Highlight
Nitration and S-nitrosylation, two post-translational modifications (PTMs) mediated by nitric oxide, differentially regulate MDAR and GR activities which are key components of the ascorbate–glutathione cycle.
Key words: Glutathione reductase, monodehydroascorbate reductase, nitration, nitric oxide, peroxynitrite, reactive nitrogen species, salinity, S-nitrosylation, S-nitrosoglutathione.
Abstract
The ascorbate–glutathione cycle is a metabolic pathway that detoxifies hydrogen peroxide and involves enzymatic and non-enzymatic antioxidants. Proteomic studies have shown that some enzymes in this cycle such as ascorbate peroxidase (APX), monodehydroascorbate reductase (MDAR), and glutathione reductase (GR) are potential targets for post-translational modifications (PMTs) mediated by nitric oxide-derived molecules. Using purified recombinant pea peroxisomal MDAR and cytosolic and chloroplastic GR enzymes produced in Escherichia coli, the effects of peroxynitrite (ONOO–) and S-nitrosoglutathione (GSNO) which are known to mediate protein nitration and S-nitrosylation processes, respectively, were analysed. Although ONOO– and GSNO inhibit peroxisomal MDAR activity, chloroplastic and cytosolic GR were not affected by these molecules. Mass spectrometric analysis of the nitrated MDAR revealed that Tyr213, Try292, and Tyr345 were exclusively nitrated to 3-nitrotyrosine by ONOO–. The location of these residues in the structure of pea peroxisomal MDAR reveals that Tyr345 is found at 3.3 Å of His313 which is involved in the NADP-binding site. Site-directed mutagenesis confirmed Tyr345 as the primary site of nitration responsible for the inhibition of MDAR activity by ONOO–. These results provide new insights into the molecular regulation of MDAR which is deactivated by nitration and S-nitrosylation. However, GR was not affected by ONOO– or GSNO, suggesting the existence of a mechanism to conserve redox status by maintaining the level of reduced GSH. Under a nitro-oxidative stress induced by salinity (150mM NaCl), MDAR expression (mRNA, protein, and enzyme activity levels) was increased, probably to compensate the inhibitory effects of S-nitrosylation and nitration on the enzyme. The present data show the modulation of the antioxidative response of key enzymes in the ascorbate–glutathione cycle by nitric oxide (NO)-PTMs, thus indicating the close involvement of NO and reactive oxygen species metabolism in antioxidant defence against nitro-oxidative stress situations in plants.
Introduction
The ascorbate–glutathione cycle is composed of monodehydroascorbate reductase (MDAR), glutathione reductase (GR), ascorbate peroxidase (APX), and dehydroascorbate reductase (DHAR), as well as the antioxidant metabolites ascorbate and glutathione and the reductive coenzyme NADPH. This metabolic pathway is essential for the detoxification and regulation of the cellular level of hydrogen peroxide (H2O2) in plant cells (Hossain and Asada, 1985; Asada, 1992; Noctor and Foyer, 1998; Shigeoka et al., 2002). Thus, H2O2 is reduced to water by APX using ascorbate as the electron donor. The oxidized ascorbate (monodehydroascorbate or dehydroascorbate) is then regenerated by MDAR and DHAR using reduced glutathione (GSH) with the concomitant generation of oxidized glutathione (GSSG). Finally, GSSG is reduced by GR with the aid of NADPH as the electron donor. At the subcellular level, these enzymes have been demonstrated to be located in different cellular compartments including the cytosol, chloroplasts, peroxisomes, and mitochondria (Foyer and Halliwell, 1976; Groden and Beck, 1979; Jiménez et al., 1998; Asada, 2006; Palma et al., 2006; Reumann and Corpas, 2010).
Nitric oxide (NO) belongs to a family of related molecules known as reactive nitrogen species (RNS). Although the specific source of NO in plants is still under debate (see Corpas et al., 2009), there is no doubt that plants have an endogenous NO source(s). S-Nitrosoglutathione (GSNO) is formed by the S-nitrosylation reaction of NO with GSH, and it has a significant physiological relevance because GSNO is considered to function as a mobile reservoir of NO bioactivity. On the other hand, the reaction of NO with superoxide radicals (O2·–) generates a powerful oxidant, designated peroxynitrite (ONOO–). Furthermore, these NO-derived molecules can mediate several post-translational modifications (PTMs) such as nitration and S-nitrosylation. Protein tyrosine nitration involves the addition of a nitro group (-NO2) to one of the two equivalent ortho-carbons of the tyrosine residue aromatic ring. Protein nitration is affected by various features such as the protein quaternary structure, the environment in which the protein is located, and the nitration mechanism. Consequently, these covalent changes may result in effects such as loss or gain of protein function or no change in function (Souza et al., 2008; Radi, 2013). On the other hand, S-nitrosylation involves the binding of an NO group to a protein cysteine residue and is also able to change the function of many proteins (Astier et al., 2012). Proteomic studies have identified potential plant protein candidates for nitration and S-nitrosylation which belong to diverse functional categories such as redox-related, stress-related, metabolic, and signalling/regulating proteins (Lindermayr et al., 2006; Corpas et al., 2015). These analyses have identified some enzymatic components of the ascorbate–glutathione cycle as potential PTM targets mediated by NO-derived molecules. However, little is known about the specific impact of these NO-related PTMs on the activity and structure of particular proteins involved in antioxidative systems (Holtgrefe et al., 2008; Chaki et al., 2011; Astier et al., 2012; Begara-Morales et al., 2013a , 2014). In previous studies, in order to understand the fine-tuned regulation of this key antioxidant system by NO, the effect of NO-related PTMs on the activity of cytosolic APX was analysed. Thus, APX was identified as a target of S-nitrosylation in Arabidopsis plants (Fares et al., 2011; Keyster et al., 2011), and pea cytosolic APX has very recently been demonstrated to have a dual mechanism of regulation mediated by NO-derived molecules; while the nitration of Tyr235 provoked a deactivation of APX activity, S-nitrosylation at Cys32 activated this activity (Begara-Morales et al., 2014). On the other hand, a nitroproteomic study of sunflower hypocotyls identified GR as a target for tyrosine nitration (Chaki et al., 2009) and for S-nitrosylation in rice (Lin et al., 2012). Additionally, MDAR has also been identified as a potential candidate for both S-nitrosylation and nitration in citrus plants (Tanou et al., 2012) and for S-nitrosylation in rice (Lin et al., 2012) and Arabidopsis (Hu et al., 2015). However, to the authors’ knowledge, no information is available on the effect of any PTMs mediated by NO-derived molecules on the molecular function of MDAR or GR.
Thus, the aim of the present study is to gain a more in-depth understanding of the regulation of the antioxidative ascorbate–glutathione cycle by NO-PTMs. Pea (Pisum sativum) was selected as the model plant for several reasons: (i) previous studies have allowed significant information on NO metabolism to be obtained (Barroso et al., 2006; Corpas et al., 2008, 2013b ); (ii) the isolation and characterization of a full-length genomic clone encoding the pea MDAR has been reported; and (iii) a previous study on pea APX showed dual regulation by S-nitrosylation and nitration (Begara-Morales et al., 2014). On the other hand, both chloroplastic and cytosolic isoforms of GR (Edwards et al., 1990; Stevens et al., 1997; Gill et al., 2013; Wu et al., 2013) were used to obtain the most complete information about this enzyme. Consequently, using in vitro approaches, the potential effect of NO-derived molecules such as ONOO– and GSNO, which trigger nitration and S-nitrosylation, respectively, on the molecular function of two enzymes (peroxisomal MDAR and cytosolic and chloroplastic GR) in this cycle was analysed.
As a result, peroxisomal MDAR was deactivated by both nitration and S-nitrosylation, which compromise the cycle’s antioxidant capacity. However, cytosolic and chloroplastic GR were unaffected by any of these NO-PTMs in an attempt to maintain the levels of GSH and the cellular redox state.
Materials and methods
Plant material and growth conditions
Pea (Pisum sativum L., cv. Lincoln) seeds were obtained from Royal Sluis (Enkhuizen, The Netherlands). Seeds were surface sterilized with 3% (v/v) commercial bleach solution for 3min, and then were washed with distilled water, and germinated in vermiculite for 3–4 d under the following growth chamber conditions: 24 ºC/18 °C (day/night), 80% relative humidity, a 16h photoperiod, and a light intensity of 190 μE m–2 s–1. Healthy and vigorous seedlings were selected and grown in nutrient solutions (Corpas et al., 1993). After 14 d, plants were transplanted to similar media supplemented with 150mM NaCl and were grown for 4 d (Begara-Morales et al., 2014).
Expression and purification of pea MDAR and GR
The cDNAs encoding mature pea peroxisomal MDAR (AY662655.1) and cytosolic (X98274.1) and chloroplastic (X60373.1) GR were amplified by PCR from total pea leaf RNA using the Fast Start High Fidelity polymerase (Roche) and the specific primer sets: 5′- GGATCCGATGGTGCATTCGTTCAAGTATATC-3′ (forward) and 5′-GCTCGAGTATTAAATTTTACTTGCAAA AGAAAGG-3′ (reverse) for MDAR; 5′-AGGATCCAATGAA CCAAGCAATGGCTACTC-3′ (forward) and 5′-CTCGAGTCTTA AGATCCAGCCACAGCTTTTG-3′ (reverse) for chloroplastic GR; and 5′-GGATCCGATGGCTAGAAAGATGCTTAACG-3′ (forward) and 5′-CTCGAGTTTTACAATTTGTCTT TGGCT TCAC-3′ (reverse) for cytosolic GR. The PCR products (1316bp for MDAR, 1671bp for chloroplastic GR, and 1510bp for cytosolic GR) were cloned into the pGEM-T Easy Vector (Promega). The positive clones were confirmed by sequencing and then subcloned following prior digestion with BamHI and XhoI into the pALEXb vector.
