Abstract
MicroRNAs (miRNAs) have emerged as important players in the regulation of T-cell functionality. However, comprehensive insight into the extent of age-related miRNA changes in T cells is lacking. We established miRNA expression patterns of CD45RO- naïve and CD45RO+ memory T-cell subsets isolated from peripheral blood cells from young and elderly individuals. Unsupervised clustering of the miRNA expression data revealed an age-related clustering in the CD45RO- T cells, while CD45RO+ T cells clustered based on expression of CD4 and CD8. Seventeen miRNAs showed an at least 2-fold up- or downregulation in CD45RO- T cells obtained from young as compared to old donors. Validation on the same and independent samples revealed a statistically significant age-related upregulation of miR-21, miR-223 and miR-15a. In a T-cell subset analysis focusing on known age-related phenotypic changes, we showed significantly higher miR-21 and miR-223 levels in CD8+CD45RO-CCR7- TEMRA compared to CD45RO-CCR7+ TNAIVE-cells. Moreover, miR-21 but not miR-223 levels were significantly increased in CD45RO-CD31- post-thymic TNAIVE cells as compared to thymic CD45RO-CD31+ TNAIVE cells. Upon activation of CD45RO- TNAIVE cells we observed a significant induction of miR-21 especially in CD4+ T cells, while miR-223 levels significantly decreased only in CD4+ T cells. Besides composition and activation-induced changes, we showed a borderline significant increase in miR-21 levels upon an increasing number of population doublings in CD4+ T-cell clones. Together, our results show that ageing related changes in miRNA expression are dominant in the CD45RO- T-cell compartment. The differential expression patterns can be explained by age related changes in T-cell composition, i.e. accumulation of CD8+ TEMRA and CD4+ post-thymic expanded CD31- T cells and by cellular ageing, as demonstrated in a longitudinal clonal culture model.
Introduction
Advanced age has been associated with defects of the immune system to mount appropriate antigen specific responses to pathogens. The most profound age-associated changes are observed in T cells. Due to thymus involution with age, the output of naïve T cells is reduced, while the proportion of memory T cells increases, thereby compromising the diversity of the T-cell pool.
Naïve T cells express CD45RA, while being negative for CD45RO [1]. Within the CD8+ T cell fraction, expression of CD45RA or CD45RO in combination with the C-C chemokine receptor type 7 (CCR7) is used to further define CCR7+CD45RA+(CD45RO-) naïve (TNAIVE), CCR7+CD45RA-(CD45RO+) central memory (TCM), CCR7-CD45RA-(CD45RO+) effector memory (TEM) and CCR7-CD45RA+(CD45RO)- late-stage effector memory (TEMRA) T-cell subsets [2]. Whether or not this model can also be applied to the CD4+ subset is still a matter of debate. Various age-related differences have been reported in the distribution of T-cell phenotypes in peripheral blood. For instance, the proportion of CD8+ TEMRA cells is higher in elderly than in young individuals [3]. Within the CD4+CD45RO- T-cell population, the proportion of CD31- TNAIVE cells increases with age, while the fraction of CD31+ T cells progressively decreases [4]. Kohler et al [5] characterized CD4+CD31+ T cells as recent thymic emigrants, while CD4+CD31- T cells represented central naïve peripherally expanded CD4+ T cells. Downregulation of surface expression of CD31 has been associated with homeostatic proliferation [6]. In elderly individuals, clonal expansion of memory T cells is required to preserve effective immune responses for combating antigenic re-challenges. This leads to a marked proliferative stress resulting in clonal exhaustion and senescence [7–9]. Human T-cell clones are characterized by altered cell surface and cytokine expression signatures that resemble the in vivo situation of chronic antigenic stress. Long-term cultured T-cell clones may thus represent a model for cellular ageing [10].
MiRNAs are a class of small non-coding RNAs that bind to mRNA transcripts of protein-coding genes in a sequence-specific manner. Based on the degree of sequence complementarity they induce degradation of the mRNA or repress translation [11]. A single miRNA potentially regulates up to several hundreds of target genes, thus orchestrating many pathways [12]. Differentiated cells in complex cellular systems are characterized by the expression of specific miRNA profiles. Moreover, miRNAs are fundamental to the regulation of complex cellular processes, such as those that regulate the immune system.
The contribution of changes in miRNA expression patterns to the age-associated decreased functionality of the immune system is largely unexplored. Differential miRNA expression patterns were shown in replicative in vitro and in in vivo models of CD8 T-cell ageing [13]. Some of the deregulated miRNAs were shown to be involved in the DNA damage response [14]. A role of miR-92a has been reported in age-related attrition of CD8+ TNAIVE cells [15]. In addition, a role has been proposed for miR-181a in modulation of TCR sensitivity of CD4+ T cells upon ageing [16].
There are no comprehensive studies of differences in miRNA expression patterns in primary human T cells from younger and older donors. To address this issue, we analyzed miRNA expression in CD4+ and CD8+ T cells sorted based on expression of CD45RO from peripheral mononuclear blood cells (PBMCs) of young and elderly individuals. We observed age-related differences in miRNA expression predominantly within the CD45RO- T-cell compartment. These differences could be explained by a combination of age-associated accumulation of TEMRA and CD31- T cells, T-cell activation and by cellular ageing. Finally, we assessed changes longitudinally in a clonal T cell culture model and found alterations in miRNA expression as a function of population doublings in vitro.
Material and Methods
Participants
In total 27 healthy young (≤ 30 yrs) and 24 healthy elderly (≥ 55 yrs) participants were included in this study. Demographic characteristics of all donors and miRNA analysis strategies are summarized in Table A in S1 File. Healthy young and old were recruited via advertisements and contacting the elderly association and were selected for healthiness based on the Senieur health admission criteria for immunogerontological studies [17]. None of the subjects had a history of infection, malignancy, autoimmune disease, chronic liver or kidney disease, alcohol or drug abuse, diabetes mellitus, current pregnancy or immunosuppressive therapy. An elevated blood pressure and the use of anti-hypertensive treatment were accepted.
