Abstract
Mitochondrial dysfunction has reported in several diseases including diabetes, cancer, skeletal muscle disorders and neurodegenerative diseases such as Wolfram syndrome. Several different methods have evolved to study mtDNA damage including Southern blotting, 8-oxoG damage, or a comprehensive scanning of the mitochondrial genome by RFLP or TTGE analyses. However these approaches require large amounts of DNA or are labor intensive. The use of polymerase amplification of long DNA products (LRPCR) has been described by several groups and more recently summarized by Van Houten's group. The underlying basis use of DNA polymerases capable of generating long DNA products and the rationale is that any lesion (strand breaks, base modifications, apurinic sites) will stop a thermostable DNA polymerase. In this method, band density of the PCR product is quantified either by Southern blotting or binding of a fluorescent dye. Although the latter approach still has some limited use in the study gene expression, it is semi-quantitative and realtime PCR analysis has largely supplanted it. Direct application of real-time PCR to LRPCR has been made difficult because of low processivity and polymerization rates of the DNA polymerases used and SYBR green inhibition of DNA amplification. We have modified the LRPCR protocol to use the commercially available PfuUltra™ II Fusion HS DNA Polymerase for real-time determination of mitochondrial DNA amplification as a means to simplify and improve of the accuracy for quantification of mtDNA damage.
Keywords: QPCR, mitochondrial DNA, DNA damage
Introduction
The mitochondrial genome is a circular double-stranded DNA of 16,569 basepairs in humans. It codes for 13 separate proteins that are part of the electron transport chain as well as for the synthesis of mitochondrial-specific ribosomal RNAs and transfer RNAs. Mitochondrial disorders are a heterogeneous group of diseases that may be characterized by maternal inheritance, heteroplasmy, and threshold effect. Aside from diabetes, mitochondrial dysfunction has reported in cancer, skeletal muscle disorders and neurodegenerative diseases such as Wolfram syndrome (Gokey et al. 2004; Kagan et al. 2005; Sciacco et al. 2005; Domenech et al. 2006). Mitochondrial DNA is thought to be more sensitive to oxidative stress induced damage compared to genomic DNA (Yakes et al. 1997).
Over the years, several different methods have evolved to study mtDNA mutations including, Southern blots, mtDNA copy number, 8-oxoG damage, comet assay, or approaches that permit comprehensive scanning of the mitochondrial genome including RFLP and TTGE analyses (Mellon et al. 1986; Leadon et al. 1988; Richter et al. 1988; Thomas et al. 1988; Wong et al. 2002; Wong 2004). However these latter approaches require large amounts of DNA, are labor intensive, or require optimization of a large number of primer sets. The use of DNA polymerase amplification of long DNA products (LRPCR) to study DNA damage was first described by Govan et.al and adapted by other groups, and more recently summarized by Santos et.al. (Govan et al. 1990; Kalinowski et al. 1992; Oshita et al. 1993; Ballinger et al. 1996; Yakes et al. 1997; Ballinger et al. 2000; Santos et al. 2006). This approach makes use of DNA polymerases capable of generating long DNA products and the rationale was that any lesion (strand breaks, base modifications, apurinic sites) will stop a thermostable DNA polymerase, and only those DNA templates not containing any lesions will be amplified (Govan et al. 1990; Kalinowski et al. 1992; Ballinger et al. 1996). The copy number derived from this amplification will be compared to the copy number of a short PCR product (150-250 bp) that is unlikely to contain any lesions. The DNA lesion frequency is then calculated from the relative ratio of long:short amplicons. This approach has been used not only to study genomic DNA repair mechanisms, but also to demonstrate that mtDNA damage is more extensive than genomic DNA damage and that mtDNA repair processes are slower (Govan et al. 1990; Ballinger et al. 1996; Yakes et al. 1997; Ballinger et al. 1999; Ballinger et al. 2000).
