Abstract
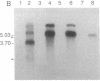
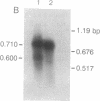
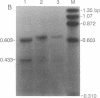
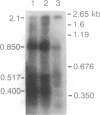
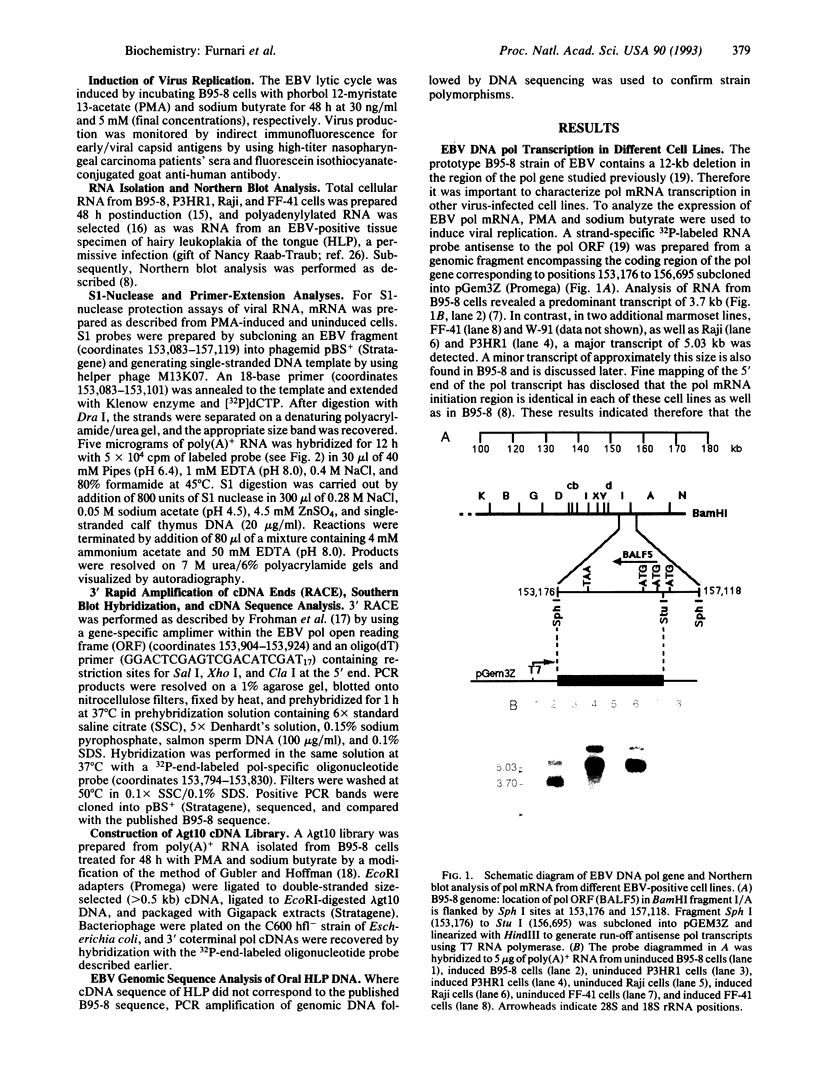
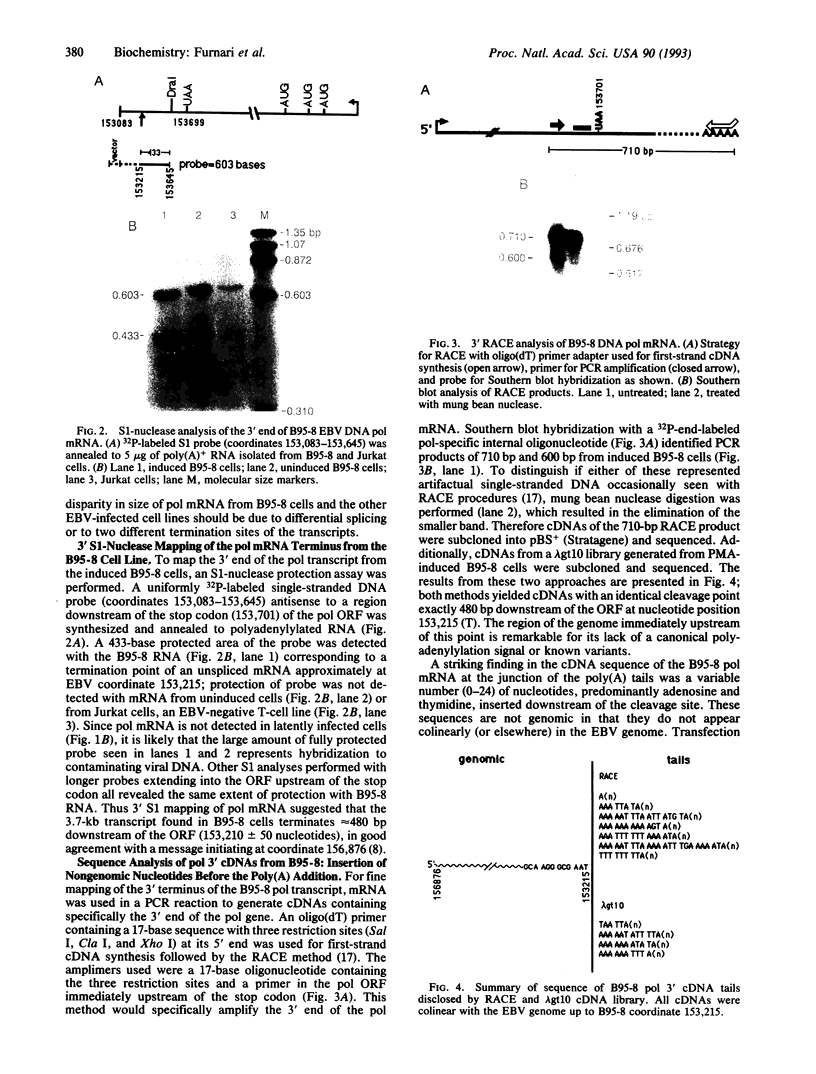
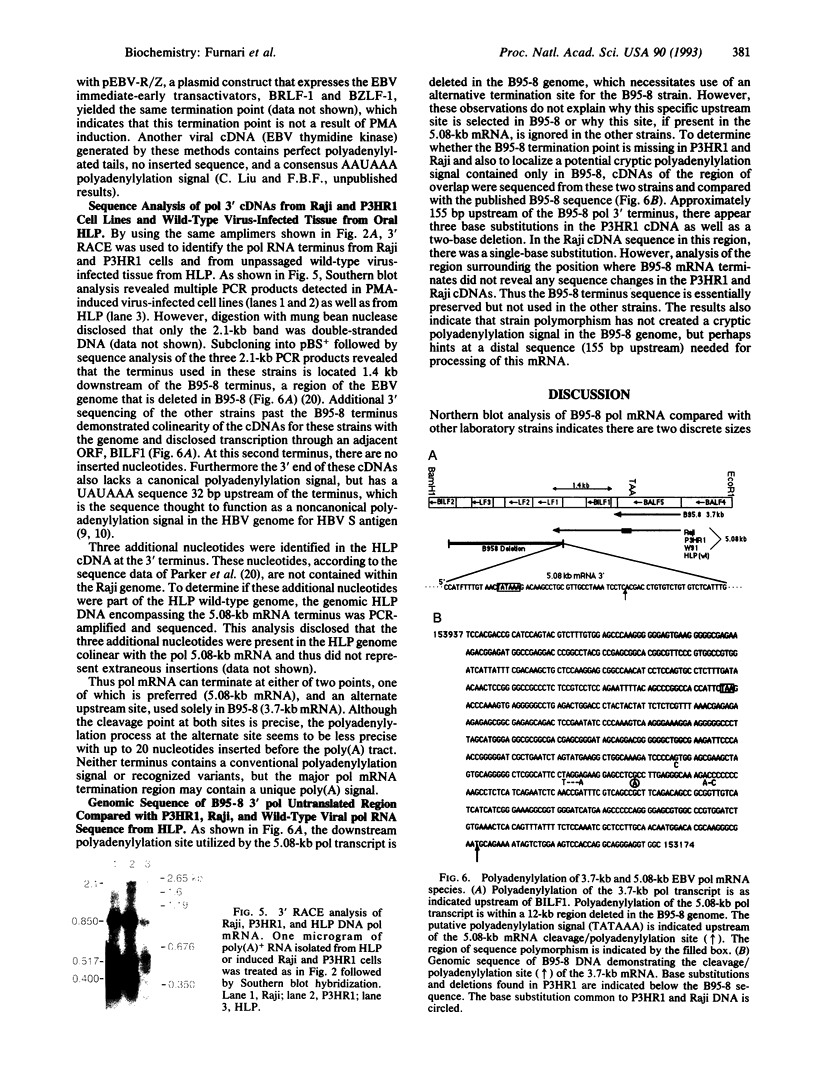
Northern blot analysis of the Epstein-Barr virus DNA polymerase mRNA identified two discrete sizes of virally encoded polymerase transcripts, 5.08 kb detected in strains P3HR1, Raji, W-91, and FF-41 and 3.7 kb detected solely in the prototype B95-8 strain. 3' S1-nuclease mapping and analysis of cDNA sequence generated by RNA-based PCR demonstrated that the 3.7-kb polymerase mRNA from B95-8 terminates 484 base pairs downstream of the open reading frame in a region of the genome remarkable for its lack of an apparent polyadenylylation signal. Moreover, between the cleavage point and the poly(A) tract of the cDNAs are a series of inserted nucleotides, mostly adenosine and uridine residues of unknown origin. A similar analysis of the 3' terminus of the 5.0-kb mRNA from the other cell lines revealed that polyadenylylation occurs 1.4 kb downstream of the B95-8 terminus. This region is deleted in B95-8, which accounts for the alternate upstream terminus used in B95-8. Like the 3.7-kb terminus, the 5.0-kb terminus lacks a canonical polyadenylylation signal, but contains a rarely used UAUAAA sequence 32 bp upstream of the poly(A) tail. These results indicate that the mRNA encoded by the Epstein-Barr virus DNA polymerase gene is polyadenylylated at two different termini without the use of canonical signals, raising the possibility of involvement of a virus-encoded factor in 3' processing of this message.