Abstract
During the first step of biofilm formation, initial attachment is dictated by physicochemical and electrostatic interactions between the surface and the bacterial envelope. Depending upon the nature of these interactions, attachment can be transient or permanent. To achieve irreversible attachment, bacterial cells have developed a series of surface adhesins promoting specific or non-specific adhesion under various environmental conditions. This chapter will review the recent advances in our understanding of the secretion, assembly and regulation of the bacterial adhesins during biofilm formation with a particular emphasis on the fimbrial, non-fimbrial and discrete polysaccharide adhesins in Gram-negative bacteria.
INTRODUCTION
The ability of bacterial cells to adhere to and interact with surfaces to eventually form a biofilm is a crucial trait for the survival of any microorganism in complex environments. As a result, different strategies aimed at providing specific or non-specific interactions between the bacterial cell and the surface have evolved. While adhesion to abiotic surfaces is usually mediated by non-specific interactions, adhesion to biotic surfaces typically requires a specific receptor-ligand interaction (1). In both cases, these interactions usually originate from the same fundamental physicochemical forces: covalent bonds, Van der Waals forces, electrostatic forces and acid-base interactions (2). Strong adhesion occurs if a bacterium and a surface are capable of forming either covalent, ionic or metallic bonds, but weaker forces, such as polar, hydrogen bonding or Van der Waals interactions can also strengthen or achieve strong interactions when a high number of contacts are involved (2, 3). Due the net negative charge of their cell envelope, bacteria are subjected to repulsive electrostatic forces when approaching surfaces. Bacterial cells also encounter repulsive hydrodynamic forces near the surface in a liquid environment. To overcome these two repulsive barriers, bacteria typically use organelles, such as flagella or pili, which act either as an active propeller or a grappling hook (4–6). Once on the surface, the cell can enhance attachment to the surface via specific and/or non-specific adhesins to eventually trigger irreversible attachment. This irreversible attachment is strongly influenced by environmental factors (i.e. pH, salinity, …) and the physicochemical properties of the surface (i.e. rugosity, hydrophobicity, charge,…), but also by the presence of the conditioning film, a layer of organic and inorganic contaminants adsorbed on the surface which changes its physicochemical properties (7). To achieve permanent adhesion under such variable conditions, bacterial cells have developed a series of adhesins able to facilitate adhesion under various environmental conditions (8, 9). In this chapter, we will focus exclusively on non-specific adhesins, which are primarily responsible for biofilm formation and bacterial adhesion to abiotic surfaces. We will review the current knowledge of fimbrial, non-fimbrial and discrete polysaccharide adhesins involved in adhesion to abiotic surfaces and cell aggregation in Gram-negative bacteria.
1. Fimbrial adhesins/Pili
Fimbrial adhesins are a varied yet ubiquitous group of adhesins in both Gram-positive and Gram-negative bacteria. Also referred to as attachment pili, these polymeric fibers are involved in an array of functions, including attachment to both biotic and abiotic surfaces, motility, DNA transfer, and biofilm formation. Visible on the cell surface using electron microscopy, fimbrial adhesins are complex appendages that often require a large number of proteins for proper assembly.
The role of pili in the biofilm is multifaceted. First, they are important for the initial stage of bacterial cell attachment to both biotic and abiotic surfaces. Pili are often involved in the transition between motility and irreversible attachment, as seen with CUP pili of Escherichia coli and Tad pili of Caulobacter crescentus (10, 11). Second, they facilitate intercellular interactions through aggregation or microcolony formation, which is demonstrated with both Tad pili and curli. Finally, they also play a role in the secondary structure of the biofilm through their function in twitching motility. The Type IVa pili of Pseudomonas aeruginosa help generate mature biofilm structure through type IV pili-mediated migration of a key subpopulation of cells within the biofilm (12–14). Many bacterial species have more than one type of pili and these can be divided into four subgroups generally defined by their secretion and assembly processes: 1) the chaperone-usher pili or CUP; 2) Type IV pili; 3) the alternative chaperone-usher pathway pili; and 4) pili assembled by the extracellular nucleation-precipitation pathway, also known as curli. The structure and the role of Gram-negative fimbrial adhesins in biofilm formation will be discussed in this section.
1.1. The Chaperone-Usher pili (CUP)
The CUP are the most ubiquitous type of pili and are generally made of either long and thin or heavier rod-like filaments. CUP are assembled by the concerted action of a periplasmic chaperone and a pore-forming protein, called the usher, which gives this type of pili its name. Although best described in Enterobacteriaceae, CUP are also found in a variety of other Gram-negative bacteria, including Acinetobacter sp., Haemophilus influenzae, Burkholderia cenocepacia, Rhizobium sp. and Xylella fastidiosa (Table 1) (15–18). Based on the phylogeny of the usher protein sequences, CUP are subdivided into six main clades: α, b, γ, κ, π, and σ (19). Although there are as many as 38 different CUP in E. coli (20), the best studied CUP are Type I (γ1 clade) and P pili (π clade), which provide a good model for CUP assembly. Here we will use the E. coli Type I pilus as the example for assembly (for greater details, see (21) or (22)).
Table 1.
Examples of fimbrial adhesins involved in biofilm formation.
| Pili type | Major Pilus Proteins | Minor Proteins and Assembly Proteins | Bacterium | reference |
|---|---|---|---|---|
| Chaperone/Us her | EcpA or MatA (ECP pili) | EcpC, EcpD, EcpE | E. coli | (197) |
| FimA, (TypeI) | FimC, FimD, FimF, FimG, FimH | E. coli, Klebsiella pneumoniae, X. fastidiosa, Enterobacter amylovora. Serratia marcescans | (21, 198–201) | |
| CsuA/B (Type I) | CsuC, CsuD, CsuE | A. baummanii | (202) | |
| MrkA (Type 3) | MrkB, MrkC, MrkD | K. pneumoniae, E. coli (UPEC), Citrobacter koseri | (203, 204) | |
| Type IV pili | ||||
| Type IVa | PilA, | PilE, PilD, PilV, PilW, PilX | P. aeruginosa | (14, 45, 46) |
| PilE | PilD, PilH, PilI, PilJ, PilK, PilX, PilV, ComP, PilD | Neiserria spp. | (205, 206) | |
| MshA (MSHA) | MshB, C, D, E, F G,I,J,K,L,M,N, O, P, and MshQ | V. parahemolyticus and V. cholerae | (49, 207) | |
| PilA (ChiRP) | PilB,PilC,PilD | V. parahemolyticus | (49, 208) | |
| TypeIVb | BfpA (bundle forming) | BfpP, BfpI, BfpJ, BfpK, | E. coli (EPEC) | (209) |
| TcpA (Tcp) | TcpB,C, D,E,F,TcpJ | V. cholerae | (50) | |
| Tad | Flp | TadA, TadB/C, TadD, TadE, TadF, TadG, TadV, RcpA, RcpB, RcpB, TadZ, | A. actinomycetemcomita ns | (58, 64) |
| PilA | CpaA, CpaB, CpaC, CpaC, CpaD, CpaE, CpaF | C. crescentus | (11, 66) | |
| Flp | TadA, TadB, TadC, TadD, TadF, TadG, FppA, RcpA, RcpC, TadZ, | P. aeruginosa | (56, 210) | |
| CtpA (common pili) | CtpA, CtpB, CtpC, CtpD, CtpE, CtpF, CtpG, CtpH, CtpI | A. tumefaciens | (70) | |
| Alternative CU | CooA (CS1) | CooB, CooC, CooD | E. coli (ETEC) | (26, 28) |
| CblA (cable pilus) | CblB, CblC, CblD | B. cepacia complex | (36) | |
| Nucleation/Pre cipitation | CsgA(Curli) | CsgB, CsgG, CsgE, CsgE, CsgF, CsgD | E. coli, Enterobacter cloacae, Citrobacter sp. | (73, 78) |
| AgfA (Tafi) | AgfB, AgfC, AgfD, AgfE, AgfF | Salmonella enteritidis | (211, 212) | |
| FapC | FapA, FapB, FapD, FapE, FapF | Pseudomonas spp. | (213) |
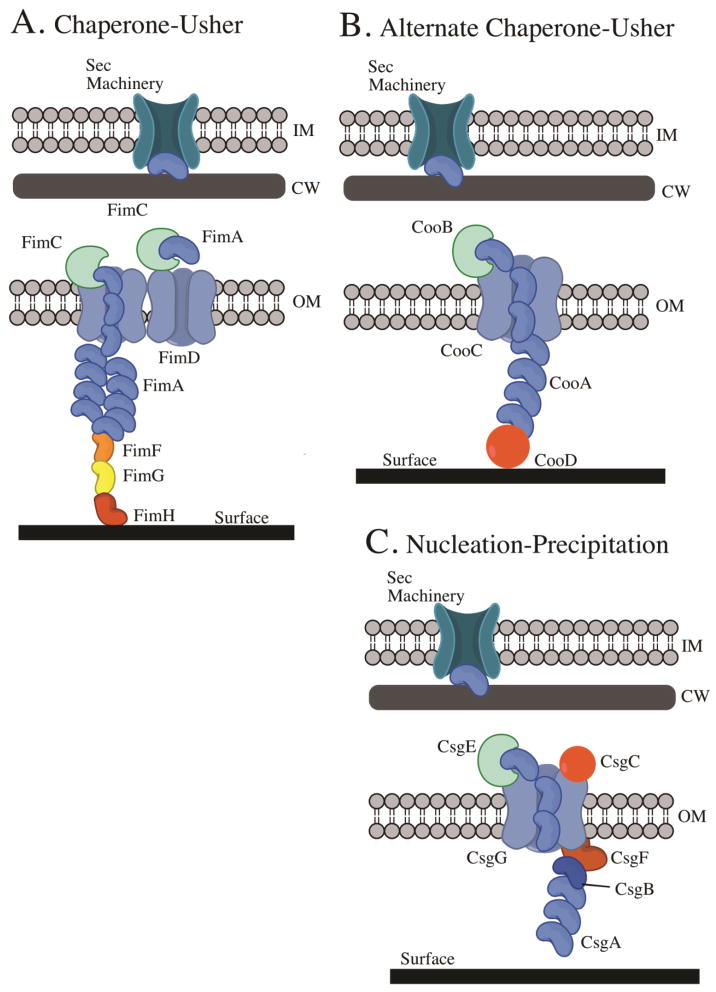
The Type I pili consists of six subunits that are first translocated into the periplasm via the Sec translocation pathway (Fig. 1A). After crossing the inner membrane, the pilus subunits associate with a periplasmic chaperone, FimC, which facilitates association with FimD, the outer-membrane usher protein. FimD, an 800 amino acid β-barrel protein, is referred to as the Type 1 pilus assembly platform. The pilus subunit, chaperone and usher proteins participate in a process called donor strand exchange and donor strand complementation to facilitate pilus filament assembly (21). Briefly, each pilus subunit has the structure of an incomplete immunoglobulin fold such that it is missing a C-terminal β-strand within the fold. Each subunit also contains an N-terminal extension. During folding and association with the chaperone subunit (FimC), the chaperone complements the missing β-strand within the pilus subunit with hydrophobic residues from the G1 strand of the chaperone. Then, the chaperone (FimC) and the pilus subunit associate with the usher (FimD) (Fig. 1A). The N-terminal extension of pilus subunit already bound to FimD moves into the groove of the pilus subunit occupied by the chaperone (FimC) G1 strand. The donor strand exchange occurs subsequently by a zip-in zip-out mechanism to mediate transfer of the incoming pilus subunit onto the growing pilus rod. The chaperone (FimC) is then recycled to bind newly formed pilus subunits (Fig. 1A). The initiation of pilus biogenesis occurs when the FimC-FimH complex associates with the usher FimD. Elongation begins with the arrival of the FimC-FimG complex. FimH and FimG comprise the 3 nm tip fibrillum, a small fiber associated with the tip of the main pilus (Fig. 1A). Then, FimF is connected to the tip adhesin and subsequently initiates assembly of the body of the main rod that is composed of FimA (Fig. 1A). P pili assembly is then terminated with the addition of PapH at the proximal end of the pilus. PapH is unable to undergo donor-strand exchange and thus terminates pilus growth. By comparison, no homologues of PapH has been identified in the Type I pilus assembly.
Figure 1.
Assembly and secretion of fimbrial adhesins. All the assembly pathways are oriented such that the inside of the cell is at the top and the surface to which the adhesin is binding, represented by the thick black line, is at the bottom. (IM) inner membrane; (CW) cell wall; (OM) outer membrane. The subunits for the three described systems are believed to be transported across the inner membrane by the Sec machinery. (A) A schematic of the chaperone-usher assembly (CUP) pathway represented by the assembly of the E. coli Type I pilus. FimC (green moon) is a chaperone. FimD (blue grey) is the outer membrane usher shown as a dimeric channel. FimA (blue beans) is the main pilus subunit. FimF (orange bean) links the tip fibrillum to the main fiber. FimG (yellow bean) is the tip fibrillum. FimH (red bean) is the mannose-specific tip fibrillum adhesin. (B) A schematic of the alternative chaperone-usher pathway using the E. coli CS1 pilus as a model. CooB (green moon) is the chaperone. CooC (blue grey) is the outer membrane usher. CooA (blue bean) is the main pilus subunit. CooD (red circle) is the pilus tip adhesin. (C) Model of E. coli curlin assembly as nucleation-precipitation pathway model. CsgE (green moon) is the chaperone. CsgG (blue grey) is the outer membrane usher. CsgA (blue beans) is the main curlin subunit. CsgB (dark blue bean) is the minor curlin subunit. CsgF (red bean) is the outer membrane protein needed for curlin polymerization and CsgB localization. CsgC (red ball) may be important for CsgG localization.
The E. coli Type I pilus is composed of 500–3000 copies of FimA resulting in a fiber that is 1 to 2 μm long and 6.9 nm wide. The absence of the tip adhesin FimH reduces adherence and biofilm formation, suggesting that FimH is the business end of the Type I pilus (10). FimH is specific for mannose-containing receptors of host cells, but can also bind to abiotic surfaces in a mannose-dependent manner (10, 13). Mannose was found to specifically inhibit biofilm formation in E. coli on a variety of abiotic surfaces, including various plastic polymers and glass (10). This specificity resulted in the development of mannose drug analogs to inhibit bacterial attachment (23). In addition to being involved in attachment to abiotic surfaces, FimH primarily mediates strong adherence to mannose receptors using catch bonds, a type of bond that becomes stronger over time or becomes activated when the receptor and ligand are pulled apart (24, 25). Catch bonds are particularly effective for organisms that experience shear stress, such as those that colonize host mucosal surfaces or air-water interfaces. A variety of other CUP have been reported to play a role in biofilm formation (Table 1).
1.2. The Alternative Chaperone-Usher pili
The alternative chaperone-usher pili (or Class 5 pilus assembly) requires “usher” and “chaperone” proteins and have been classified into the α-clade of the CU family of fimbriae (19). However, the proteins in the alternate CUP pathway have little to no similarity to those of the CUP pathway, and only four proteins are known to be required for proper assembly of the alternative CUP (19). This clade includes several coli surface antigens (CS) of Enterotoxigenic E. coli (ETEC) which are involved in the intestinal epithelium colonization or in biofilm formation on indwelling devices like bladder catheters (26), and the cable pili of Burkholderia cepacia that facilitate bacterial aggregation, microcolony formation and association with mucin, a prerequisite in the development of cystic fibrosis (Table 1) (27). In this section, the coli surface antigen 1 (CS1) pilus of ETEC will serve as our model (Fig. 1B) (Table 1) (for greater details on the alternate CU pathway see (28), (29) and (30)).
