Abstract
AIM: To evaluate the correlations between cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and multi-drug resistance 1 (MDR1) genes polymorphisms with ulcerative colitis (UC) risk.
METHODS: PubMed, EMBASE, Web of Science, Cochrane Library, CBM databases, Springerlink, Wiley, EBSCO, Ovid, Wanfang database, VIP database, China National Knowledge Infrastructure, and Weipu Journal databases were exhaustively searched using combinations of keywords relating to CTLA-4, MDR1 and UC. The published studies were filtered using our stringent inclusion and exclusion criteria, the quality assessment for each eligible study was conducted using Critical Appraisal Skill Program and the resultant high-quality data from final selected studies were analyzed using Comprehensive Meta-analysis 2.0 (CMA 2.0) software. The correlations between SNPs of CTLA-4 gene, MDR1 gene and the risk of UC were evaluated by OR at 95%CI. Z test was carried out to evaluate the significance of overall effect values. Cochran’s Q-statistic and I2 tests were applied to quantify heterogeneity among studies. Funnel plots, classic fail-safe N and Egger’s linear regression test were inspected for indication of publication bias.
RESULTS: A total of 107 studies were initially retrieved and 12 studies were eventually selected for meta-analysis. These 12 case-control studies involved 1860 UC patients and 2663 healthy controls. Our major result revealed that single nucleotide polymorphisms (SNPs) of CTLA-4 gene rs3087243 G > A and rs231775 G > A may increase the risk of UC (rs3087243 G > A: allele model: OR = 1.365, 95%CI: 1.023-1.822, P = 0.035; dominant model: OR = 1.569, 95%CI: 1.269-1.940, P < 0.001; rs231775 G > A: allele model: OR = 1.583, 95%CI: = 1.306-1.918, P < 0.001; dominant model: OR = 1.805, 95%CI: 1.393-2.340, P < 0.001). In addition, based on our result, SNPs of MDR1 gene rs1045642 C > T might also confer a significant increases for the risk of UC (allele model: OR = 1.389, 95%CI: 1.214-1.590, P < 0.001; dominant model: OR = 1.518, 95%CI: 1.222-1.886, P < 0.001).
CONCLUSION: CTLA-4 gene rs3087243 G > A and rs231775 G > A, and MDR1 gene rs1045642 C > T might confer an increase for UC risk.
Keywords: Ulcerative colitis, Cytotoxic T lymphocyte-associated antigen-4, Multi-drug resistance 1, rs3087243 G > A, rs231775 G > A, rs1045642 C > T, Polymorphism, Meta-analysis
Core tip: To evaluate the correlations between cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and multi-drug resistance 1 (MDR1) genes polymorphisms with ulcerative colitis (UC) risk. CTLA-4 gene rs3087243 G > A and rs231775 G > A, and MDR1 gene rs1045642 C > T might confer an increases for UC risk.
INTRODUCTION
Ulcerative colitis (UC) is known as an idiopathic, chronic inflammatory disease of the large intestine, frequently involving the rectum, and characterized by continuous inflammation and ulceration of intestinal mucosa and submucosa[1]. In the United States, UC affects approximately 500000 individuals with an incidence of 8-12 per 100000 populations per year and the incidence has remained relatively constant over the last five decades[2]. Crohn’s disease (CD) and UC are two forms of inflammatory bowel diseases (IBD), and while CD can impact any segment of the gastrointestinal tract, UC pathology is restricted to the colon[3]. The precise etiology of UC remains unknown, but factors such as the host immune system, other genetic factors, and environmental factors, contribute to the occurrence of UC[4,5]. Typical symptoms of UC include abdominal cramping, rectal bleeding and persistent bloody diarrhea, and other symptoms such as severe fecal urgency resulting from reduced rectal compliance, irritability, general malaise, incontinence, and weight loss are also common[6]. UC is treated in clinics with azathioprine, mesalamine, glucocorticoids, and anti-tumor necrosis factor agents (infliximab and adalimumab)[7]. Recently, single nucleotide polymorphisms (SNPs) of Cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and multi-drug resistance 1 (MDR1) genes were found to be associated with the pathogenesis of UC[8,9].
CTLA-4 plays a crucial role in the immune system inducing immune tolerance and is an essential negative regulator of T cell-mediated immune response[10]. CTLA-4 usually functions as a co-inhibitory molecule interacting with B7.1 (CD80) and B7.2 (CD86) expressed on antigen-presenting cells[11]. CTLA-4 gene encodes a 40-kDa transmembrane CTLA-4 glycoprotein and the gene is located on chromosome 2q33 in humans[12]. CTLA-4 dampens the signal transduction in T cells in the presence of antigen presenting cells, and downregulation of CTLA-4 expression is implicated in T cell associated autoimmunity and lymphoproliferative diseases[13]. MDR1, also called ATP-binding cassette subfamily B member 1 (ABCB1), is extremely important in multidrug resistance of cancer cells and therapy effectiveness in several other disorders[14]. The MDR1 gene is located on chromosome 7q21.1 and encodes a glycoprotein of 170 kDa[15]. MDR1 was originally identified as a gene amplified in multiple drug-resistant cells, and its product, P-gp, plays an important role in drug resistance[16]. Previous studies have proposed that some SNPs of CTLA-4 gene, such as rs3087243 G > A and rs231775 G > A, and SNPs of MDR1 gene rs1045642 C > T may increase the risk of UC[17,18]. However, these associations have not been confirmed, and contradictory data exists in different populations[19,20]. In order to address this relationship further, we undertook a meta-analysis based approach to evaluate the associations of SNPs of CTLA-4 and MDR1 genes with the risk of UC, by pooling all relevant published data.
MATERIALS AND METHODS
Search strategy
An extensive literature search for relevant studies was conducted on PubMed, EMBASE, Web of Science, Cochrane Library, CBM databases, Springerlink, Wiley, EBSCO, Ovid, Wanfang database, VIP database, China National Knowledge Infrastructure (CNKI), and Weipu Journal databases from their inception through to October 1st, 2014. We used the following keywords and MeSH terms: “costimulatory and inhibitory t-cell receptors” or “CTLA-4 antigen” or “cytotoxic t-lymphocyte-associated antigen 4” or “CD152 antigen” or “cytotoxic t lymphocyte antigen 4” OR “CTLA-4” and (“colitis, ulcerative” or “idiopathic proctocolitis” or “ulcerative colitis” or “Colitis gravis” or “inflammatory bowel disease, ulcerative colitis type”); “genes, MDR” or “ABCB1” or “MDR1” or “multidrug resistance 1” or “ABCB1 protein, human” or “MDR” and (“colitis, ulcerative” or “idiopathic proctocolitis” or “ulcerative colitis” or “colitis gravis” or “Inflammatory bowel disease, ulcerative colitis type”). All study types with no language restrictions were included in our search. Manual searches were carried on to retrieve other cross-references.
Inclusion and exclusion criteria
Identified articles were reviewed in their entirety and selected if they met the following inclusion criteria: (1) study type: case-control studies; (2) research topic: correlations between SNPs of CTLA-4 gene, MDR1 gene and the risk of UC; (3) subject investigated: UC patients in the case group and normal controls in the control group; (4) end indicators: all the include studies provided complete data such as: age, country, ethnicity, language, detection method, SNP site information; and (5) studies were published in Chinese or in English. The exclusion criteria were: (1) unclear diagnostic basis for subjects investigated; (2) animal research; (3) studies with insufficient data; and (4) duplicate publications.
