Abstract
Hereditary nonsyndromic hearing loss is extremely heterogeneous. Mutations in the transmembrane channel-like gene1 (TMC1) are known to cause autosomal dominant and recessive forms of nonsyndromic hearing loss linked to the loci of DFNA36 and DFNB7/11, respectively. We characterized a six-generation Chinese family (5315) with progressive, postlingual autosomal dominant nonsyndromic hearing loss (ADNSHL). By combining targeted capture of 82 known deafness genes, next-generation sequencing and bioinformatic analysis, we identified TMC1 c.1714G>A (p. D572N) as the disease-causing mutation. This mutation co-segregated with hearing loss in other family members and was not detected in 308 normal controls. In order to determine the prevalence of TMC1 c.1714G>A in Chinese ADNSHL families, we used DNA samples from 67 ADNSHL families with sloping audiogram and identified two families carry this mutation. To determine whether it arose from a common ancestor, we analyzed nine STR markers. Our results indicated that TMC1 c.1714G>A (p.D572N) account for about 4.4% (3/68) of ADNSHL in the Chinese population.
Keywords: Autosomal dominant nonsyndromic hearing loss, targeted gene capture, massively parallel sequencing, TMC1
INTRODUCTION
Hearing loss is a common sensory defect in humans. Nonsyndromic hereditary forms, in which hearing loss is the only clinical sign, are known to be genetically heterogeneous [Friedman et al., 2003]. To date, more than 100 genetic loci for nonsyndromic hereditary hearing loss have been mapped, and 67 responsible genes have been identified (Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org/). Mutations in these genes do not occur at the same frequencies across ethnicities. Thus, the genetic heterogeneity of hearing loss will be a major challenge for traditional genetic testing and counseling.
Next Generation Sequencing (NGS) has commonly been used to identify disease genes within even a limited number of patient samples [Kalay et al., 2011; Krawitz et al., 2010; Kuhlenbaumer et al., 2011; Musunuru et al., 2010; Simpson et al., 2011b]. Because of its large capacity to survey the whole exome and genome in an unbiased manner, NGS is well suited to identifying the causative mutations of hereditary hearing loss. Novel genes and mutations for non-syndromic [Azaiez et al., 2014; Behlouli et al., 2014; Girotto et al., 2013; Imtiaz et al., 2014; Jaworek et al., 2013; Oh et al., 2014; Rehman et al., 2010; Santos-Cortez et al., 2013; Santos-Cortez et al., 2014; Schrauwen et al., 2012; Walsh et al., 2010; Xing et al., 2014; Zhao et al., 2013] and syndromic hearing loss [Pierce et al., 2010] have been identified using the NGS approach. Adding a targeted capture step prior to NGS enables the analysis of a focused subset of the whole genome, thereby reducing the required sequencing capacity and data management and analysis. Targeted deaf gene enrichment usually increases the disease-associated genes or mutations by at least 1000 fold [Brownstein et al., 2011; Shearer et al., 2010].
TMC1 encodes membrane protein with six transmembrane domains that may be involved in the functional maturation of cochlear hair cells [Labay et al., 2010; Marcotti et al., 2006]. TMC1 mutations cause nonsyndromic autosomal dominant and recessive hearing loss. More than 30 recessive mutations in TMC1 are reported worldwide [Brownstein et al., 2011; Davoudi-Dehaghani et al., 2014; Davoudi-Dehaghani et al., 2013; Gao et al., 2013; Hilgert et al., 2008; Kalay et al., 2005; Kitajiri et al., 2007b; Kurima et al., 2002; Meyer et al., 2005; Santos et al., 2005; Tlili et al., 2008]. However, only four dominant mutations have been reported, in which two dominant mutations (c.1714G>A and c.1714G>C) affect the same nucleotide and are predicted to encode the amino acid missense substitutions p.D572N and p.D572H. These mutations were found in three unrelated North American families with progressive autosomal dominant nonsyndromic hearing loss (ADNSHL) at the DFNA36 locus, suggesting that they play a critical and specific role in the normal function and pathology of TMC1. Hilgert et al. proposed that c.G1714 could be a “hotspot” for mutational events supported by the observation of several differences in linked haplotypes of STR markers of two family segregating p.D572N [Hilgert et al., 2009]. Another two dominant mutations, p.G417R and p.M418K, were identified in large DFNA36 families from Iran and China, separately [Yang et al., 2010; Zhao et al., 2014]. p.M418K in TMC1 is orthologous to murine p.M412, which is replaced by lysine in the Bth mouse mutant [Zhao et al., 2014]. The hearing loss in these dominant-inherited families is postlingual with an onset in the first or second decade of life and progression to profound deafness.
Here we identified three Chinese families with ADNSHL caused by the c.1714G>A (p.D572N) mutation of TMC1. We have assessed the prevalence of this mutation in a large cohort of controls. To determine whether this recurrent mutation have a common founder or be the result of a mutational hot spot, we investigated the origin of the TMC1 p.D572N by haplotype analysis of the three families.