It is believed that the CS1 pilus proteins rely on the Sec pathway for translocation across the inner membrane, as also observed for the proteins of the CUP system. The 15.2-kDa CooA is the major pilin subunit. CooA can spontaneously form multimers in the periplasm but the multimerization is greatly improved in presence of CooB, which acts as a chaperone for CooA pilin subunits (Fig. 1B) (28, 31, 32). CooC is a 94-kDa integral outer membrane protein (33) predicted to be the usher protein required for transport and assembly of the pili on the cell surface (Fig. 1B). CooD is a 38-kDa minor pilin associated with the tip of the CS1 pili (Fig. 1B) (33). The ratio of CooD to CooA is 1:1800, indicating that one CooD could be present at the tip of the pilus. CS1 pili are not assembled in the absence of CooD, suggesting that CooD could initiate pilus assembly as observed for the FimH tip adhesin (34). Similar to the CUP pilin subunits, the pilin of the alternative CUP pathway also undergoes donor strand exchange to facilitate transfer of the pilin subunits from the chaperone to the growing pilus fiber.
ETEC is a leading cause of diarrhea in children of developing countries. These bacteria use a variety of surface adhesins to bind to the intestinal epithelium (35). ETEC can also form biofilms on indwelling devices like bladder catheters. Both an ETEC CS2 cotD mutant and a colonization factor antigen II (CFA/II) cfaE mutant, had reduced biofilm formation relative to the wild-type parental strains (26), suggesting that alternative CUP are important for adhesion.
Burkholderia cenocepacia is an important pathogen associated with cystic fibrosis. The cable pili of B. cenocepacia facilitate bacterial cell-cell interactions or microcolony formation and association with mucin (Table 1) (27). However, these cell-cell interactions actually prevent non-specific aggregation that can result in clearance by the host. A pilin subunit (cblA) mutant had increased aggregation relative to wild-type (36).
1.3. Type IV pili
The Type IV pili are multifunctional organelles, involved in diverse functions such as bacterial twitching motility, auto-aggregation, attachment and DNA uptake (37). They are generally long thin filaments (6–8 nm wide and 1–4 μm long) composed of pilin subunits that in some cases assemble into bundles.
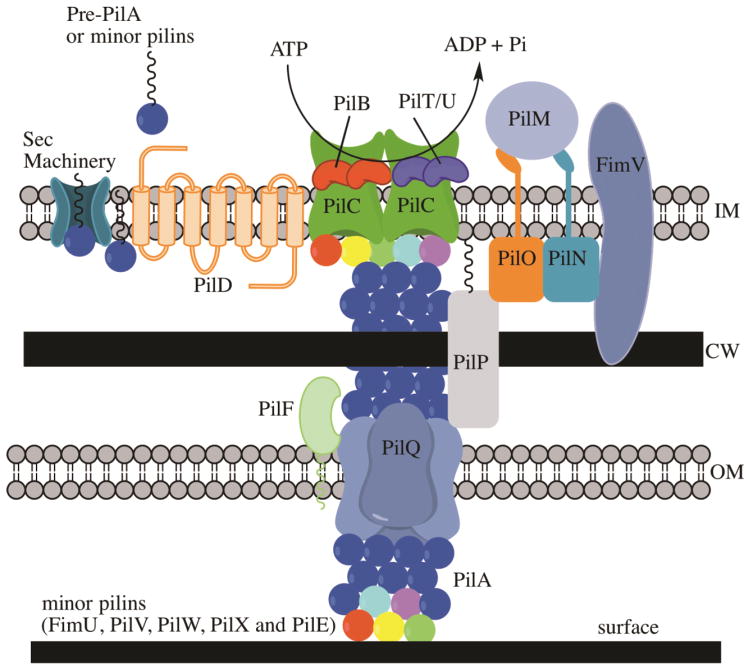
The Type IV pili are often homopolymers of a 15–20 kDa pilin protein, and they can also have an additional adhesive tip subunit. Both the CU and Type IV pilus assembly requires the general secretory pathway to translocate subunits across the inner membrane as well as a variety of outer membrane components to assemble the pilus on the cell surface. However, by comparison with CU pili, Type IV pili also require additional inner membrane components to facilitate fiber assembly. The dynamic assembly/disassembly of type IV pilin into a fiber is powered by ATP and requires at least 10 proteins: a) the major pilin subunit and sometimes minor pilin subunits, b) an assembly-specific ATPase, c) a prepilin peptidase that cleaves the N-terminal signal peptide, d) an assembly platform, e) an outer membrane protein that recruits the ATPase and f) an outer-membrane secretin that is necessary for the presentation of the pili on the cell surface (Fig. 2) (13). Based on the length of the prepilin signal peptides, Type IV pili can be subdivided into two categories: the Type IVa, characterized by prepilins with a short signal peptide (5–10 amino acids) and the Type IVb, characterized by prepilins with a long signal peptide (15–30 amino acids) (13). In this section we will focus on individual examples of the Type IVa, IVb and a subset of the Type IVb called the tight adherence pili or Tad Pili.
Figure 2.
Type IV assembly and secretion pathway. Given that the TypeIV pili have similar elements, we are using the P. aeruginosa TypeIVa pilus as the model for biogenesis. (IM) inner membrane; (CW) cell wall; (OM) outer membrane. Many TypeIVa proteins utilize the Sec machinery to translocate the inner membrane (aqua pore). PilA (blue sphere) is the main pilus subunit. FimU, PilE, PilX, PilW and PilV are minor pilins (red, yellow, light blue, green and purple spheres, respectively). The prepilins are processed by PilD (orange integral IM protein), the pre-pilin protease. PilB (red bean) is the ATPase that supplies energy for pilus assembly and PilU/PilT (purple beans) is the ATPase for pilus retraction. PilC (green porin) is an inner membrane protein of the motor complex for assembly of the pilus. PilM, PilN, PilO, PilP and FimV are the alignment complex. PilQ is the multimeric secretin in the outer membane that translocates the pilus outside the cell. PilF is a pilotin needed for localization of the PilQ in the OM. FimV is a peptidoglycan binding protein needed for multimerization of PilQ.
1.3.1. Type IVa pili
We will use P. aeruginosa as a model for Type IVa pilus assembly and secretion (Fig. 2, Tables 1 and 2.) (for greater details see (14)). The pilus itself is primarily composed of the 15-kDa main pilin PilA (Fig. 2) and the minor pilins FimU, PilV, PilW, PilX and PilE. All The pilin subunits have signal peptides and are processed by PilD, a prepilin peptidase, which cleaves pilin signal peptides and is bound to the inner-membrane. The minor pilins are incorporated throughout the pilus structure and are important in the initiation of pilus assembly (38). There have been several other roles suggested for the minor Type IVa pilins. PilX, PilY1 and PilW have been shown to be involved in cyclic diguanosine monophosphate (c-di-GMP) mediated suppression of swarming motility and biofilm formation which is facilitated through a decrease in cellular c-di-GMP levels in cells grown on an agar surface compared to liquid medium (39). In addition, PilY1 is needed for both twitching and swarming motility and facilitates attachment to host cells in a calcium-dependent manner (40).
Table 2.
Type IV pilus components
| Type IVa | Type IVb | Tad or Flp | |
|---|---|---|---|
| Bacteria | P. aeruginaosa | V. cholerae | A. actinomycetemcomitans |
| Components | |||
| Pilin subunit | PilA | TcpA | Flp-1 |
| Minor pilins | FimU, PilV, PilW, PilX, and PilE | TcpB | Flp-2,TadE,TadF |
| Prepilin peptidase | PilD | TcpJ | TadV |
| Assembly ATPase | PilB | TcpT | TadA |
| Retraction ATPase | PilT/PilU | NFa | NFa |
| Secretin | PilQ | TcpC | RcpA |
| Platform proteins | PilC, | TcpE | TadB, TadC, TadG |
| Pilotin | PilF | TpcQ | TadD |
| Alignment complex | PilM, PilN, PilO, PilP, FimV | TcpD, TcpR | RcpB, RcpC |
| Secreted Proteins | TcpF | ||
| Localization proteins | TadZ |
NF, not found
The inner membrane components of the Type IVa pilus include the motor and alignment subcomplexes. The motor complex is composed of PilB, PilC, PilT and PilU (Fig. 2, Table 2). PilB, PilT and PilU are putative ATPases required for either pilus assembly (PilB) or disassembly (PilT and PilU). PilC is an integral inner membrane protein thought to play a role in regulation of pilus assembly and depolymerization (41). The alignment complex bridges the inner membrane motor and the outer membrane secretion complex and is composed of PilM, PilN, PilO and PilP (Fig. 2). PilM is an actin-like cytoplasmic protein that is associated with PilN, an inner membrane protein (42). PilN also associates with PilO and the lipoprotein PilP to subsequently interact with PilQ, the outer membrane secretin. PilN also associates with FimV, a peptidoglycan binding protein that affects the multimerization of PilQ in the outer membrane (Fig. 2) (43). Finally, PilQ forms a large 1 MDa multimeric secretin that facilitates the presentation of the pilus on the cell surface. PilQ is associated with PilF via several tetratricopeptide repeats (TPR). PilF is an outer membrane lipoprotein required for the proper localization of PilQ in the outer membrane and referred to as a pilotin (44).
While Type IVa pili are usually associated with twitching motility, they also play an important role in attachment and biofilm structure. Twitching motility or crawling involves the ability of bacteria to move over semisolid surfaces using Type IV pili. Cells extend individual pili that attach to a surface, then retract the pili pulling the cell forward (14). Type IVa pilus-mediated twitching motility of P. aeruginosa is important for not only the initial attachment of the bacteria, but also for the formation of microcolonies and 3-D structures in the biofilm architecture (45, 46). Hyperpiliated strains of P. aeruginosa, where twitching motility is inhibited, form dense flat monolayers (47). Type IVa pili, and more specifically the MSHA (mannose sensitive hemagglutinating) pilus of Vibrio cholerae and Vibrio parahaemolyticus, facilitates proper biofilm formation on biotic and abiotic surface including both chitin and non-chitinaceaous surfaces, but remains dispensable for the process of initial attachment (48, 49).
1.3.2. Type IVb pili
While most Type IVa pili are associated with twitching motility, the Type IVb and Tad pili are more often associated with attachment, since the PilT ATPase homolog required for pilus retraction is absent. There are seven Type IVb pili that have been characterized in Gram-negative bacteria including the Tad pili. To examine the structure and assembly of Type IVb pili, we will look at the V. cholerae toxin co-regulated pilus (TCP) (Tables 1 and 2). TCP plays a role in microcolony formation and mediates bacterial interactions on chitinaceous surfaces (50) that are abundant as part of the shellfish in aquatic environments. The TCP result in a more stable and fit biofilm on chitin-rich surfaces (50). V. cholerae can degrade chitin and utilize it as a source of food, which can facilitate survival and spread of V. cholerae in the environment (51).
To be properly assembled, TCP require about 10 to 14 genes that are usually organized into one long operon called the TCP locus (52). TcpA and TcpB are the main and minor pilin of TCP pili, respectively (Table 2). TcpC is an outer membrane secretin interacting with a small protein, TcpQ, which provides proper TcpC localization and stability (53). TcpJ is the prepilin peptidase, belonging to the aspartic acid protease family, which cleaves the prepilin signal sequence between a charged and hydrophobic region within the N-terminus. The Type IVb prepillin signal sequences are approximately 25 residues (Fig. 2) (54). TcpT, the putative ATPase required for pili biogenesis, is localized to the inner membrane in a TcpR-dependent manner (Table 2) (55).
1.3.3. Tight adherence pili (Tad)
The tight adherence (Tad) loci of bacteria are a subset or distinct clade of the Type IVb pili and are found in both Gram-negative and Gram-positive bacteria. Tad pili are very important for initial adhesion, cellular aggregation and biofilm formation (56). Due to their small size, pili of the Tad system are also referred to as Flp (fimbrial-low-molecular weight protein) (Table 2) (56). We will review the Flp pili of Aggregatibacter actinomycetemcomitans as an example of Tad Pili. Many proteins of the tad loci in A. actinomycetemcomitans share similarities with proteins involved in the type II, type III and the type IV secretion pathways (Flp1, TadV, RcpA, TadA, TadB, TadC, TadE, and TadF), but some of the Tad proteins (RcpB, RcpC, TadZ, TadD, and TadG) remain exclusively associated with the Tad system (Table 2). The pilin Flp1 is the major structural component of the Flp pili (57). TadE and TadF are important for the Flp pili biogenesis (58), but are classified as pseudopilins, as they have conserved prepilin processing sites, and have never been found associated with the pilus (59). Thus far, it is believed that Tad pili are not retracted, as no homolog of PilT, the ATPase required for pilus retraction, has been identified thus far (Table 2). As observed for the P. aeruginosa Type IVa ATPase PilB, the putative ATPase TadA forms hexomeric complexes to power pilus assembly (60). The localization of TadA is facilitated by TadZ, a cytoplasmic multi-domain protein localized to the cell pole (61). TadB and TadC share similarity with PilC-like proteins and may associate to facilitate the passage of other Tad components across the inner membrane. RcpA is strongly suspected to form a channel required for the translocation of the pilus to the cell surface, but the ability of RcpA to function as a secretin are not well-characterized (62, 63) (Table 2). RcpB is an outer membrane protein that may function as a gating protein (63). TadD is a predicted lipoprotein and may be important for the assembly and targeting of the other outer membrane proteins (63).
A. actinomycetemcomitans is a strong biofilm former on a variety of surfaces and is a causative agent of infective endocarditis. The Tad pili of A. actinomycetemcomitans form bundles of fibers about 5 nm in diameter, composed of the 6.5-kDa main pilin Flp1, involved in adhesion to abiotic surfaces (58) and deletion of pilus components (rcpA and rcpB), lack of processing of pre-Flp1, pre-TadE or pre-TadF or loss of pili results in decreased cell aggregation and biofilm formation (59, 64, 65).
Unlike A. actinomycetemcomitans, the Tad pili in C. crescentus are not bundled, but are expressed as individual pili at the flagellar pole (66). The Tad pili in C. crescentus are important for the initiation of adherence and biofilm formation (Table 1). They are crucial for the initial reversible attachment to surfaces in conjunction with flagella since the loss of pili decreases initial adherence by about 50% (67). In addition, pili are involved in surface contact stimulation of holdfast secretion. Holdfast is the powerful polysaccharide adhesin responsible for irreversible adhesion and biofilm formation, as described below (see section 3.3.1 on polysaccharide adhesins) (68). Pili also play an important role in biofilm formation and maturation, as they are responsible for the integrity of the biofilm architecture (11)
P. aeruginosa makes a Tad or Flp pilus in addition to Type IVa and Type IVb pili (Table 1). The Flp pilus of P. aeruginosa has also been shown to be involved in biofilm formation and facilitates attachment to epithelial cells (69).
The plant pathogen Agrobacterium tumefaciens has a Tad pilus, encoded by the ctp locus. Mutations in ctpA, ctpB or ctpG resulted in decreased adherence on abiotic surfaces in both short-term assays and biofilm formation relative to wildtype. However, no difference was readily observed between a ctpA mutant and wildtype during root biofilm formation (70).
1.4. The Nucleation-Precipitation pili
The final group of fimbrial adhesins is assembled via the nucleation-precipitation pathway. The curli of E. coli and Salmonella are the best characterized (Table 1). This group of adhesins is covered in depth in the chapter on E. coli curli in biofilms in this book so it will only be touched on briefly here. Curli are members of a growing group of proteins called functional amyloids. Aggregation is the basis of amyloid formation. A protein is in an “amyloid state” when it forms bundles of fibers made of β-sheets. Uncontrolled aggregation of proteins can result in damage or disease, such as may be the case for Alzheimer’s disease. Functional amyloids in bacteria arise from aggregation producing an ordered β-sheet structure in a controlled process used to generate fibrils, spore coats or surface coats (71). Curli systems are present in a wide variety of bacteria (72). Curli are thin fimbriae (6–12 nm wide) with variable lengths (73). The main curlin monomer is CsgA, which when secreted via CsgG, self-assembles into fibers on the cell surface in association with the minor curlin nucleator CsgB (Fig. 1C). CsgE and CsgF function as chaperones and CsgD is a positive regulator of the curlin system. For an in-depth understanding of curli biogenesis and regulation, see the chapter on E. coli curli or the review by Evans and Chapman (74).
