Abstract
Background
Buruli ulcer (BU) is a skin infection caused by Mycobacterium ulcerans. The wounds of most BU patients are colonized with different microorganisms, including Staphylococcus aureus.
Methodology
This study investigated possible patient-to-patient transmission events of S. aureus during wound care in a health care center. S. aureus isolates from different BU patients with overlapping visits to the clinic were whole-genome sequenced and analyzed by a gene-by-gene approach using SeqSphere+ software. In addition, sequence data were screened for the presence of genes that conferred antibiotic resistance.
Principal Findings
SeqSphere+ analysis of whole-genome sequence data confirmed transmission of methicillin resistant S. aureus (MRSA) and methicillin susceptible S. aureus among patients that took place during wound care. Interestingly, our sequence data show that the investigated MRSA isolates carry a novel allele of the fexB gene conferring chloramphenicol resistance, which had thus far not been observed in S. aureus.
Author Summary
Buruli ulcer (BU) is a skin infection caused by Mycobacterium ulcerans. The wounds of most BU patients are colonized with different microorganisms, including Staphylococcus aureus. This study investigated patient-to-patient transmission events during wound care in a health care center. S. aureus isolates from patients who visited the health center at the same time points were analyzed using whole-genome sequencing. Analysis of sequence data confirmed transmission of methicillin resistant S. aureus and methicillin susceptible S. aureus among patients that took place during wound care.
Introduction
Buruli ulcer (BU) is a neglected necrotizing skin disease caused by Mycobacterium ulcerans, with the highest burden of the disease in West Africa, particularly in Benin, Cote d’Ivoire and Ghana [1]. The disease usually starts as a nodule, plaque, oedema or papule and progresses to form large ulcers with undermined edges if left untreated. It was previously shown that wounds of most BU patients are heavily colonized by many microorganisms, including Staphylococcus aureus [2,3].
S. aureus can be part of the human microbiota colonizing the skin and mucosal membranes without any clinical manifestations. However, once it crosses the skin barrier, or when the host immune system is compromised, this bacterium is able to cause a wide range of diseases, such as skin and soft tissue infections, osteomyelitis, pneumonia, meningitis, or bacteremia [4,5]. Therefore, S. aureus is considered a dangerous pathogen in both community-acquired and nosocomial infections. Colonization of healthy individuals with multi-drug resistant S. aureus is regarded as a risk factor for future development of S. aureus infections that are difficult to treat [6]. The S. aureus colonization of patients with a serious breach of skin barrier, such as patients with BU, burn wounds or the group of hereditary mechanobullous diseases epidermolysis bullosa (EB), was previously shown to be very high [2,3,7–9]. Molecular typing of S. aureus isolated from the wounds of BU and EB patients has shown that their wounds often harbor multiple genotypes of this pathogen [3,10].
Recently, several S. aureus clones have been reported in health care institutions in Ghana with the sequence types (ST) 15, 121 and 152 being the most prevalent as determined by multi-locus sequence typing (MLST) [11]. Notably, health care-associated infections (HAIs) caused by S. aureus impose a significant burden on patient care as a result of prolonged hospital stays, increased cost of treatments and high morbidity and mortality rates. Current practices implemented to reduce HAIs include cleaning of the hospital environment, hand hygiene and screening and decolonization of patients and health care workers [12–15]. Epidemiological data and molecular typing methods, such as pulsed-field gel electrophoresis (PFGE), MLST, spa-typing, multiple-locus variable number tandem repeat fingerprinting (MLVF), and whole-genome sequencing (WGS) of the infecting strains can be used to trace transmission events [16–19]. Each of these typing methods has particular advantages [20]. For example, MLVF is fast, cheap and highly discriminatory [21], while WGS provides additional information on the genetic makeup of investigated isolates on top of a highly discriminatory typing result.
BU patients may be at risk of hospital-associated colonization with S. aureus due to their frequent visits to particular health care centers for wound care. This represents an additional health risk for these patients, even if they are already colonized with community-acquired S. aureus. Therefore, the present study was aimed at uncovering possible S. aureus transmission events among BU patients using MLVF and WGS. Furthermore, WGS was applied to identify antimicrobial resistance (AMR) genes and to screen for mutations in genes that confer certain resistance phenotypes. The results obtained underpin the potential of the combined use of MLVF and WGS for the surveillance of S. aureus outbreaks in hospital settings.
Materials and Methods
Ethics statement
The ethical committee of the Noguchi Memorial Institute for Medical Research (NMIMR) (FEDERAL WIDE ASSURANCE FWA 00001824) approved the use of clinical samples for this investigation. Samples were collected upon written informed consent from adult subjects and a parent or guardian of any child participant on their behalf.
Bacterial isolates and genomic DNA extraction
A subset of the S. aureus isolates from BU patients that were previously collected and grouped by MLVF into thirteen clusters (A-M) [3] were selected for WGS. For the present study, isolates were selected from each of the thirteen MLVF clusters including two clusters suspected of patient-to-patient transmission events during wound care (clusters H and F). Screening of BU patients for the presence of S. aureus had been repeated every two weeks for a period of seven months, which defined the sampling time points t1 to t13 in this study (Table 1). Patients involved in this screening were at different stages of the disease and treatment for BU. All presently investigated S. aureus isolates were obtained from positive anterior nares and wound cultures of eleven BU patients who attended the Pakro Health Center in the Eastern region of Ghana for antimicrobial therapy (Table 1).