MATERIALS AND METHODS
Clinical data
Family 5315 is a six-generation Chinese family with autosomal dominant, late-onset, progressive, nonsyndromic sensorineural hearing loss (ADNSHL) from Harbin in north-east China. To screen for candidate mutations, we used 308 ethnicity-matched controls and 67 affected DNA samples obtained from the Department of Otolaryngology. Sixty-seven affected individuals were from families presenting with ADNSHL and in whom mutations of GJB2 and SLC26A4 had been excluded previously. Fully informed written consent was attained from each subject or their guardians. The study was approved by the Chinese PLA General Hospital ethics of research committees. Clinical information was gathered through multiple interviews with all participating members of the family. Medical history was obtained using a questionnaire regarding the following aspects of this condition: subjective degree of hearing loss, age at onset, evolution, symmetry of the hearing loss, use of hearing aids, presence of tinnitus, medication, noise exposure, pathological changes in the ear and other relevant clinical manifestations. Otoscopy, physical examination and pure tone audiometric examination (at frequencies from 250 to 8000 Hz) were performed to identify the phenotype. Immittance testing was applied to evaluate middle-ear pressure, ear canal volume and tympanic membrane mobility. Unaffected phenotype status was defined by a threshold lower than age- and gender-matched 50th centile values for all frequencies measured. Physical examination of all members revealed no signs of systemic illness or dysmorphic features. CT scans of the temporal bone in the index patients were performed. The diagnosis of profound sensorineural hearing loss was made according to the WHO criteria based on audiometric examination. Tandem gait and Rhomberg tests were performed to evaluate balance.
Deafness gene capture and Illumina library preparation
DNA quality was examined based on the optical density ratio (260/280 ratio) and by gel electrophoresis imaging. High-molecular-weight gDNA (~3 μg) was fragmented ultrasonically using a Covaris E210 DNA-shearing instrument (Covaris, Inc., Woburn, MA) to an average size of 300 base pairs (bps). The Covaris protocol was set at 3 min total duration, duty cycle 10%, intensity 5, and 200 cycles per burst.
Exons and their flanking 50 bps of 82 known human deafness genes (Supplementary Table I) were selected for capture and NGS sequencing by Illumina HiSeq2000. Hybridization probes at sizes of 0.5 to 1.6 kilobase pairs (kbps) targeting these genes were generated from either cDNA clones of the genes or by polymerase chain reaction (PCR) amplifications from targeted gDNA regions. To ensure reliable capture of shorter exons, we specifically generated longer hybridization probes from gDNA for exons shorter than 50 bps by including about 100 bps genomic DNA flanking the exons on both sides. All PCR products (10 ng each) were cleaned using the QIAquick PCR Purification Kit (Qiagen, Valencia, CA) before use. More details of capture probe validation and preparation can be found in our previous study [Tang et al., 2011].
Fragmented gDNA libraries for Illumina GAII sequencing were prepared with the NEBNext™ DNA Sample Prep Master Mix set (E6040, NEB Biolab, Ipswich, MA). End repair of DNA fragments, addition of a 3′ adenine (A), adaptor ligation and reaction cleanup were performed following the manufacturer’s protocol. The libraries were cleaned and size-selected using the AMPure DNA Purification kit (Beckman Agencourt, Danvers, MA). The ligated product (20 ng) was amplified for 14 PCR cycles with Illumina PCR primers InPE1.0, InPE2.0 and indexing primers following the manufacturer’s instructions.
For targeted enrichment of deafness genes, the Illumina library DNA was purified with QIAquick MinElute column and eluted into 50 μL of hybridization buffer (HB, Roche NimbleGen, Madison, WI). The barcoded Illumina gDNA libraries (5 μg) were incubated in 16 μL of HB on the surface of hybridization glass slides on a hybridization station (BioMicro Systems, Inc., Salt Lake City, UT) at 42°C for 72 hrs. Nonspecific DNA fragments were removed after a series of six washing steps in a washing buffer (Roche NimbleGen, Madison, WI). The DNA bound to the probes was eluted by incubating with NaOH (425 mL, 125 mM) for 10 min. The eluted solution was transferred to a 1.5-mL Eppendorf tube containing 500 μL neutralization buffer (Qiagen PBI buffer). The neutralized DNA was desalted and concentrated on a QIAquickMinElute column and eluted into 30 μL in EB buffer. To increase the yield, we typically amplified 5 μL of eluted solution by 12 PCR cycles using the Illumina PCR primers InpE1.0 and 2.0. Enrichment of targeted deafness genes was examined using quantitative PCR (qPCR) by comparing the growth curves of captured and noncaptured samples [Tang et al., 2011]. Twelve barcoded libraries of captured samples were pooled and paired-end Illumina sequencing was performed using the Illumina HiSeq system (Illumina, San Diego, CA). Details of bioinformatics analysis methods have been published previously [Tang et al., 2011].
