Highlight
This study provides the first characterization of an R2R3 family MYB transcription factor involved in cuticle and epicuticular wax deposition, whose action is confined to maize embryogenesis and juvenile phase.
Key words: Cuticle, embryogenesis, epicuticular waxes, fused leaves1, shoot development, ZmMYB94.
Abstract
The fdl1-1 mutation, caused by an Enhancer/Suppressor mutator (En/Spm) element insertion located in the third exon of the gene, identifies a novel gene encoding ZmMYB94, a transcription factor of the R2R3-MYB subfamily. The fdl1 gene was isolated through co-segregation analysis, whereas proof of gene identity was obtained using an RNAi strategy that conferred less severe, but clearly recognizable specific mutant traits on seedlings.
Fdl1 is involved in the regulation of cuticle deposition in young seedlings as well as in the establishment of a regular pattern of epicuticular wax deposition on the epidermis of young leaves. Lack of Fdl1 action also correlates with developmental defects, such as delayed germination and seedling growth, abnormal coleoptile opening and presence of curly leaves showing areas of fusion between the coleoptile and the first leaf or between the first and the second leaf. The expression profile of ZmMYB94 mRNA—determined by quantitative RT-PCR—overlaps the pattern of mutant phenotypic expression and is confined to a narrow developmental window. High expression was observed in the embryo, in the seedling coleoptile and in the first two leaves, whereas RNA level, as well as phenotypic defects, decreases at the third leaf stage. Interestingly several of the Arabidopsis MYB genes most closely related to ZmMYB94 are also involved in the activation of cuticular wax biosynthesis, suggesting deep conservation of regulatory processes related to cuticular wax deposition between monocots and dicots.
Introduction
Seedling architecture is established during maize embryogenesis, leading to the formation of a well developed embryo axis consisting of the embryonic primary root and the embryonic shoot separated by the scutellar node and encapsulated by two protective structures, the coleoriza for the root and the coleoptile for the shoot. The shoot includes the first internode, called the mesocotyl, located between the scutellar node and the coleoptilar node, and—depending on the genetic background—five or six short internodes located above the coleoptilar node, with a leaf primordium at each node. Each leaf primordium encircles those below, thus forming a conical structure that is encapsulated in the coleoptile, which develops as a sheathing structure enveloping the stem tip and the embryonic leaves (Abbe and Stein, 1954). Coleoptile and leaf primordia, which are initiated from 8 days after pollination (DAP) onwards (Clark, 1996), derive from distinct origins, the former arising as a ring on the surface of the scutellum and the latter from the shoot apical meristem (SAM) (Abbe and Stein, 1954; Bommert and Werr, 2001). The coleoptile is the first organ to grow when seed germination occurs. It appears as a conical structure that elongates and pierces the soil, enclosing and thus protecting the young leaves in the shoot apex. In this initial phase leaf elongation keeps pace with that of the coleoptile. Later on, the coleoptile arrests its growth and opens at its apex, wherefrom leaves emerge. The coleoptile dies soon after leaf emergence.
This study describes the maize fused leaves1-1 (fdl1-1) mutant, which exhibits abnormalities during embryo development and the first phases of seedling growth, between coleoptile emergence and disclosure of the third leaf. The main features of mutant plants are irregular coleoptile opening and the presence of regions of adhesion between the coleoptile and the first leaf and between the first and second leaves. In the adherent regions the cuticle is not deposited. In addition epicuticular wax deposition on the epidermis of young leaves is irregular.
MYB (myeloblastosis) family proteins possess a conserved DNA-binding domain (DBD), which is homologous to the DBD of animal c-Myb (Klempnauer et al., 1982) and is composed of up to three imperfect repeats (named sequentially as R1, R2 and R3, respectively) (Dubos et al., 2010). The R2R3 subclass of MYB factors contains two repeats and is the most common type in plants (Du et al., 2013). 157 genes encoding R2R3-MYB proteins have been identified in the maize genome and classified into 37 subgroups, according to their structure and phylogenetic relationships (Du et al., 2013). However, functional studies have been performed for only a few maize myb genes that are involved in the control of the phenylpropanoid metabolic pathway (Paz-Ares et al., 1987; Marocco et al., 1989; Grotewold et al., 1991; Fornalé et al., 2006; Heine et al., 2007).
Here we show that the fdl1-1 mutant phenotype is attributable to the insertion of an Enhancer/Suppressor mutator (En/Spm) element in ZmMYB94, an R2R3 family MYB transcription factor that appears to be part of a monocot-specific subfamily expansion. Its functional analyses indicate that during maize embryo and seedling development, cuticle deposition is required to establish correct organ morphogenesis and the formation of boundaries between organs, while phylogenetic reconstructions suggest conservation of roles for MYB94-like proteins in the regulation of wax deposition between monocots and dicots.
Materials and methods
Mutant isolation and propagation
The fdl1-1 mutation, previously designated as fdl347, was first identified by Professor Jane A. Langdale (Department of Plant Science, University of Oxford, Oxford, UK) in the selfed progeny of a maize (Zea mays) stock crossed as female to an En/Spm line. The F2 progeny segregated for normal and defective seedlings in a 3:1 ratio, showing that fdl1-1 behaves as a monogenic recessive mutation. The fdl1-1 allele was maintained in homozygosis as well as in heterozygosis. Heterozygous plants were obtained by outcrossing plants homozygous for the mutation to B73 and H99 maize inbred lines.
Wild-type and mutant maize seeds were germinated in a growth chamber at 25°C on wet filter paper. Seeds were kept in the dark for 4–5 d and then transferred to a 14h light photoperiod and photon fluence of 70 µmol m-2 s-1.
For embryo-rescue experiments, 12 and 18 DAP immature embryos were excised and cultivated in vitro, as described by Dolfini et al., (1999).
fdl1-1 mapping
Crosses of heterozygous +/fdl1-1 females to stocks carrying the TB-A translocation (Maize Genetics Cooperation Stock Center) were used to position fdl1 on the long arm of chromosome 7. Individuals bearing a B-A translocation produce hypoploid sperms, lacking an extended portion of a specific chromosome arm, which can be used to fertilize a female donor plant carrying the mutation under study. The recovery of hemizygous mutant embryos allows localization of a recessive gene to the chromosome arm involved (Beckett, 1978).
fdl1-1 was then mapped in segregating F2 populations using simple sequence repeat (SSR) markers. F1 seedlings were obtained from the cross between the homozygous fdl1-1 stock and the H99 inbred line. F1 plants were then self-pollinated to produce F2 progenies. The genomic DNA was extracted from parental and F1 plants, and from individual mutant and wild-type F2 seedlings, and was amplified with primers for SSR markers on chromosome 7L. DNA extraction, reaction conditions and gel electrophoresis were carried out as described in the SSR protocols available from the Maize Genetics and Genomic Database (http://www.maizegdb.org). Mapping data were analysed with the mapmaker/EXP3.0 software (Lander et al., 1987).
Light and electron microscopy
For transmission electron microscopy and light microscopy, immature 18 DAP embryos, germinating seedlings and young leaves from wild type and fdl1-1 mutants were fixed overnight at 4°C in 6% glutaraldehyde in 0.1M cacodylate buffer (pH 6.9) and post-fixed for 2h in 1% osmium tetroxide in the same buffer. The fixed samples were dehydrated in a series of ethyl alcohol and propylene oxide solutions and embedded in araldite.
For transmission electron microscopy, ultrathin sections (600nm thick) were cut with an ultramicrotome (Ultracut, Reichert-Jung, Leica Wetzlar, Germany), stained with uranyl acetate and lead citrate and examined with an electron microscope (TEM 300, Hitachi, Japan) operating at 75kV.
For light microscopy, thin sections (1 µm thick), cut with the same ultramicrotome, were stained with toluidine blue [1% toluidine blue 1% sodium tetraborate (1:1, v/v)] and examined with a light microscope (Ortholux, Leitz, Germany).
The fluorescence emission of unstained thin sections was tested using an epifluorescence microscope (AXIOSCOP, Zeiss, Germany) fitted with an excitation filter (BP 365/12), a chromatic beam splitter (FT 395) and a barrier filter (LP 397). Cell walls excited with UV light can exhibit blue autofluorescence due to the presence of phenolic components (Harris and Hartley, 1976).