Data extraction and quality assessment
Literature screening was performed by two independent reviewers, based on a predetermined data collection table. Relevant information including first author, published year, country, ethnicity, sample, disease, source of controls, gender, genotype method, gene, SNP were extracted from the eligible literature. Any disagreements in study selection between the two reviewers were resolved through discussion by all authors until consensus was obtained. The quality assessment for each eligible study was conducted using Critical Appraisal Skill Program (CASP) (http://www.casp-uk.net/#!casp-tools-checklists/c18f8).
Statistical analysis
The statistical methods of this study were reviewed by Zhou LP from Clinical Laboratory and Department of Clinical Epidemiology, the First Affiliated Hospital of China Medical University, See Supplement S1.Comprehensive Meta-analysis 2.0 software (Biostatic Inc., Englewood, New Jersey, United States) was used for data analysis in present meta-analysis. The correlations between SNPs of CTLA-4 gene, MDR1 gene and the risk of UC were evaluated by OR at 95%CI. Z test was carried out to evaluate the significance of overall effect values[21]. Forest plots were drawn to reflect the comparisons of OR and 95%CI among the study groups. Cochran’s Q-statistic (P < 0.05 was considered significant) and I2 tests were applied to quantify heterogeneity among studies[22,23]. In order to calculate the pool ORs, fixed/random effects model were used. When significant heterogeneity was observed (P < 0.05 or I2 > 50%), a fixed effect model was used, otherwise, the random effect model was employed[24]. Univariate and multivariate meta-regression analysis was utilized to identify potential sources of heterogeneity, and further confirmed by Monte Carlo method[25,26]. One-way sensitivity analysis was performed to assess whether the results had significant influences on the overall outcomes by deleting single studies one by one. Funnel plots, classic fail-safe N and Egger’s linear regression test were inspected for indication of publication bias, and confirmed the reliability of original analysis results[27,28]. A bilateral test was conducted with P value of less than 0.05 considered to be significant.
RESULTS
Baseline characteristics of included studies
The initial database and manual search retrieved a total of 107 relevant articles. After excluding duplicates (n = 8), non-human studies (n = 6), letters, reviews (n = 8) and studies unrelated to topic (n = 15), 70 full-text articles remained. Twelve studies finally met the inclusion criteria after we eliminated studies that were not case-control (n = 14), irrelevant to CTLA-4 (n = 22), not irrelevant MDR1 (n = 12), irrelevant to UC (n = 6) and insufficient information (n = 4). The twelve selected studies[17,19,20,29-37], published between 2003 and 2013, contained a total of 1860 UC patients and 2663 healthy controls. Sample size varied between 195-900 patients. Of the 12 studies, 5 studies were conducted in Asians (2 from Iran, 1 from China and 2 from Japan), 1 study was performed in Africa (Tunisia), and the remaining 6 studies were in Caucasians with one study each in Croatia, Slovenia, Hungary, United Kingdom, Netherlands and Germany. SNP detection methods included polymerase chain reaction with the restriction fragment length polymorphism (PCR-RFLP) and TaqMan assay. The genotype distributions of all included studies conformed to Hardy Weinberg Equilibrium (HWE) (P > 0.05) except for MDR1 rs1045642 C > T in one study. The baseline characteristics of the included studies and CASP for eligible studies are shown in Table 1 and Figure 1, respectively.
Table 1.
Baseline characteristics of all eligible studies
| First author | Year | Ethnicity | Genotype method |
Number |
Gender (M/F) |
Gene | SNP | ||
| UC | Control | UC | Control | ||||||
| Brinar et al[29] | 2013 | Caucasian | PCR-RFLP | 52/57 | 29.9 (26.0-35.70) | - | MDR1 | C3435T | |
| Repnik et al[35] | 2010 | Caucasian | PCR-RFLP | - | 119/147 | - | 37.2 | CTLA4 | rs3087243 G/A |
| Ben Alaya et al[17] | 2009 | Africa | PCR-RFLP | 18/47 | 52/48 | 39.9 (25-74) | 32.4 (24-55) | CTLA4 | rs231775 G/A |
| Farnood et al[30] | 2007 | Asian | PCR-RFLP | - | - | - | - | MDR1 | C3435T |
| Magyari et al[19] | 2007 | Caucasian | PCR-RFLP | 63/87 | 49/121 | 46.1 ± 1.3 | 57.7 ± 1.3 | CTLA4 | rs231775 G/A |
| Lankarani et al[32] | 2006 | Asian | PCR-RFLP | 48/52 | 48/52 | 31.6 ± 13.4 | 35.36 ± 11.94 | CTLA4 | rs231775 G/A |
| Wang et al[37] | 2006 | Asian | PCR-RFLP | 41/38 | 92/72 | 44.2 ± 16.2 | 37.8 ± 14.1 | CTLA4 | rs3087243 G/A |
| Osuga et al[34] | 2006 | Asian | PCR-RFLP | 37/29 | 97/76 | 42.7 ± 14.2 | 19-76 | MDR1 | C3435T |
| Ho et al[31] | 2006 | Caucasian | PCR-RFLP | - | - | - | - | MDR1 | C3435T |
| Rueda et al[20] | 2005 | Caucasian | TaqMan | - | - | - | - | CTLA4 | rs3087243 G/A |
| Machida et al[33] | 2005 | Asian | PCR-RFLP | 57/51 | 125/75 | 44.0 ± 16.9 | 32.5 ± 11.1 | CTLA4 | rs3087243 G/A |
| Machida et al[33] | 2005 | Asian | PCR-RFLP | 57/51 | 125/75 | 44.0 ± 16.9 | 32.5 ± 11.1 | CTLA4 | rs231775 G/A |
| Schwab et al[36] | 2003 | Caucasian | TaqMan | 86/63 | 86/63 | 34 ± 11 | - | MDR4 | C3435T |
PB: Population-based; HB: Hospital-based; PCR-RFLP: Polymerase chain reaction restriction fragment length polymorphism; UC: Ulcerative colitis; MDR1: Multi-drug resistance 1; CTLA4: Cytotoxic T lymphocyte-associated antigen-4; SNP: Single nucleotide polymorphisms.
Figure 1.
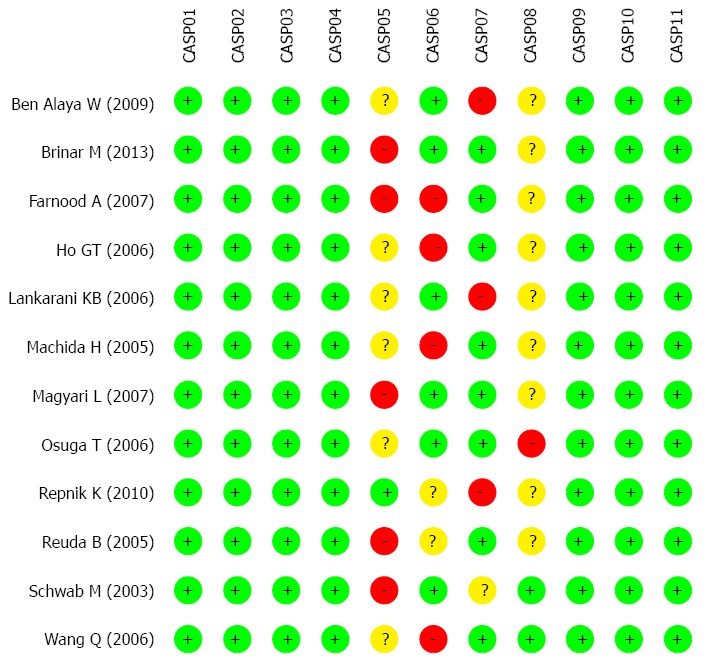
Methodological quality of included studies was evaluated by Critical Appraisal Skill Program criteria. CASP: Critical Appraisal Skill Program.
