Abstract
Background
Bovine viral diarrhoea virus (BVDV) is a member of the genus Pestivirus that belongs to the family Flaviviridae. BVDV is found worldwide in cattle population and causes significant economic losses to the dairy and beef industries. Two distinct genotypes of BVDV exist: BVDV type 1 (BVDV-1) and BVDV type 2 (BVDV-2).
Objective:
The aim of the present study was to investigate retrospectively the presence of BVDV-2 in Spain.
Results
With this objective, 47 blood samples that had tested positive in an ELISA for BVDV antigen were selected. Samples had been submitted by practitioners to the Diagnostic Service of NEIKER. The 18 herds of origin were all located in the northern half of Spain. BVDV positive samples were genotyped by reverse transcription-PCR. BVDV-1 was detected with the highest frequency (46/47), in contrast to BVDV-2 (2/47). In one blood sample, both pestivirus genotypes, BVDV-1 and BVDV-2, were detected. Sequencing of a viral genomic region, 5′ untranslated region, confirmed the identity of the BVDV-2 isolate.
Conclusions
So far as the authors know, this is the first reported presence of BVDV-2 in cattle herds in Spain. This finding may have important implications for the epidemiology, diagnosis and control of BVDV infection in the country.
Keywords: Bovine viral diarrhoea virus (BVDV), Cattle, Pestiviruses
Introduction
Bovine viral diarrhoea virus (BVDV) is included in the genus Pestivirus of the family Flaviviridae. BVDV is found worldwide in cattle population and causes substantial economic losses, mainly due to its impact on health and reproduction (Fray and others 2000). Early embryonic death, mummification, congenital defects or abortion are possible consequences of infection during pregnancy. Moreover, fetuses infected during the first 120 days of gestation with non-cytopathic strains can develop immunotolerance and become lifelong carriers of the virus. These persistently infected (PI) animals are a continuous source of virus and play a major role in the spread of the disease. In those regions with a high prevalence of BVDV antibodies, over 1–2 per cent of the newborn calves may be PI (Houe 1999). Mucosal disease, a severe and fatal clinical form of BVDV infection, may develop in these PI animals in case of superinfection with cytopathic (cp) strains. Infection of a healthy adult is usually subclinical, and the main consequences are decrease in reproductive function and immunosuppression. Nevertheless, non-specific symptoms such as pyrexia or diarrhoea may be reported. A decrease in milk yield has also been described in lactating cows. In addition, the immunosuppressive effect of the virus probably plays an important role in calf respiratory disease complex (Howard 1990).
Two genotypes of BVDV (BVDV-1 and BVDV-2) have been differentiated using serology (monoclonal and polyclonal antisera) and molecular biology (Ridpath 2003). Additionally, subtypes of the two genotypes have also been described (Vilcek and others 2001, Ridpath 2003). Recently published reverse transcription-PCR (RT-PCR) methods facilitate typing BVDV at the genotype level directly from blood samples of PI cattle (Gilbert, and others 1999, Letellier and Kerkhofs 2003, Baxi and others 2006).
BVDV-2 has been associated with severe clinical disease in adult cattle and with a haemorrhagic syndrome in young animals (Carman and others 1998). This pestivirus genotype would appear to be more prevalent in North America (Fulton and others 2005), although it has been also described in other European (Letellier and others 1999, Luzzago and others 2001, Tajima and others 2001, Jackova and others 2008) and Asiatic (Nagai and others 1998) countries. In the past two years, a severe form of BVDV-2 with high mortality has been reported initially in Germany and later in the Netherlands (Schirrmeier 2014). Previous studies showed that the BVDV-1b subtype was predominantly circulating in the cattle population in Spain; at that time no evidence for the presence of BVDV-2 was found (Arias and others 2003, Hurtado and others 2003).
Methods
A total of 47 bovine blood samples BVDV positive by ELISA (BVD Ag Serum Plus, IDEXX) were included in the study. Samples had been submitted to the diagnostic service of NEIKER between 2012 and 2013 from 18 different cattle herds located in Northern Spain. Some clinical and epidemiological data were also recorded by the practitioners. ELISA results were confirmed with a highly sensitive real-time RT-PCR assay (Hofmann and others 2006).
