Abstract
Introduction
Border disease virus (BDV) causes congenital disorders in sheep and results in severe, but underestimated, economic losses worldwide. However, information about BDV strains affecting several ruminants worldwide is scarce. Therefore, antigenic and genetic classification of isolates from different geographical regions is important to enhance the knowledge of the epidemiology of BDV.
Materials and methods
Five pestiviruses isolated from lambs in an epidemic outbreak with an unusually high mortality in Spain in 1997 were characterised antigenically with a panel of monoclonal antibodies and genetically by sequencing within the 50 untranslated (50UTR) region of the genome.
Results
All the isolates were classified as BDV and showed a high homology with the Aveyron strain (Av), which was associated with an epidemic reported in sheep from the Aveyron region of France in 1984.
Conclusions
Classification of the isolates from this study provides valuable information on the molecular epidemiology of BDV.
Introduction
Border disease virus (BDV, genus Pestivirus, Family Flaviviridae) is the causative agent of an important congenital disease in sheep. Sheep flocks with Border disease are characterised by barren ewes, abortions, stillbirths, births of persistently infected lambs with tremors, ataxia, hairy fleece, brain malformations and poor growth (Nettleton and others 1998). Clinical manifestations of Border disease in acutely infected healthy sheep are mild or unapparent. However, an unusually virulent BDV isolate, Aveyron strain (Av), was reported in sheep from the Aveyron region (France) in 1984 and was associated with an outbreak of disease with high mortality (Chappuis and others 1986). In 1997, an epidemic outbreak in sheep associated with horizontal BDV infection and characterised by high mortality and clinical signs compatible with the Aveyron disease was observed in lambs in a flock in north east Spain. Since 1997, no other similar outbreaks have been reported in Europe. Although these two epizootic episodes were separated in time, the relatively low amount of data about BDV isolates and the possibility of future outbreaks of BDV infection in ruminants or swine means further knowledge is required about highly pathogenic BDV isolates. The objective of the present study was the antigenic and phyllogenetic analysis of five pestiviruses isolated from diseased sheep during the 1997 outbreak, one of the few reported episodes of sheep mortality associated with horizontal BDV infection.
Materials and methods
Five pestiviruses (ESP97-1 to ESP97-5) were isolated from lambs from Catalonia (north east Spain) in an epidemic outbreak in 1997. The flock comprised 250 Lacaune lambs reared intensively from two months of age. Some of these animals were imported from the Aveyron region (France) a few weeks before the outbreak was detected. The outbreak was characterised by high mortality (70 per cent of the lambs present in the flock died), anorexia, depression, diarrhoea, pyrexia and respiratory clinical signs. Spleen samples from dead animals were sent to the veterinary school of the Universidad Complutense de Madrid in order to confirm the pestivirus infection and to characterise the isolate. Spleen homogenates from dead lambs were inoculated in Madin-Darby bovine kidney (MDBK) cells in order to isolate the virus. Single passage was performed and the isolates were stored at −80°C.
The five pestivirus isolates were antigenically characterised with monoclonal antibodies directed at one of three viral proteins: E2 (gp53), Erns (gp48), and NS2-3 (p80/125) (Deregt and others 1991). Preparation and characterisation of the monoclonal antibodies (mAbs) used was previously described (Edwards and others 1988, Edwards and Sands 1990, Paton and others 1991, 1994). They were raised against the NADL (National Animal Disease Laboratory) and Oregon C24V strains of bovine viral diarrhoea virus (BVDV), the 87/6 strain of BDV, the Baker A and 86/2 strains of classical swine fever virus (CSFV), and the Vosges and 59386 strains of atypical (BVDV-II) pestiviruses. Protein specificity and parental virus of the mAbs used in this study are shown in Table 1. Each isolate was grown in bovine turbinate cell line cultures infected with a virus innoculum of 300 TCID50 (median tissue culture infective dose)/well. Immunostaining was performed by means of the peroxidase-linked assay (OIE 2008).