Association of CTLA-4 gene rs3087243 G > A and the risk of UC
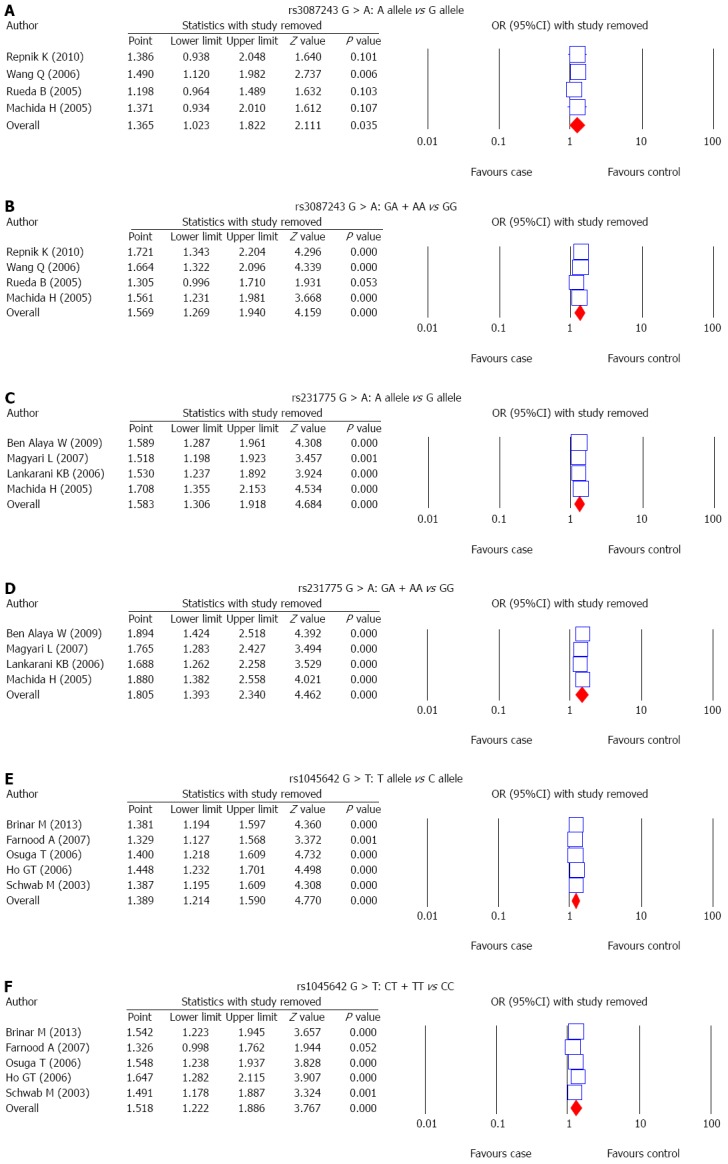
The correlations between SNP of CTLA-4 gene, rs3087243 G > A, and UC risk were reported in 4 studies. A random-effects model was used since there was evidence of heterogeneity under the allele model and a fixed-effects model was used for the absence of heterogeneity under the dominant model (P = 0.03; P = 0.121). The results of meta-analysis showed that rs3087243 G > A is associated with increased risk of UC (allele model: OR = 1.365, 95%CI: 1.023-1.822, P = 0.035; dominant model: OR = 1.569, 95%CI: 1.269-1.940, P < 0.001) (Figure 2A and B and Table 2). Subgroup analysis based on ethnicity showed no significant association between rs3087243 G > A and the risk of UC in Asians (allele model: OR = 1.153, 95%CI: 0.860-1.546, P = 0.340; dominant model: OR = 1.377, 95%CI: 0.963-1.970, P = 0.079). However, in Caucasians, rs3087243 G > A is associated with an increased risk of UC (allele model: OR = 1.563, 95%CI: 1.056-2.313, P = 0.026; dominant model: OR = 1.685, 95%CI: 1.294-2.194, P < 0.001) (Figure 3A and B; Table 2).
Figure 2.
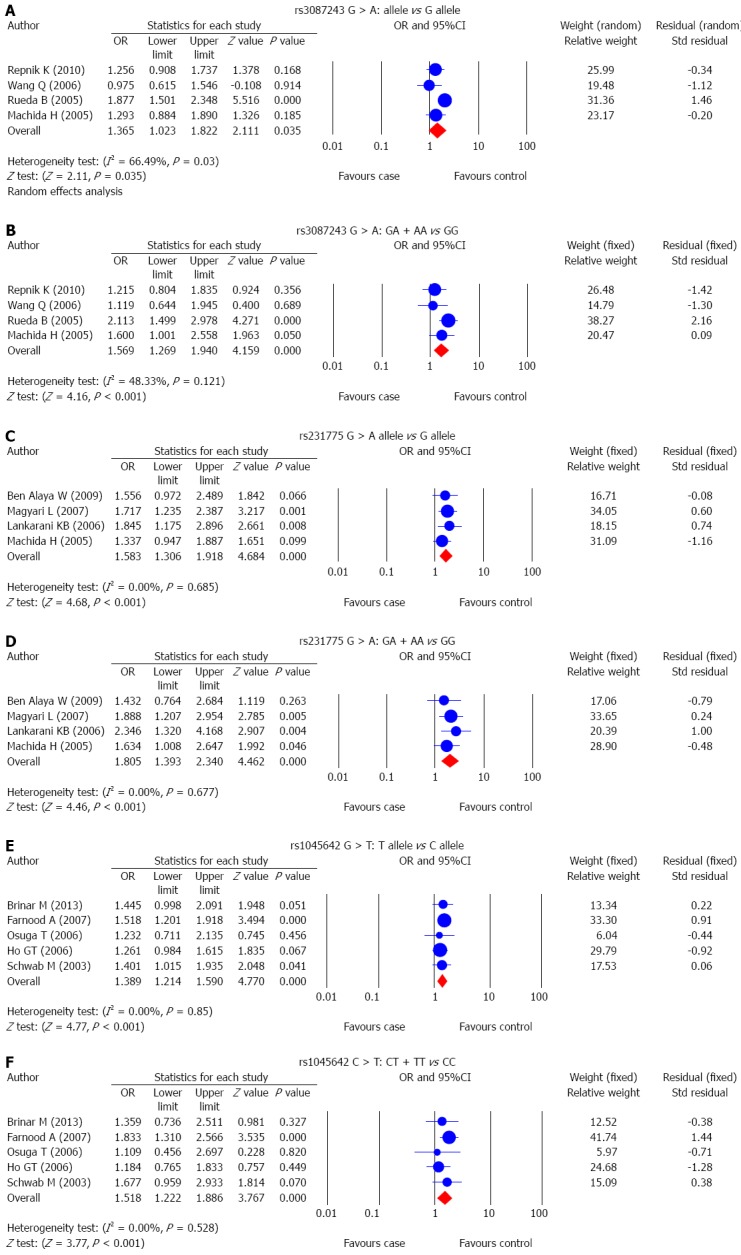
Forest plots for the correlations between single nucleotide polymorphisms of CTLA-4 and MDR1 with ulcerative colitis risk.