The LRPCR approach described by these groups quantifies DNA band density of PCR products by either 32P-Southern blotting (Govan et al. 1990; Jennerwein et al. 1991; Kalinowski et al. 1992; Yakes et al. 1997) or determination of fluorescent dye binding (Ballinger et al. 1996; Ballinger et al. 2000; Santos et al. 2006). The LRPCR approach requires DNA amplification to be carried out, such that measurements are made within the linear range of amplification. This requires optimization to find the optimal starting concentration of DNA template (Govan et al. 1990; Jennerwein et al. 1991; Yakes et al. 1997), or identify the range in which DNA amplification is linear (step-analysis), both approaches require a significant amount of optimization (Kalinowski et al. 1992; Ballinger et al. 1996; Yakes et al. 1997; Ballinger et al. 1999; Santos et al. 2006). Although this latter approach still has some limited use in the study of gene expression and is accepted as a semi-quantitative method, that realtime PCR analysis has largely supplanted for the determination of mRNA levels in gene expression studies.
Two significant problems occur in the direct application of realtime PCR to LRPCR; 1) the low processivity and polymerization rates of the DNA polymerases used in comparison to the length of the transcripts, 2) SYBR green inhibition of DNA amplification (Gudnason et al. 2007). More recently, modified DNA polymerases and optimization protocols have improved both the rate of polymerization as well as increasing processivity. We have modified the LRPCR approach to use the commercially available PfuUltra™ II Fusion HS DNA polymerase for real-time determination of mitochondrial DNA amplification. In comparison to the earlier semi-quantitative protocols this represents a significant improvement in both the ease of data acquisition and the precision for quantification of mtDNA damage.
Methods
H9c2 myoblasts (ATCC, Manassas, VA) were used as the source of DNA for all experiments, and cells were grown in DMEM/10% FBS. Total DNA was isolated using DirectPCR lysis reagent (Viagen Biotech Inc., Los Angeles, CA). To induce mtDNA damage, the media was removed and the cells washed once with PBS just prior to UV exposure. Ultraviolet (UV) damage was induced by exposure to 254nm light using a Stratalinker model 1800 (Stratagene, La Jolla, CA) and the cells were harvested immediately thereafter.
Temperature annealing optimization was performed using an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany), and realtime DNA amplification was performed using a MX3000P (Stratagene, La Jolla, CA). Rat primer sequences used are listed in Table 1; the primer set for the short range PCR (SRPCR) was located within the 12S rRNA region. Temperature annealing optimization was performed for each primer set using the enzyme/buffer combination used for data collection; SRPCR was done using a Brilliant QPCR Master Mix, while LRPCR was done using PfuUltra™ II Fusion HS DNA Polymerase (Stratagene, La Jolla, CA). The final LRPCR cycling parameters followed manufacturer's recommendations: hot start of 2min@92°C followed by; 15sec@92°C, 30sec@50°C, 8:00min@68°C for 40 cycles. Crossing points were automatically generated by the software (version 1.2) and calculation of mtDNA damage and mitochondrial copy number were made using the Δ2Ct method. mtDNA lesion frequency was calculated as the amplification of damaged mtDNA relative to amplification of control mtDNA and used the Δ2Ct method to normalize the LRPCR:SRPCR for each group for the Poisson transformation as described by Ballinger et.al (Ballinger et al. 1999). DNA band images were captured and where applicable band density was quantified using AlphaEaseFC software (AlphaInnotech, San Leando CA).
Table 1.
