Abstract
Introduction
Brucella suis is the causative agent of brucellosis in suidae and is differentiated into five biovars (bv). Biovars 1 and 3 possess zoonotic potential and can infect humans, whereas biovar 2 represents the main source of brucellosis in feral and domestic pigs in Europe. Both aspects, the zoonotic threat and the economic loss, emphasize the necessity to monitor feral and domestic pig populations. Available serological or PCR based methods lack sensitivity and specificity.
Results
Here a bioinformatics approach was used to identify a B. suis specific 17 bp repeat on chromosome II (BS1330_II0657 locus). This repeat is common for B. suis bv 1 to 4 and was used to develop a TaqMan probe assay. The average PCR efficiency was determined as 95% and the limit of detection as 12,5 fg/µl of DNA, equally to 3.7 bacterial genomes. This assay has the highest sensitivity of all previously described B. suis specific PCR assays, making it possible to detect 3-4 bacterial genomes per 1 µl of sample. The assay was tested 100% specific for B. suis and negative for other Brucella spp. and closely related non-Brucella species.
Conclusions
This novel qPCR assay could become a rapid, inexpensive and reliable screening method for large sample pools of B. suis 1 to 4. This method will be applicable for field samples after validation.
Keywords: Brucellosis, Diagnostics, Disease surveillance, Food safety, Infectious diseases, RT-PCR techniques
Introduction
Brucellosis is a zoonotic infectious disease caused by different species of the genus Brucella. One of the prevalent species in feral pigs and hares is Brucella suis with five different biovars. The biovars (bv) 1–3 infect mainly suidae, bv 4 infects reindeers and bv 5 is rarely found in rodents (Alton and others 1988, Forbes 1991). All of them possess a zoonotic potential, but only bv 1 and 3 seem to cause severe diseases in humans (Munoz and others 2010). In Europe the most common cause of porcine brucellosis is due to infections with B. suis bv 2, whereas bv 1 and 3 are reported in Croatia only once (Garcia-Yoldi and others 2007, Cvetnic and others 2009). In most European countries feral pigs and hares seem to be the main reservoir for B. suis bv 2 (Garcia-Yoldi and others 2007, Leuenberger and others 2007, Melzer and others 2007, Munoz and others 2010, Gregoire and others 2012, Wu and others 2012). Transmission to domestic pigs may occur through direct contact, sexual transmission or shared water reservoirs (Risco and others 2014). The disease then causes infertility and abortion in female pigs and orchitis in male pigs. Furthermore, lymphadenitis, arthritis and subcutaneous abscesses may occur. Studies show that interactions between feral pigs and domestic pigs occur quite often (Wu and others 2012). Regarding the growing populations of feral pigs in Europe this could explain the increase of infections in domestic pigs with brucellosis since 1990 (Godfroid and Kasbohrer 2002). Therefore, brucellosis as a zoonotic disease is of great public health importance in many countries (Melzer and others 2007, Gerst and others 2010, Von Roost and others 2010). Monitoring of Brucella spp. infections in domestic livestock and wild animals is mainly based on detection of antibodies specific for smooth lipopolysaccharide. This includes the risk of false positive reactions, especially for porcine brucellosis due to cross-reacting antibodies to the highly similar O-antigen of Yersinia enterocolitica O:9 (Hurvell 1973). Additionally, non-reactors or animals which have not yet developed an antibody level can cause false negative results. During recent years PCR-based methods became more and more important as fast and reliable methods to detect Brucella-specific DNA from colony material or directly from field samples. On one hand conventional or real-time Brucella genus-specific PCR methods are used to identify brucellae from colony material or field samples (Bricker 2002, Probert and others 2004). On the other hand species-specific multiplex PCR techniques are implemented to differentiate on species level (Lopez-Goni and others 2011). Since real-time PCR is much more sensitive compared with conventional PCR it is often used to detect specific DNA in field samples directly. Obviously, a positive result in a genus-specific real-time PCR should be confirmed by a second method which ideally is a species-specific method. So far published real-time PCR methods are able to distinguish B. abortus, B. melitensis and B. suis bv 1 (Al Dahouk and others 2007a). In central Europe B. suis bv 2 is endemic in wild boar and hare populations and therefore influences monitoring of brucellosis at least in pig farms(Garcia-Yoldi and others 2007, Leuenberger and others 2007, Munoz and others 2010, Gregoire and others 2012, Wu and others 2012). The last outbreaks of brucellosis in pigs in Germany were B. suis bv 2 infections in outdoor holdings which have been confirmed by isolation of the infectious agent (Melzer and others 2007).