T-cell clones were generated from phytohaemagglutinin (PHA)-stimulated peripheral blood mononuclear cells (PBMC) by limiting dilution in the presence of IL-2 using irradiated PBMCs as feeder cells [10,18]. In total, we included 16 CD4+ T-cell clones for each clone a sample with a low and a sample with a high number of population doublings. The T-cell clones were initially isolated from 5 individual donors (Table B in S1 File).
Ethics statement
All participants provided written informed consent according to the Declaration of Helsinki to participate in this study that was approved by The Medical Ethical Committee (METC) (project number: 2009.118) of the University Medical Center Groningen UMCG.
PBMC isolation and fluorescence-activated cell sorting (FACS) of human primary lymphocyte subsets
Peripheral blood was collected in heparin-containing vacutainer tubes (Becton Dickinson, Franklin Lakes, USA) and peripheral blood mononuclear cells (PBMC) were freshly isolated by density gradient centrifugation using Lymphoprep (Axis-Shield, Oslo, Norway) according to the manufacturer’s protocol. CD4 T cells were sorted by fluorescence-activated cell sorting (FACS) as CD3+CD4+ and CD8 T cells as CD3+CD4-. Within the CD4 and CD8 T-cell subsets, naïve (CD45RO-) and memory (CD45RO+), truly TNAIVE (CD45RO-CCR7+) and terminally differentiated (TEMRA) cells (CD45RO-CCR7-); as well as CD3+ and CD31- populations were isolated using combinations of the following anti-human monoclonal antibodies: CD3-e450, CD4-A647 (eBioscience, Vienna, Austria), CD45RO-FITC, CCR7-PE, CCR7-PECY7 and CD31-PE (BD Bioscience, Breda, The Netherlands). See Figure A in in S1 File for sorting schemes.
RNA extraction and purification
Total RNA was extracted using the miRNeasy Mini Kit (Qiagen, Venlo, The Netherlands) following the manufacturer’s instructions. Micro Bio-SpinTM chromatography columns, supplied with Bio-Gel P-6 polyacrylamide gel matrices, were applied for efficient purification of RNA samples (Bio-Rad laboratories B.V. Veenendaal, The Netherlands). RNA concentration was measured on a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, USA). RNA samples with 260 / 280 and 260 / 230 ratio of ≥ 1.90 were used for further analysis.
MiRNA profiling
Equal amounts of RNA samples from FACS sorted CD3+CD4+CD45RO+/- and CD3+CD4-CD45RO+/- T cells from healthy young (n = 5) and elderly (n = 5) donors were pooled for miRNA expression profiling. Microarray profiling was performed using the SurePrint Human miRNA Array Kit with 8x15K format (V2, sequence source from miRBase 10.1) from Agilent (G447013) according to manufacturer’s protocol (Agilent Technologies, Santa Clara, California, USA). Arrays were scanned using an Agilent scanner according to the manufacturer's instructions (Agilent Technologies). Array images were analyzed using Agilent feature extraction software. For data analysis, raw data were log2 transformed and normalized using a 90th percentile shift using GeneSpring software (v.11.5.1). We excluded control probes and probes detecting viral miRNAs. MiRNAs detected in at least 7 out of 8 samples were included for further analysis. Visualization of normalized data and hierarchical clustering was performed with Pearson correlation distance and complete linkage clustering parameters using Genesis software (v.1.7.6) (Graz University of Technology, Graz, Austria) [19]. The microarray data were deposited in Gene Expression Omnibus and are accessible with the following series record number: GSE69191.
Quantitative RT-PCR
MiRNA expression levels were determined by quantitative (q)RT-PCR. Multiplexed cDNA synthesis for up to 6 specific miRNAs was performed using Taqman MicroRNA Reverse transcription kit with a multiplexed reverse transcription primers of TaqMan microRNA Assays (Life Technologies, Carlsbad, USA): for miR-21 (000397), miR-223 (002295), miR-451 (001141), miR-22 (000398), miR-15a (000389), miR-197 (000497), miR-766 (001986), miR-574-3p (002349), miR-328 (000543), miR-483-3p (CSS07FR), miR-885-5p (241451_mat), miR-28-5p (000411) and RNU49 (001005) as described earlier [20]. RNU49 served as a reference gene to normalize miRNA expression levels.
All PCR reactions were run in triplicate. Mean cycle threshold (Ct) values were quantified with the Sequence Detection Software (SDS, version 2.3, Life Technologies, Amsterdam, The Netherlands). Relative expression levels were quantified using the 2–ΔCt (Δ Ct = Ct gene—Ct reference gene).
T cell stimulation with αCD3/αCD28 monoclonal antibodies (Abs)
Culture plates were coated with goat-anti-mouse IgG2a Ab (Cat. No. 1080–01, Southern Biotechnology, Uden, The Netherlands) and mouse anti-human CD3 hybridoma supernatant (clone WT32, IgG2a). CD3+CD4+CD45RO- and CD3+CD4-CD45RO- T cells were seeded at a density of 0,5x106 cells/mL in RPMI medium (Lonza, Breda, The Netherlands) supplemented with 5% V/V mouse anti-human CD28 hybridoma (clone 20–4669, IgG1), 2% HSA (Sanquin, Amsterdam, The Netherlands). Cells were split and placed in fresh medium supplemented with αCD3/αCD28 on day 3, 5 and 7. On day 10 cells were harvested and stained for FACS analysis or lysed for RNA isolation.
