Highlight
A brown planthopper resistance recessive gene, BPH29, was cloned which contained a B3 DNA-binding domain and conferred resistance by a mechanism that was similar to plant defence against pathogens.
Key words: B3 domain protein, BPH29, brown planthopper resistance, map-based cloning, Oryza sativa L., plant–insect defence, recessive gene.
Abstract
Rice (Oryza sativa L.) production, essential for global food security, is threatened by the brown planthopper (BPH). The breeding of host-resistant crops is an economical and environmentally friendly strategy for pest control, but few resistance gene resources have thus far been cloned. An indica rice introgression line RBPH54, derived from wild rice Oryza rufipogon, has been identified with sustainable resistance to BPH, which is governed by recessive alleles at two loci. In this study, a map-based cloning approach was used to fine-map one resistance gene locus to a 24kb region on the short arm of chromosome 6. Through genetic analysis and transgenic experiments, BPH29, a resistance gene containing a B3 DNA-binding domain, was cloned. The tissue specificity of BPH29 is restricted to vascular tissue, the location of BPH attack. In response to BPH infestation, RBPH54 activates the salicylic acid signalling pathway and suppresses the jasmonic acid/ethylene-dependent pathway, similar to plant defence responses to biotrophic pathogens. The cloning and characterization of BPH29 provides insights into molecular mechanisms of plant–insect interactions and should facilitate the breeding of rice host-resistant varieties.
Introduction
Rice (Oryza sativa L.), an important staple food for >3 billion people worldwide (Khush, 1997), is currently beset by multiple threats. Plant domestication has presumably narrowed crop genetic diversity and decreased resistance to abiotic and biotic stresses, leading to potential ‘broad susceptibility’ (Chaudhary, 2013).
The brown planthopper (BPH), Nilaparvata lugens Stål (Homoptera: Delphacidae), is a monophagous herbivore of rice and causes heavy economic losses throughout Asia. These insects feed mainly on stems, and account for 28% of total plant dry matter reduction (Sōgawa, 1994). Feeding by low-density BPH populations can reduce rice yields (Watanabe et al., 1997), while heavy infestation causes ‘hopperburn’ (Sōgawa, 1982). The BPH is also a vector of viruses responsible for ragged stunt and grassy stunt diseases. Control of BPHs using chemical pesticides is expensive, harmful to natural predators, and conducive to resistance build up in pests (Tanaka et al., 2000). As such, development of insect-resistant crop varieties is of interest as an economical, environmentally friendly alternative strategy (Khush, 2001).
Beginning with the study of Pathak et al. (1969), considerable effort has been expended in the search for rice host resistance to BPHs. To date, 28 BPH resistance genes have been detected (Wu et al., 2014) and mapped to six of the 12 rice chromosomes (2, 3, 4, 6, 11, and 12) (Cheng et al., 2013). Among them, only Bph14 has been cloned (Du et al., 2009). Resistant rice varieties inhibit BPH oviposition and lower nymph survival rates, with little or no physiological stress or ensuing yield loss (Alam and Cohen, 1998b ; Alagar et al., 2007). Additional BPH resistance resources are consequently needed to support sustained control of these pests. Furthermore, studies of novel BPH resistance genes may provide more details of the molecular mechanisms underlying plant resistance to these insects.
In response to insect predation, plants have evolved a sophisticated system of defence mechanisms to deter herbivores. Two layers of the plant immune system perceive various invaders through different classes of immune receptors. Resistance (R) proteins which function in the second layer can effectively recognize specific pathogens that break through the first layer, and activate effector-triggered immunity (ETI) (Jones and Dangl, 2006). Most R genes which have been identified encode polymorphic ‘nucleotide-binding site plus leucine-rich repeat’ (NB-LRR) domains (Dangl and Jones, 2001). In addition, plant innate immunity involves the activation of expression changes to hormones such as abscisic acid, jasmonic acid (JA), salicylic acid (SA), and ethylene that mediate signalling cross-talk (Thompson and Goggin, 2006; Chen and Ronald, 2011; Consales et al., 2012) and play a role in defence regulation against different pathogens (Thomma et al., 1998). Pathogen response is usually regulated by an SA-dependent pathway and is associated with systemic acquired resistance (SAR), whereas the wounding response is usually controlled by a JA/ethylene-dependent pathway (Felton and Korth, 2000).
Previously, two BPH resistance genes, bph20(t) and bph21(t), were identified in RBPH54, a rice introgression line derived from wild rice Oryza rufipogon (Yang et al., 2011). Genetic segregation in the F2 generation showed a ratio of 1:15, implying that the resistance was governed by recessive alleles at two loci. Examination of the BC1 generation also confirmed the duplicate interaction between bph20(t) and bph21(t). bph20(t) was mapped on the short arm of chromosome 6, and bph21(t) on the short arm of chromosome 10. The gene designations, bph20(t) and bph21(t), conflict with those of Rahman et al. (2009), who used the same names for genes that mapped to definitely different loci. To avoid confusion, the gene names BPH29 and BPH30 are proposed as replacements for bph20(t) and bph21(t), respectively, in accordance with the new CGSNL nomenclature system for rice (McCouch, 2008).
In this study, it was sought to fine-map further and clone BPH29 using a map-based cloning approach. Seedling tests were also used to confirm the crucial function of BPH29 in RBPH54 resistance to BPH. BPH29 encodes a B3 DNA-binding domain-containing protein and is specifically restricted to vascular tissue, the site of BPH predation. In a BPH infestation experiment, RBPH54 defence responses included activation of the SA signalling pathway and suppression of the JA/ethylene-dependent pathway. The results provide a BPH resistance gene resource and offer valuable information regarding plant responses to herbivore pressure.
Materials and methods
Plant materials and growth conditions
Three rice sources, RBPH54, Taichuang Native 1 (TN1), and TR539, were used in this study. RBPH54 is an indica introgression line with BPH resistance derived from the wild rice species O. rufipogon Griff.; it has proved to be highly resistant to BPH biotypes 1 and 2 and resistant to biotype Bangladesh (Yang et al., 2011). TN1, used as a control, is an international BPH-susceptible indica variety. TR539, containing BPH29, BPH30, and a few non-target background introgressions, is a line selected from a BC3F2 population constructed using TN1 as the recurrent parent and RBPH54 as the donor parent. All rice materials were grown in a greenhouse at 28 °C under 14h light/10h dark conditions or in test fields in Shanghai (31°11′N, 121°29′E) and Sanya (18°14′N, 109°31′E), China.
Mapping population development and marker design
Near isogenic lines (NILs) were constructed by backcrossing TR539 to TN1 for two generations, followed by selfing to eliminate non-target introgressed genomic regions. From these NILs, one NIL heterozygous for the target locus BPH29 (identified by markers BYL7 and BYL8), and recessive homozygous for BPH30 (identified by markers RM222 and RM244) was selected and selfed to generate an NIL-F2 segregating population. NIL-F2 progeny were screened using insertion deletion (InDel) markers between BYL7 and BYL8, and recombinant plants were selected for further fine mapping of BPH29. Recombinant lines were evaluated for BPH resistance at the seedling stage by infestation with BPH biotype 2.
InDel markers for mapping were designed on the basis of sequence differences between Japonica rice Nipponbare (http://rgp.dna.affrc.go.jp/) and indica cultivar 9311 (http://rise.genomics.org.cn), using the online primer design tool Primer3 version 4.0 (http://primer3.ut.ee/).