A previously published one-step multiplex real-time RT-PCR for BVDV was used to characterise positive samples (Baxi and others 2006). This protocol allows a discrimination of the two genotypes BVDV-1 and BVDV-2 in the same assay. To set up the method BVDV-1 positive controls were obtained from the Veterinary Laboratory Agencies (UK), and those of the BVDV-2 were kindly supplied by Dr Meyers from the Friedrich-Loeffler Institute (Germany).
Briefly, total nucleic acid was extracted from blood samples (200 µl) using the QIAamp DNA Blood Mini Kit (Qiagen) following the manufacturer's recommendations but with the following modification: 1 µg of RNA carrier (Qiagen) was added to the buffer AL. Extracted RNA was eluted with 80 µl of RNase free water and stored at −70°C until needed. Real-time RT-PCR testing was performed in a final volume of 25 µl containing 1X One Step RT-PCR Buffer III, 0.5 µl of TaKaRa Ex Taq HS (HotStart Taq DNA polymerase), 0.5 µl of Primer Script RT enzyme Mix II (recombinant MMLV enzyme), 0.5 µl of Rox Dye II, 400nM of each PCR primer and 100 nM of the fluorescent probes (FAM-BVDV-1 and Cy5-BVDV-2). Before amplification, the RNA was transcribed at 42°C for 5 minutes. This was followed by one cycle of 94°C for 10 seconds for activation of the TaKaRa Ex Taq HS and inactivation of reverse transcriptase. Cycle times were as follows: 45 cycles of denaturation at 94°C for 5 seconds and of annealing and extension at 62°C for 34 seconds.
For genomic confirmation of BVDV-2 isolates, a 288 bp fragment from the 5′ untranslated region (5′ UTR) of the genome was amplified using primers 324 and 326 as described previously (Vilcek and others 1994). Purified amplicons were sequenced in both directions using the Big Dye Terminator V.3.1 Cycle Sequencing Kit (Applied Biosystems) and the ABI 3130 sequencer, as per manufacturer’s recommendations.
Sequence consensus was assembled using BioEdit V.7.2.5 software (Hall 1999). Primer sequences were clipped from each consensus before phylogenetic analysis conducted using MEGA 6 program (Tamura and others 2013). The sequence obtained was compared with those deposited in GenBank using the BLAST software. 5′ UTR sequences were imported into MEGA 6 and sequence alignments performed using ClustalW. Phylogenetic tree was constructed using the Neighbour-joining Kimura 2 parameter method (Kimura 1980). Bootstrap analysis of the resultant tree was performed using 1000 replicates. Bootstrap values of over 70 per cent are shown (Hillis and Bull 1993). The sequences included in the phylogenetic analysis were as follows: BVDV-1a (NADL-M31182, SD1-M96751); BVDV-1b (Osloss-M96687); BVDV-2b (Soldan-U94914, 17237-EU747875); and BVDV-2a (890-U18059, 1373-AF145967, 4 5174-AF298063, 15 103-AF298055, 37Gr-EU327594, 104/98-AJ304381, B91/05-EU224242, HI749-AY379543, HI917-AY379545, SH28-HQ258818, i61380-AF417986, 53100-FJ431190, 108KL/13-KJ616408, 111KL/13-KJ613410).
Results
The 18 herds included in the study were located in the northern half of Spain. A higher number of samples came from dairy herds (35) than from beef herds (12). Clinical problems reported by practitioners in the 18 affected herds were as follows: reproductive disease (8); mucosal disease (3); calf mortality (2); no disease, samples taken as part of a control programme for BVDV (4) and not reported (1). Thrombocytopenia, haemorrhages or other clinical signs attributed to highly virulent strains of BVDV were not reported in any of the cases.
