Abstract
The intracellular localization and colocalization of a fluorescently labeled G3 amine-terminated cationic polyamidoamine (PAMAM) dendrimer and its biotin–pyridoxal (BC-PAMAM) bioconjugate were investigated in a concentration-dependent manner in normal human fibroblast (BJ) and squamous epithelial carcinoma (SCC-15) cell lines. After 24 hours treatment, both cell lines revealed different patterns of intracellular dendrimer accumulation depending on their cytotoxic effects. Cancer cells exhibited much higher (20-fold) tolerance for native PAMAM treatment than fibroblasts, whereas BC-PAMAM was significantly toxic only for fibroblasts at 50 µM concentration. Fibroblasts accumulated the native and bioconjugated dendrimers in a concentration-dependent manner at nontoxic range of concentration, with significantly lower bioconjugate loading. After reaching the cytotoxicity level, fluorescein isothiocyanate-PAMAM accumulation remains at high, comparable level. In cancer cells, native PAMAM loading at higher, but not cytotoxic concentrations, was kept at constant level with a sharp increase at toxic concentration. Mander’s coefficient calculated for fibroblasts and cancer cells confirmed more efficient native PAMAM penetration as compared to BC-PAMAM. Significant differences in nuclear dendrimer penetration were observed for both cell lines. In cancer cells, PAMAM signals amounted to ~25%–35% of the total nuclei area at all investigated concentrations, with lower level (15%–25%) observed for BC-PAMAM. In fibroblasts, the dendrimer nuclear signal amounted to 15% at nontoxic and up to 70% at toxic concentrations, whereas BC-PAMAM remained at a lower concentration-dependent level (0.3%–20%). Mitochondrial localization of PAMAM and BC-PAMAM revealed similar patterns in both cell lines, depending on the extracellular dendrimer concentration, and presented significantly lower signals from BC-PAMAM, which correlated well with the cytotoxicity.
Keywords: PAMAM G3 dendrimer, bioconjugate, normal and cancer cells, nuclei, mitochondria, confocal microscopy, colocalization
Introduction
Dendrimers are involved in numerous pharmaceutical and biomedical research applications and act as ubiquitous carriers for targeted drugs, nucleic acids, and diagnostic agents. Growing research interest over the last 2 decades has resulted in a number of publications concerning dendrimers. In particular, the amine-terminated cationic polyamidoamine (PAMAM) dendrimers of various generations and their bioconjugates have been intensively studied because of their wide range of possible applications as carriers of antibodies and contrast molecules,1,2 in protection against nonenzymatic modifications of biomacromolecules that cause various metabolic disorders,3 in therapies for cancer,4–8 and genetic and immune diseases.9,10 The unique molecular architecture of dendrimers allows for the precise control of their size, shape, charge and targeting of ligands, and therapeutic compounds attached to surface groups, which determine their function and application.11,12 The design and synthesis of highly effective carrier systems depend on understanding the mechanisms of delivery and internalization of modified dendrimers inside the targeted cells. Because of the possible localization of dendrimer bioconjugates in intracellular domains (cytosol, endosomes, lysosomes, endoplasmic reticulum, Golgi apparatus, mitochondria, and nucleus) as well as the direct interaction with cellular membranes and specificity of various cells and tissues (normal and pathological), interdisciplinary studies are required to achieve the desired effects.13 To assess the safety of dendrimers, the possible toxic and immunogenic effects on cells and organisms in vivo and in vitro must be considered.14–17 An important property of cationic dendrimers is the binding and aggregation of DNA; thus, these dendrimers are excellent tools for gene delivery, but they also have the genotoxic potential under different conditions.18,19 Therefore, it is essential to understand the movement of delivered carriers within the cells, particularly nuclear penetration.
The interactions of PAMAM dendrimers with cell membranes and their internalization have been extensively investigated in vitro in various cell lines using specific markers; indirectly through flow cytometry and directly through scanning electron microscopy,20,21 atomic force microscopy, and fluorescence microscopy.22 The application of fluorescence imaging in living cells by confocal laser scanning microscopy and recently by two-photon imaging microscopy can monitor the dynamic aspects of cellular trafficking and colocalization of dendrimers with high spatial and temporal resolution.23–25 The cellular internalization and trafficking of dendrimers depend on their size, shape, charge, and surface functionality as well as the cell type.26,27 Many studies have revealed that cationic PAMAM dendrimers can cross cell membranes by various endocytic pathways, including clathrin-dependent pathways in Caco-2 cells,28,29 clathrin-dependent and macropinocytosis pathways in HeLa cells,30 and non-clathrin, non-caveolae, cholesterol-dependent pathways in A549 lung epithelial cells and B16F10 melanoma cells.31,32 The conjugation of small bioactive molecules to PAMAM cationic dendrimers increases their transport and decreases toxicity, and it has been demonstrated in vitro and in vivo by numerous investigators as reviewed by Sadekar and Ghandehari.33 A potential carrier system is created through PAMAM dendrimers modified with vitamins and their precursors, which are exogenous molecules that can penetrate cells by various transport mechanisms and participate in a variety of cellular functions. Water-soluble group B vitamins are indispensable for a number of vital metabolic pathways in mammals. One member of this group, biotin, is required for anabolic carboxylation and carboxylic group transfer.34,35 Several authors have shown that rapidly dividing cells, such as found in malignant tumors, develop significantly increased biotin uptake through the increased expression of cell surface receptors and activation of endocytosis.36 Biotin conjugates are therefore considered to be biocompatible carriers for delivering anticancer drugs, and they can be transferred through the blood–brain barrier.37–39 Pyridoxal (PL) and its 5′ phosphorylated derivative are active forms of vitamin B6 that play a critical role in cellular metabolism and stress response.40,41 Recent studies have highlighted the importance of the protective role of vitamin B6 in carcinogenesis, especially colon carcinogenesis, by reducing cell proliferation, oxidative stress, NO production, and angiogenesis.42,43 Our previous study revealed that after internalization, biotin–PL-conjugated PAMAM G3 dendrimers had lower cytotoxicity than the nonconjugated forms of the dendrimer in both normal human fibroblasts (BJ) and squamous epithelial carcinoma (SCC-15) cancer cells.44 Although the various dendrimer transcellular transport mechanisms were the subjects of a wide range of investigation, including quantitative studies,45 less is known of the nuclear or mitochondrial localization of native PAMAM dendrimers and their bioconjugates, particularly when located inside normal cells as opposed to cancer cells. Published reports on the intracellular distribution of dendrimers, that are not designed for nuclear gene delivery or as mitochondria-targeted carriers, vary depending on the dendrimer surface modification, concentration, cell line, and time of observation. Such variations are particularly important when considering potential genomic DNA damage18 and the deleterious effects of dendrimers on mitochondrial function in normal and cancer cells.46 The main objective of this study was to investigate the nuclear and mitochondrial penetration of tertiary bioconjugate biotin–PL–PAMAM G3 (BC-PAMAM) in relation to that of native PAMAM G3 dendrimers in human fibroblasts (BJ) and epithelial cancer (SCC-15) cell lines at nontoxic and toxic concentrations. Dendrimer accumulation and localization in the nuclei and mitochondria were investigated by laser scanning confocal microscopy imaging. The effect on cell viability and proliferation was also investigated and discussed.
Materials and methods
Materials
BJ (ATCC-CRL 2522) and SCC-15 (ATCC-CRL-1623) cell lines and Dulbecco’s Modified Eagle’s Medium (DMEM-F12), Eagle’s minimum essential medium (EMEM), fetal bovine serum (FBS), penicillin, streptomycin, trypsin/ethylenediaminetetraacetic acid solution, and phosphate-buffered saline (PBS; with and without magnesium and calcium ions) were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA). Hydrocortisone, Trypan blue, and fluorescein isothiocyanate (FITC) were provided by Sigma-Aldrich Co. (St Louis, MO, USA). The fluorescent markers 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI), MitoTracker® Deep Red FM and Rhodamine Red™-X (succinimidyl ester, 5-isomer), ATP Determination Kit, and CyQUANT® NF Cell Proliferation Assay Kit were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Cell culture sterile plastics were purchased from Corning Incorporated (Corning, NY, USA), Sigma-Aldrich Co., and Brand GMBH + CO KG (Wertheim, Germany).