For scanning electron microscopy, immature 18 DAP embryos and seedlings were fixed and post-fixed with the same procedure used for transmission electron microscopy and dehydrated in a graded ethyl alcohol series. They were then dried at critical point, coated with gold and palladium and examined with a scanning electron microscope (Stereoscan 260, Cambridge, UK) operating at 25kV.
For the analysis of epicuticular waxes, leaf pieces of wild-type, fdl1-1 mutant and RNAi transformed plants, were dried and processed according to La Rocca et al., (2014). The specimen surfaces were examined with the same scanning electron microscope.
Toluidine blue test
The toluidine blue test for detecting defects in the leaf cuticle (Tanaka et al., 2004) was carried out on fdl1-1 mutant and wild-type seedlings from F2 segregating progenies. All the samples were submerged in an aqueous solution of 0.05% (w/v) toluidine blue (Sigma). After 3min the solution was removed and the samples were washed gently with distilled water to remove excess dye. Images were taken by using a digital camera.
Co-segregation analysis and SSR mapping
For Southern analysis, maize genomic DNA was extracted from 7-day-old seedlings and leaf tissues using the urea extraction method (Chen and Dellaporta, 1994). Then 10 μg of DNA were digested with the BamH1 endonuclease, which cuts once inside the En/Spm element, and separated on 0.8% agarose gels. DNA fragments were transferred to Hybond N+ membranes (Amersham), and membranes were subsequently probed with α-32P-labelled probes, as described in Gutierrez-Marcos et al., (2007). Hybridization probes were as follows. The En/Spm-specific probe (pBX1), corresponding to a 457bp BanII-XbaI fragment internal to the En/Spm element, was isolated by restriction of the plasmid containing the cloned element (Pereira et al., 1986) followed by gel purification. Probe1, whose length is 78bp, was amplified from the genomic clone containing the 2.6kb fragment co-segregating with the mutant phenotype, by using two primers (5′-ACTAGTGGATCCTCGGACG-3′; 5′-TGTAGTGCCCACCACAAGC-3′), corresponding to the end of the cloning vector and of the En/Spm element respectively.
For PCR analysis, 50ng of genomic DNA was subjected to 35 cycles of amplification with 1.25U of GoTaq® Flexi DNA Polymerase (Promega), 1× Colourless GoTaq®Flexi Buffer, 2.5mM MgCl2, 0.2mM dNTPs, 0.2 µM primers and 1M betaine. Annealing temperatures were 58–62°C (optimized for each primer pair, as listed in Supplementary Table S1) with an extension time of 60 s.
Primers used for co-segregation analysis included two primer sets specific for the mutant allele, which comprised a En/Spm primer and a gene specific primer (Consfdl2-F, Spm1-R; and Spm3-F, AW-R), and a primer set specific for the wild-type allele (ConsFdl2, AWR), spanning the region containing the En/Spm insertion (Supplementary Fig. S3A). PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Simple sequence repeat (SSR) markers were recovered from the MaizeGDB database (www.maizegdb.org/probe.php).
Genomic cloning and cDNA synthesis
The Spm-BamH1 2.6kb polymorphic restriction fragment co-segregating with the mutant phenotype was cloned from a subgenomic library prepared from size-fractionated BamH1 fragments of fdl1-1/fdl1-1 genomic DNA in the pBluescript SK vector, according to the manufacturer’s instructions (Stratagene). Four independent clones were selected and the presence of the En/Spm element and DNA flanking the insertion was determined by sequencing.
For cDNA synthesis, total RNA was extracted with 1ml TRI reagent (Molecular Research Center), according to the supplier’s instructions and, after treating RNA samples with DNase I, (Invitrogen) for 30min at 25°C, cDNA was synthesized using SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), starting from 5 µg of total RNA primed with oligo(dT).
For wild-type and mutant gene structure analysis, PCR were conducted on both genomic DNA and cDNA using gene specific primers (GSP) and En/Spm specific primers. Primer sequences, listed in Supplementary Table S1, were designed using the Primer3 software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
PCR products were electrophoresed on agarose gels and purified with the Illustra GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare), according to the instructions of the manufacturer, and sequenced in both orientations at the CRIBI Biotechnology Center (BMR Genomics, University of Padova, Padova, Italy, http://www.bmr-genomics.it).
RNAi
A 481bp ZmMYB94 fragment was amplified from genomic DNA of genotype B73 with primers attB1-FDL-RNAi and attB2-FDL-RNAi (Supplementary Table S1) and recombined in inverted orientation and under the control of the constitutive rice Actin promoter into a derivative of the integrative plasmid pSB11 (Ishida et al., 1996), which contained a Basta resistance cassette and a GFP cassette, to yield plasmid L1258. Transformation of genotype A188 was performed as described previously (Pouvreau et al., 2011). To evaluate the molecular efficiency of the RNAi construct, ZmMYB94 expression was assayed in the third leaf of primary transformants by quantitative RT-PCR in technical duplicate.
Reverse transcription and quantitative reverse transcription PCR analysis
Tissue samples were from plants of the B73 inbred line and are listed in Supplementary Table S2. For each sample, total RNA was obtained from two biological replicates to assess the repeatability of the data.
cDNAs were diluted 100-fold, and 2 μl were used in a final volume of 20 μl containing 10 μl of iQ SYBR Green Supermix and 0.25 μM of each primer. Amplifications were carried out using an iCycler thermocycler equipped with the MyiQ detection system (Bio-Rad, Milano, Italy) in 96-well optical reaction plates sealed with optical tapes (Bio-Rad). The reaction conditions were as follows: 96°C for 30 s, 40 cycles 96°C for 30 s, 63°C for 30 s, 72°C for 30 s. All primers used are listed in Supplementary Table S1. Data are presented as the mean of two biological replicates and three technical replicates, with the exception of bracts, ear, anthers and silks, which are presented as the mean of three technical replicates.
Each reaction was performed in triplicate. No-template controls and not retro-transcribed samples were included in the experimental set. The absence of aspecifically-amplified products was checked with derivative melting curves, obtained by progressive heating at 0.3°C every 15 s. Raw data were collected and analysed with the iQ5 software (Bio-Rad, Milano, Italy) with the following parameters: baseline from the 2nd to the 10th cycle and threshold calculated automatically by the software for every reaction. Differences in gene expression were calculated by the comparative delta–delta CT method (Schmittgen and Livak, 2008) with a dedicated Microsoft Excel macro created by Bio-Rad. All quantifications were normalized to the housekeeping gene 18S.
Phylogenetic analyses
All annotated R2R3 Myb domain-containing protein sequences were recovered by sequence similarity searches from the Arabidopsis thaliana, Vitis vinifera, Brachypodium distachyon, Oryza sativa (all from www.phytozome.net), and Zea mays B73 (www.maizegdb.org/) genome sequences. Protein sequences were aligned using Muscle (Edgar, 2004) and unambiguously aligned regions (102 amino acid sites) were selected with GBlocks (Castresana, 2000). The ProtTest software (Darriba, et al., 2011) selected the JTT substitution model (Jones et al., 1992) with four gamma distributed site rate categories as the best fit to the data. Initial distance analyses were performed using the programs Seqboot, Protdist, Neighbor and Consense from the PHYLIP package (Felsenstein, 1989). Subsequently, only sequences from the well-supported clade containing the ZmMYB94 gene were realigned and 166 unambiguously alignable sites were selected for maximum likelihood bootstrap analysis using PHYML (Guindon et al., 2010).
Results
The fdl1-1 mutation causes abnormal seedling development
The fused leaves1-1 (fdl1-1) recessive mutation, originally isolated in the F2 generation of a cross between a En/Spm line and a maize inbred line, was maintained by outcrossing fdl1-1 homozygous plants to different inbred lines (A188, W64A, H99 and B73) for two or three generations. In all genetic backgrounds F2 progeny carrying the fdl1-1 mutation segregated 3:1 for seedlings with abnormal morphology, which could be macroscopically distinguished from wild-type seedlings (Fig. 1).