Recombinant proteins carrying an N-terminal choline-binding domain were produced using Escherichia coli strain BIVU0811, which were routinely cultured overnight at 37 ºC in LB with kanamycin (25mg l–1) and ampicillin (100mg l–1). Gene expression was induced by the addition of 1mM salicylate and 10mM 3-methyl benzoate in a 250ml culture grown at 20 ºC overnight in order to produce a higher proportion of soluble protein. Cells were harvested by centrifugation and re-suspended in 20ml of phosphate-buffered saline (PBS; pH 7.0) containing 25U ml–1 DNAse I, 10mM MgCl2, and commercial protease inhibitor (Complete, Roche). Cells were lysed with a Niro Soavi NS1001L Panda High-Pressure homogenizer at a pressure of 800–900 bar. The cell lysate was then centrifuged at 10 000 g at 4 °C for 15min, and the supernatant was used for the purification of recombinant proteins with the aid of a 1ml LYTRAP column (Biomedal). The column was washed with 20ml of 20mM K phosphate buffer (pH 7.0) containing 300mM NaCl and 5mM choline. The protein was eluted in 1ml fractions using a discontinuous gradient of choline prepared in the same buffer with 100mM NaCl and 20mM choline (fraction E1), 50mM choline (E2), 75mM choline (E3), 100mM choline (E4), 150mM choline (E5), 200mM choline (E6), 250mM choline (E7), and 500mM choline (E8). The samples were analysed by 10% SDS–PAGE and stained with Coomassie blue dye. Supplementaryy Fig. S1 available at JXB online shows the SDS–PAGE analysis of the purification of recombinant peroxisomal MDAR (Supplementary Fig. S1A) and chloroplastic and cytosolic GR (Supplementary Fig. S1B, C).
MDAR and GR activity assays: treatment with a peroxynitrite donor (SIN-1) and a nitric oxide donor (GSNO)
MDAR (EC 1.6.5.4) activity was determined spectrophotometrically by measuring the reduction of absorbance at 340nm according to the technique described by Hossain et al. (1984) with some modifications. The 1.0ml assay mixtures contained 50mM TRIS-HCl (pH 7.8), 0.2mM NADH, 1mM ascorbate, and sample. The reaction was initiated by adding 0.2U of ascorbate oxidase (EC 1.10.3.3 from Cucurbita; Sigma-Aldrich, St. Louis, MO. USA), and the decrease in A 340 due to NADH oxidation was monitored. One milliunit of activity was defined as the amount of enzyme required to oxidize 1 nmol NADH min–1 at 25 ºC. GR (EC 1.6.4.2) activity was assayed by monitoring NADPH oxidation coupled with the reduction in GSH (Edwards et al., 1990). The reaction rate was corrected for the slight non-enzymatic oxidation of NADPH by glutathione disulphide (GSSG).
The molecule SIN-1 (3-morpholinosydnonimine) has been shown to generate peroxynitrite (ONOO–), a protein-nitrating compound (Daiber et al., 2004). Recombinant proteins were therefore incubated at 37 °C for 1h with increasing concentrations (0–5mM) of SIN-1 (Calbiochem) freshly made up before use (Begara-Morales et al., 2014). For treatments with GSNO (NO donor), recombinant proteins were incubated at room temperature for 30min with 0.5mM and 2mM GSNO (Begara-Morales et al., 2014). As a control, proteins were also incubated with 0.5mM and 2mM GSH. The protein concentration was determined with the aid of the Bio-Rad protein assay using bovine serum albumin (BSA) as standard.
Identification of nitrated tyrosine residues in recombinant pea MDAR using mass spectrometric techniques
Purified recombinant pea MDAR was processed according to a protocol involving reduction with dithiothreitol (DTT), derivatization with iodoacetamide (IAA), and enzymatic digestion with trypsin (37 ºC, 8h). The sample was purified using solid-phase extraction cartridges to eliminate choline interference. The resulting peptide mixture was analysed using a MALDI-TOF/TOF (matrix-assisted laser desorption ionization-time of flight/time of flight) mass spectrometer (4800, AB Sciex) to evaluate the quality of the sample. MALDI-TOF spectra were interpreted using a peptide mass fingerprinting (PMF) database search (Protein Prospector program). The database used for identification was UniProt (release 2011_02). The sample was then analysed by liquid chromatography–tandem mass spectometry (LC-MS/MS) using a Velos-LTQ mass spectrometer equipped with a micro-ESI ion source (ThermoFisher, San Jose, CA, USA). The sample was evaporated to dryness and diluted up to 40 μl with water containing 5% methanol and 1% formic acid. The sample was then loaded in a chromatographic system consisting of a C18 pre-concentration cartridge (Agilent Technologies, Santa Clara, CA, USA) connected to a 10cm long, 150 μm id Vydac C18 column (Vydac, IL, USA). Separation was carried out at 1 μl min–1 with a 3–40% acetonitrile gradient for 30min (solvent A, 0.1% formic acid; solvent B, acetonitrile with 0.1% formic acid). The high-performance liquid chromatography (HPLC) system contained an Agilent 1200 capillary pump, a binary pump, a thermostated microinjector, and a micro switch valve.
The Velos-LTQ instrument was operated in positive ion mode with a spray voltage of 2kV. The scan range of each full MS scan was m/z 400–2000. After each MS scan, a collection of targeted MS/MS spectra was obtained in order to identify both the unmodified and nitrated form of the predicted tyrosine-containing peptides. The parent mass list of the targeted scan was selected to ensure maximum coverage of the tyrosine-containing tryptic peptides for MDAR. The list of targeted m/z values was obtained after in silico digestion of the proteins using nitrated tyrosine as a dynamic modification. The resulting list of predicted peptides (in both nitrated and unmodified form) was filtered to exclude all peptides not containing tyrosine residues.
The MS/MS spectra were searched using Proteome Discoverer software (ThermoFisher) on the basis of the following parameters: peptide mass tolerance 2Da, fragment tolerance 0.8Da, enzyme set as trypsin, and no missed cleavages. The dynamic modifications were cysteine carbamidomethylation (+57Da), methionine oxidation (+16Da), and tyrosine nitration (+45). The searches were carried out using a database containing all the peptides listed in Table 1. Identifications were filtered with XCorr >3, P(pep) <15%. The MS/MS spectra of the nitrated tyrosines were manually validated by comparing the spectra obtained for the unmodified peptide and the nitrated peptide.
Table 1.
List of pea MDAR peptides scanned and identified by LC-MS/MS
| Peptides identifieda | Peptides scanned | Length (no. of amino acids) | M r (Da) | No. of tyrosine residues | |
|---|---|---|---|---|---|
| Not nitrated | Nitrated | ||||
| AKPAVEDVNQLAEEGLSFASK | 21 | 2203 | 0 | ||
| AVVVGGGYIGLELSAVLK | AVVVGGGYIGLELSAVLK | 18 | 1745 | 1 | |
| AYLFPESPAR | AYLFPESPAR | 10 | 1150 | 1 | |
| EAVAPYERPALSK | EAVAPYERPALSK | 13 | 1431 | 1 | |
| FGTYWIK | 7 | 914 | 1 | ||
| GIQLYLSTEIVSADLAAK | GIQLYLSTEIVSADLAAK | 18 | 1892 | 1 | |
| LFTSEIAAFYEGYYANK | LFTSEIAAFYEGYYANK | 17 | 1987 | 2032 | 3 |
| LLPEWYSEK | 9 | 1164 | 1 | ||
| LNDLDVTMVYPEPWCMPR | 18 | 2180 | 1 | ||
| LPGFHTCVGSGGER | 14 | 1417 | 0 | ||
| NIFYLR | 6 | 825 | 1 | ||
| SANGEHFDYQTLVIATGSAVIR | SANGEHFDYQTLVIATGSAVIR | 22 | 2350 | 1 | |
| SFDLSWQFYGDNVGETVLFGDNDPASSKPK | SFDLSWQFYGDNVGETVLFGDNDPASSKPK | 30 | 3322 | 1 | |
| SVEEYDYLPYFYSR | SVEEYDYLPYFYSR | 14 | 1831 | 1876 | 4 |
| TSVPDVYAVGDVATFPLK | TSVPDVYAVGDVATFPLK | 18 | 1879 | 1924 | 1 |
| YILIGGGVSAGYAAR | YILIGGGVSAGYAAR | 15 | 1468 | 2 | |
Some peptides detected do not contain tyrosines. These peptides were not included in the targeted MS/MS detection. They were detected and identified as their molecular weight coincides with that of predicted peptides.
Site-directed mutagenesis
Conversion of the tyrosine codon (TAT) to phenylalanine (TTT) in the pea cDNA of peroxisomal MDAR (accession no, AY662655) was accomplished by oligonucleotide-directed mutagenesis. The template for PCR mutagenesis was also the pALEXb expression vector carrying the entire P. sativum pMDAR I gene obtained as described above. The site-directed mutant of MDAR Tyr345Phe was obtained by using the following primers: 5′-CTTTGATCTTTTCCAATCCAC-3′ (MDAR Tyr345Phe forward) and 5′-GTGGATTGGAAAAGATCAAAG-3′ (MDAR Tyr1345Phe reverse) where the mutated nucleotides are underlined. The mutation was introduced using the QuikChange kit following the manufacturer’s protocol. The mutant plasmid encoding nucleotide sequence was confirmed by DNA sequencing. The plasmids containing the mutation were transformed into XL1-Blue supercompetent cells and stored at –80 °C in 85% glycerol. The expression and purification of the recombinant mutant MDAR protein were obtained as previously described for the wild-type MDAR protein.
In silico analysis of MDAR
The three-dimensional structure of pea peroxisomal MDAR was modelled at the Geno3D server (Combet et al., 2002) using as template the structures of the putidaredoxin reductase from Pseudomonas putida (PDB code access 1q1w and 1q1r) (Sevrioukova et al., 2004) and the ferredoxin reductase from Rhodopseudomonas palustris (PDB code access 3fg2) (Xu et al., 2009) with an identity of 26.8% and 30.1%, respectively. The analysis of the quality of the models was carried out at the Structural Analysis and Verification Server (SAVES) in terms of atomic non-local environment assessment (ANOLEA) (Melo and Feytmans, 1998), three-dimensional profiles (Verify3D) (Eisenberg et al., 1997), and Procheck (Laskowski et al., 1993). The co-ordinates of FAD and NAD were calculated by superposition of the model on those of the X-ray structure 2YVG, which shares 30% identity with the primary structure of the pea MDAR and 1.55 Å rms (backbone atoms) with the pea MDAR model.
Docking of the model of pea MDAR with GSH was carried out at the SwissDock sever (Grosdidier et al., 2011) in accurate mode and without defining the region of interest (blind docking) but allowing flexibility for the side chains within 5 Å of any atom of the ligand in its reference binding mode. Analysis of the results was carried out with the help of Swiss PdbViewer (Guex and Peitsch, 1997) and UCSF Chimera (Pettersen et al., 2004)
Molecular evolution studies were carried out at the Evolutionary Trace server (Mihalek et al., 2004) using the model of the tertiary structure as input. The phylogenetic significance and evolutionary conservation were explored by BLASTP searches (Altschul et al., 1997) on the subsection Viridiplantae of UniProtKB release 2013_05 (UniProt Consortium, 2011). The phosphorylation score was computed at the NetPhos 2.0 Server (Blom et al., 1999), and the solvent-accessible area by DSSP (Kabsch and Sande, 1983). The estimation of the pKa and the analysis of the interactions was carried out with Propka 3.1 (Olsson et al., 2011). The algorithm was originally described by Li et al. (2005) as a very fast empirical method for structure-based pKa prediction that relies on the estimation of desolvation effects and intraprotein interactions to account for the variation in the standard pKa of ionizable groups.