Gene transformation in rice
The binary plasmid vector pCAMBIA1304 (Center for the Application of Molecular Biology to International Agriculture) was assembled for the gene transformation. The pCAMBIA1304 vector carried kanamycin and hygromycin resistance for bacterial and transformed plant selection, respectively. Dehusked seeds of rice (RBPH54) were surface-sterilized by soaking in 70% (v/v) ethanol for 10min followed by 0.1% (w/v) HgCl2 for 20min, washed with sterile distilled water, and sown on gel medium for embryogenic callus induction. After bombardment with a gene gun, calli were selected after a 2 week exposure to 50mg l−1 hygromycin. Hygromycin-resistant calli regenerated and grew into transformed T0 lines. For each gene transformation, 10–20 independent T0 lines were collected. Genomic DNAs were extracted from these transformed lines as templates for PCR, and the hygromycin gene was then amplified to confirm the transformation.
BPH resistance bioassay
BPHs were collected from fields in Nanning (22°48′N, 108°22′E) and females were reared separately. F1 lines were evaluated for BPH biotype. Insects of BPH biotype 2 were selected and maintained on TN1 under greenhouse conditions (25–30 °C) at the Plant Protection Research Institute, Guangxi Academy of Agricultural Sciences.
The BPH resistance of rice plants was evaluated by the modified standard seed-box screening technique (Brookes and Barfoot, 2003). About 20 seeds of each rice line were sown in a row in a metal box (80×55×8cm) along with resistant control RBPH54 and susceptible control TN1. When seedlings reached the three-leaf stage, they were infested with first- to second-instar BPHs (biotype 2) at a density of 10 insects per seedling. When all the TN1 seedlings had died, damage to seedlings was scored as 0, 1, 3, 5, 7, or 9 according to International Rice Research Institute (IRRI) guidelines (IRRI, 1988). Lower scores indicate higher resistance to BPHs.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNAs of different rice lines and tissues were isolated using an RNAprep plant kit (Tiangen, Beijing, China). First-strand cDNA was generated using a Primescript RT reagent kit (Perfect Real Time; Takara, Otsu, Shiga, Japan). qRT-PCR analysis was carried out using the SYBR Premix Ex Taq (Takara) on a CFX96 Real-Time system (Bio-Rad). Samples were amplified by two-step qRT-PCR, which was 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Melting curves and standard curves were calculated and analysed, with relative mRNA expression levels normalized using OsActin1 as a reference. Primers used in this study are shown in Supplementary Table S2 available at JXB online.
Subcellular localization
The full-length coding region of BPH29 was cloned from RBPH54 cDNA using G5 Vector primer (Supplementary Table S2 at JXB online), assembled into the vector pCAMBIA1304, and fused in-frame with green fluorescent protein (GFP) under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. The modified pCAMBIA1304 construct and control were transiently transformed into rice protoplasts. The rice protoplasts were prepared from three-leaf stage plant leaves and transformed using 40% polyethylene glycol solution (Yoo et al., 2007). After 16–24h incubation at 28 °C in darkness, the protoplast location of the fusion protein was observed by confocal fluorescence microscopy (LSM A710; Zeiss).
β-Glucuronidase expression analysis
A 1.5kb 5′-upstream (−1500 to −1 for BPH29-ATG and 29 818 to 31 317 for P0514G12) segment of the BPH29 promoter region was amplified with the G5 promoter primer (Supplementary Table S2 at JXB online) and cloned into a promoterless pCAMBIA1301 vector followed by the β-glucuronidase (GUS) reporter gene. This PBPH29::GUS fusion vector was introduced into RBPH54, and 30–40 independent positive T0 lines were collected for subsequent experiments. GUS activity was detected by histochemical GUS staining (Jefferson et al., 1987) and observed by light microscopy.
Hormone measurements
About 40 seeds of each rice line were sown in soil and grown to the three-leaf stage, and then the seedlings were infected with first- to second-instar BPHs (biotype 2) at a density of 10 insects per seedling. Rice leaves were harvested 0, 24, and 48h after BPH infestation, frozen in liquid nitrogen, and ground to a fine powder for hormone measurements. Quantification of endogenous SA and JA was performed as described in Chen et al. (2012).
Accession numbers
Sequence data from this article can be found in the EMBL/Gen Bank data libraries under accession numbers: LOC_Os06g01820, LOC_Os06g01830, LOC_Os06g01840, LOC_Os06g01850, LOC_Os06g01860, LOC_Os06g01870, KC019172, KC019173, Os03g0718100, D38170, X87946, X89859, AY923983, AY062258, D14000, and AY396568.
Results
Fine mapping of BPH29
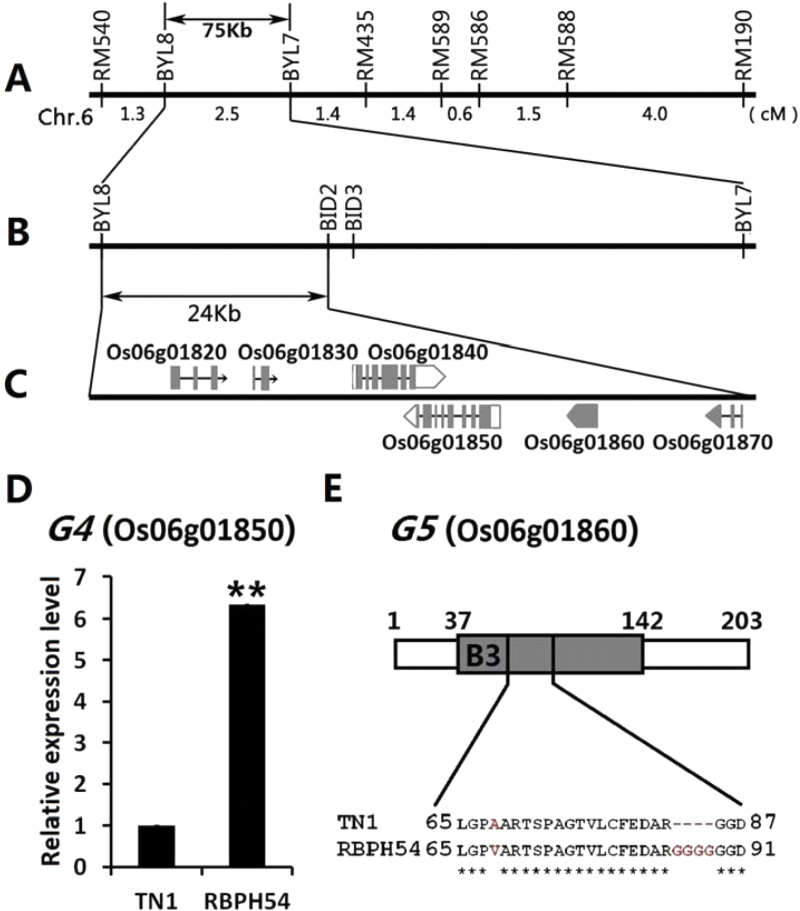
To narrow down the BPH29 locus region, a NIL-F2 segregating population which is heterozygous for the target locus BPH29 and recessive homozygous for BPH30 was developed. A total of 3435 NIL-F2 progeny were subjected to InDel analysis with markers between BYL7 and BYL8 (Fig. 1A). Out of five InDel markers, BID2 and BID3 exhibited polymorphism between the introgression line parents. As a result, 32 recombinants were identified between BYL8 and BID2; 18 of these restricted BPH29 to the region downstream of BYL8, while the remaining 14 localized the gene to the region upstream of BID2 (Table 1). For each of the 32 recombinant families, 20 homozygous selfed progeny (F2:3 family) were genotyped by the appropriate segregating markers and analysed for resistance to BPH. BLAST analysis revealed that a PAC (P1-derived artificial chromosome) clone, P0514G12, was anchored by BYL8 and BID2. In this fashion, BPH29 was confined to a 24kb region flanked by BYL8 and BID2 (Fig. 1B).