Primer Sequences used for PCR amplification.
| Primers | Genbank Reference/Sequence | Fragment Length | Annealing Temperature |
|---|---|---|---|
| mtDNA | X14848 | ||
| LRPCR/f: | ATTTTCTCCCAGTTACGAAAG | 14958 bp | 50°C |
| LRPCR/r: | CTTGGTAAGTAAATTTCTTTCTCC | ||
| SRPCR/f: | ATGCACGATAGCTAAGACCCAA | 210 bp | 60°C |
| SRPCR/r: | GCTGAATTAGCGAGAAGGGGTA | ||
| D-Loop/f | CCCTACACCTGAAACTTCAATGCCA | 207 bp | 58°C |
| D-Loop/r: | GTGGAATTTTCTGAGGGTAGG | ||
| Cyto B/f: | CCGACGCAGACAAAATCCCA | 112 bp | 59°C |
| Cyto B/r: | TAGTAGGTCTGGGAAGAATAGTAC | ||
| 16S RNA/f: | AGGGACAGCTCTTTAGGAAACG | 235 bp | 63°C |
| 16S RNA/r: | CTTGTGCTTGGTGGATTGGTTC | ||
| COX1/f: | CACAGTAGGGGGCCTAACAG | 199 bp | 58°C |
| COX1/r: | CAAAGTGGGCTTTTGCTCAT | ||
| COX3/f: | TCAGGAGTCTCAATTACATG | 244 bp | 58°C |
| COX3/r: | CGTAGTAGACAGACAATTAGG | ||
| β-actin | V01217 | ||
| SRPCR/f: | GCGGTGACCATAGCCCTCTTT | 191 bp | 60°C |
| SRPCR:r: | TGCCACTCCCAAAGTAAAGGGTCA |
The structure of SYBR® Green (Invitrogen, Carlsbad CA) has been described by Zipper et. al. and they reported an extinction coefficient of 73,000 M−1cm−1, more recently, Mao et.al corrected this to be 58,000 M−1cm−1 and it is this latter value that was used to calculate SYBR® Green concentrations (Zipper et al. 2004; Mao et al. 2007). EvaGreen™ was obtained from Biotium (Biotium Hayward CA).
Results
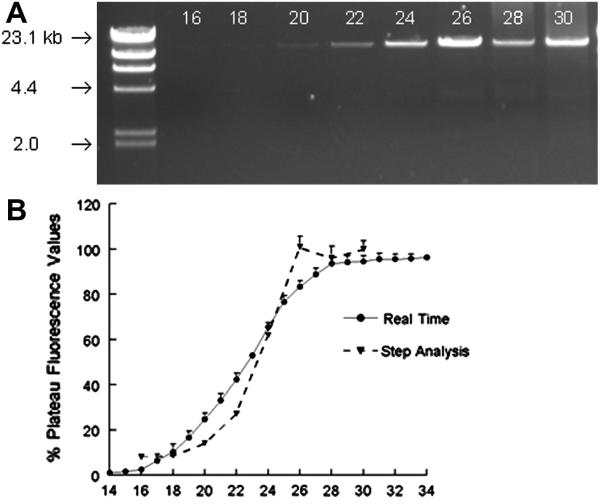
Both mitochondrial primer sets were initially optimized for annealing temperatures using the buffers and DNA polymerase utilized in data collection. As shown in Figure 1A, annealing of LRPCR/f and LRPCR/r primers was decreased above 52°C. The mitochondrial SRPCR primer set amplified a portion of the 12S rRNA region and annealing temperatures as high as 68°C could be used (Figure 1B). A comparison of methods was made between detection of amplification using the realtime protocol and a conventional protocol where samples were removed at selected time points and band density quantified. Similar to what others have observed, amplification/detection was exponential only over a few of the cycles (cycles 18:24) and above that a plateau was evident. Although detection of amplification was similar, real-time detection of amplification appeared slightly more sensitive. It should also be noted the real-time approach used 2 μl (n=4) of DNA polymerase compared to 12 μl of DNA polymerase (n=3 for each step) for the conventional approach (Figure 2).
Figure 1.