Table 1. Frequency of S. aureus with different spa-types isolated during patient visits for wound care.
| 12-12-2012 | 9-1-2013 | 23-1-2013 | 6-2-2013 | 20-2-2013 | 6-3-2013 | 20-3-2013 | 3-4-2013 | 17-4-2013 | 9-5-2013 | 23-5-2013 | 20-6-2013 | 4-7-2013 | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient No. | Start date of treatment | t1 | t2 | t3 | t4 | t5 | t6 | t7 | t8 | t9 | t10 | t11 | t12 | t13 | |
| 2 | 1-12-2012 | p | t786 | t786 | x | p | x | x | t786 | t786 | x | x | x | x | |
| ST88 | 7 | 26-12-2012 | p | p | t355 | t786 | t084, t939 | x | t084, t939, t1096 | x | x | t002, t084 | x | x | |
| 19 | 5-1-2013 | p | t786 | x | x | x | x | x | x | x | x | x | x | ||
| ST152 | 7 | 26-12-2012 | p | p | t355 | t786 | t084, t939 | x | t084, t939, t1096 | x | x | t002, t084 | x | x | |
| 5 | 22-12-2012 | p | p | p | p | x | x | x | x | x | t355 | x | x | ||
| 6 | 12-12-2012 | t7835 | x | x | x | x | x | x | x | x | x | t355 | x | x | |
| 3 | 6-12-2012 | p | t084 | p | p | x | x | t084 | t084, t355 | t084, t314 | t084 | t084 | p | p | |
| 10 | 12-12-2012 | p | t355 | t355 | t355 | t355 | x | x | t355 | x | x | x | x | x | |
| 11 | 19-12-2012 | t355 | x | x | t355 | x | x | x | x | x | x | x | x | ||
| 18 | 7-1-2013 | p | p | p | t355 | p | p | p | x | x | x | x | x | ||
| 24 | 14-2-2013 | t355 | x | x | x | x | x | x | x | x |
‘p’ indicates that a patient visited the health center, but no S. aureus was detected.
‘x’ indicates that a patient did not visit the health center at the respective time point of sampling.
‘t1’ to ‘t13’ refers to the time points at which samples were collected.
Cells with bold formatting indicate involvement of the respective S. aureus isolates in transmission events. spa-types with italic formatting are isolates that were sequenced.
Genomic DNA was extracted from S. aureus isolates grown overnight on blood agar by using the Ultraclean microbial DNA isolation kit (mo bio laboratories, Inc, Carlsbad, California, USA) according to the manufacturers’ instructions.
Whole-genome sequencing, sequence assembly and data analyses
DNA libraries were prepared using the Nextera XT v2 kit (Illumina, San Diego, CA, USA) according to the manufacturers’ instructions and then run on a Miseq (Illumina) for generating paired-end 250-bp reads. De novo sequence assembly was performed using CLC Genomics Workbench v7.0.4 (CLC bio A/S, Aarhus, Denmark) after quality trimming (Qs > 28) with optimal word sizes based on the maximum N50 value. The assembled files were imported as Fasta files into SeqSphere+ software version 1.1 (Ridom GmbH). The sequence reads were submitted to the National Center for Biotechnology Information GenBank and are available under the BioProject PRJNA283747 and accession numbers: LGAE00000000, LFTW00000000, LFTV00000000, LFTU00000000, LFTT00000000, LFOH00000000, LFOG00000000, LFNS00000000, LFNR00000000, LFNQ00000000, LFNP00000000, LFNO00000000, LFNN00000000, LFNM00000000, LFNL00000000, LFNK00000000, LFNJ00000000, LFNI00000000, LFNH00000000, LFMH00000000, LFMG00000000. The sequence data of the 21 isolates were characterized by using the core genome multilocus sequence typing (cgMLST) consisting of 1,861 genes and 706 S. aureus accessory genes. The complete sequence of each isolate was analyzed based on gene-by-gene comparison with the reference S. aureus strain COL (GenBank accession no. NC_002951) and S. aureus cgMLST target definer function with the default parameters of the software as previously described [22]. Each allele was assigned a number and an allelic typing profile based on the combination of all alleles for each isolate by the software. A dendrogram of the sequenced isolates and two additional reference genomes that represent different sequence types (ST5 [N315 GenBank accession no. BA000018.3] and ST8 [COL]) was generated using an unweighted-pair group method using average linkages (UPGMA). The concordance between the two typing methods was calculated with the Ridom EpiCompare software version 1.0 as described previously [19].