The aim of this study was to develop a quantitative real-time PCR (qPCR) assay as a sensitive diagnostic tool for detection of Brucella suis bv 1–4. These biovars are the most prevalent and are of special epidemiological importance.
Materials and methods
Bacterial strains and growth conditions
Bacteria used in this study are listed in Tables 1 and 2. A collection of 25 Brucella spp. reference strains, 75 B. suis field isolates and 30 closely related and clinically relevant non-Brucella species were obtained from the in house strain collection of the national reference laboratory for brucellosis (Friedrich-Loeffler-Institut, Germany). Brucella spp. strains were maintained on Brucella agar (Becton Dickinson) or Brucella agar, containing polymyxin B (2500 IE) bacitracin (12,500 IE), cyclohexymid (50 mg/l), nalidixic acid (2.5 mg/l), nystatin (50,000 IE) and vancomycin (10 mg/l), respectively (Farrell 1974). Brucella cultures were incubated with 5–10 per cent of CO2 at 37°C for 72 hours (Alton and others 1988). All other bacterial strains were routinely cultivated on 5 per cent sheep blood agar (Becton Dickinson) or Luria Bertani agar (Fisher Scientific) overnight at 37°C.
TABLE 1:
Bacterial strains used in this study
| Bacterial species | Reference | Bacterial species | Reference |
|---|---|---|---|
| Brucella spp. | Non-Brucella species | ||
| B. abortus 544 | ATCC 23448 | Actinobacillus pleuropneumoniae | ATCC 27088 |
| B. abortus S99 | Bacillus brevis | ATCC 8246 | |
| B. abortus S19 | Bacillus cereus | ATCC 10876 | |
| B. abortus 86/8/59 | ATCC 23449 | Bacillus megaterium | ATCC 14581 |
| B. abortus Tulya | ATCC 23450 | Bacillus subtilis | ATCC 6633 |
| B. abortus 292 | ATCC 23451 | Bacillus thuringiensis | ATCC 10792 |
| B. abortus B3196 | ATCC 23452 | Bordetella bronchiseptica | ATCC 19395 |
| B. abortus 870 | ATCC 23453 | Burkholderia cepacia | DSM7288 |
| B. abortus C68 | ATCC 23455 | Burkholderia mallei | ATCC 23344 |
| B. melitensis 16M | ATCC 23456 | Burkholderia pseudomallei | ATCC 23343 |
| B. melitensis Elberg | Escherichia coli | DSM 30083 | |
| B. melitensis 63/9 | ATCC 23457 | Lactobacillus ruminis | DSM 20403 |
| B. melitensis Ether | ATCC 23458 | Mannheimia haemolytica | ATCC 33396 |
| B. suis 1330 (bv 1) | ATCC 23444 | Ochrobactrum anthropi | CCUG 1047 |
| B. suis Thomson (bv 2) | ATCC 23445 | Oligella urethralis | DSM 7531 |
| B. suis 686 (bv 3) | ATCC 23446 | Pasteurella multocida | DSM 5281 |
| B. suis 40 (bv 4) | ATCC 23447 | Pasteurella multocida subsp. multocida | ATCC 43137 |
| B. suis 513 (bv 5) | NCTC 11996 | Proteus mirabilis | ATCC 29906 |
| B. canis RM6/66 | ATCC 23365 | Pseudomonas aeruginosa | ATCC 9027 |
| B. ovis 63/290 | ATCC 25840 | Pseudomonas alcaligenes | ATCC 14909 |
| B. neotomae 5K33 | ATCC 23459 | Pseudomonas fluorescens | ATCC 13525 |
| B. ceti B1/94 | NCTC 12891 | Pseudomonas polymyxa | ATCC 842 |
| B. pinnipedialis B2/94 | NCTC 12890 | Rhodococcus equi | DSM 20307 |
| B. microti CCM4915 | BCCN 07-01 | Staphylococcus aureus subsp. aureus | ATCC 25178 |
| B. inopinata BO1 | BCCN 09-01 | Stenotrophomonas maltophilia | ATCC 13637 |
| Streptococcus agalactiae | ATCC 27956 | ||
| Streptococcus equinus | ATCC 9812 | ||
| Streptococcus parauberis | DSM 6631 | ||
| Taylorella equigenitalis | DSM 10668 | ||
| Yersinia enterocolitica subsp. enterocolitica | ATCC 9610 |
ATCC, American Type Culture Collection; bv, biovar; BCCN, Brucella Culture Collection from Nouzilly; CCUG, Culture Collection, University of Göteborg; DSM, German collection of microorganisms (Deutsche Sammlung von Mikroorganismen); NCTC, National Collection of Type Culture
TABLE 2:
B. suis field isolates used in this study
| Biovar | Source of isolates | No. of isolates |
|---|---|---|
| 1 | Feral pig | 1 |
| 2 | Hare | 20 |
| Feral pig | 32 | |
| Domestic pig | 21 | |
| 5 | Human | 1 |
Preparation of DNA templates
DNA was extracted from single colonies and resuspended in 200 µl of sterile distilled water after heat inactivation for two hours at 80°C. DNA isolation was carried out using the HighPure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany) according to the manufacturer's instructions for bacteria. DNA quantity and purity was determined using a NanoDrop™ 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, USA). Serial 1:10 dilutions ranging from 100 ng to 1 fg/µl of DNA were prepared for qPCR efficiency and specificity testing. DNA was also directly isolated from one Brucella culture-positive spleen sample from a hare with the same extraction kit according to the manufacturer's instructions for mammalian tissue. This was done as an example to show suitability of this assay for field samples.