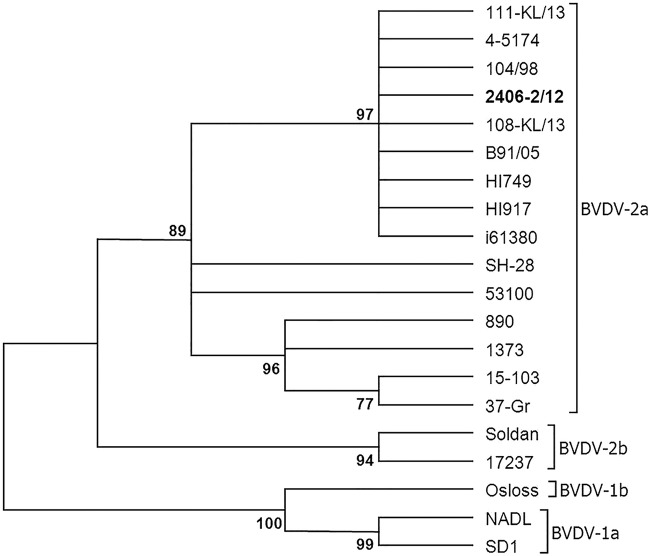
All 47 samples selected for being positive in the antigen ELISA were confirmed by the highly sensitive RT-PCR assay. As regards RT-PCR typing results, 45 of the 47 samples were typed as BVDV-1, 1 as BVDV-2 (strain 2406-2/12; Ct value: 33.9), and in the one remaining sample both pestivirus genotypes were detected (Ct value: 37.4). Sequencing of the PCR product of strain 2406-2/12 confirmed the identity of BVDV-2 (Fig 1).
FIG 1:
Phylogenetic analysis of 5′ untranslated region (5′ UTR) nucleotide sequences of selected bovine viral diarrhoea virus (BVDV) isolates
The 5′ UTR sequence for sample 2406-2/12 was submitted for a BLAST search using the blast algorithm of National Center for Biotechnology Information. It shared a 100 per cent identity to the strains 104/98 and 111-KL/13 isolated in Germany and Poland, respectively.
The phylogenetic tree constructed from 5′-UTR nucleotide sequences of selected BVDV-1 and BVDV-2 strains indicated that the strain 2406-2/12 was classified as BVDV-2a together with some isolates from Germany, France and Poland (Fig 1). The nucleotide sequence identity of the strain 2406-2/12 was closer related to the strain 890 (93 per cent) than to the BVDV-1 reference NADL and Osloss strains (76 per cent and 77 per cent, respectively).
Discussion
BVDV is found worldwide and causes considerable economic losses to the cattle industry, mainly due to its impact on reproduction. In a serological survey conducted in Asturias (North Spain), 86 per cent of the herds and 21 per cent of the animals were BVDV antibody positive (Mainar-Jaime and others 2001). More recently, 95 and 41 BVDV isolates were obtained from Spanish cattle herds, thus confirming that the virus is widespread in this country (Arias and others 2003, Hurtado and others 2003). In Spain BVDV-1b is the most prevalent subtype, as occurs in the USA and other European countries (Kuta and others 2013, Yan and others 2011). No evidence of BVDV-2 was found, suggesting either a very low prevalence or its absence. In the present study, 2 of the 47 isolates were identified as BVDV-2. As far as the authors are aware, this is the first detection of BVDV-2 in Spain. BVDV-2 was first reported in the USA (Ridpath, and others 1994), and later in other European countries, although at a lower rate than BVDV-1, except in Germany (Wolfmeyer and others 1997) where a prevalence >10 per cent was reported. Recently, the first report of BVDV-2 in Poland has been published (Polak and others 2014). Due to the low number of isolates analysed in the present study, it is not possible to determine with exactitude the prevalence of BVDV-2 in Spain. In consequence, further work is needed to establish the real extension of this pestivirus in Spanish cattle herds and the possible clinical consequences. Interestingly, one of the animals harboured both genotypes BVDV-1 and BVDV-2, indicating a mixed infection. In this case, sequencing was not performed due to the interference of the two peaks in the chromatogram. Dual infections have also been reported by others (Yan and others 2011). A superinfection of a PI animal with a different genotype could be an explanation for this finding.
The clinical symptoms reported in the herds of origin are consistent with BVDV infection. Reproductive signs were reported with a higher frequency. This is scarcely surprising as BVDV infection in pregnant cattle can result in early embryonic death, abortion, mummification or congenital defects. No clinical signs suggestive of an infection with highly virulent strains of BVDV were reported by any of the practitioners. This is consistent with the fact that most acute infections with either pestivirus genotype, BVDV-1 or BVDV-2, result in mild or subclinical disease (Ridpath 2003).