Transmission events
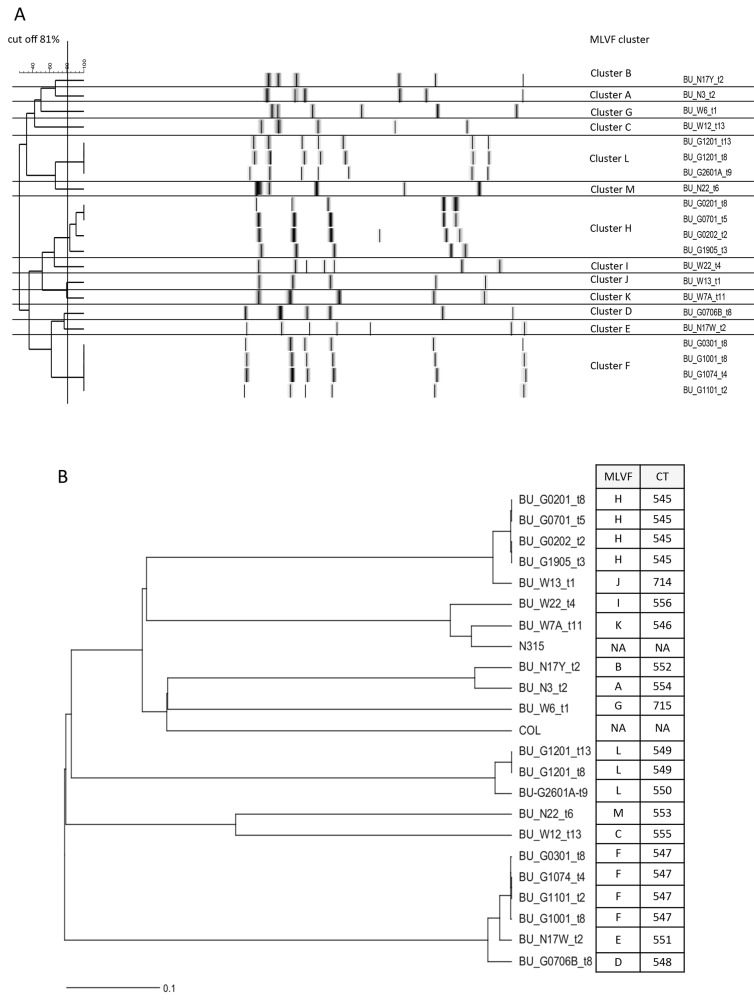
In this study a transmission event is defined to have occurred if the wound of a patient, previously not containing a particular S. aureus genotype, becomes colonized over time by an S. aureus with a genotype that is identical with the genotype of an S. aureus isolate collected from the wound of another patient. Here we investigated whether transmission events had indeed occurred during wound care of patients treated in the Pakro Health Center, using the SeqSphere+ scheme which assigned each S. aureus isolate an allelic typing profile as previously described. The typing profile will, subsequently, be known as cluster type (CT). Hence a transmission event would have occurred if S. aureus isolates from different BU patients are grouped within the same CT. In our previous study, S. aureus isolates from BU out-patients, who visited the health center for wound care, were suspected to be involved in patient-to-patient transmission events [3]. These isolates were initially grouped by MLVF into clusters H and F (Fig 1A).
Fig 1. MLVF and SeqSphere+ dendrograms of 21 S. aureus isolates from BU patients.
The dendrograms were generated using the UPGMA algorithm. (A), Dendrogram showing the previously identified MLVF clusters A-M. (B), SeqSphere+ dendrogram showing the 14 CTs identified in the present study. The reference strains COL (GenBank accession no. NC_002951) and N315 (GenBank accession no. BA000018.3) were included in the SeqSphere+ dendrogram. Isolates (BU_G0201_t8 and BU_G0202_t2 belonging to CT 545 and BU_G1074_t4 and BU_G1001_t8 belonging to CT 547) originating from patients 2 and 10 at two different time points are included in both transmission events. NA means ‘Not Applicable’.
Screening for antimicrobial resistance
De novo assembled genome sequences of S. aureus isolates were queried against specific previously identified sequence features, or compared to complete S. aureus reference genomes with associated annotated genes (S1 Table) using blastN in the WebACT comparison tool with default settings (http://www.webact.org/WebACT/prebuilt#). Further detailed analyses were performed with the Artemis Comparison Tool (ACT) software [23]. Specifically sequence data were queried for the presence of SCCmec elements and AMR genes. Similarity matches were filtered based on their length and percentage similarity scores, and only the filtered hits with at least 80% sequence similarity were then displayed by ACT and analyzed in detail. The AMR genes that were screened confer resistance to chloramphenicol, clindamycin, erythromycin, fusudic acid, kanamycin, lincosamide, methicillin, mupirocin, penicillin, rifampicin, streptogramin A and B, tetracycline, trimethoprim, tobramycin, and/or vancomycin. Antibiotic resistance profiles of the sequenced isolates were previously determined using vitek according to the EUCAST guidelines [3].
Results
Phylogeny of sequenced S. aureus isolates found in BU patients in Ghana based on a gene-by-gene comparison
From a total of 13 BU patients who visited the Pakro Healthcare Center for wound care 21 S. aureus isolates from the anterior nares (n = 4) and wounds (n = 17) were sequenced. These included six methicillin resistant MRSA and 15 methicillin susceptible S. aureus (MSSA) isolates. These isolates have been previously characterized by MLVF and spa-typing as shown in Table 2 [3].