Biotin switch method
For in vitro S-nitrosylation, peroxisomal MDAR and chloroplastic and cytosolic GR were incubated with GSNO as the NO donor for 30min at room temperature. S-Nitrosylated recombinant proteins were subjected to the biotin switch method as described by Begara-Morales et al. (2014). The non-nitrosylated free cysteine residue was blocked by incubation with 30mM methyl methane thionsulphonate and 2.5% SDS at 50 °C for 20min with frequent vortexing. Residual methyl methane thionsulphonate was removed by precipitation with 2 vols of –20 °C acetone, and the proteins were resuspended in 0.1ml of HENS buffer (25mM HEPES pH 7.7 buffer containing 1mM EDTA, 0.1mM neocuproine, and 1% SDS) per milligram of protein. Biotinylation was obtained by adding 1mM N-[6-(biotinamido) hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP) and 0.1mM ascorbate, and incubating at room temperature for 1h. The proteins were then precipitated with 2 vols of –20 ºC acetone. Biotin-labelled proteins were separated by non-reducing 10% SDS–PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Immobilon P, Millipore, Bedford, MA, USA) using a semi-dry transfer system (Bio-Rad Laboratories). PVDF membranes were blocked using TRIS-buffered saline (TBS)+1% BSA. The blots were incubated with anti-biotin antibody at a dilution of 1:20 000 for 1h, and the immunoreactive bands were detected using a photographic film (Hyperfilm, Amersham Pharmacia Biotech) with an enhanced chemiluminescence kit (ECL-PLUS, Amersham Pharmacia Biotech).
Purification of biotinylated proteins and immunodetection of MDAR
Purification of biotinylated proteins from control and NaCl-treated pea plant leaves was carried out as described by Begara-Morales et al. (2014). Briefly, biotinylated proteins and 30 μl of neutravidin agarose 50% (w/v) slurry (high capacity neutravidin agarose resin, Thermo Scientific) per milligram of protein were equilibrated with a neutralization buffer [20mM HEPES pH 7.7 containing 100mM NaCl, 1mM EDTA, and 0.5% (v/v) Triton X-100]. Proteins were added to the neutravidin agarose matrix and were incubated 1h at room temperature with gentle shaking. The matrix with bound proteins was washed several times with washing buffer [20mM HEPES pH 7.7 containing 600mM NaCl, 1mM EDTA, and 0.5% (v/v) Triton X-100] and was transferred to an empty column. Finally, biotinylated proteins were eluted after incubation for 30min with elution buffer (20mM HEPES pH 7.7 containing 0.1M NaCl, 1mM EDTA, and 100mM β-mercaptoethanol) at room temperature. Purified biotinylated proteins were separated by 12% SDS–PAGE and transferred to PVDF membranes as described above.
For MDAR immunodetection, the membrane was incubated with a rabbit polyclonal antibody against cucumber MDAR (Sano and Asada, 1994) diluted 1:3000. Immunoreactive bands were detected using a photographic film (Hyperfilm, Amersham Pharmacia Biotech) with an enhanced chemiluminescence kit (ECL-PLUS, Amersham Pharmacia Biotech).
Real-time quantitative RT–PCR
Real-time quantitative reverse transcription–PCR (RT–PCR) was performed in 20 μl of reaction mixture, composed of 1 μl of cDNA, master mix IQ™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), and 10 pmol gene-specific forward and reverse oligonu cleotides (5′-AGAAGAATGCGAAAGCTGTGGTTGTTGGAG-3′ and 5′-TGCTTCCAGGACCCTACCATCCTTTAGTTT-3′, respec tively) for pea MDAR using a iCycler iQ system (Bio-Rad). Amplifications were performed under the following conditions: initial polymerase activation, 95 ºC for 5min; then 35 cycles of 30 s at 95ºC, 30 s at 62.2 ºC, and 1min at 72ºC; with a final extension at 72ºC for 7min. An internal control of 18S rRNA was used for the normalization with the following forward and reverse oligonucleotides: 5′-GTGCAACAAACCCCGACTTTTGAAGGATG-3′ and 5′-GTGGTAGCCGTTTCTCAGGCTCCCTCTC-3′.
Results
Expression and purification of recombinant pea MDAR and GR proteins: effect of peroxynitrite
In order to increase the knowledge of the regulatory mechanism of the ascorbate–glutathione cycle involved in the decomposition of H2O2, the recombinant proteins of two of its components, the MDAR and GR isoforms, were obtained by sequencing the pea clones and overexpression in E. coli (see the Materials and methods). Supplementary Fig. S1 at JXB online shows the electrophoretic analysis of the different fractions obtained after LYTRAP affinity column chromatography of recombinant MDAR and GR proteins. Recombinant MDAR (Supplementary Fig. 1A) showed a molecular mass of 68.6kDa, which is within the range of the theoretical value predicted for the peroxisomal MDAR protein (47.3kDa) with the Ly-tag (21.3kDa). The fractions E3–E5 with an MDAR activity of 1200 nmol NADH min–1 mg–1 protein showed an adequate grade of purity for this protein which was used for subsequent experiments. On the other hand, the recombinants chloroplastic and cytosolic GR (Supplementary Fig. 1B, C) showed a molecular mass of ~81kDa and 75kDa, respectively, which is within the range of the theoretical value predicted for both GR proteins (59.7kDa with the Ly-tag 21.3kDa for the chloroplastic isoform, and 53.7kDa with the Ly-tag 21.3kDa for the cytosolic isoform). The fractions E2–E4 with GR activities of 18 μmol NADPH min–1 mg–1 protein were used for subsequent experiments.
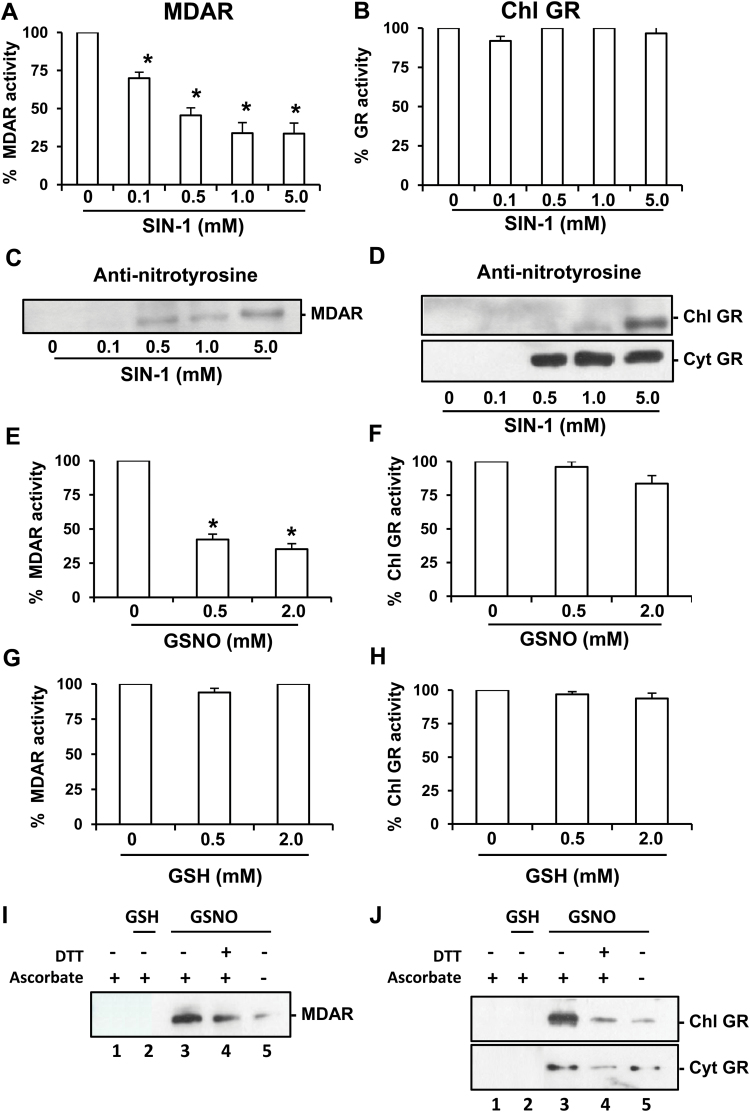
In order to evaluate the potential action of different NO-derived molecules, an in vitro assay was carried out in the presence of ONOO– using SIN-1 as the peroxynitrite donor (Chaki et al., 2009; Begara-Morales et al., 2014). Figure 1A depicts the inhibitory effect of ONOO– on MDAR activity in a dose-dependent manner that ranges from 30% with 0.1mM SIN-1 to 67% with 1mM and 5mM SIN-1, respectively. On the other hand, Fig. 1B shows that chloroplastic GR activity was not affected by any assayed concentration of ONOO–. Similar behaviour was observed for cytosolic GR (results not shown).
Fig. 1.
Effect of nitration and S-nitrosylation on recombinant monodehydroascorbate reductase (MDAR) and glutathione reductase (GR). Effect of SIN-1 (peroxynitrite donor) on recombinant MDAR (A) and GR (B) activities. Representative immunoblot showing the grade of tyrosine nitration of MDAR (C) and chloroplastic and cytosolic GR (D), treated with different concentrations of SIN-1 and detected with an antibody against 3-nitrotyrosine (dilution 1:2500). A 5 μg aliquot of protein was used per line. Effect of S-nitrosoglutathione (GSNO) on recombinant MDAR (E) and chloroplastic GR (F). Effect of glutathione (GSH) on recombinant MDAR (G) and chloroplastic GR (H). Purified MDAR and GR proteins were incubated at different concentrations of SIN-1 at 37 ºC for 60min, GSNO at 25 ºC for 30min, or GSH at 25 ºC for 30min. The specific activity of the recombinant MDAR was 1200 nmol NADH min–1 mg–1 and for GR proteins it was 18 μmol NADPH min–1 mg–1. S-Nitrosylation of recombinant MDAR (I) and chloroplastic and cytosolic GR (J). A 5 μg aliquot of purified recombinants proteins was treated with 2mM GSH and 2mM GSNO and was subjected to the biotin switch method. Control treatments were carried out with water (lane 1) and 2mM GSH (lane 2). Additionally, recombinants proteins were S-nitrosylated with 2mM GSNO (lane 3) and reduced again with 50mM DTT (lane 4). Furthermore, GSNO-treated recombinant proteins underwent the biotin switch method without ascorbate (lane 5). Proteins were separated under non-reducing conditions by SDS–PAGE and blotted onto a PVDF membrane. Biotinylated proteins were detected using an anti-biotin antibody. Data are means ±SEM of at least three replicates. *Differences from control values were significant at P<0.05.