Bioinformatic analyses
For identification of a B. suis-specific target whole genome alignments were performed using the Geneious software (Geneious 6.0.3, Biomatters Ltd., Auckland, New Zealand). Genome sequences from various Brucella spp. (Table 3) were obtained from the NCBI GenBank database and analysed for B. suis bv 1 and 2 conserved regions. Identified targets were then tested for specificity using BLAST search with a Discontiguous Megablast against the NCBI GenBank database.
TABLE 3:
Brucella spp. genome sequences used for whole genome alignment analysis
| Brucella spp. | Accession no.* |
|---|---|
| B. abortus | NC_016777 |
| NC_006933 | |
| NC_007624 | |
| B. melitensis | NC_003318 |
| NC_012442 | |
| NC_017247 | |
| NC_017245 | |
| NC_017283 | |
| B. ovis | NC_009504 |
| B. canis | NC_010104 |
| B. microti | NC_013118 |
| B. pinnipedialis | NC_015858 |
| B. suis bv 1 | NC_017250 |
| B. suis bv 2 | NC_010167 |
| B. suis bv 3 | NZ_DS999731 |
| B. suis bv 4 | NZ_GG703794 |
| B. suis bv 5 | NZ_DS999712 |
*NCBI GenBank accession number
Quantitative real time PCR
Oligonucleotides B. suis for 5′-GCC AAA TAT CCA TGC GGG AAG-3′ and B. suis rev 5′-TGG GCA TTC TCT ACG GTG TG-3′ targeting a 106 bp fragment of the BS1330_II0657 locus encoded on chromosome 2 of B. suis bv 1 with the FAM-labelled hydrolysis probe B. suis probe 6-carboxyfluorescein (FAM)-TTG CGC TTT TGT GAT CTT TGC GCT TTA TGG-BHQ1 were used for the qPCR detection assay (Jena Bioscience, Germany). Primer and probe were designed with Geneious software (Geneious 6.0.3, Biomatters Ltd., Auckland, New Zealand). For qPCR analysis the Stratagene Mx3000P QPCR System (Agilent Technologies, Santa Clara, USA) was used. The qPCR was performed in a 25 µl format using the 2× TaqMan Universal PCR Mastermix (Applied Biosystems, Foster City, California, USA), 0.3µM of each primer, 0.1µM probe and 2 µl of template DNA. The qPCR was carried out on an Mx3000P QPCR Instrument (Agilent Technologies, Santa Clara, California, USA) using the following cycling conditions: 50°C for 2 minutes, 95°C for 10 minutes and 50 cycles of 95°C for 15 seconds and 60°C for 1 minute. Data were analysed with the Mx3000Pro software. Raw data were normalised using an adaptive baseline correction and a smoothing with a moving average of three amplification points. Initially cycle threshold (Ct) was set with background-based threshold algorithm. For better comparison of the results a threshold of 0.05 (dRn) was applied to all qPCR signals, as the highest of all computed thresholds from all experiments. Fluorescence signals above this value were considered as positive, below as negative results.
