Abstract
The advent of next-generation sequencing technologies has not only accelerated findings on various novel non-coding RNA (ncRNA) species but also led to the revision of the biological significance and versatility of fundamental RNA species with canonical function, such as transfer RNAs (tRNAs). Although tRNAs are best known as adapter components of translational machinery, recent studies suggest that tRNAs are not always end products but can further serve as a source for short ncRNAs. In many organisms, various tRNA-derived ncRNA species are produced from mature tRNAs or their precursor transcripts as functional molecules involved in various biological processes beyond translation. In this review, we focus on the tRNA-derived ncRNAs associated with Argonaute proteins and summarize recent studies on their conceivable biogenesis factors and on their emerging roles in gene expression regulation as regulatory RNAs.
Keywords: transfer RNA, tRNA fragment, microRNA, Argonaute family protein, AGO protein
Introduction
The advent of next-generation sequencing (NGS) technologies has unveiled the entire picture of the cellular transcriptome, wherein various non-coding RNA (ncRNA) are widely expressed from a large part of the genome.1,2 The functional significance of ncRNAs is particularly evident for the short 20- to 31-nucleotide (nt) ncRNAs bound to the family of Argonaute proteins.3–5 MicroRNAs (miRNAs), the best-studied class of such RNAs, are approximately 22 nt in length and posttranscriptionally regulate the expression of complementary target mRNAs. While nearly 2,800 species of human miRNAs are currently annotated in the miRBase database,6 computational analyses of massive RNA-sequencing (RNA-Seq) data has kept increasing the number of miRNA species.7,8 miRNAs are estimated to regulate the expression of most protein-coding genes,9 and a wide variety of other short and long functional ncRNA species coexist in the cellular transcriptome,10,11 exhibiting a tremendous impact of ncRNA-orchestrated regulatory mechanisms on normal developmental and physiological processes and diseases.12,13 NGS studies have accelerated findings on not only novel ncRNA repertoires but also well-characterized fundamental RNA species with canonical functions, such as transfer RNAs (tRNAs), which stimulated us to revisit their significance and versatility.
tRNAs are 70- to 90-nt ncRNAs that fold into a cloverleaf secondary structure and L-shaped tertiary structure. Since their discovery in the 1950s, tRNAs have been best known as fundamental adapter components of translational machinery, where they translate codons in mRNA into amino acids in protein.14 The human genome encodes >500 tRNA genes,15 along with numerous tRNA-lookalikes resembling nuclear and mitochondrial tRNAs.16,17 The multitude of tRNA genes in the genome, along with high stability,18 places tRNAs among the most abundant RNA molecules in the cellular transcriptome. Because of their abundance and well-defined role in translation, RNA fragments derived from tRNAs, found