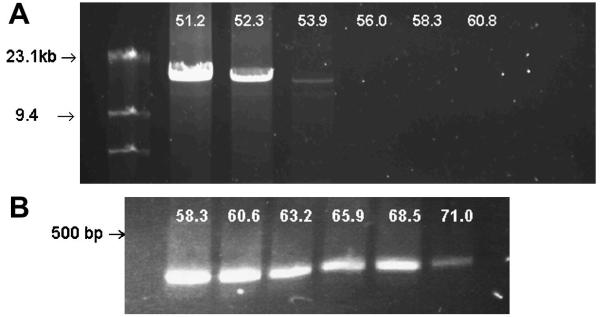
Temperature gradient amplification of mtDNA. A. Long Range amplification of mtDNA. mtDNA was annealed at the temperatures indicated above each band and amplification carried out as described in Methods. B. Short range PCR (SRPCR) of mtDNA from the cytochrome oxidase subunit 3 region. The annealing temperature used for DNA amplification is shown above each band.
Figure 2.
Comparison of realtime LRPCR and step cycle amplification of mtDNA. A. Representative picture of step LRPCR; 100 ng of total DNA was amplified using the LRPCR protocol and the reaction tube was removed at the cycle number indicated above each band. 10% of the LRPCR product was run on a 1% agarose gel. B. Realtime determination of DNA fluorescence (n=4) was determined as described in Methods. For the step cycle, the integrated density value (IDV) of the bands (n=3 for each step) was determined using AlphaInnotech/AlphaEaseFC software. All individual reactions contained 1:1250 dilution of EvaGreen. Values represented are mean±SEM of fluorescence normalized to the plateau values.
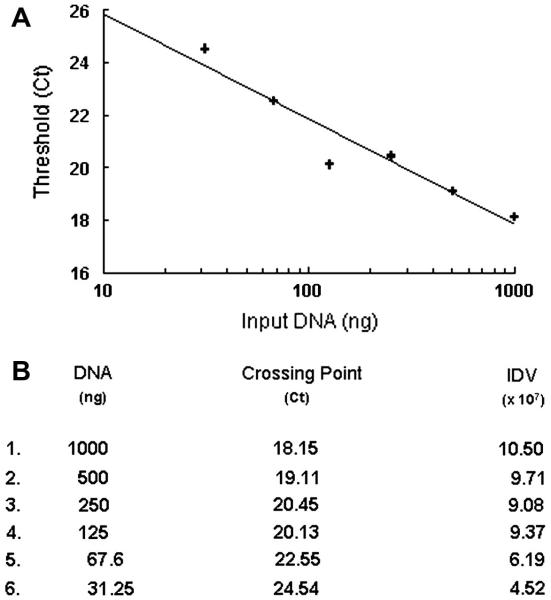
The processivity of PfuUltra™ II Fusion HS DNA Polymerase is greater than Pfu or EXL DNA polymerases (Stratagene 2006). The recommended extension times are 15-30 second per kb of amplification. This permitted reactions to be completed in less than six hours compared to other enzymes that required overnight incubations. SYBR green inhibition of DNA amplification has been know for sometime, and our initial attempts to directly amplify long DNA fragments in the presence of SYBR (1:10000 to 1:40000 dilution) were unsuccessful (Karsai et al. 2002; Arezi et al. 2003). However decreasing the concentration to 156 nM (1:56250 dilution), some amplification of the 15kb fragment was observed (Figure 3B). Further decreasing the SYBR green concentration enhanced DNA yields. The 39-78 nM range appeared to be the best compromise between signal generation and DNA yield as indicated by producing the lowest Ct (Figure 3A). The impact of input DNA over the range of 31 to 1000 ng was examined and it was observed that similar to amplification of short PCR products (data not shown), the amplification of long mtDNA products was logarithmic over the range of input DNA used, the standard curve is shown in Figure 4.
Figure 3.
LRCPR of mtDNA using increasing concentrations of SYBR Green. A. Crossing point analysis of the LRPCR using increasing concentrations of SYBR Green. B. 10% of the LRPCR product after 40 cycles of amplification was electrophoresed on a 1% agarose gel. The integrated density value (IDV) of the bands was determined using AlphaInnotech/AlphaEaseFC software following 40 cycles of amplification. NTC: non-template control
Figure 4.