Analysis of cell surface markers
Cell surface markers on in vitro activated T cells were assessed using mAbs against human CD25-PE (BC96) (eBioscience, Vienna, Austria) and CD45RO-FITC (UCHL1) (BD Biosciences, Breda, The Netherlands). Cells were analyzed on BD LSR-II Flow Cytometer by Diva software (BD Biosciences). Data analysis was performed on Kaluza Flow Analysis Software (1.2) (Beckman Coulter, Woerden, The Netherlands).
Statistical analysis
Results obtained from qRT-PCR are expressed as median respectively. Unpaired samples were compared using the Mann-Whitney test and paired samples using the Wilcoxon signed-rank test. For comparisons of paired unstimulated and stimulated CD4+ and CD8+ T cells, we applied the Wilcoxon matched pairs test. T-cell clones with low and high PD were compared using a paired Wilcoxon signed-rank test. Statistical analyses were performed with GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA) and SPSS Statistics version 22.0 (IBM Corp. Armonk, NY, USA). P < 0.05 was considered statistically significant.
Results
Age-related miRNA expression differences are restricted to CD45RO- T cells
To identify age related differences in T cells, we compared miRNA expression profiles from sorted T cells from peripheral blood of 5 healthy young (median age 27) and 5 healthy old (median age 64) donors. CD4 and CD8 T cells were sorted from the CD45RO- and CD45RO+ gates (Figure A in S1 File). Purity of the T-cell subsets are shown in Figure B in S1 File. Equal amounts of RNA were pooled for each T-cell subset resulting in a total of 8 T-cell subset pools. MiRNA signatures were generated from these pools by miRNA expression arrays. Unsupervised hierarchical clustering of the 166 miRNAs that were detected in at least 7 out of 8 samples revealed a clustering into two main groups, i.e. CD45RO- and CD45RO+ T-cells (Table C and Figure C in S1 File). In the CD45RO- T cell subset, the clustering was based on age, irrespective of CD4 and CD8 expression status (Fig 1A). In the CD45RO+ T-cell subset, the clustering was between CD4+ and CD8+ T cells, irrespective of age. Based on this marked difference we focused our further analysis on the CD45RO- T-cell subset.
Fig 1. Age-related differences in miRNA expression are predominantly found in CD45RO- T cells.
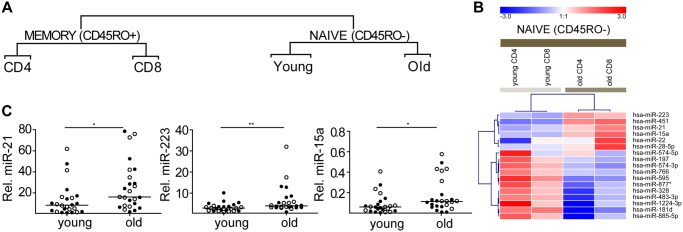
(A) Unsupervised hierarchical cluster analysis of 166 miRNAs revealed a clear distinction between CD45RO- and CD45RO+ T cells. Within the CD45RO- subgroup the next separation was based on age, whereas in CD45RO+ T cells the next clustering was based on expression of CD4/CD8 (the complete heatmap can be seen in Figure C in S1 File). (B) Heatmap of miRNAs differentially expressed in young versus old CD45RO- T cells (n = 17) using a 2-fold expression change cut-off (≥ 2 fold) between young and old CD45RO- T cells. The blue to red gradient on the heatmap indicates relative miRNA expression levels varying from low (blue) to high (red). (C) Relative expression levels in CD45RO- T cells as determined by qRT-PCR of miR-21, miR-223 and miR-15a in CD45RO- T cells from young and old donors. MiRNA expression levels were normalized to the expression levels of RNU49 (lines indicates median). Filled circles indicate CD4 T cells and open circles indicate CD8 T cells, *p ≤ 0.05, **p ≤ 0.01.
Seventeen miRNAs showed a ≥ 2-fold difference in expression level between young and old CD45RO- T cells (Fig 1B). Based on miRNA expression level and availability of qRT-PCR assays, 13 out of the 17 miRNAs were selected for validation on the individual samples of the pools used for the array analysis combined with a second independent cohort (young; n = 12, median age 27; old; n = 13, median age 65) (Table A and Table D in S1 File). Array results of miR-21, miR-223 and miR-15a were validated on the combined cohort (Fig 1C). MiR-28-5p expression levels were increased in old compared to young subjects, but this difference did not reach significance (Figure D in S1 File). Of the other selected miRNAs, we could not validate differences between young and old donors for miR-197, miR-766, miR-328 and miR-451 (Figure D in S1 File). We next studied if the observed changes could be assigned to ageing related changes in the T-cell composition. We focused these analyses on miR-21 and miR-223, as the miR-15a levels were very low.
Increased numbers of CD8+ TEMRA cells in elderly contributes to the higher miR-21 and miR-223 levels in CD45RO- T cells
Loss of CCR7 expression is a hallmark of differentiation from central memory to effector and late effector CD8+ TEMRA cells. To explore if the observed changes in miR-21 and miR-223 levels are related to the increase of TEMRA cells in elderly, we sorted CD45RO-CCR7+ TNAIVE and CD45RO-CCR7- TEMRA cells from peripheral blood of 7 healthy young (median age 27) and 7 healthy old (median age 67) donors (Figure A in S1 File). Purity of the T-cell subsets are shown in Figure B in S1 File. The percentage of CD45RO-CCR7+ TNAIVE cells within the CD4+ gate was high for both young (median is 93,6%) and old donors (median is 87,1%) (Fig 2A). The yield of CD4+CD45RO-CCR7- TEMRA cells was insufficient for the qRT-PCR analysis. For CD8+ T cells we observed a significantly lower percentage of CD45RO-CCR7+ TNAIVE cells in elderly donors (median of 33% in elderly vs 59% in young) with a concomitant significant increase in the percentage of TEMRA cells (median percentage 67% in elderly vs 41% in young) (Fig 2B). QRT-PCR analysis revealed significant higher expression of both miR-21 and miR-223 in the TEMRA populations when compared to TNAIVE cells (Fig 2C and 2D). Hence, increased expression of both miRNAs in the CD45RO- T-cell subset upon ageing can at least in part be explained by the age-associated accumulation of CD8 TEMRA cells.