Synthesis of biotin and PL-conjugated and fluorescent-labeled dendrimers
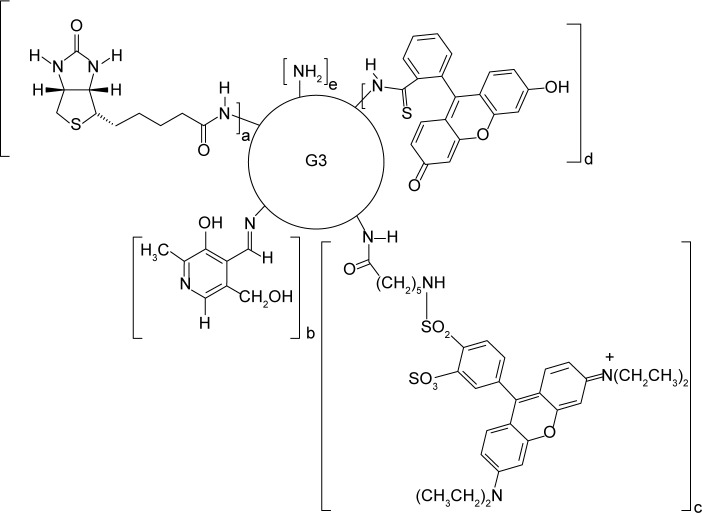
The bioconjugate of the PAMAM G3 dendrimer with nine biotin (linked via amide bond) and ten PL (linked via aldimine bond) G39B10P was obtained as previously described.44 The PAMAM G3 (PAMAM) and G39B10P (BC-PAMAM) were fluorescently labeled with FITC as described by Filipowicz and Wołowiec,47 whereas G3 was labeled with Rhodamine Red (Figure 1) as follows: 5 mg (6.5 µmol) Rhodamine Red™-X (succinimidyl ester, 5-isomer, molecular weight 768.9 Da) was added to 45 mg PAMAM G3 dissolved in methanol and stirred for 2 hours in darkness until all of the N-hydroxysuccinimidyl (NHS)-rhodamine dissolved. The methanol was vacuum evaporated, and the residue was dissolved in water and dialyzed against water for 48 hours (membrane cutoff 2,000 Da) in darkness. Next, the solvents were removed under vacuum to obtain G3R (Figure 1), where R = rhodamine linked via amide bond. The G39B bioconjugate and ternary bioconjugate G39B10P (BC-PAMAM) were identified by proton nuclear magnetic resonance (1H NMR) spectroscopy as previously described.44 PAMAM G3 is >32–36 Å in diameter depending on the calculation approach and experimental method.48
Figure 1.
Schematic presentation of the formulas of the conjugates.
Notes: BC-PAMAM (a=10, b=9, c=0, d=0, e=13), FITC-BC-PAMAM (a=9, b=10, c=0, d=1, e=12), RR-PAMAM (a=0, b=0, c=1, d=0, e=31) and FITC-PAMAM (a=0, b=0, c=0, d=1, e=31). a – biotin, b – pyridoxal, c – Rhodamine Red™-X, d – fluorescein isothiocyanate (FITC).
Abbreviations: BC-PAMAM, biotin–polyamidoamine; FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate.
Cell cultures
Two human cell lines were investigated, such as normal fibroblasts BJ (ATCC CRL-2522) and squamous cell carcinoma SCC-15 (ATCC CRL-1623), both growing in layers. BJ cells (population doubling 1.9 days) were cultured in EMEM supplemented with nonessential amino acids, 10% heat-inactivated FBS, 100 U/mL penicillin, and 100 µg/mL streptomycin. SCC-15 cells (population doubling 5 days) were grown in DMEM containing 15 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, and 400 ng/mL hydrocortisone. The cells were grown at 37°C in an atmosphere of 5% CO2 and 95% humidity, and the medium was changed every 2–3 days and passaged at 70%–85% confluence using 0.25% trypsin–0.03% ethylenediaminetetraacetic acid in PBS (calcium and magnesium free). The cell morphology was verified under a Nikon TE2000S Inverted Microscope (Tokyo, Japan) with phase contrast, and the cell numbers and viability were estimated by a Trypan blue exclusion test using an Automatic Cell Counter TC10™ (Bio-Rad Laboratories Inc., Hercules, CA, USA) or hemocytometer. All experiments were performed in triplicate. No Ethics Committee permission was required for the use of human cell lines because only certificated cell lines provided by ATCC were used.
Cytotoxicity assay
The CyQUANT NF Cell Proliferation Assay Kit (Molecular Probes®) is a sensitive assay for the quantitation of cell number using a microplate reader with excitation at 485 nm and emission detection at 530 nm. The dye is cell permeable, develops an intense fluorescence signal after binding to cellular DNA, and has linearity over a broad range of cell densities (50–5×104 cells/well).49 BJ and SCC-15 cells were seeded (1×104 cells/well) into 96-well plates and preincubated (24 hours, 37°C, 5% CO2, 95% humidity). Subsequently, 1 mM dendrimer stock solution in water was filter sterilized and then added. Working solutions were prepared in culture media and delivered at 200 µL/well in concentrations ranging from 0.05 µM to 50 µM. After 24 hours, the media were exchanged for new media without dendrimers (200 µL/well), and the microplates with BJ and SCC-15 cells were returned to the incubator for 2 days and 5 days, respectively. After removing the medium, CyQUANT NF/HBSS working solution was added (100 µL/well) according to the manufacturer’s protocol. After the mixture was incubated for 30 minutes at 37°C in the dark the fluorescence was measured using a microplate reader (FilterMax F5 Multi-Mode Microplate Reader; Molecular Devices LLC, Sunnyvale, CA, USA) at 485 nm/530 nm wavelength. The fluorescence signal corresponds to the number of adherent, viable cells. The results are expressed as the % of the control, nontreated cell samples.
Cellular accumulation of PAMAM and BC-PAMAM
The evaluation of the cellular dendrimer accumulation was performed as previously described.50 The cells were seeded into 96-well microtiter plates at a density of 2×104 cells/well and preincubated (37°C, 5% CO2, 95% humidity) for 24 hours, and then various concentrations (0.05–50 µM) of the FITC-conjugated native PAMAM or BC-PAMAM were added. After an additional 24 hours incubation, the medium was removed and plates were extensively washed with PBS using an automatic cell washer (ELx50; BioTek Instruments, Inc., Winooski, VT, USA) to ensure the complete removal of unbound dendrimers. Fluorescence was read at 485 nm/530 nm (excitation/emission [exc/em]) using a FilterMax F5 Multi-Mode Microplate Reader (Molecular Devices LLC). The median of the triplicate sample measurements was calculated after the background values (samples incubated with fluorescent-labeled PAMAM or BC-PAMAM, containing no cells) were subtracted. The fluorescence intensity was calibrated using FITC-labeled dendrimers diluted in water.
PAMAM and BC-PAMAM colocalization analysis
BJ and SCC-15 cells (5×104/well) were seeded on 0.1% gelatin-coated glass coverslips and placed in 6-well cell culture plates. After 24 hours incubation (37°C, 5% CO2, 95% humidity), the culture medium was removed and cells were treated for 24 hours with Rhodamine Red™-X-labeled PAMAM G3 (RR-PAMAM) and FITC-labeled BC-PAMAM G3 (FITC-BC-PAMAM) dendrimers at concentrations ranging from 1 µM to 50 µM, extensively washed with PBS, and fixed with 3.7% formaldehyde (2 mL/well) for 10 minutes at room temperature (RT). The dried coverslips were embedded in polyvinyl alcohol mounting medium (Fluka) and covered with a glass slide. After 3 hours, images were collected with a Carl Zeiss Axio Observer Z1 LSM 710 confocal microscope (Carl Zeiss Jena GmbH, Germany) with a 63×/1.4 plan-apochromat oil immersion lens with its pinhole set to 1.0 airy units (AU) in both the red (RhodamineRed™-X) and green (FITC) channels. Lasers with 488 nm and 555 nm excitation wavelengths were used for FITC and rhodamine, respectively. Green and red fluorescence were separated by a dichroic filter.
The colocalization of signals from two channels was evaluated using ImageJ software (US National Institute of Health, Bethesda, MD, USA) and the JACoP plugin.51 This plugin provides Pearson and Mander coefficient values. Both coefficients determine the degree of overlap between pixels from two different channels expressed in a range from 0 to 1, where 0 means no overlap (no correlation) and 1 represents total overlap (full correlation). The M1 coefficient estimates the degree of green signal overlapping red, and the M2 coefficient estimates the degree of red signal overlapping green.52
PAMAM and BC-PAMAM penetration into nuclei
Cells were plated into a 96-well black-wall, thin-bottom clear plate at 1×104 cells/well in 200 µL culture medium in three replicates and treated with FITC-labeled dendrimers (PAMAM or BC-PAMAM) at the conditions described earlier. Following extensive washing with PBS, the cells were fixed in 3.7% formaldehyde (200 µL/well, 10 minutes, RT), and permeabilized with 0.1% Triton X-100 in PBS (200 µL/well, 30 minutes, RT). Nuclear visualization of the permeabilized cells was performed by DAPI staining of DNA with 300 nM of DAPI/PBS (200 µL/well, 30 minutes, RT with mixing).