Fig. 1.
Fine mapping and prediction of the BPH29 gene. (A) BPH29 location in a 75kb region between BYL8 and BYL7 on the short arm of chromosome 6 as determined previously. (B) Further refinement of the locus region to a 24kb location between BYL8 and BID2. (C) The six predicted genes (LOC_Os06g01820, LOC_Os06g01830, LOC_Os06g01840, LOC_Os06g01850, LOC_Os06g01860, and LOC_Os06g01870) annotated in this region. (D) Quantitative real-time PCR (qRT-PCR) analysis of G4 (LOC_Os06g01850) expression in rice lines Taichuang Native 1 (TN1) and RBPH54. Student’s t-test: **P<0.01. Data are means ±SD (n=3 individuals), and expression levels are shown relative to that of OsActin1. (E) Protein structure of G5 (LOC_Os06g01860) annotated by Pfam (http://pfam.xfam.org/). The B3 DNA-binding domain is indicated by a grey box. Differences in G5 between TN1 and RBPH54 amino acid sequences include a single mutation and a four-glycine insertion. Identical amino acids are marked with a single asterisk.
Table 1.
Genotypes of key recombinants from the NIL-F2 population
| Flanked marking | Individuala | Chromosome | Markersb | |||
|---|---|---|---|---|---|---|
| RM540 | BYL8 | BID2 | BYL7 | |||
| BYL8 | BPH-132 | 6 | A | A | B | B |
| BPH-133 | 6 | A | A | B | B | |
| BPH-134 | 6 | A | A | B | B | |
| BPH-135 | 6 | A | A | B | B | |
| BPH-136 | 6 | A | A | B | B | |
| BPH-138 | 6 | A | A | B | B | |
| BPH-139 | 6 | A | A | B | B | |
| BPH-140 | 6 | A | A | B | B | |
| BPH-141 | 6 | A | A | B | B | |
| BPH-142 | 6 | A | A | B | B | |
| BPH-416 | 6 | A | A | B | B | |
| BPH-417 | 6 | A | A | B | B | |
| BPH-419 | 6 | A | A | B | B | |
| BPH-1097 | 6 | A | A | B | B | |
| BPH-1100 | 6 | A | A | B | B | |
| BPH-1103 | 6 | A | A | B | B | |
| BPH-1104 | 6 | A | A | B | B | |
| BPH-1106 | 6 | H | H | B | B | |
| BID2 | BPH-49 | 6 | B | B | H | H |
| BPH-91 | 6 | B | B | A | A | |
| BPH-110 | 6 | B | B | A | A | |
| BPH-143 | 6 | B | B | H | H | |
| BPH-151 | 6 | B | B | H | H | |
| BPH-159 | 6 | B | B | H | H | |
| BPH-162 | 6 | B | B | H | H | |
| BPH-172 | 6 | B | B | H | H | |
| BPH-183 | 6 | B | B | H | H | |
| BPH-211 | 6 | B | B | H | H | |
| BPH-246 | 6 | B | B | H | H | |
| BPH-251 | 6 | B | B | A | A | |
| BPH-254 | 6 | B | B | A | A | |
| BPH-259 | 6 | B | B | H | H | |
a For each of the 32 recombinant families, 20 homozygous selfed progeny (F2:3 family) were evaluated for BPH resistance at the seedling stage by infestation with BPH biotype 2 and identified as resistant to BPH.
b A, TN1 homozygous genotype; B, RBPH54 homozygous genotype; H, heterozygous genotype.
Analysis of candidate genes
Six genes (LOC_Os06g01820, LOC_Os06g01830, LOC_Os06g01840, LOC_Os06g01850, LOC_Os06g01860, and LOC_Os06g01870) were predicted in the 24kb target region based on annotations in the TIGR Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/) (Kawahara et al., 2013) (Fig. 1C). LOC_Os06g01850 and LOC_Os06g01860 showed functional differences between the susceptible parent TN1 and the resistant parent RBPH54, and were designated as G4 and G5, respectively.
An 11bp insertion was present in the 505bp upstream region of the G4 start codon of RBPH54 compared with TN1. This regulatory region difference may have an impact on G4 expression levels, and remarkably high levels of G4 expression were detected in RBPH54 by qRT-PCR (Fig. 1D). Differences of G5 were found in its coding regions. These differences included three single nucleotide polymorphism, resulting in one single amino acid mutation from alanine to valine, and a 12bp insertion corresponding to four glycines in RBPH54 (Fig. 1E; Supplementary Fig. S1 at JXB online). These G5 sequence discrepancies between the two mapping parents may affect the structure and function of the encoded protein. On the basis of these observations, G4 and G5 were selected as candidates for the BPH resistance gene BPH29.
Complementation tests
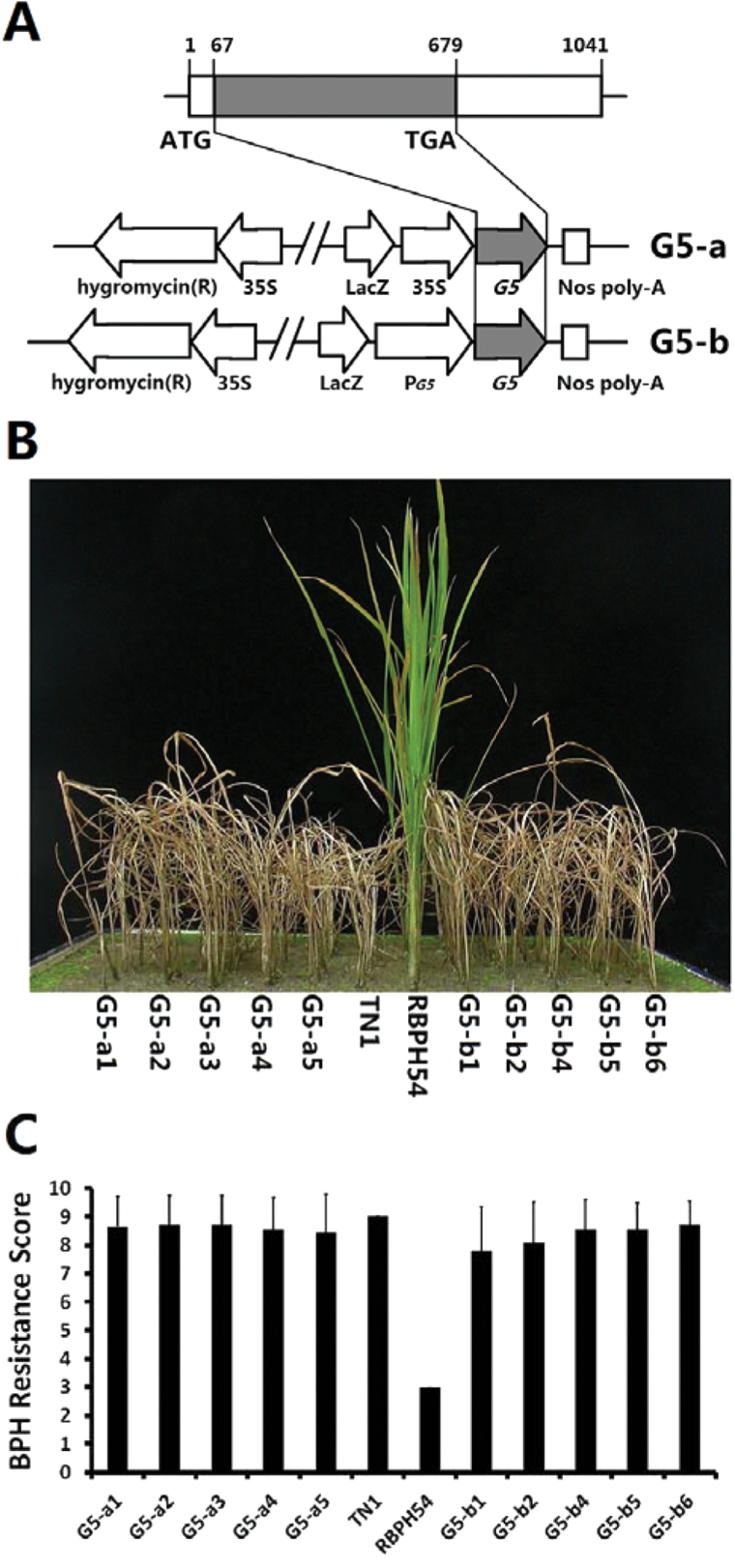
To investigate which candidate gene corresponded to BPH29, complementation tests were performed. Different strategies were adopted based on discrepancies in G4 and G5. An antisense transformation experiment was conducted to repress the expression of endogenous G4 in RBHP54, and the dominant G5 allele was transferred from TN1 into RBPH54 to mask the phenotype of the recessive G5 allele. An antisense-G4 vector was generated by fusion of the CaMV 35S promoter with the antisense G4 coding sequence (Supplementary Fig. S2A at JXB online). For G5, the dominant allele was cloned from TN1 and assembled downstream of either the CaMV 35S promoter (35S::G5) or the G5 1.5kb (−1500 to −1 for G5-ATG, and 29 818 to 31 317 for P0514G12) native promoter (PG5::G5) (Fig. 2A). These transformations were all performed in the resistant parent RBPH54 using a pCAMBIA1304 vector.
