Abstract
The infection of Cryptococcus neoformans is acquired through the inhalation of desiccated yeast cells and basidiospores originated from the environment, particularly from bird’s droppings and decaying wood. Three environmental strains of C. neoformans originated from bird droppings (H4, S48B and S68B) and C. neoformans reference clinical strain (H99) were used for intranasal infection in C57BL/6 mice. We showed that the H99 strain demonstrated higher virulence compared to H4, S48B and S68B strains. To examine if gene expression contributed to the different degree of virulence among these strains, a genome-wide microarray study was performed to inspect the transcriptomic profiles of all four strains. Our results revealed that out of 7,419 genes (22,257 probes) examined, 65 genes were significantly up-or down-regulated in H99 versus H4, S48B and S68B strains. The up-regulated genes in H99 strain include Hydroxymethylglutaryl-CoA synthase (MVA1), Mitochondrial matrix factor 1 (MMF1), Bud-site-selection protein 8 (BUD8), High affinity glucose transporter 3 (SNF3) and Rho GTPase-activating protein 2 (RGA2). Pathway annotation using DAVID bioinformatics resource showed that metal ion binding and sugar transmembrane transporter activity pathways were highly expressed in the H99 strain. We suggest that the genes and pathways identified may possibly play crucial roles in the fungal pathogenesis.
Introduction
Over the last decade, opportunistic fungal pathogens such as Cryptococcus neoformans have become increasingly important due to AIDS pandemic and the emergence of drug resistant strains [1]. C. neoformans is an encapsulated yeast-like fungus which causes life-threatening diseases of the pulmonary and central nervous system. More than 900,000 cases of cryptococcal meningitis were reported each year, resulting in approximately 600,000 deaths worldwide [2]. Cryptococcal infection is acquired by the infectious propagules [3] derived from desiccated yeast cells and basidiospores from birds excreta and decaying wood [4, 5]. Primary cryptococcal infection occurs in the lung through inhalation, causing cryptococcal pneumonia, while secondary infection due to dissemination can cause fatal meningitis/encephalitis [6].
Many studies have focused on identifying and examining the virulence factors of C. neoformans [7]. The polysaccharide capsule which increased in size during in vivo infection, or in vitro stimulation with the presence of low iron, mammalian serum, and physiological concentrations of carbon dioxide [7, 8], is a known virulence factor of C. neoformans. Additionally, growth at 37°C [9], α-mating type [10], and the production of melanin [11], phospholipase [11, 12], protein kinases [12], urease [13], cell wall integrity enzymes [14] and superoxide dismutases [15] have been documented to play a role in the invasion and survival of C. neoformans in vivo [16]. In vivo experiments were commonly carried out in mice model through intranasal inoculation of live yeast cells, which can result in lesions in the lung and brain tissues.
In this study, we aimed to identify novel genes which may contribute to the virulence of the C. neoformans. We first investigated the infectivity of three environmental C. neoformans isolates (H4, S48B and S68B), and the H99 reference strain, using C57BL/6 mice. Microaray analysis was then conducted to identify the differentially expressed genes among the strains, which may be responsible for cryptococcal pathogenicity.
Materials and Methods
2.1. Yeasts and media
C. neoformans var. grubii (serotype A) H99, an isolate derived from a meningitis patient, was obtained from American Type Culture Collection (ATCC) and used as the control strain for the experiment. Three environmental strains H4, S48B and S68B were isolated from bird droppings at different locations in Klang valley, a densely populated city in the central region of Malaysia [17]. Similar to the H99 strain, all environmental strains were C. neoformans serotype A, genotype VNI with an α-mating type [18], the predominant type of Cryptococcus isolated worldwide [19]. All four C. neoformans strains demonstrated genetic similarity of >92.9% (Supporting Information S1 Fig).
Yeast strains were maintained at -80°C prior to the study. Cultures were streaked on the Sabouraud’s dextrose agar (SDA) and incubated at 37°C for 48 hours. To prepare cell suspension, 2 to 3 single colonies from freshly prepared plate were inoculated into Sabouraud’s dextrose broth (SDB) and incubated at 37°C for 48 hours.
2.2. Growth curve analysis
Three single colonies from different C. neoformans strains were inoculated separately into SDB. Cells were cultured at 37°C with agitation at 150 rpm for 108 hours. The optical density (OD) at 600 nm was measured at 6 hours intervals using a Spectronic 20 Genesys spectrophotometer (Thermo Scientific, Milford, MA). The average OD readings (mean ± SD) for each strain at different time points were calculated.
2.3. In vivo infection study
C57BL/6 mice were purchased from Jackson Laboratory (Bar Harbor, ME) and maintained in individually ventilated cages under specific pathogen free condition. Fresh culture of cryptococcal strains were washed and harvested by centrifugation at 1800 × g for 10 min. Cells were then adjusted to 107 cells/ml in PBS using a heamocytometer. Mice were first anesthetized with intraperitoneal injection of a mixture ketamine (90 mg/kg) and xylazine (10 mg/kg) before inoculated with intranasal pipetting of 20 μl (2 × 105 cells) yeast suspension. For fungal burden assay, mice (n = 4) were sacrificed at 20-day post-infection. Organs (lung and brain) were excised aseptically and homogenized using two glass slides in 1 ml PBS and then diluted to 10, 100 and 1000 folds. A volume of 20 μl serially diluted homogenates was plated on SDA plates and cultured at 37°C for 48 hours. Colony forming unit (CFU) per ml was determined by calculating yeast colonies on each plate. For survival study, a total of 6–9 mice were infected. All mice were examined for up to 42 days post infection. Mice were euthanized with CO2 inhalation to minimize suffering and distress if they exhibited severe signs including hunched posture, fur ruffling, weakness, increased respiratory rate and difficulty breathing. This study was approved by the Faculty of Medicine Ethics Committee for Animal Experimentation at the University of Malaya.
2.4. Antimicrobial susceptibility testing
Antimicrobial susceptibility to fluconazole (FLU) and amphotericin B (AMB) were tested using E-test strips (Liofilchem, Italy). Freshly cultured cells were adjusted to OD reading 75% transmittancy at 530 nm before spread onto SDA plate using a sterile swap. E-test strips containing AMB at concentration of 0.002 to 32 mg/l or FLU at 0.016 to 256 mg/l were then applied. The plates were incubated at 37°C for 48 hr. Minimal inhibitory concentration (MIC) values were read where the edge of the inhibition ellipse intersected with the strip.
2.5. Capsule induction
Yeast cells were incubated at 37°C, for 24 hours in 2 ml phosphate buffer saline (PBS) with or without 10% heat-inactivated fetal bovine serum (FBS) (Life Technologies, Rockville, MD) in six-well plates. Inactivation of sera was performed by incubation at 56°C for 30 min. To visualize the size of the capsule, a drop of India ink was added to the cell suspension on a glass slide and viewed under light microscope.
2.6. RNA Isolation
Yeast RNA was isolated using RNeasy mini kit (Qiagen, Valencia, CA) according to the manual instruction with minor modifications. Fresh yeast cultures at approximately 109 cells were prepared and washed with cold water and harvested at 1000 ×g, 4°C for 5 minutes. The pellet was resuspended in 350 μl of lysis buffer, and distributed into two 1.5-ml microcentrifuge tubes. Then, 200 μl of 0.5 mm diameter glass beads (MoBio Laboratories, Carlsbad, CA) were added and the cells were disrupted mechanically with one cycle of agitation at 6800 rpm for 40 seconds using a homogenizer (Precellyse 24 lysis, Berlin technology, France). Samples were incubated on ice for 5 minutes and centrifuged in a microcentrifuge (Hermle, Germany) for 2 minutes at 14,000 rpm. The supernatants were transferred to a new microcentrifuge tube, mixed with 1 volume of 70% ethanol before filtering through RNeasy spin column. Column was washed and RNA was eluted using 30 μl RNase free water. The quality and quantity of the RNA were analyzed using a bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). All samples showed RNA integrity number (RIN)>7. For each sample, two biological replicates (rep1 and rep2) were prepared from two independent yeast cultures for microarray analysis.
2.7. Microarray
Microarray analysis was performed using customized chips (Design ID:G4102A-066930). Each microarray chip was prepared in an 8-array slide with 22,313 features per array, inclusive of the control probes. The array features were customized based on the transcript sequences available at the online BROAD Institute database (www.broadinstitute.org/annotation/genome/Cryptococcus_neoformans) [20]. A total of 7813 transcript sequences in fasta format were uploaded into the eArray webtool (Agilent Technologies) to design specific probes against each sequence. Three probes for each transcript were successfully designed for a total number of 7419 transcript sequences.
For microarray assay, all samples were run in duplicate and were prepared as previously described [21]. Briefly, RNA isolated from yeast cells were first labeled using a Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies). Total RNA (100 ng) was converted to double-stranded cDNA by priming with an oligo-dT primer and in vitro transcribed using T7 RNA polymerase to produce cyanine 3-CTP labeled cRNA. The cRNA (600 ng) was then hybridized on the slide at 10 rpm at 65°C in a hybridization oven for 17 hours. The slide was washed and scanned on a High Resolution Microarray Scanner C-model (Agilent Technologies). Raw signal data was extracted using Feature Extraction Software (V107.1.1) and data analysis was performed using a GeneSpring GX version 12.6.1 (Agilent Technologies). During data analysis, the expression filter were performed whereby probes with low raw signal intensities (<20) were excluded to eliminate noise. After filtration, data from 21,069 probes were successfully retrieved.
Hierachrical clustering was done with Euclidean distance metric and Ward’s Linkage rule clustering on both Genelist and Conditions. Predicted gene set was compared to gene ontology database by using Blast2GO software [22]. Non-matched genes were annotated using Basic Local Alignment Search Tool (BLAST) to select genes with the highest percentage of similarities. Pathway annotations of significant genes were analyzed using DAVID software [23].
2.8. Quantitative RT-PCR
RNA (5 μg) was reverse transcribed as previously described [24]. Quantitative real time PCR (qRT-PCR) was performed using SsoAdvanced SYBR Green Supermix (Biorad, Hercules, CA) in a Real-Time PCR 7500 (Applied Biosystems, Foster City, CA). Table 1 shows the primers used in this study. The fold change of each gene was calculated using formula (2-ΔΔC T). All samples were run in triplicates and the results were presented as mean ± SD.