Cell imaging was performed using a laser scanning Carl Zeiss Axio Observer Z1 LSM 710 confocal microscope with pinhole set to 1.0 AU in each channel. To visualize the dendrimers, the fluorescence emission was measured using 488 nm/530 nm exc/em for FITC and 405 nm/461 nm exc/em for DAPI to visualize the nuclei.
Images were collected in the z-axis position to estimate the area with the largest nuclear cross section. Confocal images were processed using ImageJ software. The green (dendrimers) and blue (nuclei) channels were thresholded and binarized, and the nuclear areas were marked using the wand tool. The nuclear penetration by dendrimers was expressed as follows: to compare the nuclear permeability of normal and cancer cells, the ratio of the fluorescence signal of the nuclear area occupied by dendrimers to the total nuclear area was calculated and expressed as percentage.
PAMAM or BC-PAMAM colocalization in mitochondria
The fibroblasts and carcinoma cells were plated into a 96-well black-wall, thin-bottom clear plate at 1.5×104 cells/well in 200 µL culture medium in three replicates and then preincubated for 24 hours, and subsequently, FITC-labeled dendrimers (PAMAM and BC-PAMAM) at concentrations from 1 µM to 50 µM and 24 hours incubation (37°C) were added. Following triple washing with PBS, 100 nM MitoTracker Deep Red FM working solution was added (100 µL/well), according to the manufacturer’s protocols, and the cells were incubated for 30 minutes in 37°C, 5% CO2, and 95% humidity. After extensive washing with PBS on a shaking platform, the cells were fixed with formaldehyde as described earlier and nuclei were stained with 300 nM DAPI/PBS (200 µL/well, 30 minutes, RT, with mixing).
Images were obtained with a 40× oil immersion lens and pinhole set to 1.0 AU. Lasers with 405 nm, 488 nm, and 644 nm excitation wavelengths were used for DAPI, FITC-labeled dendrimers and MitoTracker, respectively, and fluorescence signals were read at 461 nm, 530 nm, and 665 nm, respectively. An analysis of the images with the JACoP plugin to determine the Mander’s coefficient was performed as described earlier.
Statistical analysis
A statistical analysis was performed using the Kruskal–Wallis test to estimate the differences between the dendrimer-treated and -nontreated control samples and paired Mann–Whitney U-test to evaluate the differences between cells treated with PAMAM and BC-PAMAM. P<0.05 was considered statistically significant.53
Results and discussion
Cytotoxicity of PAMAM and BC-PAMAM dendrimers
A CyQUANT NF Cell Proliferation Assay was conducted to compare the cytotoxicity of PAMAM and BC-PAMAM in both cell lines (Figure 2).
Figure 2.
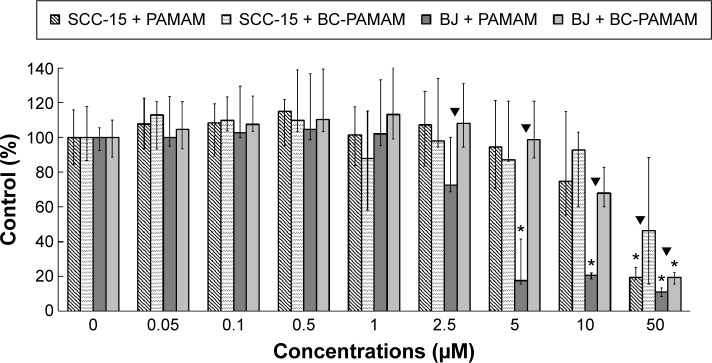
Concentration-dependent effects of PAMAM or BC-PAMAM treatment (24 hours) on the survival of normal human fibroblasts (BJ) and squamous carcinoma cells (SCC-15) measured with CyQUANT® NF dye.
Notes: Results (median of triplicate assays from four independent experiments) are expressed as a % of the nontreated controls. The whiskers are lower (25%) and upper (75%) quartile ranges. *P<0.05; Kruskal–Wallis test (against nontreated control), ▼P<0.05; Mann–Whitney U-test (PAMAM against BC-PAMAM).
Abbreviation: BC-PAMAM, biotin–polyamidoamine.
In BJ cells, PAMAM exhibited significant cytotoxicity at 5 µM and higher concentrations, decreasing the viable cell population to ~10% of the control number at 50 µM. BC-PAMAM produced no significant cytotoxic effects at concentrations up to 50 µM, at which only 20% of the cells remained viable. According to Fischer et al,54 15 µM PAMAM G3 resulted in >80% viability of L929 mouse fibroblasts after 24 hours. In our experiments, BC-PAMAM was significantly less toxic than the native dendrimer at concentrations >2.5 µM (except at 50 µM). Yellepeddi et al39 reported that biotinylated PAMAM dendrimers G3 decreased the viability of human embryonic kidney HEK293T cells by 50% at 30 µM, which was similar to that of the native dendrimers.
A different response to PAMAM was observed in the SCC-15 cells in these experiments, which is consistent with our previous studies.44 Cancer cells exhibited much higher tolerance to both PAMAM and BC-PAMAM at concentrations from 2.5 µM to 10 µM. At 50 µM, BC-PAMAM decreased the viable cell population to ~50% (nonsignificant), whereas PAMAM yielded high toxicity, decreasing cell viability to 20% compared with that of the control (Figure 2).
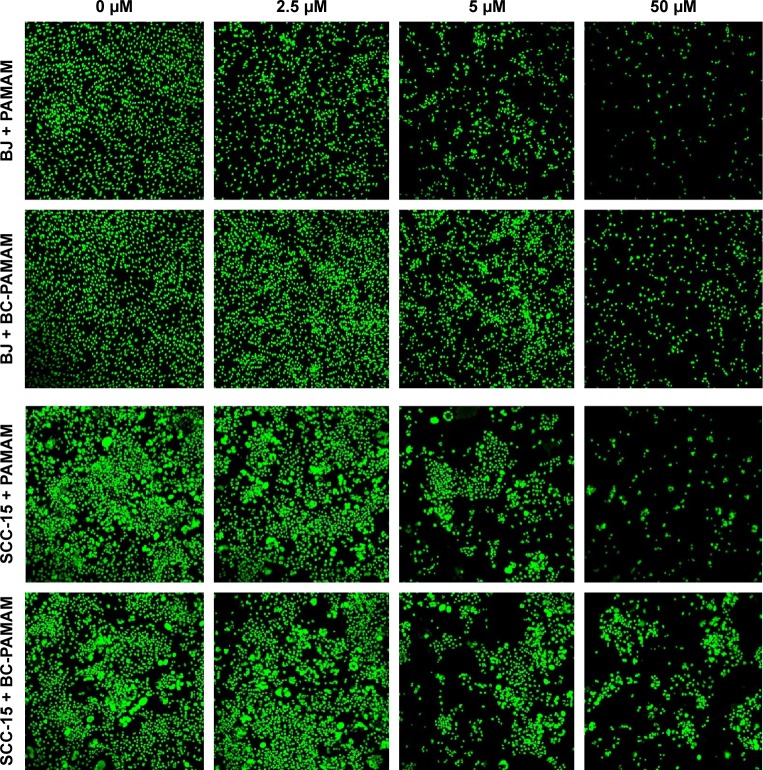
Similar results were obtained by other researchers, who reported that PAMAM was nontoxic to cancer cells in the concentration range of 70–100 µM in Caco-2, A549,55 MCF-7, and MDA-MB-23156 cell lines. In contrast, B16f10 melanoma cells are sensitive to cationic G3 and G4 PAMAM, showing a high toxicity after 72 hours treatments at concentrations >2 µM.32 The cytotoxicity experiments were confirmed by the confocal microscopy images of BJ and SCC-15 cell cultures maintained under the same conditions as described in Figure 2. After 24 hours treatment with PAMAM or BC-PAMAM dendrimers, decreased cell populations were clearly visible at concentrations that were estimated to be cytotoxic for both cell lines (Figure 3).
Figure 3.
Confocal microscopy images of BJ and SCC-15 cell nuclei after 24 hours treatment with PAMAM or BC-PAMAM at concentrations from 2.5 µM to 50 µM or without dendrimers (control).
Notes: After treatment, the cells were incubated 2 days or 5 days in medium (BJ or SCC-15, respectively) and stained with CyQUANT® NF Cell Proliferation Assay Kit.
Abbreviations: BC-PAMAM, biotin–polyamidoamine; PAMAM, polyamidoamine.
The signs of bioconjugate cytotoxicity were absent at the same concentrations of native PAMAM, which has been reported by others.30,57 The effective positive charge of PAMAM G3 at (physiological) pH 7.7 is +20.58 Thus, the ratio of effective charge to the number of primary amine groups in G3 (equal to 32) results in a coefficient of 0.625. If this value is considered a constant, then the effective positive charge of the G39B10P PAMAM bioconjugate (where an average of 13 amine groups remained unsubstituted) equals 13×0.625=+8 at physiological conditions. The bioconjugate therefore remains cationic, although the charge is reduced from +20 to +8 in relation to the native PAMAM G3.