Fig. 2.
Complementation test for BPH29. (A) Gene and vector structure of G5. G5 has only one exon, with the open reading frame indicated by a grey box. Two linear maps of pCAMBIA1304 are shown: G5-a and G5-b represent the vector assembled with the 35S and G5 native promoter, respectively. (B) BPH resistance bioassay of G5 transgenic lines at the seedling stage. G5-a1 to G5-a5, 35S::G5 transgenic T1 lines; G5-b1 to G5-b6, PG5::G5 transgenic T1 lines; TN1, susceptible parent control; RBPH54, resistant parent control. (C) Brown planthopper (BPH) resistance scores of G5 transgenic lines. Both G5-a and G5-b transgenic lines showed higher scores, indicating their high susceptibility to BPH. Data are means ±SD (n=40 plants).
After seedling tests in the transgenic population, it was found that loss of BPH resistance in RBPH54 co-segregated with the G5 dominant allele gene transformations. In the BPH resistance bioassay, both 35S::G5 and PG5::G5 transformation lines showed high susceptibility to BPH, while RBPH54 seedlings remained healthy (Fig 2B, C). cDNA sequencing confirmed that the G5 dominant allele had been expressed successfully in these BPH-susceptible transformation lines (Supplementary Fig. S3 at JXB online). The 35S::anti-G4 positive transformants were still highly resistant to BPH (Supplementary Fig. S2B, C, D). It was therefore concluded that G5 was BPH29. Scores of G5 transgenic lines in the bulked seedling test revealed a significant loss of resistance, quite similar to levels of the susceptible control TN1 (Fig. 2C), indicating that the recessive allele of BPH29 is crucial to the BPH resistance of RBPH54. Sequence data of both dominant and recessive alleles have been deposited in GenBank under accession numbers KC019172 and KC019173, respectively.
Analysis of BPH29 protein
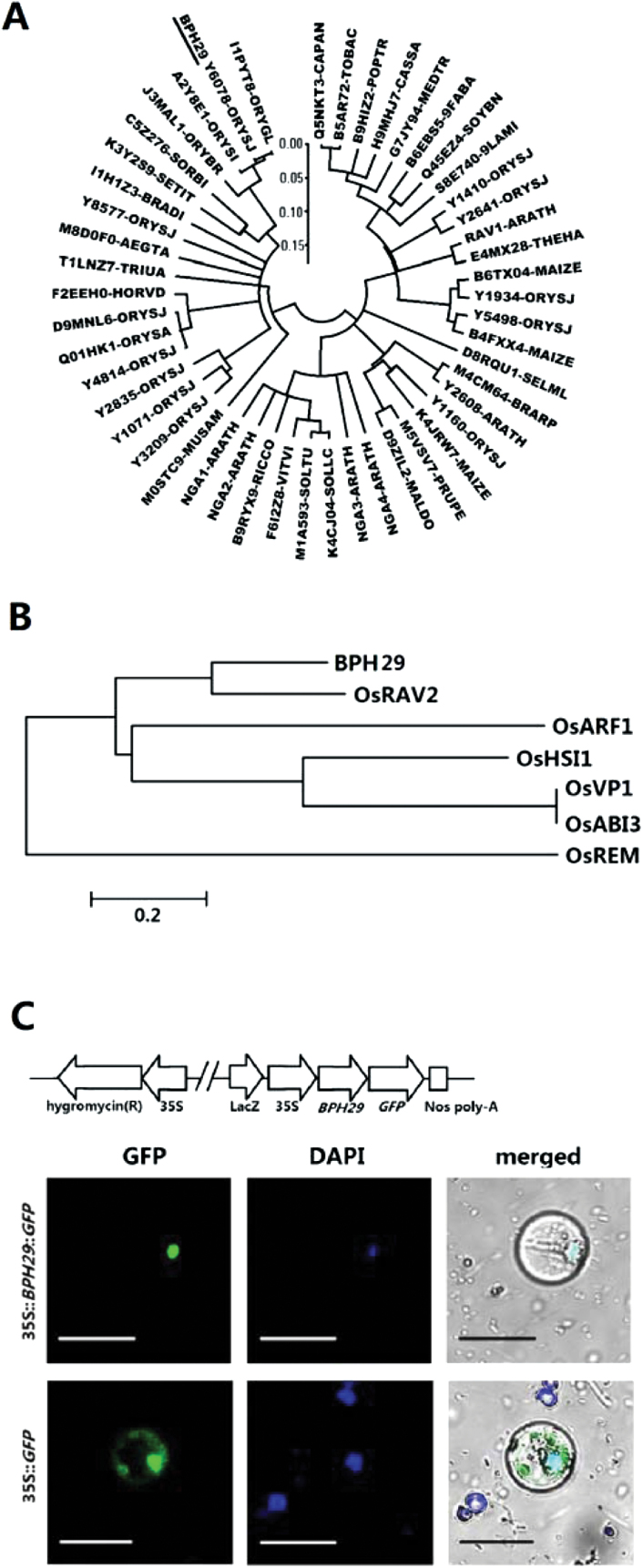
Amino acid sequence alignment analysis showed that BPH29 homologues are extensively distributed among various species of higher plants, such as rice, Arabidopsis, tomato, pepper, soybean, potato, wheat, maize, sorghum, and tobacco (Fig. 3A). BPH29 in Oryza sativa shares 96% and 77% sequence identity with its homologues in Oryza glaberrima and Oryza brachyantha, respectively. The widespread presence and high similarity of BPH29 reveals that the BPH29 gene is highly conserved in the plant kingdom and may have a critical function.