Fig 2. Age-related upregulation of miR-21 and miR-223 can be partly explained by the accumulation of TEMRA (CD8 T cells) in the CD45RO- T-cell compartment.
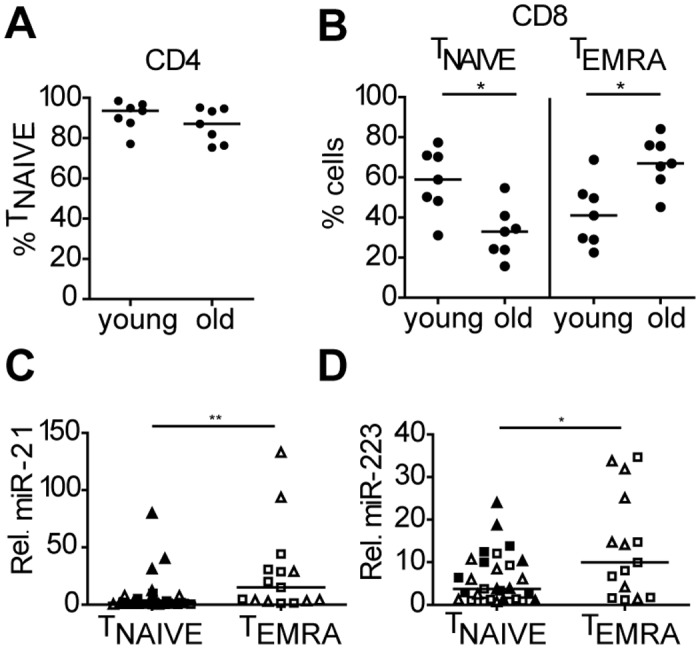
Percentage of (A) TNAIVE CD4+CD45RO-CCR7+ and (B) TERMA CD4-CD45RO-CCR7- T cells in young and old donors. Relative expression of (C) miR-21 and (D) miR-223 in TNAIVE (CD4 and CD8) and TERMA (CD8) T cells in young vs old donors. MiRNA expression was normalized to the expression of RNU49 (Lines indicate median). Squares indicate cells from young and triangles cells from elderly donors. Filled symbols indicate CD4+ T cells and open symbols indicate CD8+ T cells. *p ≤ 0.05, **p ≤ 0.01.
Increased numbers of CD31- central naïve T cells contributes to the increased levels of miR-21 in CD45RO- T cells
As a next step, we studied expression patterns of miR-21 and miR-223 in relation to CD31 expression as it is known that CD31- central naïve T cells increase with age especially in the CD4+ T cell subset. CD31+ and CD31- T cells were sorted from the CD45RO- compartment for both CD4 and CD8 from PBMCs of 8 healthy young (median age 28) and 7 healthy old (median age 75) donors (Figure A in S1 File). Purity of the T-cell subsets are given in Figure B in S1 File. The percentage of CD4+CD31- T cells was significantly higher in elderly donors (median is 45%) when compared to younger individuals (median is 19%). In the CD8 compartment, we observed a slightly higher, albeit not significant, percentage of CD31- T cells in elderly (median is 17%) compared to young donors (median is 9,3%)(Fig 3A and 3B). The expression level of miR-21 was significantly increased in CD45RO-CD31- T cells compared to CD45RO-CD31+ T cells, whereas the miR-223 levels were similar in CD31+ and CD31- T cells (Fig 3C and 3D). Thus, the accumulation of CD31- T cells contributes to the observed age related increase of miR-21 levels, especially in the CD4+CD45RO- T cell compartment in which CD31 loss is most pronounced.
Fig 3. MiR-21 is upregulated in CD31- T cells in the CD45RO- compartment.
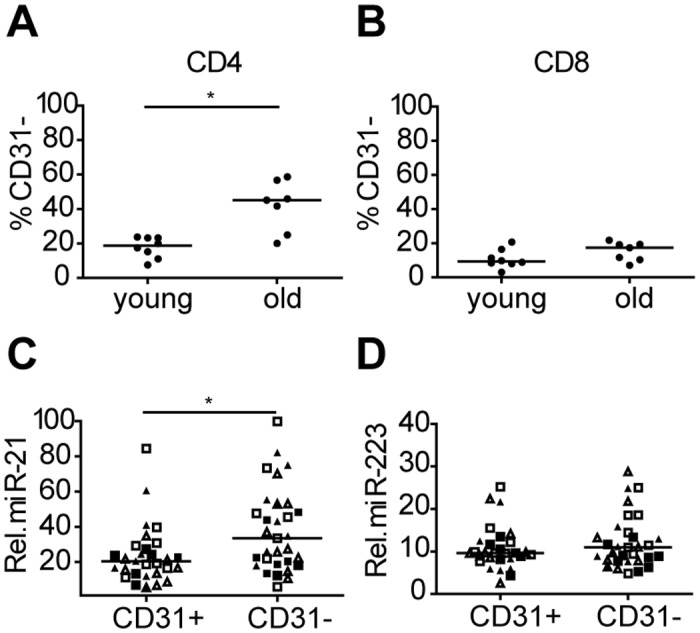
Percentage of CD31+ and CD31- in (A) CD4+CD45RO- and (B) CD4-CD45RO- T cells in young and old donors. Relative expression of (C) miR-21 and (D) miR-223 in CD31-and CD31+ T-cell subset sorted from the CD45RO- T-cell compartment. MiRNA expression was normalized to the expression of RNU49 (line indicates median). Squares indicate cells from young and triangles indicate cells from old donors. Filled symbols indicate CD4+ T cells and open symbols indicate CD8+ T cells. *p ≤ 0.05.