in early NGS studies, were often disregarded as nonfunctional degradation products. However, recent biochemical and bioinformatic evidence has generated a previously unsuspected concept that tRNAs are not always end products but are further able to serve as a source for short ncRNAs. In many organisms, specific tRNA-derived ncRNAs are produced from mature tRNAs or their precursor transcripts, not as random degradation products, but as functional molecules involved in many biological processes beyond translation.19–23
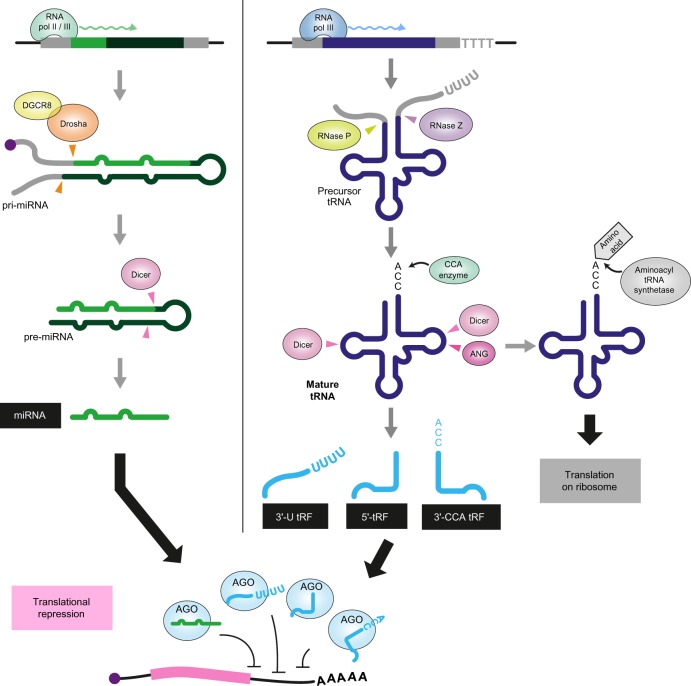
tRNA-derived ncRNAs identified to date can be classified into two groups: tRNA halves and tRNA-derived fragments (tRFs).19–23 tRNA halves are composed of 30–35 nt fragments derived from either the 5′- or the 3′-part of mature tRNAs. tRFs are shorter than tRNA halves, ranging from 13 to 32 nt in length and are currently subclassified into four subgroups: 5′-tRFs, 3′-CCA tRFs, 3′-U tRFs, and 5′-leader-exon tRFs. The 5′-tRFs and 3′-CCA tRFs correspond to the 5′- and 3′-parts of mature tRNAs containing processed mature 5′- and 3′-CCA termini, respectively (Fig. 1). The 3′-U tRFs are derived from the 3′-leader sequence of precursor tRNAs (pre-tRNAs) that harbor 3′-terminal uridine stretches, which result from transcriptional termination by RNA polyme rase III at thymidine tracts (Fig. 1). The 5′-leader-exon tRFs include the 5′-leader sequence of pre-tRNAs and the 5′-part of mature tRNAs. Although several excellent reviews have already described the repertoire, biogenesis, and functional significance of each class of the tRNA-derived ncRNAs,19–23 in this review, we particularly focus on the tRFs bound to Argonaute proteins and summarize their conceivable biogenesis factors and emerging roles in gene expression regulation as miRNAs.
Figure 1.
Schematic representation of the biogenesis of miRNAs and tRFs associated with AGO proteins.
tRFs Associated with AGO Proteins
In humans, the Argonaute protein family has eight members: four of the AGO clade (AGO1–4), which are ubiquitously expressed in numerous tissues, and four of the PIWI clade, whose expressions are restricted to the germline.3–5 All four AGO members associate with miRNAs to form effector complexes for their posttranscriptional regulatory roles, while only AGO2 shows endonucleolytic cleavage activity of target mRNAs. Biochemical purification and identification of miRNAs and their target mRNAs have been accomplished by purifying AGO proteins using immunoprecipitation (AGO-IP) followed by investigation of the AGO-bound RNAs in the IP fraction.24,25 Similar sizes of tRFs with miRNAs allowed researchers to reason that tRFs may be AGO-bound RNA species. Although early studies have already observed tRNA-derived sequences from AGO-IP fraction, as described below, more recent reports have demonstrated an evident association of the tRNA-derived ncRNAs with AGO proteins. Hereafter, we will use tRF amino acid/anticodon to denote a tRF derived from the tRNA corresponding to the amino acid and anticodon sequences.
