaz C., Legarra A., 2012. Joint genomic evaluation of French dairy cattle breeds using multiple-trait models. Genet. Sel. Evol. 44: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehermeier C., Krämer N., Bauer E., Bauland C., Camisan C., et al. , 2014. Usefulness of multiparental populations of maize (Zea mays L.) for genome-based prediction. Genetics 198: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makgahlela M. L., Mäntysaari E. A., Strandén I., Koivula M., Nielsen U. S., et al. , 2013. Across breed multi-trait random regression genomic predictions in the Nordic Red dairy cattle. J. Anim. Breed. Genet. 130: 10–19. [DOI] [PubMed] [Google Scholar]
- Olson K. M., VanRaden P. M., Tooker M. E., 2012. Multibreed genomic evaluations using purebred Holsteins, Jerseys, and Brown Swiss. J. Dairy Sci. 95: 5378–5383. [DOI] [PubMed] [Google Scholar]
- Pérez P., de los Campos G., 2014. Genome-wide regression and prediction with the BGLR statistical package. Genetics 198: 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., et al. , 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2014. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Schulz-Streeck T., Ogutu J. O., Karaman Z., Knaak C., Piepho H. P., 2012. Genomic selection using multiple populations. Crop Sci. 52: 2453–2461. [Google Scholar]
- Wenzl P., Carling J., Kudrna D., Jaccoud D., Huttner E., et al. , 2004. Diversity Arrays Technology (DArT) for whole-genome profiling of barley. Proc. Natl. Acad. Sci. USA 101: 9915–9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer V., Lehermeier C., Albrecht T., Auinger H.-J., Wang Y., et al. , 2013. Genome-wide prediction of traits with different genetic architecture through efficient variable selection. Genetics 195: 573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windhausen, V. S., G. N. Atlin, J. M. Hickey, J. Crossa, J.-L. Jannink et al., 2012 Effectiveness of genomic prediction of maize hybrid performance in different breeding populations and environments. G3 (Bethesda) 2: 1427–1436. [DOI] [PMC free article] [PubMed]
- Wu B., Nianjun L., Zhao H., 2006. PSMIX: an R package for population structure inference via maximum likelihood method. BMC Bioinformatics 7: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., et al. , 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42: 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C.-W., Eizenga G. C., Wright M. H., Ali M. L., et al. , 2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Lund M. S., Wang Y., Su G., 2014. Genomic predictions across Nordic Holstein and Nordic Red using the genomic best linear unbiased prediction model with different genomic relationship matrices. J. Anim. Breed. Genet. 131: 249–257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All three data sets used in this study have already been published. The rice data can be downloaded from http://www.ricediversity.org/data/. The genotypic data of the maize lines can be downloaded from http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE50558 and the phenotypic data are available as Supporting Information, File S1 of Lehermeier et al. (2014). The wheat data set is available within the R package BGLR (http://cran.r-project.org/web/packages/BGLR/index.html). Details on the R code for fitting the models are given in the Supporting Information, File S1 of this article.