Both BVDV-2 infected animals originated from Asturias, an area with a high density of dairy herds. Apparently, both animals were clinically normal at the time of sampling. Trade in animals is one of the main risk factors for the entrance of BVDV infection (Houe 1999) and could also be the source in these cases. Control programmes based exclusively on testing and culling of PI animals were initiated in Nordic countries some years ago (Houe and others 2006). In other European countries, vaccination with killed or modified-live vaccines is a common practice to control BVDV infection. At present, there is not a countrywide control programme of BVDV in Spain. A globalised world with trade in animals, embryos and semen can favour the spread of viruses such as BVDV if biosecurity measures are not taken. In this scenario, knowledge of BVDV diversity is the first step towards understanding the epidemiology of the infection and towards the design of efficient diagnostic tests and vaccines (Hamers and others 2001). As for diagnostic tests, the two tests used in this study, antigen ELISA and RT-PCR, demonstrated perfect concordance (47/47). Therefore, both appear adequate for detecting BVDV infections in blood samples. BVDV vaccines available in Spain only contain BVDV-1. It has been reported that there are differences in the antigenic structure of both pestivirus genotypes BVDV-1 and BVDV-2, even at the subtype level (Ridpath and others 2010). Although several studies have demonstrated some degree of cross-immunity, better fetal protection is to be expected when a strain of the same genotype is used (Walz 2009). In a recent study, a vaccine containing BVDV-1 did not protect against a BVDV-2 infection (Polak and others 2014). The development of safe and efficacious vaccines with viruses from all antigenic groups has been identified as a priority (Hamers and others 2001) in order to implement control programmes of BVDV based on the vaccination of naive cattle.
Conclusion
It is reported in the present study, for the first time, the identification of BVDV-2 in Spanish cattle herds. Further work is required to determine the real prevalence of this pestivirus and the practical implications with regard to the diagnosis and control of BVDV infection in Spanish cattle population.
Acknowledgments
The authors thank Dr Gregor Meyers (Friedrich-Loeffler Institute, Germany) for supplying BVDV-2 RNA and for critically reading the manuscript. The authors are grateful to Itziar del Pozo, Gloria Rubio, Argiñe Zubieta and Ainara Badiola for the excellent technical assistance. Many thanks also to the practitioners and farmers who participated in the study.
Footnotes
Contributors: GA, RA and NC: participated in the design of the study and discussion of results. GA: drafted the manuscript. NC: performed the PCR analysis and genotyping. All the authors read and approved the final manuscript.
Funding: Viceconsejería de Agricultura, Pesca y Política Alimentaria of the Basque Government & Boehringer-Ingelheim Spain.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- Arias P., Orlich M., Prieto M., Cedillo-Rosales S., Thiel H. -J., Álvarez M., Becher P. (2003) Genetic heterogeneity of bovine viral diarrhoea viruses from Spain. Veterinary Microbiology 96, 327–336 doi:10.1016/j.vetmic.2003.09.009 [DOI] [PubMed] [Google Scholar]
- Baxi M., McRae D., Baxi S., Greiser-Wilke I., Vilcek S., Amoako K., Deregt D. (2006) A one-step multiplex real-time RT-PCR for detection and typing of bovine viral diarrhoea viruses. Veterinary Microbiology 116, 37–44 doi:10.1016/j.vetmic.2006.03.026 [DOI] [PubMed] [Google Scholar]
- Carman S., van Dreumel T., Ridpath J., Hazlett M., Alves D., Dubovi E., Tremblay R., Bolin S., Godkin A., Anderson N. (1998) Severe acute bovine viral diarrhea in Ontario, 1993–1995. Journal of Veterinary Diagnostic Investigation 10, 27–35 doi:10.1177/104063879801000106 [DOI] [PubMed] [Google Scholar]