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Cattaneo R., Will H., Hernandez N., Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983 Sep 22;305(5932):336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- Feighny R. J., Henry B. E., 2nd, Datta A. K., Pagano J. S. Induction of DNA polymerase activity after superinfection of Raji cells with Epstein-Barr virus. Virology. 1980 Dec;107(2):415–423. doi: 10.1016/0042-6822(80)90308-6. [DOI] [PubMed] [Google Scholar]
- Fischer D. K., Miller G., Gradoville L., Heston L., Westrate M. W., Maris W., Wright J., Brandsma J., Summers W. C. Genome of a mononucleosis Epstein-Barr virus contains DNA fragments previously regarded to be unique to Burkitt's lymphoma isolates. Cell. 1981 May;24(2):543–553. doi: 10.1016/0092-8674(81)90345-7. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari F. B., Adams M. D., Pagano J. S. Regulation of the Epstein-Barr virus DNA polymerase gene. J Virol. 1992 May;66(5):2837–2845. doi: 10.1128/jvi.66.5.2837-2845.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Herman R. C., Schubert M., Keene J. D., Lazzarini R. A. Polycistronic vesicular stomatitis virus RNA transcripts. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4662–4665. doi: 10.1073/pnas.77.8.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinuma Y., Konn M., Yamaguchi J., Wudarski D. J., Blakeslee J. R., Jr, Grace J. T., Jr Immunofluorescence and herpes-type virus particles in the P3HR-1 Burkitt lymphoma cell line. J Virol. 1967 Oct;1(5):1045–1051. doi: 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney S., Kamine J., Holley-Guthrie E., Lin J. C., Mar E. C., Pagano J. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J Virol. 1989 Apr;63(4):1729–1736. doi: 10.1128/jvi.63.4.1729-1736.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka J., Kallin B., Klein G. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology. 1979 Apr 15;94(1):228–231. doi: 10.1016/0042-6822(79)90455-0. [DOI] [PubMed] [Google Scholar]
- Miller G., Coope D., Niederman J., Pagano J. Biological properties and viral surface antigens of Burkitt lymphoma- and mononucleosis- derived strains of Epstein-Barr virus released from transformed marmoset cells. J Virol. 1976 Jun;18(3):1071–1080. doi: 10.1128/jvi.18.3.1071-1080.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller G., Shope T., Lisco H., Stitt D., Lipman M. Epstein-Barr virus: transformation, cytopathic changes, and viral antigens in squirrel monkey and marmoset leukocytes. Proc Natl Acad Sci U S A. 1972 Feb;69(2):383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. L., Glaser R., Rapp F. Studies of an Epstein-Barr virus-induced DNA polymerase. Virology. 1977 Feb;76(2):494–502. doi: 10.1016/0042-6822(77)90232-x. [DOI] [PubMed] [Google Scholar]
- PULVERTAFT J. V. A STUDY OF MALIGNANT TUMOURS IN NIGERIA BY SHORT-TERM TISSUE CULTURE. J Clin Pathol. 1965 May;18:261–273. doi: 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker B. D., Bankier A., Satchwell S., Barrell B., Farrell P. J. Sequence and transcription of Raji Epstein-Barr virus DNA spanning the B95-8 deletion region. Virology. 1990 Nov;179(1):339–346. doi: 10.1016/0042-6822(90)90302-8. [DOI] [PubMed] [Google Scholar]