MiR-21 is upregulated upon activation of CD45RO- TNAIVE cells
Prolonged exposure of T cells to pathogens in elderly increases the number of TCR primed antigen-experienced memory T cells. To explore a relation between the increased miR-21 and miR-223 levels in elderly and T cell activation, we sorted both CD4+ and CD8+ CD45RO- T cells from peripheral blood of 7 healthy young donors (median age 27) (Table A in S1 File) and stimulated these cells with αCD3/αCD28. Stimulation of CD45RO- T cells in vitro induced an activated memory phenotype with a marked induction of both CD25 and CD45RO (Figure E in S1 File). A significant induction of miR-21 was observed in CD4+ T cells and a borderline significant induction in CD8+ T cells after stimulation for 10 days (Fig 4A and 4B). Expression of miR-223 was significantly decreased upon αCD3/αCD28 triggering in CD4+ T cells, whereas no difference was observed in CD8+ T cells (Fig 4C and 4D). Thus, our data show that in vitro activation of CD4+ T cells is associated with the transgression to the memory phenotype (CD45RO+), induction of miR-21 and reduction of miR-223. In vitro activation of CD8+ T cells is also associated with acquisition of the memory phenotype (CD45RO+) and the induction of miR-21, whereas the miR-223 levels remain constant.
Fig 4. T-cell activation induces upregulation of miR-21 and downregulation of miR-223.
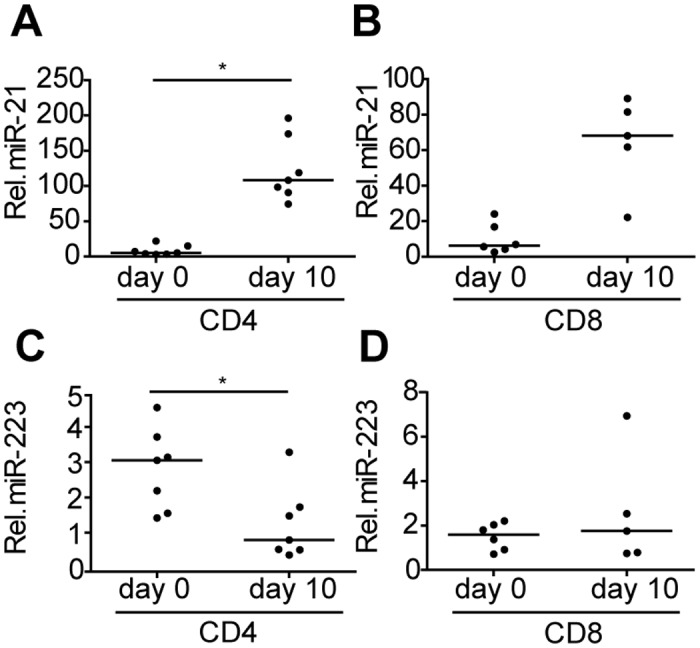
Relative expression of miR-21 in stimulated (A) CD4+CD45RO- and (B) CD4-CD45RO- T cells. Relative expression of miR-223 in stimulated (C) CD4+CD45RO- and (D) CD4-CD45RO- T cells. MiRNA expression was normalized to the expression of RNU49 (line indicates median). *p<0.05.
MiR-21 is increased in aged CD4+ T cell clones
We next tested, if besides T-cell composition and activation, we could also identify an age-related component that could contribute to the enhanced miR-21 and miR-223 levels. As an in vitro model of T-cell aging, we studied expression of these two miRNAs in 16 CD4+ T-cell clones harvested at a lower (<40) (median population doubling (PD) is 28) and a higher (>40) (median PD is 55) number of PDs. Expression of miR-21 tended to be increased in high vs low PD T-cell clones (p = 0.0525), whereas the miR-223 levels did not differ in T-cell clones with low and high population doublings (Fig 5A and 5B). Thus, based on this in vitro model of T-cell ageing, the age-related increase of miR-21 may be associated with increased numbers of cell divisions and continuous activation of the T-cell clones.
Fig 5. MiR-21 is upregulated with increasing population doublings in T-cell clones.
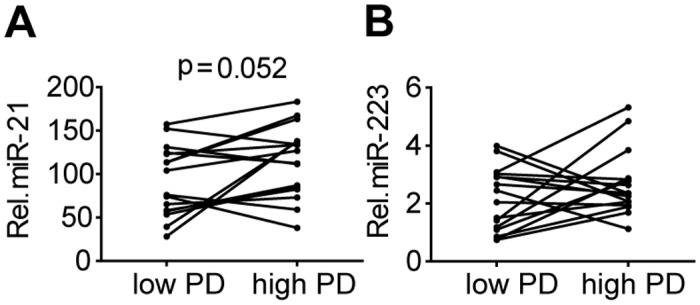
Relative expression of (A) miR-21 and (B) miR-223 in T cell clones harvested at lower and higher number of population doublings (PD). MiRNA expression was normalized to the expression of RNU49.
Discussion
In order to decipher age-related differences in miRNA expression patterns we studied CD45RO+ and CD45RO- T cells of healthy young and elderly individuals. We observed clear differences in miRNA expression patterns between CD45RO- and CD45RO+ T cells, consistent with previous studies [21,22]. The next clustering step indicated an age-related difference in CD45RO- T cells and a CD4/CD8 based clustering in CD45RO+ T cells. The pronounced age-related clustering in CD45RO- T cells has not been described previously.