Table 1. Primer sequences for qRT-PCR analysis.
List of the forward and reverse primer sequences (5’-3’) of the up- and down-regulated genes selected for qRT-PCR analysis.
| Gene Symbol | Forward primer | Reverse primer |
|---|---|---|
| MMF1 | CCCTACCCAACAGTCCTCAA | CCAGCCGATTTGGAGTGTAT |
| MPH2/3 | CAACAGGGTAAGTGCGGAAT | GCAGAGGACTCCAAGTCCAG |
| MVA1 | TGTATGGTGAGCATGGCTGT | TGAGGAGTAGCCGAGCAAAT |
| PUT3 | CGCGAGTGTTTCGCATACTA | GCGCGAGTTGATACACTTCA |
2.9. Statistical analysis
Microarray data was analyzed using one-way analysis of variance (ANOVA) followed by Bonferroni’s post hoc test. Unpaired student’s t-test was used for comparison between two groups of data. Data were considered statistical significance if P < 0.05.
Results
3.1. Environmental strains of C. neoformans are less virulent compared to H99
To compare the degree of virulence between C. neoformans H99 strain and three locally isolated environmental strains, we administrated wildtype C57BL/6 mice intranasally with yeast cells, as the respiratory tract is the portal of entry for C. neoformans. A total number of 2 × 105 yeasts cells were inoculated because previous report showed that cryotococcal dissemination to the brain occurred in 100% of animals when infected at approximately 104 or 105 organisms, but not at lower numbers of organisms [25]. Mice infected with C. neoformans H99 strain were found dead starting from day 18, while the remaining of the mice infected with environmental strains survived throughout the end of observation period of 42 days (Fig 1A). This was in consistent with a previous study suggesting that most of the environmental isolates of C. neoformans serotype A were non-lethal to the mice [26].
Fig 1. In vivo virulence study among different C. neoformans strains.
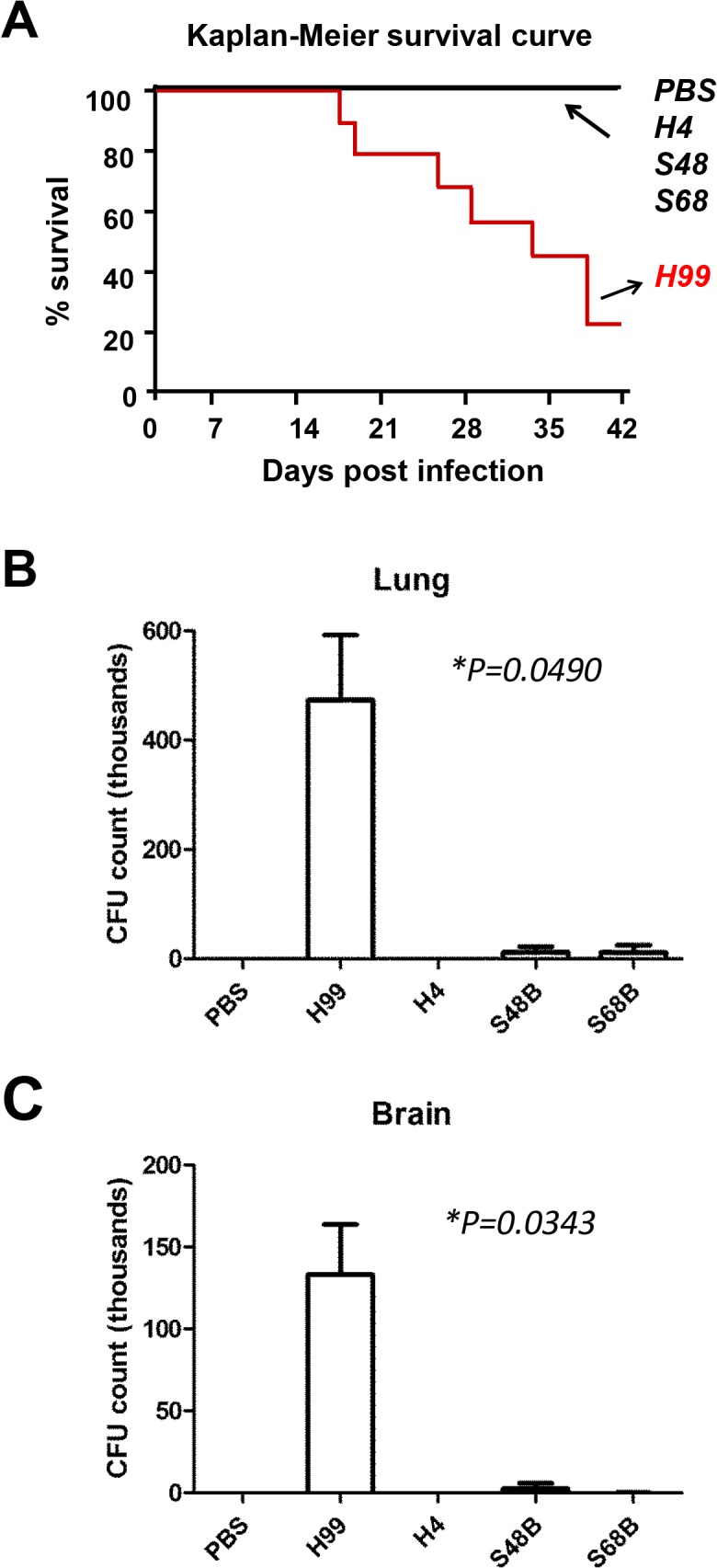
(A) Kaplan-Meier survival curve. C57BL/6 mice at age 8–12 weeks old were administrated intranasally with 2 × 105 yeast cell suspension. Mice were infected with C. neoformans clinical strain H99 or environmental strains H4, S48B and S68B, and observed for a period of 42 days. (n = 6–9). (B and C) CFU assay. Lung (B) or brain (C) homogenates from C. neoformans-infected mice (n = 4) were serially diluted and plated onto agar plates in duplicates. Numbers of colonies (CFU/ml) were counted after 48 hours. CFU formations on the plates were measured and shown as mean ± SD. Group statistical significance measured was by two-way ANOVA analysis (*P<0.05).
At 20-day post-infection, the lung (primary infection site) and brain (secondary infection site) of the infected mice were excised, homogenized and plated for CFU counts. The fungal load in both lung and brain tissues was higher for mice infected with the H99 strain, comparing to those infected with the environmental strains (Fig 1B and 1C). The lung homogenates from the C. neoformans H99-infected mice showed an average of 4.73±1.19 × 105 CFU/ml (P = 0.049), approximately forty folds higher than those obtained from S48B and S68B-infected mice (0.11±0.09 × 105 and 0.11±0.13 × 105 CFU/ml, respectively) (Fig 1B). Consistently, the brain homogenates from the C. neoformans H99-infected mice demonstrated higher CFU counts (1.33±0.31 × 105 CFU/ml, P = 0.034) compared to those obtained from S48B and S68B-infected mice (0.02±0.03 × 105 CFU and 0.01±0.006 × 105 CFU/ml, respectively) (Fig 1C). No growth was obtained from the homogenates of organs excised from those mice infected with H4 environmental strain, suggesting that H4 strain was probably a non-virulent strain. Additionally, inflammation and edema were only observed from the lung tissues of mice infected with H99 strain. No specific sign of illness or pathological abnormality was observed for mice infected with the environmental strains. Hence, based on the findings obtained from the mice virulence study, we proposed that the degree of virulence for the four strains tested were H99>S48B/S68B>H4.
3.2. Pathogenicity of C. neoformans H99 strain was not due to extensive cell growth
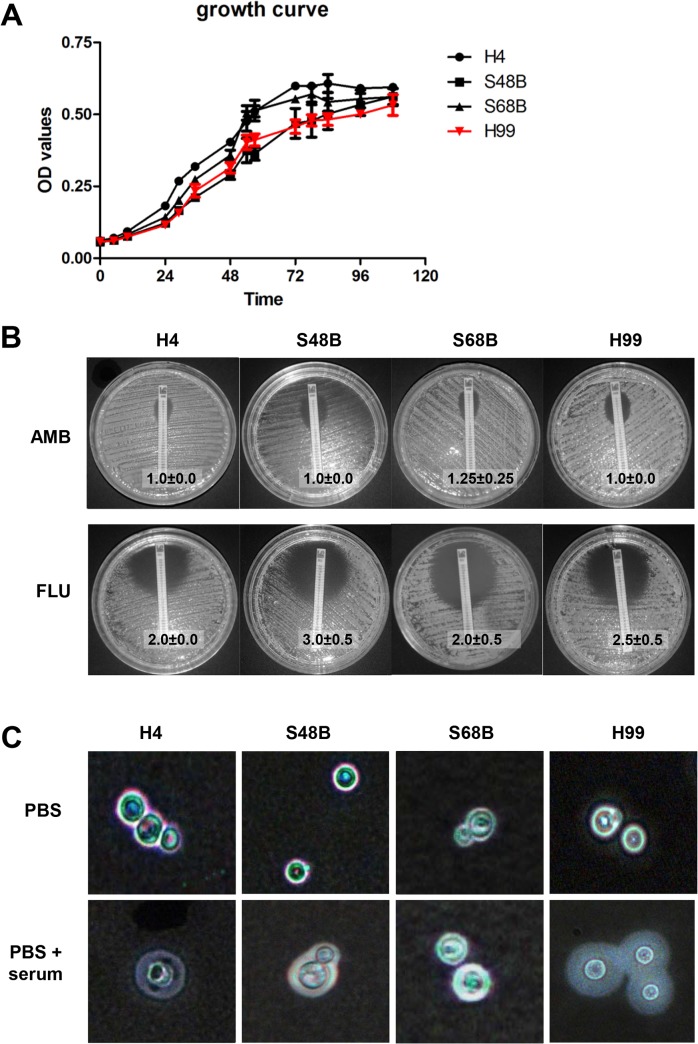
An experiment was performed to assess the growth rate of each cryptococcal strain as the virulence of an organism may be affected by its growth rate. The real-time cell growths of the H99 compared to H4, S68B and S48 strains were determined (Fig 2A). No significant changes were observed in the growth rates of the cryptococcal strains throughout the observation period of 108 hours. However, the least virulent H4 strain demonstrated a relatively higher growth rate compared to the rest of the strains tested, followed by S48B. Meanwhile, S68B and H99 strains showed almost identical growth curve throughout the observation period. The result suggests that there is no direct link between in vitro growth activity in growth media and the in vivo pathogenicity results performed in C57BL/6 mice.