Table 2.
Comparisons of genotype and allele frequencies between the case and the control groups
| SNP |
rs3087243 G/A |
rs231775 G/A |
rs1045642 C/T |
||||||
| OR | 95%CI | P value | OR | 95%CI | P value | OR | 95%CI | P value | |
| Allele model | |||||||||
| Asians | 1.15 | 0.86-1.55 | 0.340 | 1.51 | 1.15-1.98 | 0.003 | 1.47 | 1.19-1.82 | < 0.001 |
| Caucasians | 1.56 | 1.06-2.31 | 0.026 | 1.72 | 1.24-2.39 | 0.001 | 1.34 | 1.13-1.59 | 0.001 |
| Africas | - | - | - | 1.56 | 0.97-2.49 | 0.066 | - | - | - |
| Overall | 1.37 | 1.02-1.82 | 0.035 | 1.58 | 1.31-1.92 | < 0.001 | 1.39 | 1.21-1.59 | < 0.001 |
| Dominant model | |||||||||
| Asians | 1.38 | 0.96-1.97 | 0.079 | 1.90 | 1.31-2.75 | 0.001 | 1.72 | 1.26-2.36 | 0.001 |
| Caucasians | 1.69 | 1.29-2.19 | < 0.001 | 1.89 | 1.21-2.95 | 0.005 | 1.35 | 1.00-1.83 | 0.048 |
| Africas | - | - | - | 1.43 | 0.76-2.68 | 0.263 | - | - | - |
| Overall | 1.57 | 1.27-1.94 | < 0.001 | 1.81 | 1.39-2.34 | < 0.001 | 1.52 | 1.22-1.89 | < 0.001 |
| Homozygous model | |||||||||
| Overall | 2.41 | 1.67-3.48 | < 0.001 | 2.21 | 1.45-3.37 | < 0.001 | 1.92 | 1.44-2.56 | < 0.001 |
| Heterozygous model | |||||||||
| Overall | 0.62 | 0.44-0.89 | 0.008 | 0.76 | 0.50-1.15 | 0.190 | 0.67 | 0.53-0.86 | 0.001 |
| Recessive model | |||||||||
| Overall | 1.98 | 1.42-2.76 | < 0.001 | 1.73 | 1.16-2.57 | 0.007 | 1.62 | 1.29-2.05 | < 0.001 |
OR: Odds ratio.
Figure 3.
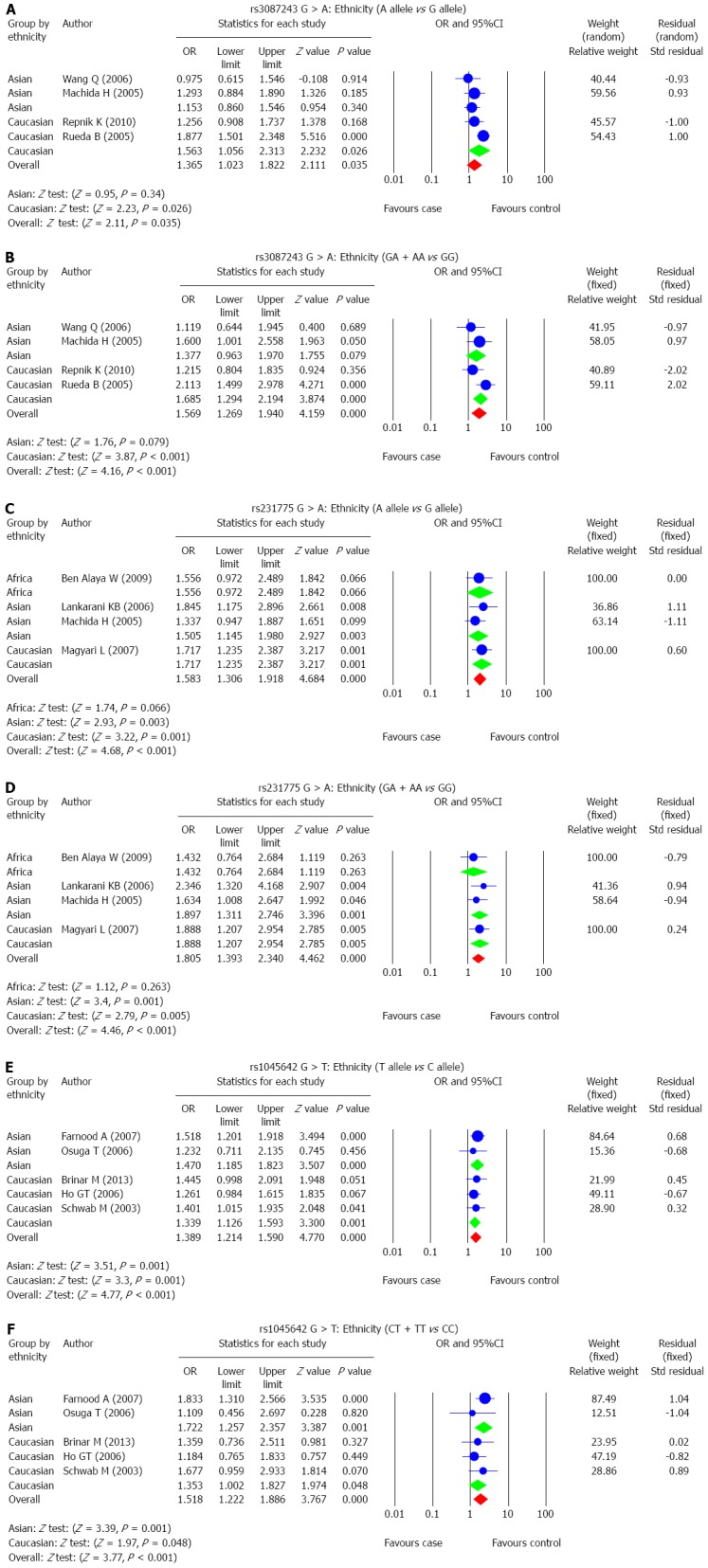
Subgroup analysis for the correlations between single nucleotide polymorphisms of CTLA-4 and MDR1 with ulcerative colitis risk.
Association of CTLA-4 gene rs231775 G > A and the risk of UC
The correlation between SNP of CTLA-4 gene rs231775 G > A, and UC risk was reported in 4 studies. The results of the heterogeneity test showed no significant heterogeneity under the allele model and dominant model, thus the fixed-effects model was applied in this meta-analysis (all P > 0.05). Pooled data in this meta-analysis showed that rs231775 G > A is associated with significantly increased risk of UC (allele model: OR = 1.583, 95%CI: 1.306-1.918, P < 0.001; dominant model: OR = 1.805, 95%CI: 1.393-2.340, P < 0.001) (Figure 2C and D; Table 2). Subgroup analysis based on ethnicity showed no significant association between rs231775 G > A and the risk of UC in Africans (allele model: OR = 1.556, 95%CI: 0.972-2.489, P = 0.066; dominant model: OR = 1.432, 95%CI: 0.764-2.684, P = 0.263), but in both Asians and Caucasians, rs231775 G > A was strongly associated with significant increases in the incidence of UC (Asians: allele model: OR = 1.505, 95%CI: 1.145-1.980, P = 0.003; dominant allele: OR = 1.897, 95%CI: 1.311-2.746, P = 0.001; Caucasians: allele model: OR = 1.717, 95%CI: 1.235-2.387, P = 0.001; dominant model: OR = 1.888, 95%CI: 1.207-2.954, P = 0.005) (Figure 3C and D; Table 2).