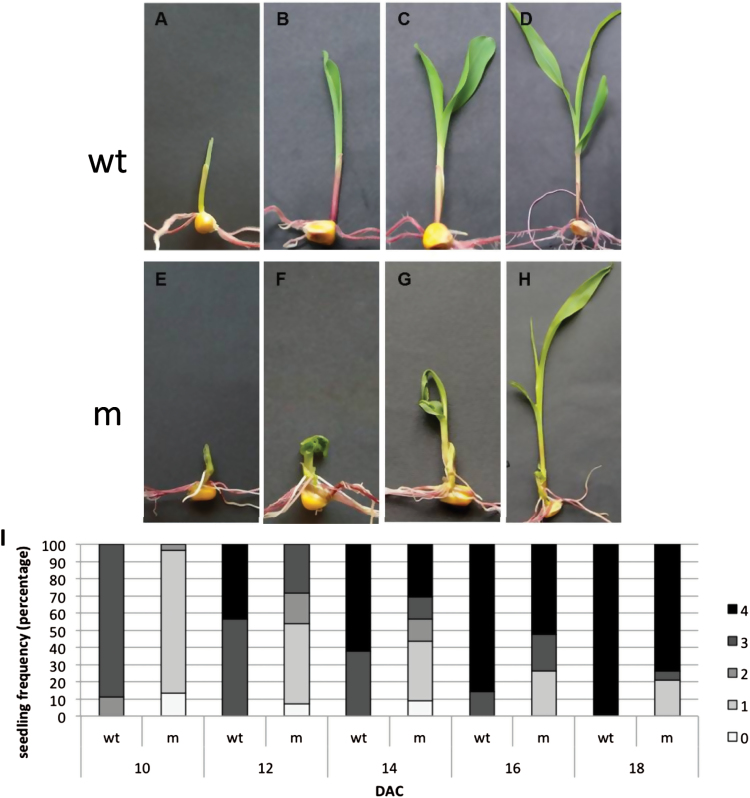
Fig. 1.
Seedling phenotypes and developmental profile of fdl1-1. Representative wild-type (A–D) and mutant (E–H) seedlings were photographed at successive stages of development: coleoptile (A, E), first leaf (B, F), second leaf (C, G) and third leaf (D, H). The developmental profile of fdl1-1 mutant (m) and wild-type (wt) seedlings is presented as percentage of seedlings in each stage at a specific time after initiation of cultures (I). Between 30 and 40 seedlings were analysed for each time point. 0, coleoptile presence; 1, first leaf presence; 2, second leaf presence, 3, third leaf presence; 4, fourth leaf presence; DAC, days after initiation of cultures.
The fdl1-1 mutation specifically affected the early stages of seedling development, from germination to the 3–4 leaf stage, causing a general delay in germination and seedling growth as well as phenotypic abnormalities (Fig. 1E–G). An evident trait in mutant seedlings was the presence of a slightly thicker coleoptile (Fig. 1E). In contrast with wild-type leaves that elongated straight following their emergence from the coleoptile tip (Fig. 1A, B), mutant leaves emerged from the coleoptile in a warped manner, and appeared rolled and twisted (Fig. 1F, G). Moreover, regions of adhesion between the coleoptile and the first leaf, and between the first and the second leaf were observed and interfered with leaf expansion, causing bending of seedlings (Fig. 1G). After the third leaf stage, most of the mutant plants resumed normal phenotypic development and were indistinguishable from the wild-type plants (Fig. 1D, H).
The phenotypes of the fdl1-1 mutants were rather variable. The most severe phenotypes exhibited extremely stunted growth and heavily rolled (Supplementary Fig. S1A) or fractured (Supplementary Fig. S1B) leaves. In most of the mutant seedlings the first and second leaves appeared fused (Supplementary Fig. S1C) and showed remnants of the fusion at later stages (Supplementary Fig. S1D, E). To compare mutant and wild-type growth rate, seedlings belonging to the same F2 progeny were allowed to germinate on wet filter paper and, at succeeding time points, classified according to their stage of development into specific classes: presence of coleoptile (stage 0, Fig. 1A, E), first leaf (stage 1, Fig. 1 B, F), second leaf (stage 2, Fig. 1 C, G), third leaf (stage 3, Fig. 1D, H) and fourth leaf (stage 4). Ten days after culture initiation (DAC), the majority of wild-type plants had reached stage three, whereas the majority of the mutant seedlings were in stage one (Fig.1 I). Similarly, at later times (12 and 14 DAC), a substantial percentage of wild-type plants had already reached stage four, whereas mutant plants were at earlier stages. At 16 and 18 DAC, the discrepancy between wild-type and mutant was less consistent.
The strongest phenotypes, i.e. seedlings at stage one after 18 days of culture, representing 21.05 % of the mutant seedlings, arrested their growth, whereas the rest of the mutants, after the emergence of the third/fourth leaf, resumed almost normal growth, except for a reduction in their height. They could be grown to maturity and were fertile.
To better examine the effect of the mutation on seedling organization, mutant germinating shoots were analysed by scanning electron microscopy. Mutant coleoptiles appeared thicker and variably misshapen (Fig. 2B–F) compared to wild-type (Fig. 2A). Soon after emergence wild-type coleoptiles exhibited an aperture at the top from which leaves easily emerged (Fig. 2A); in contrast, mutant coleoptiles often showed a lateral subapical slit (Fig. 2B). Mutant leaves did not emerge from the tip but from the side of the coleoptile by forcing and breaking its tissues (Fig. 2C, D), apparently because the upper leaf region was joined with the coleoptile apex (Fig. 2E). Finally, leaf detachment occurred through apical tissue tearing. In some greatly deformed mutant seedlings, coleoptile and first leaf were not easily distinguishable (Fig. 2F).
Fig. 2.
Scanning electron microscope micrographs of wild-type (A) and fdl1-1 mutant (B–F) germinating seedlings. (A) The first leaf emerges from the aperture at the top of a wild-type coleoptile. (B) A mutant coleoptile exhibiting a long subapical cleft. (C) A mutant coleoptile fractured by forcing leaves. (D) Leaves slipping laterally from the broken coleoptile; the leaf apical region is inside the closed top of the coleoptile. (E) An emerged leaf still joined with the coleoptile top. (F) A thick and greatly malformed mutant coleoptile. co, coleoptile; lf, leaf. Bars, 2mm.
The fdl1-1 mutation affects embryonic shoot development and organ separation
To ascertain if the fdl1-1 phenotype was detectable prior to germination, both SEM and histological analysis were carried out on wild-type and homozygous mutant immature seeds from segregating F2 ears.
SEM analysis revealed that in immature 18 DAP wild-type seeds (Fig. 3A) the embryo shoot was enveloped by the scutellum, from which only the apical region of the coleoptile emerged. By contrast, in mutant seeds (Fig. 3B) the scutellum is less curved, leaving the embryo shoot uncovered. Moreover, the embryo shoot exhibited external malformations with incomplete surfaces and irregular openings of coleoptile and first leaf. At the same stage light microscope observations of thin transversal sections disclosed the presence of wide areas of coleoptile and first leaf fusion in the mutant embryo (Fig. 3E, F) that were absent in wild-type embryos (Fig. 3C, D). In the fused regions the two organs were not clearly distinguishable, since attribution of cells to the coleoptile or leaf was not feasible (Fig. 3F). In addition, the sections showed that, contrary to wild type, the mutant scutellum did not extend towards the adaxial face of the embryo shoot and that the coleoptile and first leaf had incomplete and opened surfaces.
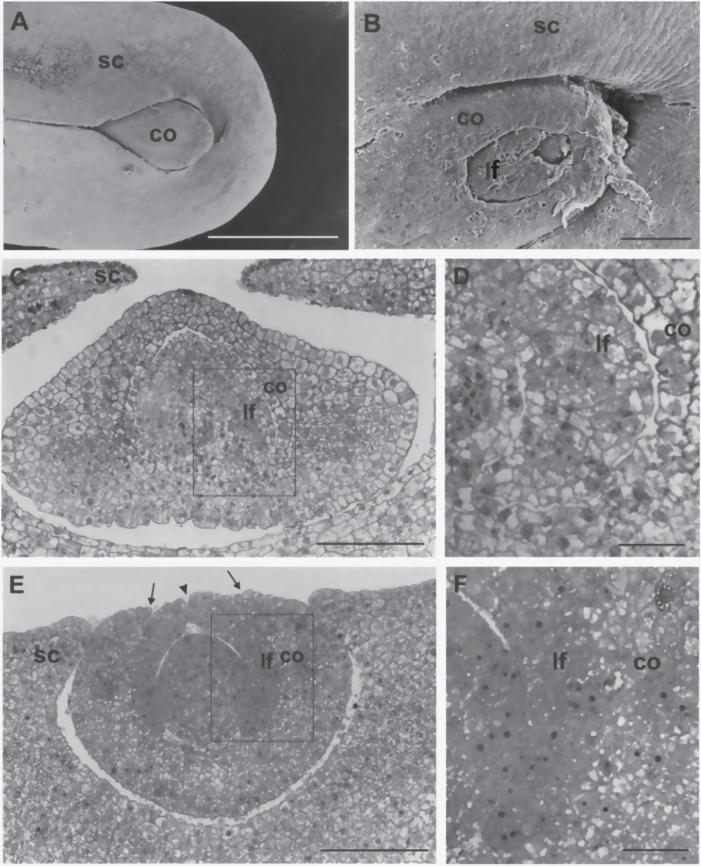
Fig. 3.