Cellular accumulation of FITC-labeled PAMAM and BC-PAMAM
The total fluorescence of internalized FITC-labeled PAMAM G3 and BC-PAMAM dendrimers was plotted against the dendrimer concentrations after treatment for 24 hours in both cell lines (Figure 4). The 24 hours dendrimer treatment was selected based on observations by other groups. Pai et al59 showed that the time course of FITC PAMAM G4 uptake into HaCaT cells reached saturation at 5 hours incubation, and Albertazzi et al30 reported the stabilization of PAMAM G4-Alexa647 dendrimer localization in HeLa cells after 12 hours treatment.
Figure 4.
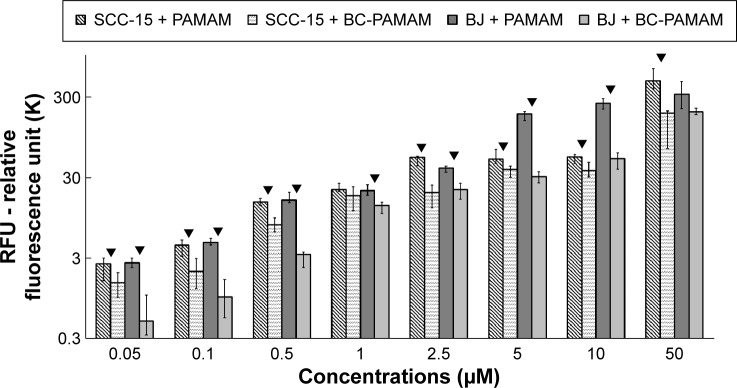
The total fluorescent intensity of the FITC-labeled PAMAM and BC-PAMAM in SCC-15 and BJ cells as a function of dendrimer concentration.
Notes: Cells were incubated with FITC-labeled dendrimers at various concentrations for 24 hours. Results are presented as median of triplicate assays from four independent experiments and whiskers are lower (25%) and upper (75%) quartile ranges. ▼P<0.05; Mann–Whitney U-test (PAMAM against BC-PAMAM).
Abbreviations: FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate; BC-PAMAM, biotin–polyamidoamine.
Based on the viability of both cell populations as assessed by CyQUANT NF dye (Figure 2), the concentration-dependent FITC-PAMAM accumulation was observed at nontoxic 0.05–2.5 µM concentration range in BJ fibroblast cells, whereas at cytotoxic 5–50 µM concentrations, dendrimer loading remained at increased, similar level. The fluorescence of FITC-PAMAM in cancer cells increased up to 2.5 µM concentration and remained at constant level up to 10 µM, with a sharp increase at toxic 50 µM concentration (Figure 4). The intracellular loading of FITC-PAMAM dendrimers is comparable at low concentrations up to 2.5 µM in both cell lines, whereas the native dendrimer fluorescence signal was higher at 5 µM and 10 µM in BJ cells (Figure 4).
The bioconjugated dendrimer FITC-BC-PAMAM accumulated at a significantly lower amount compared to that of the native FITC-PAMAM at all concentrations in both SCC-15 cells and fibroblasts. These results confirmed that the internalization of cationic PAMAM dendrimers depends on the surface charge, as it was documented by the others.30,31 Our results, however, differ from those obtained by Yellepeddi et al who reported that the biotinylated PAMAM dendrimers of different generations were internalized at a significantly higher extent in human ovarian cancer cells (OVCAR-3) compared to that of human embryonic kidney cells (HEK 293T).27 The internalization of the FITC-PAMAM G3 dendrimer involves energy and may involve caveole-dependent and clathrin-dependent endocytosis pathways.28,29,60 However, a cholesterol-dependent pathway was also observed in B16f10 melanoma cells.32 Yellepeddi et al showed that biotin-conjugated PAMAM dendrimers of the G1–G4 generations enter OVCAR-3 cells by both absorptive endocytosis and biotin/SMVT receptor-mediated endocytosis.39 Observed at cytotoxic concentrations, increased dendrimer loading in both cell lines may be due to cell membrane damage and simple diffusion process.
Notably, PAMAM accumulation in SCC-15 cells at nontoxic 2.5–10 µM concentrations was kept at similar level. This may be explained by increased dendrimer exocytosis mechanisms. This, however, requires further studies, as so far only a few studies on this phenomenon are available in B16f10 cells32 and in Capan-1 pancreatic cancer cells.45 Various degrees of internalization of early generations (up to G4) of cationic PAMAM dendrimers were reported by investigators for different cell types, including HeLa, HepG2, PC-12 cancer cells, MRC5 fetal lung fibroblasts and primary astrocytes,30 B16f10 melanoma,32 Caco-2,28 and HT-29 adenocarcinoma cell lines.60 The analysis of these results clearly indicates that the various effectiveness of cationic PAMAM internalization observed here may depend on specific differences in cellular membrane composition and mechanisms of endocytosis as well as on cell type, normal versus cancer.
PAMAM and BC-PAMAM colocalization
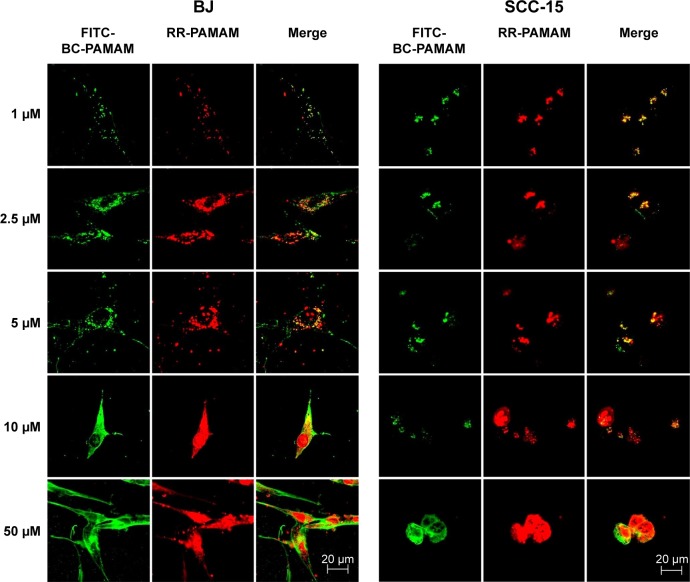
The degree of colocalization of RR-PAMAM and FITCBC-PAMAM in BJ and SCC-15 cells is shown in Figure 5.
Figure 5.
Intracellular colocalization of RR-PAMAM (red) and FITC-BC-PAMAM (green) in BJ and SCC-15 cells after 24 hours incubation with dendrimers at various concentrations (1–50 µM).
Notes: Colocalization is shown as a yellow signal. Cell imaging was performed on a Carl Zeiss Axio Observer Z1 LSM 710 inverted confocal microscope equipped with a 63× oil immersion objective.
Abbreviations: RR-PAMAM, Rhodamine Red™-X-labeled PAMAM G3; FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate.
A high degree of dendrimer colocalization was observed in both cell lines at all applied concentrations as demonstrated by Pearson coefficient values between 0.53 and 0.87 (Table 1). However, the higher value of the M1 Mander coefficients compared with the M2 coefficients indicate that nonconjugated RR-PAMAM more efficiently penetrates both cell lines (Table 1). This indicates that in areas where RR-G3 PAMAM was observed, FITC-G3 BC-PAMAM was also recorded.
Table 1.
Pearson’s coefficients and Mander’s coefficients (M1, M2) for RR-PAMAM colocalization with FITC-BC-PAMAM
| Cell line | PAMAM and BC-PAMAM concentration (µM) | Pearson’s coefficient | M1 | M2 |
|---|---|---|---|---|
| BJ | 1 | 0.63±0.15 | 0.66±0.22 | 0.49±0.23 |
| 2.5 | 0.65±0.04 | 0.69±0.08 | 0.49±0.12 | |
| 5 | 0.60±0.07 | 0.79±0.04 | 0.41±0.05 | |
| 10 | 0.63±0.04 | 0.64±0.04 | 0.50±0.06 | |
| 50 | 0.78±0.03 | 0.73±0.04 | 0.66±0.05 | |
| SCC-15 | 1 | 0.87±0.04 | 0.73±0.06 | 0.69±0.07 |
| 2.5 | 0.66±0.07 | 0.70±0.18 | 0.49±0.12 | |
| 5 | 0.65±0.05 | 0.75±0.14 | 0.47±0.15 | |
| 10 | 0.53±0.06 | 0.59±0.20 | 0.30±0.23 | |
| 50 | 0.77±0.09 | 0.72±0.12 | 0.73±0.07 |
Note: Results are the mean of triplicate measurements of four independent experiments ± the standard deviation.
Abbreviations: RR-PAMAM, Rhodamine Red™-X-labeled polyamidoamine G3; FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate.