Association of MDR1 gene rs1045642 C > T and the risk of UC
The correlation between SNP of MDR1 gene, rs1045642 C > T, and UC risk was discussed in 5 studies. A fixed-effects model was used for the absence of heterogeneity under the allele model and the dominant model (all P > 0.05). The results of meta-analysis showed that rs1045642 C > T is linked to increased risk of UC (allele model: OR = 1.389, 95%CI: 1.214-1.590, P < 0.001; dominant model: OR = 1.518, 95%CI: 1.222-1.886, P < 0.001) (Figure 2E and F and Table 2). Subgroup analysis based on ethnicity showed a significant association between rs1045642 C > T and the risk of UC in both Asians and Caucasians (Asians: allele model: OR = 1.470, 95%CI: 1.185-1.823, P < 0.001; dominant model: OR = 1.722, 95%CI: 1.257-2.357, P = 0.001; Caucasians: allele model: OR = 1.339, 95%CI: 1.126-1.593, P = 0.001; dominant model: OR = 1.353, 95%CI: 1.002-1.827, P = 0.048) (Figure 3E and F; Table 2).
Sensitivity analysis and publication bias
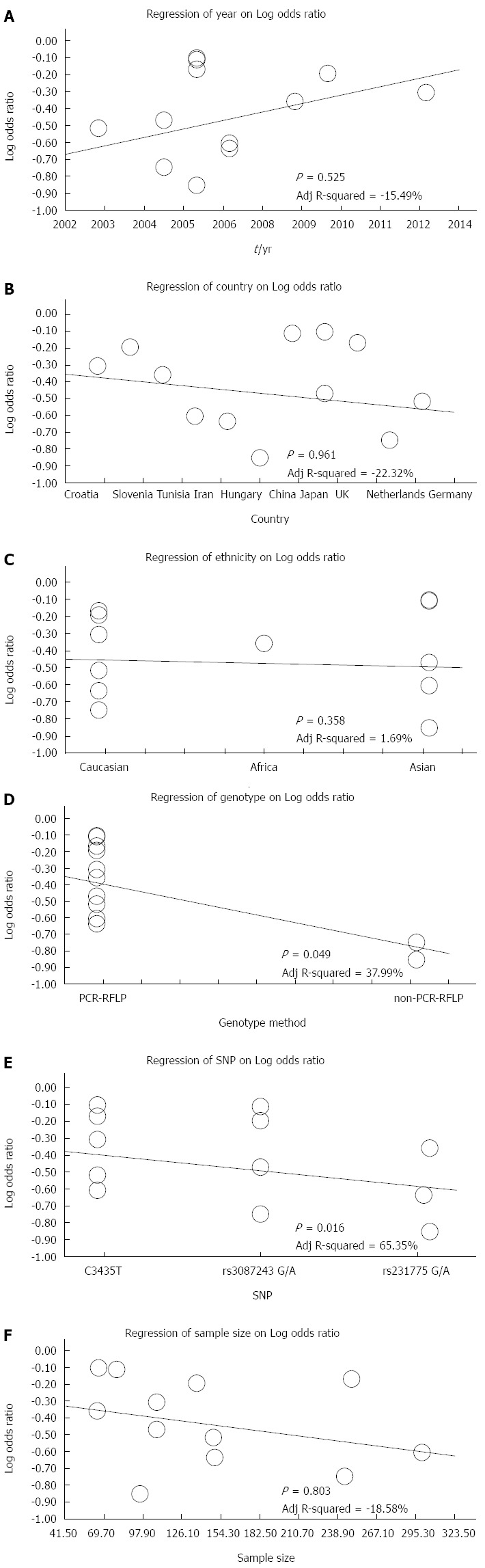
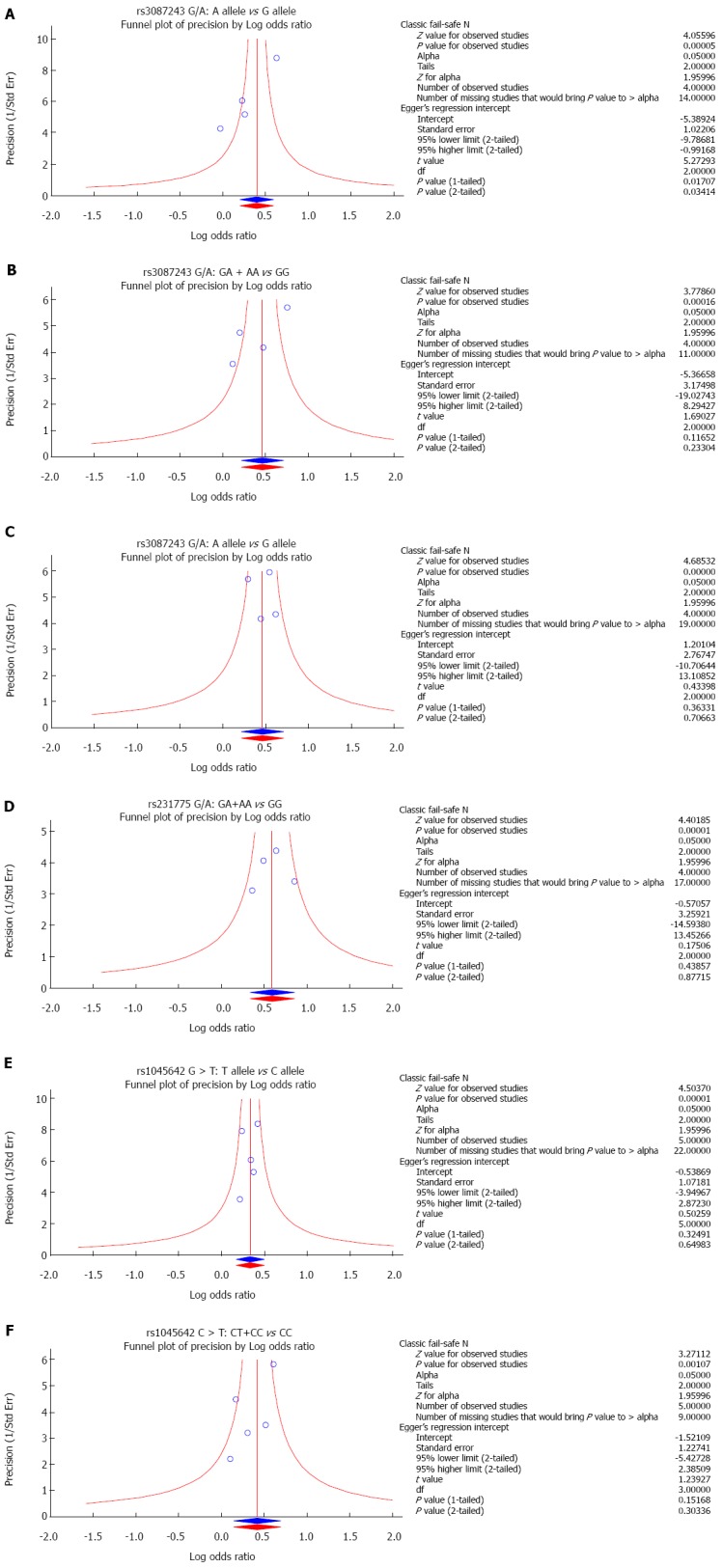
The results of sensitivity analysis demonstrated that any single study had no significant effect on pooled ORs of correlations between SNPs of CTLA-4 gene rs3087243 G > A, rs231775 G > A and MDR1 gene rs1045642 C > T, and the risk of UC (Figure 4). Univariate meta-regression analysis suggested that publication year, country, ethnicity, sample size, SNP, and detection methods were not the main sources for heterogeneity or the key factors affecting the overall effect values (P > 0.05). Multivariate meta-regression analysis further confirmed that the published year, country, ethnicity, sample size, SNP, and detection methods are not the sources of heterogeneity (Figure 5 and Table 3). Funnel plots for rs3087243 G > A under the allele model were asymmetric, suggesting the existence of publication bias. Classic fail-safe N and Egger’s linear regression tests further confirmed there was publication bias. However, funnel plots for rs3087243 G > A under the dominant model, rs231775 G > A and rs1045642 C > T both under the allele model and dominant model were symmetrical, revealing no significant publication bias. Classic fail-safe N and Egger’s linear regression tests further confirmed there was no publication bias (Figure 6).