LRCPR of mtDNA using increasing input total DNA. A. The standard curve was derived from the crossing point analysis using MX3000P software. The log fit values: Y= −3.975*log(X) + 29.98, R2= 0.92 and calculated efficiency was 78%. B. Crossing point analysis of the LRPCR using increasing amounts of total DNA. The integrated density value (IDV) of the bands was determined using AlphaInnotech/AlphaEaseFC software following 40 cycles of amplification.
Although numerous DNA binding dyes are available, more recently EvaGreen™ has become available from Biotium (Biotium Hayward CA). Several different applications have already been described for EvaGreen™ including DNA quantification, methylation-sensitive melting curve analysis, and QPCR (Ihrig et al. 2006; Mao et al. 2007; White et al. 2007). In comparison to SYBR Green, it is reported to be less inhibitory of DNA amplification and have a similar emission spectra that permits quantification using the FAM channel available on many real-time thermo-cyclers (Mao et al. 2007). Although the structure and extinction coefficient have not been reported, the recommended dilution of the commercially available solution is 1:20 for typical QPCR use. Using that concentration no amplification of the 15kb mitochondrial DNA fragment was observed (data not shown). However, when the EvaGreen concentration was decreased to final dilutions of 1:625 to 1:2500, comparable amplification of the 15kb DNA fragment was observed (Figure 5).
Figure 5.

LRCPR of mtDNA using increasing concentrations of EvaGreen. A. Crossing point analysis of the LRPCR using increasing concentrations of EvaGreen. 100ng of total DNA was used as described in Methods. B. 10% of the LRPCR product after 40 cycles of amplification was electrophoresed on a 1% agarose gel. The integrated density value (IDV) of the bands was determined using AlphaInnotech/AlphaEaseFC software following 40 cycles of amplification.
Several lines of evidence indicate that mitochondrial DNA is more susceptible to damage compared to genomic DNA (Richter et al. 1988; Yakes et al. 1997). In the present study we have used UV exposure as an example of a known agent that will cause DNA damage (Govan et al. 1990; Kalinowski et al. 1992; Yakes et al. 1997). Using the real-time detection of LRPCR we examined the impact of UV light on mtDNA. Exposure to low doses of UV light induced a significant amount of damage to the mtDNA (Figure 6A) and mtDNA lesion frequency was increased relative to controls (Figure 6C). In comparison, beta-actin as a marker of genomic DNA was used to calculate the ratio of mtDNA to genomic DNA. UV light did not significantly alter this ratio (Figure 6B) indicating that no significant changes in mitochondrial copy number occurred. Using a similar does of 10 J/m2, Kalinowski et.al observed an approximately 20% decrease in amplification which was less than the 73% observed in the present study. These differences may reflect that their mitochondrial DNA target was only 2.3 kb in length, compared to the 15 kb fragment in the present study. There a linear response to a doubling of the UV dose. Other studies have reported greater decreases in amplification in response to the lower range of dosages of UV or H2O2 used (Govan et al. 1990; Kalinowski et al. 1992; Yakes et al. 1997).
Figure 6.
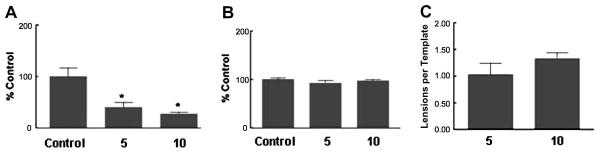
UV light induces mtDNA damage. H9c2 myoblasts were exposed to 5 or 10 J/m2 ultraviolet light. Total DNA was isolated for LRPCR and SRPCR analysis using mitochondrial and genomic probes as indicated in Methods. A. mtDNA damage calculated using the Δ2Ct method; damage expressed as a % of control. B. Mitochondrial copy number was the ratio of mtDNA: genomic DNA calculated using the Δ2Ct as described in Methods, expression expressed as a percentage of control. C. lesion frequency was calculated as described in Methods. Values are mean±SEM, * p<.05 compared to respective control.