The first focused detection of AGO-bound tRNA-derived ncRNAs in humans was reported in 2009.26 Cole et al. identified numerous tRFs in RNA-Seq data from HeLa cells.26 Majority of the identified tRFs were 5′-tRFs, and 5′-tRFGlnCUG, selected for the analyses, was shown to be present in AGO1- and AGO2-IP fractions. This result suggests the loading of the tRF in both of the AGO proteins, although 5′-tRFGlnCUG was also found in the fraction where AGO proteins were absent and therefore it seems that not all tRFs are loaded onto the AGO proteins. Haussecker et al. detected 3′-CCA tRFs and 3′-U tRFs in Human Embryonic Kidney 293 (HEK293) cells by northern blots.27 Interestingly, the identified tRF was selectively loaded onto different AGO proteins; 3′-U tRFSerUGA was found to be loaded onto AGO3 and AGO4 but not to AGO1 and AGO2.27 Burroughs and Ando et al. performed AGO1-3 IPs using THP-1 cells followed by NGS of the RNA contents of the IP fractions, and identified several 5′-tRFs that were specifically loaded onto AGO1, but slightly or not at all onto AGO2 and AGO3.28,29 They also observed the presence of 3′-CCA tRFs in their AGO-bound RNA libraries. Li et al. identified numerous 5′-tRFs and 3′-CCA tRFs in RNA-Seq libraries from BCP1 and HEK293 cells and demonstrated that both of the selected tRFs, 3′-CCA tRFHisGUG and 3′-CCA tRFLeuCAG, were loaded onto AGO2 but not onto AGO1.30 Maute et al. focused on 3′-CCA tRFGlyGCC whose expression levels are regulated by differentiation stages in mature B cells.31 The tRF was enriched in the AGO4-IP fraction, followed by fractions from AGO1- and AGO3-IPs, but less enriched in the AGO2-IP fraction. Kumar et al. analyzed many available short RNA libraries and observed numerous 5′-tRFs, 3′-CCA tRFs, and 3′-U tRFs, whose expression profiles varied in different cell lines and tissues.32 Their analyses of HEK293 AGO-IP fractions obtained from Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP)33 suggested that 5′-tRFs and 3′-CCA tRFs associate with AGO1, AGO3, and AGO4, but not with AGO2,32 whereas 3′-U tRFs were absent in all four AGO-IP fractions. Positional analyses of crosslink-induced mutations in PAR-CLIP data indicated that tRFs were loaded onto AGO proteins in a manner similar to miRNAs.32 Taken together, it could be concluded that the attempts to connect tRFs, at least partially, with the miRNA pathway have been successful. As miRNAs, some tRF species of the three classes (5′-tRFs, 3′-CCA tRFs, and 3′-U tRFs) are the interacting partners of AGO proteins.
The presence of tRFs in the AGO-IP fraction has also been reported for many other organisms such as mouse,34 Drosophila,35 and Bombyx,36 as well as in plant.37 Along the same lines, PIWI proteins, the other Argonaute family subclade that associates with PIWI-interacting RNAs (piR-NAs) for germline development,3–5 have also been reported to interact with tRNA-derived ncRNAs in various organisms such as human,38 marmoset,39 Drosophila,40 Trypanosoma,41 and Tetrahymena.42,43 Functional significance of the PIWI-bound tRFs in RNA metabolism has been shown in Tetrahymena.43 These findings suggest that tRNA-derived ncRNAs are widely-conserved functional factors across phyla that interact with Argonaute family proteins.
Biogenesis Factors for AGO-Bound tRFs
Asymmetric loading of tRFs onto the different AGO proteins could be due to coupling of tRF biogenesis with AGO loading mechanism and/or preferences of the AGO proteins and other biogenesis factors for the particular structures of tRFs or their precursors, namely mature tRNAs and pre-tRNAs. Mature tRNAs are produced from pre-tRNAs, which undergo several maturation steps44 (Fig. 1). Pre-tRNAs are first transcribed by RNA polymerase III, and their 5′-leader and 3′-trailer sequences are removed by endonucleolytic cleavage of RNase P and RNase Z, respectively. The “CCA” trinucleotides are then attached to the 3′-termini by a CCA-adding enzyme, and various different non-canonical base modifications are attached by modification enzymes. The resultant mature tRNAs are finally aminoacylated by their cognate aminoacyl-tRNA synthetases. Judging from the regions and sequences that the tRFs originate from, both pre-tRNAs and mature tRNAs must serve as substrates for the production of tRFs.