Curli are associated with both initial adherence and biofilm formation in Shigatoxin producing E. coli (75). Gastrointestinal commensal E. coli express curli that were shown to contribute to biofilm formation (76). The curli and cellulose of Enterohemorragic E. coli and ETEC work in concert in adherence and biofilm formation (77). As a consequence of curli protein self-assembly on the cell surface, cross-seeding of curli proteins has been documented between multiple species, including E. coli, Salmonella, Citrobacter. Enhanced adherence and pellicle biofilm formation upon cross-seeding indicates that curli can facilitate multispecies biofilm formation (78).
2. Non-fimbrial adhesins
Unlike the long, polymeric adhesins discussed above, the non-fimbrial adhesins are short monomeric or trimeric structures. This type of adhesin is widely spread among bacteria and is involved in cell attachment to abiotic surfaces and/or host cells. In this section, only non-fimbrial adhesins involved in biofilm formation will be presented. The different roles in non-specific adhesion played by these adhesins will be discussed: non-fimbrial adhesins are directly anchored to the outer cell membrane via covalent or non-covalent interactions and, due to their relatively short size, are usually involved in close contact between the bacterial cell and the substrate. Non-fimbrial adhesins are also involved in cell-cell interactions and aggregation. Furthermore, this type of adhesin is also shown to interact with various components of the biofilm extracellular matrix, linking the bacteria to the matrix and maintaining the biofilm architecture.
Although highly diverse in terms of structure and/or adhesives properties, non-fimbrial adhesins in Gram-negative bacteria are usually grouped into two main categories: the non-fimbrial adhesins secreted through a type 1 secretion system (T1SS) and the non-fimbrial adhesins secreted through one of the type 5 secretion system (T5SS). Detailed examples of non-fimbrial adhesins from both categories will be discussed in this section (for greater details on non-fimbrial adhesins see(79) and(80)).
2.1. Adhesin secreted by the type 1 secretion system (T1SS)
In Gram-negative bacteria, a very large group of non-fimbrial adhesins is secreted by the T1SS. This secretion system is one of the simplest described to date in Gram-negative bacteria. It is a heteromeric complex composed of three components: 1) an inner-membrane ABC (ATP binding cassette) transporter; 2) a membrane fusion protein in the periplasmic space; and 3) an outer-membrane pore (Fig. 3). These three proteins interact and form a channel that specifically transports T1SS substrates through the bacterial envelope in a single step, directly from the cytoplasm to the extracellular space. Proteins targeted by the T1SS share a C-terminal domain of lightly conserved secondary structure that is not cleaved off during the secretion process. Proteins are secreted in an unfolded state and first interact with the ABC transporter, triggering a conformational change of the channel and subsequent ATP hydrolysis, followed by the transport of the protein through the entire secretion system and subsequent folding of the secreted protein (reviewed in (81)).
Figure 3.
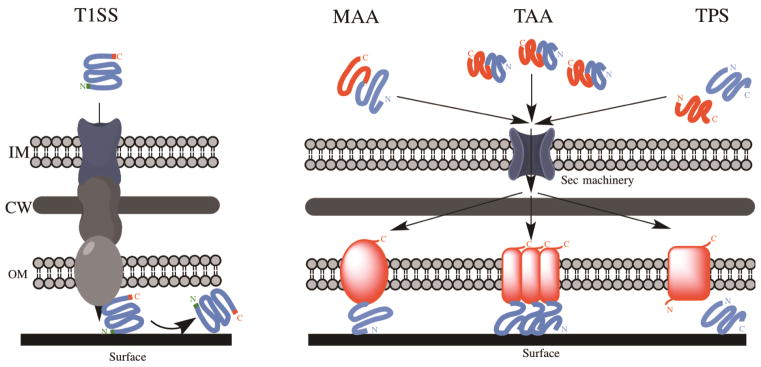
Schematic overview of the different secretion systems of non-fimbrial adhesins. Type 1 Secretion System (T1SS) and three classes of Type 5 Secretion System (T5SS) (Monomeric Autotransporters (MAA); Trimeric Autotransporters (TAA) and Two-Partner Secretion (TPS) systems) are represented. In T1SS, the adhesin is exported directly from the cytoplasm to the extracellular milieu via a pore constituted of three proteins. In T5SS, the adhesin is translocated from the cytoplasm to the periplasm by the Sec machinery and auto-assembled in the outer membrane. See text for more details. (IM) inner membrane; (CW) cell wall; (OM) outer membrane.
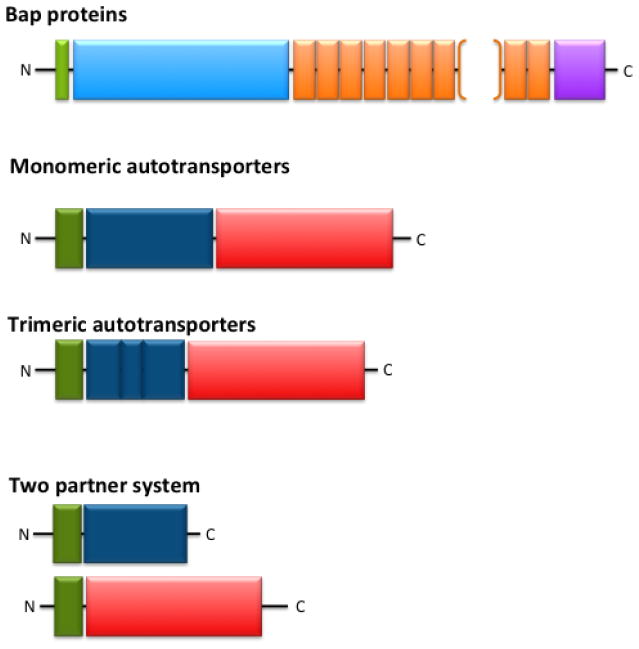
The most studied example of non-fimbrial adhesins secreted via T1SS is the Bap (biofilm-associated protein) family of proteins. The adhesins belonging to this group are high molecular weight multi-domain proteins and contain a core domain of long repeated units of variable length and sequence. All Baps share three distinct features (Fig. 4): 1) an N-terminal secretion signal for transport from the cytoplasm to the periplasm; 2) a core domain composed of highly repeated motifs; and 3) a glycine-rich C-terminal domain. The first Bap was identified in Staphylococcus aureus (82) and, since then, numerous other Bap family members have been shown to be involved in cell adhesion to abiotic surfaces and biofilm formation in both Gram-positive and Gram-negative bacteria (for a review, see (83)). A computational study revealed that large adhesins sharing the Bap family features are widespread among bacteria (84). However, only a few of these proteins have been characterized experimentally. Table 3 shows the Bap proteins that have been reported to be involved in biofilm formation by Gram-negative bacteria.
Figure 4.
Non-fimbrial adhesin organization. See text for further information. Green, signal sequence for proper localization and processing; Turquoise Blue, core domain; Orange, glycine-rich repeated domain (brackets depict the variable number of repeats); Magenta, Serine-rich C-terminal region; Navy Blue, passenger domain; Red, translocator domain.
Table 3.
Selected examples of non-fimbrial adhesins experimentally shown to be involved in biofilm formation by Gram-negative bacteria.
| Protein | Organism | Size (aa) | Reference |
|---|---|---|---|
| Biofilm associated proteins (Bap) – T1SS | |||
| LapA | P. putida | 8682 | (85) |
| BapA / AdhA | B. cenocepacia | 2924 | (214) |
| LapA | P. fluorescens | 4920 | (86) |
| BapA | S. enterica | 3825 | (215) |
| YeeJ | E. coli | 2358 | (103) |
| Bap | A. baumannii | 8621 | (216) |
| LapF | P. putida | 6310 | (90) |
| BfpA | Shewanella oneidensis | 2768 | (217) |
| MRP | Pectobacterium atrosepticum | 4558 | (218) |
| BfpA | Shewanella putrefaciens | 4220 | (219) |
| Cat-1 | Psychrobacter articus | 6715 | (220) |
| Monomeric autotransporter adhesins – T5SS | |||
| Ag43 | E. coli | 1039 | (105) |
| Cah | E. coli | 2850 | (221) |
| AIDA | E. coli | 1237 | (108) |
| TibA | E. coli | 989 | (222) |
| YfaL/EhaC | E. coli | 1250 | (103) |
| YpjA/EhaD | E. coli | 1526 | (103) |
| YcgV | E. coli | 955 | (103) |
| Hap | H. influenzae | 1392 | (128) |
| EhaA | E. coli | 1328 | (223) |
| EhaB | E. coli | 980 | (224) |
| UpaH | E. coli | 2845 | (225) |
| UpaC | E. coli | 996 | (226) |
| UpaI | E. coli | 1254 | (227) |
| MisL | S. enterica | 955 | (228) |
| Trimeric autotransporter adhesins – T5SS | |||
| YadA | Y. pseudotuberculosis | 434 | (120) |
| UspA1 | Moraxella catarrhalis | 955 | (229) |
| Hap/MID | M. catarrhalis | 2090 | (229) |
| UpaG | E. coli | 1779 | (121) |
| SadA | S. enterica | 1461 | (122) |
| AtaA | Acinetobacter sp. Tol5 | 3630 | (230) |
| EhaG | E. coli | 1589 | (231) |
| BbfA | B. pseudomallei | 1527 | (123) |
| Hemagglutin-like adhesins – T5SS | |||
| HxfB | X. fastidiosa | 3376 | (232) |
| HxfA | X. fastidiosa | 3458 | (232) |
| HMW1 | H. influenzae | 1536 | (128) |
| HMW2 | H. influenzae | 1477 | (128) |
| XadA | X. fastidiosa | 763 | (126) |
| YapH | Xanthomonas fuscans | 3397 | (125) |
| FhaB | X. fuscans | 4490 | (125) |
| XacFhaB | Xanthomonas axonopodis | 4753 | (233) |
| CdrA | P. aeruginosa | 2154 | (130) |
| FHA | B. pertussis | 3590 | (129) |
| BcpA | Burkholderia thailandensis | 3147 | (234) |
In Pseudomonas fluorescens and P. putida, the Bap protein LapA (large adhesion protein A) is crucial for biofilm formation (85, 86). This surface-associated protein, the largest one expressed by both organisms, is responsible for the transition between reversible adhesion via a single pole to irreversible adhesion along the entire cell length (85, 86). Indeed, lapA or the ABC transporter for LapA (required for the export of LapA to the cell surface) mutants are able to first attach to surfaces via their pole, but fail to undergo later irreversible adhesion and to develop the typical biofilm architecture (86). LapA is conserved between many P. fluorescens and P. putida strains, but the length of the protein in different strains is highly variable due to the flexible number of amino acid repeats (87). LapA mediates cell adhesion to a wide array of abiotic surfaces, from various plastics to glass or quartz, suggesting that the interactions between this protein and the surface are non-specific (85, 86). Single Cell Force Spectroscopy (SCFS) experiments recently showed that different domains of LapA are involved in different adhesion processes: while the repeated units in the core domain are mainly responsible for adhesion on hydrophobic surfaces, the C-terminal domain is involved in adhesion to hydrophilic surfaces, allowing LapA adhesion to a wider range of substrates and increasing the versatility of P. fluorescens colonization in diverse environments (88). In addition, SCFS analysis of the footprint left behind by P. fluorescens cells detached from a surface reveals a local accumulation of LapA occurring at the cell-surface interface and the presence of multiple adhesion peaks with extended rupture lengths, highlighting the critical role of LapA in mediating the irreversible cell adhesion to surfaces (89).
Another large adhesion protein found in P. putida that participates in biofilm formation is LapF (90). LapF is a surface associated protein and shares the key features of Baps (90). While LapA is responsible for irreversible adhesion of single cells to the surface, LapF is believed to meditate the cell-cell interaction in the biofilm and to play a major role during later development phases in biofilm maturation and stabilization (87, 90). It is worth noting that LapA is not present in pathogenic pseudomonads, such as P. aeruginosa and P. syringae (86, 91), suggesting that, while LapA is crucial for biofilm formation in environmental pseudomonads, pathogenic strains have developed other mechanisms for adhesion. P. aeruginosa strains have a lapF gene, whereas no LapF ortholog has been found in P. syringae (91). A similar observation is reported for Bap in S. aureus, where bap genes are absent from the genomes of human pathogenic strains (92).
Once secreted via the T1SS, all Baps remain loosely anchored to the outer membrane and are frequently released into the extracellular space. The nature of the interaction between the cell envelope and Baps remains unknown, but various hypotheses have been proposed: 1) Baps could transiently interact with the T1SS apparatus upon secretion; 2) Baps could interact with an unknown surface-exposed outer membrane protein; or 3) released Baps could eventually engage homotypic interaction with the residual surface-associated Baps (79, 93). Recent studies on SiiE from Salmonella enterica revealed that the retention of SiiE on the cell surface is modulated by pH, ionic strength, and osmolarity and is probably due to the interaction of the SiiE coil-coiled domains with the proteins forming the T1SS pore (94). In addition, extensive studies on the Baps LapA and LapF from P. putida show that these proteins are important components of the biofilm extracellular matrix and could be a link between the cells and matrix exopolysaccharides (95–97). During biofilm dispersal, LapA is cleaved from the cell surface by the specific protease LapG and released into the extracellular medium, freeing the bacteria from the biofilm matrix (95). Thus, the balance between retention on the cell surface and the release of Baps into the extracellular space may play a role in the regulation of cell adhesion to surfaces and eventually biofilm formation.
2.2. Type 5 secretion system (T5SS) autotransporter adhesins
The other major class of non-fimbrial adhesins is secreted by the T5SS. One of the largest group of secreted proteins in Gram-negative bacteria, T5SS proteins display a two-step process: proteins are first transported from the cytoplasm to the periplasm via the Sec machinery and are then secreted across the outer membrane through a channel formed by a β-barrel pore (Fig. 3) (98). Originally thought to be self-sufficient for proper assembly on the cell surface, this type of protein was called “autotransporter”. However, more recent studies suggest that the Bam complex, the machinery responsible for proper folding and proper insertion of outer membrane proteins into the outer membrane, is required for proper assembly of T5SS (99, 100).
All T5SS proteins share common structural and functional features (Figure 4): 1) an N-terminal Sec-dependent signal peptide; 2) a passenger domain providing the protein function; and 3) a C-terminal β-barrel domain that allows secretion of the passenger domain. Once the protein is processed, the β-barrel domain integrates into the outer membrane to form a pore through which the passenger domain is secreted to the cell surface (for recent reviews, see (80, 101)).
The T5SS autotransporter family is represented by three distinct groups: 1) the monomeric autotransporters; 2) the trimeric autotransporters; and 3) the two-partner secretion systems.
2.2.1. Monomeric autotransporters
This group is the largest group of T5SS proteins. In monomeric autotransporters, passenger and secretion domains are integrated into a single multidomain protein (Fig. 3). The secretion β-barrel domain (250–300 aa) has high sequence similarity among monomeric autotransporters and is predicted to form a pore comprised of 14 antiparallel β-strands (98). Conversely, the passenger regions consist of repetitive amino acid motifs, highly variable in sequence and size, responsible for the difference in specificity of each protein (102). The properties of autotransporters are very diverse, as they can be virulence factors, toxins or adhesins, among other functions. The majority of the studies on these autotransporter adhesins focuses on specific interaction and host cell-bacterial adhesion, and only a few monomeric autotransporter adhesins have been shown to play a role in biofilm formation and non-specific adhesion to abiotic surfaces (Table 3).