Figure 4.
Sensitivity analysis for the correlations between single nucleotide polymorphisms of CTLA-4 and MDR1 with ulcerative colitis risk. SNP: Single nucleotide polymorphism.
Figure 5.
Meta-regression analysis on the correlations between single nucleotide polymorphisms of CTLA-4 and MDR1 with ulcerative colitis risk.
Table 3.
Meta-regression analyses of potential source of heterogeneity
| Heterogeneity factors | Coefficient | SE | t | P value |
95%CI |
|
| (adjusted) | LL | UL | ||||
| Year | -0.061 | 0.035 | -1.76 | 0.359 | -0.15 | 0.028 |
| Country | -0.054 | 0.03 | -1.78 | 0.352 | -0.131 | 0.024 |
| Ethnicity | 0.001 | 0.042 | 0.03 | 1 | -0.106 | 0.109 |
| Method | 0.203 | 0.092 | 2.2 | 0.219 | -0.034 | 0.44 |
| SNP | 0.045 | 0.057 | 0.79 | 0.85 | -0.101 | 0.191 |
| Sample | 0 | 0 | 0.09 | 1 | 0 | 0 |
SE: Standard error; LL: Lower limit; UL: Upper limit. SNP: Single nucleotide polymorphism.
Figure 6.
Funnel plot of publication biases on the correlations of single nucleotide polymorphisms of CTLA-4 and MDR1 with ulcerative colitis risk.
DISCUSSION
UC is a non-specific chronic inflammatory disorder which, together with CD, is known as IBD, and it is characterized by diffuse mucosal inflammation confined to the colon[8]. Evidence has revealed that genetic factors and immune dysregulation may be two main important components in the etiology and pathogenesis of UC[38]. Recent genome-wide association studies (GWAS) have discovered multiple genes and loci for UC risk factors; for example, GWAS meta-analyses have established more than 30 loci in CD, and several of these loci have also been found to be correlated to UC[39]. The GWAS that evaluated the correlation between UC and the variants of the CTLA-4 gene and MDR1 genes produced contradictory or inconclusive results; we found that associations have been found in some, but not all populations[40]. Therefore, in order to investigate the correlations between SNPs of the CTLA-4 gene, MDR1 gene and the risk of UC, a meta-analysis was conducted.
Our meta-analysis demonstrated that CTLA-4 gene polymorphisms, rs3087243 G > A and rs231775 G > A, are closely associated with the increased risk of UC. CTLA-4 is primarily expressed in activated T cells, and it also plays an inhibitory role in the regulation of self-tolerance and T-cell functions, suggesting that CTLA-4, by virtue of its influence on T-cell regulation, may potentially influence UC disease susceptibility[17]. CTLA-4 is described as a vital downregulator of T-cell activation resulting in peripheral tolerance, and is also a negative regulator of T/B, T-cell activation and T/monocyte-macrophage cognate interaction. Thus, CTLA-4 has been recognized as a good candidate gene for the susceptibility to UC[41]. In this context, downregulation of CTLA-4 may contribute to an exaggerated T cell response, along with a persistent inflammation response in the gastrointestinal mucosa, possibly initiating the development of UC[42]. Data from a previous study showed that the CTLA-4 gene is involved in the pathogenesis of IBD and UC among Slovenian patients and that CT60 (rs3087243 G > A) polymorphism is an important target regulating CTLA-4 gene expression[35].
Our meta-analysis also revealed a significant association between MDR1 gene polymorphism, rs1045642 C > T, and the risk of UC. As an energy-dependent efflux pump, MDR1 (P-gp) plays a crucial role in the bioavailability and cytotoxicity of a large number of drugs, substances, and xenobiotics including sugars, glycans, ions, proteins, phospholipids, antibiotics, corticosteroids, anticancer drugs, immunosuppressants, calcium-channel blocker agents and anti- (human immunodeficiency virus) HIV protease inhibitors[43]. UC patients frequently exhibit reduced P-gp expression levels, and MDR1 mRNA expression might also be reduced in the colonic tissue of UC patients. G2677T/A (Ala893Ser/Thr, rs2032582) and C3435T (Ile1145Ile, rs1045642) are two common polymorphisms (variants) in MDR1 gene, and correlate with the function and activity of P-gp[44]. Data from a previous study revealed a 2-fold increased OR for the development of UC in patients with the MDR1 rs1045642 C > T genotype, supporting the notion that P-gp expression plays an essential role in defense against intestinal bacteria and low P-gp expression, as a result of an rs1045642 C > T genotype, contributes to the development of UC[34]. Based on our results and previous studies supporting our conclusions, we propose that individuals with the rs1045642 C > T have a reduced intestinal barrier function, and thus are at a significantly higher risk for developing UC.
To further explore the effect of other influential factors like ethnicity on the correlation between SNPs of CTLA-4 gene, MDR1 gene and the risk of UC, a subsequent subgroup meta-analysis was conducted. Subgroup analysis based on ethnicity showed that in Asians, there was no significant association between rs3087243 G > A and the risk of UC. However, in Caucasians, rs3087243 G > A may increase the risk of UC. In Africans, no significant association was found between rs231775 G > A and the risk of UC. But both in Asians and Caucasians, rs231775 G > A was likely to increase the incidence of UC. Furthermore, both in Asians and Caucasians, there was a significant association between rs1045642 C > T and the risk of UC. Our overall results are consistent with previous studies, which suggested a significant association of SNPs of the CTLA-4 gene and the MDR1 gene with the pathogenesis of UC.
Limitations of this meta-analysis need to be addressed. First, the sample size is relatively small. Second, all eligible studies were written in English and Chinese indexed by the selected databases. It is possible that published studies in other languages or unpublished studies could be missed, which might bias the results. Third, the genotyping methods were not uniform and might increase the deviation of outcomes. Therefore, more studies with larger sample sizes are still needed to provide a more accurate statistical analysis.
This meta-analysis provides strong evidence that SNPs of CTLA-4 gene rs3087243 G > A and rs231775 G > A, and MDR1 gene polymorphism rs1045642 C > T significantly increase the risk of UC, and the polymorphisms can be used as important biological indicators for early diagnosis of UC.