In vertebrates, canonical miRNA biogenesis (Fig. 1) starts from transcription of the miRNA gene by RNA polymerase II to generate a primary transcript (pri-miRNA), which is subsequently cleaved by Drosha, an endonuclease belonging to the RNase III superfamily, forming a complex with DGCR8.3–5 The resultant ∼70-nt precursor miRNA (pre-miRNA) with a hairpin-shaped structure is further processed by Dicer, another RNase III endonuclease, leading to the production of RNA duplex, one strand of which is then loaded onto AGO proteins.3–5 Considering the expression of tRFs complexed with AGO proteins, one may infer Drosha and Dicer endonucleases are involved in the production of such AGO-loaded tRFs. Indeed, the biogenesis of some tRFs belonging to 5′-tRFs, 3′-CCA tRFs, or 3′-U tRFs have been reported to be dependent on Dicer in humans,26,27,31 mice,45 and Drosophila35 (Fig. 1). Furthermore, Dicer is also reported to cleave tRNAGlnCUG to generate 5′-tRFGlnCUG in vitro.26 Although the cloverleaf secondary structure of tRNAs does not meet the criteria for canonical Dicer substrates, some precursor and mature tRNA molecules may alternatively form a long hairpin structure, particularly before modifications are attached, which may make tRNA structures suitable for Dicer processing as predicted in the case of mice tRNAIluUAU.45
Some tRFs seem to be generated via Dicer-independent biogenesis mechanism.27,32 Because several non-canonical miRNA biogenesis pathways are independent of Drosha or Dicer and utilize precursor RNAs forming various structures other than canonical hairpin-loop structures, such as single-stranded RNAs, AGO-bound tRFs may be produced through such non-canonical pathways.46,47 Alternatively, the other enzymes, which are not involved in miRNA biogenesis would be involved in the production of AGO-bound tRFs. RNase Z cleavage has been shown to produce 3′-U tRFs from pre-tRNAs in a Dicer-independent manner.27,48 Moreover, Angiogenin (ANG), a member of the RNase A superfamily, is the candidate enzyme responsible for AGO-bound tRFs. In mammalian cells, ANG was first discovered to be the enzyme that cleaves the anticodon loops of mature tRNAs to produce tRNA halves upon stress stimuli.49,50 However, it has also been shown that ANG is able to cleave the T-loop of tRNAs to produce 3′-CCA tRFLysUUU in vitro (Fig. 1),30 although this has yet to be confirmed in vivo.
It remains to be elucidated how the biogenesis factors are able to access and process the rigid structure of mature tRNAs, because, in the cytoplasm, mature aminoacylated tRNAs form an L-shaped tertiary structure and are tightly bound by an elongation factor to be transported to the ribosome. Because tRFs and tRNA halves coexist (e.g., 5′-tRFs were purified together with 5′-tRNA halves in Dicer-immunoprecipitates),36,51–54 the tRNA halves may be direct precursors of tRFs in some instances. Because the anticodon loop is exposed and accessible in animoacylated tRNA–elongation factor complexes, mature tRNAs may first be subjected to anticodon cleavage by ANG or other nucleases and then the generated tRNA halves dissociated from an elongation factor may be further processed for the production of tRFs. Further analyses are required to fully unveil the biogenesis factors and mechanisms for AGO-bound tRFs.
Functional Significance of AGO-Bound tRFs Acting as miRNAs
miRNAs deposit AGO proteins onto target RNAs by recognizing complementary sequences generally located in their 3′-UTR.3–5 Imperfect miRNA base-pairing with target mRNAs appears to induce translational silencing, whereas extensive base-pairing triggers exonucleolytic decay of the target mRNAs. Considering their association with AGO proteins and their similar size to miRNAs, it is not surprising that AGO-bound tRFs act as miRNAs and silence the expression of their complementary target mRNAs.
Research on viruses has shown the first demonstration of AGO-bound tRFs functioning as miRNAs. It has long been known that retroviruses use host tRNA as a primer for reverse transcription during the first step of retroviral replication. The 3′-part of such tRNAs is complementary and binds to the primer-binding site (PBS) of the virus sequence. In human MT4 T-cells infected with human immunodeficiency virus type 1 (HIV-1), Yeung et al. observed the abundant expression of an 18-nt 3′-CCA tRFLysUUU which is complementary to the PBS of HIV-1.55 Moreover, the 3′-CCA tRFLysUUU was loaded onto AGO2 and induced AGO2-mediated cleavage of the PBS, thereby silencing PBS-containing reporter gene and a HIV-1 gene. These results suggest important roles of AGO-loaded tRFs on repression of virus replication. Because the human genome contains many endogenous retroviral sequences and 3′-tRFs are highly complementary to these,30 tRF-directed pathways may also play ubiquitous roles in silencing endogenous viral elements. As another example of the involvement of tRFs in viral expression, Wang et al. reported that infection with human respiratory syncytial virus (RSV) led to accumulation of 5′-tRFs derived from various cytoplasmic tRNAs.56 One chosen 5′-tRF, 5′-tRFGluCUC, was shown to be generated by ANG-mediated cleavage at sites adjacent to the 5′-end of the anticodon loop. Because the 5′-tRFGluCUC showed activity in repressing complementary reporter gene expression, it is highly plausible that the tRF is loaded onto the AGO proteins. The tRF was further shown to have functional significance in regulating RSV replication.56