Determination of qPCR efficiency
Standard curves were done with serial dilutions 1:10 from 106 to 100 fg/µl of bacterial DNA from B. suis bv 2. The efficiency of the PCR was calculated from the slope of the logarithmic regression of Ct values plotted against DNA concentrations by E=e(−1/slope)−1. For statistical analysis and assessment of reproducibility, standard curve experiments were done in quadruplicate and repeated five times on five different days. Additionally, qPCR efficiencies for B. suis bv 1–4 were compared by analysing serial dilutions 1:10 of genomic DNA from 106 to 102 fg/µl. Dilutions were chosen within the linear dynamic range of the assay and expected concentrations of DNA within possible specimens. Samples were analysed in duplicate.
Determination of the qPCR limit of detection
The limit of detection (LOD) was defined as the DNA concentration with 95 per cent positive qPCR results. Therefore we used 100, 50, 25, 12.5 and 6.25 fg/µl of B. suis bv 2 DNA spiked into DNA preparations from anti-Brucella antibody and Brucella DNA-free swine sera. To proof Brucella DNA free status of the sera a published real-time PCR (Probert and others 2004) was used as described. LOD samples were analysed in octuplicate for each concentration and repeated three times on three different days.
Capillary gel electrophoresis
Real time PCR products were analysed by capillary gel electrophoresis using the QIAxcel DNA high resolution kit (1200) with an analytical range of 15 bp to 10 kb on the QIAxcel Advanced system (QIAGEN, Hilden, Germany). Briefly, 10 µl of each PCR product were added to the template tray and a standardised volume of 0.1 μl was injected automatically into the QIAxcel cartridge for analysis. For accurate fragment size determination the QX alignment marker 15 bp/3 kb and the QX DNA size marker FX174/HaeIII with a range of 50–1350 bp was used. Electrophoresic separation of amplicons was then carried out by applying the OM500 program. For data analysis and generation of a virtual gel image the Biocalculator QIAxcel software was used.
Results
Identification of a B. suis bv 1–4 specific qPCR target by whole genome comparison.
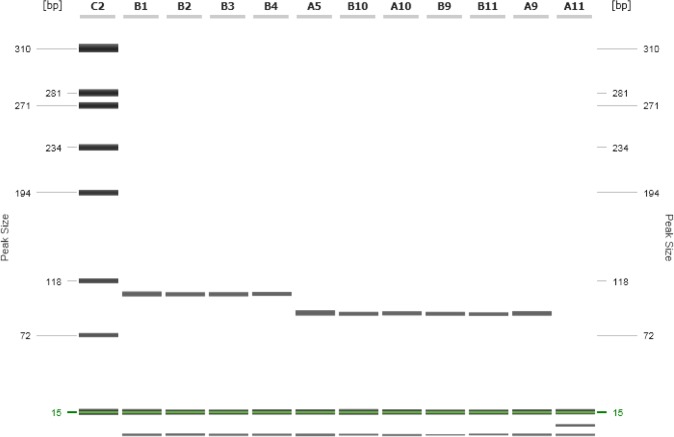
Annotated genome sequences from 12 Brucella spp. and B. suis bv 1 and 2 (Table 3) were analysed by whole genome comparison with B. suis bv 2 as reference genome. A region within the 3′ end of BS1330_II0657 locus was identified to be highly specific for B. suis with a 17 bp repeat exclusively found in B. suis bv 1–4 (Fig 1). All other Brucella spp. and B. suis bv 5 lack this repeat. Designed real-time primer were located up- and downstream of this repeat generating a 106 bp fragment for B. suis bv 1–4 and a 89 bp fragment for all other Brucella spp. and B. suis bv 5 (Fig 2). BLAST search performed for the 106 bp amplicon revealed 100 per cent complementarity for B. suis bv 1–4. The probe spanning the 17 bp repeat in B. suis bv 1–4 shows a 7 bp mismatch within the 5′-end in all other Brucella spp., lacking this repeat. However, this mismatch lowers the Tm to 57°C and prevents binding of the probe at the assay-specific annealing temperature of 60°C.