The consistency of nitration by SIN-1 was confirmed by immunoblot analysis of the recombinant proteins using an antibody against 3-nitrotyrosine. Figure 1C and D show that the degree of nitration of both MDAR and GR isoforms increases as a function of the SIN-1 concentration.
Effect of S-nitrosylation of recombinant pea MDAR and chloroplastic and cytosolic GR
In order to gain additional insight into the regulation of pea MDAR and GR proteins, the effect of increasing concentrations of GSNO, a well-known NO donor (Begara-Morales et al., 2014), on the enzymatic activities was evaluated. As shown in Fig. 1E, 0.5mM and 2.0mM GSNO considerably inhibited MDAR activity by between 58% and 65%, respectively. In contrast, chloroplastic GR activity is not significantly affected (Fig. 1F). When both activities were assayed in the presence of 0.5mM and 2mM GSH to determine whether this effect was due to the release and binding of NO to the protein, GSH was found not to affect either MDAR or GR activities (Fig. 1G and H, respectively). Similar behaviour was observed for cytosolic GR (results not shown). This indicates that none of these enzymes is affected by S-glutathionylation. Furthermore, to show that GSNO treatment of recombinant MDAR and GR isoforms causes S-nitrosylation, the biotin switch assay method (Begara-Morales et al., 2014) was specifically used to detect S-nitrosylated proteins. Figure 1I and J shows that MDAR and chloroplastic and cytosolic GR are S-nitrosylated after treatment with 2mM GSNO (lane 3), whereas treatment with GSH does not produce any signal in the biotin switch assay (lane 2). As expected, S-nitrosylation of these proteins is reversible and can be down-regulated by adding a reducing agent such as DTT to the S-nitrosylated proteins (lane 4) or in the absence of ascorbate (lane 5) which is used as an SNO-specific reducing agent, further demonstrating S-nitrosylation.
Mapping tyrosine nitration sites and spectral characterization of nitrated pea peroxisomal MDAR
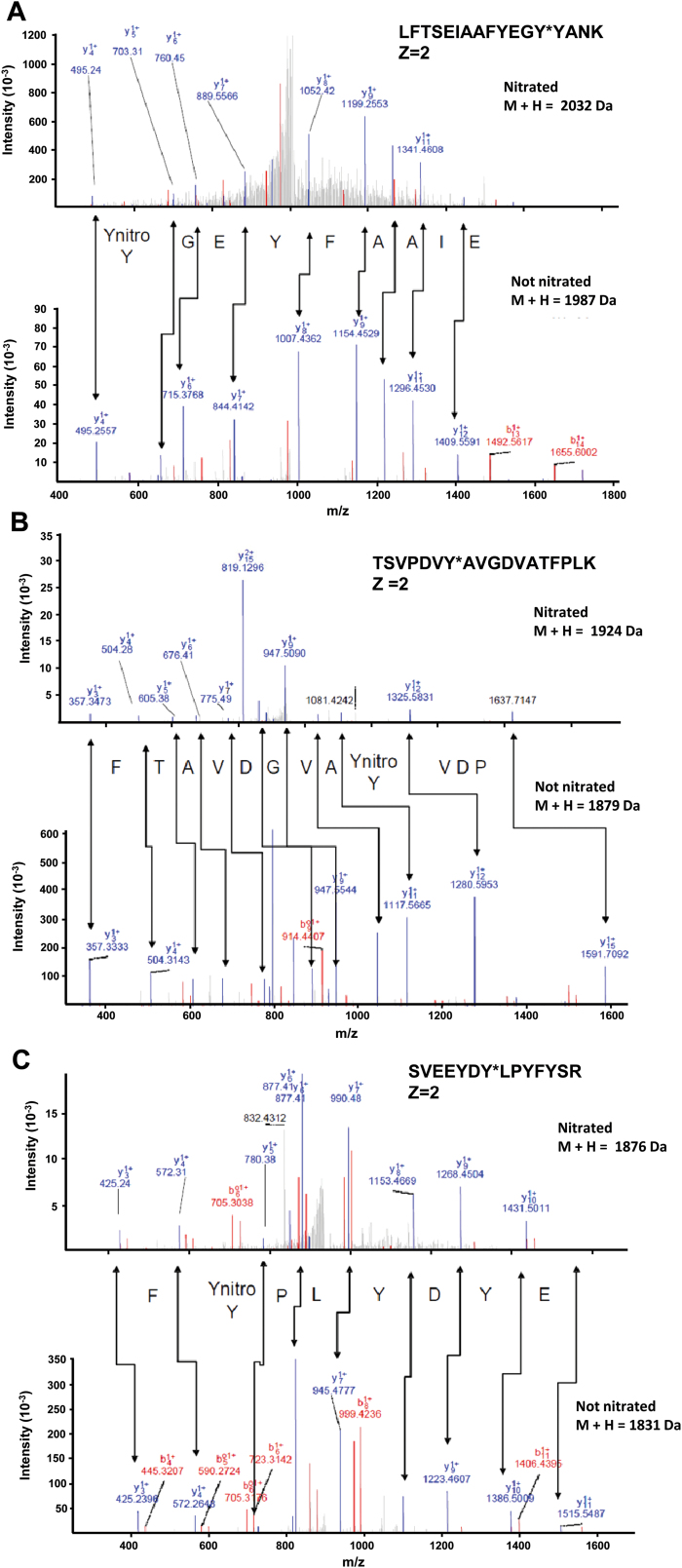
Given that GR is not affected by any of the PTMs mediated for NO-derived molecules, further study was conducted on MDAR. With the aim of identifying which of the 22 tyrosines present in the pea MDAR is(are) target(s) of this PTM, peroxynitrite-treated recombinant MDAR was subjected to trypsin digestion followed by MALDI-TOF/TOF mass spectrometry examination. Table 1 shows the list of peptides scanned and those identified by LC-MS/MS. Among the peptides identified, only three contained a nitrated tyrosine. Figure 2 shows the comparison of the nitrated (top) and unmodified (bottom) MS/MS spectra of these identified peptides from the pea MDAR. The nitrated peptide LFTSEIAAFYEGY*YANK (Z=2) has a total of 17 amino acids and a mass of 2032Da (1987Da plus 45Da) which is compatible with the acquisition of a nitro group in Tyr213 (Fig. 2A). The nitrated peptide TSVPDVY*AVGDVATFPLK (Z=2) has a total of 18 amino acids and a mass of 1924Da (1879Da plus 45Da) which is also compatible with the acquisition of a nitro group in Tyr292 (Fig. 2B). The nitrated peptide SVEEYDYLPY*FYSR (Z=2) has a total of 14 amino acids and a mass of 1876Da (1831Da plus 45Da) which is also compatible with the acquisition of a nitro group in Tyr345 (Fig. 2C).
Fig. 2.
Comparison of the nitrated (top) and unmodified (bottom) MS/MS spectra of the peptides identified from the pea peroxisomal MDAR in the corresponding panels: (A) LFTSEIAAFYEGY*YANK, (B) TSVPDVY*AVGDV ATFPLK, and (C) SVEEYDY*LPYFYSR. Peptide fragment ions are indicated by ‘b’ if the charge is retained on the N-terminus and by ‘y’ if the charge is maintained on the C-terminus. The subscript indicates the number of amino acid residues in the fragment studied from either the N-terminus or the C-terminus. The superscript indicates the charge (1+ or 2+) of the backbone fragmentation. (This figure is available in colour at JXB online.)
Modelling of pea MDAR and identification of potential residues affected by peroxynitrite and GSNO
The low identity of pea MDAR with the structures available from the PDB made homology modelling a difficult task. The best co-ordinates were obtained from Geno3D server (Combet et al., 2002) using as template the PDB entries 1q1w, 1q1r (26.8% identity), and 3fg2 (30.1% identity) (Sevrioukova et al., 2004; Xu et al., 2009). The model was refined to –18225.1 kcal mol–1 and comprises from residue 6 to residue 433 (98.8% coverage). The analysis of the quality of the model yields an Errat overall quality factor of 88.496, 81.82% of the residues with averaged 3D-1D score larger than 0.2, and 64.6% at the most favoured regions in the Ramachandran plot, 1.9% (i.e. seven residues) being at disallowed regions.