Table 2. Genotypic and phenotypic characteristics of 21 S. aureus isolates from BU patients.
| Patient No. | Sample ID | Start of treatment | End of wound care | Week of sampling | MLVF | CT | ST | spa-type | SCCmec | CXT | OXA | CIP | TET | CHL | TRI | STM | RIF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | BU_G0201_t8 | 1-12-2012 | Not yet | 8 | H | 545 | 88 | t786 | IVa | + | mecA | tetM, tetL | fexB | ||||
| 7 | BU_G0701_t5 | 26-12-2012 | 25-6-2013 | 5 | H | 545 | 88 | t786 | IVa | + | mecA | tetM, tetL | fexB | ||||
| 2 | BU_G0202_t2 | 1-12-2012 | Not yet | 2 | H | 545 | 88 | t786 | IVa | + | mecA | tetM, tetL | fexB | ||||
| 19 | BU_G1905_t3 | 5-1-2013 | 22-7-2013 | 3 | H | 545 | 88 | t786 | IVa | + | mecA | tetM, tetL | fexB | ||||
| 13 | BU_W13_t1 | 12-12-2012 | 26-4-2013 | 1 | J | 714 | 88 | t186 | IVa | + | mecA | tetM, tetL | fexB | ||||
| 22 | BU_W22_t4 | 12-12-2012 | Not yet | 4 | I | 556 | 5 | t2724 | tetK | drfG | |||||||
| 7 | BU_W7A_t11 | 26-12-2012 | 25-6-2013 | 11 | K | 546 | 5 | t002 | none | + | + | gyrA * | |||||
| 17 | BU_N17Y_t2 | 28-12-2012 | N/A | 2 | B | 552 | 15 | t346 | drfG | ||||||||
| 3 | BU_N3_t2 | 6-12-2012 | N/A | 2 | A | 554 | 15 | t084 | tetM | ||||||||
| 6 | BU_W6_t1 | 12-12-2012 | 26-4-2013 | 1 | G | 715 | 1 | t7835 | tetK | fexB | |||||||
| 12 | BU_G1201_t13 | 19-12-2012 | 25-6-2013 | 13 | L | 549 | 121 | t314 | tetK | catA | drfG | ||||||
| 12 | BU_G1201_t8 | 19-12-2012 | 25-6-2013 | 8 | L | 549 | 121 | t314 | tetK | catA | drfG | ||||||
| 26 | BU_G2601A_t9 | 6-3-2013 | 21-6-2013 | 9 | L | 550 | 121 | t314 | tetK | drfG | rpoB * | ||||||
| 22 | BU_N22_t6 | 12-12-2012 | N/A | 6 | M | 553 | 3019 | t939 | catA | ||||||||
| 12 | BU_W12_t13 | 19-12-2012 | 25-6-2013 | 13 | C | 555 | 508 | t12836 | |||||||||
| 3 | BU_G0301_t8 | 6-12-2012 | Not yet | 8 | F | 547 | 152 | t355 | tetK | catA | str | ||||||
| 10 | BU_G1074_t4 | 12-12-2012 | 26-4-2013 | 4 | F | 547 | 152 | t355 | tetK | catA | str | ||||||
| 11 | BU_G1101_t2 | 19-12-2012 | 5-3-2013 | 2 | F | 547 | 152 | t355 | tetK | ||||||||
| 10 | BU_G1001_t8 | 12-12-2012 | 26-4-2013 | 8 | F | 547 | 152 | t335 | tetK | catA | str | ||||||
| 17 | BU_N17W_t2 | 28-12-2012 | N/A | 2 | E | 551 | 152 | t11375 | |||||||||
| 7 | BU_G0706B_t8 | 26-12-2012 | 25-6-2013 | 8 | D | 548 | 152 | t1096 | tetK |
CXT—cefoxitin, OXA—oxacillin, CIP—ciprofloxacillin, TET—tetracycline, CHL–chloramphenicol, TRI–trimethoprim, STM–streptomycin and RIF—rifampicin.
The ‘*’ symbol indicates a mutation in a core genome gene that is involved in antibiotic resistance. All genes are involved in specific resistance phenotypes.
A dendrogram was generated after SeqSphere+ analysis of the 21 sequenced isolates, which revealed 14 cluster types (CTs) (Figs 1B and 2) denoted as 545 (n = 4), 714 (n = 1), 556 (n = 1), 546 (n = 1), 552 (n = 1), 554 (n = 1), 715 (n = 1), 549 (n = 2), 550 (n = 1), 553 (n = 1), 555 (n = 1), 547 (n = 4), 551 (n = 1) and 548 (n = 1). This clustering by SeqSphere+ seemed to match well with the previous clustering of isolates by MLVF. To calculate the concordance between the SeqSphere+ and MLVF typing data, the Ridom epicompare software 1.0 (Ridom GmbH) was used with the Rands Adjusted co-efficient. This revealed a concordance of 0.924.