Fig 2. Different traits of C. neoformans strains.
(A) Growth curve. C. neoformans strains (H99, H4, S48B and S68B) cells were cultured in triplicates at 37°C with gentle shaking. OD reading for each sample was measured throughout a time course of 108 hours. (B) E-test. Fungal strains were inoculated evenly onto SDA plate. An AMB or FLU-containing MIC strip was then placed on each plate and incubated at 37°C for 48 hours. Eclipse sizes which intersected with the MIC strips were recorded. Shown were mean±SD from 2 duplicate plates. Data were representative of two independent experiments. (C) Capsule formation. Fungal cells were incubated in PBS with or without the presence of 10% serum, and incubated for 24 hours at 37°C. Cells were then applied on glass slide for India ink staining.
In addition to growth curve, we examined other traits of the cryptococcal strains i.e. anti-fungal resistance and capsule formation. For comparison of anti-fungal resistance among strains, an E-test was performed (Fig 2B). We observed that all 4 strains showed comparative MIC values, at 1.0 to 1.25 for AMB, and 2.0 to 3.0 for FLU, indicating strong susceptibility and no significant difference of anti-fungal resistance among H99 and environmental strains.
Capsule size is known to increase during in vivo infection and is associated with virulence. To examine capsule formation ability among all strains, we incubated cells in the presence of FBS at 37°C (Fig 2C). Interestingly, we observed all environmental strains were able to form capsule. Among all strains, H99 showed most prominently enlarged capsule size which may confer to strong virulence in animal study.
3.3. Differential expression pattern of H99 strain versus the environmental strains
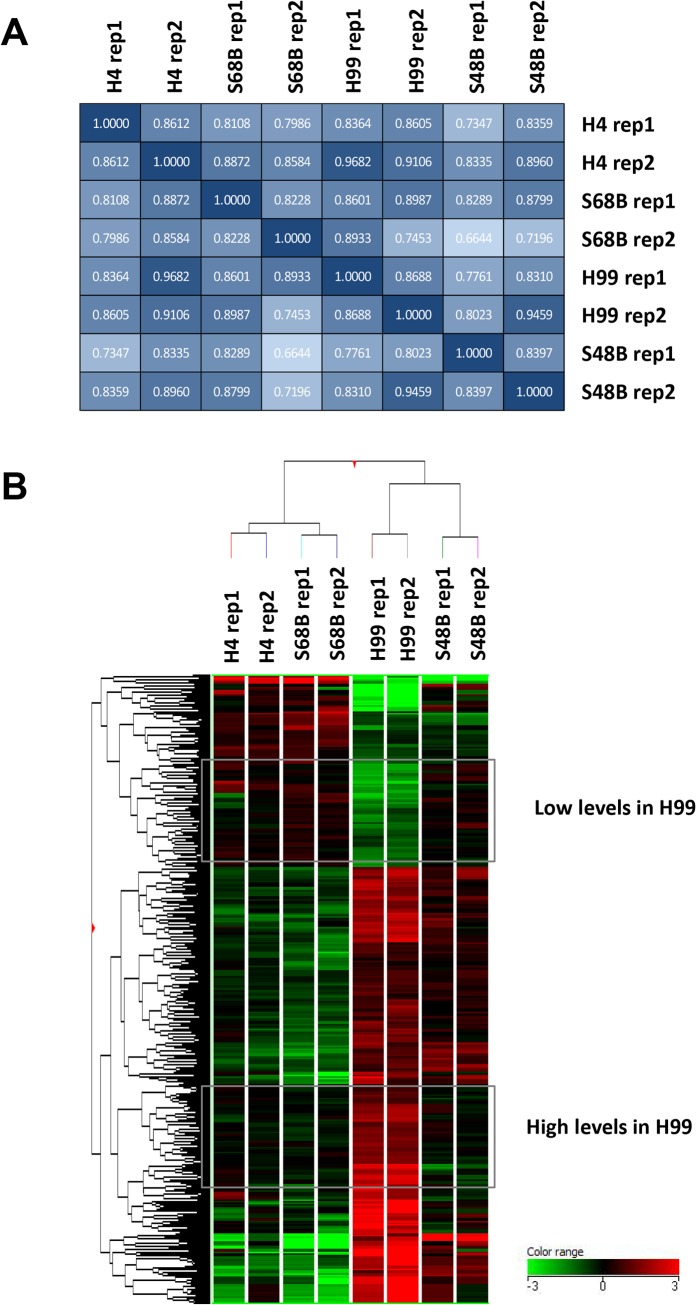
To examine total gene expression in H99 and environmental C. neoformans strains, we isolated RNA from the fungal cells cultured at 37°C, 150 rpm (mimics the condition during in vivo infection) and processed for microarray analysis. Because the C. neoformans microarray chip was not commercially available, microarray slide on an 8-array chip was costumed-made. The total numbers of features analyzed on each array were 22,257 probes (7,419 genes) in triplicates. Pairwise correlation coefficient matrixes among all samples tested were as shown in Fig 3A. Duplicates of each strain (rep1 and rep2) showed correlation values which ranged from 0.8 to 1.0. The correlation among different strains were at an average of 0.8584 (0.6644 to 0.9682), suggesting that all strains shared a relatively close expression patterns. To identify differentially expressed genes, fold change (FC) of each probe was calculated by normalizing the expression level in H99 strain to the expression in environmental strains. After microarray result was filtered with FC<-2 or FC>2 (P<0.05 by Post-Hoc testing), a total of 871 probes (435 genes) were found to be significantly up- or down-regulated. Note that the number of probes was not exactly three times of the number of genes because some probes were excluded during stringent filtration process. The heatmap generated from these 871 significant probes revealed a distinct expression pattern of the H99 strain in comparison to other environmental strains (Fig 3B). Hierachrical clustering based on Euclidean distance metric and Ward’s Linkage showed closer distance of H4 to S68B, and H99 to S48B strains, consistent with molecular typing of the strains into Gr1 and Gr2 subtypes, respectively [18].
Fig 3. Microarray analysis.
(A) Pairwise correlation matrix. Four C. neoformans strains (H99, H4, S48B and S68B) were compared. Two independent samples were prepared for each fungal strain. Rep1–2: replicates of samples. Numbers in the box represents correlation coefficient values among groups. Dark box: high correlation; light box: low correlation. (B) Gene expression heat map. Dendrogram represents the colour-coded expression levels of the significantly regulated genes. The groups of genes which showed differential expressions in H99 versus other strains were as indicated. Low levels in H99 category indicates the genes with low expression levels in H99 (green) but high expression in the environmental strains (red). In contrast, high levels in H99 category indicates the genes with high expression levels in H99 (red) but show low expression in the environmental strains (green). Hierachrical cluster was performed with Euclidean distance metric and Ward’s Linkage rule clustering. Colour range represents log10 (FC) of the microarray intensities.
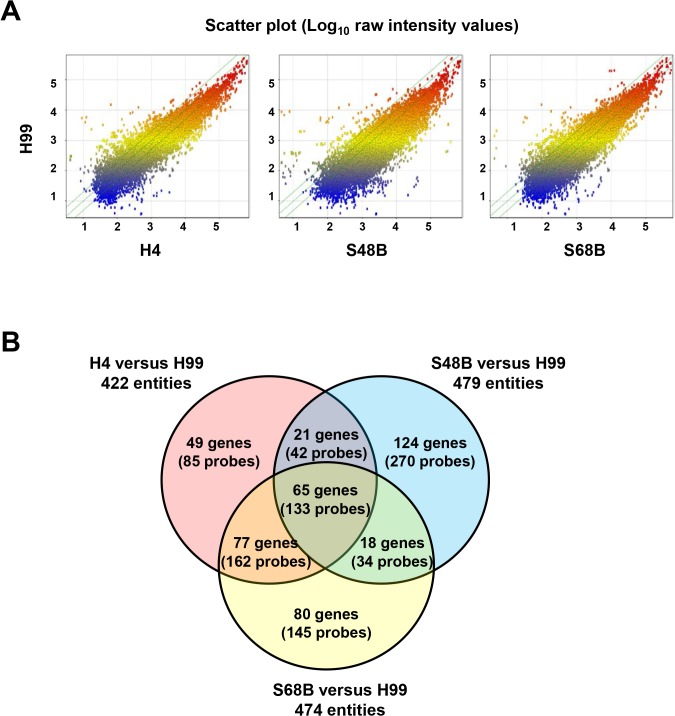
Scatter plots were used to compare the gene expression of H4, S48B and S68B with H99 strain (Fig 4A). Majority of the plots were scattered around the medium line, indicating that no differential expressions between two strains. Those plots exceeding upper and lower border lines were determined as up- or down-regulated probes. A Venn diagram was schemed using significantly expressed genes (FC<-2 or >2, P<0.05) to show the distribution of the differentially regulated probes/genes in each group of comparisons (Fig 4B). Some overlapping genes were detected between the strains. Remarkably, 133 probes (65 genes) were overlapped among all groups of comparisons, which suggest that these 65 genes were differentially expressed in the H99 strain relative to all H4, S48B and S68B environmental strains. These genes may have association with the virulence of Cryptococcus since H99 strain demonstrated stronger virulence in the intranasal infection of C57BL/6 mice. A list of 65 significant genes was as shown in Table 2.
Fig 4. Gene expression of H99 versus environmental strains.
(A) Scatter plots show the expressions of total probes in the H99 versus H4, S48B or S68B. X and Y axis show log10 raw intensity values. (B) Venn diagram. Significantly regulated probes (FC>2 or FC<-2, P<0.05) were selected from the comparisons of H99 versus each environmental strain. Venn diagram shows the distribution and the number of the genes (probes) in each fraction.
Table 2. 65 significant up- and down-regulated genes in C. neoformans H99 relative to non-virulent strains.