TABLE 1:
Protein specificity and parental virus of the monoclonal antibodies used in this study and reactivity of the Border disease virus isolates included in this study with these mAbs
| mAb | Parental virus | Protein specificity | Reactivity* |
|---|---|---|---|
| WB105 | BVDV-I Oregon C24V | NS2-3 | + |
| WB115 | BVDV-I Oregon C24V | E2 | − |
| WB162 | BVDV-I Oregon C24V | E2 | − |
| WB163 | BVDV-I Oregon C24V | E2 | − |
| WB165 | BVDV-I Oregon C24V | E2 | − |
| WB215 | BVDV-I Oregon C24V | E2 | − |
| WB158 | BVDV-I NADL | E2 | − |
| WB160 | BVDV-I NADL | NS2-3 | + |
| WB166 | BVDV-I NADL | E2 | + |
| WB170 | BVDV-I NADL | E2 | − |
| WB212 | BVDV-I NADL | NS2-3 | + |
| WB214 | BVDV-I NADL | E2 | − |
| WH216 | CSFV Baker A | Erns | + |
| WH299 | CSFV 86/2 | NS2-3 | − |
| WH303 | CSFV 86/2 | E2 | − |
| WH304 | CSFV 86/2 | E2 | + |
| WS363 | BDV 87/6 | Erns | + |
| WS368 | BDV 87/6 | Erns | + |
| WS369 | BDV 87/6 | Erns | + |
| WS371 | BDV 87/6 | Erns | + |
| WS373 | BDV 87/6 | Erns | + |
| WS381 | BDV 87/6 | E2 | + |
| WS384 | BDV 87/6 | E2 | + |
| WV433 | BVDV-II Vosges | Erns | + |
| WV437 | BVDV-II Vosges | NS2-3 | + |
| WV438 | BVDV-II Vosges | E2 | − |
| WV443 | BVDV-II Vosges | NS2-3 | + |
| WV459 | BVDV-II Vosges | NS2-3 | + |
| WV461 | BVDV-II Vosges | NS2-3 | + |
| WA536 | BVDV-II 59386 | Erns | − |
| WA538 | BVDV-II 59386 | E2 | + |
| WA539 | BVDV-II 59386 | E2 | − |
| WA548 | BVDV-II 59386 | NS2-3 | − |
| WA576 | BVDV-II 59386 | E2 | − |
*The patterns of monoclonal reactivity were identical for the five BDV isolates studied
BDV, Border disease virus; BVDV, bovine viral diarrhoea virus; CSFV, classical swine fever virus; mAb, monoclonal antibody
Phylogenetic study of the five isolates was performed. Viral RNA was extracted directly from spleen homogenates using a commercial kit (Macherey Nagel Nucleospin Viral RNA Isolation) according to the manufacturer's instructions. Reverse transcriptase-PCR was performed to detect pestiviral RNA (5′ untranslated region, 5′UTR) using previously described pan-pestivirus primers 324 and 326 (Vilcek and others 1994). Amplified DNA was purified (Minelute Gel Extraction Kit, Qiagen Inc) and sequenced. Purified amplicons were analysed with the Big Dye Terminator V.3.1 Kit (Applied Biosystems) and the ABI 3130xl Genetic Analyser (Applied Biosystems). The phylogenetic tree was calculated by the neighbour-joining method (Saitou and Nei 1987) using an automatic root location. To test the reliability of the branches in the tree a bootstrap analysis of 1000 replicates was performed by creating a series of randomly selected bootstrap samples.