In BJ cells, RR-PAMAM accumulated in the cytoplasm and nuclei at concentrations >10 µM. FITC-BC-PAMAM was distributed mainly in the cytoplasm at concentrations between 10 µM and 50 µM, and it was also present in the nuclei but in lower degree than that of RR-PAMAM. In cancer cells, a similar pattern of dendrimer localization was observed; however, FITC-BC-PAMAM penetrated nuclei only at the 50 µM concentration.
More effective penetration of the cells by PAMAM may occur because of its higher effective positive charge compared to that of BC-PAMAM, or as result of different mechanisms of endocytosis of native and bioconjugated dendrimers. Clathrin-mediated endocytosis and macropinocytosis are involved in G2, G4, and G6 dendrimer internalization in HeLa cells.28 Moreover, the amine-modified cationic dendrimers were shown to utilize multiple routes of entry into Caco-2 and B16f10 melanoma cells,28,32 and a cholesterol-dependent route was also recognized in B16f10 cells.31 After internalization, clathrin-coated vesicles may be transferred to lysosomes for degradation,61 whereas the fate of the dendrimers internalized by macropinocytosis depends on the cell type.62 It was also established that positively charged nanoparticles can escape from endosomes after internalization, and they exhibit perinuclear localization because of the “proton sponge” effect.63
Dendrimer penetration into nuclei
To estimate the nuclear penetration by dendrimers, the fluorescence signal of the nuclei was recorded in both cell lines treated with either FITC-PAMAM or FITC-BC-PAMAM dendrimers at increasing concentrations ranging from 1 µM to 50 µM for 24 hours at 37°C. Nuclear visualization was performed by staining the DNA with DAPI. Figures 6 and 7 show the BJ and SCC-15 cell images, respectively.
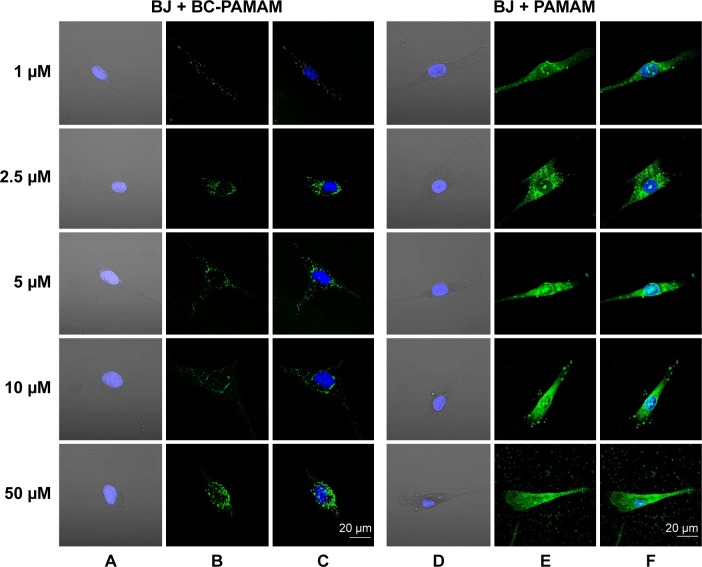
Figure 6.
Localization of FITC-BC-PAMAM (green) (A–C) and FITC-PAMAM (green) (D–F) in BJ cells.
Notes: Nuclei (blue) stained with DAPI. Images collected after 24 hours incubation at various concentrations of dendrimers.
Abbreviations: FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate; DAPI, 4′,6-diamidino-2-phenylindole, dihydrochloride.
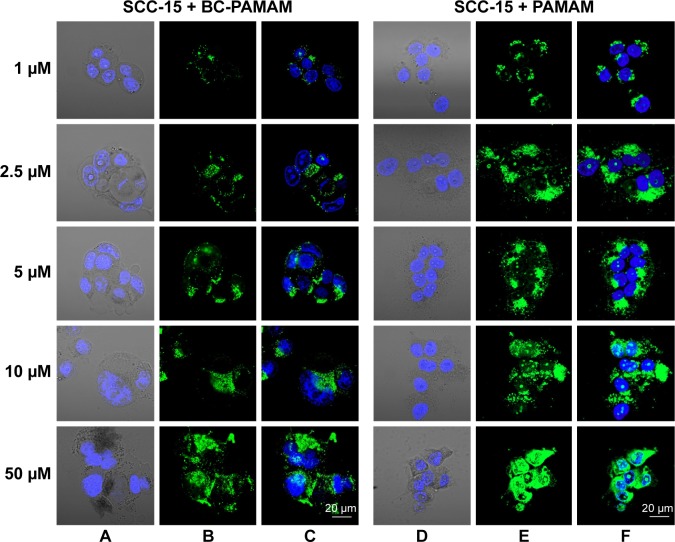
Figure 7.
Localization of FITC-BC-PAMAM (green) (A–C) and FITC-PAMAM (green) (D–F) in SCC-15 cells.
Notes: Nuclei (blue) stained with DAPI. Images collected after 24 hours incubation with various concentrations of dendrimers.
Abbreviations: FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate; DAPI, 4′,6-diamidino-2-phenylindole, dihydrochloride.
Increasing intense fluorescence signals were observed in the cytoplasmic compartments as well as in the nuclei of fibroblasts treated for 24 hours with FITC-PAMAM at 1–50 µM, whereas much lower signals were observed for the FITC-BC-PAMAM bioconjugate internalization. At toxic FITC-PAMAM concentrations of 5–50 µM, the destruction of cells and condensed nuclei was seen (Figure 6D).
In cancer cells, the penetration of nuclei by either FITC-BC-PAMAM or FITC-PAMAM was observed at 10 µM and 50 µM, despite dense perinuclear accumulation of FITC-labeled dendrimers at lower concentrations. At 50 µM of the native dendrimer, swollen and sparsely condensed nuclei are visible.
The results of the earlier experiments are summarized in Figure 8, which illustrates the dendrimer nuclear content in both cell lines.
Figure 8.
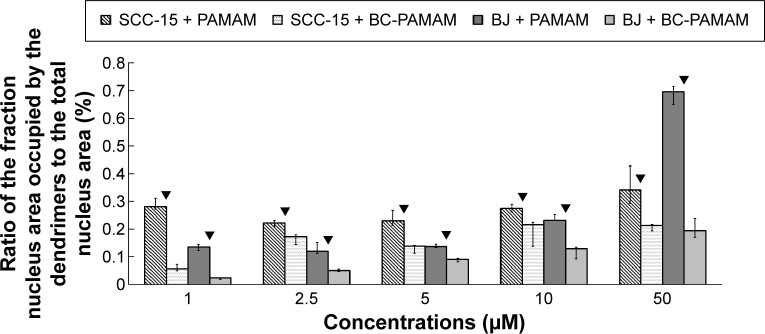
Ratio of the fraction of nuclear area occupied by the dendrimers (FITC-PAMAM or FITC-BC-PAMAM) to the total nuclear area (%) in BJ and SCC-15 cells after 24 hours treatment with various concentrations of dendrimers.
Notes: Results are the median of triplicates of four independent experiments, whiskers are lower (25%) and upper (75%) quartile ranges. ▼P<0.05; Mann–Whitney U-test (PAMAM against BC-PAMAM).
Abbreviation: FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate.
Fibroblasts accumulated FITC-PAMAM dendrimers in the nuclei at a constant level (~15% of the nuclear area) at 1–5 µM, but at toxic dendrimer concentrations (10–50 µM) the nuclear content dramatically increased (Figure 8). This indicates the possible interruption of controlled transport mechanisms when the FITC-G3 dendrimers enter the nuclei at toxic concentrations. Bioconjugate dendrimers accumulated in the nuclei at significantly lower levels at all concentrations, and the accumulation was dependent on the total BC-PAMAM accumulation in the cells (Figure 4). Considering the similar levels of total cellular FITC-BC-PAMAM accumulation (Figure 8), the controlled transport of bioconjugated dendrimers into the nucleus might only proceed at nontoxic cellular dendrimer concentrations. At higher toxic concentrations, the direct interaction of cationic dendrimers with elements of the nuclear envelope may cause lipid membrane perforation as previously described22,64,65 and concentration-dependent dendrimer entry into nucleus by simple diffusion.