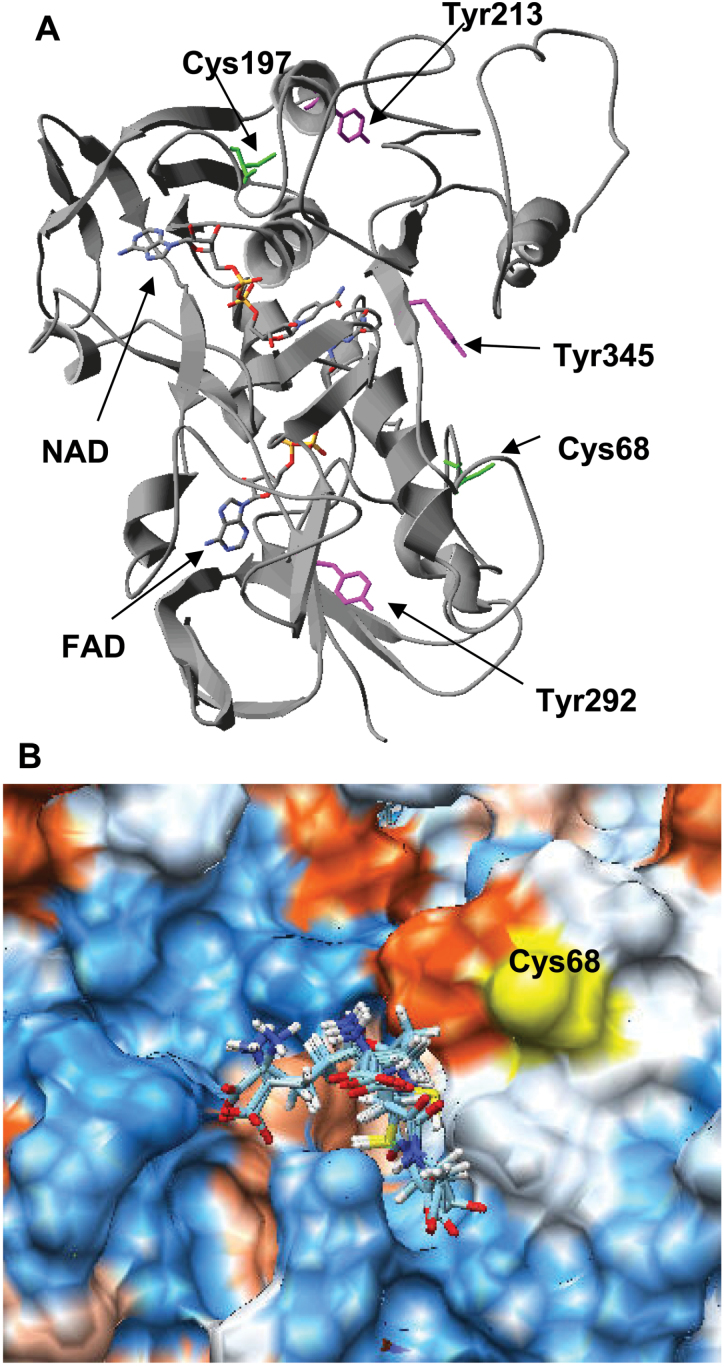
The location on the model of the pea MDAR of the three tyrosine residues identified as nitrated by LC-MS/MS and the two cysteine residues that are potential targets for GSNO did not provide any conclusive information regarding the modulation of the enzyme by tyrosine nitration and/or for S-nitrosylation (Fig. 3A). However, since MDAR plays an important role in homeostasis, it is reasonable to assume that its regulation has been preserved through evolution. On the other hand, the fact that the modulation takes place via the modification of only a few residues points to a particular reactivity of those residues. Bearing in mind both ideas, residues identified as potential targets of the PTM were further analysed. The results are summarized in Supplementary Table S1 at JXB online and reveal that Tyr292 is absolutely preserved and shows the lowest estimated pKa, Tyr213 is prone to phosphorylation, and Tyr345 is the most accessible. Cys197 is buried and less preserved that Cys68. Molecular docking with GSNO failed due to problems with the parameterization of the S–O bond, but when the calculations were carried out with GSH an area close to Cys68 was spotted where GSH fits with an estimated K d of 6.5 μM (Fig. 3B).
Fig. 3.
(A) Location of the tyrosine residues and cysteine residues susceptible to being responsible for the modulation of the enzymatic activity of pea MDAR by peroxynitrite and GSNO. (B) GSH binding site close to Cys68 located by blind docking. (This figure is available in colour at JXB online.)
Effect of SIN-1 on recombinant mutant pea MDAR (Tyr345Phe) obtained by site-directed mutagenesis analysis
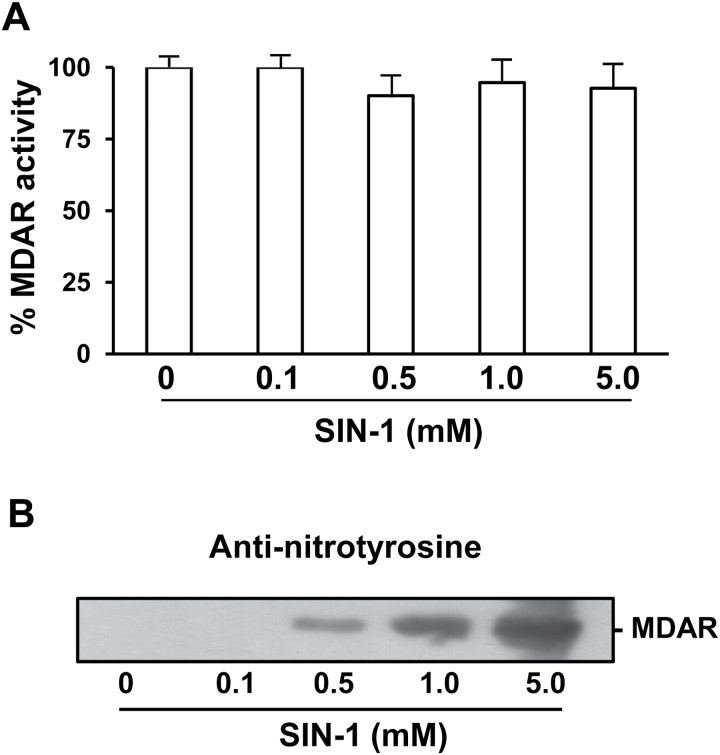
Of the nitrated tyrosines, in silico analysis outputs Tyr345 as the most likely candidate responsible for the observed inhibition by ONOO– since this residue is located at the active site and closely interacts with the cofactor. To corroborate this hypothesis, the Tyr345 residue in MDAR was replaced with phenylalanine by site-directed mutagenesis to obtain mutant MDAR Tyr345Phe. The overexpression and purification of the mutant were obtained as the recombinant wild type which yielded a functional enzyme resistant to inhibition by 0.5mM and 5.0mM ONOO– (Fig. 4A). Additionally, the nitration by SIN-1 of the mutant MDAR Tyr345Phe was confirmed by immunoblot analysis using an antibody against 3-nitrotyrosine (Fig. 4B) which shows that the degree of nitration increases as a function of the SIN-1 concentration. This allows the confirmation that the nitration of Tyr345 is the residue responsible for the inhibition of MDAR activity.
Fig. 4.
Effect of SIN-1 (peroxynitrite donor) on the recombinant mutant pea MDAR I (Tyr345Phe). (A) Effect of SIN-1 on recombinant MDAR activity. (B) Representative immunoblot showing the grade of tyrosine nitration of recombinant mutant pea MDAR and detected with an antibody against 3-nitrotyrosine (dilution 1:2500). Recombinant mutant pea MDAR I (Y345F) protein was incubated at different concentrations of SIN-1 at 37 ºC for 1h. Data are means ±SEM of at least three replicates.
Analysis of protein and gene expression of MDAR under salt-induced oxidative stress
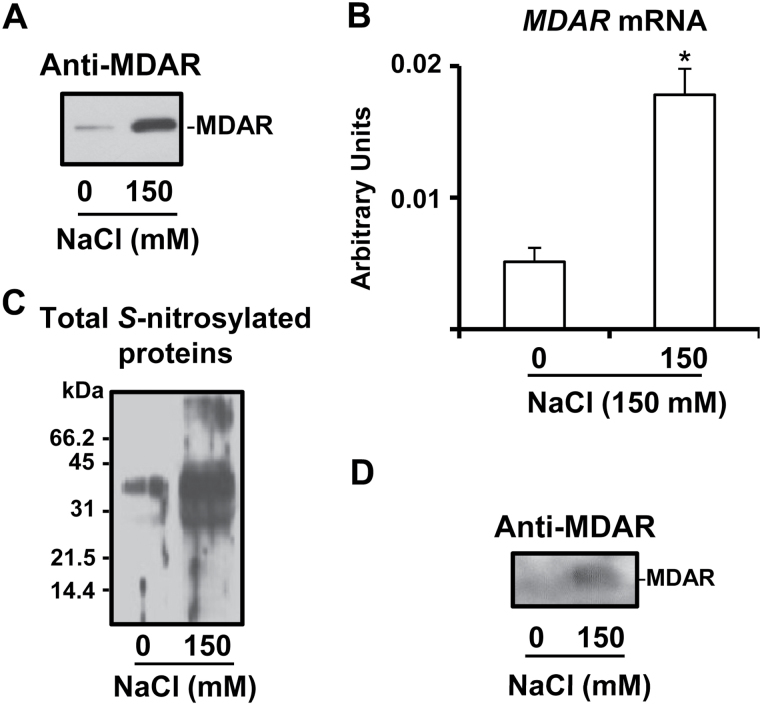
In order to gain additional insight into the physiological relevance of MDAR under an oxidative stress situation, it was analysed in leaves of pea plants grown in the presence of 150mM NaCl as was previously reported (Begara-Morales et al., 2014). Figure 5A shows by immunoblot analysis the MDAR protein expression which was found to increase clearly under salinity conditions. MDAR gene expression was also analysed and a similar behaviour to the protein expression was observed, with an increase of 3.5-fold under salt stress (Fig. 5B).
Fig. 5.
Protein and gene expression of MDAR and analysis of S-nitrosylated MDAR in leaves of pea plants under salinity (150mM NaCl) stress conditions. (A) Immunoblotting analysis of MDAR protein expression using an antibody against cucumber MDAR (dilution 1:3000). A 10 μg aliquot of protein was used per lane. (B) Real-time quantitative RT–PCR transcript analysis (arbitrary units) of the MDAR gene. Data are means ±SEM of at least four independent RNA samples. *Differences from control values were significant at P>0.05. (C) Detection of total S-nitrosylated proteins separated under non-reducing conditions by 12% SDS–PAGE and blotted onto a PVDF membrane. Biotinylated proteins were detected using anti-biotin antibodies as described in the Materials and methods. (D) Immunoblot of total S-nitrosylated proteins probed with an antibody against cucumber MDAR (dilution 1:3000). A 5 μg aliquot of protein was used per lane.
Purification of total S-nitrosylated proteins under salinity stress and detection of S-nitrosylated MDAR
To evaluate if MDAR under salinity stress conditions undergoes a process of S-nitrosylation, total S-nitrosylated proteins were purified from leaves of pea plants grown under control and salinity stress conditions and then the presence of MDAR protein among these S-nitrosylated protein was evaluated by immunoblotting. Figure 5D depicts the electrophoretic analysis of total S-nitrosylated proteins. Thus, under salinity stress, the pattern of S-nitrosylated proteins showed an increase in the number and in the intensity of some specific bands. Figure 5D shows the immunoblot analysis of the total S-nitrosylated proteins probed with an antibody against cucumber MDAR where an increase under salinity stress was also observed. Taken together, the results indicate that MDAR is S-nitrosylated in vivo and this process is increased under salinity conditions, which supports the data observed in in vitro conditions (Fig. 1I).
Discussion
PTMs such as nitration and S-nitrosylation mediated by NO-derived molecules are now considered to be crucial elements in the fine-tuned regulation of the function of their protein targets. In higher plants, proteomic analyses of nitration/S-nitrosylation have shown that a certain number of proteins are targets of PTMs mediated by NO-derived molecules (Lindermayr et al., 2005; Chaki et al., 2009; Álvarez et al. 2011; Lozano-Juste et al., 2011; Tanou et al., 2012; Begara-Morales et al., 2013a , b , 2014; Corpas et al., 2007, 2015). However, information on the specific impact of NO-PTMs on key proteins involved in antioxidative systems and the consequences for their functionality and protein structure is scarce (Yu et al., 2014).