Results
Pestiviruses were isolated from MDBK cells from all the spleen homogenates analysed. The patterns of monoclonal reactivity against the BDV isolates in this study were identical for the five isolates studied (Table 1). The five isolates were recognised by all mAbs raised against the BDV-87/6 strain. The isolates also reacted with mAbs raised against other pestivirus species. Analyses of the 5′UTR of the isolates showed that the isolates ESP97-1 to ESP97-5 (GenBank Acc. Numbers FR714860, HE999869, HE999870, HE999871 and HE999872) were almost identical and were classified as a BDV-5 genogroup, in agreement with mAb reactivity.
Discussion
BDV causes congenital disorders in sheep and severe economic losses worldwide. Although Spain is one of the most important countries of the EU for sheep meat production (Valdazo-González and others 2006), knowledge of circulating BDV strains in domestic small ruminants is scarce. For several years, all of the BDV strains sequenced from domestic small ruminants in Spain have been isolated from farms with ‘classical’ clinical signs of Border disease, that is, abortions and diseased newborns (Hurtado and others 2003, Valdazo-González and others 2006). This work provides information about the only outbreak of disease with high mortality caused by a BDV postnatal horizontal infection in lambs in Spain.
The BDV analysed in this study (ESP97) was characterised by causing an unusually high mortality rate in lambs and abortions in sheep. This epidemiological feature was only previously described in the Aveyron region in France. The Aveyron outbreak was also characterised by high mortality rates in horizontally infected lambs (Chappuis and others 1986). Interestingly, several lambs from the Aveyron region were introduced to the flock where the ESP97 pestivirus was isolated.
The patterns of monoclonal reactivity against the BDV isolates in this study were identical for the five isolates studied and they were identified as BDV. In reference to the cross-reaction observed in the mAb panel used in this study, this was observed previously in other BDV antigenic characterisations (Giammarioli and others 2011).
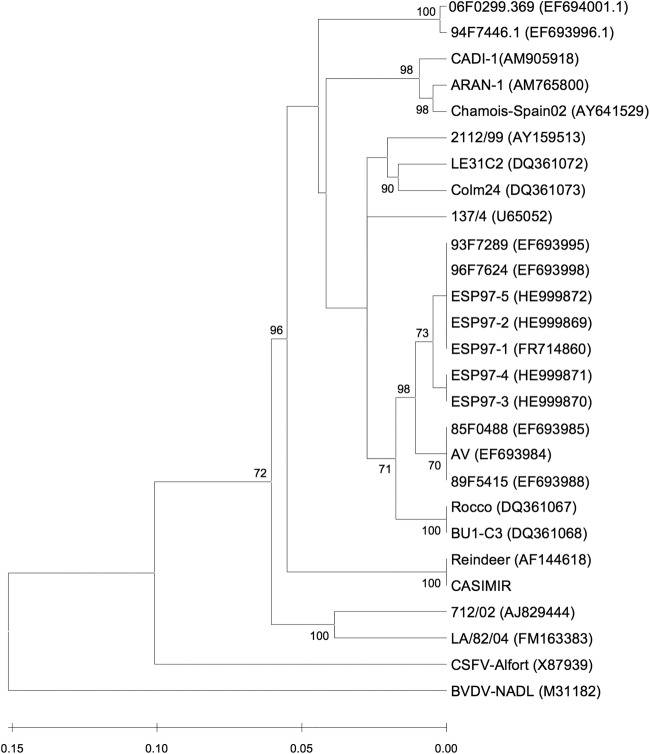
Analyses of the 5′UTR classified the five isolates within the BDV-5 genogroup, in agreement with mAb reactivity. Also, phylogenetic analysis of the ESP97 isolates showed a high homology between ESP97 and Av (Fig 1). Classification of BDV isolates has been widely developed in recent years. In this sense, the molecular analysis of the 5′UTR has proved to be useful to classify pestivirus strains (Vilcek and others 1994, 1996). The Av strain has recently been classified on the basis of the 5′UTR into the genotype BDV-5 (Giammarioli and others 2011). However, isolates classified by Giammarioli and others (2011) as BDV-5 were grouped together with BDV isolated from Spain by Valdazo-González and others (2006) and classified as BDV-4b. Thus, we observed that ESP97 grouped with several previously studied BDVs. However, the nomenclature of BDV isolates should be homogenised.