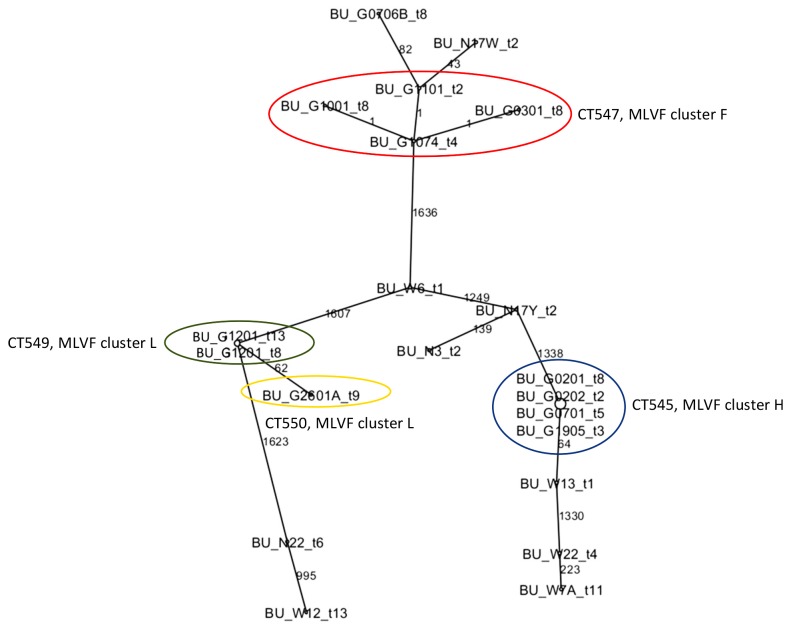
Fig 2. Minimum spanning tree showing the allelic difference between 21 S. aureus isolated from BU patients.
The tree was generated using the SeqSphere+ software. The node size in the tree is proportional to the frequency of genotype occurrence. The allelic difference between S. aureus isolates is indicated as numbers between each node. Isolates with blue and red circles belonging to CTs 545 (MLVF cluster H) and 547 (MLVF cluster F) confirm transmission events. Isolates with green and yellow circles belonging to CTs 549 and 550 were initially grouped in MLVF cluster L and do not confirm transmission events.
The 21 sequenced isolates were assigned to eight MLST types, namely ST1, ST5, ST15, ST88, ST121, ST152, ST508, and the new ST3019. The ST3019 is a single-locus variant of ST45 at the yqiL locus.
Evidence of patient transmission events
A first transmission event was identified for four MRSA isolates belonging to ST88, which were previously grouped in the MLVF cluster H (Fig 1A) [3]. These isolates were classified by SeqSphere+ as CT 545 (Fig 1B). Within CT 545 the allelic profiles were identical (Fig 2). Of note, the four isolates were obtained from three different patients visiting the healthcare center over a period of seven months. The medical care for these patients involved antibiotic treatment and wound dressing changes (Tables 1 and 2). This particular MRSA was first identified in the wound of patient 2, who tested negative at the first sampling time point (t1). Patient 2 was the first to start treatment in this study and was found to carry this particular S. aureus genotype at several sampling time points during treatment (i.e. at t2, t3, t8 and t9). Patients 7 and 19 started treatment 25 and 35 days later, respectively. They both visited the health care center for wound care at time point t2. The wounds of patients 7 and 19 tested positive for S. aureus with the genotype of CT 545 at the sampling time points t5 (patient 7) and t3 (patient 19; Table 1), which is indicative of transmission events.
A second suspected transmission event was initially identified by MLVF typing (cluster F) and involved eight BU patients [3]. To investigate this possible transmission event in more detail four of the 25 isolates obtained from three patients were randomly selected and sequenced. These four MSSA ST152 isolates were assigned to CT 547 (Fig 1B). The allelic profiles within this cluster differed by one (Fig 2). Patients 10 and 11 tested positive for S. aureus with this particular genotype at the same sampling time point (t2). Patient 10 remained positive for S. aureus with the CT 547 until sampling time point t8, and patient 11 until time point t5 (Table 1). A third patient (patient 3) was found to be positive for S. aureus with the CT 547 at sampling time point t8 (Table 1). Patients 5, 6, 7, 18 and 24 became positive for S. aureus with this genotype at later time points than patients 10 and 11. The patients 5, 6, 7, 18 and 24 paid at least one visit at the health center for wound care that overlapped with visits by three other patients, which were found to be positive for the S. aureus genotype with the CT 547 (Table 1).
It is noteworthy to mention that in each of the transmission events, the gene allele variation between isolates was not higher than one. This implies that the isolates were nearly identical with respect to their core genome.