- Pourcel C., Louise A., Gervais M., Chenciner N., Dubois M. F., Tiollais P. Transcription of the hepatitis B surface antigen gene in mouse cells transformed with cloned viral DNA. J Virol. 1982 Apr;42(1):100–105. doi: 10.1128/jvi.42.1.100-105.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. S., Schubert M., Lazzarini R. A. Polyadenylation sites for influenza virus mRNA. J Virol. 1981 Apr;38(1):157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C. C., Levinson A. D. Analysis of processing and polyadenylation signals of the hepatitis B virus surface antigen gene by using simian virus 40-hepatitis B virus chimeric plasmids. Mol Cell Biol. 1983 Dec;3(12):2250–2258. doi: 10.1128/mcb.3.12.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair A. J., Brimmell M., Shanahan F., Farrell P. J. Pathways of activation of the Epstein-Barr virus productive cycle. J Virol. 1991 May;65(5):2237–2244. doi: 10.1128/jvi.65.5.2237-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urier G., Buisson M., Chambard P., Sergeant A. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 1989 May;8(5):1447–1453. doi: 10.1002/j.1460-2075.1989.tb03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling D. M., Edmiston S. N., Sixbey J. W., Abdel-Hamid M., Resnick L., Raab-Traub N. Coinfection with multiple strains of the Epstein-Barr virus in human immunodeficiency virus-associated hairy leukoplakia. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6560–6564. doi: 10.1073/pnas.89.14.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz J., Pettine S. M., Shenk T. Functional analysis of point mutations in the AAUAAA motif of the SV40 late polyadenylation signal. Nucleic Acids Res. 1989 May 25;17(10):3899–3908. doi: 10.1093/nar/17.10.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]