The different patterns of dendrimer accumulation in cancer cell nuclei are seen. Approximately 25%–30% of the nuclei are occupied at all tested concentrations (Figure 8), in a manner independent of their total cellular content (Figure 4). BC-PAMAM accumulated at significantly lower levels depending on the cellular bioconjugate concentration (Figures 4 and 8). These results clearly indicate different mechanisms of nuclear penetration in normal and cancer cells, with the normal cells maintaining a controlled nuclear penetration at noncytotoxic concentrations of both native and bioconjugated dendrimers and cancer cells demonstrating a lack of such limitations. The observed nuclear penetration by FITC-PAMAM dendrimers in cancer cells is inconsistent with observations in other studies. Albertazzi et al reported that G4 Alexa647 dendrimer conjugates accumulated only in the perinuclear region within HeLa cells at treatments up to 48 hours.30 No nuclear localization was observed by these authors after treatment for 3 hours in HepG2 and MRC5 cancer cells or in normal PC-12 and primary astrocyte cultures. No entry of FITC-G4 PAMAM into human keratinocyte HaCaT cell nuclei was observed after 24 hours and 48 hours treatments by Pai et al,59 or after 24 hours in primary neuronal cultures.25 The localization of later generation G6 PAMAM dendrimers conjugated with estrogen was observed in the membrane and cytoplasm, but not in the nuclei of estrogen receptor-positive MCF-7 human breast cancer cells.66 At the nondividing phase, the nuclear envelope acts as a molecular permeability barrier, enabling the free diffusion of small hydrophilic molecules (up to 9 nm size) through the nuclear pore complex. Larger molecules (up to 40 kDa) undergo active transport processes involving the interaction of multiple cellular components, including oligopeptide sequences containing lysine and arginine, which is known as a nuclear localization signal.67 The theoretical size of the PAMAM G3 dendrimer is ~ nm. However, dendrimers and their conjugates can aggregate, which was found for PAMAM G4 dendrimers.21,68 Presumably, the increased size and decreased cationic charge of bioconjugates determine their entry into the nuclei of normal cells. However, in cancer cells, changes in nuclear morphology and laminin expression were often observed, and these changes caused damage to and the collapse of the nuclear envelope function.69 This may explain the extensive penetration observed in the SCC-15 cell nuclei by both types of investigated dendrimers.
PAMAM and BC-PAMAM localization within mitochondria
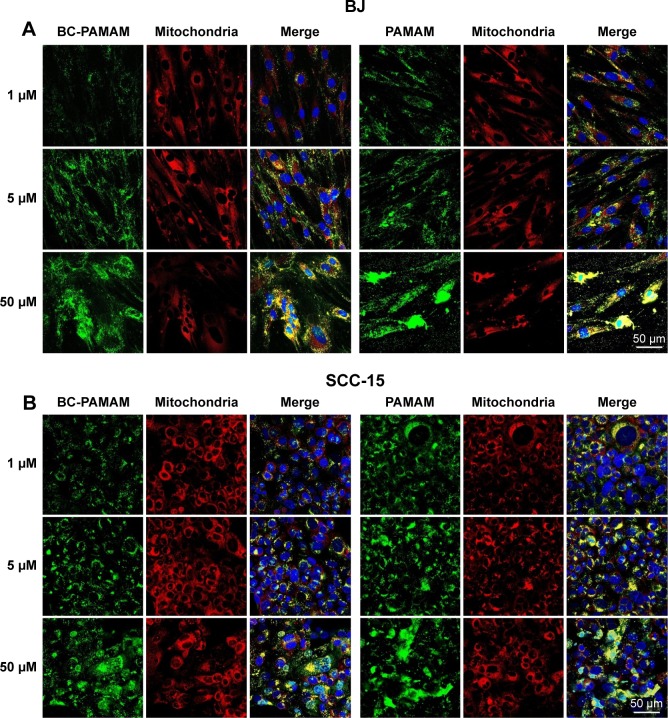
Colocalization of MitoTracker-labeled mitochondria and FITC-labeled native and bioconjugated dendrimers is shown in Figure 9A and B.
Figure 9.
FITC-PAMAM or FITC-BC-PAMAM (green) colocalization with mitochondria (red) in (A) fibroblasts and (B) squamous cell carcinoma after 24 hours incubation with dendrimers (1–50 µM).
Notes: Mitochondria (red) were stained with MitoTracker® Deep Red FM and nuclei were stained with DAPI (blue). Colocalization is shown as a yellow signal. Cell imaging was performed on a Carl Zeiss Axio Observer Z1 LSM 710 inverted confocal microscope equipped with a 40× oil immersion objective.
Abbreviations: FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate; DAPI, 4′,6-diamidino-2-phenylindole, dihydrochloride.
Data expressed as M2 coefficients (mitochondrial signal overlapping FITC-PAMAM or FITC-BC-PAMAM signals) are shown in Figure 10. The colocalization of cationic PAMAM dendrimers with mitochondrial markers has been observed by other researchers.21,59 Investigations of the interaction between cationic dendrimers and artificial and cellular lipid bilayers revealed electrochemical as well as hydrophobic interactions that promote membrane disruption and impair transport mechanisms.70,71 The formation of pores in the outer mitochondrial membrane promotes apoptotic cell death.72
Figure 10.
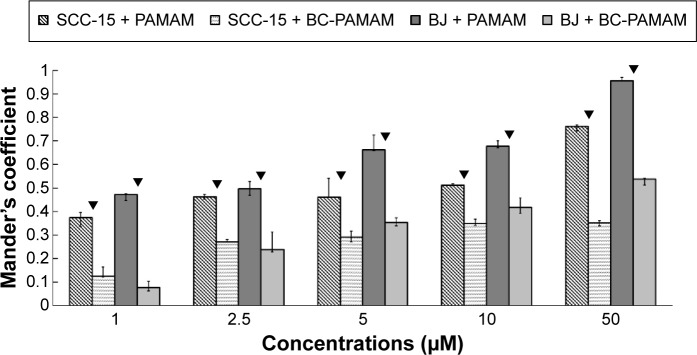
Mitochondrial localization of native FITC-PAMAM or FITC-BC-PAMAM expressed as Mander’s coefficient M2 (fraction of mitochondrial signal overlapping dendrimers) in BJ and SCC-15 cells after 24 hours dendrimer treatment (1–50 µM).
Notes: Results are the median of triplicates of four independent experiments, whiskers are lower (25%) and upper (75%) quartile ranges. ▼P<0.05; Mann–Whitney U-test (PAMAM against BC-PAMAM).
Abbreviations: FITC-PAMAM, fluorescein isothiocyanate–polyamidoamine; FITC-BC-PAMAM, fluorescein isothiocyanate-labeled biotin–polyamidoamine bioconjugate.
In our experiments, the localization of native and bioconjugate dendrimers in the mitochondria correlated well with their cytotoxicity. In fibroblasts, the Mander’s coefficient of PAMAM colocalization in mitochondria increased from 0.6 to almost 1.0 at higher dendrimer concentrations, which is consistent with their cytotoxic effect (Figures 2 and 10). In cancer cells, an M2 coefficient >0.5 was only observed at the toxic 50 µM concentration. In both cell lines, a lower BC-PAMAM dendrimer signal colocalized with the mitochondria and was correlated with the much lower cytotoxicity threshold. This result is consistent with our previous results concerning mitochondrial metabolism, which were estimated with the tetraziolium salt assay test (XTT), and the proapoptotic effects of both native and bioconjugated dendrimers assayed under identical conditions.44 Lee et al documented that in human lung cells (WI-26 VA4) exposed to G4 PAMAM, dendrimers colocalized with mitochondria, disrupted the mitochondrial membrane potential and released cytochrome c, promoting apoptosis. Moreover, the downregulation of mitochondrial DNA-encoded genes involved in the maintenance of mitochondrial membrane potential was observed.46
Conclusion
Our results provide evidence that the intracellular localization of native and bioconjugated dendrimers of early generations depends on their physical–chemical characteristics (size, surface, and substituted functional groups) as well as on the type of targeted cell (normal or transformed). The comparison of internalization and intracellular localization of the PAMAM G3 dendrimer and its biotin–PL bioconjugate BC-PAMAM in normal fibroblast (BJ) and cancer cells (SCC-15) as well as their cytotoxicity revealed significant differences in the investigated parameters.
In particular, our results do not confirm observations made by other authors, that biotin increases dendrimer entry to cancer cells, pointing out the importance of individual cell line evaluations. Of interest is also increased penetration of nuclei by native and conjugated dendrimers in cancer cells.
Thus, to determine the mechanisms of various dendrimer effects on target cells, careful estimations of numerous biological parameters and their spatial and temporal dynamics must be performed because the results cannot be generalized for different cell populations. Therefore, introducing common protocols for various assays, including in vivo and in vitro assays, is required to construct safe multifunctional carriers for therapy and imaging in living organisms.