Fig. 3.
Analysis of BPH29 protein. (A) Amino acid sequence homologue analysis of BPH29. BPH29 homologues are extensively distributed among higher plants, such as rice, Arabidopsis, tomato, pepper, soybean, potato, wheat, maize, sorghum, and tobacco. Analysis was run by UniProt (http://www.uniprot.org/). The tree was generated by the Neighbor–Joining method (scale bar=0.05 estimated amino acid substitutions per residue). (B) Homology analysis of the BPH29 B3 domain. B3 domains of BPH29, OsABI3 (NM_001051697), OsVP1 (D16640), OsRAV2 (AK241984), OsARF1 (AK065936), OsHSI1 (AK241199), and OsREM (NM_001185617) were analysed by MEGA 4. The tree was generated by the Neighbor–Joining method (scale bar=0.2 estimated amino acid substitutions per residue). (C) Subcellular localization of BPH29. The 35S::BPH29::GFP fusion protein was transiently transformed into rice protoplasts. Green fluorescent protein (GFP) fluorescence was detected co-localized with 4′,6-diamidino-2-phenylindole (DAPI), a nucleus-specific fluorescent stain, in the nucleus. GFP fluorescence in control protoplasts (35S::GFP) was detected throughout the cytosol. Scale bar=30 μm.
According to information in GenBank, BPH29 is a single-copy gene that encodes a 203 amino acid (recessive allele) putative uncharacterized protein with no introns. Amino acid sequence analysis has shown that BPH29 contains only one B3 DNA-binding domain (Pfam accession 02362) (Punta et al., 2012) (Fig. 1E), a highly conserved domain found exclusively in transcription factors that interacts with the major groove of DNA (Yamasaki et al., 2004). Considering this characteristic, BPH29 should be localized to the nucleus. To confirm the subcellular localization of the BPH29 protein, the BPH29 coding region was fused to the GFP gene under the control of the 35S promoter (35S::BPH29::GFP). After the activity was confirmed by gene transformation and BPH resistance bioassay in rice (Supplementary Fig. S4 at JXB online), this BPH29–GFP fusion protein was transiently transformed into rice protoplasts, and its fluorescence was detected in the nucleus by co-localization with nuclear-specific 4′,6-diamidino-2-phenylindole (DAPI) (Fig. 3C). This result indicates that BPH29 is a nuclear-localized protein.
Expression analysis of BPH29
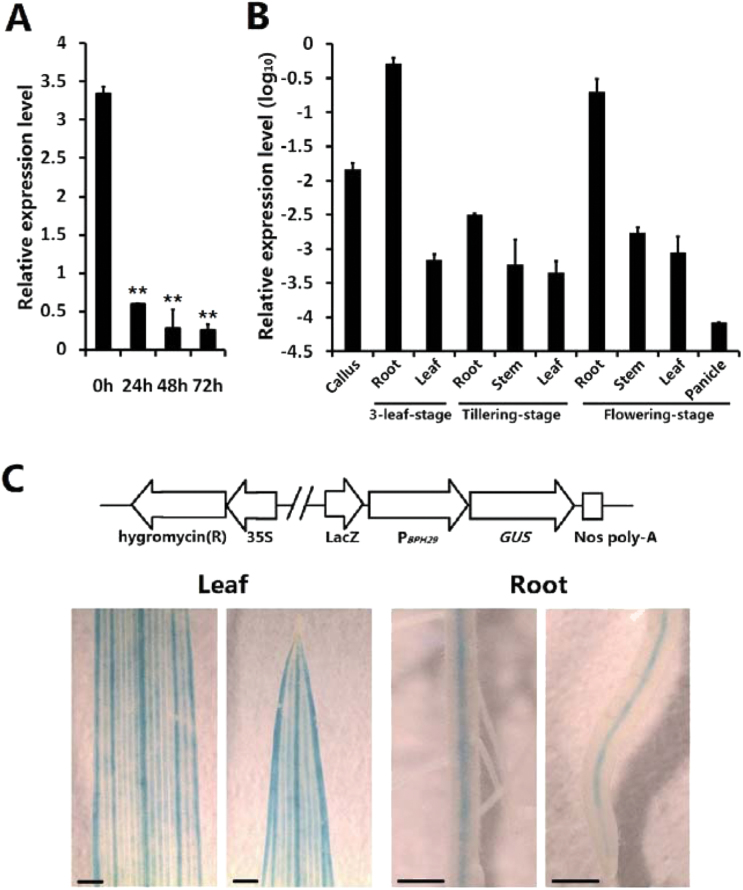
Expression level changes in BPH29 following the BPH infestation were detected. RBPH54 seedlings on the three-leaf stage were infested with first- to second-instar BPHs, and whole plants were harvested 0, 24, 48, and 72h later. The qRT-PCR analysis revealed that BPH29 decreased rapidly 24h after BPH infestation, and remained low afterwards (Fig. 4A). This expression pattern suggests that BPH29 is suppressed following BPH infestation.
Fig. 4.
Expression analysis of BPH29. (A) qRT-PCR analysis of the BPH29 expression pattern under BPH infestation. RBPH54 plants were collected 0, 24, 48, and 72h after infestation. Expression levels revealed that BPH29 is suppressed by BPH infestation. Student’s t-test: **P<0.01. Data are means ±SD (n=3 individuals). Expression levels are relative to OsActin1. (B) qRT-PCR analysis of BPH29 spatial- and temporal-specific expression. Different tissues from RBPH54 callus, three-leaf, tillering, and flowering stages were detected. BPH29 showed relatively high levels in callus and in roots at the three-leaf and flowering stages. Data are means ±SD (n = 3 individuals). Expression levels are relative to OsActin1 and log10 transformed. (C) Histochemical localization of BPH29 revealed by β-glucuronidase (GUS). The BPH29 promoter was fused with the GUS reporter gene (PBPH29::GUS), and introduced into RBPH54. GUS activity is indicated in blue. It showed that GUS stained the vascular system of leaf and root. Scale bar=1mm.
Based on the CREP rice gene expression database (http://crep.ncpgr.cn/), BPH29 showed extremely low expression levels in most tissues throughout the entire rice life cycle, except for roots of seedlings with two tillers (Supplementary Table S1 at JXB online). A qRT-PCR analysis was carried out to confirm this expression pattern using various RBPH54 tissue materials obtained from callus, three-leaf, tillering, and flowering stages. The qRT-PCR analysis confirmed that BPH29 was expressed at low basal levels, with marked increases during the callus stage and in roots at the three-leaf and flowering stages (Fig. 4B). These plant parts are located near the region of ingestion during BPH predation on rice plants.
To characterize tissue specificity of BPH29 expression, GUS histochemical localization of the BPH29 gene was examined. The BPH29 promoter (−1500 to −1 for BPH29-ATG) was fused to the GUS reporter gene and introduced into RBPH54. GUS activity was strongly detected in the vascular system of leaf and root (Fig. 4C), where BPH sucked the sap by using its stylet. Above all, this specific expression pattern of BPH29 is consistent with the location of BPH attack.
Defence-related gene and hormone modulation in BPH29-mediated insect resistance
Plant responses to herbivores involve global changes in gene expression mediated by multiple signalling pathways, including plant hormone SA and JA/ethylene signalling pathways (Walling, 2000). BPH infestation experiments were performed on three-leaf stage rice to investigate the changes of defence-related genes transcript level as well as the corresponding hormones level during infestation (Qiu et al., 2007) and to compare differences between BPH-resistant RBPH54 and loss-of-resistance G5 transformation lines.