COMMENTS
Background
Ulcerative colitis (UC) is known as an idiopathic, chronic inflammatory disease of the large intestine, frequently involving the rectum, and characterized by continuous inflammation and ulceration of intestinal mucosa and submucosa.
Research frontiers
Recently, single nucleotide polymorphisms (SNPs) of Cytotoxic T lymphocyte-associated antigen-4 (CTLA-4) and multi-drug resistance 1 (MDR1) genes were found to be associated with the pathogenesis of UC.
Innovations and breakthroughs
The authors undertook a meta-analysis based approach to evaluate the associations of SNPs of CTLA-4 and MDR1 genes with the risk of UC, by pooling all relevant published data.
Applications
This meta-analysis provides strong evidence that SNPs of CTLA-4 gene rs3087243 G > A and rs231775 G > A, and MDR1 gene polymorphism rs1045642 C > T significantly increase the risk of UC, and the polymorphisms can be used as important biological indicators for early diagnosis of UC
Peer-review
The authors address, with their meta-analysis, the risk of ulcerative colitis in the presence of genetic polymorphisms of CTLA and MDR1.
Footnotes
Conflict-of-interest statement: The declaration of the authors reveals that they have not received any financial payments or other benefits from any commercial entity associated with the subject of this article.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: January 7, 2015
First decision: January 22, 2015
Article in press: May 4, 2015
P- Reviewer: Kapischke M, Karatapanis S, Pessi T S- Editor: Qi Y L- Editor: O’Neill M E- Editor: Liu XM
References
- 1.Reinisch W, Sandborn WJ, Hommes DW, D’Haens G, Hanauer S, Schreiber S, Panaccione R, Fedorak RN, Tighe MB, Huang B, et al. Adalimumab for induction of clinical remission in moderately to severely active ulcerative colitis: results of a randomised controlled trial. Gut. 2011;60:780–787. doi: 10.1136/gut.2010.221127. [DOI] [PubMed] [Google Scholar]
- 2.Kornbluth A, Sachar DB. Ulcerative colitis practice guidelines in adults: American College Of Gastroenterology, Practice Parameters Committee. Am J Gastroenterol. 2010;105:501–523; quiz 524. doi: 10.1038/ajg.2009.727. [DOI] [PubMed] [Google Scholar]
- 3.Krishnan K, Arnone B, Buchman A. Intestinal growth factors: potential use in the treatment of inflammatory bowel disease and their role in mucosal healing. Inflamm Bowel Dis. 2011;17:410–422. doi: 10.1002/ibd.21316. [DOI] [PubMed] [Google Scholar]
- 4.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 5.Salim SY, Söderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:362–381. doi: 10.1002/ibd.21403. [DOI] [PubMed] [Google Scholar]
- 6.Colombel JF, Rutgeerts P, Reinisch W, Esser D, Wang Y, Lang Y, Marano CW, Strauss R, Oddens BJ, Feagan BG, et al. Early mucosal healing with infliximab is associated with improved long-term clinical outcomes in ulcerative colitis. Gastroenterology. 2011;141:1194–1201. doi: 10.1053/j.gastro.2011.06.054. [DOI] [PubMed] [Google Scholar]
- 7.Sandborn WJ, Ghosh S, Panes J, Vranic I, Su C, Rousell S, Niezychowski W. Tofacitinib, an oral Janus kinase inhibitor, in active ulcerative colitis. N Engl J Med. 2012;367:616–624. doi: 10.1056/NEJMoa1112168. [DOI] [PubMed] [Google Scholar]
- 8.Jiang T, Ge LQ, Chen ZT, Li C, Zhou F, Luo Y, Xia B. Effect of cytotoxic T lymphocyte-associated molecule 4 1661 gene polymorphism on its expression and transcription in ulcerative colitis. J Dig Dis. 2010;11:369–375. doi: 10.1111/j.1751-2980.2010.00462.x. [DOI] [PubMed] [Google Scholar]
- 9.Herrlinger KR, Koc H, Winter S, Teml A, Stange EF, Fellermann K, Fritz P, Schwab M, Schaeffeler E. ABCB1 single-nucleotide polymorphisms determine tacrolimus response in patients with ulcerative colitis. Clin Pharmacol Ther. 2011;89:422–428. doi: 10.1038/clpt.2010.348. [DOI] [PubMed] [Google Scholar]
- 10.Gao JW, Guo YF, Fan Y, Qiu JX, Bao ED, Liu Y, Qin Y, Zhang F. Polymorphisms in cytotoxic T lymphocyte associated antigen-4 influence the rate of acute rejection after renal transplantation in 167 Chinese recipients. Transpl Immunol. 2012;26:207–211. doi: 10.1016/j.trim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Li M, Zheng H, Li T, Gao P, Zhang XL, Liu DW. Cytotoxic T-lymphocyte associated antigen-4 gene polymorphisms and primary biliary cirrhosis: a systematic review. J Gastroenterol Hepatol. 2012;27:1159–1166. doi: 10.1111/j.1440-1746.2012.07118.x. [DOI] [PubMed] [Google Scholar]
- 12.Wells AD, Walsh MC, Bluestone JA, Turka LA. Signaling through CD28 and CTLA-4 controls two distinct forms of T cell anergy. J Clin Invest. 2001;108:895–903. doi: 10.1172/JCI13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang ST, Tang HQ, Zhang Q, Wang CJ, Wang YM, Peng WJ. Association of cytotoxic T-lymphocyte associated antigen 4 gene polymorphism with type 1 diabetes mellitus: a meta-analysis. Gene. 2012;508:165–187. doi: 10.1016/j.gene.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 14.Ni LN, Li JY, Miao KR, Qiao C, Zhang SJ, Qiu HR, Qian SX. Multidrug resistance gene (MDR1) polymorphisms correlate with imatinib response in chronic myeloid leukemia. Med Oncol. 2011;28:265–269. doi: 10.1007/s12032-010-9456-9. [DOI] [PubMed] [Google Scholar]
- 15.Bodor M, Kelly EJ, Ho RJ. Characterization of the human MDR1 gene. AAPS J. 2005;7:E1–E5. doi: 10.1208/aapsj070101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takakura Y, Hinoi T, Oue N, Sasada T, Kawaguchi Y, Okajima M, Akyol A, Fearon ER, Yasui W, Ohdan H. CDX2 regulates multidrug resistance 1 gene expression in malignant intestinal epithelium. Cancer Res. 2010;70:6767–6778. doi: 10.1158/0008-5472.CAN-09-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben Alaya W, Sfar I, Aouadi H, Jendoubi S, Najjar T, Filali A, Gorgi Y, Ben Abdallah T, Mouelhi L, Matri S, et al. Association between CTLA-4 gene promoter (49 A/G) in exon 1 polymorphisms and inflammatory bowel disease in the Tunisian population. Saudi J Gastroenterol. 2009;15:29–34. doi: 10.4103/1319-3767.43285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zintzaras E. Is there evidence to claim or deny association between variants of the multidrug resistance gene (MDR1 or ABCB1) and inflammatory bowel disease? Inflamm Bowel Dis. 2012;18:562–572. doi: 10.1002/ibd.21728. [DOI] [PubMed] [Google Scholar]