One of the most studied monomeric autotransporters is Ag43 from E. coli. Based on sequence similarity with Ag43, several other monomeric autotransporter adhesins have been identified in E. coli, and a few have been shown experimentally to be involved in cell aggregation and biofilm formation (Table 3) (103). Ag43 is present in the majority of commensal and pathogenic E. coli strains (104) and promotes autoaggregation and biofilm formation (105, 106). The exact role of monomeric autotransporters in biofilm formation has not been entirely elucidated yet, but recent experiments suggest that Ag43, as with other related autotransporters including AIDA (adhesin involved in diffuse adherence), may mediate cell-cell aggregation by homotypic interactions that could eventually play a role in biofilm formation (107, 108). In addition, it has been shown that different monomeric autotransporter adhesins, like Ag43 and AIDA, can cross-interact, leading to the formation of mixed cell aggregates (80, 102, 108). It has been shown that many autotransporters are O-glycosylated proteins, like Ag43, AIDA and TibA in E.coli (109–111). However, this glycosylation is not playing a role in cell-cell aggregation or biofilm formation (110, 112). Ag43 expression is controlled by phase variation and switches from Ag43on to Ag43off at a high rate (113). A recent study highlighted the importance of Ag43 phase variation during biofilm formation mediated by both Ag43 and other adhesins in E. coli (114). Indeed, Ag43on bacteria are physically selected for in the biofilm, probably due to their inherent property to auto-aggregate, which may subsequently modulate interaction with surfaces via the production of other adhesins (114). The recent resolution of the crystal structure of Ag43 reveals a “Velcro-like” organization similar to the self-oligomerization of the Hap autotransporter from H. influenzae (115), mediating Ag43-Ag43 interactions and probably Ag43 interactions with other autotransporters, promoting cell-cell aggregation (116).
2.2.2. Trimeric autotransporters (TAA)
All the members of the trimeric autotransporter (TAA) family described so far are non-fimbrial adhesin proteins (93). Numerous TAAs have been reported to specifically bind to biotic surfaces on host cells, or host extracellular matrix components including fibronectin or collagen, and only a few studies have focused on the role of these adhesins in biofilm formation and attachment to abiotic surfaces (Table 3).
While the overall organization of these autotransporters is similar to the monomeric autotransporters, TAA usually have a shorter C-terminal secretion domain (50–100 aa) with four β-strands. The formation of a full size 12 β-strand pore is achieved by trimerization (117). The passenger domain contains conserved elements designated as head, connector, and stalk domains (118). The head harbors the adhesive properties, while the connector and stalk primarily function to extend the head domain away from the bacterial cell body (118). Unlike the monomeric autotransporters described above, TAA adhesins are not cleaved or released into the extracellular space (107).
The most studied TAA in the multifunctional non-fimbrial adhesins is YadA from Yersinia spp.(119). This adhesin has been linked to virulence, adhesion to host cells, and autoaggregation (119). However, the putative role of YadA in adhesion to abiotic surfaces and biofilm formation is poorly understood. A specific 31-aa motif, present only in the N-terminal sequence of YadA from Y. pseudotuberculosis, is required for cell aggregation and biofilm formation, but the mechanism of adhesion is unknown (120). Only a few other TAAs have been reported to play a role in biofilm formation (Table 3). UpaG is involved in cell-cell interactions, promoting biofilm formation to abiotic surfaces in E. coli (121). SadA, an ortholog of UpaG in S. enterica and S. Typhimirium, has been shown to mediate cell aggregation and biofilm formation in cells unable to express long O-antigen on the cell surface (122), suggesting that bacteria need to be in close proximity for SadA to mediate cell-cell interactions. Recently, BbfA (Burkolderia biofilm factor A) has been shown to play a major role in biofilm formation in B. pseudomallei, probably by mediating cell aggregation (123).
2.3. Two-partners secretion system (TPS)
Two-partner secretion system (TPS) proteins form another group of autotransporters utilizing T5SS. In this family, the secretion mechanism is very similar to the ones described above, but the passenger and membrane anchor domains are encoded by two distinct genes, usually organized in a single operon. One gene encodes the secretion signal and the adhesin domain, and the other encodes the outer membrane β-barrel pore (80) (Fig. 3). The pores are highly conserved proteins of typically 500–800 aa (124). The effectors are large proteins and can be highly variable, but they all contain two common features: 1) a β-helix or β-solenoid secondary structure, and 2) a conserved N-terminal domain, the TPS domain (124). The TPS domain provides specific recognition of the appropriate pore for translocation through the outer membrane (117).
Filamentous haemagglutinin proteins are typical adhesins exported by the TPS (Table 3) and are usually involved in tight interaction between the bacterium and a specific receptor on the host cell. Their involvement in biofilm formation and non-specific adhesion to abiotic surfaces has been studied only sparsely. Filamentous haemagglutinins have been shown to play a role in biofilm formation by different plant pathogens, including X. fastidiosa or different Xanthomonas spp. (125–127). Unlike the two other types of autrotransporter adhesins described above, it seems that filamentous haemagglutinins from these species do not mediate cell-cell interactions and autoaggregation, but rather attach cells to abiotic surfaces, thereby facilitating their interaction with each other (126). Filamentous haemagglutinins also play a role in biofilm formation in the human pathogens H. influenzae (128) and Bordetella pertussis (129). FHA (filamentous haemagglutinin) from B. pertussis is a critical virulence factor and is involved in interactions between bacterial cells, as well as interaction between the bacteria and abiotic surfaces (129). In addition, it has been shown that the haemagglutinins HMW (high molecular weight protein) 1 and HMW 2 are found around H. influenzae cells grown in a biofilm, as well as interacting with the biofilm extracellular matrix (128), strongly suggesting that these proteins play an important role in adhesion to abiotic surfaces by linking the bacteria to the biofilm matrix and stabilizing the overall architecture.
Another example of a non-fimbrial adhesin belonging to the TPS group is CdrA (c-di-GMP regulated TPS A) in P. aeruginosa. This adhesin is transported by CdrB (c-di-GMP regulated TPS B) and is expressed when the level of c-di-GMP in the cell is high, or in biofilm cultures (130). Once secreted outside the cell, CdrA is proposed to bind to the exopolysaccharide Psl to promote cell aggregation and biofilm formation. This adhesin is part of the biofilm matrix and plays a role in maintaining biofilm structural integrity, probably by bundling individual Psl strands together and/or by anchoring Psl fibers to the bacteria in the biofilm (130).
3. Polysaccharide Adhesins
In addition to the different protein adhesins described above, many bacterial species produce extracellular polysaccharides that promote adhesion. These polysaccharides can be firmly associated with the cell surface, forming a capsule (capsular polysaccharide, CPS), or be loosely associated or even released from the cell surface (extracellular polysaccharide, EPS). The difference between CPS and EPS is usually experimentally defined and may have limited physiological relevance. From an adhesive point of view, it is more appropriate to distinguish protective from aggregative polysaccharides. While protective polysaccharides provide a protective barrier around the cell, the aggregative polysaccharides have adhesive and/or cohesive properties that are exploited by bacteria to mediate adherence to surfaces and/or reinforce structural integrity of the biofilm (131, 132). Although protective polysaccharides could also contribute to the adhesive properties of bacterial cells, only the aggregative polysaccharides that serve as adhesins will be discussed in this section. For simplicity, aggregative polysaccharides will be referred to as EPS.
3.1. Structure and Function
EPS often have long chains (MW>106 Da) composed of a repetition of the same sugar residue (homopolysaccharide) or a mixture of neutral and/or charged sugar residues (heteropolysaccharide). These polymers are mostly negatively charged, but neutral and even positively charged EPS also exist (6, 133–135). The adhesiveness of EPS strongly depends on chain conformation and is greatly influenced by substituents that modify inter-chain and intra-chain interactions (136). The polar and hydrogen bonding functional groups of polysaccharides, such as ethers and hydroxyls, provide good adhesion to polar surfaces. The adhesiveness of EPS can be strongly influenced by the presence carboxylic acid groups that promote the association of divalent cations such as Ca2+ and Mg2+ (6, 133–135, 137).
The degree of acetylation of a polysaccharide can drastically modify its cohesiveness and adhesiveness (138–142). Partial deacetylation of the well-characterized poly-β-1,6-N-acetyl-D-glucosamine EPS (PGA) is required to initiate biofilm formation by E. coli, S. aureus, and S. epidermidis (143–146). Similarly, the selective O-acetylation of mannuronate residues on alginate, a polysaccharide secreted by P. aeruginosa, seems to improve biofilm adhesion and increase adhesion to host surfaces (138, 140, 147). Furthermore, partial deacetylation of E. coli PGA is required to facilitate the secretion of the polymer through the outer membrane (144). On the other hand, partial deacetylation of holdfast in C. crescentus and Asticcacaulis biprosthecum, appears to be essential to maintain the structure, the adhesiveness, and the retention of the polymer on the cell surface in both species (142). In this case, the deacetylation of polysaccharides might facilitate the conformational transition of the polymer chain from random coils to ordered helices, promoting better inter-chain interactions and higher stiffness of the polymer (139).
3.2. Biosynthesis, secretion, and anchoring
Although chemically diverse, the molecular mechanisms by which EPS are assembled and exported to the cell surface can be categorized into three distinct mechanisms: 1) the Wzx/Wzy transporter-dependent pathway; 2) the ATP-binding cassette (ABC) transporter-dependent pathway; and 3) the synthase-dependent pathway (Fig. 5). These three mechanisms have been extensively reviewed and only a brief overview will be presented in this section (for greater details see (132))
Figure 5.
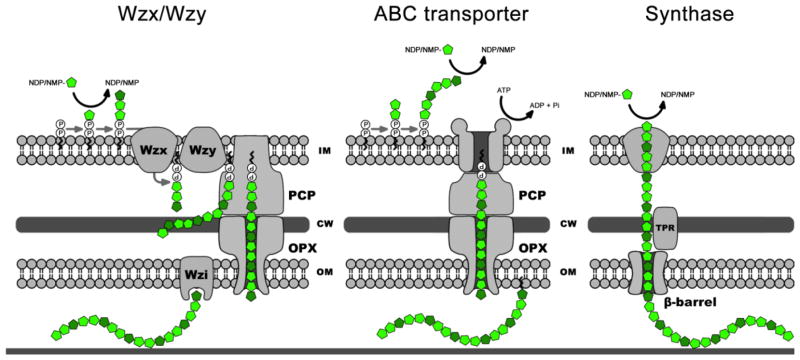
Polysaccharide Biosynthesis pathways. Overview of the Wzx/Wzy-, ABC-transporter-, and synthase-dependent exopolysaccharide biosynthesis pathways. Only the key components for each pathway are indicated on the diagram. In the Wzx/Wzy-dependent pathway, the polysaccharide repeat unit assembly is initiated on an undecaprenyl phosphate acceptor moiety located in the inner leaflet of the inner membrane, which is then transported across the inner membrane by the flippase, Wzx. The polymerization into high-molecular-weight polysaccharide occurs in the periplasm by the action of the polymerase Wzy. The export and secretion of the polysaccharide through the outer membrane are facilitated by the outer membrane polysaccharide export (OPX) and the polysaccharide co-polymerase (PCP) protein families. Depending on the polysaccharide being synthesized, the nascent polymer could be anchored to the outer-membrane via a specific protein, such as Wzi. In the ABC transporter-dependent pathway, the entire polysaccharide chain is assembled into the cytoplasm on a lipid acceptor that is then transported across the inner membrane by the ABC transporter. As observed for the Wzx/Wzy-dependant pathway, the export and secretion of the polysaccharide through the outer membrane also involve OPX and PCP protein families. In the synthase-dependent pathway, both the polymerization and the transport of the polymer across the inner membrane are carried out by the same membrane-embedded glycosyl transferase. The export and secretion of the polysaccharide through the outer membrane is facilitated by a molecular chaperone and a β-barrel porin. (IM) inner membrane; (CW) cell wall; (OM) outer membrane.
In the Wzx/Wzy-dependent pathway, each polysaccharide repeat unit is first assembled on an undecaprenyl phosphate acceptor moiety in the cytoplasm, which is then transported across the inner membrane before being polymerized into a high-molecular weight polysaccharide in the periplasm. In contrast, the entire polysaccharide chain is assembled in the cytoplasm on a lipid acceptor in the ATP-binding cassette (ABC) transporter-dependent pathway. Although mechanistically different, these two pathways use a similar mechanism to facilitate EPS export across the periplasm and through the outer membrane. Both pathways use a large protein complex spanning the periplasmic space, formed by an outer membrane polysaccharide export pore and a polysaccharide co-polymerase. In the synthase-dependent pathway, both the polymerization and the transport of the polymer across the inner membrane are carried out by the same membrane-embedded glycosyl transferase (132, 148). The polymer is protected by a molecular chaperone, a TPR repeat containing protein, and then exported across the outer membrane through a β-barrel porin (148).
When associated with the cell surface, the exact connection between EPS and outer membrane is not always known, and usually involves a linker, which can be a sugar, lipid or protein. For EPS exported through the ABC transporter-dependent pathway, the conserved phospholipid terminus is attached at the reducing end of the polysaccharide via a poly-3-deoxy-D-manno-oct-2-ulosonic acid (also known as KDO) linker that is responsible for proper translocation and contributes to the attachment of the polymer to the cell surface (149), although ionic interactions between the core region of the LPS and the polymer may be also involved (150). As observed for LPS O-antigen, carbohydrate chains forming the capsular or K-antigen (derived from the German word Kapsel (151)) may be also linked to the lipid A core of a LPS molecule, but the distinction from a traditional O-antigen remains subtle and purely operational (152, 153). Finally, polysaccharides can be anchored to the cell surface via dedicated proteins (154–156). In E. coli and Klebsiella pneumoniae, the outer-membrane protein Wzi is a critical factor in the anchoring of the K30 type I capsule to the cell surface (154, 156). Recent 3D-structure of Wzi suggest that Wzi may act as a lectin binding to the nascent polymer once translocated through the Wza translocase, but other cell-surface components may also play a role (154). In C. crescentus, the HfaA, HfaB and HfaD proteins provide an extremely strong anchor for the holdfast polysaccharide to the cell envelope, as discussed below (155). Alternatively, polysaccharides can be anchored to the cell surface by glycosylation of proteins, a system primarily utilized to decorate flagellar proteins and protein adhesins (for review see (157, 158)).
3.3. Polysaccharide diversity
The composition and the structure of EPS can vary both between and within species (149) and it is now well accepted that bacteria are able to produce different types of EPS to provide adhesive adaptability under varying conditions (Table 4). Most of these carbohydrate polymers generate a highly hydrated and extensive layer surrounding the cell, but a few bacterial species adhere to surfaces by using a clearly defined, discrete patch of polysaccharide (159–161). We will focus on the discrete aggregative polysaccharides in this section.
Table 4.
Selected examples of aggregative polysaccharides experimentally shown to be involved in biofilm formation by Gram-negative bacteria.
| Polysaccha ride | Organism | Composition/Structure | Reference |
|---|---|---|---|
| Alginate | P. aeruginosa | β-1,4-linked mannuronic acids and guluronic acids | (235) |
| Cellulose | Gluconacetobacter xylinus, A. tumefaciens, R. leguminosarum bv. Trifolii, Sarcina ventriculli, Salmonella spp., E. coli, K. pneumoniae | β-1,4-linked D-Glucose | (236–240) |
| Holdfast | Caulobacter spp., A. biprosthecum, Hyphomonas adherens, Hyphomonas rosenbergii, Hyphomicrobium zavarzinii, Maricaulis maris, Oceanicaulis alexandrii | Suspected to contain β-1,4-linked N-acetyl-D-glucosamine, but the exact composition and structure remain unknown. | (161, 164, 167, 241–244) |
| PGA | E. coli, Yersinia pestis, Bordetella spp., Actinobacillus spp., P. fluorescens | β-1,6-linked N-Acetyl-D-glucosamine | (245, 246) |
| Psl | P. aeruginosa | Repeating pentasaccharide of 3 mannose, 1 rhamnose, and 1 glucose. | (247–249) |
| Pel | P. aeruginosa, P. fluorescens | Unknown, but reported to be a glucose-rich polysaccharide polymer. | (247, 250, 251) |
| Slime | M. xanthus | Suspected to contain α-D-mannose or α-D-glucose residues, but the exact composition and structure remain unknown. | (193) |
| UPP | A. tumefaciens | Suspected to contain N-acetyl-D-glucosamine residues, but the exact composition and structure remain unknown. | (68, 180) |
There are diverse instances of discrete adhesive polysaccharide mediating surface attachment among these are the well described unipolar attachment to surfaces of Alphaproteobacteria, such as C. crescentus or Agrobacterium tumefaciens and the less common discrete slime deposition by the motile Deltaproteobacterium, Myxococcus xanthus (Fig. 6). In both cases, the discrete polysaccharide provides a specific function for different phases of the life cycle, supporting a transient or an irreversible attachment with the surface. In this section, the unipolar polysaccharide adhesin produced by C. crescentus and by some Rhizobiales, and the adhesive slime produced by M. xanthus will be discussed.