- Fray M. D., Paton D. J., Alenius S. (2000) The effects of bovine viral diarrhoea virus on cattle reproduction in relation to disease control. Animal Reproduction Science 60–61, 615–627 doi:10.1016/S0378-4320(00)00082-8 [DOI] [PubMed] [Google Scholar]
- Fulton R. W., Ridpath J. F., Ore S., Confer A. W., Saliki J. T., Burge L. J., Payton M. E. (2005) Bovine viral diarrhoea virus (BVDV) subgenotypes in diagnostic laboratory accessions: Distribution of BVDV1a, 1b, and 2a subgenotypes. Veterinary Microbiology 111, 35–40 doi:10.1016/j.vetmic.2005.10.002 [DOI] [PubMed] [Google Scholar]
- Gilbert S. A., Burton K. M., Prins S. E., Deregt D. (1999) Typing of bovine viral diarrhea viruses directly from blood of persistently infected cattle by multiplex PCR. Journal of Clinical Microbiology 37, 2020–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symposium Series 41, 95–98 [Google Scholar]
- Hamers C., Dehan P., Couvreur B., Letellier C., Kerkhofs P., Pastoret P. P. (2001) Diversity among bovine pestiviruses. The Veterinary Journal 161, 112–122 doi:10.1053/tvjl.2000.0504 [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Bull J. J. (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42, 182–192 doi:10.1093/sysbio/42.2.182 [Google Scholar]
- Hofmann B., Depner K., Schirrmeier H., Beer M. (2006) A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. Journal of Virological Methods 136, 200–209 doi:10.1016/j.jviromet.2006.05.020 [DOI] [PubMed] [Google Scholar]
- Houe H. (1999) Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Veterinary Microbiology 64, 89–107 doi:10.1016/S0378-1135(98)00262-4 [DOI] [PubMed] [Google Scholar]
- Houe H., Lindberg A., Moening V. (2006) Test strategies in bovine viral diarrhea virus control and eradication campaigns in Europe. Journal of Veterinary Diagnostic Investigation 18, 427–436 doi:10.1177/104063870601800501 [DOI] [PubMed] [Google Scholar]
- Howard C. J. (1990) Immunological responses to bovine virus diarrhoea virus infections. Revue scientifique et technique Office international des Épizooties 9, 95–103 [DOI] [PubMed] [Google Scholar]
- Hurtado A., García-Pérez A. L., Aduriz G., Juste R. A. (2003) Genetic diversity of ruminant pestiviruses from Spain. Virus Research 92, 67–73 doi:10.1016/S0168-1702(02)00315-5 [DOI] [PubMed] [Google Scholar]
- Jackova A., Novackova M., Pelletier C., Audeval C., Gueneau E., Haffar A., Petit E., Rehby L., Vilcek S. (2008) The extended genetic diversity of BVDV-1: Typing of BVDV isolates from France. Veterinary Research Communications 32, 7–11 doi:10.1007/s11259-007-9012-z [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16, 111–120 doi:10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Kuta A., Polak M. P., Larska M., Zmudzinski J. F. (2013) Predominance of bovine viral diarrhea virus 1b and 1d subtypes during eight years of survey in Poland. Veterinary Microbiology 166, 639–644 doi:10.1016/j.vetmic.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Letellier C., Kerkhofs P. (2003) Real-time PCR for simultaneous detection and genotyping of bovine viral diarrhoea virus. Journal of Virological Methods 114, 21–27 doi:10.1016/j.jviromet.2003.08.004 [DOI] [PubMed] [Google Scholar]
- Letellier C., Kerkhofs P., Wellemans G., Vanopdenbosch E. (1999) Detection and genotyping of bovine diarrhoea virus by reverse transcription-polymerase chain amplification of the 5′ untranslated region. Veterinary Microbiology 64, 155–167 doi:10.1016/S0378-1135(98)00267-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzzago C., Bandi C., Bronzo V., Ruffo G., Zecconi A. (2001) Distribution pattern of bovine viral diarrhoea virus strains in intensive cattle herds in Italy. Veterinary Microbiology 83, 265–274 doi:10.1016/S0378-1135(01)00429-1 [DOI] [PubMed] [Google Scholar]