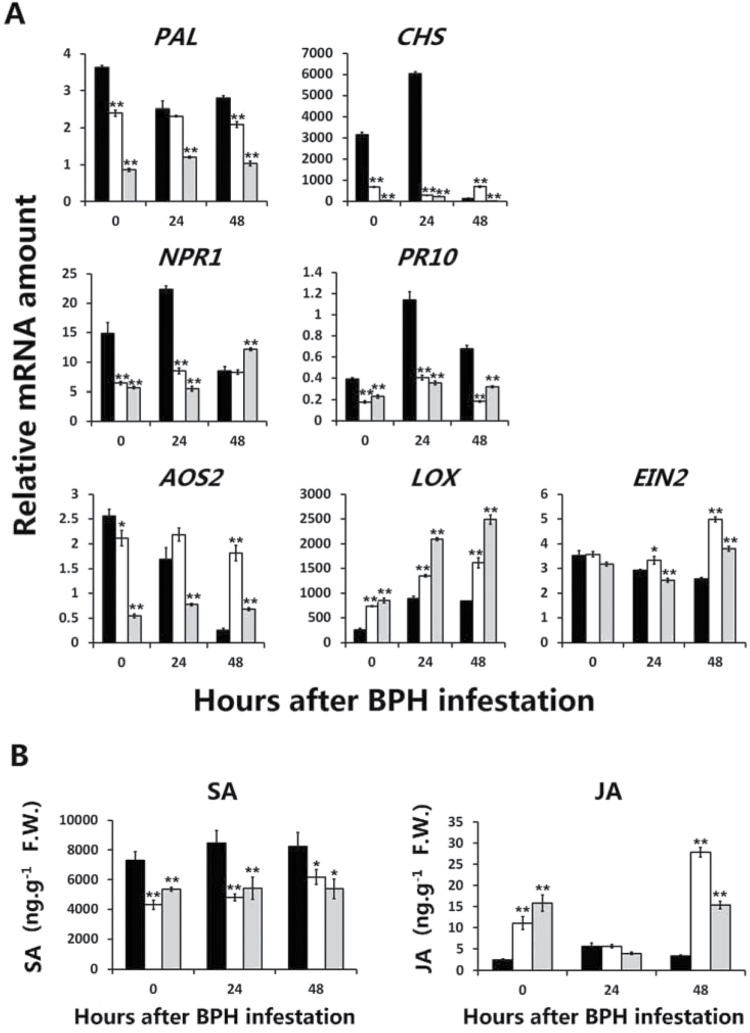
SA synthesis can be accomplished via either the isochorismate or phenylpropanoid pathways (Lee et al., 1995; Mauch et al., 2001). Two genes in the phenylpropanoid pathway, PAL (phenylalanine ammonia-lyase) and CHS (chalcone synthase), showed significant high transcript levels in RBPH54. CHS, in particular, showed pronounced background transcription in RBPH54 and rapid accumulation 24h after infestation. NPR1 (homologue of Arabidopsis non-expressor of pathogenesis-related genes 1), a key regulator of SAR in Arabidopsis that functions downstream of SA signalling pathways (Durrant and Dong, 2004), was transcribed at high levels in RBPH54. One of the PR (pathogenesis-related) genes PR10 was also higher in RBPH54 than in G5 lines (Fig. 5A). LOX and AOS are two important enzymes in the JA synthesis pathway (Zhao et al., 2005). After BPH infestation, AOS2 (allene oxide synthase 2) decreased rapidly in RBPH54, with no significant change observed in BPH-susceptible G5 lines. LOX (lipoxygenase) transcript levels were generally lower in RBPH54 as well. Additionally, expression of the ethylene signalling pathway receptor gene EIN2 (ethylene insensitive 2) (Jun et al., 2004) was higher in loss-of-resistance G5 lines than in RBPH54 at 48h after infestation (Fig. 5A).
Fig. 5.
Quantification of plant defence-related genes and hormone amounts in BPH29-mediated insect resistance. (A) Expression analysis of plant defence-related genes. PAL and CHS are two salicylic acid (SA) synthesis-related genes in the phenylpropanoid SA synthesis pathway. NPR1 is a key regulator of systemic acquired resistance (SAR) and operates downstream of the SA pathway. PR10 is one of the pathogenesis-related (PR) genes. AOS2 and LOX are two important genes in the jasmonic acid (JA) synthesis pathway. EIN2 is the ethylene signalling pathway receptor gene. OsActin1 was used as a reference control. (B) Hormone measurements of SA and JA levels. For (A) and (B), black bars represent BPH-resistant RBPH54, white bars correspond to the BPH-susceptible transgenic line G5-a1, and grey bars to G5-a2. Samples were obtained 24h and 48h after BPH infestation; 0h corresponds to no insect treatment. Student’s t-test: *P<0.05; **P<0.01. Data are means ±SD (n=3).
Quantification results of hormone amounts were consistent with the gene transcript levels above (Fig. 5B). The amount of SA showed a higher level in RBPH54, and quantification of JA confirmed the lower JA level in RBPH54. These results indicate that BPH-resistant RBPH54 activate SA synthesis after BPH feeding, and may defend against insects through SA-dependent SAR. In the meanwhile, the JA/ethylene-dependent pathway was suppressed during RBPH54 defence against BPH feeding.
Discussion
BPH29, a new recessive BPH resistance gene
Owing to climate change, pest frequency has become a critical problem that threatens global food security. To date, 28 BPH resistance genes have been detected, and only one (Bph14) has been cloned and published (Du et al., 2009). In this study, the BPH29 locus region was narrowed down to 24kb on the short arm of chromosome 6, and the location of BPH29 was finally confirmed by genetic analysis in this target region (Figs 1, 2; Supplementary Fig. S2 at JXB online). Thus a new BPH resistance gene, BPH29, has been cloned.
BPH resistance is known to be monogenically controlled in most resistant sources, and little progress has been made on recessive resistance genes compared with dominant ones (Huang et al., 2013). RBPH54 exhibits sustainable resistance to BPHs that is governed by recessive alleles at two loci (Yang et al., 2011). Generally, mechanisms of plant resistance to insects can be categorized into antixenosis, antibiosis, and tolerance (Painter, 1951). Antixenosis refers to a quality that repels or disturbs insects (Alam and Cohen, 1998a ). In the present study, the recessive BPH29 allele that contains a DNA mutation in the B3 domain (Fig. 1E; Supplementary Fig. S1 at JXB online) might lose the function of a dominant allele which was required for the settling of insects, and confer an antixenosis resistance in conjunction with another recessive locus. Ultimately, formal proof of the role of BPH29 in antixenosis resistance should be tested in the future, and the mechanism of co-operation of the two recessive loci remains to be explored. Cloning of BPH29 provides valuable information on molecular mechanisms of recessive resistance genes and offers a chance to understand the uncommon resistance sources conferred by polygenes.
The interaction between rice and the BPH reflects the co-evolutionary arms race between plants and herbivorous pests (Cheng et al., 2013) and provides an ideal system for studying the molecular mechanisms underlying plant defences against phloem-feeding insects, which are still fairly unclear. Further efforts related to the identification of herbivore-specific signal molecules, their recognition, and signal transduction might result in major breakthroughs in the future (Zebelo and Maffei, 2015). The information gleaned from BPH29 offers further insights into the field of plant–insect interactions and plant defence response.