Seventeen miRNAs were found to be ≥2-fold differentially expressed. For miR-15a, miR-21, miR-22 and miR-223 a relation with age was shown previously in studies conducted in humans, mice and rhesus monkeys [23,24]. RNA sequencing of skeletal muscles of old rhesus monkeys revealed age-related upregulation of miR-15a, of which functional involvement in ageing yet remains to be identified [25]. An age-related elevated expression of miR-21 has been described in plasma samples of healthy elderly and geriatric patients with cardiovascular disease and, as such, miR-21 has been established as a circulating marker of inflammageing [26]. Expression of miR-22 was prominently upregulated during cardiac ageing of C57/BI6N mice and overexpression was shown to induce cellular senescence of cardiac fibroblasts [27]. MiR-223 levels were increased in bone marrow derived dendritic cells from aged C57BL/6 mice [28]. Several previously reported age-related miRNA expression changes in T cells were confirmed by our study. Expression levels of the miR-17, miR-19, miR-20a and miR-106a were downregulated in a mix of naïve and memory CD8+CD28+ T cells [13]. We did not find downregulation of these miRNAs in naive CD45RO- T-cell subsets. However, in memory CD8+CD45RO+ T cells we observed a similar age-related downregulation for miR-17, miR-19 and miR-20a, whereas the levels of miR-106a were below detection limit in most samples. In CD4+ TNAIVE cells an age-associated decline of miR-181a expression has been reported [16]. Consistent with this finding we also observed a decreased expression of miR-181a expression with age although this did not reach our 2-fold cut off criterion (1.5 fold). Thus our data substantially increase the list of currently known age-related miRNA expression changes in specific T-cell subsets.
We next assessed if the observed age related differences in miRNA expression patterns are caused by heterogeneity in the composition of the CD45RO- T cells or are truly associated with age. In the CD8 T-cell compartment we showed that the expression levels of miR-21 and miR-223 were strongly upregulated in CD45RO-CCR7- TEMRA vs CD45RO-CCR7+ TNAIVE CD8+ cells. CCR7 is a predicted target for miR-21 and we previously demonstrated that miR-21 regulates expression of CCR7 in CD4+CD45RO- TNAIVE cells following activation [29]. The increased expression of miR-21 in CD8+CD45RO-CCR7- TEMRA cells is thus consistent with its CCR7 targeting potential. There are currently no studies showing a direct link between miR-223 and CCR7. MiRNA target gene prediction algorithms such as TargetScan, PicTar and miRanda also showed no predicted miR-223 binding sites in the CCR7 transcript. In CD4+ T cells, ageing has also been shown to alter T-cell composition, i.e. loss of CD31 a characteristic marker of post-thymic central naïve CD4+ T cells [5]. We observed higher levels of miR-21 in central naive CD4+CD31- T cells compared to CD31+ TNAIVE cells. As miR-21 expression levels are induced upon activation [29] and central naïve CD4+CD31- T cells have a history of antigenic stimulation [30], it might be anticipated that stimulation of CD31+ and CD31- CD4+ T-cell subsets will result in differential induction of miR-21 levels. Target gene prediction programs show no predicted target sites for miR-21 in the 3’-UTR of the CD31 mRNA transcript. Thus, it remains unclear if and how increased miR-21 levels affect CD31 loss. Overall, our data show that ageing-related changes in miRNA expression levels are at least partly induced by changes in T-cell subset composition.
Stimulation of T cells with αCD3/αCD28 results in enhanced miR-21 levels [31]. Consistent with this we showed that miR-21 is highly induced upon TCR triggering, with the most pronounced effects in CD4+ T cells. As the increase of both TEMRA and CD31- T cells in elderly is at least in part related to prolonged stimulation, this might explain the accumulation of miR-21 in these T-cell subsets. The vast majority of CD4 and CD8 CD45RO- T cells in young individuals are CD28+ [32]. Nonetheless, we cannot exclude the possibility that differences in CD28 expression between the CD4 and CD8 populations could influence the effectiveness of the αCD3/CD28 stimulation and thus the degree of miRNA expression increase or decrease.
TEMRA cells clonally expand as a result of chronic antigenic stimulation and their numbers increase upon ageing [33]. CD8+ TEMRA cells re-express CD45RA and lose expression of CD45RO, CD27 and CD28. Hence, TEMRA cells are considered senescent T cells with impaired T-cell functionality [2,34]. Our data show that age-related differences in miR-21 and miR-223 expression levels in CD45RO- T cells may partly reflect the accumulation of CD8+ TEMRA cells in elderly. Post-thymic proliferating central naïve CD31- T cells have undergone TCR stimulation, which might explain the increased miR-21 levels in central naïve T cells. Induction of miR-21 in response to TCR engagement has been reported previously and the levels of miR-21 are increased in inflammatory conditions, such as rheumatoid arthritis [29,35]. Moreover, miR-21 is well characterized in terms of its anti-apoptotic and pro-survival properties in T cells [36,37]. In line with this, miR-21 was shown to act as an oncomiR in a conditional mouse model [38]. Another study showed increased miR-21 levels upon replicative and stress-induced senescence in endothelial cells, suggesting a role for miR-21 in senescence-induced growth arrest [39]. MiR-21 mediates suppression of apoptosis in part by targeting Tipe2 [37]. These data support a role of the TCR-induced increase of miR-21 in expansion and survival of both TEMRA and central naïve T cells in aged individuals. Expression of miR-223 was decreased in naïve CD4+ T cells following αCD3/αCD28 stimulation, whereas levels remained constant in CD8+ T cells. This is in line with the previously reported reduction of miR-223 upon activation of TNAIVE cells [40].