Scanning electron microscope (A, B) and light microscope (C–F) micrographs of the shoot region of 18 DAP embryos. (A) A wild-type embryo with the coleoptile enveloped by the scutellum. (B) A malformed fdl1-1 mutant embryo. Note the less curved scutellum, the uncovered shoot and the openings in coleoptile and first leaf surfaces. (C) The wild-type scutellum encloses the coleoptile, which contains the first leaf. (D) Higher magnification of the area framed in C shows the distinct coleoptile and leaf. (E) Laterally expanded scutellum, open coleoptile surface (arrows) and cleft leaf (arrowhead) are visible in the fdl1-1 mutant embryo shoot. (F) Higher magnification of the area framed in E. Note the lack of tissue identity between coleoptile and leaf in the fused region. co, coleoptile; lf, first leaf; sc, scutellum. Bars: 1mm (A); 100 μm (B, C, E); 25 μM (D, F).
Taken together, these observations indicate that the effect of the fdl1-1 mutation is exerted prior to germination. Accordingly, mutant seedlings cultivated through embryo rescue exhibited morphological defects. Seedlings showing all the typical fdl1-1 traits were detected, with the expected segregation ratio, in cultures obtained from immature embryos deriving from the same F2 ear (Supplementary Fig. S1F). Moreover, mutant seedling growth was reduced and delayed in comparison with that of wild-type siblings (Supplementary Fig. S1G). After 20 days of in vitro growth, most of the mutant seedlings (90%) from 12 DAP old embryos were at stage zero, whereas the majority of wild-type seedlings (80%) had already reached stage one. In cultures initiated from 18 DAP embryos the developmental profiles were less divergent. Among wild-type seedlings, stage two and three were the most represented (37.5% and 57.5% respectively); among mutant seedlings 55.2% reached stage three, while the growth of the remaining seedlings was at earlier stages.
Analysis of tissues identities and cuticle and epicuticular wax deposition in mutant seedlings
To better examine the effect of the mutation on the early phases of plant development, apical and basal cross-sections of wild-type (Fig. 4 A, B) and mutant (Fig. 4C, D) seedlings at the coleoptile stage were compared through light microscopy analysis. In the wild-type sections, leaves were enclosed and regularly rolled within the coleoptile and always appeared as distinct structures (Fig. 4A, B). In contrast, in the mutant apical sections, the inner surface of the coleoptile was fused with masses of a presumed first leaf tissue, lacking organization and exhibiting unclear cell identity (Fig. 4C). In the basal sections wide regions of fusion between coleoptile and first leaf epidermises and between epidermises of adjacent leaves were present (Fig. 4D).
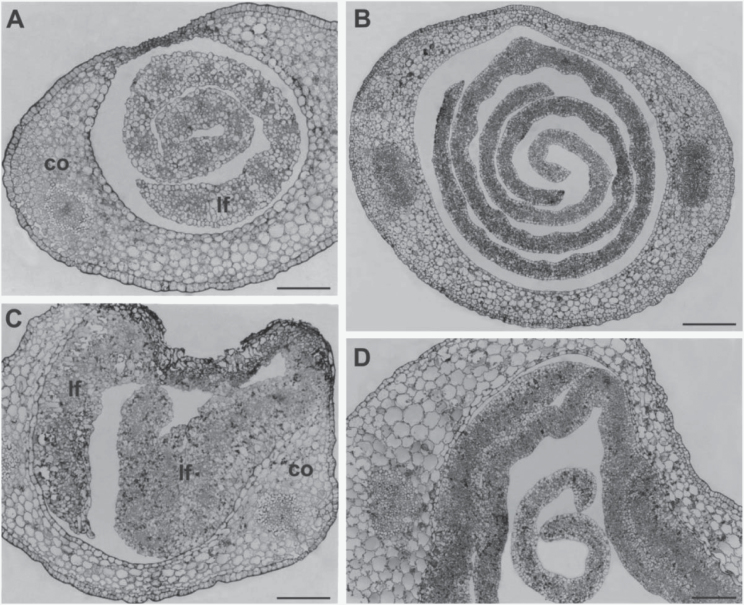
Fig. 4.
Light microscope micrographs of thin transversal sections of apical (A, C) and basal (B, D) regions of germinating seedling. (A) The apical region of a wild-type shoot shows a well-organized leaf in the hollow of the still closed coleoptile. (B) Young leaves independently enrolled are inside the coleoptile in the basal region of the wild-type shoot. (C) In the apical region of the mutant shoot masses of disorganized leaf tissues are fused with the coleoptile. (D) Fused areas between coleoptile and leaf surfaces are visible in the basal region of the mutant shoot. A young free leaf is also present. co, coleoptile, lf, leaf. Bars: 100 μm (A, C, D); 200 μm (B).
At higher magnification, areas of merged surfaces alternating with areas of free surfaces were visible (Fig. 5A). In areas involving coleoptile and first leaf fusion, epidermal cell shapes were frequently altered, whereas fused epidermal cells between two leaves assumed a more regular shape.
Fig. 5.
Light and fluorescence microscope micrographs of merged coleoptile and leaf blades in the fdl1-1 mutant. (A) At the light microscope some epidermal cell shapes are altered in areas involving coleoptile and first leaf fusion. Fused epidermal cells between two leaves look more regularly shaped. (B) At the fluorescence microscope severely misshaped cells can be seen in merged zone between undistinguishable leaf blade and edge of coleoptile lateral opening. (C) The tissue organization of fused blades from succeeding leaves does not show significant alterations. In the more differentiated leaf, xylem vessels are regularly distributed in the parallel bundles, the epidermises are recognizable and have stomata (arrow head) in the free regions. co, coleoptile; lf, leaf. Bars, 50 μm.
Fluorescence microscopy analysis of fused regions revealed that in the merged zone between leaf blade and edge of coleoptile lateral opening, cells of the two organs were indistinguishable and the boundary was no longer clearly recognizable (Fig. 5B). By contrast, the blades of adjacent leaves had recognizable epidermises with fused areas and free regions. The latter also showed well-differentiated stomata as evidenced by the intense autofluorescence of their cell walls (Fig 5C).
TEM analysis revealed that, within zones of fusion between two organs, mutant cells were joined by an apparent single cell wall (Fig. 6A), the cell wall proper of each of them being undistinguishable. While a thick, continuous cuticle layer could be seen on the cell walls of separated epidermises in adjacent zones (Fig. 6B, D, F) covering the free surfaces of the two organs even in the initial point of separation (Fig. 6D, E), cuticular material was not detectable between joined cell wall zones even at higher magnification (Fig. 6C). Moreover, the cuticle was always detectable along free epidermises (Fig. 6F) and fusion was never found between cutinized surfaces of leaves or other plant organs.
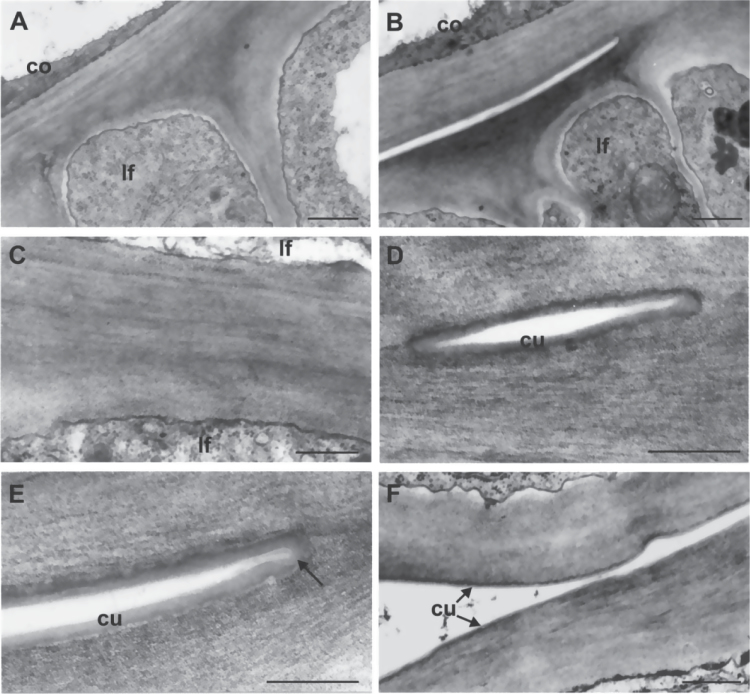
Fig. 6.