In addition to the viral research, Maute et al. reported a tRF functioning as miRNA in mature human B cells.31 A 22-nt AGO-bound 3′-CCA tRFGlyGCC showed activity in repressing the expression of target mRNAs in a sequence-specific manner. RPA1, an essential gene for DNA dynamics and repair, was identified as one of the endogenous targets containing complementary sequences of the tRF in its 3′-UTR. Stable expression of the tRF suppressed cell proliferation and modulated the response to DNA damage in an RPA1-dependent manner, indicating the clear biological importance of 3′-CCA tRFGlyGCC.31 Haussecker et al. used a reporter assay to confirm that AGO-bound 3′-CCA tRFs and 3′-U tRFs repressed complementary target RNAs.27 These results suggested that the tRFs were able to function as miRNAs, although further identification of endogenous tRF targets using CLIP25,33 and degradome RNA analysis57 will be required to fully understand their biological significance. One of the tRFs used in their analysis, a 3′-U tRFSerUGA, was reported to promote cell proliferation in the other study by Lee et al.48, validating that at least this tRF has an important cellular role. In addition to their roles in translational repression in the cytoplasm, AGO proteins have been reported to play distinct roles in the nucleus, such as modulation of histone methylation, mRNA splicing, and DNA damage repair, which is guided by miR-NAs.58,59 AGO-bound tRFs may be involved in such nuclear functions of AGO proteins. Accumulation of further evidence of the functionality and endogenous biological roles of AGO-bound tRFs would lead to clearcut designation of these tRFs as regulatory RNA molecules.
Perspectives
The number of reports characterizing expression and function of tRNA-derived ncRNAs has been growing, which enforces the conceptual consensus that cells utilize mature and precursor tRNAs as a source for short, functional ncRNAs. Although this review detailed Argonaute-bound tRFs, there are various other classes of tRNA-derived ncRNAs that are functional through mechanisms without involvement of Argonaute proteins. A representative example is the tRNA halves, termed tRNA-derived stress-induced RNAs (tiRNAs),49 whose production through ANG cleavage of the tRNA anticodon are triggered by various stress stimuli.19–23 tiRNAs promote the stress response by stress granule formation60 and global inhibition of translation by YB-1-involved pathway,49,61,62 thereby associating with normal biological processes and diseases.19–23,52 As another example of functional tRNA halves, sex hormone-dependent tRNA-derived RNAs (SHOT-RNAs) have been recently identified in breast and prostate cancers.63 SHOT-RNAs are also produced by ANG cleavage of the tRNA anticodon, which is promoted by sex hormones and their receptors, and these enhance cell proliferation.63
Despite the findings described in this review, information regarding expression profiles, biogenesis pathways, and biological meanings of AGO-bound tRFs is still fragmented. One of the obstacles for comprehensive identification of the tRFs would be the extensive presence of posttranscriptional modifications in tRNAs. Over 100 posttranscriptional modifications are present in tRNAs, many of which play crucial roles in tRNA folding and function such as codon recognition.64–67 Because it is highly plausible that many tRNA-derived ncRNAs are produced from modified tRNAs and because many modifications inhibit Watson–Crick base paring and thus arrest reverse transcription,68 heavily modified tRNA-derived ncRNAs would not be accurately captured by RT-PCR based sequencing methods. Modifications would also influence the biogenesis and function of tRNA-derived ncRNAs. For example, the presence of base-pairing-inhibitory modification inside the AGO-bound tRF would affect the efficiency of targeting and regulation of mRNAs. These points based on tRNA biological properties should be borne in mind for analyses of tRNA-derived ncRNAs.
As another considerable point, analyses of tRNAs and their fragments should be executed by proper bioinformatics. Genomes encode many identical tRNA genes and their slightly different variants, and identical tRNA fragment sequences can be present in non-tRNA regions in the genome.69 Very recently, during revising this manuscript, exhaustive analyses of massive RNA-Seq data have newly identified the fifth class of tRF, i-tRFs, which are wholly derived from internal regions of mature tRNAs and are loaded onto AGO proteins.70 tRFs including i-tRFs were reported to be differentially expressed in different cell types, tissues, disease states and human race.70 As in the effort to generate a tRNA fragment database by Kumar et al.71, cataloging the expression of tRNA fragments in various cell types and tissues, which should be based on appropriate bioinformatics approach and uniform nomenclatures will accelerate future investigations of tRNA-derived ncRNAs.