To study whether CD4+ T cell changes in miR-21 and miR-223 could also be related to cellular ageing we used T-cell clones with low and high population doublings as an in vitro model of ageing [10]. Extensive inter-clonal heterogeneity made it difficult to establish a clear correlation for miR-223, but there was a borderline significant increase in miR-21 with increasing PD when averaging the results from all the clones. This heterogeneity is not unexpected among monoclonal T-cell populations of different individuals. Next to the proposed role of miR-21 on survival of TEMRA and central naïve T cells, we propose that an age-related increase in miR-21 expression levels may also contribute to the increased miR-21 levels in ageing T cells. MiR-21 was found to induce replicative and stress-induced senescence in human endothelial cells via targeting p21(CIP) and CDC25A cell cycle genes [39]. These T-cell clone experiments thus provide a first indication that changes in levels of miR-21 and potentially also miR-223 measured longitudinally may be related to cellular ageing, but further experiments are required to substantiate this notion.
Conclusions
Taken together, our results provide an evidence for an age-related miRNA expression pattern especially in CD45RO- T cells. These changes are most likely defined by a combination of ageing-related changes in T-cell composition, activation and the number of cell divisions. MiR-223 changes are associated specifically with an accumulation of TEMRA cells, whereas increase in miR-21 levels are related to the number of cell divisions, the accumulation of TEMRA and CD31- central naïve T cells, the latter likely reflecting post-thymic TCR stimulation.
Supporting Information
Figure A-Representative scatter plots from T-cell subsets sorting strategies. Figure B- T-cell subsets purity following cell sorting. Figure C-Heatmap of the differentially expressed miRNAs in naïve (CD45RO-) and memory (CD45RO+) T cells from young and old donors. Figure D-Validation of microRNA array. Figure E-Activated T cell phenotype. Table A-Characteristics of donors. Table B-Characteristics of T cell clones. Table C-Normalized expression values of miRNAs. Table D-Differentially expressed miRNAs in CD45RO- (CD4/CD8) T cells.
(DOC)
Acknowledgments
We thank all healthy young and elderly volunteers for participating in the study. We are grateful for the outstanding support of the flow cytometry team, Geert Mesander and Henk Moes.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by unrestricted funds from Abbot, Jan Kornelis de Cock Foundation and the Graduate School of Medical Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Michie CA, McLean A, Alcock C, Beverley PC. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 1992. November 19;360(6401):264–265. [DOI] [PubMed] [Google Scholar]
- 2. Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999. October 14;401(6754):708–712. [DOI] [PubMed] [Google Scholar]
- 3. Fagnoni FF, Vescovini R, Mazzola M, Bologna G, Nigro E, Lavagetto G, et al. Expansion of cytotoxic CD8+ CD28- T cells in healthy ageing people, including centenarians. Immunology 1996. August;88(4):501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kimmig S, Przybylski GK, Schmidt CA, Laurisch K, Mowes B, Radbruch A, et al. Two subsets of naive T helper cells with distinct T cell receptor excision circle content in human adult peripheral blood. J Exp Med 2002. March 18;195(6):789–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohler S, Thiel A. Life after the thymus: CD31+ and CD31- human naive CD4+ T-cell subsets. Blood 2009. January 22;113(4):769–774. 10.1182/blood-2008-02-139154 [DOI] [PubMed] [Google Scholar]
- 6. Kohler S, Wagner U, Pierer M, Kimmig S, Oppmann B, Mowes B, et al. Post-thymic in vivo proliferation of naive CD4+ T cells constrains the TCR repertoire in healthy human adults. Eur J Immunol 2005. June;35(6):1987–1994. [DOI] [PubMed] [Google Scholar]
- 7. Effros RB. Replicative senescence: the final stage of memory T cell differentiation? Curr HIV Res 2003. April;1(2):153–165. [DOI] [PubMed] [Google Scholar]
- 8. Goronzy JJ, Weyand CM. Aging, autoimmunity and arthritis: T-cell senescence and contraction of T-cell repertoire diversity—catalysts of autoimmunity and chronic inflammation. Arthritis Res Ther 2003;5(5):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Effros RB. The role of CD8 T cell replicative senescence in human aging. Discov Med 2005. June;5(27):293–297. [PubMed] [Google Scholar]
- 10. Pawelec G, Rehbein A, Haehnel K, Merl A, Adibzadeh M. Human T-cell clones in long-term culture as a model of immunosenescence. Immunol Rev 1997. December;160:31–42. [DOI] [PubMed] [Google Scholar]
- 11. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009. January 23;136(2):215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004. January 2;303(5654):83–86. [DOI] [PubMed] [Google Scholar]
- 13. Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C, et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 2010. April;9(2):291–296. 10.1111/j.1474-9726.2010.00549.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brunner S, Herndler-Brandstetter D, Arnold CR, Wiegers GJ, Villunger A, Hackl M, et al. Upregulation of miR-24 is associated with a decreased DNA damage response upon etoposide treatment in highly differentiated CD8(+) T cells sensitizing them to apoptotic cell death. Aging Cell 2012. August;11(4):579–587. 10.1111/j.1474-9726.2012.00819.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ohyashiki M, Ohyashiki JH, Hirota A, Kobayashi C, Ohyashiki K. Age-related decrease of miRNA-92a levels in human CD8+ T-cells correlates with a reduction of naive T lymphocytes. Immun Ageing 2011. November 15;8(1):11-4933-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li G, Yu M, Lee WW, Tsang M, Krishnan E, Weyand CM, et al. Decline in miR-181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med 2012. October;18(10):1518–1524. 10.1038/nm.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ligthart GJ, Corberand JX, Fournier C, Galanaud P, Hijmans W, Kennes B, et al. Admission criteria for immunogerontological studies in man: the SENIEUR protocol. Mech Ageing Dev 1984. November;28(1):47–55. [DOI] [PubMed] [Google Scholar]