Very recently, in vitro assays of several recombinant Arabidopsis superoxide dismutases (MnSOD1, FeSOD3, and CuZnSOD3) have shown that these SOD activities were not altered upon GSNO treatment but were inhibited to different degrees by ONOO– (Holzmeister et al., 2015). In this context, the ascorbate–glutathione pathway is one of the key antioxidant systems involved in the regulation of H2O2 levels in plant development and under abiotic stress conditions. In this respect, it has very recently been demonstrated that pea cytosolic APX, a key enzyme system in the antioxidant ascorbate–glutathione cycle, presents dual regulation by both tyrosine nitration and S-nitrosylation (Begara-Morales et al., 2014). Furthermore, several proteomic studies have identified GR and MDAR, other important components in the ascorbate–glutathione cycle, as targets of tyrosine nitration/S-nitrosylation processes (Chaki et al., 2009; Lin et al., 2012; Tanou et al., 2012). To elucidate the molecular mechanism and physiological relevance of these NO-PTMs to GR and MDAR functionality and their effect on the potential operation of the cycle, the present study analyses the regulation of MDAR and GR activities at the molecular level by using NO-derived molecules such as peroxynitrite and S-nitrosoglutathione that have the capacity to mediate tyrosine nitration and S-nitrosylation, respectively.
GR is an important enzyme in the antioxidative defense system that converts oxidized glutathione (GSSG) to reduced glutathione (GSH) using NADPH as cofactor. This reaction enables the GSH/GSSG ratio to be maintained at a high level, which is very important given that GSH is considered to be the most abundant soluble antioxidant in plants. This enzyme system has different isoenzymes located in a diverse range of cell compartments (Edwards et al., 1990; Romero-Puertas et al., 2006; Wu et al., 2013) and plays an important physiological role in maintaining and regenerating GSH in response to biotic and abiotic stresses in plants (Creissen et al., 1992; Foyer and Noctor, 2011; Leterrier et al., 2012; Gill et al., 2013; Signorelli et al., 2013). Under the present experimental conditions, the activities of either isoform tested was unaffected by any NO-PTM assayed and mediated by ONOO– and GSNO, suggesting that this could be a mechanism for maintaining GSH regeneration in order to sustain antioxidant capacity of the ascorbate–glutathione cycle against nitro-oxidative cell conditions. Moreover, it must be pointed out that, in the case of tyrosine nitration mediated by peroxynitrite, this behaviour is unusual as, to the authors’ knowledge, the pea GR is the first case of a nitrated protein found to be unaffected by this NO-PTM in higher plants as, up to now, most analyses have shown that nitration causes loss of function in all proteins identified in higher plants (Astier and Lindermayr, 2012; Corpas et al., 2013b ).
Given that pea GR was unaffected by NO-PTMs either for peroxynitrite or for GSNO, this study was focused on MDAR which is the enzyme in the ascorbate–glutathione cycle involved in the regeneration of reduced ascorbate. The pea (P. sativum) plant has only one gene encoding MDAR (Murthy and Zilinskas, 1994) whose corresponding protein has been immunolocalized in the different subcellular compartments including chloroplasts, peroxisomes, mitochondria, and the cytosol (Leterrier et al., 2005). As part of the ascorbate–glutathione cycle, MDAR also plays an important role under environmental stresses in which nitro-oxidative stress could be a significant component. In pea, although MDAR activity increased under high light intensity and cadmium, it was reduced by the herbicide 2,4-D (Leterrier et al., 2005). However, during the natural senescence of pea leaves, a simultaneous decrease in MDAR and APX activities has been reported (Jiménez et al., 1998). In tomato, MDAR activity is also increased by salinity (Mittova et al., 2003) and high light intensity (Gechev et al., 2003), in rice by low temperature (Oidaira et al., 2000), and in Arabidopsis by UV-B radiation (Kubo et al., 1999). However, in Arabidopsis, stresses such as high temperature (30 ºC), enhanced light intensity (200 μE m–2 s–1), water deficiency (water deprivation for 2 d), and low temperature (5 ºC) did not affect MDAR activity (Kubo et al., 1999).
As mentioned above, several proteomic studies have identified MDAR as a potential target for both S-nitrosylation and nitration (Lin et al., 2012; Tanou et al., 2012). However, the specific effects of these NO-PTMs on MDAR functions are not known. Thus, the present data show that both processes cause loss of MDAR function. Three putative candidates have been identified for tyrosine nitration and two cysteine residues are present in pea MDAR, but the fact that none of them is located in a relevant position from a functional point of view makes it difficult to understand the mechanism of inhibition and whether all of them are equally relevant. However since MDAR plays an important role in homeostasis and only a few residues are modified, it is reasonable to assume that those residues have been preserved through evolution and that they show a particular reactivity (Sano et al., 2004). For the particular case of tyrosine nitration, it is accepted that it proceeds though a radical mechanism where tyrosine cannot react directly with peroxynitrite but rather with carbon dioxide or a metal centre to yield a secondary oxidizing species that reacts with tyrosine to form the tyrosyl radical (Alvarez and Radi, 2003). Thus, from both evolutionary and chemical points of view, Tyr292 should be the strongest candidate for nitration, Tyr345 the weakest, and Tyr213 may be discarded and considered as a good candidate to undergo phophorylation. However, an analysis of the interaction network of the protein with its cofactors shows that only Tyr345 is involved (Table 2). Thus, the second interaction sphere of Tyr345 includes residues that directly interact with the atoms N3 (His313) and N10 (Glu45 and Asp296) from FAD (Table 2). Interestingly the prediction of the pKa values output by PropKa 3.1 for the ionizable groups present in FAD and NAD included in the model of pea MDAR reveals that atoms N3 and N10 from FAD show extreme anomalous values, the former being –1.72 (expected 5) and the latter 4.35 (expected 10). This analysis led to the hypothesis that nitration of Tyr345 should influence the functionality of MDAR. This hypothesis was confirmed by site-directed mutagenesis and, as expected, the functionality of the mutant MDAR Tyr345Phe was not affected by 5.0mM ONOO–.
Table 2.
Analysis of the first and second interaction spheres (in italics) of the three tyrosine residues identified as nitrated
The contribution of hydrogen bonding (involving side chains and backbone), coulombic interactions, and desolvation effects (regular, which is calculated according to Coulomb’s law, and RE, which includes all interactions between the ionizable residue and the remaining protein, apart from the Coulomb energy, that affects the deprotonation energy of the residue). Residues of the second interaction sphere that interact with any of the cofactors are shown in bold.
| Residue | pKa | Buried (%) | Desolvation effects | Hydrogen bond | Coulombic interaction | Atoms cofactor | ||
|---|---|---|---|---|---|---|---|---|
| Regular | RE | Side chain | Backbone | |||||
| Tyr345 G | 11.99 | 56 % | 0.90 438 | 0.00 0 | 0.21 Asp315 G | –0.02 Arg318 0.90 Asp315 |
||
| Asp315 | 2.63 | 97 | 3.82 554 | 1.17 0 |
–1.60 His313 G
–0.85 Lys319 G –0.20 Tyr342 G –0.21 Tyr345 G |
–0.08 Tyr345 G |
–0.10 Arg318
–0.32 His316 –1.13 His313 –1.66 Lys319 |
=>N3 FAD |
| Arg318 | 11.00 | 92 | –1.93 540 | 0.00 0 |
0.21 Glu45
0.04 Asp296 0.10 Asp315 0.02 Tyr345 |
=>N10 FAD =>N10 FAD |
||
| Tyr292 | 9.95 | 38 | 1.12 387 | 0.00 0 | –0.77 Lys285 G | 0.00 Xxx0 X | 0.08 Asp281 G 0.10 Asp290 –0.01 Lys102 –0.58 Lys285 |
|
| Asp281 | 3.91 | 32 | 1.47 370 | 0.13 0 |
–0.18 Asp281 G
–0.19 Lys285 G |
-0.08 Lys279
0.02 Glu 339 |
||
| Asp290 | 3.07 | 3 | 0.53 290 | 0.02 0 | –0.85 Lys102 G | –0.02 Asp290 G |
–0.03 Lys 285
–0.38 Lys102 |
|
| Lys102 | 11.07 | 0 | –0.67 268 | 0.00 0 | 0.85 Asp290 G |
0.01 Tyr292 G
0.38 Asp 290 |
||
| Lys285 | 10.17 | 45 | –2.04 406 | 0.00 0 | 0.77 Tyr 292 G |
0.34 Asp281 GX
0.03 Asp290 G 0.58 Tyr292 |
||
| Tyr213 | 11.13 | 23 | 0.86 347 | 0.00 0 | 0.27 Asp352 | |||
| Asp352 | 3.50 | 5 | 0.42 295 | 0.01 0 |
-0.52 Asp352 G
–0.09 Leu353 G |
–0.02 Lys217
–0.10 Arg349 |
||
The enzymatic activity of MDAR is also regulated by GSNO, and a link between MDAR and NO metabolism throughout iron metabolism has been reported. Specifically, plant oxyhaemoglobin (Hb) can act as an NO scavenger with the concomitant production of nitrate and Fe3+-Hb which is reduced to Fe2+-Hb in the presence of ascorbate and NADH, and MDAR activity is a key element in this process because it facilitates the regeneration of ascorbate (Igamberdiev et al., 2006; Wang and Hargrove, 2013). The sequence of MDAR comprises two cysteine residues and, according to Sano et al. (1995), one cysteine is more reactive towards DNTBA and it seems to participate in the reduction of the enzyme by NADH. As depicted in Supplementary Table S1 at JXB online, Cys68 seems to be the best candidate for S-nitrosylation because it is better preserved and more accessible. The identification was approached taking into account that S-nitrosylation by GSNO is a transnitrosylation reaction, an affinity between GSNO and the target proteins is expected, and hypothesizing that it may be detected in docking experiments. In fact it has been demonstrated that such an affinity is strong enough to be exploited to isolate targets proteins (Begara-Morales et al., 2013a ). Blind docking experiments with GSH found a region close to Cys68 where GSH fits with an estimated K d of 6.5 μM (Fig. 3B). This result is significant because amino acid sequence alignment reveals that Cys68 is equivalent to Cys117 of chloroplastic MDAR that has been reported to be involved in the activity and structural stability of chloroplastic MDAR (Li et al., 2010)
Additionally, both ONOO– and GSNO showed concentration-dependent inhibitory effects on MDAR activity, which suggests that these NO-derived molecules can have a fine-tuned regulatory effect depending on their cellular production and physiological and stress conditions. It is well known that peroxisomes have a remarkable oxidative metabolism. However, it must also be pointed out that peroxisomes are subcellular compartments where the presence of l-arginine-dependent nitric oxide synthase activity (Barroso et al., 1999), NO generation, and other NO-related species including ONOO– and GSNO has been demonstrated (Barroso et al., 2013a ; Corpas and Barroso, 2013). Therefore, peroxisomal MDAR could be modulated through loss of function when these molecules are overproduced under adverse conditions and could consequently contribute to a nitro-oxidative stress. In order to assess the in vivo relevance of the present study, salt stress (150mM NaCl) was applied to plants as an inducer of nitro-oxidative stress (Begara-Morales et al., 2014). It was found that MDAR expression (mRNA, protein, and activity) increased, which may reflect a mechanism to compensate the inhibitory effect of S-nitrosylation and nitration upon the enzyme in salt-stressed pea leaves. To date, other peroxisomal enzymes such as catalase (Clark et al., 2000) and NADH-dependent hydroxypyruvate reductase (Corpas et al., 2013a ) have been demonstrated to be targets of NO, which confirms the importance of NO in the peroxisomal metabolism.