BPH29 encodes a B3 domain-containing resistance protein
An essential layer of the plant immune system is based on highly polymorphic R proteins and is effective against specialized pathogens (Jones and Dangl, 2006). Most R proteins are multidomain NB-LRR proteins (Takken and Tameling, 2009). Mi-1.2, Vat, and Bph14 (Rossi et al., 1998; Pauquet et al., 2004; Du et al., 2009), the three plant insect resistance genes that have been cloned, all encode NB-LRR R proteins (Bruce, 2015). BPH29 is a single-exon gene that encodes a 203 amino acid protein which only contains one B3 DNA-binding domain (Fig. 1E), that is a novel structure for the R proteins.
The B3 domain is a highly conserved domain found only in vascular plants (Woo et al., 2010). Five major gene classes containing the B3 domain have been identified: ABI3/VP1 (Abscisic acid insensitive3/Viviparous1) (Giraudat et al., 1992; Suzuki et al., 1997), HSI (high-level expression of sugar-inducible gene) (Tsukagoshi et al., 2005), RAV (related to ABI3/VP1) (Kagaya et al., 1999), ARF (auxin response factor) (Ulmasov et al., 1997), and REM (reproductive meristem) (Franco-Zorrilla et al., 2002) gene families. Among these genes, the B3 domain of BPH29 is most similar to that of the RAV family (Fig. 3B). With regard to function, genes from the different subfamilies are involved in similar issues, such as hormone signalling pathways, flowering time control, organ growth, and polarity (Swaminathan et al., 2008). A previous study has shown that the RAV1 gene plays an important role in bacterial disease resistance (Sohn et al., 2006). Identification of BPH29 offers a unique example of the B3 domain’s role in plant insect resistance function.
Similarity of plant BPH resistance responses mediated by BPH29 and plant defences against pathogens
Molecular responses of plants against herbivores are mainly correlated with insect feeding modes and degree of plant tissue damage. In contrast to chewing insects that cause extensive damage to plant foliage and activate wound response pathways, BPHs are typical phloem-feeding insects that suck sap using their stylets, and therefore cause minimal physical injury to the host, but the interaction between insect stylets and plant cells is prolonged and intimate (Du et al., 2009). In some respects, these features are similar to attacks arising from fungal pathogens (Jones and Dangl, 2006; Wang et al., 2008). Additionally, BPHs also act as virus vectors, causing insect feeding to be accompanied by plant pathogen-related defences. As a consequence, the resistance factors are thought to occur within the phloem (Walling and Thompson, 2012), and host responses to phloem-feeding insects are thought to mirror responses to fungal or bacterial pathogens (Walling, 2000, 2008).
The present results revealed that the tissue-specific expression of BPH29 was restricted to plant vascular tissue, the location of BPH attack (Fig. 4C). Plant hormone pathway responses to BPH suggest the activation of SA-dependent SAR, and the JA/ethylene-dependent pathway was suppressed during RBPH54 defence against BPHs (Fig. 5). These characteristics are consistent with molecular responses that occur during plant–pathogen interaction (Felton and Korth, 2000). These facts indicate that BPH29-mediated resistance against the BPH is similar to defensive molecular responses that plants apply to biotrophic pathogens.
A valuable host resistance gene resource for crop breeding development
During crop breeding improvement, the elimination of insect damage to broadly susceptible domesticated modern crops is desired, with an economical and environmentally friendly strategy strongly preferred. Genetic resources from natural wild germplasm may be able to meet these demands (Bruce, 2012). Common cultivated rice possesses an AA genome. The host genetic background is an important factor that influences the function of resistance genes (Cao et al., 2007). RBPH54 resistance is derived from the wild rice O. rufipogon, which has a close evolutionary relationship with Oryza sativa and possesses the same AA genome as Asian rice cultivars. This background suggests that BPH29 is able to maintain a fine interaction with the cultivated rice genome. In addition, a single BPH resistance gene has reportedly been quickly overcome by insects under natural conditions (Khush and Brar, 1991), with the pyramiding of two or three genes generally found to provide greater resistance (Sharma et al., 2004; Hu et al., 2012). Considering the frequency of resistant BPH outbreaks and the need to pyramid multiple resistance genes for greater resistance, the identification of the BPH resistance gene BPH29 should greatly facilitate the breeding of rice host-resistant varieties.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Differences in G5 between the susceptible parent TN1 and the resistant parent RBPH54.
Figure S2. Complementation test for G4.
Figure S3. Expression of G5 alleles in transgenic lines.
Figure S4. BPH29–GFP fusion protein activity test.
Table S1. Transcript levels of BPH29 and OsActin1 at different rice developmental stages.
Table S2. Primers used in this work.
Acknowledgments
We thank Biqiu Wu, Chi Liu, Dahui Huang, and Zengfeng Ma (Guangxi Academy of Agricultural Sciences) for planting and managing the mapping population materials. This research was supported by grants from the Genetically Modified Organisms Breeding Major Projects (2009ZX08009-047B), Talent Development Program of Shanghai Academy of Agricultural Sciences (Pg09), and the Scientific Research Foundation of Guangxi University (XDZ110082).
References
- Alagar M, Suresh S, Samiyappan R, Saravanakumar D. 2007. Reaction of resistant and susceptible rice genotypes against brown planthopper (Nilaparvata lugens). Phytoparasitica 35, 346–356. [Google Scholar]
- Alam SN, Cohen MB. 1998. a Detection and analysis of QTLs for resistance to the brown planthopper, Nilaparvata lugens, in a doubled-haploid rice population. Theoretical and Applied Genetics 97, 1370–1379. [Google Scholar]
- Alam SN, Cohen MB. 1998. b Durability of brown planthopper, Nilaparvata lugens, resistance in rice variety IR64 in greenhouse selection studies. Entomologia Experimentalis et Applicata 89, 71–78. [Google Scholar]
- Brookes G, Barfoot P. 2003. GM rice: will this lead the way for global acceptance of GM crop technology? ISAAA Briefs No. 28. Ithaca, NY: ISAAA. [Google Scholar]
- Bruce TJA. 2012. GM as a route for delivery of sustainable crop protection. Journal of Experimental Botany 63, 537–541. [DOI] [PubMed] [Google Scholar]
- Bruce TJA. 2015. Interplay between insects and plants—dynamic and complex interactions that have coevolved over millions of years but act in milliseconds. Journal of Experimental Botany 66, 455–465. [DOI] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. 2007. The expression pattern of a rice disease resistance gene Xa3/Xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics 177, 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B. 2013. Plant domestication and resistance to herbivory. International Journal of Plant Genomics 2013, 572784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Fu XM, Liu JQ, Ye TT, Hou SY, Huang YQ, Yuan BF, Wu Y, Feng YQ. 2012. Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. Journal of Chromatography B 905, 67–74. [DOI] [PubMed] [Google Scholar]
- Chen X, Ronald PC. 2011. Innate immunity in rice. Trends in Plant Science 16, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Zhu L, He G. 2013. Towards understanding of molecular interactions between rice and the brown planthopper. Molecular Plant 6, 621–634. [DOI] [PubMed] [Google Scholar]
- Consales F, Schweizer F, Erb M, Gouhier-Darimont C, Bodenhausen N, Bruessow F, Sobhy I, Reymond P. 2012. Insect oral secretions suppress wound-induced responses in Arabidopsis . Journal of Experimental Botany 63, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J, Jones J. 2001. Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Du B, Zhang W, Liu B, et al. 2009. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proceedings of the National Academy of Sciences, USA 106, 22163–22168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W, Dong X. 2004. Systemic acquired resistance. Annual Review of Phytopathology 42, 185–209. [DOI] [PubMed] [Google Scholar]