It is not known why the cells utilize tRNAs for ncRNA production. One potential answer may be the regulatory mechanism of tRNA abundance and heterogeneity. tRNA abundance varies greatly among different human cells and tissues, and the translational regulation by tRNA heterogeneity has been suggested.72–74 The expression of tRNA halves is also variable in different cells and tissues and further regulated by tRNA modification states, various stressors, and hormones.19–23,63 The uniquely regulated mechanism in controlling the quality and quantity of tRNAs and tRNA halves may consequently result in the opportunity for cells to gene rate, from these precursor RNAs, specific class of ncRNAs whose species and abundance would then also be uniquely controlled. In that regards, as shown in the case of various types of ncRNAs,11 tRNA-derived ncRNAs may emerge as a source of biomarkers and targets for therapeutics. Indeed, use of tRNA-derived ncRNAs as indicators for biological states has been suggested.63,70,75,76 Research on tRNA-derived ncRNAs is still in the initial stage and their biological functions may extend beyond our expectations. Further efforts to understand the expression profiles, biogenesis, and function of tRNA-derived ncRNAs will advance our knowledge regarding the expanding tRNA world and may provide significant insights into the pathophysiology of diseases.
Acknowledgments
We are grateful to Drs Shozo Honda and Keisuke Morichika for helpful discussions.
Footnotes
ACADEMIC EDITOR: James Willey, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 761 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived the concepts: MS, YK. Wrote the first draft of the manuscript: MS, YK. Jointly developed the structure and arguments for the paper: MS, YK. Made critical revisions: MS, YK. Both authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135(7):1201–14. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- 4.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedländer MR, Lizano E, Houben AJ, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol. 2014;15(4):R57. doi: 10.1186/gb-2014-15-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Londin E, Loher P, Telonis AG, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci USA. 2015;112(10):E1106–15. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10(12):833–44. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 11.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31(5):239–51. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet. 2014;15(6):423–37. doi: 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RajBhandary UL, Soll D. Transfer RNA in its fourth decade. In: Soll D, RajBhandary UL, editors. tRNA: Structure, Biosynthesis and Function. Washington, DC: American Society for Microbiology; 1995. pp. 1–4. [Google Scholar]
- 15.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37(Database issue):D93–7. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Telonis AG, Loher P, Kirino Y, Rigoutsos I. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front Genet. 2014;5:344. doi: 10.3389/fgene.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Telonis AG, Kirino Y, Rigoutsos I. Mitochondrial tRNA-lookalikes in nuclear chromosomes: could they be functional? RNA Biol. 2015;12(4):375–80. doi: 10.1080/15476286.2015.1017239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nwagwu M, Nana M. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J Embryol Exp Morphol. 1980;56:253–67. [PubMed] [Google Scholar]
- 19.Shigematsu M, Honda S, Kirino Y. Transfer RNA as a source of small functional RNA. J Mol Biol Mol Imaging. 2014;1(2):1–8. [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588(23):4297–304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol. 2013;10(12):1798–806. doi: 10.4161/rna.27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA. 2011;2(6):853–62. doi: 10.1002/wrna.96. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Silva MR, Cabrera-Cabrera F, Guida MC, Cayota A. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes (Basel) 2012;3(4):603–14. doi: 10.3390/genes3040603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azuma-Mukai A, Oguri H, Mituyama T, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA. 2008;105(23):7964–9. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cole C, Sobala A, Lu C, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15(12):2147–60. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–95. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burroughs AM, Ando Y, de Hoon MJ, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20(10):1398–410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burroughs AM, Ando Y, de Hoon MJ, et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8(1):158–77. doi: 10.4161/rna.8.1.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40(14):6787–99. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maute RL, Schneider C, Sumazin P, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci USA. 2013;110(4):1404–9. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hafner M, Landthaler M, Burger L, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanada T, Weitzer S, Mair B, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495(7442):474–80. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawamura Y, Saito K, Kin T, et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453(7196):793–7. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- 36.Nie Z, Zhou F, Li D, et al. RIP-seq of BmAgo2-associated small RNAs reveal various types of small non-coding RNAs in the silkworm, Bombyx mori. BMC Genomics. 