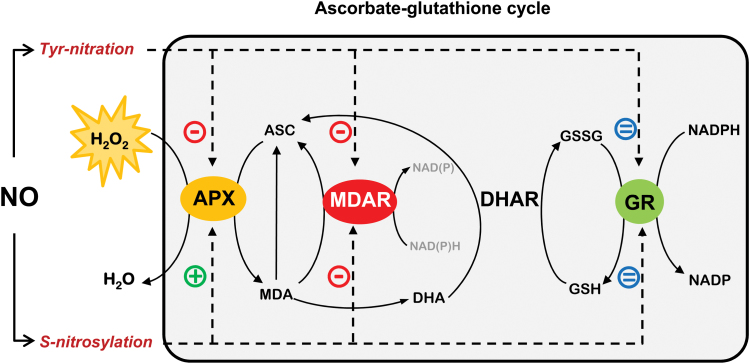
In summary, the present results provide new insights into the molecular mechanism involved in regulating MDAR and GR through PTMs mediated by NO-derived molecules and confirm the close involvement of NO and ROS metabolism in the antioxidant defence against nitro-oxidative stress situations in plants. These data, together with previous findings on the dual regulation of APX by S-nitrosylation/nitration (Begara-Morales et al., 2014), are summarized in Fig. 6. It shows the modulation of the antioxidative response of key enzymes in the ascorbate–glutathione cycle by NO-PTMs, where MDAR was deactivated by nitration and S-nitrosylation, which could compromise the cycle’s antioxidant capacity. However, GR was not affected by any of these NO-PTMs in an attempt to maintain the levels of GSH and the cellular redox state, suggesting that this could be a crucial mechanism to sustain the antioxidant capacity of the ascorbate–glutathione cycle against nitro-oxidative cell conditions.
Fig. 6.
Regulation of the ascorbate–glutathione cycle by nitric oxide (NO). NO modulates the ascorbate–glutathione cycle throughout post-translational modifications (PTMs) as tyrosine nitration and S-nitrosylation of APX and MDAR proteins. MDAR activity is reduced after both modifications, with APX activity also being reduced by tyrosine nitration. Under nitro-oxidative stress conditions, these modifications could compromise the antioxidant capacity of the cycle. However, APX activity is enhanced by S-nitrosylation while GR activity is not significantly affected by these NO-related PTMs. This behaviour suggests that APX and GR try to detoxify hydrogen peroxide and maintain regeneration of GSH, respectively, and consequently the cellular redox state to maintain the antioxidant resistance of the ascorbate–glutathione cycle against nitro-oxidative cell conditions. (This figure is available in colour at JXB online.)
Supplementary data
Supplementary data are available at JXB online.
Figure S1. SDS–PAGE analysis of the purification of the recombinant proteins.
Figure S2. Multiple alignment of the deduced amino acid sequences of MDAR in different plant species.
Table S1. Characterization in terms of evolutionary conservation, likelihood of tyrosine phosphorylation, solvent-accessible surface area, and estimated pKa of the three tyrosine and two cysteine residues susceptible to being responsible for the modulation of the enzymatic activity of pea MDAR by peroxynitrite and GSNO.
Acknowledgements
JBM would like to thank the Ministry of Science and Innovation for funding his PhD fellowship (F.P.U.). The study was supported by an ERDF grant co-financed by the Ministry of Economy and Competitiveness (BIO2012-33904) and the Junta de Andalucía (groups BIO286 and BIO192) in Spain. LC/MS/MS analyses were carried out at the Laboratorio de Proteómica LP-CSIC/UAB, a member of ProteoRed network.
Glossary
Abbreviations:
- NO
nitric oxide
- PTM
post-translational modification
- ROS
reactive oxygen species
- RNS
reactive nitrogen species.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25, 3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, Lozano-Juste J, Romero LC, García I, Gotor C, León J. 2011. Inhibition of Arabidopsis O-acetylserine(thiol)lyase A1 by tyrosine nitration. Journal of Biological Chemistry 286, 578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez B, Radi R. 2003. Peroxynitrite reactivity with amino acids and proteins. Amino Acids 25, 295–311. [DOI] [PubMed] [Google Scholar]
- Asada K. 1992. Ascorbate peroxidase: a hydrogen peroxide-scavenging enzyme in plants. Physiologia Plantarum 85, 235–241. [Google Scholar]
- Asada K. 2006. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiology 141, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier J, Kulik A, Koen E, Besson-Bard A, Bourque S, Jeandroz S, Lamotte O, Wendehenne D. 2012. Protein S-nitrosylation: what’s going on in plants? Free Radical Biology and Medicine 53, 1101–1110. [DOI] [PubMed] [Google Scholar]
- Astier J, Lindermayr C. 2012. Nitric oxide-dependent posttranslational modification in plants: an update. International Journal of Molecular Sciences 13, 15193–15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupiáñez JA, del Río LA. 1999. Localization of nitric-oxide synthase in plant peroxisomes. Journal of Biological Chemistry 274, 36729–36733. [DOI] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, et al. 2006. Localization of S-nitrosoglutathione and expression of S-nitrosoglutathione reductase in pea plants under cadmium stress. Journal of Experimental Botany 57, 1785–1793. [DOI] [PubMed] [Google Scholar]
- Begara-Morales JC, Chaki M, Sánchez-Calvo B, Mata-Pérez C, Leterrier M, Palma JM, Barroso JB, Corpas FJ. 2013. a Protein tyrosine nitration in pea roots during development and senescence. Journal of Experimental Botany 64, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begara-Morales JC, López-Jaramillo FJ, Sánchez-Calvo B, Carreras A, Ortega-Muñoz M, Santoyo-González F, Corpas FJ, Barroso JB. 2013. b Vinyl sulfone silica: application of an open preactivated support to the study of transnitrosylation of plant. BMC Plant Biology 13, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begara-Morales JC, Sánchez-Calvo B, Chaki M, Valderrama R, Mata-Pérez C, López-Jaramillo J, Padilla MN, Carreras A, Corpas FJ, Barroso JB. 2014. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. Journal of Experimental Botany 65, 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S, Brunak S. 1999. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. Journal of Molecular Biology 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernández-Ocaña AM, et al. 2009. Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. Journal of Experimental Botany 60, 4221–4234. [DOI] [PubMed] [Google Scholar]
- Chaki M, Valderrama R, Fernández-Ocaña AM, et al. 2011. High temperature triggers the metabolism of S-nitrosothiols in sunflower mediating a process of nitrosative stress which provokes the inhibition of ferredoxin-NADP reductase by tyrosine nitration. Plant, Cell and Environment 34, 1803–1818. [DOI] [PubMed] [Google Scholar]
- Clark D, Durner J, Navarre DA, Klessig DF. 2000. Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Molecular Plant-Microbe Interactions 13, 1380–1384. [DOI] [PubMed] [Google Scholar]
- Combet C, Jambon M, Deléage G, Geourjon C. 2002. Geno3D an automated protein modelling Web server. Bioinformatics 18, 213–214. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB. 2013. Nitro-oxidative stress vs oxidative or nitrosative stress in higher plants. New Phytologist 199, 633–635. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Begara-Morales JC, Sánchez-Calvo B, Chaki M, Barroso JB. 2015. Nitration and S-nitrosylation: two posttranslational modifications (PTMs) mediated by reactive nitrogen species and their role in signaling processes of plant cells. In: Gupta KJ, Igamberdiev AU, eds. Reactive oxygen and nitrogen species signaling and communications in plants. Signaling and communication in plants, Vol. 23 . Berlin: Springer, 267–281. [Google Scholar]
- Corpas FJ, Chaki M, Fernández-Ocaña A, Valderrama R, Palma JM, Carreras A, Begara-Morales JC, Airaki M, del Río LA, Barroso JB. 2008. Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant and Cell Physiology 49, 1711–1722. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, del Río LA, Barroso JB. 2007. Need of biomarkers of nitrosative stress in plants. Trends in Plant Science 12, 436–438. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Gómez M, Hernández JA, del Río LA. 1993. Metabolism of activated oxygen in peroxisomes from two Pisum sativum cultivars with different sensitivity to sodium chloride. Journal of Plant Physiology 141, 160–165. [Google Scholar]
- Corpas FJ, Leterrier M, Begara-Morales JC, et al. 2013. a Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochimica et Biophysica Acta 1830, 4981–4989. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, del Río LA, Barroso J. 2009. Evidence supporting the existence ofl-arginine-dependent nitric oxide synthase activity in plants. New Phytologist 184, 9–14. [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Palma JM, del Río LA, Barroso J. 2013. b Protein tyrosine nitration in higher plants grown under natural and stress conditions. Frontiers in Plant Science-Plant Proteomics 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creissen G, Edwards EA, Enard C, Wellburn A, Mullineaux P. 1992. Molecular characterization of glutathione reductase cDNAs from pea (Pisum sativum L.). The Plant Journal 2, 129–131. [PubMed] [Google Scholar]
- Daiber A, Bachschmid M, Beckman JS, Munzel T, Ullrich V. 2004. The impact of metal catalysis on protein tyrosine nitration by peroxynitrite. Biochemical and Biophysical Research Communications 317, 873–881. [DOI] [PubMed] [Google Scholar]
- Edwards EA, Rawsthonem S, Mullineaux PM. 1990. Subcellular distribution of multiple forms of glutathione reductase in leaves of pea (Pisum sativum L.). Planta 180, 278–284. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Lüthy R, Bowie JU. 1997. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods in Enzymology 277, 396–404. [DOI] [PubMed] [Google Scholar]
- Fares A, Rossignol M, Peltier JB. 2011. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochemical and Biophysical Research Communications 416, 331–336. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Halliwell B. 1976. The presence of glutathione and glutathione reductase in chloroplasts: a proposed role in ascorbic acid metabolism. Planta 133, 21–25. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2011. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology 155, 2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T, Willekens H, Van Montagu M, Inze D, Van Camp W, Toneva V, Minkov I. 2003. Different responses of tobacco antioxidant enzymes to light and chilling stress. Journal of Plant Physiology 160, 509–515. [DOI] [PubMed] [Google Scholar]