Transmission electron microscope micrographs of epidermal cell walls of coleoptile and first two leaves from the fdl1-1 mutant seedlings. (A) The cell wall proper of coleoptile and first leaf cannot be distinguished in a fused zone. (B) Distinct cell walls are visible in the free area adjacent to the fused one. (C) The higher magnification reveals the absence of any cuticular materials between the fused cell walls of two leaf surfaces. (D) A cuticle layer can be seen on the cell walls of the small free zone. (E) An evident continuous cuticle covers the free surfaces of the epidermal cells, also bordering the point of fusion (arrow). (F) The cuticle overlays the surfaces of two unfused leaf epidermises. co, coleoptile; cu, cuticle; lf, leaf. Bars: 1 μm (A, B); 0.5 μm (C, F); 0.25 μm (D, E).
Toluidine blue stained roots and seeds (which are devoid of cuticular material), whereas it did not stain aerial parts of wild-type or mutant seedlings, with the exception of mutant coleoptile fractured edges (Supplementary Fig. S2) since this dye permeates epidermal surfaces only if the cuticle is absent or defective. According to this assay (Tanaka et al., 2004), the cuticle covering the free surfaces of mutant leaves displayed a waterproofing property.
It was also observed that, if mutant seedlings are misted with water, they retain water beads on their surface (Fig. 7D). SEM analysis suggested that this trait could be attributable to the alterations in epicuticular wax deposition, which was less homogenous on the mutant (Fig. 7E), than wild-type (Fig. 7B) surfaces. Indeed, mutant leaf surfaces exhibited a patchy distribution of epicuticular waxes, with some areas evenly covered and others completely devoid (Fig. 7E). This trait, similarly to the other fdl1-1 traits, was confined to the first two leaves, since later leaves show a regular distribution of epicuticular waxes that was indistinguishable from that of wild-type leaves (data not shown).
Fig. 7.
Second leaf of wild-type (A) and mutant (D) misted with water and micrographs of epicuticular waxes at scanning electron microscopy (SEM). The epicuticular waxes are regularly distributed on the upper surface of the second leaf of wild-type (B) while they show a patchy deposition on the same leaf of fdl1-1 (E). An irregular distribution of epicuticular waxes can be seen on the second leaf of an RNAi transformed seedling (F) if compared with the second leaf of an untransformed seedling (C). (This figure is available in colour at JXB online.)
The fdl1-1 mutant phenotype is linked to the insertion of a En/Spm element in a R2R3-MYB transcription factor
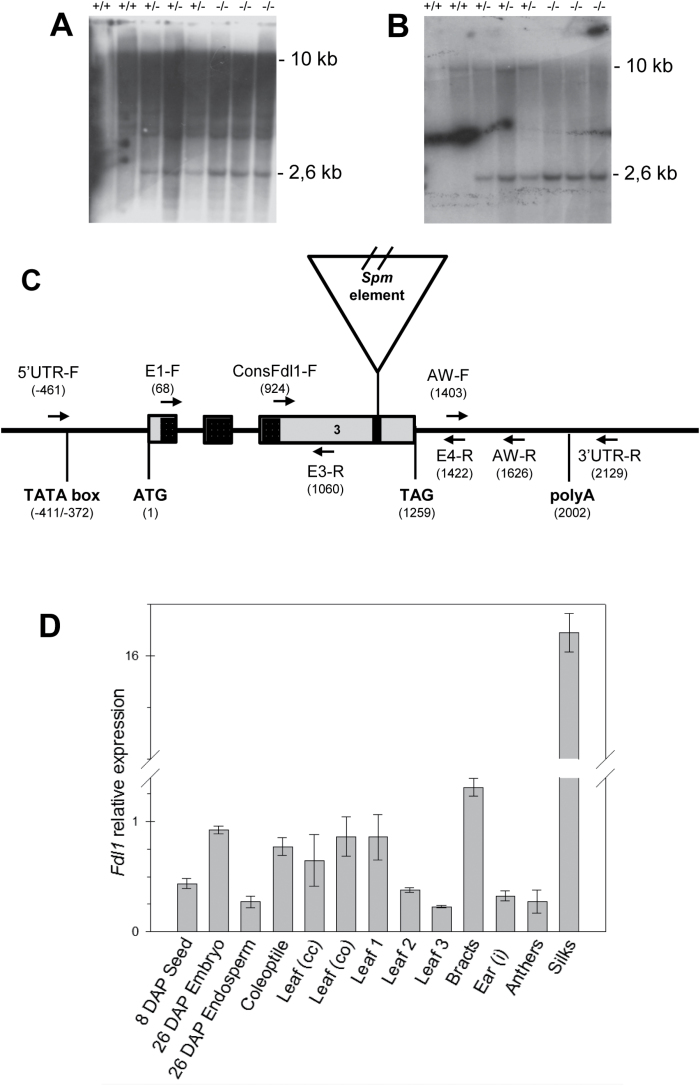
Chromosomal arm location of fdl1-1 was first established by means of TB-A translocations. Heterozygous +/fdl1-1 females were crossed with a set of heterozygous or hyperploid B-A translocation males. Only the F1 obtained from crosses involving the TB-7L male parent uncovered the mutant phenotype, indicating that fdl1 lies on the long arm of chromosome 7. A more refined position for fdl1 was achieved through analysis of SSR marker distribution in a segregating F2 population obtained by crossing homozygous fdl1-1/fdl1-1 females with H99 inbred male parents. Polymorphism for the markers UMC1125 and UMC1342 allowed the positioning of fdl1 at 6.3 cM from the former and 5.4 cM from the latter. Since no similar mutants have so far been located in this genomic region, it was assumed that the fdl1 phenotype defines a novel gene. The fdl1-1 mutant had been isolated from an active En/Spm line, therefore a co-segregation analysis was performed to identify whether the insertion of an En/Spm element in the fdl1 locus was causing the mutant phenotype. Genomic DNA was extracted from single individuals from F1 and F2 segregating progenies, including homozygous fdl1-1/fdl1-1 mutants, fdl1-1/+ heterozygous and +/+ homozygous wild-types and compared by gel blot analysis. Hybridization of genomic DNAs with the En/Spm-specific probe pbx1, revealed a BamH1 restriction fragment length polymorphism (RFLP) of approximately 2.6kb co-segregating with the mutant phenotype (Fig. 8A). Cloning of the RFLP highlighted the presence of 78bp of genomic sequence flanking the En/Spm element. When used as a probe against the BamH1 blot, this fragment identified the same 2.6kb mutant RFLP in fdl1-1/+ and fdl1-1/fdl1-1 individuals (Fig. 8B) as well as a 10kb restriction fragment ascribable to the wild-type allele of fdl1-1/+ and +/+ individuals (Fig. 8B). Co-segregation analysis, performed with both the En/Spm probe and the genomic probe on a total of 50 individuals (data not shown) indicated that the element was inserted either in the fdl1 gene itself or in a closely linked locus. Alignment of the 78bp flanking sequence with the maize genome sequence (www.maizesequence.org) placed the En/Spm insertion in the GRMZM2G056407 gene model corresponding to the ZmMYB94 gene, which is located on chromosome 7L in the region delimited by the UMC1125 and UMC1342 markers where the fdl1 gene was mapped.
Fig. 8.
ZmMYB94 gene structure and expression analysis. Southern analysis of fdl1-1 and wild-type alleles (A, B). DNAs from homozygous wild-type (+/+), heterozygous (+/-) and homozygous mutant (-/-) seedlings were cleaved with BamH1. Filters were hybridized with the En/Spm-specific probe (A) or the 78bp fragment flanking the En/Spm insertion (B). (C) Schematic representation of the ZmMYB94 gene showing the position of the three exons, indicated as grey rectangles, and the two introns, indicated as black lines. The En/Spm element inserted in the mutant allele is designated as a triangle, the 78bp genomic DNA flanking the insertion site, which was cloned after co-segregation analysis, is designed as a black rectangle and the region containing the MYB domain is highlighted in grey. The putative translational start (ATG) and stop (TAG) codons and polyA signal are reported. Exact positions are indicated with respect to the putative transcription initiation site. Names and positions of primers used for gene sequence analysis and qRT-PCR are also reported. The En/Spm element is not drawn to scale. (D) Relative expression level in different wild-type tissue samples from plants of the B73 inbred line. Expression level was established by qRT-PCR and the 18S gene was used as internal control. Bars represent means ±SD. DAP, days after pollination; cc, inside the closed coleoptile; co, emerging from the open coleoptile; i, immature. Tissue samples are listed in Supplementary Table S2.