FIG 1:

Alignment of the targeted 3′ end of BS1330_II0657 locus from different Brucella spp. strains and B. suis bv 1–5. The alignment shows the 17 bp repeat specific for B. suis bv 1–4 and the resulting 7 bp mismatch of the probe (indicated by asterisks) for all other Brucella spp
FIG 2:
High-resolution capillary gel electrophoresis of the B. suis-targeted quantitative real-time PCR amplicons. Primer B. suis for and B. suis rev were used for amplification of the targeted region within BS1330_II0657 locus. Lack of the 17 bp repeat in Brucella spp. or B. suis bv 5 are visualised and resulted in a fragment size of 106 bp for B. suis bv 1–4 and 89 bp for all other strains. Lane C2: Size marker; lane B1: B. suis bv 1; lane B2: B. suis bv 2; lane B3: B. suis bv 3; lane B4: B. suis bv 4; lane A5: B. suis bv 5; lane B10: B. abortus 544; lane A10: B. melitensis 16M; lane B9: B. ovis; lane B11: B. canis; lane A9: B. microti; lane A11: no template control
PCR efficiency
For PCR efficiency determination genomic DNA standards from 106 to 100 fg/µl of B. suis bv 2 DNA were analysed. Efficiency was calculated from the slope and ranged between 90.8 and 97.6 per cent with a y-interception between 38.88 and 40.15 and a R2 value between 0.9821 and 0.9965 (Fig 3). Analysis of B. suis bv 1, 3 and 4 DNA standards revealed similar efficiencies of 94.3–95.5 per cent with y-intercepts between Ct- value 44.24 and 45.24 and an R2 value between 0.9567 and 0.9965 (Fig 4). In general we observed a decreasing accuracy, assessed as an increasing standard error (se) for the resulting Ct values with decreasing DNA concentrations. The se increased from 1 ng/µl to 10 fg/µl from 0.20 to 0.83 during the PCR efficiency testing and from 100 fg/µl to 12.5 fg/µl from 0.36 to 1 for the LOD determination (Fig 5).
FIG 3:
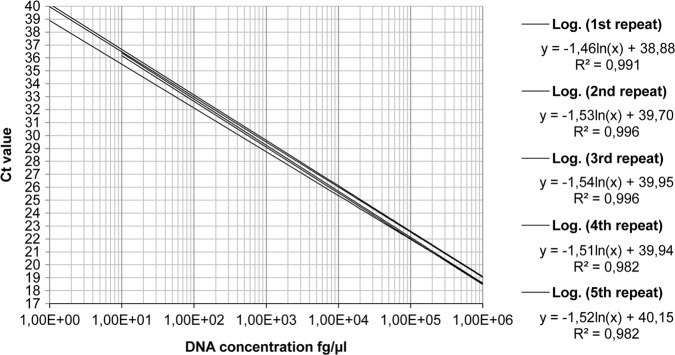
Logarithmic regression of quantitative real-time PCR efficiency testing experiments. Formulas show the slope values used for the efficiency calculation (slope ln). The values at the end of each formula show the y interceptions, which represent the theoretical maximal Ct values
FIG 4:
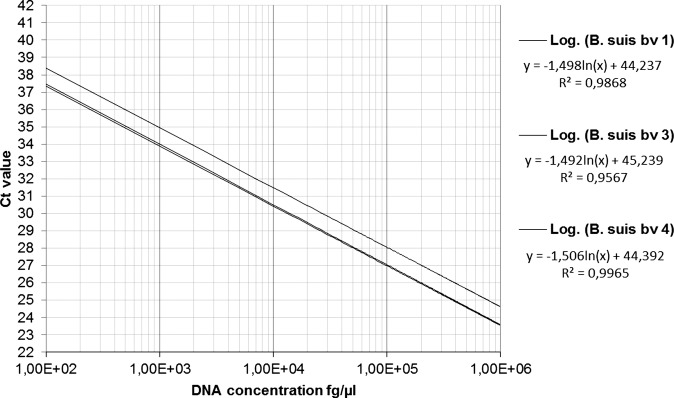
Logarithmic regression of the B. suis bv 1–4 genomic DNA standards. Formulas show the slope values used for the efficiency calculation (slope ln). The values at the end of each formula show the y-interceptions, which represent the theoretical maximal Ct values
FIG 5:
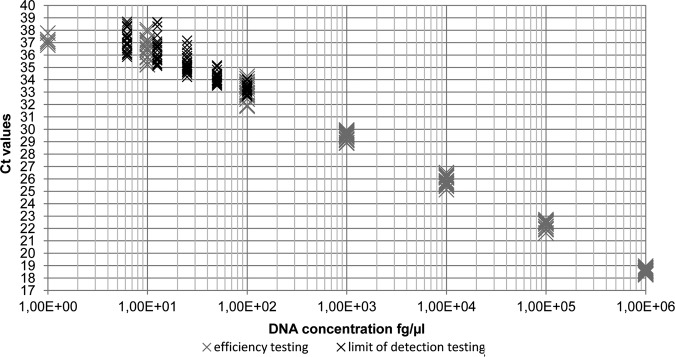
Scatter plot showing the increasing variation of Ct values with decreasing concentrations of B. suis bv 2 genomic DNA
Limit of detection
The developed qPCR assay reproducibly detected 12.5 fg/µl of B. suis bv 2 DNA (3.7 bacterial genomes). At this DNA concentration all qPCR assays were 100 per cent positive. However, at a DNA concentration of 6.25 fg/µl (1.8 bacterial genomes), still 83 per cent of performed reactions were positive (Fig 5).
