Abstract
Ubiquitin-dependent proteolysis is a major proteolytic pathway in the cytoplasm and nucleus of eukaryotic cells. We introduced a gene encoding a substrate for this pathway into the genome of Arabidopsis thaliana. The transgene codes for a hybrid protein consisting of dihydrofolate reductase (DHFR, EC 1.5.1.3) fused to a degradation signal that is specifically recognized by components of the ubiquitin-dependent proteolysis pathway. Elevated concentrations of the DHFR protein confer resistance to the drug methotrexate, but rapid degradation prevents accumulation of the protein in the plant. Therefore, transgenic A. thaliana lines expressing the DHFR fusion protein are methotrexate-sensitive. Selection for mutants resistant to methotrexate resulted in plants impaired in degradation of the DHFR model substrate, as shown by an increase in protein level in the mutants.
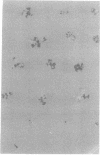
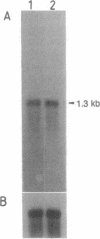
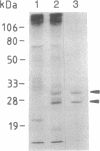
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armon T., Ganoth D., Hershko A. Assembly of the 26 S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J Biol Chem. 1990 Dec 5;265(34):20723–20726. [PubMed] [Google Scholar]
- Bachmair A., Becker F., Masterson R. V., Schell J. Perturbation of the ubiquitin system causes leaf curling, vascular tissue alterations and necrotic lesions in a higher plant. EMBO J. 1990 Dec;9(13):4543–4549. doi: 10.1002/j.1460-2075.1990.tb07906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989 Mar 24;56(6):1019–1032. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- Bartel B., Wünning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990 Oct;9(10):3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989 Mar 24;243(4898):1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Dohmen R. J., Madura K., Bartel B., Varshavsky A. The N-end rule is mediated by the UBC2(RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll J., Goldberg A. L. The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J Biol Chem. 1990 Mar 25;265(9):4789–4792. [PubMed] [Google Scholar]
- Finley D., Chau V. Ubiquitination. Annu Rev Cell Biol. 1991;7:25–69. doi: 10.1146/annurev.cb.07.110191.000325. [DOI] [PubMed] [Google Scholar]
- Glotzer M., Murray A. W., Kirschner M. W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991 Jan 10;349(6305):132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gonda D. K., Bachmair A., Wünning I., Tobias J. W., Lane W. S., Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989 Oct 5;264(28):16700–16712. [PubMed] [Google Scholar]
- Hershko A. The ubiquitin pathway for protein degradation. Trends Biochem Sci. 1991 Jul;16(7):265–268. doi: 10.1016/0968-0004(91)90101-z. [DOI] [PubMed] [Google Scholar]
- Hough R., Pratt G., Rechsteiner M. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem. 1987 Jun 15;262(17):8303–8313. [PubMed] [Google Scholar]
- Kane S. E., Pastan I., Gottesman M. M. Genetic basis of multidrug resistance of tumor cells. J Bioenerg Biomembr. 1990 Aug;22(4):593–618. doi: 10.1007/BF00762963. [DOI] [PubMed] [Google Scholar]
- Koncz C., Martini N., Mayerhofer R., Koncz-Kalman Z., Körber H., Redei G. P., Schell J. High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Mayerhofer R., Koncz-Kalman Z., Nawrath C., Reiss B., Redei G. P., Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990 May;9(5):1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbrandt J. D., Heintz N. H., White W. C., Rothman S. M., Hamlin J. L. Methotrexate-resistant Chinese hamster ovary cells have amplified a 135-kilobase-pair region that includes the dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6043–6047. doi: 10.1073/pnas.78.10.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. C., Finley D., Szostak J. W. A strategy for the generation of conditional mutations by protein destabilization. Proc Natl Acad Sci U S A. 1992 Feb 15;89(4):1249–1252. doi: 10.1073/pnas.89.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M. Natural substrates of the ubiquitin proteolytic pathway. Cell. 1991 Aug 23;66(4):615–618. doi: 10.1016/0092-8674(91)90104-7. [DOI] [PubMed] [Google Scholar]
- Richter-Ruoff B., Heinemeyer W., Wolf D. H. The proteasome/multicatalytic-multifunctional proteinase. In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett. 1992 May 11;302(2):192–196. doi: 10.1016/0014-5793(92)80438-m. [DOI] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. In vivo function of the proteasome in the ubiquitin pathway. EMBO J. 1992 Aug;11(8):3077–3080. doi: 10.1002/j.1460-2075.1992.tb05379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. L., Vierstra R. D. Cloning of a 16-kDa ubiquitin carrier protein from wheat and Arabidopsis thaliana. Identification of functional domains by in vitro mutagenesis. J Biol Chem. 1991 Dec 15;266(35):23878–23885. [PubMed] [Google Scholar]
- Töpfer R., Matzeit V., Gronenborn B., Schell J., Steinbiss H. H. A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acids Res. 1987 Jul 24;15(14):5890–5890. doi: 10.1093/nar/15.14.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D., Van Montagu M., Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. Naming a targeting signal. Cell. 1991 Jan 11;64(1):13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule. Cell. 1992 May 29;69(5):725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]