To experimentally validate the gene model, overlapping amplification products were obtained with gene specific primer sets reported in Fig. 8C (5′UTR-F and E3-R, ConsFdl1 and 3′UTR-R), from wild-type genomic DNA and cDNA and sequenced. Sequences were assembled to generate a contig showing 100% identity with the GRMZM2G056407 gene prediction comprising three exons separated by two introns (Fig. 8C), whose putative product consists of 330 amino acids. Sequence analysis of mutant DNA confirmed that the En/Spm element insertion was within the GRMZM2G056407 coding sequence (data not shown).
To verify the results obtained from the Southern-based co-segregation analysis, a PCR-based analysis was performed on 100 mutant and 40 wild-type F2 individuals, whose genotypes were ascertained by selfing. The two products, of 309bp and 625bp respectively, which are specific for the mutant allele (Supplementary Fig. S3C, D), were always and exclusively present in all mutant samples, whereas the third amplification product of 692bp, was only yielded by wild-types (Supplementary Fig. S3B). No recombination was detected between the fdl1-1 allele and the En/Spm insertion in the ZmMYB94 gene, indicating a tight linkage between this insertion and the fdl1-1 mutation.
Mutant cDNAs were also amplified by using the same primer sets (Consfdl2-F, Spm1-R; and Spm3-F, AW-R) and produced amplicons of the same size as those obtained from genomic DNA (data not shown). Sequence analysis showed that the fdl1-1 mutant transcript retains the En/Spm element and is predicted to encode a non-functional protein carrying a frameshift mutation at amino acid position 278 and a premature stop codon at position 298.
Seed progeny obtained from crossing fdl1-1 with a waxy (wx) endosperm line homozygous for the wx-m8 allele (Shure et al., 1983), which contains a defective En/Spm element, did not highlight any Waxy revertant sector, suggesting that the element inserted in the fdl1 locus is a non-autonomous element. A similar conclusion arose from the PCR based analysis of DNA extracted from homozygous entire mutant leaves fractionated in sectors. The primer set specific for the mutant allele, i.e. ConsFdl2-F and Spm1-R, gave the expected product, whereas the primer set specific for the wild-type allele, i.e. ConsFdl2-F and AW-R, did not produce an amplicon (data not shown).
Attempts to generate germinal revertants by crossing the mutant line with a line containing an autonomous En/Spm element, as well as to isolate novel transposon-induced alleles through Mutator-targeted mutagenesis were unfruitful (data not shown). Proof of gene identity was thus obtained using an alternative approach consisting of an RNAi experiment, in which ZmMYB94 as well as the closely related paralogue ZmMYB70 (GRMZM2G139284), were targeted. Six independent transformants were obtained and T1 progenies, which showed 50% segregation of transgenic individuals, as expected for single locus insertion, were visually scored for mutant phenotypes. Although the mutant phenotype was very mild and rather sensitive to environmental conditions, transgenic seedlings of two transformants grown at 23°C showed some typical fdl1-1 mutant traits consisting of curly leaves and regions of adhesions between the first two leaves (Supplementary Fig. S4B, C, D). In these two transformants the relative expression level of ZmMYB94 in the third leaf was 4% and 5% respectively of that observed in wild-type.
Moreover, SEM analysis of a transformed mutant second leaf revealed an uneven epicuticular wax distribution on its epidermal surface (Fig. 7F) resembling that of the fdl1-1 mutant epidermal surface (Fig. 7E). This pattern was not detected on the second leaf of an untransformed sibling plant (Fig.7C).
The expression profile of the ZmMYB94 gene in the seedling overlaps with the pattern of mutant phenotype expression
The spatial and temporal expression profile of the ZmMYB94 gene was analysed by qRT-PCR in different maize tissues (Fig. 8D; Supplementary Table S2), using the AW-F and AW-R ZmMYB94 specific primers. The ZmMYB94 mRNA was detected in all organ/tissues analysed, except in the primary root, and the expression pattern obtained (Fig. 8D) did not deviate from that described in the work of Sekhon et al., (2013). The highest expression (5-fold higher than other tissues) was observed in ear silks. Within the seed, ZmMYB94 was more expressed in the embryo than the endosperm. In the seedling, higher expression levels were observed in the coleoptile and in the first leaf, with a progressive decrease of expression in the second and third leaf. We note that in seedlings, the expression profile detected by qRT-PCR analysis matches the pattern of fdl1-1 phenotypic expression observed (Fig. 1).
Phylogenetic analysis
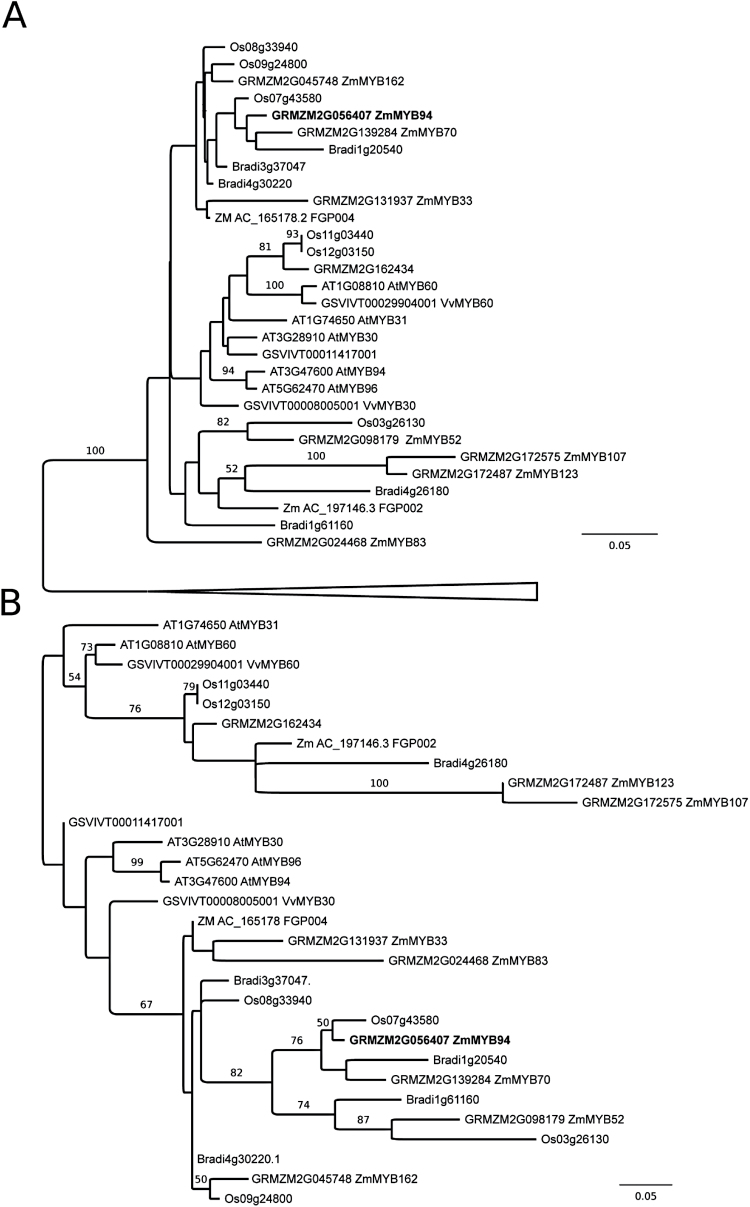
Preliminary phylogenetic reconstructions considering 104 unambiguously alignable amino acid residues from the highly conserved MYB domains of all annotated R2R3-MYB proteins from Z. mays, O. sativa, B. distachyon, A. thaliana and V. vinifera indicate that ZmMYB94 falls within a well supported clade containing representatives of all species considered (Fig. 9A), but did not allow confident assignment of an exact phylogenetic placement. Consideration of only sequences within this clade allowed a subsequent phylogenetic reconstruction including 166 aligned amino acid sites (Fig. 9B), which, while failing to unambiguously resolve all relationships within the MYB subclade, provided strong support for co-orthology of ZmMyb94 and ZmMyb70 with functionally uncharacterized Brachypodium and rice genes and as part of a monocot-specific gene family expansion. Our analyses recover no support for direct orthology between ZmMyb94 or ZmMyb70 and any dicot homologue. Indeed, the most closely related dicot MYBs were V. vinifera MYB30, an additional uncharacterized V. vinifera MYB and a clade of recently duplicated Arabidopsis MYBs (AtMYB30/94/96). The absence of direct dicot orthologues of ZmMyb94 notwithstanding, it is interesting to note that MYB30, MYB94 and MYB96 have all been implicated in the regulation of cuticular wax biosynthesis in Arabidopsis (Shepherd et al., 2006; Raffaele et al., 2008; Seo et al., 2011; Lee et al., 2014).