- Felton GW, Korth KL. 2000. Trade-offs between pathogen and herbivore resistance. Current Opinion in Plant Biology 3, 309–314. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Cubas P, Jarillo JA, Fernández-Calvín B, Salinas J, Martínez-Zapater JM. 2002. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiology 128, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. The Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Cheng M, Gao G, Zhang Q, Xiao J, He Y. 2012. Pyramiding and evaluation of three dominant brown planthopper resistance genes in the elite indica rice 9311 and its hybrids. Pest Management Science 69, 802–808. [DOI] [PubMed] [Google Scholar]
- Huang D, Qiu Y, Zhang Y, Huang F, Meng J, Wei S, Li R, Chen B. 2013. Fine mapping and characterization of BPH27, a brown planthopper resistance gene from wild rice (Oryza rufipogon Griff.). Theoretical and Applied Genetics 126, 219–229. [DOI] [PubMed] [Google Scholar]
- IRRI. 1988. Standard evaluation system for rice . Manila, The Philippines: International Rice Research Institute. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jun SH, Han MJ, Lee S, Seo YS, Kim WT, An G. 2004. OsEIN2 is a positive component in ethylene signaling in rice. Plant and Cell Physiology 45, 281–289. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T. 1999. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Research 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS. 1997. Origin, dispersal, cultivation and variation of rice. Plant Molecular Biology 35, 25–34. [PubMed] [Google Scholar]
- Khush GS. 2001. Green revolution: the way forward. Nature Reviews Genetics 2, 815–822. [DOI] [PubMed] [Google Scholar]
- Khush GS, Brar DS. 1991. Genetics of resistance to insects in crop plants. Advances in Agronomy 45, 223–274. [Google Scholar]
- Lee HI, León J, Raskin I. 1995. Biosynthesis and metabolism of salicylic acid. Proceedings of the National Academy of Sciences, USA 92, 4076–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C. 2001. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. The Plant Journal 25, 67–77. [DOI] [PubMed] [Google Scholar]
- McCouch SR. 2008. Gene nomenclature system for rice. Rice 1, 72–84. [Google Scholar]
- Painter RH. 1951. Insect resistance in crop plants. Soil Science 72, 481. [Google Scholar]
- Pathak MD, Cheng CH, Fortuno ME. 1969. Resistance to Nephotettix impicticeps and Nilaparvata lugens in varieties of rice. Nature 223, 502–504. [Google Scholar]
- Pauquet J, Burget E, Hagen L, et al. 2004. Map-based cloning of the Vat gene from melon conferring resistance to both aphid colonization and aphid transmission of several viruses. In: Lebeda A, Paris HS, eds. Progress in cucurbit genetics and breeding research. Proceedings of Cucurbitaceae 2004, the 8th EUCARPIA Meeting on Cucurbit Genetics and Breeding, Olomouc, Czech Republic, 12–17 July, 2004 . Olomouc, Czech Republic: Palacký University in Olomouc, 325–329. [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J. 2012. The Pfam protein families database. Nucleic Acids Research 40, D290–D301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. 2007. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Molecular Plant-Microbe Interactions 20, 492–499. [DOI] [PubMed] [Google Scholar]
- Rahman ML, Jiang W, Chu SH, et al. 2009. High-resolution mapping of two rice brown planthopper resistance genes, Bph20(t) and Bph21(t), originating from Oryza minuta . Theoretical and Applied Genetics 119, 1237–1246. [DOI] [PubMed] [Google Scholar]
- Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. 1998. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proceedings of the National Academy of Sciences, USA 95, 9750–9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma PN, Torii A, Takumi S, Mori N, Nakamura C. 2004. Marker-assisted pyramiding of brown planthopper (Nilaparvata lugens Stål) resistance genes Bph1 and Bph2 on rice chromosome 12. Hereditas 140, 61–69. [DOI] [PubMed] [Google Scholar]
- Sōgawa K. 1982. The rice brown planthopper: feeding physiology and host plant interactions. Annual Review of Entomology 27, 49–73. [Google Scholar]
- Sōgawa K. 1994. Feeding behaviour and damage mechanism of the rice planthoppers. In: Elings A, Rubia EG, eds. Analysis of damage mechanisms by pests and diseases and their effects on rice yield . Wageningen, The Netherlands: SARP Research Proceedings, 143–154. [Google Scholar]
- Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK. 2006. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Molecular Biology 61, 897–915. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR. 1997. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. The Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan K, Peterson K, Jack T. 2008. The plant B3 superfamily. Trends in Plant Science 13, 647–655. [DOI] [PubMed] [Google Scholar]
- Takken FLW, Tameling WIL. 2009. To nibble at plant resistance proteins. Science 324, 744–746. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Endo S, Kazano H. 2000. Toxicity of insecticides to predators of rice planthoppers: spiders, the mirid bug and the dryinid wasp. Applied Entomology and Zoology 35, 177–187. [Google Scholar]
- Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, Cammue BP, Broekaert WF. 1998. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences, USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL. 2006. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. Journal of Experimental Botany 57, 755–766. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K. 2005. Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiology 138, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. The Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. 2000. The myriad plant responses to herbivores. Journal of Plant Growth Regulation 19, 195–216. [DOI] [PubMed] [Google Scholar]
- Walling LL. 2008. Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiology 146, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL, Thompson GA. 2012. Behavioral and molecular-genetic basis of resistance against phloem-feeding insects. In: Thompson GA, van Bel AJE, eds. Phloem: molecular cell biology, systemic communication, biotic interactions . Oxford: Wiley-Blackwell, 328–351. [Google Scholar]
- Wang Y, Tang M, Hao P, Yang Z, Zhu L, He G. 2008. Penetration into rice tissues by brown planthopper and fine structure of the salivary sheaths. Entomologia Experimentalis et Applicata 129, 295–307. [Google Scholar]
- Watanabe T, Fabellar LT, Almazan LP, Rubia EG, Heong KL, Sogawa K. 1997. Quantitative evaluation of growth and yield of rice plants infested with rice planthoppers. In: Kropff MJ, Teng PS, Aggarwal PK, Bouma J, Bouman BAM, Jones JW, van Laar HH, eds. Applications of systems approaches at the field level . Dordrecht, The Netherlands: Springer, 365–382. [Google Scholar]
- Woo HR, Kim JH, Kim J, Kim J, Lee U, Song I-J, Kim J-H, Lee H-Y, Nam HG, Lim PO. 2010. The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis . Journal of Experimental Botany 61, 3947–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu Y, He J, Liu Y, Jiang L, Liu L, Wang C, Cheng X, Wan J. 2014. Fine mapping of brown planthopper (Nilaparvata lugens Stål) resistance gene Bph28(t) in rice (Oryza sativa L.). Molecular Breeding 33, 909–918. [Google Scholar]
- Yamasaki K, Kigawa T, Inoue M, et al. 2004. Solution structure of the B3 DNA binding domain of the Arabidopsis cold-responsive transcription factor RAV1. The Plant Cell 16, 3448–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Li RB, Li YR, et al. 2011. Genetic mapping of bph20(t) and bph21(t) loci conferring brown planthopper resistance to Nilaparvata lugens Stål in rice (Oryza sativa L.). Euphytica 183, 161–171. [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zebelo SA, Maffei ME. 2015. Role of early signalling events in plant–insect interactions. Journal of Experimental Botany 66, 435–448. [DOI] [PubMed] [Google Scholar]
- Zhao J, Davis LC, Verpoorte R. 2005. Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnology Advances 23, 283–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.