#No hit were obtained using C. neoformans mRNA sequence in NCBI blast database and a closest hit from Saccharomyces cerevisiae was selected. Only known genes or genes which matched blast database are described. vs: versus. The P value is determined by an ANOVA analysis, which compares among all sample pairs.
| ProbeName | Blast_Hits | Yeast Gene | Yeast Description | H4 vs H99 | S48B vs H99 | S68B vs H99 | P value |
|---|---|---|---|---|---|---|---|
| Up-regulated genes in H99 | |||||||
| CNAG_03084T0_334 | NP_012213# | MMF1 | Protein MMF1, mitochondrial | 336.6 | 133.1 | 453.4 | 0.0008 |
| CNAG_06956T0_399 | NP_192919.1 | MVA1 | Hydroxymethylglutaryl-CoA synthase | 318.5 | 36.1 | 1151.7 | 0.0017 |
| CNAG_04901T0_584 | NP_013457.1# | BUD8 | Bud-site-selection protein 8 | 40.5 | 37.5 | 62.5 | 0.0267 |
| CNAG_03433T0_1016 | 15.3 | 15.4 | 10.7 | 0.0116 | |||
| CNAG_03772T0_1438 | NP_010087 | SNF3 | High-affinity glucose transporter SNF3 | 15.2 | 8.0 | 5.5 | 0.0113 |
| CNAG_03477T0_50 | NP_010667.1 | RGA2 | Rho GTPase-activating protein 2 | 13.6 | 7.2 | 11.9 | 0.0150 |
| CNAG_05426T0_545 | NP_013064.1# | PRP19 | E3 ubiquitin-protein ligase PRP19 | 13.1 | 5.2 | 10.2 | 0.0072 |
| CNAG_01995T0_522 | NP_013452.1# | DIC1 | Mitochondrial dicarboxylate transporter | 11.6 | 12.1 | 13.4 | 0.0004 |
| CNAG_00826T0_1147 | NP_013641 | DAK1 | Dihydroxyacetone kinase 1 | 11.5 | 2.6 | 10.7 | 0.0006 |
| CNAG_06337T0_1243 | NP_010872.1# | GDA1 | Guanosine-diphosphatase | 7.6 | 17.5 | 6.3 | 0.0020 |
| CNAG_07387T0_1849 | NP_011816 | ARN2 | Siderophore iron transporter ARN2 | 6.7 | 10.2 | 11.4 | 0.0109 |
| CNAG_04507T0_826 | 6.4 | 4.1 | 6.6 | 0.0018 | |||
| CNAG_01494T1_640 | 6.0 | 5.3 | 5.2 | 0.0011 | |||
| CNAG_03400T0_1015 | NP_014490 | GRE2 | NADPH-dependent methylglyoxal reductase GRE2 | 5.9 | 3.7 | 7.9 | 0.0009 |
| CNAG_01498T0_1754 | NP_013090.1# | SOF1 | Protein SOF1http://www.ncbi.nlm.nih.gov/blast/Blast.cgi-alnHdr_6323018 | 5.7 | 7.7 | 8.1 | 0.0053 |
| CNAG_02011T0_1027 | NP_010883.3# | SPF1 | Ion-transporting P-type ATPase SPF1 | 5.4 | 2.6 | 5.0 | 0.0019 |
| CNAG_07428T0_625 | NP_015525.1# | ARR1 | Arsenicals resistance | 4.8 | 6.1 | 5.9 | 0.0165 |
| CNAG_02083T0_1719 | NP_011823 | ARN1 | Siderophore iron transporter ARN1 | 4.7 | 8.1 | 5.5 | 0.0020 |
| CNAG_00838T0_310 | NP_009353.1# | AIM1 | Altered inheritance of mitochondria protein 1 | 4.5 | 3.1 | 4.7 | 0.0141 |
| CNAG_04967T0_1021 | NP_011608.1# | VAS1 | Valine-tRNA ligase | 4.5 | 4.2 | 3.7 | 0.0156 |
| CNAG_06653T1_22 | NP_014127.1# | SEC2 | Rab guanine nucleotide exchange factor | 4.0 | 3.3 | 6.6 | 0.0046 |
| CNAG_04541T0_1287 | NP_011471 | RNA15 | mRNA 3'-end-processing protein RNA15 | 3.9 | 3.6 | 4.0 | 0.0016 |
| CNAG_06411T0_1614 | NP_116685.1# | PTR3 | SPS-sensor componenthttp://www.ncbi.nlm.nih.gov/blast/Blast.cgi-alnHdr_14318552 | 3.8 | 4.9 | 4.9 | 0.0036 |
| CNAG_04618T1_2110 | NP_010111 | UGA3 | Transcriptional activator protein UGA3 | 3.8 | 3.0 | 3.3 | 0.0226 |
| CNAG_05644T0_925 | NP_012683 | YJR149W | Putative 2-nitropropane dioxygenase | 3.1 | 2.7 | 3.9 | 0.0001 |
| CNAG_03393T0_741 | NP_012553 | TES1 | Peroxisomal acyl-coenzyme A thioester hydrolase 1 | 3.0 | 4.6 | 5.3 | 0.0031 |
| CNAG_02439T0_493 | NP_009974.1# | CSG2 | Mannosylinositol phosphorylceramide synthase regulatory subunit | 3.0 | 3.2 | 2.7 | 0.0001 |
| CNAG_02439T1_409 | NP_009974.1# | Rrt12p | Subtilase type proteinase | 3.0 | 3.2 | 2.6 | 0.0002 |
| CNAG_00484T0_1473 | 2.9 | 3.0 | 2.4 | 0.0186 | |||
| CNAG_04346T0_968 | 2.9 | 3.4 | 2.9 | 0.0076 | |||
| CNAG_06540T1_1679 | NP_012550 | ILV3 | Dihydroxy-acid dehydratase, mitochondrial | 2.8 | 2.0 | 3.9 | 0.0038 |
| CNAG_04122T0_1086 | NP_013755 | ARA2 | D-arabinose 1-dehydrogenase | 2.7 | 2.7 | 2.4 | 0.0100 |
| CNAG_02336T0_1615 | NP_013031 | VBA5 | Vacuolar basic amino acid transporter 5 | 2.6 | 2.6 | 4.7 | 0.0096 |
| CNAG_07980T0_45 | NP_010066.1# | GDH2 | Glutamate dehydrogenase (NAD(+)) | 2.5 | 2.4 | 3.2 | 0.0034 |
| CNAG_04034T0_839 | NP_013086.1 | BPT1 | ATP-binding cassette bilirubin transporter BPT1 | 2.4 | 2.2 | 3.1 | 0.0123 |
| CNAG_03819T0_544 | NP_013706 | ERG6 | Sterol 24-C-methyltransferase | 2.4 | 2.7 | 3.2 | 0.0187 |
| CNAG_04363T0_2449 | NP_011853.1# | ETP1 | Ring finger proteinhttp://www.ncbi.nlm.nih.gov/blast/Blast.cgi-alnHdr_6321777 | 2.4 | 2.5 | 3.0 | 0.0199 |
| CNAG_06239T0_1499 | NP_013580.1 | ERG13 | Hydroxymethylglutaryl-CoA synthase | 2.2 | 2.7 | 2.2 | 0.0151 |
| CNAG_06806T0_862 | NP_015329 | AIM45 | Probable electron transfer flavoprotein subunit alpha, mitochondrial | 2.1 | 2.7 | 2.3 | 0.0078 |
| CNAG_05784T1_1057 | NP_012234 | SSM4 | E3 ubiquitin-protein ligase Doa10 | 2.1 | 2.0 | 2.2 | 0.0008 |
| Down-regulated genes in H99 | |||||||
| CNAG_05332T0_622 | NP_012694 | MPH2 | Alpha-glucosides permease MPH2/3 | 57.4 | 189.8 | 69.0 | 0.0001 |
| CNAG_05333T0_1673 | NP_012910 | PUT3 | Proline utilization trans-activator | 51.3 | 69.7 | 32.0 | 0.0179 |
| CNAG_03087T0_1523 | NP_009857 | MAL31 | Maltose permease MAL31; Maltose permease MAL61 | 44.8 | 13.6 | 23.3 | 0.0064 |
| CNAG_01858T0_1311 | NP_010021 | HMRA1 | Mating-type protein A1 | 21.3 | 17.6 | 12.1 | 0.0086 |
| CNAG_07313T0_320 | 16.2 | 151.2 | 16.3 | 0.0003 | |||
| CNAG_04030T0_242 | NP_014029.1 | ADE4 | amidophosphoribosyltransferase | 12.6 | 5.8 | 5.0 | 0.0117 |
| CNAG_06868T0_730 | NP_012044 | ENO2 | Enolase 2 | 11.2 | 9.4 | 15.3 | 0.0063 |
| CNAG_07707T0_1685 | NP_015447.1# | AXL1 | Putative protease | 7.2 | 4.9 | 9.4 | 0.0068 |
| CNAG_05973T0_1159 | NP_013769.1# | STB2 | Sin3 complex subunit | 6.2 | 15.8 | 8.2 | 0.0109 |
| CNAG_05872T0_1058 | NP_015171 | PEP4 | Saccharopepsin | 6.1 | 2.5 | 4.6 | 0.0014 |
| CNAG_04185T0_667 | NP_015414.2# | YPR089W | hypothetical protein YPR089W | 5.4 | 3.5 | 4.8 | 0.0158 |
| CNAG_06016T0_1557 | NP_012789.2# | YKL133C | hypothetical protein YKL133C | 5.2 | 5.9 | 3.8 | 0.0044 |
| CNAG_02992T0_2056 | NP_010414 | SAC6 | Fimbrin | 5.0 | 5.0 | 4.8 | 0.0303 |
| CNAG_05786T1_1058 | NP_015338 | YPR013C | Zinc finger protein YPR013C | 4.6 | 2.4 | 3.6 | 0.0037 |
| CNAG_02351T0_1732 | 3.8 | 3.9 | 4.4 | 0.0145 | |||
| CNAG_04978T0_794 | NP_011884.1# | YHR020W | proline—tRNA ligase | 3.7 | 4.2 | 3.5 | 0.0233 |
| CNAG_06334T0_1554 | NP_009405.1# | SEN34 | tRNA-splicing endonuclease subunit | 3.6 | 2.6 | 3.2 | 0.0037 |
| CNAG_01533T0_2123 | NP_015210 | BEM3 | GTPase-activating protein BEM3 | 3.4 | 5.2 | 3.4 | 0.0004 |
| CNAG_04737T0_2461 | NP_013895.1# | HOT1 | Highosmolarity-induced transcription protein 1 | 3.2 | 5.3 | 5.5 | 0.0046 |
| CNAG_06359T0_714 | NP_013372 | DCS1 | Scavenger mRNA-decapping enzyme | 3.1 | 3.0 | 2.3 | 0.0130 |
| CNAG_03136T0_1325 | 2.8 | 2.3 | 2.8 | 0.0166 | |||
| CNAG_01082T0_3293 | NP_010550 | AKR1 | Palmitoyltransferase AKR1 | 2.6 | 2.4 | 2.9 | 0.0240 |