- Mainar-Jaime R. C., Berzal-Herranz B., Arias P., Rojo-Vázquez F. A. (2001) Epidemiological pattern and risk factors associated with bovine viral diarrhoea virus (BVDV) infection in non-vaccinated dairy cattle population from the Asturias region of Spain. Preventive Veterinary Medicine 52, 63–73 doi:10.1016/S0167-5877(01)00239-2 [DOI] [PubMed] [Google Scholar]
- Nagai M., Sato M., Nagano H., Pang H., Kong X., Murakami T., Ozawa T., Akashi H. (1998) Nucleotide sequence homology to bovine viral diarrhea virus 2 (BVDV 2) in the 5′ untranslated region of BVDVs from cattle with mucosal disease or persistent infection in Japan. Veterinary Microbiology 60, 271–276 doi:10.1016/S0378-1135(98)00158-8 [DOI] [PubMed] [Google Scholar]
- Polak M. P., Kuta A., Rybałtowski W., Rola J., Larska M., Żmudziński J. F. (2014) First report of bovine viral diarrhoea virus-2 infection in cattle in Poland. The Veterinary Journal 202, 643–645 doi:doi:10.1016/j.tvjl.2014.09.026 [DOI] [PubMed] [Google Scholar]
- Ridpath J. F. (2003) BVDV genotypes and biotypes: practical implications for diagnosis and control. Biologicals, 31, 127–131 doi:10.1016/S1045-1056(03)00028-9 [DOI] [PubMed] [Google Scholar]
- Ridpath J. F., Bolin S. R., Dubovi E. J. (1994) Segregation of bovine viral diarrhoea virus into genotypes. Virology 205, 66–74 doi:10.1006/viro.1994.1620 [DOI] [PubMed] [Google Scholar]
- Ridpath J. F., Fulton R. W., Kirkland P. D., Neill J. D. (2010) Prevalence and antigenic differences observed between Bovine viral diarrhea virus subgenotypes isolated from cattle in Australia and feedlots in the south-western United States. Journal of Veterinary Diagnostic Investigation 22, 184–191 doi:10.1177/104063871002200203 [DOI] [PubMed] [Google Scholar]
- Schirrmeier U. (2014) Three years of mandatory BVD control in Germany – lessons to be learned. Proceedings of the XXVIII World Buiatrics Congress Cairns, Australia: pp 245–248 [Google Scholar]
- Tajima M., Frey H. R., Yamato O., Maede Y., Moenning V., Scholz H., Greiser-Wilke I. (2001) Prevalence of genotypes 1 and 2 of bovine viral diarrhea virus in Lower Saxony, Germany. Veterinary Microbiology 76, 31–42 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729 doi:10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilcek S., Herring A. J., Herring J. A., Nettleton P. F, Lowings J. P., Paton D. J. (1994) Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Archives of Virology 136, 309–323 doi:10.1007/BF01321060 [DOI] [PubMed] [Google Scholar]
- Vilcek S., Paton D. J., Durkovic B., Strojny L., Ibata G., Moussa A., Loitsch A., Rossmanith W., Vega S., Scicluna M. T., Palfi V. (2001) Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Archives of Virology 146, 99–115 doi:10.1007/s007050170194 [DOI] [PubMed] [Google Scholar]
- Walz P. H. (2009) Bovine viral diarrhea virus. In: Current Veterinary Therapy 5th edition: Food Animal Practice. Eds Anderson D. E., Rings D. M.. Missouri, USA: Saunders, Elsevier; pp 96–106 [Google Scholar]
- Wolfmeyer A., Wolf G., Beer M., Strube W., Hehnen H. R., Schmeer N., Kaaden O. R. (1997) Genomic (5′-UTR) and serological differences among German BVDV field isolates. Archives of Virology 142, 2049–2057 doi:10.1007/s007050050222 [DOI] [PubMed] [Google Scholar]
- Yan L., Zhang S., Pace L., Wilson F., Wan H., Zhang M. (2011) Combination of reverse transcription real-time polymerase chain reaction and antigen capture enzyme-linked immunosorbent assay for the detection of animals persistently infected with bovine viral diarrhea virus. Journal of Veterinary Diagnostic Investigation 23, 16–25 doi:10.1177/104063871102300103 [DOI] [PubMed] [Google Scholar]