Fig. 9.
Phylogenetic placement of ZmMYB94. (A) Distance tree of all annotated R2R3-MYB protein sequences from Zea mays, Oryza sativa, Brachypodium distachyon, Arabidopsis thaliana and Vitis vinifera (104 unambiguously aligned amino acid positions). The tree is arbitrarily rooted at the base of the well-supported clade that contains ZmMYB94 and all relationships outside this clade collapsed. (B) Arbitrarily rooted maximum likelihood phylogenetic reconstruction of relationships within the ZmMYB94 clade (166 amino acid positions) from the same taxonomic sampling employed in panel A. For both panels, bootstrap support values for internal branches are marked where greater than 50%.
Discussion
Seed germination and seedling emergence represent crucial events in the plant life cycle, and rapid and robust emergence allows plants to take advantage of favourable environments and to compete with neighbours. Moreover synchrony in seedling emergence is a profitable character for cultivated plants because it facilitates the weed control practices (Buhler et al., 2000). The study of the fd11-1 mutant carried out in this work adds knowledge on the molecular mechanisms underlying shoot formation during embryogenesis, germination and early seedling development in Z. mays. The fdl1-1 seedling phenotype is characterized by curly leaves with regions of adhesion between the first leaf and the coleoptile or between the first and the second leaves. In these regions suture zones devoid of any intervening cuticular material between juxtaposed cell walls are present (Fig. 6), which are adjacent to free zones, where cell wall surfaces are covered by a continuous cuticle layer. Indeed, while most of the young epidermis is covered by a well-organized cuticle making them waterproof (Supplementary Fig. S2), an additional feature of the mutant seedling is a patchy distribution of epicuticular waxes on the epidermis of the second leaf, where areas devoid of epicuticular waxes are clearly visible (Fig. 7E).
The fdl1-1 mutant, located on the long arm of chromosome seven, has been ascribed to the insertion of an En/Spm transposable element inside the third exon of a maize R2R3-MYB transcription factor whose gene model has been named either ZmMYB94 in the last version of the maize genome sequence (http://www.maizesequence.org) or ZmMYB110 in the paper of Du et al., (2013). The decision to adopt the first designation in this work is based on the relationship, mentioned hereafter, with AtMYB94.
ZmMyb94, and its close paralogue ZmMyb70, are members of a monocot-specific local expansion of the MYB family, within which ZmMyb94 is the first member to be functionally analysed. The function of ZmMYB94 recalls that of AtMYB96, AtMYB94 and AtMYB30, which are its closest Arabidopsis members. AtMYB96 is involved in the activation of cuticular wax biosynthesis (Seo et al., 2011). AtMYB30 acts as a transcriptional activator of genes encoding the fatty acid elongase complex, being responsible for the biosynthesis of very long-chain fatty acids (VLCFA), which are leaf epidermal wax components (Shepherd et al., 2006; Raffaele et al., 2008). AtMYB94 has recently been implicated in regulation of wax deposition through direct binding of promoters of various genes involved in wax biosynthesis (Lee et al., 2014). The action of these Arabidopsis genes has been correlated with environmental responses (Shepherd et al., 2006; Raffaele et al., 2008; Seo et al., 2011; Lee et al., 2014) but not, to our knowledge, with developmental processes.
The relationship between alterations in epidermis covering material and developmental defects, in particular organ fusion, has been reported (Nawrath, 2006; Bernard and Joubès, 2013). For example, the maize adherent1 mutant exhibits defective cell wall and epicuticular wax deposition and shows organ fusion (Sinha and Linch, 1998). In Arabidopsis, organ fusions were observed in mutants such as fiddlehead-1 (Lolle et al., 1992, 1997; Yephremov et al., 1999; Pruitt et al., 2000), bodyguard (Kurdyukov et al., 2006a) and hothead (Kurdyukov et al., 2006b), which are defective in the biosynthesis of cuticle components as well as in the dso (DESPERADO) mutant, defective in a transporter required for cutin and wax secretion (Panikashvili et al., 2007). Postgenital fusion also occurred in the lacerata mutant of Arabidopsis in which cutine biosynthesis is impaired (Wellesen et al., 2001), and in the cer3/wax2 mutant, specifically impaired in the elongation of very long-chain (VLC) alkanes, which are major components of cuticular waxes (Chen et al., 2003; Rowland et al., 2007). Organ fusion has also been observed in the lacs1/lacs2 double mutant whose corresponding genes are involved in cuticular wax and cutin biosynthetic pathways, respectively (Weng et al., 2010).
Particular analogies can be recognised between fdl1-1 and bodyguard (Kurdyukov et al., 2006a) as both exhibit direct adhesion of cell walls in the suture zones that are devoid of intervening cuticular membrane (Fig. 6), and between fdl1-1 and lacerata, the latter forming suture zones in which the cuticle was not particularly conspicuous, and wherein contacting cell walls adhere directly to each other (Wellesen et al., 2001). Our functional study of the fdl1-1 mutant provides further confirmation that an intact cuticle layer is necessary for establishing organ boundaries by preventing post-genital organ fusion and is consistent with analyses of Arabidopsis plants expressing a fungal cutinase and exhibiting leaf fusions, in which cell walls of both epidermal cells came into direct contact (Sieber et al., 2000).
Additional developmental defects in the fdl1-1 mutant include delays in germination and seedling elongation (Fig. 1) as well as the presence of a thicker coleoptile, with impaired opening (Fig. 2). Mutant leaves are forced to start growing when still within the coleoptile and instead of emerging through a clear-cut hole at the apex of the coleoptile, emerge from the lateral side of this organ (Figs 1, 2). Fdl1-1 mutant seedling defects can be traced back to abnormalities in the embryogenetic process. At 18 DAP (Fig. 3E), as well as at later developmental stages (data not shown), both scutellum and coleoptile are not properly developed in correspondence with the adaxial side of the mutant embryo. Leaf primordia are abnormal and not regularly rolled and coleoptile and first leaf primordia are fused (Fig. 3F). It is thus conceivable that the fdl1 gene begins to exert its influence when coleoptile and first leaf primordia are initiated.
Seedling phenotypes display variability in phenotypic expression at early stages of seedling growth (Supplementary Fig. S1). However after stage three (presence of third leaf), most homozygous individuals, bearing either mild or severe phenotypes, resumed a wild-type phenotype (Fig. 1). This observation, together with the mRNA expression profile (Fig. 8D), suggests that fdl1 gene action is exerted during a strict developmental window. In maize, juvenile and adult leaves differ since the former are coated with epicuticular waxes and lack specialized cells, whereas the latter lack epicuticular waxes, bear trichomes and bulliform cells and have crenulated epidermal cell walls (Sylvester et al., 2001). There is evidence that the juvenile and adult phases are genetically separated. The action of glossy15, an APETALA2-like gene, is required to maintain juvenile leaf identity and its down-regulation promotes phase change (Moose and Sisco, 1996; Lauter et al., 2005). The glossy appearance of the first fdl1-1 leaves and the stronger expression of ZmMYB94 in the first leaves are indications that ZmMYB94 may be another actor in phase change and it would be interesting to investigate regulatory links between ZmMYB94 and Gl15.
In conclusion, ZmMYB94, whose action is specific for embryogenesis and early phase of seedling development, is required for determining a regular pattern of cuticle deposition in the young shoot, which is shown to be a prerequisite for the establishment of organ boundaries and identities. As outlined in the study of the maize OUTER CELL LAYER1 (OCL1) gene, cuticle deposition implies the coordinated activity of different regulatory factors (Javelle et al., 2011). Further analysis of the MYB94 action will contribute to the understanding of this regulatory network.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. List of primers used in this study.