| CNAG_06223T0_1780 | NP_014799 | NFI1 | E3 SUMO-protein ligase | 2.5 | 2.1 | 2.2 | 0.0013 |
| CNAG_03689T0_1787 | NP_013860 | SIP5 | Protein SIP5 | 2.5 | 2.4 | 2.4 | 0.0268 |
| CNAG_07939T0_65 | 2.1 | 2.6 | 3.1 | 0.0084 |
3.4. Significant up-regulated genes in C. neoformans H99 compared to non-virulent strains
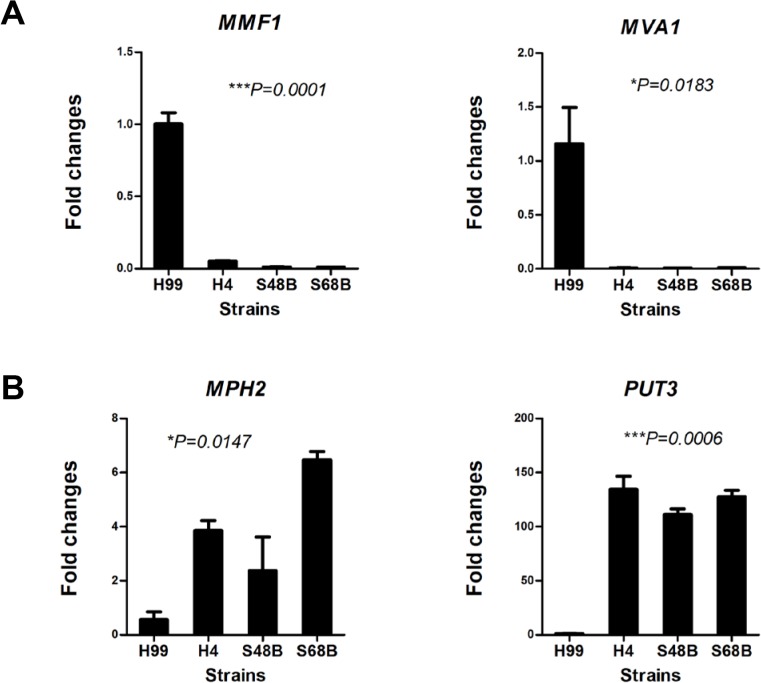
Among top of the genes that were expressed at an intensely high level in C. neoformans H99 relative to environmental strains included MVA1, MMF1, BUD8, SNF3 and RGA2. In contrast, genes that showed significant lower expression in H99 strain included MPH2, PUT3, MAL31, HMRAI and ENO2. The potential functions for significantly up-regulated genes in causing fungal virulence are discussed in the Discussion. To validate the microarray data, expression of selected top differentially regulated genes including MVA1, MMF1, MPH2 and PUT3 were verified using qRT-PCR (Fig 5). Consistent with the microarray data, qRT-PCR showed that expression levels of MVA1 and MMF1 were significantly increased in H99 relative to other environmental strains at >142 and >19 folds, respectively. On the other hand, the MPH2 and PUT3 were reduced in H99 at >4.2 and >25 folds, respectively.
Fig 5. qRT-PCR verification.
RNAs were extracted from H99, H4, S48B and S68B for qRT-PCR analysis. Two independent samples were prepared for each fungal strain. All samples were run in triplicates and data were shown as mean ± SD. Group statistical significance measured was by one-way ANOVA analysis (*P<0.05, **P<0.01, ***P<0.001).
3.5. Pathway analysis
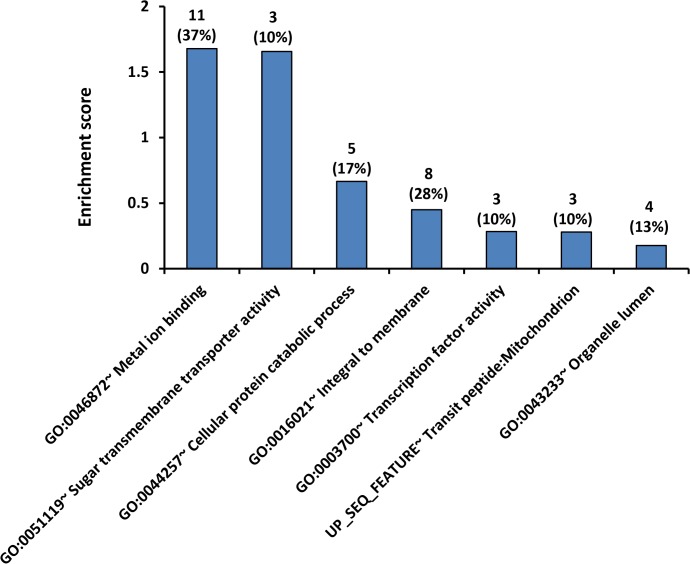
The over-represented pathway categories of the 65 significant genes in H99 strain were analyzed using DAVID bioinformatics resource (Fig 6). Representative annotation of each cluster and gene details were as listed in Table 3. The top in the list of the over-represented pathways was metal ion binding (GO:0045872) with enrichment score (ES) at 1.68 (P = 0.015). A total of eleven genes (out of the 65 significant genes) were found to be listed in the metal ion binding pathway database, suggesting that considerable numbers of the H99-specific genes were associated with metal ion binding process (Table 3). In addition, three genes were found to be associated with sugar transmembrane transporter activity in fungal cells (GO:0051119) (ES = 1.65, P = 0.010). Other relevant pathways included cellular catabolic process (GO:0044257), integral to membrane (GO:0016021), transcription factor activity (GO:0003700), transit peptide:mitochondrion (Up_Seq) and organelle lumen (GO:0043233), however these five pathways were not statistically significant.
Fig 6. Pathway analysis.
Histogram shows a summary of DAVID pathway annotation analysis. The 65 genes which were commonly regulated in H99 versus three environmental strains were selected and analyzed. The number of significant genes and their percentages of genes in the database are as indicated.
Table 3. Pathway annotation by DAVID bioinformatics resource.
The 65 significant genes (133 overlapped probes) which were differentially regulated in H99 compared to environmental strains were analyzed. Shown were representative of each annotation cluster detected. Count represents number of genes which match the pathway database, and % represents the percentage of gene hits among the total genes in the pathway database. Enrichment score (ES) of each group was measured by the geometric mean of the EASE Scores (modified Fisher Exact) associated with the enriched annotation terms that belong to this gene group. Population hit (Pop Hits) represents how many have the function name in your gene list of interest, and population total (Pop Total) represents how many genes in overall population has that function name in the background genome (all genes in the species of interest in DAVID database). False discovery rate (FDR) represents the percentages of test which might be false positive. P values were analyzed using Fisher exact score to identify which sub-populations are over- or under-represented in a sample. Data were considered significant if *P<0.05.
| Category | Term | Genes | ES | Count | % | Pop Hits | Pop Total | FDR | P |
|---|---|---|---|---|---|---|---|---|---|
| GOTERM_MF_FAT | GO:0046872~ Metal ion binding | UGA3, SSM4, PUT3, YPR013C, SAC6, ENO2, AKR1, ARN1, ILV3, ARN2, NFI1 | 1.67 | 11 | 37.9 | 830 | 4190 | 16.7 | 0.0159* |
| GOTERM_MF_FAT | GO:0051119~ Sugar transmembrane transporter activity | MAL31, SNF3, MPH2 | 1.65 | 3 | 10.3 | 26 | 4190 | 10.9 | 0.0101* |
| GOTERM_BP_FAT | GO:0044257~ Cellular protein catabolic process | SIP5, PEP4, SSM4, DCS1, NFI1 | 0.66 | 5 | 17.2 | 351 | 4870 | 82.9 | 0.1255 |
| GOTERM_CC_FAT | GO:0016021~ Integral to membrane | SSM4, MAL31, SNF3, MPH2, AKR1, VBA5, ARN1, ARN2 | 0.44 | 8 | 27.5 | 1488 | 4595 | 99.7 | 0.4214 |
| GOTERM_MF_FAT | GO:0003700~ Transcription factor activity | UGA3, PUT3, HMRA1 | 0.28 | 3 | 10.3 | 135 | 4190 | 91.1 | 0.1917 |
| UP_SEQ_FEATURE | UP_SEQ_FEATURE~ Transit peptide:Mitochondrion | AIM45, MMF1, ILV3 | 0.27 | 3 | 10.3 | 325 | 6448 | 99.7 | 0.415 |
| GOTERM_CC_FAT | GO:0043233~ Organelle lumen | PEP4, AIM45, RNA15, MMF1 | 0.17 | 4 | 13.7 | 783 | 4595 | 99.9 | 0.6522 |
Discussion
The mice virulence study showed that the fungal loads in both lung and brain tissues of H99-infected mice were significantly higher relatively to the mice infected with Malaysian environmental strains (S48B, S68B or H4). The environmental strains of C. neoformans can be found ubiquitously in the soil, certain trees and bird guano. Most of the people are exposed to C. neoformans during childhood through inhalation because of the ubiquitous existence of environmental strains [27]. However, a previous study suggests that most of the environmental strains isolated are non-lethal to the mice and only one out of eleven strains tested is virulent [26]. Our study showed that H99 clinical strain was highly virulent in mice, while S48B and S68B strains were less virulent with detectable numbers of CFU in lung and brain homogenates. The H4 strain was considered non-virulent as no CFU could be observed in both tissue homogenates. Because the four strains used in this study were all C. neoformans serotype A, molecular type VNI, and shared ~93% genetic similarities (Supporting Information S1 Fig) [18], the mechanism underlying their difference degrees of infectivity remains elusive. We began this study by hypothesizing that this difference could plausibly be a resultant of their distinct gene transcriptional programs, which renders invasive or non-invasive characteristics. Microarray data analysis was conducted to substantiate our hypothesis.