Figure 6.
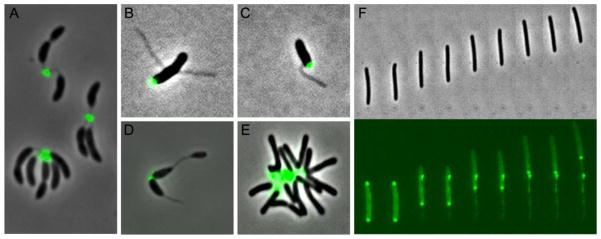
Selected examples of discrete polysaccharides. AF488-conjugated WGA lectin labelling of the holdfast in C. crescentus (A), A. biprosthecum (B) (Courtesy of Chao Jiang), A. excentricus (C) (Courtesy of Chao Jiang), Hyphomicrobium vulgare (D) (Courtesy of Ellen Quardokus). AF488-conjugated WGA lectin labelling of the UPP in A. tumefaciens (E). FITC-conjugated ConA lectin labelling of the slime in M. xanthus (F).
3.3.1. Holdfast
Caulobacter spp. and other stalked Alphaproteobacteria (162) synthesize a polar polysaccharide called holdfast (163) (Fig. 6). Only a small amount of holdfast is produced at a discrete site on the cell surface and can be observed either at one of the cell poles or at the tip of a thin cylindrical extension of the cell envelope referred to as the stalk. The holdfast is responsible for permanent adhesion to a surface and can maintain attachment for many generations, presumably to provide better access to limited nutrients under flow (164). The holdfast is also crucial for biofilm formation (163). C. crescentus holdfast secretion is regulated both developmentally and by contact-dependence (68). C. crescentus exhibits a dimorphic life cycle that begins as a motile swarmer cell. Unless they come in contact with a surface, swarmer cells are able to swim for a period equivalent to one-third of the cell cycle, after which they differentiate into stalked cells. During this developmentally programmed transition, swarmer cells release their flagellum, retract their pili, and synthesize a holdfast and a stalk at the same pole. The small protein regulator HfiA is crucial for holdfast production at the correct time during the cell cycle, as well as under the correct nutritional conditions (165). In addition to the developmental timing of holdfast secretion, if a swarmer cell encounters the surface, the coordinated action of the flagellum and pili stimulates early holdfast secretion, thereby driving the rapid, just-in-time transition from reversible to irreversible attachment (68). The addition of crowding agents that impede the rotation of the flagellum also stimulates the production of holdfast, suggesting that increased load on the flagellum triggers holdfast synthesis (68).
Two main gene clusters in C. crescentus have been identified that are required for holdfast synthesis (hfs) and holdfast anchoring to the tip of the stalk (hfa). The hfs cluster encodes proteins involved in the biosynthesis and secretion of the polysaccharide in a process similar to the Wzx/Wzy transporter-dependent pathway (161, 166). The hfa cluster encodes proteins crucial for attaching the holdfast to the cell envelope. Mutations in the hfa locus result in inefficient attachment of the holdfast to the tip of the stalk, causing holdfast shedding into the culture supernatant (155). The hfa locus is a single operon comprised of hfaA, hfaB and hfaD. The HfaA, HfaB and HfaD proteins are associated with the outer membrane and are polarly localized. HfaA and HfaD form high-molecular-weight complexes sharing properties with amyloid proteins, and HfaB plays a role in the secretion of both proteins (155). Although the exact anchoring mechanism between the polysaccharide and the high-molecular-weight complexes formed by HfaA and HfaD remains unknown, the HfaA, HfaB and HfaD proteins provide an extremely strong anchor for the holdfast polysaccharide to the cell envelope of C. crescentus (155).
Holdfast is bound specifically by wheat germ agglutinin (WGA), a lectin that specifically recognizes N-acetyl-D-glucosamine (GlcNAc)-containing polymers. In addition, holdfast is sensitive to lysozyme, a glycolytic enzyme specific for cleavage of β-1,4 linkages in oligomers of GlcNAc, suggesting that the holdfast is mostly comprised of β-1,4-linked GlcNAc (164, 167). The exact composition and structure of the holdfast is still unknown, due in good part to the small quantity synthesized by cells and its insoluble nature. The holdfast of C. crescentus has a gel-like nature and exhibits an impressive adhesive force in the μNewton range, the equivalent of 10,000 psi, making holdfast one of the strongest known biological adhesives yet characterized (167, 168). GlcNAc polymers have been proposed to provide elastic properties to the holdfast (167). Recent force spectroscopy experiments suggest that additional adhesive components within the holdfast might be responsible for the bulk of adhesion (169). The measure of time-dependence of holdfast rupture force on a variety of substrates suggests the existence of discrete and cooperative events of single adhesin molecules that may act in concert with the GlcNAc polymers (169). As mentioned earlier, the degree of deacetylation strongly influences the physical properties of the holdfast polysaccharide (142). The deletion of hfsH, a polysaccharide deacetylase, strongly affects the adhesiveness and the cohesiveness of holdfast (142).
Although C. crescentus holdfast remains the best-studied, other stalked bacteria including Hyphomonas, Hyphomicrobium, Asticcacaulis, other freshwater Caulobacter and a variety of marine bacteria also produce a discrete holdfast polysaccharide (Fig. 6) (for a review see (170)). These polysaccharides localize to the pole of the cell and provide strong adhesion to surfaces in marine or freshwater environments. The holdfast biosynthesis gene clusters of the other species often contain additional genes for gycosyltransferases or polysaccharide modification enzymes as compared to C. crescentus (170), suggesting that holdfast composition may vary depending on the environment in which the different species attach.
3.3.2. Polar surface polysaccharide from Rhizobiales
In addition to the Caulobacterales holdfast, other members of the Alphaproteobacteria exhibit similar polar polysaccharides that are involved in adhesion. The Rhizobiales A. tumefaciens, Rhizobium leguminosarum and Bradyrhizobium japonicum, can form a pathogenic or endosymbiotic association with certain plants (171). This association starts with the attachment of the bacteria to the host plant tissues, a multi-step process mediated by bacteria-secreted polysaccharides, bacteria-secreted adhesins, or in some cases, by plant-produced lectins (171–174). Once in contact with the plant surface, bacteria may produce cellulose fibrils to form a stable biofilm around the tips of root hairs or the plant tissues (175). Several different surface polysaccharides, including LPS and capsular acidic polysaccharides have been shown to play a role in plant infection and colonization. In addition, A. tumefaciens, R. leguminosarum and B. japonicum produce an unipolar polysaccharide that appears to be essential for the initial step of adhesion (171, 175–177).
A. tumefaciens forms complex biofilms on abiotic surfaces and plant roots that eventually cause “crown gall” disease. Unlike R. leguminosarum and B. japonicum, A. tumefaciens are pathogenic and do not benefit the plant (178). The infection involves first the attachment of the bacteria to the plant surface and then the transfer of a DNA segment carried on the tumor-inducing (Ti) plasmid into the host plant genome. It has been shown that A. tumefaciens produces several different EPS: succinoglycan, cellulose, β1–2 glucans and β1–3 glucans. However, more recent studies indicate that the initial attachment to the plant root surface and abiotic surfaces is mediated by a coordinated action of flagella and the unipolar polysaccharide (UPP) (68, 179, 180). UPP is crucial for permanent attachment, but by comparison with the holdfast of C. crescentus, UPP is only observed upon surface contact (68). More recently, it has been proposed that secretion of UPP is stimulated by an increase of the intracellular level of c-di-GMP, suggesting that surface contact may stimulate the increase of c-di-GMP through the activity of diguanylate cyclases or phosphodiesterases by an as yet unknown mechanism (180).
The secretion of UPP is abolished by deletion of the uppABCDEF locus (179, 180). The genes within the uppABCDEF locus shares similarity with several within the hfs locus in C. crescentus, most notably UppC with HfsD, the predicted Wza outer membrane export porin (176). No homologues of the hfa genes, which are involved in holdfast anchoring in C. crescentus, have been identified in agrobacterial genome sequences (176), suggesting that UPP and holdfast are anchored to the cell surface via different mechanisms. As observed for holdfast, UPP reacts specifically with WGA, suggesting that the UPP also contains GlcNAc residues, but its exact composition and structure remain unknown (68, 180).
R. leguminosarum forms an endosymbiotic association with certain leguminous plants such as Trifolium (clover), Pisum (pea), Vicia (vetch) and Phaseolus (bean) to potentially nodulate and differentiate into nitrogen fixing bacteroids (181). This bacterium also synthesizes a discrete polar polysaccharide that is responsible for specific adhesion to plant receptors. The primary attachment of R. leguminosarum to host plant root hairs involves bacteria-secreted polysaccharides and plant-produced lectins (172, 173). The plant-produced lectins, such as the Pea (P. sativum) lectin and the vetch (V. sativa) lectin are hypothesized to recognize and bind to a polarly localized glucomannan exopolysaccharide present at the surface of R. leguminosarum, promoting bacterial attachment at one pole (172–174). The glucomannan exopolysaccharide is mainly composed of mannose and glucose with minor amounts of galactose and rhamnose (172). Terminal mannose and glucose were identified, in addition to 1,2-mannose, 1,4-glucose and 1,3,4-linked glucose, but the exact structure remains unknown (172). At least one gene (gmsA), whose deletion abolishes glucomannan secretion and biofilm formation on root hairs, has been identified as being involved in glucomannan biosynthesis (175). Interestingly, gmsA belongs to a conserved gene locus among other rhizobia and more specifically A. tumefaciens. This locus shares sequence similarity with the uppABCDEF locus in A. tumefaciens, suggesting that glucomannan could be biosynthesized through a similar pathway as UPP (179). Glucomannan is required for initial attachment only under moderate acidic condition. Under neutral, or alkaline conditions, attachment to root hairs is mediated by Rhicadhesin, a calcium-binding protein, or rhizobium adhering proteins, named as “Rap” (172, 174, 182).
B. japonicum forms an endosymbiotic association with soybean plants. It was proposed that initial attachment was mediated by the plant lectin SBA (soybean agglutinin) but another carbohydrate binding protein, called BJ38, has been subsequently identified (177). Both SBA and BJ38 bind to a polarly localized surface polysaccharide produced by B. japonicum, but surprisingly, SBA and BJ38 recognize polysaccharides at opposite ends of the cell (177). It was suggested that the pole bound by BJ38 is probably involved in cell-cell and cell-host attachment since BJ38 labeled the point of bacterial attachment (177, 183). However, the exact nature of this interaction and the composition of this polar polysaccharide remain to be determined (177, 183). More recently, SBA has been shown to promote biofilm formation even in absence of plant roots, suggesting that SBA also contributes to adhesion on abiotic surfaces (184).
In conclusion, the use of a polar adhesin appears to be a common feature shared by some Alphaproteobacteria to mediate adhesion of the bacterial cell to biotic or abiotic surfaces. The conservation of this feature suggests that the resulting asymmetric adhesion is important for the fitness of these species. However, future studies will be needed to identify the adhesive mechanism behind these strongly adhesive unipolar polysaccharides, their regulation and biosynthesis.
3.3.3. Slime
By comparison with the discrete unipolar polysaccharide of C. crescentus, the Deltaproteobacterium M. xanthus exhibits an unusual patchy localization of an adhesive polysaccharide referred to as slime (Fig. 6). M. xanthus moves across solid surfaces by both twitching and gliding motility. While twitching motility results from polar retractile type IV pili, gliding motility results from the action of internal molecular motors that exert traction against the substrate through a large envelope-spanning complex (185–188). Under low nutrient conditions, M. xanthus cells coordinate their motility to merge into multi-cellular structures called fruiting bodies where cells follow a developmental program that eventually triggers their differentiation into stress-resistant spores. Under high nutrient conditions, spores germinate to form predatory swarms that secrete degradative enzymes to lyse prey bacteria (189). While moving over surfaces, slime is observed in the wake of motile bacteria. Slime has been observed for more than three decades (190) and has been proposed to be directly responsible for gliding motility (191), but recent evidence suggest the slime most likely promotes adhesion of the gliding motors to the substrate by a “paste-release” mechanism (192).
When observed at high contrast by a non-invasive technique called Wet-SEEC, slime trails appear to be secreted inconsistently behind the cell as extended continuous segments separated by patchy and localized deposits (192, 193). On cells, slime is unevenly distributed, forming conspicuous patches distributed at regular intervals on the surface of the cell body (193). Careful examination suggests that slime is locally concentrated and deposited underneath the cell body at fixed and discrete positions that co-localize with the gliding machinery. It has been suggested that the gliding machinery itself may produce its own adhesion substrate, enhancing Myxococcus substrate adhesion and allowing cell gliding on a larger range of substrates, but the exact anchoring mechanism between the polymer and the tip of the machinery remains to be elucidated (193).
Although slime reacts with Concanavalin-A, a lectin specific to internal and non-reducing terminal α-D-mannose or α-D-glucose, its exact composition remains unknown (193). Moreover, slime secretion remains unaffected by the deletion of genes involved in the secretion of exopolysaccharides through the Wzx/Wzy transporter-dependent pathway or in the biosynthesis of the O-antigen, suggesting that slime is secreted by an alternative secretion pathway. More recently, slime has been shown to contain embedded outer membrane vesicles, which might be involved in cell-cell communication or cell-to-cell transfer of outer-membrane proteins between Myxococcus cells (192). Slime may play multiple roles by not only providing specific adhesion for the gliding machinery, but also by potentially supporting a form of cell–cell communication between distant Myxoccocus cells sharing the same track.
CONCLUSION
During the last decade a large repertoire of bacterial adhesins have been identified. The recent advances in our understanding of the secretion, assembly and regulation of these bacterial adhesins demonstrate that, even if they share some key structural and regulatory factors that determine their expression at the surface of cells, their level of specificity varies significantly depending on the targeted surface. This characteristic clearly illustrates the ability of bacteria to successfully adapt and adhere to virtually all-natural and man-made materials. This book chapter reviewed major protein and localized discrete polysaccharide adhesins, but large variety of adhesins suggests that virtually any biological molecule made by the bacterium can be used to support or modulate its adhesion to surfaces. Even previously unsuspected biological molecules, such as extracellular DNA or lipids, have been shown to play a role in the process of adhesion or biofilm maturation (194, 195). Recent discoveries in the regulation and the modulation of adhesion suggest that this process is much more complex and dynamic than originally anticipated. We have just begun to understand the factors that govern the temporal regulation of the expression of adhesin molecules, but many questions concerning the nature of signals triggering the secretion of the proper adhesin or the interaction between the different types of adhesins remain unanswered. The recent advent of single-cell approaches will yield a better understanding of the detailed roles of each adhesive factor and the environment during this transition from reversible to irreversible attachment (196). Surprisingly, even with decades of research, the exact molecular mechanisms that support or modulate the interaction between adhesins and surfaces have seldom been characterized. Even if we can grasp the huge versatility of bacterial adhesives, we are lacking the most important knowledge about how this adhesion occurs.
A better understanding of the adhesive properties of these adhesins holds promise for future methods to control bacterial surface attachment, promoting it when beneficial and eradicating it when detrimental. In addition, an improved knowledge in this field will allow us to use certain bacterial adhesins as models of biological adhesives, which could offer an alternative strategy to their synthetic counterparts in industrial and medical applications. Indeed, biological adhesives demonstrate impressive performance in their natural context: they enable attachment to a broad variety of surfaces in aqueous environments, and share desirable properties, such as sustainability, biodegradability and biocompatibility, resulting in a much-reduced impact on the environment.