Antibiotic resistance genes
The assembled genomes of the 21 S. aureus isolates were used in blast comparisons to detect the presence of AMR genes, and the results are shown in Table 2. Among the investigated isolates none was found to carry genes involved in resistance to erythromycin, fusidic acid, kanamycin, mupirocin, or vancomycin. Antibiotic resistance of the sequenced isolates was previously most often found against penicillin, chloramphenicol, tetracycline and trimethoprim [3]. Consistent with their penicillin resistance, all sequenced isolates carried various types of blaZ operons, which were located on chromosomally integrated transposons or plasmids. Specifically, the blaZ gene was found in 16 isolates that belonged to ST1, ST5, ST15, ST88, ST152 and ST3019, while the blaZ-B variant was found in five isolates representing ST5, ST508 and ST121. Fourteen sequenced S. aureus isolates were chloramphenicol resistant of which six (ST121, ST3019 and ST152) carried various plasmids with a catA gene. Six other isolates (ST88 and ST1) carried a novel allele of fexB that was not previously reported in S. aureus. In the case of one isolate, the phenotypic resistance for chloramphenicol could not be confirmed at the genomic level, which was potentially due to the loss of the resistance gene. Resistance to rifampicin was identified in one isolate belonging to ST121 where the rpoB gene was found to encode an amino acid substitution that changed Asp471 into Gly. Resistance to tetracycline was identified in 16 isolates, which was confirmed by the identification of resistance genes, such as tetK, tetL and tetM. The tetK gene was located on plasmid pT181, which was found in 10 isolates representing different STs. Five isolates of ST88 contained the tetL and tetM genes located on identical mobile genetic elements integrated into their genomes, while one isolate of ST15 contained a transposon with tetM. The presence of a plasmid or transposon carrying the drfG gene responsible for trimethoprim resistance was detected in five isolates that belonged to ST5, ST15 and ST121. Resistance to streptomycin was limited to three isolates of ST152 where the str gene was present. Of the six methicillin resistant isolates, five belonging to ST88 contained the mecA gene, whereas one ST5 isolate contained neither mecA nor mecC. The latter isolate was termed borderline oxacillin resistant S. aureus (BORSA). Intriguingly, the BORSA isolate contained no mutations in the genes for the penicillin-binding proteins PBP1, PBP2, and PBP3 or the YjbH protein, which were previously proposed to be involved in BORSA phenotypes [24]. However, sequence comparisons revealed that the PBP2 protein of the BORSA isolate contains a Tyr residue at position 197, while the PBP2 protein of S. aureus N315 contains a Cys residue at this position. Furthermore, the BORSA isolate showed resistance to fluoroquinolones, which may be due to a specific mutation in the gyrase A gene (Ser84Leu).
Discussion
In the present study, we have investigated S. aureus transmission events in BU patients during wound care by implementing a WGS-based gene-by-gene typing approach using SeqSphere+. The SeqSphere+ scheme grouped the 21 sequenced S. aureus isolates into eight different STs. Sequenced S. aureus that belonged to ST88 isolates shared identical characteristics (spa-types t186/t786, SCCmec type IVa, PVL-negative) with isolates collected from out-patients in Egypt and Angola, indicating a larger geographic distribution on the African continent [25,26]. The new ST3019 (spa-type t939) identified in this study belongs to the same clonal complex (CC45) as ST45 and ST508. Compared to ST45 a single locus variation was observed at the yqiL locus for ST3019 and in the aroE locus for ST508.
Using SeqSphere+, we identified two major clusters of S. aureus isolates from different BU patients, which may reflect transmission events that occurred during overlapping visits to the Pakro Healthcare Center where these patients received wound care. None of these patients carried S. aureus with the CTs 545 or 547 on their first visit to the Pakro Healthcare Center, strongly suggesting that they acquired the respective S. aureus types upon wound care. Interestingly, the majority of S. aureus isolates from BU patients belong to lineages characterized by spa-types t786 and t355 that have been already reported in health care settings in Ghana [11]. This suggests the nosocomial acquisition of these S. aureus types by patient-to-patient transmission between BU patients and healthcare workers that may have occurred due to inadequate hygiene. Indeed, it has been reported in a recent study that 8 of 11 MRSA transmission events among patients in intensive care settings were potentially due to poor hand hygiene [16]. This could probably be avoided by wearing gloves and protective gowns, and strict implementation of hand hygiene [27–29]. Basic preventive measures, such as adherence to aseptic techniques may further reduce the risk of infection thereby improving wound care of patients, provided that gloves, gowns, adequate dressing materials, running water and hand rub alcohol are made available. With a steady supply and stock of equipment and disposables, routine screening of patients and healthcare workers for S. aureus may be less critical.
Genotypic data of the isolates sequenced confirmed the results of the antimicrobial resistance profiles described previously [3]. Interestingly, the chloramphenicol resistance of some isolates was conveyed by the fexB gene (Table 2), which was thus far not encountered in S. aureus. On the other hand, fexB was previously reported in Enterococcus faecium EFM-1 and Enterococcus hirae EH-1 isolates from pigs [30]. As Enterococci were previously identified in the wounds of BU patients, it is conceivable that the MRSA isolates acquired the fexB gene by horizontal gene transfer from such Enterococci [3]. Furthermore, a BORSA phenotype was identified in an isolate belonging to ST5. Such a BORSA phenotype was previously reported for S. aureus isolates with ST1, ST8 and ST15 that were implicated in wound infections in Scotland [24]. The presence of specific mutations in the genes coding for four proteins, namely PBP1, PBP2, PBP3 and YjbH, was proposed to be involved in the BORSA phenotype. However, in the genome sequence of the presently investigated BORSA isolate from a BU patient, none of these mutations was found. After genomic comparison of the BORSA isolate with the N315 reference genome, the only difference was observed for PBP2, where at position 197 a cysteine residue was replaced by a tyrosine residue. However, this PBP2 amino acid substitution is encoded by the majority of S. aureus genomes available in the NCBI database and, therefore, it may not explain the BORSA phenotype observed. Further comparative genome analyses revealed about 300 additional non-synonymous SNPs, which could contribute to the observed BORSA phenotype.