- Gill SS, Anjum NA, Hasanuzzaman M, Gill R, Trivedi DK, Ahmad I, Pereira E, Tuteja N. 2013. Glutathione and glutathione reductase: a boon in disguise for plant abiotic stress defense operations. Plant Physiology and Biochemistry 70, 204–212. [DOI] [PubMed] [Google Scholar]
- Groden D, Beck E. 1979. H2O2 destruction by ascorbate-dependent systems from chloroplasts. Biochimica et Biophysica Acta 546, 426–435. [DOI] [PubMed] [Google Scholar]
- Grosdidier A, Zoete V, Michielin O. 2011. SwissDock, a protein-small molecule docking web service based on EADOC DSS. Nucleic Acids Research 39, W270–W277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Holzmeister C, Gaupels F, Geerlof A, Sarioglu H, Sattler M, Durner J, Lindermayr C. 2015. Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. Journal of Experimental Botany 66, 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R. 2008. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiologia Plantarum 133, 211–228. [DOI] [PubMed] [Google Scholar]
- Hossain MA, Asada K. 1985. Monodehydroascorbate reductase from cucumber is a flavin adenine dinucleotide enzyme. Journal of Biological Chemistry 260, 12920–12926. [PubMed] [Google Scholar]
- Hossain MA, Nakano Y, Asada K. 1984. Monodehydroascorbate reductase in spinach chloroplast and its participation in regeneration of ascorbate for scavenging of hydrogen peroxide. Plant and Cell Physiology 25, 385–395. [Google Scholar]
- Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Wang Y, Zuo J. 2015. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiology 167, 1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Bykova NV, Hill RD. 2006. Nitric oxide scavenging by barley hemoglobin is facilitated by a monodehydroascorbate reductase-mediated ascorbate reduction of methemoglobin. Planta 223, 1033–1040. [DOI] [PubMed] [Google Scholar]
- Jiménez A, Hernández JA, Pastori G, del Rio LA, Sevilla F. 1998. Role of the ascorbate–glutathione cycle of mitochondria and peroxisomes in the senescence of pea leaves. Plant Physiology 118, 1327–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sande C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Keyster M, Klein A, Egbichi I, Jacobs A, Ludidi N. 2011. Nitric oxide increases the enzymatic activity of three ascorbate peroxidase isoforms in soybean root nodules. Plant Signaling and Behaviour 6, 956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Aono M, Nakajima N, Saji H, Tanaka K, Kondo N. 1999. Differential responses in activity of antioxidant enzymes to different environmental stresses in Arabidopsis thaliana. Journal of Plant Research 112, 279–290. [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography 26, 283–291. [Google Scholar]
- Leterrier M, Barroso JB, Valderrama R, Palma JM, Corpas FJ. 2012. NADP-dependent isocitrate dehydrogenase from Arabidopsis roots contributes in the mechanism of defence against the nitro-oxidative stress induced by salinity. Scientific World Journal 2012, 694740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leterrier M, Corpas FJ, Barroso JB, Sandalio LM, del Río LA. 2005. Peroxisomal monodehydroascorbate reductase. Genomic clone characterization and functional analysis under environmental stress conditions. Plant Physiology 138, 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Wu QY, Sun YL, Ma NN, Wang XY, Meng QW. 2010. Evidence that the amino acid residue Cys117 of chloroplastic monodehydroascorbate reductase is involved in its activity and structural stability. International Journal of Biological Macromolecules 46, 350–355. [DOI] [PubMed] [Google Scholar]
- Li H, Robertson AD, Jensen JH. 2005. Very fast empircal prediction and rationalization of protein pKa values. Proteins 61, 704–721. [DOI] [PubMed] [Google Scholar]
- Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, Hu B, Yang F, Loake GJ, Chu C. 2012. Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiology 158, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Saalbachm G, Bahnweg G, Durner J. 2006. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. Journal of Biological Chemistry 281, 4285–4291. [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J. 2005. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiology 137, 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J, Colom-Moreno R, León J. 2011. In vivo protein tyrosine nitration in Arabidopsis thaliana . Journal of Experimental Botany 62, 3501–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F, Feytmans E. 1998. Assessing protein structures with a non-local atomic interaction energy, Journal Molecular Biology 227, 1141–1152. [DOI] [PubMed] [Google Scholar]
- Mihalek I, Res I, Lichtarge O. 2004. A family of evolution–entropy hybrid methods for ranking protein residues by importance. Journal of Molecular Biology 336, 1265–1282. [DOI] [PubMed] [Google Scholar]
- Mittova V, Tal M., Volokita M, Guy M. 2003. Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii . Plant, Cell and Environment 6, 845–856. [DOI] [PubMed] [Google Scholar]
- Murthy SS, Zilinskas BA. 1994. Molecular cloning and characterization of a cDNA encoding pea monodehydroascorbate reductase. Journal of Biological Chemistry 269, 31129–31133. [PubMed] [Google Scholar]
- Noctor G, Foyer CH. 1998. Ascorbate and glutathione: keeping active oxygen under control. Annual Review of Plant Physiology and Plant Molecular Biology 49, 249–279. [DOI] [PubMed] [Google Scholar]
- Oidaira H, Sano S, Koshiba T, Ushimaru T. 2000. Enhancement of antioxidative enzyme activities in chilled rice seedlings. Journal of Plant Physiology 156, 811–813. [Google Scholar]
- Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JJ. 2011. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. Journal of Chemical Theory and Computation 7, 525–537. [DOI] [PubMed] [Google Scholar]
- Palma JM, Jiménez A, Sandalio LM, Corpas FJ, Lundqvist M, Gómez M, Sevilla F, del Río LA. 2006. Antioxidative enzymes from chloroplasts, mitochondria, and peroxisomes during leaf senescence of nodulated pea plants. Journal of Experimental Botany 57, 1747–1758. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Radi R. 2013. Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Accounts of Chemical Research 46, 550–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S, Corpas FJ. 2010. The peroxisomal ascorbate–glutathione pathway: molecular identification and insights into its essential role under environmental stress conditions. In: Anjum NA, Chan M-T, Umar S, eds. Ascorbate–glutathione pathway and stress tolerance in plants . Dordrecht, The Netherlands: Springer, 387–404. [Google Scholar]
- Romero-Puertas MC, Corpas FJ, Sandalio LM, Leterrier M, Rodríguez-Serrano M, del Río LA, Palma JM. 2006. Glutathione reductase from pea leaves: response to abiotic stress and characterization of the peroxisomal isozyme. New Phytologist 170, 43–52. [DOI] [PubMed] [Google Scholar]
- Sano S, Asada K. 1994. cDNA cloning of monodehydroascorbate radical reductase from cucumber: a high degree of homology in terms of amino acid sequence between this enzyme and bacterial flavoenzymes. Plant and Cell Physiology 35, 425–437. [PubMed] [Google Scholar]
- Sano S, Kang YN, Shigemizu H, Morishita N, Yoon HJ, Saito K, Asada K, Mikami B. 2004. Crystallization and preliminary crystallographic analysis of monodehydroascorbate radical reductase from cucumber. Acta Crystallographica Section D: Biological Crystallography 60, 1498–1499. [DOI] [PubMed] [Google Scholar]
- Sano S, Miyake C, Mikami B, Asada K. 1995. Molecular characterization of monodehydroascorbate radical reductase from cucumber highly expressed in Escherichia coli . Journal of Biological Chemistry 270, 21354–21361. [DOI] [PubMed] [Google Scholar]
- Sevrioukova IF, Li H, Poulos TL. 2004. Crystal structure of putidaredoxin reductase from Pseudomonas putida, the final structural component of the cytochrome P450cam monooxygenase. Journal of Molecular Biology 336, 889–902. [DOI] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. 2002. Regulation and function of ascorbate peroxidase isoenzymes. Journal of Experimental Botany 53, 1305–1319. [PubMed] [Google Scholar]
- Signorelli S, Corpas FJ, Borsani O, Barroso JB, Monza J. 2013. Water stress induces a differential and spatially distributed nitro-oxidative stress response in roots and leaves of Lotus japonicus . Plant Science 201–202, 137–146. [DOI] [PubMed] [Google Scholar]
- Souza JM, Peluffo G, Radi R. 2008. Protein tyrosine nitration–functional alteration or just a biomarker? Free Radical Biology and Medicine 45, 357–366. [DOI] [PubMed] [Google Scholar]
- Stevens RG, Creissen GP, Mullineaux PM. 1997. Cloning and characterisation of a cytosolic glutathione reductase cDNA from pea (Pisum sativum L.) and its expression in response to stress. Plant Molecular Biology 35, 641–654. [DOI] [PubMed] [Google Scholar]
- Tanou G, Filippou P, Belghazi M, Job D, Diamantidis G, Fotopoulos V, Molassiotis A. 2012. Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. The Plant Journal 72, 585–599. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. 2011. Ongoing and future developments at the universal protein resources. Nucleic Acids Research 39, D214–D219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hargrove MS. 2013. Nitric oxide in plants: the roles of ascorbate and hemoglobin. PLoS One 8, e82611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TM, Lin WR, Kao YT, Hsu YT, Yeh CH, Hong CY, Kao CH. 2013. Identification and characterization of a novel chloroplast/mitochondria co-localized glutathione reductase 3 involved in salt stress response in rice. Plant Molecular Biology 83, 379–390. [DOI] [PubMed] [Google Scholar]
- Xu F, Bell SG, Peng Y, Johnson EO, Bartlam M, Rao Z, Wong LL. 2009. Crystal structure of a ferredoxin reductase for the CYP199A2 system from Rhodopseudomonas palustris . Proteins 88, 867–880. [DOI] [PubMed] [Google Scholar]
- Yu M, Lamattina L, Spoel SH, Loake GJ. 2014. Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytologist 202, 1142–1156. http://www.ncbi.nlm.nih.gov/pubmed/24611485 [DOI] [PubMed] [Google Scholar]
- Yun BW, Feechan A, Yin M, et al. 2011. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.