Acknowledgments
The authors want to thank the members of the Brun lab and Clay Fuqua for their fruitful discussions and critical reading of the manuscript. Work on bacterial adhesion in the Brun lab is supported by National Institutes of Health Grant GM102841 and a grant from the Indiana METACyt Initiative of Indiana University (funded in part through a major grant from the Lilly Endowment, Inc.).
References
- 1.Dunne WM. Bacterial adhesion: seen any good biofilms lately? Clinical microbiology reviews. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Oss CJ. Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specific and aspecific interactions. J Mol Recognit. 2003;16:177–190. doi: 10.1002/jmr.618. [DOI] [PubMed] [Google Scholar]
- 3.Stewart RJ. Protein-based underwater adhesives and the prospects for their biotechnological production. Appl Microbiol Biotechnol. 2011;89:27–33. doi: 10.1007/s00253-010-2913-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annual Reviews in Microbiology. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 5.Monds RD, O’Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends in microbiology. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geng J, Henry N. Bacterial Adhesion. Springer; 2011. Short time-scale bacterial adhesion dynamics; pp. 315–331. [DOI] [PubMed] [Google Scholar]
- 8.Beloin C, Roux A, Ghigo J-M. Bacterial Biofilms. Springer; 2008. Escherichia coli biofilms; pp. 249–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiology and Molecular Biology Reviews. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 11.Entcheva-Dimitrov P, Spormann AM. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J Bacteriol. 2004;186:8254–8266. doi: 10.1128/JB.186.24.8254-8266.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol. 2005;156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria -structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrows LL. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 15.St Geme JW, Pinkner JS, 3rd, Krasan GP, Heuser J, Bullitt E, Smith AL, Hultgren SJ. Haemophilus influenzae pili are composite structures assembled via the HifB chaperone. Proc Natl Acad Sci U S A. 1996;93:11913–11918. doi: 10.1073/pnas.93.21.11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gohl O, Friedrich A, Hoppert M, Averhoff B. The thin pili of Acinetobacter sp. strain BD413 mediate adhesion to biotic and abiotic surfaces. Appl Environ Microbiol. 2006;72:1394–1401. doi: 10.1128/AEM.72.2.1394-1401.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inhulsen S, Aguilar C, Schmid N, Suppiger A, Riedel K, Eberl L. Identification of functions linking quorum sensing with biofilm formation in Burkholderia cenocepacia H111. Microbiologyopen. 2012;1:225–242. doi: 10.1002/mbo3.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ormeno-Orrillo E, Menna P, Almeida LG, Ollero FJ, Nicolas MF, Pains Rodrigues E, Shigueyoshi Nakatani A, Silva Batista JS, Oliveira Chueire LM, Souza RC, Ribeiro Vasconcelos AT, Megias M, Hungria M, Martinez-Romero E. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.) BMC Genomics. 2012;13:735. doi: 10.1186/1471-2164-13-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wurpel DJ, Beatson SA, Totsika M, Petty NK, Schembri MA. Chaperone-usher fimbriae of Escherichia coli. PLoS One. 2013;8:e52835. doi: 10.1371/journal.pone.0052835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busch A, Waksman G. Chaperone-usher pathways: diversity and pilus assembly mechanism. Philos Trans R Soc Lond B Biol Sci. 2012;367:1112–1122. doi: 10.1098/rstb.2011.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Z, Pinkner JS, Ford B, Obermann R, Nolan W, Wildman SA, Hobbs D, Ellenberger T, Cusumano CK, Hultgren SJ, Janetka JW. Structure-based drug design and optimization of mannoside bacterial FimH antagonists. J Med Chem. 2010;53:4779–4792. doi: 10.1021/jm100438s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hertig S, Vogel V. Catch bonds. Curr Biol. 2012;22:R823–825. doi: 10.1016/j.cub.2012.08.035. [DOI] [PubMed] [Google Scholar]
- 25.Rakshit S, Sivasankar S. Biomechanics of cell adhesion: how force regulates the lifetime of adhesive bonds at the single molecule level. Phys Chem Chem Phys. 2014;16:2211–2223. doi: 10.1039/c3cp53963f. [DOI] [PubMed] [Google Scholar]
- 26.Liaqat I, Sakellaris H. Biofilm formation and binding specificities of CFA/I, CFA/II and CS2 adhesions of enterotoxigenic Escherichia coli and Cfae-R181A mutant. Braz J Microbiol. 2012;43:969–980. doi: 10.1590/S1517-838220120003000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ammendolia MG, Bertuccini L, Iosi F, Minelli F, Berlutti F, Valenti P, Superti F. Bovine lactoferrin interacts with cable pili of Burkholderia cenocepacia. Biometals. 2010;23:531–542. doi: 10.1007/s10534-010-9333-1. [DOI] [PubMed] [Google Scholar]
- 28.Sakellaris H, Scott JR. New tools in an old trade: CS1 pilus morphogenesis. Mol Microbiol. 1998;30:681–687. doi: 10.1046/j.1365-2958.1998.01088.x. [DOI] [PubMed] [Google Scholar]
- 29.Starks AM, Froehlich BJ, Jones TN, Scott JR. Assembly of CS1 pili: the role of specific residues of the major pilin, CooA. J Bacteriol. 2006;188:231–239. doi: 10.1128/JB.188.1.231-239.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galkin VE, Kolappan S, Ng D, Zong Z, Li J, Yu X, Egelman EH, Craig L. The structure of the CS1 pilus of enterotoxigenic Escherichia coli reveals structural polymorphism. J Bacteriol. 2013;195:1360–1370. doi: 10.1128/JB.01989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Casal J, Swartley JS, Scott JR. Gene encoding the major subunit of CS1 pili of human enterotoxigenic Escherichia coli. Infect Immun. 1990;58:3594–3600. doi: 10.1128/iai.58.11.3594-3600.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voegele K, Sakellaris H, Scott JR. CooB plays a chaperone-like role for the proteins involved in formation of CS1 pili of enterotoxigenic Escherichia coli. Proc Natl Acad Sci U S A. 1997;94:13257–13261. doi: 10.1073/pnas.94.24.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakellaris H, Balding DP, Scott JR. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- 34.Froehlich BJ, Karakashian A, Melsen LR, Wakefield JC, Scott JR. CooC and CooD are required for assembly of CS1 pili. Mol Microbiol. 1994;12:387–401. doi: 10.1111/j.1365-2958.1994.tb01028.x. [DOI] [PubMed] [Google Scholar]
- 35.Macfarlane S, Dillon JF. Microbial biofilms in the human gastrointestinal tract. J Appl Microbiol. 2007;102:1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x. [DOI] [PubMed] [Google Scholar]
- 36.Tomich M, Mohr CD. Adherence and autoaggregation phenotypes of a Burkholderia cenocepacia cable pilus mutant. FEMS Microbiol Lett. 2003;228:287–297. doi: 10.1016/S0378-1097(03)00785-7. [DOI] [PubMed] [Google Scholar]
- 37.Giltner CL, Nguyen Y, Burrows LL. Type IV pilin proteins: versatile molecular modules. Microbiol Mol Biol Rev. 2012;76:740–772. doi: 10.1128/MMBR.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giltner CL, Habash M, Burrows LL. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J Mol Biol. 2010;398:444–461. doi: 10.1016/j.jmb.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 39.Kuchma SL, Griffin EF, O’Toole GA. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J Bacteriol. 2012;194:5388–5403. doi: 10.1128/JB.00899-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson MD, Garrett CK, Bond JE, Coggan KA, Wolfgang MC, Redinbo MR. Pseudomonas aeruginosa PilY1 binds integrin in an RGD-and calcium-dependent manner. PLoS One. 2011;6:e29629. doi: 10.1371/journal.pone.0029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takhar HK, Kemp K, Kim M, Howell PL, Burrows LL. The platform protein is essential for type IV pilus biogenesis. J Biol Chem. 2013;288:9721–9728. doi: 10.1074/jbc.M113.453506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tammam S, Sampaleanu LM, Koo J, Manoharan K, Daubaras M, Burrows LL, Howell PL. PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J Bacteriol. 2013;195:2126–2135. doi: 10.1128/JB.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wehbi H, Portillo E, Harvey H, Shimkoff AE, Scheurwater EM, Howell PL, Burrows LL. The peptidoglycan-binding protein FimV promotes assembly of the Pseudomonas aeruginosa type IV pilus secretin. J Bacteriol. 2011;193:540–550. doi: 10.1128/JB.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koo J, Tang T, Harvey H, Tammam S, Sampaleanu L, Burrows LL, Howell PL. Functional mapping of PilF and PilQ in the Pseudomonas aeruginosa type IV pilus system. Biochemistry. 2013;52:2914–2923. doi: 10.1021/bi3015345. [DOI] [PubMed] [Google Scholar]
- 45.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 46.Klausen M, Aaes-Jorgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 47.Chiang P, Burrows LL. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J Bacteriol. 2003;185:2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watnick PI, Fullner KJ, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shime-Hattori A, Iida T, Arita M, Park KS, Kodama T, Honda T. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol Lett. 2006;264:89–97. doi: 10.1111/j.1574-6968.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- 50.Reguera G, Kolter R. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J Bacteriol. 2005;187:3551–3555. doi: 10.1128/JB.187.10.3551-3555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lutz C, Erken M, Noorian P, Sun S, McDougald D. Environmental reservoirs and mechanisms of persistence of. Front Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manning PA. The tcp gene cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/s0378-1119(97)00036-x. [DOI] [PubMed] [Google Scholar]
- 53.Bose N, Taylor RK. Identification of a TcpC-TcpQ outer membrane complex involved in the biogenesis of the toxin-coregulated pilus of Vibrio cholerae. J Bacteriol. 2005;187:2225–2232. doi: 10.1128/JB.187.7.2225-2232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaPointe CF, Taylor RK. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem. 2000;275:1502–1510. doi: 10.1074/jbc.275.2.1502. [DOI] [PubMed] [Google Scholar]
- 55.Tripathi SA, Taylor RK. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J Bacteriol. 2007;189:4401–4409. doi: 10.1128/JB.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomich M, Planet PJ, Figurski DH. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol. 2007;5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 57.Inoue T, Tanimoto I, Ohta H, Kato K, Murayama Y, Fukui K. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol Immunol. 1998;42:253–258. doi: 10.1111/j.1348-0421.1998.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 58.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 2001;9:429–437. doi: 10.1016/s0966-842x(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 59.Tomich M, Fine DH, Figurski DH. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J Bacteriol. 2006;188:6899–6914. doi: 10.1128/JB.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–5936. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perez-Cheeks BA, Planet PJ, Sarkar IN, Clock SA, Xu Q, Figurski DH. The product of tadZ, a new member of the parA/minD superfamily, localizes to a pole in Aggregatibacter actinomycetemcomitans. Mol Microbiol. 2012;83:694–711. doi: 10.1111/j.1365-2958.2011.07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Haase EM, Zmuda JL, Scannapieco FA. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:2901–2908. doi: 10.1128/iai.67.6.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clock SA, Planet PJ, Perez BA, Figurski DH. Outer membrane components of the Tad (tight adherence) secreton of Aggregatibacter actinomycetemcomitans. J Bacteriol. 2008;190:980–990. doi: 10.1128/JB.01347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Inoue T, Shingaki R, Sogawa N, Sogawa CA, Asaumi J, Kokeguchi S, Fukui K. Biofilm formation by a fimbriae-deficient mutant of Actinobacillus actinomycetemcomitans. Microbiol Immunol. 2003;47:877–881. doi: 10.1111/j.1348-0421.2003.tb03454.x. [DOI] [PubMed] [Google Scholar]
- 65.Saito T, Ishihara K, Ryu M, Okuda K, Sakurai K. Fimbriae-associated genes are biofilm-forming factors in Aggregatibacter actinomycetemcomitans strains. Bull Tokyo Dent Coll. 2010;51:145–150. doi: 10.2209/tdcpublication.51.145. [DOI] [PubMed] [Google Scholar]
- 66.Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bodenmiller D, Toh E, Brun YV. Development of surface adhesion in Caulobacter crescentus. J Bacteriol. 2004;186:1438–1447. doi: 10.1128/JB.186.5.1438-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol Microbiol. 2012;83:41–51. doi: 10.1111/j.1365-2958.2011.07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ruer S, Stender S, Filloux A, de Bentzmann S. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol. 2007;189:3547–3555. doi: 10.1128/JB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang YCHHCF. submitted for publication [Google Scholar]
- 71.Bednarska NG, Schymkowitz J, Rousseau F, Van Eldere J. Protein aggregation in bacteria: the thin boundary between functionality and toxicity. Microbiology. 2013;159:1795–1806. doi: 10.1099/mic.0.069575-0. [DOI] [PubMed] [Google Scholar]
- 72.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One. 2012;7:e51274. doi: 10.1371/journal.pone.0051274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans ML, Chapman MR. Curli biogenesis: Order out of disorder. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbamcr.2013.09.010. S0167-4889(13)00332-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cookson AL, Cooley WA, Woodward MJ. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int J Med Microbiol. 2002;292:195–205. doi: 10.1078/1438-4221-00203. [DOI] [PubMed] [Google Scholar]
- 76.Bokranz W, Wang X, Tschape H, Romling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- 77.Saldana Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Giron JA. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol. 2009;11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou Y, Smith D, Leong BJ, Brannstrom K, Almqvist F, Chapman MR. Promiscuous cross-seeding between bacterial amyloids promotes interspecies biofilms. J Biol Chem. 2012;287:35092–35103. doi: 10.1074/jbc.M112.383737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 80.Chagnot C, Zorgani MA, Astruc T, Desvaux M. Proteinaceous determinants of surface colonization in bacteria: bacterial adhesion and biofilm formation from a protein secretion perspective. Front Microbiol. 2013;4:303. doi: 10.3389/fmicb.2013.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Delepelaire P. Type I secretion in gram-negative bacteria. Biochim Biophys Acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 82.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lasa I, Penades JR. Bap: a family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 84.Yousef F, Espinosa-Urgel M. In silico analysis of large microbial surface proteins. Res Microbiol. 2007;158:545–550. doi: 10.1016/j.resmic.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 85.Espinosa-Urgel M, Salido A, Ramos JL. Genetic analysis of functions involved in adhesion of Pseudomonas putida to seeds. J Bacteriol. 2000;182:2363–2369. doi: 10.1128/jb.182.9.2363-2369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hinsa SM, Espinosa-Urgel M, Ramos JL, O’Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 87.Fuqua C. Passing the baton between laps: adhesion and cohesion in Pseudomonas putida biofilms. Mol Microbiol. 2010;77:533–536. doi: 10.1111/j.1365-2958.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Kirat-Chatel S, Beaussart A, Boyd CD, O’Toole GA, Dufrêne YF. Single-Cell and Single-Molecule Analysis Deciphers the Localization, Adhesion, and Mechanics of the Biofilm Adhesin LapA. ACS Chemical Biology. 2013 doi: 10.1021/cb400794e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.El-Kirat-Chatel S, Boyd CD, O’Toole GA, Dufrêne YF. Single-Molecule Analysis of Pseudomonas fluorescens Footprints. ACS nano. 2014 doi: 10.1021/nn4060489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martinez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol. 2010;77:549–561. doi: 10.1111/j.1365-2958.2010.07249.x. [DOI] [PubMed] [Google Scholar]
- 91.Duque E, de la Torre J, Bernal P, Molina-Henares MA, Alaminos M, Espinosa-Urgel M, Roca A, Fernandez M, de Bentzmann S, Ramos JL. Identification of reciprocal adhesion genes in pathogenic and non-pathogenic Pseudomonas. Environ Microbiol. 2013;15:36–48. doi: 10.1111/j.1462-2920.2012.02732.x. [DOI] [PubMed] [Google Scholar]