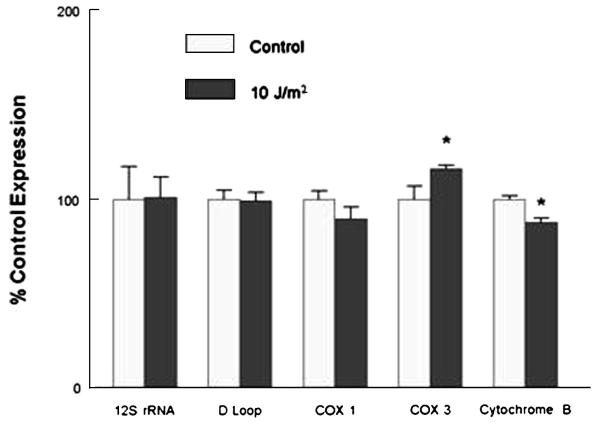
An underlying assumption is that no deletions occur within the SRPCR mtDNA region used for quantifying copy number. Using different primer sets that included the 12S rRNA, cytochrome oxidase subunit 1 region, and the D-Loop region no significant differences in copy number between control and 10 J/m2 group were observed, while the cytochrome oxidase subunit 3 region was significantly increased by 16% and the cytochrome B region decreased by 12% Figure 8. The nearest neighbor to the cytochrome B was the D-Loop, which was not altered by the UV exposure and it is unclear why these two regions may have been influenced. These findings suggest that care needs to be taken when designating the SRPCR region for any study examining mtDNA damage in response to an experimental treatment.
Summary
These protocols improve on the quality and precision in the quantification of mtDNA damage. In addition, the approach described is simpler than previously reported protocols. In comparison to 8-oxoG protocols that measure oxidative damage in the nucleus and mitochondria, the LRPCR protocols can selectively examine damage of mitochondrial DNA. Use of recently developed DNA Polymerases and protocols permits the rapid amplification of very long mtDNA fragments. Adaptation of real-time determination protocols allows for direct detection of DNA amplification of the long PCR products and its application here permits a fairly simple and reproducible determination of mitochondrial DNA damage.
Figure 7.
Regional copy number test. H9c2 DNA (control and 10 J/m2 UV light) were amplified and copy number normalized using β-actin. Three of the regions found no differences, while the cytochrome oxidase subunit 3 region was significantly increased by 16% and the cytochrome B region decreased by 12%. Values are mean±SEM expression expressed as a percentage of control; * p<.05 compared to respective control.
Acknowledgement
This work was supported in part by the PO1HL43023 and Castle-Krob Award of the New York Medical College Endowment Fund. There are no competing interests to report.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arezi B, Xing W, et al. Amplification efficiency of thermostable DNA polymerases. Anal Biochem. 2003;321(2):226–35. doi: 10.1016/s0003-2697(03)00465-2. [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Bouder TG, et al. Mitochondrial genome damage associated with cigarette smoking. Cancer Res. 1996;56(24):5692–7. [PubMed] [Google Scholar]
- Ballinger SW, Patterson C, et al. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86(9):960–6. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- Ballinger SW, Van Houten B, et al. Hydrogen peroxide causes significant mitochondrial DNA damage in human RPE cells. Exp Eye Res. 1999;68(6):765–72. doi: 10.1006/exer.1998.0661. [DOI] [PubMed] [Google Scholar]
- Domenech E, Gomez-Zaera M, et al. Wolfram/DIDMOAD syndrome, a heterogenic and molecularly complex neurodegenerative disease. Pediatr Endocrinol Rev. 2006;3(3):249–57. [PubMed] [Google Scholar]
- Gokey NG, Cao Z, et al. Molecular analyses of mtDNA deletion mutations in microdissected skeletal muscle fibers from aged rhesus monkeys. Aging Cell. 2004;3(5):319–26. doi: 10.1111/j.1474-9728.2004.00122.x. [DOI] [PubMed] [Google Scholar]
- H. L. Govan, 3rd, Valles-Ayoub Y, et al. Fine-mapping of DNA damage and repair in specific genomic segments. Nucleic Acids Res. 1990;18(13):3823–30. doi: 10.1093/nar/18.13.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudnason H, Dufva M, et al. Comparison of multiple DNA dyes for real-time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35(19):e127. doi: 10.1093/nar/gkm671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihrig J, Lill R, et al. Application of the DNA-specific dye EvaGreen for the routine quantification of DNA in microplates. Anal Biochem. 2006;359(2):265–7. doi: 10.1016/j.ab.2006.07.043. [DOI] [PubMed] [Google Scholar]