- 19.Magyari L, Faragó B, Bene J, Horvatovich K, Lakner L, Varga M, Figler M, Gasztonyi B, Mózsik G, Melegh B. No association of the cytotoxic T-lymphocyte associated gene CTLA4 +49A/G polymorphisms with Crohn’s disease and ulcerative colitis in Hungarian population samples. World J Gastroenterol. 2007;13:2205–2208. doi: 10.3748/wjg.v13.i15.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rueda B, Zhernakova A, López-Nevot MA, Gomez-Garcia M, Ortega E, Piñero A, Correro F, Brieva JA, Nieto A, Koeleman BP, et al. CTLA4/CT60 polymorphism is not relevant in susceptibility to autoimmune inflammatory intestinal disorders. Hum Immunol. 2005;66:321–325. doi: 10.1016/j.humimm.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Manning AK, Dupuis J. A method of moments estimator for random effect multivariate meta-analysis. Biometrics. 2012;68:1278–1284. doi: 10.1111/j.1541-0420.2012.01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson D, White IR, Riley RD. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med. 2012;31:3805–3820. doi: 10.1002/sim.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–680. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 24.Zintzaras E, Ioannidis JP. Heterogeneity testing in meta-analysis of genome searches. Genet Epidemiol. 2005;28:123–137. doi: 10.1002/gepi.20048. [DOI] [PubMed] [Google Scholar]
- 25.Huizenga HM, Visser I, Dolan CV. Testing overall and moderator effects in random effects meta-regression. Br J Math Stat Psychol. 2011;64:1–19. doi: 10.1348/000711010X522687. [DOI] [PubMed] [Google Scholar]
- 26.Ferrenberg AM, Swendsen RH. New Monte Carlo technique for studying phase transitions. Phys Rev Lett. 1988;61:2635–2638. doi: 10.1103/PhysRevLett.61.2635. [DOI] [PubMed] [Google Scholar]
- 27.Wikstrom EA, Naik S, Lodha N, Cauraugh JH. Balance capabilities after lateral ankle trauma and intervention: a meta-analysis. Med Sci Sports Exerc. 2009;41:1287–1295. doi: 10.1249/MSS.0b013e318196cbc6. [DOI] [PubMed] [Google Scholar]
- 28.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinar M, Cukovic-Cavka S, Bozina N, Ravic KG, Markos P, Ladic A, Cota M, Krznaric Z, Vucelic B. MDR1 polymorphisms are associated with inflammatory bowel disease in a cohort of Croatian IBD patients. BMC Gastroenterol. 2013;13:57. doi: 10.1186/1471-230X-13-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farnood A, Naderi N, Moghaddam SJ, Noorinayer B, Firouzi F, Aghazadeh R, daryani NE, Zali MR. The frequency of C3435T MDR1 gene polymorphism in Iranian patients with ulcerative colitis. Int J Colorectal Dis. 2007;22:999–1003. doi: 10.1007/s00384-007-0270-6. [DOI] [PubMed] [Google Scholar]
- 31.Ho GT, Soranzo N, Nimmo ER, Tenesa A, Goldstein DB, Satsangi J. ABCB1/MDR1 gene determines susceptibility and phenotype in ulcerative colitis: discrimination of critical variants using a gene-wide haplotype tagging approach. Hum Mol Genet. 2006;15:797–805. doi: 10.1093/hmg/ddi494. [DOI] [PubMed] [Google Scholar]
- 32.Lankarani KB, Karbasi A, Kalantari T, Yarmohammadi H, Saberi-Firoozi M, Alizadeh-Naeeni M, Taghavi AR, Fattahi MR, Ghaderi A. Analysis of cytotoxic T lymphocyte associated antigen 4 gene polymorphisms in patients with ulcerative colitis. J Gastroenterol Hepatol. 2006;21:449–453. doi: 10.1111/j.1440-1746.2005.03956.x. [DOI] [PubMed] [Google Scholar]
- 33.Machida H, Tsukamoto K, Wen CY, Narumi Y, Shikuwa S, Isomoto H, Takeshima F, Mizuta Y, Niikawa N, Murata I, et al. Association of polymorphic alleles of CTLA4 with inflammatory bowel disease in the Japanese. World J Gastroenterol. 2005;11:4188–4193. doi: 10.3748/wjg.v11.i27.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osuga T, Sakaeda T, Nakamura T, Yamada T, Koyama T, Tamura T, Aoyama N, Okamura N, Kasuga M, Okumura K. MDR1 C3435T polymorphism is predictive of later onset of ulcerative colitis in Japanese. Biol Pharm Bull. 2006;29:324–329. doi: 10.1248/bpb.29.324. [DOI] [PubMed] [Google Scholar]
- 35.Repnik K, Potocnik U. CTLA4 CT60 single-nucleotide polymorphism is associated with Slovenian inflammatory bowel disease patients and regulates expression of CTLA4 isoforms. DNA Cell Biol. 2010;29:603–610. doi: 10.1089/dna.2010.1021. [DOI] [PubMed] [Google Scholar]
- 36.Schwab M, Schaeffeler E, Marx C, Fromm MF, Kaskas B, Metzler J, Stange E, Herfarth H, Schoelmerich J, Gregor M, et al. Association between the C3435T MDR1 gene polymorphism and susceptibility for ulcerative colitis. Gastroenterology. 2003;124:26–33. doi: 10.1053/gast.2003.50010. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Xia B. Association between the cytotoxic T lymphocyte associated antigen 4/CT60 gene polymorphism and ulcerative colitis in the Chinese. Zhonghua Weichangbingxue Yu Ganbingxue Zazhi. 2006;15:158–160. [Google Scholar]
- 38.Vermeire S, Rutgeerts P. Current status of genetics research in inflammatory bowel disease. Genes Immun. 2005;6:637–645. doi: 10.1038/sj.gene.6364257. [DOI] [PubMed] [Google Scholar]
- 39.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Brant SR, Li C, Shrestha UK, Jiang T, Zhou F, Jiang Y, Shi X, Zhao Y, Li J, et al. CTLA4 -1661A/G and 3’UTR long repeat polymorphisms are associated with ulcerative colitis and influence CTLA4 mRNA and protein expression. Genes Immun. 2010;11:573–583. doi: 10.1038/gene.2010.16. [DOI] [PubMed] [Google Scholar]
- 42.Berman D, Parker SM, Siegel J, Chasalow SD, Weber J, Galbraith S, Targan SR, Wang HL. Blockade of cytotoxic T-lymphocyte antigen-4 by ipilimumab results in dysregulation of gastrointestinal immunity in patients with advanced melanoma. Cancer Immun. 2010;10:11. [PMC free article] [PubMed] [Google Scholar]
- 43.Yang QF, Chen BL, Zhang QS, Zhu ZH, Hu B, He Y, Gao X, Wang YM, Hu PJ, Chen MH, et al. Contribution of MDR1 gene polymorphisms on IBD predisposition and response to glucocorticoids in IBD in a Chinese population. J Dig Dis. 2015;16:22–30. doi: 10.1111/1751-2980.12205. [DOI] [PubMed] [Google Scholar]
- 44.Krupoves A, Mack D, Seidman E, Deslandres C, Amre D. Associations between variants in the ABCB1 (MDR1) gene and corticosteroid dependence in children with Crohn’s disease. Inflamm Bowel Dis. 2011;17:2308–2317. doi: 10.1002/ibd.21608. [DOI] [PubMed] [Google Scholar]