In summary, WGS of S. aureus isolates from BU patients and the subsequent analysis of sequencing data using the SeqSphere+ scheme revealed likely patient-to-patient transmission events in a healthcare setting in Ghana. This indicates a need for the implementation of improved hygiene protocols in healthcare settings where BU patients receive wound care. Apart from the detection of transmission events, WGS has the advantage that it also provides information on antimicrobial resistance. Related to the antimicrobial resistance pheno- and genotypes identified in S. aureus isolates from BU patients, it is important to bear in mind that antimicrobial pressure has the potential to aggravate resistance, with an inherent risk for transmission of resistant organisms. Therefore, even in low-resource settings, antimicrobial stewardship programs are likely to have added value, with more restrictive antimicrobial use than currently practiced [2].
Supporting Information
(DOCX)
Acknowledgments
The authors express their gratitude to the Buruli ulcer health team from the Pakro Health Center (Ghana) for their help in sample collection and treatment of the patients.
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
NAA was supported by fellowships from the Graduate School for Medical Sciences of the University of Groningen. YS was supported by a VENI grant from the Netherlands Organisation for Scientific Research and the Gratama foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jassens PG; Pattyn SR; Meyers WM; Portaels F (2005) Buruli ulcer: An historical overview with updating to 2005. Bull Séanc Acad R Sci O-m 51: 165–199. [Google Scholar]
- 2. Barogui YT, Klis S, Bankole H, Sopoh GE, Mamo S, et al. (2013) Towards Rational Use of Antibiotics for Suspected Secondary Infections in Buruli Ulcer Patients. PLoS Negl Trop Dis 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amissah NA, Glasner C, Ablordey A, Tetteh CS, Kotey NK, et al. (2015) Genetic Diversity of Staphylococcus aureus in Buruli Ulcer. PLoS Negl Trop Dis 9: e0003421 10.1371/journal.pntd.0003421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Belkum A, Melles D, Nouwen J, van Leewen W, van Wamel W, et al. (2009) Co-evolutionary aspects of human colonisation and infection by Staphylococcus aureus. Infect Genet Evol 9: 32–47. http://www.sciencedirect.com/science/article/pii/S1567134808001779. Accessed 30 April 2014. 10.1016/j.meegid.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 5. Lowy FD (1998) Staphylococcus aureus infections. N Engl J Med 339: 520–532. [DOI] [PubMed] [Google Scholar]
- 6. Kluytmans J, van Belkum A, Verbrugh H (1997) Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10: 505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aiken AM, Mutuku IM, Sabat AJ, Akkerboom V, Mwangi J, et al. (2014) Carriage of Staphylococcus aureus in Thika Level 5 Hospital, Kenya: a cross-sectional study. Antimicrob Resist Infect Control 3: 22 http://www.ncbi.nlm.nih.gov/pubmed/25057351. 10.1186/2047-2994-3-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tigist Alebachew, Gizachew Y, Derabe A, Sisay Z (2012) Staphylococcus aureus burn wound infection among patients attending Yekatit 12 Hospital burn unit, Addis Ababa, Ethiopia. Ethiop J Health Sci 22: 209–213. [PMC free article] [PubMed] [Google Scholar]
- 9. Van der Kooi-Pol MM, Veenstra-Kyuchukova YK, Duipmans JC, Pluister GN, Schouls LM, de Neeling AJ, Grundmann H, Jonkman MF van DJ (2012) High genetic diversity of Staphylococcus aureus strains colonizing patients with epidermolysis bullosa. Exp Dermatol 21: 463–466. 10.1111/j.1600-0625.2012.01502.x [DOI] [PubMed] [Google Scholar]
- 10. Van der Kooi-Pol MM, Sadaghian Sadabad M, Duipmans JC, Sabat AJ, Stobernack T, et al. (2013) Topography of Distinct Staphylococcus aureus Types in Chronic Wounds of Patients with Epidermolysis Bullosa. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egyir B, Guardabassi L, Sørum M, Nielsen SS, Kolekang A, et al. (2014) Molecular Epidemiology and Antimicrobial Susceptibility of Clinical Staphylococcus aureus from Healthcare Institutions in Ghana. PLoS One 9: e89716 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3934920&tool=pmcentrez&rendertype=abstract. 10.1371/journal.pone.0089716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McBryde ES, Bradley LC, Whitby M, McElwain DLS (2004) An investigation of contact transmission of methicillin-resistant Staphylococcus aureus. J Hosp Infect 58: 104–108. [DOI] [PubMed] [Google Scholar]
- 13. Otter JA, Yezli S, French GL (2011) The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect Control Hosp Epidemiol 32: 687–699. 10.1086/660363 [DOI] [PubMed] [Google Scholar]