FIG 1:
Unrooted neighbour-joining phylogenetic tree based on the 5′UTR (5′ untranslated region) sequence among pestiviruses. ESP97 strains are enclosed in BDV-4. The numbers on the branches indicate the bootstrap values (in percent; 1000 replicates). Sequences of strains taken from GenBank with the following accession numbers: Chamois-Spain02 (AY641529), ARAN-1 (AM765800), CADI-1 (AM905918), Aveyron (EF693984), BU1-C3 (DQ361068), Rocco (DQ361067), 2112/99 (AY159513), LE31C2 (DQ361072), 96F7624 (EF693998), 93F7289 (EF693995), 89F5415 (EF693988), 85F0488 (EF693985), Colm24 (DQ361073), 137/4 (U65052), Moredun (U65023), CASIMIR (AB122085), Reindeer (AF144618), Alfort (X87939) and NADL (M31182). BDV, Border disease virus; BVDV, bovine viral diarrhoea virus; CSFV, classical swine fever virus; NADL, National Animal Disease Laboratory
The high homology between ESP97 and Av is of interest because, as mentioned, these two isolates caused unusual mortalities in horizontally infected lambs. Reported phylogenetic analyses of BDV have reported a single circulating genogroup of BDV in sheep flocks in Spain, the BDV-4 (Hurtado and others 2003, Valdazo-González and others 2006). This study reports the first identification of the BDV-5 genogroup in the Iberian Peninsula. Interestingly, all the BDV-4 isolates reported in Spain have been associated with the classical clinical presentation of Border disease in ruminant flocks with reproductive disorders (Hurtado and others 2003, Valdazo-González and others 2006). Our results lead us to hypothesise that ESP97, Av and related BDV-5 could have high pathogenic effects in sheep. This situation is of interest because the two BDV-5 outbreaks occurred in different sheep races and were separated by 14 years. Thabti and others (2002) reported that Av infection in different lamb races under experimental conditions did not reproduce the high mortality observed in the field. One of the reasons for the absence of illness in the challenged lambs was that the environmental conditions of the Aveyron region outbreak were not reproduced experimentally. Therefore, we hypothesise that it is possible that BDV-5 can maintain its pathogenic potential in the field, be undetected, and under certain conditions may reproduce a high pathogenic outbreak.
Interestingly, since 2001, high mortality rates associated with a horizontal BDV-4 infection have been reported in the wild ruminant Pyrenean chamois (Rupicapra pyrenaica) populations in Catalonia (Marco and others 2009). Phylogenetic analysis (Fig 1) shows that ESP97 is not included in the group of BDV isolated from diseased chamois and excludes the possibility that ESP97 caused the high mortalities observed in this wild ruminant.
In the authors’ opinion, this study provides valuable information about the BDV-5 ESP-97 isolate, close to Av, associated with one of the few reported cases of severe horizontal BDV infection in small ruminants.
Acknowledgments
We are most grateful to Jenny Sands and Steven Edwards for permission to use their mAbs in this study.
Footnotes
Competing interests: None declared.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Authors were asked to include a data sharing statement for their original research article.