PCR specificity
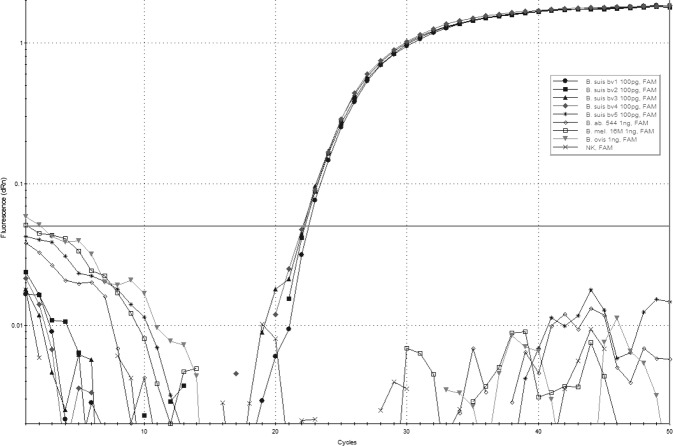
To evaluate the specificity of the B. suis qPCR, DNA preparations from 30 non-Brucella species (Table 1), 25 Brucella spp. reference strains including five B. suis reference strains (Table 1), representing all five biovars were tested at a concentration of 100 pg/µl. For all 30 non-Brucella species negative results were obtained with Ct values not exceeding the calculated y-intercept from qPCR efficiency testing of 40 for any DNA concentration used (data not shown). Similar results were obtained for all Brucella spp. analysed. Only DNA from B. suis bv 1–4 strains showed positive results with Ct values of approximately 22. There were no suspicious or undefined signals for non-B. suis bv 1–4 samples (Fig 6). Furthermore we analysed 75 field isolates (Table 2) from confirmed B. suis cases from various animal species and geographical regions which were all qPCR positive for B. suis bv 1–4. One sample, which was a confirmed human case of B. suis bv 5 was tested negative.
FIG 6:
Amplification curves of B. suis bv 1–4 (100 pg/µl DNA) with Ct values from 22.08 to 22.52 and of B. suis bv 5, B. abortus, B. melitensis and B. ovis (1 ng/µl DNA). Raw data were processed with an adaptive baseline correction and a moving average of three amplification points. The threshold was set to 0.05 (dRn)
Isolated DNA of the true positive spleen sample from the field was tested positive by B. suis qPCR also.
Discussion
The qPCR is a fast and reliable tool with still growing importance for direct detection of pathogens in clinical samples. For the diagnosis of brucellosis it has been already established and shown its usefulness. But till now and to the knowledge of the authors there is no description of a single probe qPCR that is able to detect all practically relevant B. suis bv 1–4. A test panel of overall 100 Brucella and 30 non-Brucella strains showed 100 per cent specificity of the new assay for B. suis bv 1–4. Our B. suis qPCR has an average efficiency of 95 per cent. This value shows its suitability for quantification of clinical samples even at very low concentrations of isolated DNA. A comparison of B. suis bv 1–4 showed no significant differences in Ct-values for a serial dilution 1:10 from 1 ng/µl to 1 pg/µl of DNA. For the efficiencies calculated from standard curves only B. suis bv 4 showed a decreased value of 88 per cent whereas B. suis bv 1–3 had efficiencies of 93–94.5 per cent.
The limit of detecting was set at a concentration of 12.5 fg/µl of template DNA. This means the assay is able to detect three to four bacterial genomes in 1 µl of DNA isolated from clinical samples. To our knowledge this is the highest sensitivity reached with a B. suis-specific qPCR assay (Redkar and others 2001, Bogdanovich and others 2004, Probert and others 2004, Al Dahouk and others 2007b, Fretin and others 2008). Nevertheless we observed an increased variance of Ct values for DNA concentrations of 100 fg/µl down to the LOD with 12.5 fg/µl. We think the only reason for this is the growing impact of dilution errors and the resulting variance of used DNA for a single qPCR reaction. By spiking DNA into eluates from negative swine sera we also showed that the assay could be useful for diagnosis of clinical samples. DNA directly isolated from tissue of a spleen sample of one, by isolation proofed Brucella infected hare, was tested positive in the B. suis qPCR. To use the method for clinical samples like sera or biopsy material a true validation procedure has to be done.