- 14. Dancer SJ, White LF, Lamb J, Girvan EK, Robertson C (2009) Measuring the effect of enhanced cleaning in a UK hospital: a prospective cross-over study. BMC Med 7: 28 10.1186/1741-7015-7-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simor AE, Daneman N (2009) Staphylococcus aureus Decolonization as a Prevention Strategy. Infect Dis Clin North Am 23: 133–151. 10.1016/j.idc.2008.10.006 [DOI] [PubMed] [Google Scholar]
- 16. Moore G, Cookson B, Gordon NC, Jackson R, Kearns A, Singleton J, Smyth D WA (2015) Whole-genome sequencing in hierarchy with pulsed-field gel electrophoresis: the utility of this approach to establish possible sources of MRSA cross-transmission. J Hosp Infect S0195-6701: 00032–00038. [DOI] [PubMed] [Google Scholar]
- 17. Tong SY, Holden MT, Nickerson EK, Cooper BS, Köser CU, Cori A, Jombart T, Cauchemez S, Fraser C, Wuthiekanun V, Thaipadungpanit J, Hongsuwan M, Day NP, Limmathurotsakul D, Parkhill J PS (2015) Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high transmission setting. Genome Res 25: 111–118. 10.1101/gr.174730.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long SW, Beres SB, Olsen RJ MJ (2014) Absence of patient-to-patient intrahospital transmission of Staphylococcus aureus as determined by whole-genome sequencing. MBio 5: e01692–14. 10.1128/mBio.01692-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sabat AJ, Chlebowicz MA, Grundmann H, Arends JP, Kampinga G, et al. (2012) Microfluidic-Chip-Based Multiple-Locus Variable-Number Tandem-Repeat Fingerprinting with New Primer Sets for Methicillin-Resistant Staphylococcus aureus. J Clin Microbiol 50: 2255–2262. 10.1128/JCM.00056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sabat a. J, Budimir a., Nashev D, Sá-Leão R, van Dijl J, et al. (2013) Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill 18: 20380 [DOI] [PubMed] [Google Scholar]
- 21. Karynski M, Sabat AJ, Empel J, Hryniewicz W (2008) Molecular surveillance of methicillin-resistant Staphylococcus aureus by multiple-locus variable number tandem repeat fingerprinting (formerly multiple-locus variable number tandem repeat analysis) and spa typing in a hierarchic approach. Diagn Microbiol Infect Dis 62: 255–262. 10.1016/j.diagmicrobio.2008.06.019 [DOI] [PubMed] [Google Scholar]
- 22. Leopold SR, Goering R V., Witten A, Harmsen D, Mellmann A (2014) Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52: 2365–2370. 10.1128/JCM.00262-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, et al. (2005) ACT: The Artemis comparison tool. Bioinformatics 21: 3422–3423. [DOI] [PubMed] [Google Scholar]
- 24. Ba X, Harrison EM, Edwards GF, Holden MTG, Larsen AR, et al. (2014) Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J Antimicrob Chemother 69: 594–597. 10.1093/jac/dkt418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abou Shady HM, Bakr AEA, Hashad ME, Alzohairy MA (2015) Staphylococcus aureus nasal carriage among outpatients attending primary health care centers: a comparative study of two cities in Saudi Arabia and Egypt. b raz j inf e c t di s 19: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conceição T, Coelho C, Santos-Silva I, de Lencastre H A-SM (2014) Epidemiology of methicillin-resistant and-susceptible Staphylococcus aureus in Luanda, Angola: first description of the spread of the MRSA ST5-IVa clone in the African continent. Microb Drug Resist 20: 441–449. 10.1089/mdr.2014.0007 [DOI] [PubMed] [Google Scholar]
- 27. Derde LPG, Cooper BS, Goossens H, Malhotra-Kumar S, Willems RJL, et al. (2014) Interventions to reduce colonisation and transmission of antimicrobial-resistant bacteria in intensive care units: An interrupted time series study and cluster randomised trial. Lancet Infect Dis 14: 31–39. 10.1016/S1473-3099(13)70295-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris AD, Pineles L, Belton B, Johnson JK, Shardell M, et al. (2013) Universal glove and gown use and acquisition of antibiotic-resistant bacteria in the ICU: a randomized trial. JAMA 310: 1571–1580. http://www.ncbi.nlm.nih.gov/pubmed/24097234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sopirala MM, Yahle-Dunbar L, Smyer J, Wellington L, Dickman J, et al. (2014) Infection control link nurse program: An interdisciplinary approach in targeting health care-acquired infection. Am J Infect Control 42: 353–359. 10.1016/j.ajic.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu H, Wang Y, Wu C, Schwarz S, Shen Z, et al. (2012) A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J Antimicrob Chemother 67: 322–325. 10.1093/jac/dkr481 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.