Supplementary Table S2. List of tissues used for qRT-PCR.
Supplementary Figure S1. Variable fdl1-1 seedling phenotypes.
Supplementary Figure S2. Response of wild-type and fdl1-1 mutant seedlings to the toluidine blue test.
Supplementary Figure S3. PCR based co-segregation analysis.
Supplementary Figure S4. Phenotype of RNAi transformed seedlings.
Acknowledgements
We thank Prof. Jane Langdale for donating the fdl1-1 mutant, Prof. Giuseppe Gavazzi for helpful comments on this manuscript and the Maize Genetics Cooperation Stock Center, Urbana, IL, USA, for providing various genetic stocks used in this work.
References
- Abbe EC, Stein L. 1954. The growth of the shoot apex in maize: embryogeny. American Journal of Botany 41, 173–284. [Google Scholar]
- Beckett JB. 1978. B-A translocations in maize: I. Use in locating genes by chromosome arms. The Journal of Heredity 69, 27–36. [Google Scholar]
- Bernard A, Joubès J. 2013. Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Progress In Lipid Research 52, 110–129. [DOI] [PubMed] [Google Scholar]
- Bommert P, Werr W. 2001. Gene expression patterns in the maize caryopsis: clues to decisions in embryo and endosperm development. Gene 271, 131–142. [DOI] [PubMed] [Google Scholar]
- Buhler DD, Liebman M, Obrycki JJ. 2000. Theoretical and practical challenges to an IPM approach to weed management. Weed Science 48, 274–280. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution 4, 540–552. [DOI] [PubMed] [Google Scholar]
- Chen J, Dellaporta S. 1994. Urea-based plant DNA miniprep. In: Freeling M, Walbot V, eds. The Maize Handbook . New York: Springer-Verlag, 526–527. [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA. 2003. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. The Plant Cell 15, 1170–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JK. 1996. Maize embryogenesis mutants. In: Wang TI, Cuming A, eds. Embryogenesis—the Generation of a Plant . Oxford: BIOS Scientific Publications, 89–112. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini S, Landoni M, Consonni G, Rascio N, Dalla Vecchia F, Gavazzi G. 1999. The maize lilliputian mutation is responsible of disrupted morphogenesis and minute stature. The Plant Journal 17, 11–17. [Google Scholar]
- Du H, Wang YB, Xie Y, Liang Z, Jiang SJ, Zhang SS, Huang YB, Tang YX. 2013. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Research 20, 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C. 2010. MYB transcription factors in Arabidopsis . Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1989. PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 5, 164–166. [Google Scholar]
- Fornalé S, Sonbol FM, Maes T, Capellades M, Puigdomènech P, Rigau J, Caparrós-Ruiz D. 2006. Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyltransferase genes by two new maize R2R3-MYB transcription factors. Plant Molecular Biology 62, 809–823. [DOI] [PubMed] [Google Scholar]
- Grotewold E, Athma P, Peterson T. 1991. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of Myb-like transcription factors. Proceedings of the National Academy of Sciences, USA 88, 4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 3, 307–321. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos JF, Dal Prà M, Giulini A, et al. 2007. empty pericarp4 encodes a mitochondrion-targeted pentatricopeptide repeat protein necessary for seed development and plant growth in maize. The Plant Cell 19, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PJ, Hartley RD. 1976. Detection of ferulic acid in cell walls of the Gramineae by ultraviolet fluorescence microscopy. Nature 259, 508–510. [Google Scholar]
- Heine GF, Malik V, Dias AP, Grotewold E. 2007. Expression and molecular characterization of ZmMYB-IF35 and related R2R3-MYB transcription factors. Molecular Biotechnology 37, 155–164. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. 1996. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens . Nature Biotechnology 14, 745–750. [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC. 2011. Epidermis: the formation and functions of a fundamental plant tissue. New Phytology 189, 17–39. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 3, 275–282. [DOI] [PubMed] [Google Scholar]
- Klempnauer KH, Gonda TJ, Bishop JM. 1982. Nucleotide sequence of the retroviral leukemia gene v–myb and its cellular progenitor c–myb: the architecture of a transduced oncogene. Cell 31, 453–463. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, et al. 2006. a The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis . The Plant Cell 18, 321–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bar S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A. 2006. b Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-, omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224, 315–329. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. 1987. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- La Rocca N, Pupillo P, Puppi G, Rascio N. 2014. Erythronium dens-canis L. (Liliaceae): an unusual case of change of leaf mottling. Plant Physiology and Biochemistry 74, 108–117. [DOI] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. 2005. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proceedings of the National Academy of Sciences, USA 102, 9412–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kim J, Suh MC. 2014. Overexpression of myb94 transcription factor causes activation of Arabidopsis cuticular wax biosynthesis. Plant & Cell Physiology pii: pcu142. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Cheung AY, Sussex IM. 1992. Fiddlehead—an Arabidopsis mutant constitutively expressing an organ fusion program that involves interactions between epidermal-cells. Developmental Biology 152, 383–392. [DOI] [PubMed] [Google Scholar]
- Lolle SJ, Berlyn GP, Engstrom EM, Krolikowski KA, Reiter WD, Pruitt RE. 1997. Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: A role for the epidermal cell wall and cuticle. Developmental Biology 189, 311–321. [DOI] [PubMed] [Google Scholar]
- Marocco A, Wissenbach M, Becker D, Paz-Ares J, Saedler H, Salamini F, Rohde W. 1989. Multiple genes are transcribed in Hordeum vulgare and Zea mays that carry the DNA binding domain of the myb oncoproteins. Molecular and General Genetics 216, 183–187. [DOI] [PubMed] [Google Scholar]
- Moose SP, Sisco PH. 1996. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes & Development 10, 3018–3027. [DOI] [PubMed] [Google Scholar]
- Nawrath C. 2006. Unraveling the complex network of cuticular structure and function. Current Opinion in Plant Biology 9, 281–287. [DOI] [PubMed] [Google Scholar]
- Panikashvili D, Savaldi-Goldstein S, Mandel T, Yifhar T, Franke RB, Hofer R, Schreiber L, Chory J, Aharoni A. 2007. The Arabidopsis DESPERADO/AtWBC11 transporter is required for cutin and wax secretion. Plant Physiology 145, 1345–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz–Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. 1987. The regulatory c1 locus of maize encodes a protein with homology to myb proto–oncogene products and with structural similarities to transcriptional activators. The EMBO Journal 6, 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. 1986. Molecular analysis of the En/Spm transposable element system of Zea mays . The EMBO Journal 5, 835–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouvreau B, Baud S, Vernoud V, Morin V, Py C, Gendrot G, Pichon J-P, Rouster J, Paul W, Rogowsky PM. 2011. Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiology 156, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada JP, Ploense SE, Grossniklaus U, Lolle SJ. 2000. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proceedings of the National Academy of Sciences, USA 97, 1311–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Robya D. 2008. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis . The Plant Cell 20, 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L. 2007. The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Letters 581, 3538–3544. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM. 2013. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8, e61005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. 2011. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis . The Plant Cell 23, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd T, Wynne Griffiths D. 2006. The effects of stress on plant cuticular waxes. New Phytology 171, 469–499. [DOI] [PubMed] [Google Scholar]
- Shure M, Wessler S, Fedoroff N. 1983. Molecular identification and isolation of the Waxy locus in maize. Cell 35, 225–233. [DOI] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Metraux JP, Nawrath C. 2000. Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. The Plant Cell 12, 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N, Lynch M. 1998. Fused organs in the adherent1 mutation in maize show altered epidermal walls with no perturbations in tissue identities. Planta 206, 184–195. [Google Scholar]
- Sylvester AW, Parker-Clark V, Murray GA. 2001. Leaf shape and anatomy as indicators of phase change in the grasses: comparison of maize, rice, and bluegrass. American Journal of Botany 88, 2157–2167. [PubMed] [Google Scholar]
- Tanaka T, Tanaka H, Machida C, Watanabe M, Machida Y. 2004. A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis . The Plant Journal 37, 139–146. [DOI] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A. 2001. Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different robes of fatty acid omega-hydroxylation in development. Proceedings of the National Academy of Sciences, USA 98, 9694–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H, Molina I, Shockey J, Browse J. 2010. Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis . Planta 231, 1089–1100. [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. 1999. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. The Plant Cell 11, 2187–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.