Pathway annotation analysis using DAVID bioinformatics resource revealed two significantly pathways i.e. metal ion intake (ES = 1.67, P = 0.0159) and sugar transmembrane transporter activity ES = 1.65, P = 0.0101). This finding suggests that the ability of metal ion uptake and sugar transmembrane transport might distinct between the virulent strain H99 and less/non-virulent environmental strains. Transition metals such as iron, zinc, copper, and manganese are essential elements for the growth and survival of microorganisms including fungi. These microorganisms sequester the metal ions from the host cells through expressing high affinity metal ion transporters to import the nutrient ions [28]. Evidences suggest that metal ion intake is involved in microbial pathogenesis. Iron uptake can regulate the transcription and capsule formation in C. neoformans [29] thus contribute to the fungal cell virulence. In fact, pulmonary infection using C. neoformans mutant strains lacking or defective in copper coordination exhibit reduced pulmonary colonization [30]. It has been reported that copper was required to restore the activity of defective laccase in a DVPH1 avirulent mutant of C. neoformans [31]. Hence, low expression of the environmental strains in metal ion uptake may be a cause for the lower laccase activities of our environmental strains compared to the clinical strains, as noted in a previous study [32]. Besides, mutations in Nickel transporter gene NIC1 and urease proteins attenuate the invasion of C. neoformans into mice central nervous system [33]. Up-regulation of Siderophore iron transporter ARN/SIT-associated genes are discussed below.
Another significant pathway identified was sugar transmembrane transport. C. neoformans is an encapsulated yeast with sugar-coating, in which the sugar transport regulators are essential for the production of capsular polysaccharide antigens [34]. In fact, top in the list of the up-regulated genes in the H99 comprised SNF3, a gene which is linked to the sugar transmembrane transport [35, 36]. Our data showed that SNF3 was up-regulated at 15.2-, 8.0- and 5.5-fold in H99 versus H4, S48B and S68B strains. Mutations in the yeast SNF3 gene affect glucose sensing and SNF3 mutants show defective growth on glucose [37]. In the absence of SNF3, yeast cells show reduced lifespan and caloric restriction effectiveness due to impaired mitochondrial activities [38].
Microarray data analysis showed that the expression profile of H99 differs from the rest of the environmental strains. We managed to identify a list of 65 genes that were commonly up- or down-regulated in H99 compared to H4, S48B or S68B strains. Referring to the variances of the degree of virulence among H99 and environmental strains H4, S48B and S68B, we hypothesized that the differentially regulated genes in H99 likely play roles in determining the fungal virulence. The main problem encountered in the data analysis was that not all C. neoformans genes have been fully characterized and reported. For certain genes, BLAST was carried out to find the closest matched genes from widely studied species i.e. Saccharomyces cerevisiae. Functional prediction for many genes was also reviewed based on the previous studies carried out using S. cerevisiae or Candida albicans. No suitable matches were found for some of the genes reported in Table 2.
The highest up-regulated gene in the H99 strain was Mitrochondrial matrix factor (MMF1), which was up-regulated at 336.6-, 133.1- and 453.4-fold in the C. neoformans H99 strain versus other environmental strains. MMF1 is responsible in the maintenance of intact mitochondria [39, 40]. Mitochondrial function is essential for metabolic pathways such as the glyoxylate cycle and gluconeogenesis as well as in sustaining cell survival throughout oxidative stress [41, 42]. In addition, the mitochondrial function in membrane lipid homeostasis confers drug tolerance ability in fungus [43]. C. albicans mutants which lack GOA1, a mitochondrial protein or subunits of the mitochondrial pyruvate dehydrogenase complex, show a defect in yeast-to-filamentous switching and impairment in systemic invasion [44, 45]. Besides, the hyper virulence C. gattii isolated during the outbreaks in Vancouver Island and North America [16] has been associated with more efficient mitochondrial function [46]. In addition to MMF1, four other mitochondrial associated genes were found among the 65 significant genes in H99. These genes were Mitochondrial dicarboxylate transporter DIC1, Altered inheritance of mitochondria protein 1 AIM1, Mitochondrial Dihydroxy-acid dehydratase ILV3 and Mitochondrial probable electron transfer flavoprotein subunit alpha AIM45, which were induced at >2-fold (P<0.05) in the H99 compared to environmental strain, suggesting the active involvement of mitochondrial activity in driving fungal virulence.
Another highly up-regulated gene was a novel gene with unpredicted function, which matches to S. cerevisiae MVA1 at 53% gene similarity in the NCBI database. MVA1 was upregulated at 318.5-, 26.1-, and 1151.7-fold in the H99 stain versus H4, S48B and S68B environmental strains. MVA1 is a acetyl-CoA C-acetyltransferase (also named as hydroxymethylglutaryl-CoA synthases) which changes the acetyl-CoA into hydroxymethylglutaryl-CoA in the cholesterol synthesis pathway. In fungi, acetyl-CoA is important for the synthesis of cell wall chitin and O-acetylation of the capsule [47]. Acetylation of capsule polysaccharide enables inhibition of neutrophil migration and suppression of host immune response [48]. Importantly, elevated expression of acetyl-CoA production and utilization-associated genes has been detected in C. neoformans recovered from the lung of infected mice during pulmonary infection [47].
Another significant up-regulated gene identified in this study, Bud-site-selection protein 8 (BUD8), was induced at 40.5-, 37.5- and 62.5-fold. BUD8 is a transmembrane glycoprotein which participates in the proteins complex to organize and polymerize actin and actin-associated proteins [49, 50]. The function of BUD8 is to ensure bipolar budding during cell division and polarized growth [51], important for bud initiation during invasion process [52]. Besides, another gene in the top list, RGA2, induced at 13.6-, 7.2- and 11.9-fold in H99 compared to environmental strains, was also associated with yeast budding process. RGA2 is a Rho GTPase-activating protein (GAP) of the central polarity regulator CDC42 that functions to regulate cell morphogenesis and integrity [53]. In C. albicans, hyperphosphorylation of RGA2 occurs at the bud emergence and governs different forms of polarized morphogenesis [54]. The functions of BUD8 and RGA2 in C. neoformans and their roles in fungal virulence require further investigations.
Other selected genes that were considerably up-regulated and have a reported role in virulence were as discussed below. For instance, two genes in Siderophore iron transporters family, ARN2 and ARN1 were up-regulated at 6.7-, 10.2-, and 11.4-fold, and 4.7-, 8.1-, and 5.5-fold, respectively, in H99 when compared to H4, S48B and S68B strains. Non-reductive uptake of ferric–siderophore complexes is dependent on specific siderophore transporters ARN1, ARN2, ARN3 and ARN4 [55, 56]. Some pathogenic fungi, such as S. cerevisiae, have homologues for siderophore transporters that help these fungi to utilize siderophores from competitors [55]. C. albicans ARN1/SIT1 is essential for the fungal epithelial cell invasion [57–59]. Whereas in Aspergillus fumigatus, loss of ability to synthesize siderophores appears to compromise its virulence [29, 60–62]. Recently, the transcript for a putative siderophore transporter ARN1/SIT1 which was increased in C. neoformans cells grown in low-iron medium, has been characterized [63]. Therefore, ARN1 and ARN2 may contribute to the fungal cell virulence by controlling the ability of C. neoformans to acquire siderophores.
Guanosine-diphosphatase (GDA1), a GDPase which functions to hydrolyze GDP to GMP, was induced at 7.6-, 17.5- and 6.3-fold in H99 relative to H4, S48B and S68B. The adhesive and immunomodulation properties of some fungal pathogens depend on cell wall mannoproteins [64]. GDA1 hydrolyzes the GDP-mannose complexes thus releases mannans to the cell wall. In GDA1-mutated C. albicans, defect in GDP hydrolysis, O-mannosylation can also result in impaired yeast-hypha transition [65]. Another up-regulated gene, Ino-transporting P-type ATPase (SPF1) was increased at 5.4-, 2.6- and 5.0-fold in H99 versus environmental strains. SPF1 plays essential role in calcium homeostasis, and its deletion results in calcium influx and increased cellular calcium contents, leading to expression of the calcium-dependent response elements gene CCH1 for the cell survival [66]. Besides, SPF1 null mutant shows defects in hyphal growth rate and biofilm formation, resulting in severely attenuated virulence in C. albicans [67].
NADPH-dependent methylglyoxal reductase (GRE2) was increased 5.9-, 3.7- and 7.9-fold in H99 versus H4, S48B and S68B strains. The cAMP-dependent genes GRE2 in C. neoformans is known to be induced by a variety of environmental stresses, including osmotic and oxidative stresses [68]. In addition, GRE2 involves in metabolism of ergosterol [69], a V-ATPase which functions to repress growth and attenuate fungal virulence in both S. cerevisiae and C. albicans [70]. On the other hand, NAD(+)-dependent Glutamate dehydrogenase (GDH2) was induced at 2.5-, 2.4- and 3.2-fold. GDH2 degrades the reversible oxidative deamination of glutamate to α-ketoglutarate and ammonia using NAD(H) and NADP(H) as cofactors. It also interacts with GDH3 and suppresses stress-induced apoptosis in the cells [71]. Therefore, high expression of GRE2 and GDH2 in H99 may influence cell virulence by regulating biochemistry pathways in C. neoformans.
Rab guanine nucleotide exchange factor (SEC2) was induced at 4.0-, 3.3-, 6.6-fold in H99 compared to H4, S48B and S68B. SEC2 functions in polarized directional delivery of post golgi vesicle for exocytosis activity [72, 73]. Phosphorylated form of SEC2 binds preferentially to SEC15 and a component of exocyst tethering complex which enables fusion of secretory vesicle with plasma membrane [74]. The phosphorylation of SEC2 is necessary to support hyphal in C. albicans because during hyphal extension, the distance of hyphal tip and nucleus is far, thus an effective transport machinery through SEC2 is required [75].