A real-time PCR based single nucleotide polymorphism analysis (Gopaul and others 2008) differentiates B. abortus, B. melitensis, B. ovis, B. suis, B. canis, B. neotomae and marine Brucella spp. but with lower sensitivity than what seems to be necessary to examine field samples.
Regarding the problems with specificity and sensitivity in serology and the insufficient limits of detecting in most qPCR assays, this new assay could play a key role in improving the performance of large-scale brucellosis screenings of wild boar or hare populations and surveillance in domestic pigs.
Acknowledgments
The authors gratefully thank Nadine Lemser, Michaela Ganß, Peggy Marten and Oda Schneider for excellent technical assistance.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- Al Dahouk S., Flèche P. L., Nöckler K., Jacques I., Grayon M., Scholz H. C., Tomaso H., Vergnaud G., Neubauer H. (2007a) Evaluation of Brucella MLVA typing for human brucellosis. Journal of Microbiological Methods 69, 137–145 doi:10.1016/j.mimet.2006.12.015 [DOI] [PubMed] [Google Scholar]
- Al Dahouk S., Nockler K., Scholz H. C., Pfeffer M., Neubauer H., Tomaso H. (2007b) Evaluation of genus-specific and species-specific real-time PCR assays for the identification of Brucella spp. Clinical Chemistry and Laboratory Medicine 45, 1464–1470 doi:10.1515/CCLM.2007.305 [DOI] [PubMed] [Google Scholar]
- Alton G. G., Jones L. M., Angus R. D., Verger J. M. (1988) Techniques for the Brucellosis Laboratory. Paris: INRA [Google Scholar]
- Bogdanovich T., Skurnik M., Lubeck P. S., Ahrens P., Hoorfar J. (2004) Validated 5′ nuclease PCR assay for rapid identification of the genus Brucella. Journal of Clinical Microbiology 42, 2261–2263 doi:10.1128/JCM.42.5.2261-2263.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker B. J. (2002) PCR as a diagnostic tool for brucellosis. Veterinary Microbiology 90, 435–446 doi:10.1016/S0378-1135(02)00228-6 [DOI] [PubMed] [Google Scholar]
- Cvetnic Z., Spicic S., Toncic J., Majnaric D., Benic M., Albert D., Thiebaud M., Garin-Bastuji B. (2009) Brucella suis infection in domestic pigs and wild boar in Croatia. Revue scientifique et technique 28, 1057–1067 [DOI] [PubMed] [Google Scholar]
- Farrell I. D. (1974) The development of a new selective medium for the isolation of Brucella abortus from contaminated sources. Research in Veterinary Science 16, 280–286 [PubMed] [Google Scholar]
- Forbes L. B. (1991) Isolates of Brucella suis biovar 4 from animals and humans in Canada, 1982–1990. The Canadian Veterinary Journal 32, 686–688 [PMC free article] [PubMed] [Google Scholar]
- Fretin D., Whatmore A. M., Al Dahouk S., Neubauer H., Garin-Bastuji B., Albert D., Van Hessche M., Menart M., Godfroid J., Walravens K., Wattiau P. (2008) Brucella suis identification and biovar typing by real-time PCR. Veterinary Microbiology 131, 376–385 doi:10.1016/j.vetmic.2008.04.003 [DOI] [PubMed] [Google Scholar]
- Garcia-Yoldi D., Le Fleche P., De Miguel M. J., Munoz P. M., Blasco J. M., Cvetnic Z., Marin C. M., Vergnaud G., Lopez-Goni I. (2007) Comparison of multiple-locus variable-number tandem-repeat analysis with other PCR-based methods for typing Brucella suis isolates. Journal of Clinical Microbiology 45, 4070–4072 doi:10.1128/JCM.01096-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst S., Wolf P., Uhl W., Risch K., Wolf C., Gerst K., Seelmann M. (2010) Incidence of brucella suis in wild boars in mecklenburg-western Pomerania—Pathological, bacteriological and molecular biological results within a monitoring program. Vorkommen der brucellose beim Scliwarzwild in mecklenburg-vorpommern - Pathologist Bef unde und Erreger-/Genomnachweise im Rahmen eines monitoringprogramms 65, 336–341 [Google Scholar]
- Godfroid J., Kasbohrer A. (2002) Brucellosis in the European Union and Norway at the turn of the twenty-first century. Veterinary Microbiology 90, 135–145 doi:10.1016/S0378-1135(02)00217-1 [DOI] [PubMed] [Google Scholar]