- 18. Pawelec G, Barnett Y, Mariani E, Solana R. Human CD4+ T cell clone longevity in tissue culture: lack of influence of donor age or cell origin. Exp Gerontol 2002. Jan-Mar;37(2–3):265–269. [DOI] [PubMed] [Google Scholar]
- 19. Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics 2002. January;18(1):207–208. [DOI] [PubMed] [Google Scholar]
- 20. Kluiver J, Slezak-Prochazka I, van den Berg A. Studying microRNAs in lymphoma. Methods Mol Biol 2013;971:265–276. 10.1007/978-1-62703-269-8_15 [DOI] [PubMed] [Google Scholar]
- 21. Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One 2007. October 10;2(10):e1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smigielska-Czepiel K, van den Berg A, Jellema P, van der Lei RJ, Bijzet J, Kluiver J, et al. Comprehensive analysis of miRNA expression in T-cell subsets of rheumatoid arthritis patients reveals defined signatures of naive and memory Tregs. Genes Immun 2014. March;15(2):115–125. 10.1038/gene.2013.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech Ageing Dev 2012. Nov-Dec;133(11–12):675–685. 10.1016/j.mad.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 24. Zhu S, Deng S, Ma Q, Zhang T, Jia C, Zhuo D, et al. MicroRNA-10A* and MicroRNA-21 modulate endothelial progenitor cell senescence via suppressing high-mobility group A2. Circ Res 2013. January 4;112(1):152–164. 10.1161/CIRCRESAHA.112.280016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mercken EM, Majounie E, Ding J, Guo R, Kim J, Bernier M, et al. Age-associated miRNA alterations in skeletal muscle from rhesus monkeys reversed by caloric restriction. Aging (Albany NY) 2013. September;5(9):692–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, Rippo MR, et al. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech Ageing Dev 2012. Nov-Dec;133(11–12):675–685. 10.1016/j.mad.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 27. Jazbutyte V, Fiedler J, Kneitz S, Galuppo P, Just A, Holzmann A, et al. MicroRNA-22 increases senescence and activates cardiac fibroblasts in the aging heart. Age (Dordr) 2013. June;35(3):747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park S, Kang S, Min KH, Woo Hwang K, Min H. Age-associated changes in microRNA expression in bone marrow derived dendritic cells. Immunol Invest 2013;42(3):179–190. 10.3109/08820139.2012.717328 [DOI] [PubMed] [Google Scholar]
- 29. Smigielska-Czepiel K, van den Berg A, Jellema P, Slezak-Prochazka I, Maat H, van den Bos H, et al. Dual role of miR-21 in CD4+ T-cells: activation-induced miR-21 supports survival of memory T-cells and regulates CCR7 expression in naive T-cells. PLoS One 2013. October 1;8(10):e76217 10.1371/journal.pone.0076217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fornasa G, Groyer E, Clement M, Dimitrov J, Compain C, Gaston AT, et al. TCR stimulation drives cleavage and shedding of the ITIM receptor CD31. J Immunol 2010. May 15;184(10):5485–5492. 10.4049/jimmunol.0902219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, He L, Zhang R, Liu X, Ren Y, Liu Z, et al. Regulation of T lymphocyte activation by microRNA-21. Mol Immunol 2014. June;59(2):163–171. 10.1016/j.molimm.2014.02.004 [DOI] [PubMed] [Google Scholar]
- 32. Koch S, Larbi A, Derhovanessian E, Ozcelik D, Naumova E, Pawelec G. Multiparameter flow cytometric analysis of CD4 and CD8 T cell subsets in young and old people. Immun Ageing 2008. July 25;5:6. 6-4933-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ouyang Q, Wagner WM, Zheng W, Wikby A, Remarque EJ, Pawelec G. Dysfunctional CMV-specific CD8(+) T cells accumulate in the elderly. Exp Gerontol 2004. April;39(4):607–613. [DOI] [PubMed] [Google Scholar]
- 34. Henson SM, Riddell NE, Akbar AN. Properties of end-stage human T cells defined by CD45RA re-expression. Curr Opin Immunol 2012. August;24(4):476–481. 10.1016/j.coi.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 35. van der Geest KS, Smigielska-Czepiel K, Park JA, Abdulahad WH, Kim HW, Kroesen BJ, et al. SF Treg cells transcribing high levels of Bcl-2 and microRNA-21 demonstrate limited apoptosis in RA. Rheumatology (Oxford) 2015. May;54(5):950–958. [DOI] [PubMed] [Google Scholar]
- 36. Buscaglia LE, Li Y. Apoptosis and the target genes of microRNA-21. Chin J Cancer 2011. June;30(6):371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruan Q, Wang P, Wang T, Qi J, Wei M, Wang S, et al. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis 2014. February 27;5:e1095 10.1038/cddis.2014.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010. September 2;467(7311):86–90. 10.1038/nature09284 [DOI] [PubMed] [Google Scholar]
- 39. Dellago H, Preschitz-Kammerhofer B, Terlecki-Zaniewicz L, Schreiner C, Fortschegger K, Chang MW, et al. High levels of oncomiR-21 contribute to the senescence-induced growth arrest in normal human cells and its knock-down increases the replicative lifespan. Aging Cell 2013. June;12(3):446–458. 10.1111/acel.12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fulci V, Scappucci G, Sebastiani GD, Giannitti C, Franceschini D, Meloni F, et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Hum Immunol 2010. February;71(2):206–211. 10.1016/j.humimm.2009.11.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure A-Representative scatter plots from T-cell subsets sorting strategies. Figure B- T-cell subsets purity following cell sorting. Figure C-Heatmap of the differentially expressed miRNAs in naïve (CD45RO-) and memory (CD45RO+) T cells from young and old donors. Figure D-Validation of microRNA array. Figure E-Activated T cell phenotype. Table A-Characteristics of donors. Table B-Characteristics of T cell clones. Table C-Normalized expression values of miRNAs. Table D-Differentially expressed miRNAs in CD45RO- (CD4/CD8) T cells.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