Dihydroxyacetone kinase 1 (DAK1) was found to be up-regulated in H99 for 11.5-, 2.6- and 10.7-fold. DAK1 functions to catalyze both phosphorylation of dihydroxyacetone (DHA) of glyceraldehyde. In S. cerevisiae, detoxification of dihydroxyacetone by DAK1 is suggested to be a vital part of the physiological response during diverse stress conditions [76]. Interestingly, DAK has been found to be an interacting protein for MDA-5, a pattern recognition receptor on host immune cells. DAK interacts with MDA-5 and blocks its antiviral signaling [77]. The overexpression of DAK inhibits MDA5-mediated IFN-β induction and cytoplasmic dsRNA-/virus-induced activation of the IFN-β promoter, whereas these processes can be increased by RNAi knockdown of endogenous DAK. It remains to be investigated if the DAK1 expressed in C. neoformans H99 can inhibit MDA5-mediated signaling to suppress host immune response.
Although comparative gene transcriptome profile can provide us valuable information and prediction of potential virulence genes, further investigations on C. neoformans have shown that gene regulation does not always correlate with their functional activities. For example, C. neoformans Isocitrate lyase (ICll) gene is highly upregulated during central nervous system infection nonetheless the null mutant for ICll does not exhibit an attenuated phenotype in vivo [7]. Therefore, mutagenesis studies of the genes discussed are needed in future study to confirm their functions and roles in fungal pathogenesis.
Supporting Information
Genomic DNA was isolated using MasterPure Yeast DNA Purification Kit (Epicentre, USA) and checked using a nanophotometer (Implen, Germany). PCR fingerprinting was then performed as previously described [18]. Briefly, genomic DNA was amplified using primer (GTG)5: 5’-GTGGTGGTGGTGGTG-3’ primers. PCR products were separated on 2.0% agarose gels electrophoresis and visualized under UV light. Digital images were further analyzed by the PyElph software. Cluster analysis of the cryptococcal isolates was performed with the unweighted-pair-group method using arithmetic averages (UPGMA).
(TIF)
Acknowledgments
The authors would like to thank Mr. Zhi Hui Li (Genomax Technologies, Singapore) for assistance in the design and analysis of microarray chip, and Ms. Nur Wahida Hamdan (University of Malaya) for assistance in PCR fingerprinting.
Fundings
This work was supported by University of Malaya (RG525-13HTM) and Malaysian Ministry of Higher Education (E000013-20001). EM was supported by a postgraduate research fund (PPP) from University of Malaya.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Malaysian Ministry of Higher Education (E000013-20001) and University of Malaya (RG525-13HTM). EM was supported by a postgraduate research fund (PPP) from University of Malaya.
References
- 1. Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans . Clin Microbiol Rev. 1995;8(4):515–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–30. 10.1097/QAD.0b013e328322ffac . [DOI] [PubMed] [Google Scholar]
- 3. Ellis DH, Pfeiffer TJ. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans . Lancet. 1990;336(8720):923–5. . [DOI] [PubMed] [Google Scholar]
- 4. Garcia-Hermoso D, Mathoulin-Pelissier S, Couprie B, Ronin O, Dupont B, Dromer F. DNA typing suggests pigeon droppings as a source of pathogenic Cryptococcus neoformans serotype D. J Clin Microbiol. 1997;35(10):2683–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lazera MS, Pires FD, Camillo-Coura L, Nishikawa MM, Bezerra CC, Trilles L, et al. Natural habitat of Cryptococcus neoformans var. neoformans in decaying wood forming hollows in living trees. J Med Vet Mycol. 1996;34(2):127–31. . [PubMed] [Google Scholar]
- 6. Kronstad J, Jung WH, Hu G. Beyond the big three: systematic analysis of virulence factors in Cryptococcus neoformans . Cell Host Microbe. 2008;4(4):308–10. 10.1016/j.chom.2008.09.003 . [DOI] [PubMed] [Google Scholar]
- 7. Perfect JR. Cryptococcus neoformans: a sugar-coated killer with designer genes. FEMS Immunol Med Microbiol. 2005;45(3):395–404. 10.1016/j.femsim.2005.06.005 . [DOI] [PubMed] [Google Scholar]
- 8. Ngamskulrungroj P, Himmelreich U, Breger JA, Wilson C, Chayakulkeeree M, Krockenberger MB, et al. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii . Infect Immun. 2009;77(10):4584–96. 10.1128/IAI.00565-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kraus PR, Fox DS, Cox GM, Heitman J. The Cryptococcus neoformans MAP kinase MPK1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol Microbiol. 2003;48(5):1377–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kwon-Chung KJ, Edman JC, Wickes BL. Genetic association of mating types and virulence in Cryptococcus neoformans . Infect Immun. 1992;60(2):602–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Williamson PR. Biochemical and molecular characterization of the diphenol oxidase of Cryptococcus neoformans: identification as a laccase. Journal of Bacteriology. 1994;176(3):656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wright L, Payne J, Santangelo R, Simpanya M, Chen S, Widmer F, et al. Cryptococcal phospholipases: a novel lysophospholipase discovered in the pathogenic fungus Cryptococcus gattii . Biochem. J. 2004;384:377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68(2):443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gerik KJ, Donlin MJ, Soto CE, Banks AM, Banks IR, Maligie MA, et al. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans . Mol Microbiol. 2005;58(2):393–408. 10.1111/j.1365-2958.2005.04843.x . [DOI] [PubMed] [Google Scholar]
- 15. Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, Casadevall A, et al. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71(1):173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ngamskulrungroj P, Price J, Sorrell T, Perfect JR, Meyer W. Cryptococcus gattii virulence composite: candidate genes revealed by microarray analysis of high and less virulent Vancouver island outbreak strains. PLoS One. 2011;6(1):e16076 10.1371/journal.pone.0016076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tay ST, Chai HC, Na SL, Hamimah H, Rohani MY, Soo-Hoo TS. The isolation, characterization and antifungal susceptibilities of Cryptococcus neoformans from bird excreta in Klang Valley, Malaysia. Mycopathologia. 2005;159(4):509–13. 10.1007/s11046-005-3091-6 . [DOI] [PubMed] [Google Scholar]
- 18. Tay ST, Lim HC, Tajuddin TH, Rohani MY, Hamimah H, Thong KL. Determination of molecular types and genetic heterogeneity of Cryptococcus neoformans and C. gattii in Malaysia. Med Mycol. 2006;44(7):617–22. 10.1080/13693780600857330 . [DOI] [PubMed] [Google Scholar]
- 19. Enache-Angoulvant A, Chandenier J, Symoens F, Lacube P, Bolognini J, Douchet C, et al. Molecular identification of Cryptococcus neoformans serotypes. J Clin Microbiol. 2007;45(4):1261–5. 10.1128/JCM.01839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broad Institute of Harvard and MIT. Cryptococcus neoformans var. grubii H99 Sequencing Project (http://www.broadinstitute.org). Available: http://www.broadinstitute.org.
- 21. Tan GM, Looi CY, Fernandez KC, Vadivelu J, Loke MF, Wong WF. Suppression of cell division-associated genes by Helicobacter pylori attenuates proliferation of RAW264.7 monocytic macrophage cells. Sci Rep. 2015;5:11046 10.1038/srep11046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–6. 10.1093/bioinformatics/bti610 . [DOI] [PubMed] [Google Scholar]
- 23. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. 10.1038/nprot.2008.211 . [DOI] [PubMed] [Google Scholar]
- 24. Wong WF, Looi CY, Kon S, Movahed E, Funaki T, Chang LY, et al. T-cell receptor signaling induces proximal Runx1 transactivation via a calcineurin-NFAT pathway. Eur J Immunol. 2014;44(3):894–904. 10.1002/eji.201343496 . [DOI] [PubMed] [Google Scholar]
- 25. Huffnagle GB, Yates JL, Lipscomb MF. T cell-mediated immunity in the lung: a Cryptococcus neoformans pulmonary infection model using SCID and athymic nude mice. Infect Immun. 1991;59(4):1423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Litvintseva AP, Mitchell TG. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect Immun. 2009;77(8):3188–95. 10.1128/IAI.00296-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, Niang R, et al. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics. 2001;107(5):E66 . [DOI] [PubMed] [Google Scholar]
- 28. Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Curr Opin Chem Biol. 2010;14(2):218–24. 10.1016/j.cbpa.2009.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lian T, Simmer MI, D'Souza CA, Steen BR, Zuyderduyn SD, Jones SJ, et al. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans . Mol Microbiol. 2005;55(5):1452–72. 10.1111/j.1365-2958.2004.04474.x . [DOI] [PubMed] [Google Scholar]
- 30. Ding C, Festa RA, Chen YL, Espart A, Palacios O, Espin J, et al. Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe. 2013;13(3):265–76. 10.1016/j.chom.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhu X, Gibbons J, Zhang S, Williamson PR. Copper-mediated reversal of defective laccase in a Deltavph1 avirulent mutant of Cryptococcus neoformans . Mol Microbiol. 2003;47(4):1007–14. . [DOI] [PubMed] [Google Scholar]
- 32. Chan MY, Tay ST. Enzymatic characterisation of clinical isolates of Cryptococcus neoformans, Cryptococcus gattii and other environmental Cryptococcus spp. Mycoses. 2010;53(1):26–31. 10.1111/j.1439-0507.2008.01654.x . [DOI] [PubMed] [Google Scholar]
- 33. Morrow CA, Fraser JA. Is the nickel-dependent urease complex of Cryptococcus the pathogen's Achilles' heel? MBio. 2013;4(4). 10.1128/mBio.00408-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jain N, Cordero RJ, Casadevall A, Fries BC. Allergen1 regulates polysaccharide structure in Cryptococcus neoformans . Mol Microbiol. 2013;88(4):713–27. 10.1111/mmi.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bisson LF, Neigeborn L, Carlson M, Fraenkel DG. The SNF3 gene is required for high-affinity glucose transport in Saccharomyces cerevisiae . J Bacteriol. 1987;169(4):1656–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Celenza JL, Marshall-Carlson L, Carlson M. The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. Proc Natl Acad Sci U S A. 1988;85(7):2130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Theodoris G, Bisson LF. DDSE: downstream targets of the SNF3 signal transduction pathway. FEMS Microbiol Lett. 2001;197(1):73–7. . [DOI] [PubMed] [Google Scholar]