- Jennerwein MM, Eastman A. A polymerase chain reaction-based method to detect cisplatin adducts in specific genes. Nucleic Acids Res. 1991;19(22):6209–14. doi: 10.1093/nar/19.22.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J, Srivastava S. Mitochondria as a target for early detection and diagnosis of cancer. Crit Rev Clin Lab Sci. 2005;42(5-6):453–72. doi: 10.1080/10408360500295477. [DOI] [PubMed] [Google Scholar]
- Kalinowski DP, Illenye S, et al. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain reaction assay. Nucleic Acids Res. 1992;20(13):3485–94. doi: 10.1093/nar/20.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai A, Muller S, et al. Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques. 2002;32(4):790–2. doi: 10.2144/02324st05. 794-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon SA, Snowden MM. Differential repair of DNA damage in the human metallothionein gene family. Mol Cell Biol. 1988;8(12):5331–8. doi: 10.1128/mcb.8.12.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao F, Leung WY, et al. Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 2007;7(1):76. doi: 10.1186/1472-6750-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon I, Bohr VA, et al. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci U S A. 1986;83(23):8878–82. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshita F, Eastman A. Gene-specific damage produced by cisplatin, ormaplatin and UV light in human cells as assayed by the polymerase chain reaction. Oncol Res. 1993;5(3):111–8. [PubMed] [Google Scholar]
- Richter C, Park JW, et al. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85(17):6465–7. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos JH, Meyer JN, et al. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol Biol. 2006;314:183–99. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- Sciacco M, Prelle A, et al. A case of CPT deficiency, homoplasmic mtDNA mutation and ragged red fibers at muscle biopsy. J Neurol Sci. 2005;239(1):21–4. doi: 10.1016/j.jns.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Stratagene High-Fidelity PCR Enzymes: Properties and Error Rate Determinations. Technical Notes(TN5-03/06) 2006:1–12. [Google Scholar]
- Thomas DC, Morton AG, et al. General method for quantifying base adducts in specific mammalian genes. Proc Natl Acad Sci U S A. 1988;85(11):3723–7. doi: 10.1073/pnas.85.11.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HE, Hall VJ, et al. Methylation-sensitive high-resolution melting-curve analysis of the SNRPN gene as a diagnostic screen for Prader-Willi and Angelman syndromes. Clin Chem. 2007;53(11):1960–2. doi: 10.1373/clinchem.2007.093351. [DOI] [PubMed] [Google Scholar]
- Wong LJ. Comprehensive molecular diagnosis of mitochondrial disorders: qualitative and quantitative approach. Ann N Y Acad Sci. 2004;1011:246–58. doi: 10.1196/annals.1293.024. [DOI] [PubMed] [Google Scholar]
- Wong LJ, Liang MH, et al. Comprehensive scanning of the entire mitochondrial genome for mutations. Clin Chem. 2002;48(11):1901–12. [PubMed] [Google Scholar]
- Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94(2):514–9. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipper H, Brunner H, et al. Investigations on DNA intercalation and surface binding by SYBR Green I, its structure determination and methodological implications. Nucleic Acids Res. 2004;32(12):e103. doi: 10.1093/nar/gnh101. [DOI] [PMC free article] [PubMed] [Google Scholar]