Acknowledgments
The work was supported by grant 2014/13/D/NZ3/02825 from the National Science Centre, Poland. We gratefully acknowledge Prof Grzegorz Bartosz and coworkers at the Department of Biochemistry and Cell Biology, University of Rzeszow, for kindly enabling us to perform measurements using their confocal microscope.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kobayashi H, Koyama Y, Barrett T, et al. Multimodal nanoprobes for radionuclide and five-color near-infrared optical lymphatic imaging. ACS Nano. 2007;1(4):258–264. doi: 10.1021/nn700062z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Theoharis S, Krueger U, Tan PH, Haskard DO, Weber M, George AJT. Targeting gene delivery to activated vascular endothelium using anti E/P-Selectin antibody linked to PAMAM dendrimers. J Immunol Methods. 2009;343(2):79–90. doi: 10.1016/j.jim.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Labieniec-Watala M, Karolczak K, Siewiera K, Watala C. The Janus face of PAMAM dendrimers used to potentially cure non-enzymatic modifications of biomacromolecules in metabolic disorders-a critical review of the pros and cons. Molecules. 2013;18(11):13769–13811. doi: 10.3390/molecules181113769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svenson S, Tomalia DA. Dendrimers in biomedical applications – reflections on the field. Adv Drug Deliv Revs. 2012;64(Suppl):102–115. doi: 10.1016/j.addr.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Shao N, Su Y, Hu J, Zhang J, Zhang H, Cheng Y. Comparison of generation 3 polyamidoaminedendrimer and generation 4 polypropyleniminedendrimer on drug loading, complex structure, release behavior and cytotoxicity. Int J Nanomedicine. 2011;6:3361–3372. doi: 10.2147/IJN.S27028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shcharbin D, Shakhbazau A, Bryszewska M. Poly(amidoamine) dendrimer complexes as a platform for gene delivery. Expert Opin Drug Deliv. 2013;10(12):1687–1698. doi: 10.1517/17425247.2013.853661. [DOI] [PubMed] [Google Scholar]
- 7.Wolinsky JB, Grinstaff MW. Therapeutic and diagnostic applications of dendrimers for cancer treatment. Adv Drug Deliv Rev. 2008;60(9):1037–1055. doi: 10.1016/j.addr.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Borowska K, Wołowiec S, Głowniak K, Sieniawska E, Radej S. Transdermal delivery of 8-methoxypsoralene mediated by polyamidoaminedendrimer G2.5 and G3.5 – in vitro and in vivo study. Int J Pharm. 2012;436(1–2):764–770. doi: 10.1016/j.ijpharm.2012.07.067. [DOI] [PubMed] [Google Scholar]
- 9.Daftarian P, Kaifer AE, Li W, et al. Peptide-conjugated PAMAM dendrimer as a universal DNA vaccine platform to target antigen-presenting cells. Cancer Res. 2011;71(24):7452–7462. doi: 10.1158/0008-5472.CAN-11-1766. [DOI] [PubMed] [Google Scholar]
- 10.Avti PK, Kakkar A. Dendrimers as anti-inflammatory agents. Brazilian J Pharm Sci. 2013;49:57–65. [Google Scholar]
- 11.Bai S, Thomas C, Rawat A, Ahsan F. Recent progress in dendrimer-based nanocarriers. Crit Rev Ther Drug Carrier Syst. 2006;23(6):437–495. doi: 10.1615/critrevtherdrugcarriersyst.v23.i6.10. [DOI] [PubMed] [Google Scholar]
- 12.Chai M. Unique structure and property of dendrimers in biomedical applications. Proc R Soc A. 2010;466:1441–1443. [Google Scholar]
- 13.Rajendran L, Knölker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nat Rev Drug Discov. 2010;9(1):29–42. doi: 10.1038/nrd2897. [DOI] [PubMed] [Google Scholar]
- 14.Duncan R, Izzo L. Dendrimer biocompatibility and toxicity. Adv Drug Deliv Rev. 2005;57(15):2215–2237. doi: 10.1016/j.addr.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 15.Jain K, Kesharwani P, Gupta U, Jain NK. Dendrimer toxicity: Let’s meet the challenge. Int J Pharm. 2010;394(1–2):122–142. doi: 10.1016/j.ijpharm.2010.04.027. [DOI] [PubMed] [Google Scholar]
- 16.Kwon IK, Lee SC, Han B, Park K. Analysis on the current status of targeted drug delivery to tumors. J Control Release. 2012;164(2):108–114. doi: 10.1016/j.jconrel.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruenraroengsak P, Cook JM, Florence AT. Nanosystem drug targeting: Facing up to complex realities. J Control Release. 2010;141(3):265–276. doi: 10.1016/j.jconrel.2009.10.032. [DOI] [PubMed] [Google Scholar]
- 18.Tang MX, Szoka PC. The influence of polymer structure on the interactions of cationic polymers with DNA and morphology of the resulting complexes. Gene Ther. 1997;4(8):823–832. doi: 10.1038/sj.gt.3300454. [DOI] [PubMed] [Google Scholar]
- 19.Froehlich E, Mandeville JS, Weinert CM, Kreplak L, Tajmir-Riahi HA. Bundling and aggregation of DNA by cationic dendrimers. Biomacromolecules. 2011;12(2):511–517. doi: 10.1021/bm1013102. [DOI] [PubMed] [Google Scholar]
- 20.Jevprasesphant R, Penny J, Attwood A, D’Emanuelle A. Transport of dendrimernanocarriers through epithelial cells via the transcellular route. J Control Release. 2004;97(2):259–267. doi: 10.1016/j.jconrel.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 21.Lai PS, Shich MJ, Pai CL, Wang CY, Lou PJ. Studies on Intracellular Trafficking of PAMAM Dendrimer. Nanotech. 2005;1:232–235. [Google Scholar]
- 22.Hong S, Leroueil RP, Janus KE, et al. Interaction of polycationicpolimers with supported lipid bilayers and cells: nanoscale hole formation and enhanced membrane permeability. Bioconjug Chem. 2006;17(3):728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 23.Kitchens KM, Kolhatakar RB, Swaan PW, Eddington ND. Transport of poly(amidoamine) dendrimers across Caco-2 cell monolayers: Influence of size, charge and fluorescent labeling. Pharm Res. 2006;23(12):2818–2826. doi: 10.1007/s11095-006-9122-2. [DOI] [PubMed] [Google Scholar]
- 24.Tsai HC, Imae T, Caldero G, Solans C. Two-photon confocal imaging study: cell uptake of two photon dyes-labelled PAMAM dendrons in HeLa cells. J Biomed Mat Res A. 2012;100(3):746–756. doi: 10.1002/jbm.a.33283. [DOI] [PubMed] [Google Scholar]
- 25.Albertazzi L, Gherardini L, Brondi M, et al. In vivo distribution and toxicity of PAMAM dendrimers in the central nervous system depend on their surface chemistry. Mol Pharm. 2013;10(1):249–260. doi: 10.1021/mp300391v. [DOI] [PubMed] [Google Scholar]
- 26.Satija J, Gupta U, Jain NK. Pharmaceutical and biomedical potential of surface engineered dendrimers. Crit Rev Ther Drug Carrier Syst. 2007;24(3):257–306. doi: 10.1615/critrevtherdrugcarriersyst.v24.i3.20. [DOI] [PubMed] [Google Scholar]
- 27.Yellepeddi VK, Kumar A, Palakurthi S. Surface modified poly(amido) amine dendrimers as diverse nanomolecules for biomedical applications. Expert Opin Drug Deliv. 2009;6(8):835–850. doi: 10.1517/17425240903061251. [DOI] [PubMed] [Google Scholar]
- 28.Kitchens KM, Foraker AB, Kolhatkar RB, Swaan PW, Ghandehari H. Endocytosis and interaction of poly(amidoamine) dendrimers with Caco-2 cells. Pharm Res. 2007;24(11):2138–2145. doi: 10.1007/s11095-007-9415-0. [DOI] [PubMed] [Google Scholar]
- 29.Kitchens KM, Kolhatkar RB, Swaan PW, Ghandehari H. Endocytosis inhibitors prevent poly(amidoamino) dendrimer internalization and permeability across Caco-2 cells. Mol Pharm. 2008;5(2):364–369. doi: 10.1021/mp700089s. [DOI] [PubMed] [Google Scholar]
- 30.Albertazzi L, Serresi M, Albanese A, Beltram F. Dendrimer internalization and intracellular trafficking in living cells. Mol Pharm. 2010;7(3):680–688. doi: 10.1021/mp9002464. [DOI] [PubMed] [Google Scholar]
- 31.Perumal OP, Inapagolla R, Kannan S, Kannan RM. The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials. 2008;29(24–25):3469–3476. doi: 10.1016/j.biomaterials.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 32.Seib FP, Jones AT, Duncan R. Comparison of the endocytic properties of linear and branched PEIs and cationic PAMAM dendrimers in B16f10 melanoma cells. J Control Release. 2007;117(3):291–300. doi: 10.1016/j.jconrel.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Sadekar S, Ghandehari H. Transepithelial transport and toxicity of PAMAM dendrimers: implications for oral drug delivery. Adv Drug Deliv Rev. 2012;64(6):571–588. doi: 10.1016/j.addr.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahon RJ. Biotin in metabolism and molecular biology. Annu Rev Nutr. 2002;22:221–239. doi: 10.1146/annurev.nutr.22.121101.112819. [DOI] [PubMed] [Google Scholar]