2013;14:661. doi: 10.1186/1471-2164-14-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loss-Morais G, Waterhouse PM, Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with Argonaute and identification of their putative targets. Biol Direct. 2013;8:6. doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keam SP, Young PE, McCorkindale AL, et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014;42(14):8984–95. doi: 10.1093/nar/gku620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirano T, Iwasaki YW, Lin ZY, et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA. 2014;20(8):1223–37. doi: 10.1261/rna.045310.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Silva MR, Sanguinetti J, Cabrera-Cabrera F, Franzen O, Cayota A. A particular set of small non-coding RNAs is bound to the distinctive Argonaute protein of Trypanosoma cruzi: insights from RNA-interference deficient organisms. Gene. 2014;538(2):379–84. doi: 10.1016/j.gene.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Couvillion MT, Sachidanandam R, Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24(24):2742–7. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell. 2012;48(4):509–20. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22(20):2773–85. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43(6):892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chak LL, Okamura K. Argonaute-dependent small RNAs derived from single-stranded, non-structured precursors. Front Genet. 2014;5:172. doi: 10.3389/fgene.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23(22):2639–49. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu H, Feng J, Liu Q, et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583(2):437–42. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 51.Nowacka M, Strozycki PM, Jackowiak P, Hojka-Osinska A, Szymanski M, Figlerowicz M. Identification of stable, high copy number, medium-sized RNA degradation intermediates that accumulate in plants under non-stress conditions. Plant Mol Biol. 2013;83(3):191–204. doi: 10.1007/s11103-013-0079-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanco S, Dietmann S, Flores JV, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33(18):2020–39. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013;4(5):931–7. doi: 10.1016/j.celrep.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 54.Hsieh LC, Lin SI, Shih AC, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151(4):2120–32. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37(19):6575–86. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21(2):368–79. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and degradome-Seq data. Nucleic Acids Res. 2011;39(Database issue):D202–9. doi: 10.1093/nar/gkq1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang V, Li LC. Demystifying the nuclear function of Argonaute proteins. RNA Biol. 2014;11(1):18–24. doi: 10.4161/rna.27604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schraivogel D, Meister G. Import routes and nuclear functions of Argonaute and other small RNA-silencing proteins. Trends Biochem Sci. 2014;39(9):420–31. doi: 10.1016/j.tibs.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Emara MM, Ivanov P, Hickman T, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–68. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–23. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanov P, O’Day E, Emara MM, Wagner G, Lieberman J, Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111(51):18201–6. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honda S, Loher P, Shigematsu M, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci USA. 2015;112(29):E3816–25. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRCNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009;37(Database issue):D159–62. doi: 10.1093/nar/gkn772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Limbach PA, Crain PF, McCloskey JA. Summary: the modified nucleosides of RNA. Nucleic Acids Res. 1994;22(12):2183–96. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- 67.Jackman JE, Alfonzo JD. Transfer RNA modifications: nature’s combinatorial chemistry playground. Wiley Interdiscip Rev RNA. 2013;4(1):35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kellner S, Burhenne J, Helm M. Detection of RNA modifications. RNA Biol. 2010;7(2):237–47. doi: 10.4161/rna.7.2.11468. [DOI] [PubMed] [Google Scholar]
- 69.Schopman NC, Heynen S, Haasnoot J, Berkhout B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010;7(5):573–6. doi: 10.4161/rna.7.5.13141. [DOI] [PubMed] [Google Scholar]
- 70.Telonis AG, Loher P, Honda S, et al. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015 doi: 10.18632/oncotarget.4695. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43(Database issue):D141–5. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2(12):e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37(21):7268–80. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gingold H, Tehler D, Christoffersen NR, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158(6):1281–92. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 75.Mishima E, Inoue C, Saigusa D, et al. Conformational change in transfer RNA is an early indicator of acute cellular damage. J Am Soc Nephrol. 2014;25(10):2316–26. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dhahbi JM, Spindler SR, Atamna H, et al. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]