- 92.Valle J, Latasa C, Gil C, Toledo-Arana A, Solano C, Penades JR, Lasa I. Bap, a biofilm matrix protein of Staphylococcus aureus prevents cellular internalization through binding to GP96 host receptor. PLoS Pathog. 2012;8:e1002843. doi: 10.1371/journal.ppat.1002843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wagner C, Hensel M. Adhesive mechanisms of Salmonella enterica. Adv Exp Med Biol. 2011;715:17–34. doi: 10.1007/978-94-007-0940-9_2. [DOI] [PubMed] [Google Scholar]
- 94.Wagner C, Polke M, Gerlach RG, Linke D, Stierhof YD, Schwarz H, Hensel M. Functional dissection of SiiE, a giant non-fimbrial adhesin of Salmonella enterica. Cell Microbiol. 2011;13:1286–1301. doi: 10.1111/j.1462-5822.2011.01621.x. [DOI] [PubMed] [Google Scholar]
- 95.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Molecular Microbiology. 2010;75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 96.Martinez-Gil M, Quesada JM, Ramos-Gonzalez MI, Soriano MI, de Cristobal RE, Espinosa-Urgel M. Interplay between extracellular matrix components of Pseudomonas putida biofilms. Res Microbiol. 2013;164:382–389. doi: 10.1016/j.resmic.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 97.Martínez-Gil M, Ramos-González MI, Espinosa-Urgel M. Role of c-di-GMP and the Gac System in the Transcriptional Control of the Genes Coding for the Pseudomonas putida Adhesins LapA and LapF. Journal of Bacteriology: JB. 2014:01287–01213. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Desvaux M, Parham NJ, Henderson IR. Type V protein secretion: simplicity gone awry? Curr Issues Mol Biol. 2004;6:111–124. [PubMed] [Google Scholar]
- 99.Bernstein HD. Are bacterial ‘autotransporters’ really transporters? Trends Microbiol. 2007;15:441–447. doi: 10.1016/j.tim.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 100.Leyton DL, Rossiter AE, Henderson IR. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol. 2012;10:213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 101.Leo JC, Grin I, Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Klemm P, Vejborg RM, Sherlock O. Self-associating autotransporters, SAATs: functional and structural similarities. Int J Med Microbiol. 2006;296:187–195. doi: 10.1016/j.ijmm.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 103.Roux A, Beloin C, Ghigo JM. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J Bacteriol. 2005;187:1001–1013. doi: 10.1128/JB.187.3.1001-1013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Owen P, Meehan M, de Loughry-Doherty H, Henderson I. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol Med Microbiol. 1996;16:63–76. doi: 10.1111/j.1574-695X.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 105.Hasman H, Chakraborty T, Klemm P. Antigen-43-mediated autoaggregation of Escherichia coli is blocked by fimbriation. J Bacteriol. 1999;181:4834–4841. doi: 10.1128/jb.181.16.4834-4841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Danese PN, Pratt LA, Dove SL, Kolter R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol. 2000;37:424–432. doi: 10.1046/j.1365-2958.2000.02008.x. [DOI] [PubMed] [Google Scholar]
- 107.Grijpstra J, Arenas J, Rutten L, Tommassen J. Autotransporter secretion: varying on a theme. Res Microbiol. 2013;164:562–582. doi: 10.1016/j.resmic.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 108.Sherlock O, Schembri MA, Reisner A, Klemm P. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J Bacteriol. 2004;186:8058–8065. doi: 10.1128/JB.186.23.8058-8065.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Benz I, Schmidt MA. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Molecular microbiology. 2001;40:1403–1413. doi: 10.1046/j.1365-2958.2001.02487.x. [DOI] [PubMed] [Google Scholar]
- 110.Sherlock O, Dobrindt U, Jensen JB, Vejborg RM, Klemm P. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. Journal of bacteriology. 2006;188:1798–1807. doi: 10.1128/JB.188.5.1798-1807.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lindenthal C, Elsinghorst EA. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infection and immunity. 1999;67:4084–4091. doi: 10.1128/iai.67.8.4084-4091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Côté J-P, Charbonneau M-È, Mourez M. Glycosylation of the Escherichia coli TibA self-associating autotransporter influences the conformation and the functionality of the protein. PloS one. 2013;8:e80739. doi: 10.1371/journal.pone.0080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wallecha A, Munster V, Correnti J, Chan T, van der Woude M. Dam-and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J Bacteriol. 2002;184:3338–3347. doi: 10.1128/JB.184.12.3338-3347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chauhan A, Sakamoto C, Ghigo JM, Beloin C. Did I pick the right colony? Pitfalls in the study of regulation of the phase variable antigen 43 adhesin. PLoS One. 2013;8:e73568. doi: 10.1371/journal.pone.0073568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW. Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. The EMBO journal. 2011;30:3864–3874. doi: 10.1038/emboj.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proceedings of the National Academy of Sciences. 2014;111:457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lyskowski A, Leo JC, Goldman A. Structure and biology of trimeric autotransporter adhesins. Adv Exp Med Biol. 2011;715:143–158. doi: 10.1007/978-94-007-0940-9_9. [DOI] [PubMed] [Google Scholar]
- 119.El Tahir Y, Skurnik M. YadA, the multifaceted Yersinia adhesin. Int J Med Microbiol. 2001;291:209–218. doi: 10.1078/1438-4221-00119. [DOI] [PubMed] [Google Scholar]
- 120.Heise T, Dersch P. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc Natl Acad Sci U S A. 2006;103:3375–3380. doi: 10.1073/pnas.0507749103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol. 2008;190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Raghunathan D, Wells TJ, Morris FC, Shaw RK, Bobat S, Peters SE, Paterson GK, Jensen KT, Leyton DL, Blair JM, Browning DF, Pravin J, Flores-Langarica A, Hitchcock JR, Moraes CT, Piazza RM, Maskell DJ, Webber MA, May RC, MacLennan CA, Piddock LJ, Cunningham AF, Henderson IR. SadA, a trimeric autotransporter from Salmonella enterica serovar Typhimurium, can promote biofilm formation and provides limited protection against infection. Infect Immun. 2011;79:4342–4352. doi: 10.1128/IAI.05592-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lazar Adler NR, Dean RE, Saint RJ, Stevens MP, Prior JL, Atkins TP, Galyov EE. Identification of a Predicted Trimeric Autotransporter Adhesin Required for Biofilm Formation of Burkholderia pseudomallei. PLoS One. 2013;8:e79461. doi: 10.1371/journal.pone.0079461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mazar J, Cotter PA. New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol. 2007;15:508–515. doi: 10.1016/j.tim.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 125.Darsonval A, Darrasse A, Durand K, Bureau C, Cesbron S, Jacques MA. Adhesion and fitness in the bean phyllosphere and transmission to seed of Xanthomonas fuscans subsp. fuscans. Mol Plant Microbe Interact. 2009;22:747–757. doi: 10.1094/MPMI-22-6-0747. [DOI] [PubMed] [Google Scholar]
- 126.Feil H, Feil WS, Lindow SE. Contribution of Fimbrial and Afimbrial Adhesins of Xylella fastidiosa to Attachment to Surfaces and Virulence to Grape. Phytopathology. 2007;97:318–324. doi: 10.1094/PHYTO-97-3-0318. [DOI] [PubMed] [Google Scholar]
- 127.Ryan RP, Vorholter FJ, Potnis N, Jones JB, Van Sluys MA, Bogdanove AJ, Dow JM. Pathogenomics of Xanthomonas: understanding bacterium-plant interactions. Nat Rev Microbiol. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 128.Webster P, Wu S, Gomez G, Apicella M, Plaut AG, St Geme JW., 3rd Distribution of bacterial proteins in biofilms formed by non-typeable Haemophilus influenzae. J Histochem Cytochem. 2006;54:829–842. doi: 10.1369/jhc.6A6922.2006. [DOI] [PubMed] [Google Scholar]
- 129.Serra DO, Conover MS, Arnal L, Sloan GP, Rodriguez ME, Yantorno OM, Deora R. FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PLoS One. 2011;6:e28811. doi: 10.1371/journal.pone.0028811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guo H, Yi W, Song JK, Wang PG. Current understanding on biosynthesis of microbial polysaccharides. Curr Top Med Chem. 2008;8:141–151. doi: 10.2174/156802608783378873. [DOI] [PubMed] [Google Scholar]
- 132.Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21:63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ahimou F, Semmens MJ, Haugstad G, Novak PJ. Effect of protein, polysaccharide, and oxygen concentration profiles on biofilm cohesiveness. Appl Environ Microbiol. 2007;73:2905–2910. doi: 10.1128/AEM.02420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Davey ME, O’Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sutherland I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 136.Haag AP. Biological Adhesives. Springer-Verlag; Berlin Heidelberg: 2006. Mechanical Properties of Bacterial Exopolymeric Adhesives and their Commercial Development; pp. 1–19. [Google Scholar]
- 137.Korstgens V, Flemming HC, Wingender J, Borchard W. Influence of calcium ions on the mechanical properties of a model biofilm of mucoid Pseudomonas aeruginosa. Water Sci Technol. 2001;43:49–57. [PubMed] [Google Scholar]
- 138.Franklin MJ, Ohman DE. Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J Bacteriol. 1993;175:5057–5065. doi: 10.1128/jb.175.16.5057-5065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rinaudo M. Role of substituents on the properties of some polysaccharides. Biomacromolecules. 2004;5:1155–1165. doi: 10.1021/bm030077q. [DOI] [PubMed] [Google Scholar]
- 140.Tielen P, Strathmann M, Jaeger KE, Flemming HC, Wingender J. Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol Res. 2005;160:165–176. doi: 10.1016/j.micres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 141.Villain-Simonnet A, Milas M, Rinaudo M. A new bacterial polysaccharide (YAS34). I. Characterization of the conformations and conformational transition. Int J Biol Macromol. 2000;27:65–75. doi: 10.1016/s0141-8130(99)00120-8. [DOI] [PubMed] [Google Scholar]
- 142.Wan Z, Brown PJ, Elliott EN, Brun YV. The adhesive and cohesive properties of a bacterial polysaccharide adhesin are modulated by a deacetylase. Mol Microbiol. 2013;88:486–500. doi: 10.1111/mmi.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cerca N, Jefferson KK, Maira-Litran T, Pier DB, Kelly-Quintos C, Goldmann DA, Azeredo J, Pier GB. Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-beta-(1–6)-glucosamine. Infect Immun. 2007;75:3406–3413. doi: 10.1128/IAI.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston JF, 3rd, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190:3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vuong C, Kocianova S, Voyich JM, Yao Y, Fischer ER, DeLeo FR, Otto M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279:54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 146.Pokrovskaya V, Poloczek J, Little DJ, Griffiths H, Howell PL, Nitz M. Functional characterization of Staphylococcus epidermidis IcaB, a de-N-acetylase important for biofilm formation. Biochemistry. 2013;52:5463–5471. doi: 10.1021/bi400836g. [DOI] [PubMed] [Google Scholar]
- 147.Riley LM, Weadge JT, Baker P, Robinson H, Codee JD, Tipton PA, Ohman DE, Howell PL. Structural and functional characterization of Pseudomonas aeruginosa AlgX: role of AlgX in alginate acetylation. J Biol Chem. 2013;288:22299–22314. doi: 10.1074/jbc.M113.484931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH, Howell PL. Structural basis for alginate secretion across the bacterial outer membrane. Proc Natl Acad Sci U S A. 2011;108:13083–13088. doi: 10.1073/pnas.1104984108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Willis LM, Stupak J, Richards MR, Lowary TL, Li J, Whitfield C. Conserved glycolipid termini in capsular polysaccharides synthesized by ATP-binding cassette transporter-dependent pathways in Gram-negative pathogens. Proc Natl Acad Sci U S A. 2013;110:7868–7873. doi: 10.1073/pnas.1222317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jimenez N, Senchenkova SN, Knirel YA, Pieretti G, Corsaro MM, Aquilini E, Regue M, Merino S, Tomas JM. Effects of lipopolysaccharide biosynthesis mutations on K1 polysaccharide association with the Escherichia coli cell surface. J Bacteriol. 2012;194:3356–3367. doi: 10.1128/JB.00329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gaastra W, De Graaf FK. Host-specific fimbrial adhesins of noninvasive enterotoxigenic Escherichia coli strains. Microbiological reviews. 1982;46:129. doi: 10.1128/mr.46.2.129-161.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Franco AV, Liu D, Reeves PR. A Wzz (Cld) protein determines the chain length of K lipopolysaccharide in Escherichia coli O8 and O9 strains. J Bacteriol. 1996;178:1903–1907. doi: 10.1128/jb.178.7.1903-1907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jann K, Dengler T, Jann B. Core-lipid A on the K40 polysaccharide of Escherichia coli O8:K40:H9, a representative of group I capsular polysaccharides. Zentralbl Bakteriol. 1992;276:196–204. doi: 10.1016/s0934-8840(11)80006-x. [DOI] [PubMed] [Google Scholar]
- 154.Bushell SR, Mainprize IL, Wear MA, Lou H, Whitfield C, Naismith JH. Wzi is an outer membrane lectin that underpins group 1 capsule assembly in Escherichia coli. Structure. 2013;21:844–853. doi: 10.1016/j.str.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hardy GG, Allen RC, Toh E, Long M, Brown PJ, Cole-Tobian JL, Brun YV. A localized multimeric anchor attaches the Caulobacter holdfast to the cell pole. Mol Microbiol. 2010;76:409–427. doi: 10.1111/j.1365-2958.2010.07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Rahn A, Beis K, Naismith JH, Whitfield C. A novel outer membrane protein, Wzi, is involved in surface assembly of the Escherichia coli K30 group 1 capsule. J Bacteriol. 2003;185:5882–5890. doi: 10.1128/JB.185.19.5882-5890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Iwashkiw JA, Vozza NF, Kinsella RL, Feldman MF. Pour some sugar on it: the expanding world of bacterial protein O-linked glycosylation. Mol Microbiol. 2013;89:14–28. doi: 10.1111/mmi.12265. [DOI] [PubMed] [Google Scholar]
- 158.Song MC, Kim E, Ban YH, Yoo YJ, Kim EJ, Park SR, Pandey RP, Sohng JK, Yoon YJ. Achievements and impacts of glycosylation reactions involved in natural product biosynthesis in prokaryotes. Appl Microbiol Biotechnol. 2013;97:5691–5704. doi: 10.1007/s00253-013-4978-7. [DOI] [PubMed] [Google Scholar]
- 159.Quintero EJ, Busch K, Weiner RM. Spatial and Temporal Deposition of Adhesive Extracellular Polysaccharide Capsule and Fimbriae by Hyphomonas Strain MHS-3. Appl Environ Microbiol. 1998;64:1246–1255. doi: 10.1128/aem.64.4.1246-1255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ong CJ, Wong ML, Smit J. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J Bacteriol. 1990;172:1448–1456. doi: 10.1128/jb.172.3.1448-1456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Smith CS, Hinz A, Bodenmiller D, Larson DE, Brun YV. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J Bacteriol. 2003;185:1432–1442. doi: 10.1128/JB.185.4.1432-1442.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Poindexter JS. The Prokaryotes. Springer; New York: 2006. Dimorphic Prosthecate Bacteria: The Genera Caulobacter, Asticcacaulis, Hyphomicrobium, Pedomicrobium, Hyphomonas and Thiodendron; pp. 72–90. [Google Scholar]