- 35.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells. Effects of cell proliferation. Eur J Biochem. 2001;268(20):5424–5429. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 36.Russell-Jones G, McTavish K, McEwan J. Vitamin-mediated targeting as a potential mechanism to increase drug uptake by tumors. J Inorg Biochem. 2004;98(10):1625–1633. doi: 10.1016/j.jinorgbio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Yang W, Cheng Y, Xu T, Wang X, Wen LP. Targeting cancer cells with biotin-dendrimer conjugates. Eur J Med Chem. 2009;44(2):862–868. doi: 10.1016/j.ejmech.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 38.Hemmer R, Hall A, Spaulding R, et al. Analysis of biotynylated generation 4 poly(amidoamine) PAMAM dendrimer distribution in the rat brain and toxicity in a cellular model of the blood-brain barrier. Molecules. 2013;18(9):11537–11552. doi: 10.3390/molecules180911537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yellepeddi V, Kumar A, Palakhurt S. Biotynylated poly(amido)amine (PAMAM) dendrimers as carriers for drug delivery to ovarian cancer cells in vitro. Anticancer Res. 2009;29(8):2933–2943. [PubMed] [Google Scholar]
- 40.Hellmann H, Mooney S. Vitamin B6: a molecule for human health? Molecules. 2010;15(1):442–459. doi: 10.3390/molecules15010442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mooney S, Leuendorf JE, Hendrickson C, Hellman H. Vitamin B6: a long known compound of surprising complexity. Molecules. 2009;14(1):329–351. doi: 10.3390/molecules14010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu S, Yanaka N, Matsubara K, Kato N. Antitumor effect of vitamin B6 and its mechanisms. Biochim Biophys Acta. 2003;1647(1–2):127–130. doi: 10.1016/s1570-9639(03)00076-1. [DOI] [PubMed] [Google Scholar]
- 43.Galluzzi L, Vacchelli E, Michels J, et al. Effects of vitamin B6 metabolism on oncogenesis, tumor progression and therapeutic responses. Oncogene. 2013;32(42):4995–5004. doi: 10.1038/onc.2012.623. [DOI] [PubMed] [Google Scholar]
- 44.Uram Ł, Szuster M, Gargasz K, Filipowicz A, Wałajtys-Rode E, Wołowiec S. In vitro cytotoxicity of the ternary PAMAM G3-pyridoxal-biotin bioconjugate. Int J Nanomedicine. 2013;8:4707–4720. doi: 10.2147/IJN.S53254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Opitz AW, Czymmek KJ, Wickstrom E, Wagner NJ. Uptake, efflux, and mass transfer coefficient of fluorescent PAMAM dendrimers into pancreatic cancer cells. Biochim Biophys Acta. 2013;1828(2):294–301. doi: 10.1016/j.bbamem.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH, Cha KE, Kim MS, et al. Nanosizedpolyamidoamine (PAMAM) dendrimer-induced apoptosis mediated by mitochondrial dysfunction. Toxicol Lett. 2009;190(2):202–207. doi: 10.1016/j.toxlet.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 47.Filipowicz A, Wołowiec S. Solubility and in vitro transdermal diffusion of riboflavin assisted by PAMAM dendrimers. Int J Pharm. 2011;408(1–2):152–156. doi: 10.1016/j.ijpharm.2011.01.033. [DOI] [PubMed] [Google Scholar]
- 48.Maiti PK, Cagin T, Wang G, Goddard WA., III Structure of PAMAM dendrimers: generation 1 through 11. Macromolecules. 2004;37(16):6236–6254. [Google Scholar]
- 49.Jones LJ, Gray M, Yue ST, Haugland RP, Singer VL. Sensitive determination of cell number using the CyQUANT cell proliferation assay. J Immunol Methods. 2001;254(1–2):85–98. doi: 10.1016/s0022-1759(01)00404-5. [DOI] [PubMed] [Google Scholar]
- 50.Al-Jamal KT, Ruenraroengsak P, Hartell N, Florence AT. An intrinsically fluorescent dendrimer as a nanoprobe of cell transport. J Drug Target. 2006;14(6):405–412. doi: 10.1080/10611860600834441. [DOI] [PubMed] [Google Scholar]
- 51.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 52.Manders EMM, Verbeck FJ, Aten JA. Measurement of co-localization of object in dual-colour confocal images. J Microsc. 1993;169(3):375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 53.Hollander M, Wolfe DA. Wiley Series in Probability and Statistics. 2nd ed. New York: John Wiley & Sons, Inc; 1999. Nonparametric Statistical Methods. [Google Scholar]
- 54.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 55.El-Sayed M, Ginski M, Rhodes C, Ghandehari H. Transepithelial transport of poly(amidoamine) dendrimers across Caco-2 cell monolayers. J Control Release. 2002;81(3):355–365. doi: 10.1016/s0168-3659(02)00087-1. [DOI] [PubMed] [Google Scholar]
- 56.Winnicka K, Bielawski K, Rusak M, Bielawska A. The effect of Generation 2 and 3 poly(amidoamine) dendrimers on viability of human breast cancer. J Health Sci. 2009;55:169–177. [Google Scholar]
- 57.Mukherjee SP, Lyng FM, Garcia A, Davoren M, Byrne HJ. Mechanistic studies of in vitro cytotoxicity of poly(amidoamine) dendrimers in mammalian cells. Toxicol Appl Pharmacol. 2010;248(3):259–268. doi: 10.1016/j.taap.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 58.Böhme U, Klenge A, Hänel B, Sheler U. Counterion condensation and effective charge of PAMAM dendrimers. Polymers. 2011;3(2):812–819. [Google Scholar]
- 59.Pai CL, Shieh MJ, Lou PJ, Huang FH, Wang TW, Lai PS. Characterization of the uptake and intracellular trafficking of G4 polyamidoamine dendrimers. Aust J Chem. 2011;64(3):302–308. [Google Scholar]
- 60.Saovapakhiran A, D’Emanuele A, Attwood D, Penny J. Surface modification of PAMAM dendrimers modulates the mechanism of cellular internalization. Bioconjug Chem. 2009;20(4):693–701. doi: 10.1021/bc8002343. [DOI] [PubMed] [Google Scholar]
- 61.Benmerah A, Lamaze C. Clathrin-coated pits: vive la différence? Traffic. 2007;8(8):970–982. doi: 10.1111/j.1600-0854.2007.00585.x. [DOI] [PubMed] [Google Scholar]
- 62.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PubMed] [Google Scholar]
- 63.Kou L, Sun J, Zhai Y, He Z. The endocytosis and intracellular fate of nanomedicines: Implication for rational design. Asian J Pharm Sci. 2013;8(1):1–10. [Google Scholar]
- 64.He XC, Qu ZG, Xu F, et al. Molecular analysis of interactions between dendrimers and asymmetric membranes at different transport stages. Soft Matter. 2014;10(1):139–148. doi: 10.1039/c3sm51990b. [DOI] [PubMed] [Google Scholar]
- 65.Lee H, Larson RG. Multiscale modeling of dendrimers and their interactions with bilayers and polyelectrolytes. Molecules. 2009;14(1):423–438. doi: 10.3390/molecules14010423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harrington WR, Kim SH, Funk CC, et al. Estrogen dendrimer conjugates that preferentially activate extranuclear, nongenomic versus genomic pathways of estrogen action. Mol Endocrinol. 2006;20(3):491–502. doi: 10.1210/me.2005-0186. [DOI] [PubMed] [Google Scholar]
- 67.Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386(6627):779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 68.Li J, Piehler LT, Qin D, Baker JR, Tomalia DA, Meier DJ. Visualization and characterization of poly(amidoamine) dendrimers by atomic force microscopy. Langmuir. 2000;16(13):5613–5616. [Google Scholar]
- 69.Hatch E, Hetzer M. Breaching the nuclear envelope in development and disease. J Cell Biol. 2014;205(2):133–141. doi: 10.1083/jcb.201402003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong S, Bielinska AU, Mecke A, et al. Interaction of poly(amidoamine) dendrimers with supported lipid bilayers and cells: hole formation and the relation to transport. Bioconjug Chem. 2004;15(4):774–782. doi: 10.1021/bc049962b. [DOI] [PubMed] [Google Scholar]
- 71.Smith PE, Brender JR, Dürr UH, et al. Solid-state NMR reveals the hydrophobic-core location of poly(amidoamine) dendrimers in biomembranes. J Am Chem Soc. 2010;132(23):8087–8097. doi: 10.1021/ja101524z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith DJ, Ng H, Kluck RM, Nagley P. The mitochondrial gateway to cell death. IUBMB Life. 2008;60(6):383–389. doi: 10.1002/iub.44. [DOI] [PubMed] [Google Scholar]