References
- Chappuis G., Brun A., Kato F., Dauvergne M., Reynaud G., Duret C. (1986) Études Sérologiques et Immunologiques Réalisées à la Suite de l′Isolement d′un Pestivirus dans un Foyer Ovin chez des Moutons de l′Aveyron. In Pestivirose des Ovinset des Bovins: Nouvelles Connaissances, Utilisation pour une Stratégie de Contrôle, Journées Nationales de la Société Française de Buiatrie et de son Groupe d′Étudesur la Pathologie des Ovins et des Caprins (GEPOC). Eds Espinasse J., Savey M.. Paris: pp 55–65 [Google Scholar]
- Deregt D., Masri S. A., Cho H. J. (1991) Bovine viral diarrhea virus proteins: relatedness of p175 with p80 and p125 and evidence of glycoprotein processing. Canadian Journal of Microbiology 37, 815–822 doi:10.1139/m91-141 [DOI] [PubMed] [Google Scholar]
- Edwards S., Sands J. J. (1990) Antigenic comparisons of hog cholera virus isolates from Europe, America and Asia using monoclonal antibodies. Deutsche tierarztliche Wochenschrift 97, 79–81 [PubMed] [Google Scholar]
- Edwards S., Sands J. J., Harkness J. W. (1988) The application of monoclonal antibody panels to characterise pestivirus isolates from ruminants in Great Britain. Archives of Virology 102, 197–206 doi:10.1007/BF01310825 [DOI] [PubMed] [Google Scholar]
- Giammarioli M., La Rocca S. A., Steinbach F., Casciari C., De Mia G. M. (2011) Genetic and antigenic typing of border disease virus (BDV) isolates from Italy reveals the existence of a novel BDV group. Veterinary Microbiology 147, 231–236 doi:10.1016/j.vetmic.2010.06.027 [DOI] [PubMed] [Google Scholar]
- Hurtado A., García-Pérez A. L., Adúriz G., Juste R. A. (2003) Genetic diversity of ruminant pestiviruses from Spain. Virus Research 92, 67–73 doi:10.1016/S0168-1702(02)00315-5 [DOI] [PubMed] [Google Scholar]
- Marco I., Rosell R., Cabezón O., Mentaberre G., Casas E., Velarde R., Lavín S. (2009) Border disease virus among chamois, Spain. Emerging Infectious Diseases 15, 448–451 http://dx.doi.org/10.3201/eid1503.081155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton P. F., Gilray J. A., Russo P., Dlissi E. (1998) Border disease of sheep and goats. Veterinary Research 29, 327–340 [PubMed] [Google Scholar]
- OIE (World Organisation for Animal Health) (2008) Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. www.oie.int/manual-of-diagnostic-tests-and-vaccines-for-terrestrial-animals
- Paton D. J., Sands J. J., Edwards S. (1994) Border disease virus: delineation by monoclonal antibodies. Archives of Virology 135, 241–252 doi:10.1007/BF01310011 [DOI] [PubMed] [Google Scholar]
- Paton D. J., Sands J. J., Roeche P. M. (1991) BVD monoclonal antibodies: relationship between viral protein specificity and viral strain specificity. Archives of Virology 3, 47–54 doi:10.1007/978-3-7091-9153-8_6 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Thabti F., Fronzaroli L., Dlissi E., Guibert J. M., Hammami S., Pepin M., Russo P. (2002) Experimental model of Border disease virus infection in lambs: comparative pathogenicity of pestiviruses isolated in France and Tunisia. Veterinary Research 33, 35–45 doi:10.1051/vetres:2001004 [DOI] [PubMed] [Google Scholar]
- Valdazo-González B., Álvarez-Martínez M., Greiser-Wilke I. (2006) Genetic typing and prevalence of Border disease virus (BDV) in small ruminant flocks in Spain. Veterinary Microbiology 117, 141–153 doi:10.1016/j.vetmic.2006.06.008 [DOI] [PubMed] [Google Scholar]
- Vilcek S., Belák S. (1996) Genetic identification of pestivirus strain Frijters as a border disease virus from pigs. Journal of Virological Methods 60, 103–108 doi:10.1016/0166-0934(96)02031-9 [DOI] [PubMed] [Google Scholar]
- Vilcek S., Herring A. J., Nettleton P. F., Lowings J. P., Paton D. J. (1994) Pestiviruses isolated from pig, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Archives of Virology 136, 309–323 doi:10.1007/BF01321060 [DOI] [PubMed] [Google Scholar]