- 163.Poindexter JS. Biological Properties and Classification of the Caulobacter Group. Bacteriol Rev. 1964;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Merker RI, Smit J. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl Environ Microbiol. 1988;54:2078–2085. doi: 10.1128/aem.54.8.2078-2085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fiebig A, Herrou J, Fumeaux C, Radhakrishnan SK, Viollier PH, Crosson S. A cell cycle and nutritional checkpoint controlling bacterial surface adhesion. PLoS Genet. 2014;10:e1004101. doi: 10.1371/journal.pgen.1004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Toh E, Kurtz HD, Jr, Brun YV. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J Bacteriol. 2008;190:7219–7231. doi: 10.1128/JB.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Li G, Smith CS, Brun YV, Tang JX. The elastic properties of the caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J Bacteriol. 2005;187:257–265. doi: 10.1128/JB.187.1.257-265.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Tsang PH, Li G, Brun YV, Freund LB, Tang JX. Adhesion of single bacterial cells in the micronewton range. Proc Natl Acad Sci U S A. 2006;103:5764–5768. doi: 10.1073/pnas.0601705103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Berne C, Ma X, Licata NA, Neves BR, Setayeshgar S, Brun YV, Dragnea B. Physiochemical properties of Caulobacter crescentus holdfast: a localized bacterial adhesive. J Phys Chem B. 2013;117:10492–10503. doi: 10.1021/jp405802e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Brown PJ, Hardy GG, Trimble MJ, Brun YV. Complex Regulatory Pathways Coordinate Cell-Cycle Progression and Development in Caulobacter crescentus. Advances in microbial physiology. 2008;54:1–101. doi: 10.1016/S0065-2911(08)00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rodriguez-Navarro DN, Dardanelli MS, Ruiz-Sainz JE. Attachment of bacteria to the roots of higher plants. FEMS Microbiol Lett. 2007;272:127–136. doi: 10.1111/j.1574-6968.2007.00761.x. [DOI] [PubMed] [Google Scholar]
- 172.Laus MC, Logman TJ, Lamers GE, Van Brussel AA, Carlson RW, Kijne JW. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol. 2006;59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- 173.Xie F, Williams A, Edwards A, Downie JA. A plant arabinogalactan-like glycoprotein promotes a novel type of polar surface attachment by Rhizobium leguminosarum. Mol Plant Microbe Interact. 2012;25:250–258. doi: 10.1094/MPMI-08-11-0211. [DOI] [PubMed] [Google Scholar]
- 174.Ausmees N, Jacobsson K, Lindberg M. A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology. 2001;147:549–559. doi: 10.1099/00221287-147-3-549. [DOI] [PubMed] [Google Scholar]
- 175.Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol. 2008;190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tomlinson AD, Fuqua C. Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens. Curr Opin Microbiol. 2009;12:708–714. doi: 10.1016/j.mib.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Loh JT, Ho SC, de Feijter AW, Wang JL, Schindler M. Carbohydrate binding activities of Bradyrhizobium japonicum: unipolar localization of the lectin BJ38 on the bacterial cell surface. Proc Natl Acad Sci U S A. 1993;90:3033–3037. doi: 10.1073/pnas.90.7.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Merritt PM, Danhorn T, Fuqua C. Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J Bacteriol. 2007;189:8005–8014. doi: 10.1128/JB.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol. 2012;163:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol. 2013;89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Fujishige NA, Kapadia NN, De Hoff PL, Hirsch AM. Investigations of Rhizobium biofilm formation. FEMS Microbiol Ecol. 2006;56:195–206. doi: 10.1111/j.1574-6941.2005.00044.x. [DOI] [PubMed] [Google Scholar]
- 182.Abdian PL, Caramelo JJ, Ausmees N, Zorreguieta A. RapA2 is a calcium-binding lectin composed of two highly conserved cadherin-like domains that specifically recognize Rhizobium leguminosarum acidic exopolysaccharides. J Biol Chem. 2013;288:2893–2904. doi: 10.1074/jbc.M112.411769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ho SC, Wang JL, Schindler M, Loh JT. Carbohydrate binding activities of Bradyrhizobium japonicum. III. Lectin expression, bacterial binding, and nodulation efficiency. Plant J. 1994;5:873–884. doi: 10.1046/j.1365-313x.1994.5060873.x. [DOI] [PubMed] [Google Scholar]
- 184.Pérez-Giménez J, Mongiardini EJ, Althabegoiti MJ, Covelli J, Quelas JI, López-García SL, Lodeiro AR. Soybean lectin enhances biofilm formation by Bradyrhizobium japonicum in the absence of plants. International journal of microbiology. 2009;2009 doi: 10.1155/2009/719367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T. Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet. 2011;7:e1002268. doi: 10.1371/journal.pgen.1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci U S A. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nan B, Mauriello EM, Sun IH, Wong A, Zusman DR. A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol. 2010;76:1539–1554. doi: 10.1111/j.1365-2958.2010.07184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci U S A. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang Y, Ducret A, Shaevitz J, Mignot T. From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol Rev. 2012;36:149–164. doi: 10.1111/j.1574-6976.2011.00307.x. [DOI] [PubMed] [Google Scholar]
- 190.Burchard RP. Trail following by gliding bacteria. J Bacteriol. 1982;152:495–501. doi: 10.1128/jb.152.1.495-501.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. How myxobacteria glide. Curr Biol. 2002;12:369–377. doi: 10.1016/s0960-9822(02)00716-9. [DOI] [PubMed] [Google Scholar]
- 192.Ducret A, Fleuchot B, Bergam P, Mignot T. Direct live imaging of cell-cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. Elife. 2013;2:e00868. doi: 10.7554/eLife.00868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci U S A. 2012;109:10036–10041. doi: 10.1073/pnas.1120979109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Flemming H-C, Wingender J. The biofilm matrix. Nature Reviews Microbiology. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 195.Stoodley P, Sauer K, Davies D, Costerton JW. Biofilms as complex differentiated communities. Annual Reviews in Microbiology. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 196.Wessel AK, Hmelo L, Parsek MR, Whiteley M. Going local: Technologies for exploring bacterial microenvironments. Nature Reviews Microbiology. 2013;11:337–348. doi: 10.1038/nrmicro3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Garnett JA, Martinez-Santos VI, Saldana Z, Pape T, Hawthorne W, Chan J, Simpson PJ, Cota E, Puente JL, Giron JA, Matthews S. Structural insights into the biogenesis and biofilm formation by the Escherichia coli common pilus. Proc Natl Acad Sci U S A. 2012;109:3950–3955. doi: 10.1073/pnas.1106733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kalivoda EJ, Stella NA, O’Dee DM, Nau GJ, Shanks RM. The cyclic AMP-dependent catabolite repression system of Serratia marcescens mediates biofilm formation through regulation of type 1 fimbriae. Appl Environ Microbiol. 2008;74:3461–3470. doi: 10.1128/AEM.02733-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Koczan JM, Lenneman BR, McGrath MJ, Sundin GW. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl Environ Microbiol. 2011;77:7031–7039. doi: 10.1128/AEM.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mhedbi-Hajri N, Jacques MA, Koebnik R. Adhesion mechanisms of plant-pathogenic Xanthomonadaceae. Adv Exp Med Biol. 2011;715:71–89. doi: 10.1007/978-94-007-0940-9_5. [DOI] [PubMed] [Google Scholar]
- 201.Stahlhut SG, Struve C, Krogfelt KA, Reisner A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol Med Microbiol. 2012;65:350–359. doi: 10.1111/j.1574-695X.2012.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 203.Ong CL, Beatson SA, Totsika M, Forestier C, McEwan AG, Schembri MA. Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella and Citrobacter species. BMC Microbiol. 2010;10:183. doi: 10.1186/1471-2180-10-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Di Martino P, Cafferini N, Joly B, Darfeuille-Michaud A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res Microbiol. 2003;154:9–16. doi: 10.1016/s0923-2508(02)00004-9. [DOI] [PubMed] [Google Scholar]
- 205.Lappann M, Haagensen JA, Claus H, Vogel U, Molin S. Meningococcal biofilm formation: structure, development and phenotypes in a standardized continuous flow system. Mol Microbiol. 2006;62:1292–1309. doi: 10.1111/j.1365-2958.2006.05448.x. [DOI] [PubMed] [Google Scholar]
- 206.Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol Microbiol. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- 207.Marsh JW, Taylor RK. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J Bacteriol. 1999;181:1110–1117. doi: 10.1128/jb.181.4.1110-1117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Moreira CG, Palmer K, Whiteley M, Sircili MP, Trabulsi LR, Castro AF, Sperandio V. Bundle-forming pili and EspA are involved in biofilm formation by enteropathogenic Escherichia coli. J Bacteriol. 2006;188:3952–3961. doi: 10.1128/JB.00177-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Bernard CS, Bordi C, Termine E, Filloux A, de Bentzmann S. Organization and PprB-dependent control of the Pseudomonas aeruginosa tad Locus, involved in Flp pilus biology. J Bacteriol. 2009;191:1961–1973. doi: 10.1128/JB.01330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Collinson SK, Clouthier SC, Doran JL, Banser PA, Kay WW. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Austin JW, Sanders G, Kay WW, Collinson SK. Thin aggregative fimbriae enhance Salmonella enteritidis biofilm formation. FEMS Microbiol Lett. 1998;162:295–301. doi: 10.1111/j.1574-6968.1998.tb13012.x. [DOI] [PubMed] [Google Scholar]
- 213.Dueholm MS, Sondergaard MT, Nilsson M, Christiansen G, Stensballe A, Overgaard MT, Givskov M, Tolker-Nielsen T, Otzen DE, Nielsen PH. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen. 2013;2:365–382. doi: 10.1002/mbo3.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Huber B, Riedel K, Kothe M, Givskov M, Molin S, Eberl L. Genetic analysis of functions involved in the late stages of biofilm development in Burkholderia cepacia H111. Mol Microbiol. 2002;46:411–426. doi: 10.1046/j.1365-2958.2002.03182.x. [DOI] [PubMed] [Google Scholar]
- 215.Latasa C, Roux A, Toledo-Arana A, Ghigo JM, Gamazo C, Penades JR, Lasa I. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol. 2005;58:1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x. [DOI] [PubMed] [Google Scholar]
- 216.Loehfelm TW, Luke NR, Campagnari AA. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol. 2008;190:1036–1044. doi: 10.1128/JB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Theunissen S, De Smet L, Dansercoer A, Motte B, Coenye T, Van Beeumen JJ, Devreese B, Savvides SN, Vergauwen B. The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res Microbiol. 2010;161:144–152. doi: 10.1016/j.resmic.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 218.Pérez-Mendoza D, Coulthurst SJ, Humphris S, Campbell E, Welch M, Toth IK, Salmond GP. A multi-repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a Type I pathway and is subject to complex regulation involving a non-canonical diguanylate cyclase. Molecular microbiology. 2011;82:719–733. doi: 10.1111/j.1365-2958.2011.07849.x. [DOI] [PubMed] [Google Scholar]
- 219.Wu C, Cheng YY, Yin H, Song XN, Li WW, Zhou XX, Zhao LP, Tian LJ, Han JC, Yu HQ. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci Rep. 2013;3:1945. doi: 10.1038/srep01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hinsa-Leasure SM, Koid C, Tiedje JM, Schultzhaus JN. Biofilm formation by Psychrobacter arcticus and the role of a large adhesin in attachment to surfaces. Appl Environ Microbiol. 2013;79:3967–3973. doi: 10.1128/AEM.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Torres AG, Perna NT, Burland V, Ruknudin A, Blattner FR, Kaper JB. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Molecular Microbiology. 2002;45:951–966. doi: 10.1046/j.1365-2958.2002.03094.x. [DOI] [PubMed] [Google Scholar]
- 222.Sherlock O, Vejborg RM, Klemm P. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect Immun. 2005;73:1954–1963. doi: 10.1128/IAI.73.4.1954-1963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol. 2008;10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 224.Wells TJ, McNeilly TN, Totsika M, Mahajan A, Gally DL, Schembri MA. The Escherichia coli O157:H7 EhaB autotransporter protein binds to laminin and collagen I and induces a serum IgA response in O157:H7 challenged cattle. Environ Microbiol. 2009;11:1803–1814. doi: 10.1111/j.1462-2920.2009.01905.x. [DOI] [PubMed] [Google Scholar]
- 225.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect Immun. 2010;78:1659–1669. doi: 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect Immun. 2012;80:321–332. doi: 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zude I, Leimbach A, Dobrindt U. Prevalence of autotransporters in Escherichia coli: what is the impact of phylogeny and pathotype? Int J Med Microbiol. 2013 doi: 10.1016/j.ijmm.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 228.Kroupitski Y, Brandl MT, Pinto R, Belausov E, Tamir-Ariel D, Burdman S, Sela Saldinger S. Identification of Salmonella enterica genes with a role in persistence on lettuce leaves during cold storage by recombinase-based in vivo expression technology. Phytopathology. 2013;103:362–372. doi: 10.1094/PHYTO-10-12-0254-FI. [DOI] [PubMed] [Google Scholar]
- 229.Pearson MM, Laurence CA, Guinn SE, Hansen EJ. Biofilm formation by Moraxella catarrhalis in vitro: roles of the UspA1 adhesin and the Hag hemagglutinin. Infect Immun. 2006;74:1588–1596. doi: 10.1128/IAI.74.3.1588-1596.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ishikawa M, Nakatani H, Hori K. AtaA, a new member of the trimeric autotransporter adhesins from Acinetobacter sp. Tol 5 mediating high adhesiveness to various abiotic surfaces. PLoS One. 2012;7:e48830. doi: 10.1371/journal.pone.0048830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Totsika M, Wells TJ, Beloin C, Valle J, Allsopp LP, King NP, Ghigo JM, Schembri MA. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl Environ Microbiol. 2012;78:2179–2189. doi: 10.1128/AEM.06680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Guilhabert MR, Kirkpatrick BC. Identification of Xylella fastidiosa antivirulence genes: hemagglutinin adhesins contribute a biofilm maturation to X. fastidios and colonization and attenuate virulence. Mol Plant Microbe Interact. 2005;18:856–868. doi: 10.1094/MPMI-18-0856. [DOI] [PubMed] [Google Scholar]
- 233.Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin-like protein of Xanthomonas axonopodis pv. citri, the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS One. 2009;4:e4358. doi: 10.1371/journal.pone.0004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Molecular microbiology. 2013;89:1213–1225. doi: 10.1111/mmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Evans LR, Linker A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J Bacteriol. 1973;116:915–924. doi: 10.1128/jb.116.2.915-924.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ausmees N, Jonsson H, Hoglund S, Ljunggren H, Lindberg M. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology. 1999;145 ( Pt 5):1253–1262. doi: 10.1099/13500872-145-5-1253. [DOI] [PubMed] [Google Scholar]
- 237.Matthysse AG, Thomas DL, White AR. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1076–1081. doi: 10.1128/jb.177.4.1076-1081.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 241.MacRae JD, Smit J. Characterization of caulobacters isolated from wastewater treatment systems. Appl Environ Microbiol. 1991;57:751–758. doi: 10.1128/aem.57.3.751-758.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Moore RL, Marshall KC. Attachment and rosette formation by hyphomicrobia. Appl Environ Microbiol. 1981;42:751–757. doi: 10.1128/aem.42.5.751-757.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Quintero EJ, Weiner RM. Evidence for the Adhesive Function of the Exopolysaccharide of Hyphomonas Strain MHS-3 in Its Attachment to Surfaces. Appl Environ Microbiol. 1995;61:1897–1903. doi: 10.1128/aem.61.5.1897-1903.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Yun C, Ely B, Smit J. Identification of genes affecting production of the adhesive holdfast of a marine caulobacter. J Bacteriol. 1994;176:796–803. doi: 10.1128/jb.176.3.796-803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wang X, Preston JF, 3rd, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186:2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Itoh Y, Wang X, Hinnebusch BJ, Preston JF, 3rd, Romeo T. Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms. J Bacteriol. 2005;187:382–387. doi: 10.1128/JB.187.1.382-387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ghafoor A, Hay ID, Rehm BH. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Byrd MS, Sadovskaya I, Vinogradov E, Lu H, Sprinkle AB, Richardson SH, Ma L, Ralston B, Parsek MR, Anderson EM, Lam JS, Wozniak DJ. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol. 2009;73:622–638. doi: 10.1111/j.1365-2958.2009.06795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 251.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]