- 38. Choi KM, Kwon YY, Lee CK. Disruption of SNF3/RGT2 glucose sensors decreases lifespan and caloric restriction effectiveness through Mth1/Std1 by adjusting mitochondrial efficiency in yeast. FEBS Lett. 2015;589(3):349–57. 10.1016/j.febslet.2014.12.020 . [DOI] [PubMed] [Google Scholar]
- 39. Kim JM, Yoshikawa H, Shirahige K. A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae . Genes Cells. 2001;6(6):507–17. . [DOI] [PubMed] [Google Scholar]
- 40. Oxelmark E, Marchini A, Malanchi I, Magherini F, Jaquet L, Hajibagheri MA, et al. MMF1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol Cell Biol. 2000;20(20):7784–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, et al. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. MBio. 2011;2(3):e00103–11. 10.1128/mBio.00103-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodaki A, Bohovych IM, Enjalbert B, Young T, Odds FC, Gow NA, et al. Glucose promotes stress resistance in the fungal pathogen Candida albicans . Mol Biol Cell. 2009;20(22):4845–55. 10.1091/mbc.E09-01-0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shingu-Vazquez M, Traven A. Mitochondria and fungal pathogenesis: drug tolerance, virulence, and potential for antifungal therapy. Eukaryotic cell. 2011;10(11):1376–83. 10.1128/EC.05184-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim SY, Kim J. Roles of dihydrolipoamide dehydrogenase LPD1 in Candida albicans filamentation. Fungal Genet Biol. 2010;47(9):782–8. 10.1016/j.fgb.2010.06.005 . [DOI] [PubMed] [Google Scholar]
- 45. Vellucci VF, Gygax SE, Hostetter MK. Involvement of Candida albicans Pyruvate dehydrogenase complex protein X (PDX1) in filamentation. Fungal Genet Biol. 2007;44(10):979–90. 10.1016/j.fgb.2006.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Espinel-Ingroff A, Aller AI, Canton E, Castanon-Olivares LR, Chowdhary A, Cordoba S, et al. Cryptococcus neoformans-Cryptococcus gattii species complex: an international study of wild-type susceptibility endpoint distributions and epidemiological cutoff values for fluconazole, itraconazole, posaconazole, and voriconazole. Antimicrob Agents Chemother. 2012;56(11):5898–906. 10.1128/AAC.01115-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69(6):1456–75. 10.1111/j.1365-2958.2008.06374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ellerbroek PM, Lefeber DJ, van Veghel R, Scharringa J, Brouwer E, Gerwig GJ, et al. O-acetylation of cryptococcal capsular glucuronoxylomannan is essential for interference with neutrophil migration. J Immunol. 2004;173(12):7513–20. . [DOI] [PubMed] [Google Scholar]
- 49. Taheri N, Kohler T, Braus GH, Mosch HU. Asymmetrically localized BUD8p and BUD9p proteins control yeast cell polarity and development. EMBO J. 2000;19(24):6686–96. 10.1093/emboj/19.24.6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae . Mol Cell Biol. 1996;16(4):1857–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krappmann AB, Taheri N, Heinrich M, Mosch HU. Distinct domains of yeast cortical tag proteins BUD8p and BUD9p confer polar localization and functionality. Mol Biol Cell. 2007;18(9):3323–39. 10.1091/mbc.E06-10-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harkins HA, Page N, Schenkman LR, De Virgilio C, Shaw S, Bussey H, et al. BUD8p and BUD9p, proteins that may mark the sites for bipolar budding in yeast. Mol Biol Cell. 2001;12(8):2497–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Villar-Tajadura MA, Coll PM, Madrid M, Cansado J, Santos B, Perez P. RGA2 is a Rho2 GAP that regulates morphogenesis and cell integrity in S. pombe. Mol Microbiol. 2008;70(4):867–81. 10.1111/j.1365-2958.2008.06447.x . [DOI] [PubMed] [Google Scholar]
- 54. Zheng XD, Lee RT, Wang YM, Lin QS, Wang Y. Phosphorylation of RGA2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 2007;26(16):3760–9. 10.1038/sj.emboj.7601814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Haas H. Molecular genetics of fungal siderophore biosynthesis and uptake: the role of siderophores in iron uptake and storage. Appl Microbiol Biotechnol. 2003;62(4):316–30. 10.1007/s00253-003-1335-2 . [DOI] [PubMed] [Google Scholar]
- 56. Lesuisse E, Blaiseau PL, Dancis A, Camadro JM. Siderophore uptake and use by the yeast Saccharomyces cerevisiae . Microbiology. 2001;147(Pt 2):289–98. . [DOI] [PubMed] [Google Scholar]
- 57. Ardon O, Bussey H, Philpott C, Ward DM, Davis-Kaplan S, Verroneau S, et al. Identification of a Candida albicans ferrichrome transporter and its characterization by expression in Saccharomyces cerevisiae . J Biol Chem. 2001;276(46):43049–55. 10.1074/jbc.M108701200 . [DOI] [PubMed] [Google Scholar]
- 58. Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF. The siderophore iron transporter of Candida albicans (SIT1p/ARN1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun. 2002;70(9):5246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47(5):1185–97. . [DOI] [PubMed] [Google Scholar]
- 60. Hissen AH, Chow JM, Pinto LJ, Moore MM. Survival of Aspergillus fumigatus in serum involves removal of iron from transferrin: the role of siderophores. Infect Immun. 2004;72(3):1402–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hissen AH, Wan AN, Warwas ML, Pinto LJ, Moore MM. The Aspergillus fumigatus siderophore biosynthetic gene SIDA, encoding L-ornithine N5-oxygenase, is required for virulence. Infect Immun. 2005;73(9):5493–503. 10.1128/IAI.73.9.5493-5503.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schrettl M, Bignell E, Kragl C, Joechl C, Rogers T, Arst HN Jr., et al. Siderophore biosynthesis but not reductive iron assimilation is essential for Aspergillus fumigatus virulence. J Exp Med. 2004;200(9):1213–9. 10.1084/jem.20041242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tangen KL, Jung WH, Sham AP, Lian T, Kronstad JW. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans . Microbiology. 2007;153(Pt 1):29–41. 10.1099/mic.0.2006/000927-0 . [DOI] [PubMed] [Google Scholar]
- 64. Hirschberg CB, Robbins PW, Abeijon C. Transporters of nucleotide sugars, ATP, and nucleotide sulfate in the endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem. 1998;67:49–69. 10.1146/annurev.biochem.67.1.49 . [DOI] [PubMed] [Google Scholar]
- 65. Herrero AB, Uccelletti D, Hirschberg CB, Dominguez A, Abeijon C. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryotic cell. 2002;1(3):420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bonilla M, Cunningham KW. Mitogen-activated protein kinase stimulation of Ca(2+) signaling is required for survival of endoplasmic reticulum stress in yeast. Mol Biol Cell. 2003;14(10):4296–305. 10.1091/mbc.E03-02-0113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu Q, Wang H, Xu N, Cheng X, Wang Y, Zhang B, et al. SPF1 strongly influences calcium homeostasis, hyphal development, biofilm formation and virulence in Candida albicans . Microbiology. 2012;158(Pt 9):2272–82. 10.1099/mic.0.057232-0 . [DOI] [PubMed] [Google Scholar]
- 68. Maeng S, Ko Y-J, Kim G-B, Jung K-W, Floyd A, Heitman J, et al. Comparative transcriptome analysis reveals novel roles of the Ras and cyclic AMP signaling pathways in environmental stress response and antifungal drug sensitivity in Cryptococcus neoformans . Eukaryotic cell. 2010;9(3):360–78. 10.1128/EC.00309-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Warringer J, Blomberg A. Involvement of yeast YOL151W/GRE2 in ergosterol metabolism. Yeast. 2006;23(5):389–98. 10.1002/yea.1363 . [DOI] [PubMed] [Google Scholar]
- 70. Zhang YQ, Gamarra S, Garcia-Effron G, Park S, Perlin DS, Rao R. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 2010;6(6):e1000939 10.1371/journal.ppat.1000939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee YJ, Kim KJ, Kang HY, Kim HR, Maeng PJ. Involvement of GDH3-encoded NADP+-dependent glutamate dehydrogenase in yeast cell resistance to stress-induced apoptosis in stationary phase cells. J Biol Chem. 2012;287(53):44221–33. 10.1074/jbc.M112.375360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Elkind NB, Walch-Solimena C, Novick PJ. The role of the COOH terminus of SEC2p in the transport of post-Golgi vesicles. J Cell Biol. 2000;149(1):95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Walch-Solimena C, Collins RN, Novick PJ. SEC2p mediates nucleotide exchange on SEC4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137(7):1495–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stalder D, Mizuno-Yamasaki E, Ghassemian M, Novick PJ. Phosphorylation of the Rab exchange factor SEC2p directs a switch in regulatory binding partners. Proc Natl Acad Sci U S A. 2013;110(50):19995–20002. 10.1073/pnas.1320029110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Caballero-Lima D, Hautbergue GM, Wilson SA, Sudbery PE. In Candida albicans hyphae, SEC2p is physically associated with SEC2 mRNA on secretory vesicles. Mol Microbiol. 2014;94(4):828–42. 10.1111/mmi.12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Molin M, Norbeck J, Blomberg A. Dihydroxyacetone kinases in Saccharomyces cerevisiae are involved in detoxification of dihydroxyacetone. J Biol Chem. 2003;278(3):1415–23. 10.1074/jbc.M203030200 . [DOI] [PubMed] [Google Scholar]
- 77. Diao F, Li S, Tian Y, Zhang M, Xu LG, Zhang Y, et al. Negative regulation of MDA5- but not RIG-I-mediated innate antiviral signaling by the dihydroxyacetone kinase. Proc Natl Acad Sci U S A. 2007;104(28):11706–11. 10.1073/pnas.0700544104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic DNA was isolated using MasterPure Yeast DNA Purification Kit (Epicentre, USA) and checked using a nanophotometer (Implen, Germany). PCR fingerprinting was then performed as previously described [18]. Briefly, genomic DNA was amplified using primer (GTG)5: 5’-GTGGTGGTGGTGGTG-3’ primers. PCR products were separated on 2.0% agarose gels electrophoresis and visualized under UV light. Digital images were further analyzed by the PyElph software. Cluster analysis of the cryptococcal isolates was performed with the unweighted-pair-group method using arithmetic averages (UPGMA).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.