- Gopaul K.K., Koylass M.S., Smith C.J., Whatmore A.M. (2008) Rapid identification of Brucella isolates to the species level by real time PCR based single nucleotide polymorphism (SNP) analysis. BMC Microbiol 8, 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire F., Mousset B., Hanrez D., Michaux C., Walravens K., Linden A. (2012) A serological and bacteriological survey of brucellosis in wild boar (Sus scrofa) in Belgium. BMC Veterinary Research 8, 80 doi:10.1186/1746-6148-8-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurvell B. (1973) Serological cross-reactions between different Brucella species and Yersinia enterocolitica. Biological and chemical investigations of lipopolysaccharides from Brucella abortus and Yersinia enterocolitica type IX. Acta pathologica et microbiologica Scandinavica. Section B: Microbiology and immunology 81, 105–112 [DOI] [PubMed] [Google Scholar]
- Leuenberger R., Boujon P., Thur B., Miserez R., Garin-Bastuji B., Rufenacht J., Stark K. D. (2007) Prevalence of classical swine fever, Aujeszky's disease and brucellosis in a population of wild boar in Switzerland. The Veterinary Record 160, 362–368 doi:10.1136/vr.160.11.362 [DOI] [PubMed] [Google Scholar]
- Lopez-Goni I., Garcia-Yoldi D., Marin C. M., De Miguel M. J., Barquero-Calvo E., Guzman-Verri C., Albert D., Garin-Bastuji B. (2011) New Bruce-ladder multiplex PCR assay for the biovar typing of Brucella suis and the discrimination of Brucella suis and Brucella canis. Veterinary Microbiology 154, 152–155 doi:10.1016/j.vetmic.2011.06.035 [DOI] [PubMed] [Google Scholar]
- Melzer F., Lohse R., Nieper H., Liebert M., Sachse K. (2007) A serological study on brucellosis in wild boars in Germany. European Journal of Wildlife Research 53, 153–157 doi:10.1007/s10344-006-0072-0 [Google Scholar]
- Munoz P. M., Boadella M., Arnal M., De Miguel M. J., Revilla M., Martinez D., Vicente J., Acevedo P., Oleaga A., Ruiz-Fons F., Marin C. M., Prieto J. M., De La Fuente J., Barral M., Barberan M., De Luco D. F., Blasco J. M., Gortazar C. (2010) Spatial distribution and risk factors of Brucellosis in Iberian wild ungulates. BMC Infectious Diseases 10, 46 doi:10.1186/1471-2334-10-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probert W. S., Schrader K. N., Khuong N. Y., Bystrom S. L., Graves M. H. (2004) Real-time multiplex PCR assay for detection of Brucella spp., B. abortus, and B. melitensis. Journal of Clinical Microbiology 42, 1290–1293 doi:10.1128/JCM.42.3.1290-1293.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar R., Rose S., Bricker B., Delvecchio V. (2001) Real-time detection of Brucella abortus, Brucella melitensis and Brucella suis. Molecular and Cellular Probes 15, 43–52 doi:10.1006/mcpr.2000.0338 [DOI] [PubMed] [Google Scholar]
- Risco D., Garcia A., Serrano E., Fernandez-Llario P., Benitez J. M., Martinez R., Garcia W. L., De Mendoza J. H. (2014) High-density dependence but low impact on selected reproduction parameters of Brucella suis Biovar 2 in wild boar hunting estates from South-Western Spain. Transboundary and Emerging Diseases 61, 555–62 doi:10.1111/tbed.12060 [DOI] [PubMed] [Google Scholar]
- Von Roost H., Seelmann M., Konow M., Klopries M., Melzer F., Wölk R., Kay M., Dey E., Mildner H., Heyne H. (2010) Early recognition and monitoring of brucellosis in free-range pig farms in Mecklenburg-Western Pomerania, Germany. Untersuchungen zur früherkennung und überwachung der schweinebrucellose (Brucella suis) in Freilandhaltungen in Mecklenburg-Vorpommern 65, 278–284 [Google Scholar]
- Wu N., Abril C., Thomann A., Grosclaude E., Doherr M. G., Boujon P., Ryser-Degiorgis M. P. (2012) Risk factors for contacts between wild boar and outdoor pigs in Switzerland and investigations on potential Brucella suis spill-over. BMC Veterinary Research 8, 116 doi:10.1186/1746-6148-8-116 [DOI] [PMC free article] [PubMed] [Google Scholar]