Mutational analysis of TMC1
A total of 308 negative samples and 67 ADNSHL cohorts with sloping audiogram underwent mutation screening by direct sequencing or digestion reactions (restriction-fragment-length polymorphism). Also, 67 ADNSHL cohorts were previously excluded due to GJB2 and SLC26A4 mutations. Genomic DNA was extracted from the blood using the blood DNA extraction kit (TianGen, Beijing, China). Genotyping for c.1714G>A was performed by PCR with modified primers (Forward: tcctctagccttcatacaccgaagtc and Reverse: aacctgggaggcttttctgt) to produce a 160 bp product. The forward primer was designed to introduce a Sal1 site for c.1714G only. Each PCR product (4 μl) was analyzed by electrophoresis in a 1.5% agarose gel, and 5 μL was digested with the Sal1 restriction enzyme in a 20-μL reaction at 37°C for 4 h. Digestion products were analyzed by electrophoresis on a 2.5% agarose gel. Since a Sal1 recognition site was removed by the mutation, only the wild type sequence was digested into two fragments of 130 and 30 bp. Digestion cleaves the normal allele into two fragments (130 and 30 bp), whereas the mutant allele remains uncleaved (160 bp). Therefore, the PCR product containing the heterozygous D572N mutation was cleaved into three fragments of 160, 130 and 30 bp, whereas the wild-type DNA was cleaved into two fragments of 130 and 30 bp. Digestion reactions were performed according to the manufacturer’s protocols. A total of 100 negative samples were confirmed by digestion reactions.
Another 208 negative samples were detected by bidirectional sequences of the amplified fragments and were determined using an automated DNA sequencer (ABI 3100, Applied Biosystems, USA). Nucleotide alteration(s) was identified by sequence alignment with the TMC1 GenBank sequence using the Genetool software.
Haplotype analysis
Four DNA samples including three index patients of three families and one negative control of Family 5315 were haplotyped using nine STR markers within and flanking TMC1 followed the previous literature [Hilgert et al., 2009].
RESULTS
Family and clinical evaluations
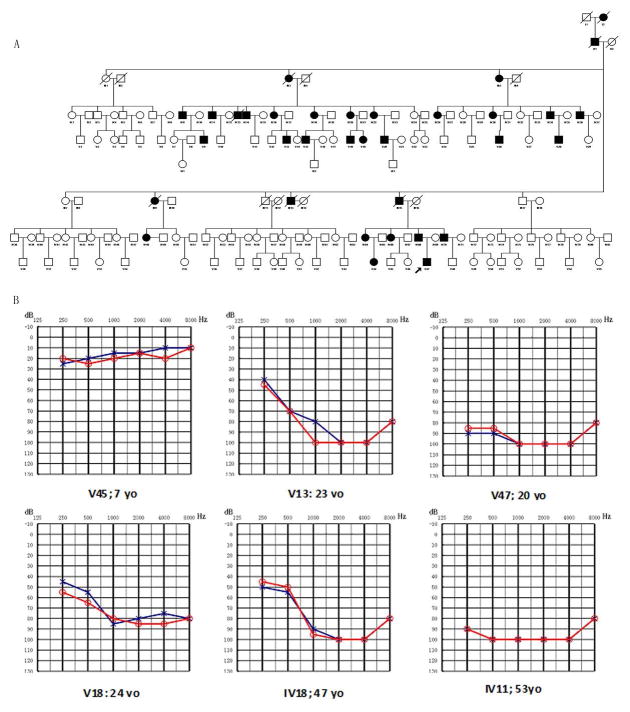
Figure 1A shows the pedigree of Family 5315 with postlingual ADNSHL. There were 34 affected individuals in the family over six generations with apparent autosomal dominant inheritance. In this study, 48 family members participated, 20 of whom were classified as affected. In affected subjects, hearing loss began in the second decade of life and varied from mild to profound on their audiograms. Affected family members had bilateral hearing loss present with a sloping audiogram at the age of onset at high frequencies. Although low-frequency hearing is initially normal, hearing deteriorates at all frequencies by the fourth or fifth decades. Audiograms of some affected members of this family are shown in Figure 1B.
Figure 1.
A. Pedigree of Chinese Family 5315 with Dominant Progressive Hearing Loss and Combined Audiogram. Affected subjects are denoted in black. The proband is indicated by an arrow. B. Audiograms of the affected subjects. The severity of hearing loss and prevalence involvement appears to be correlated with the individual’s age. Hearing loss ranged from mild to profound. Note that affected individual v45 has normal hearing sensitivity.
Hearing loss of Patient 5879-III:6 (Figure 3A upper panel, male/29 years old) was initially detected at age 14 years with mild hearing loss and the average threshold was 35dB HL. The hearing loss showed rapid progression with annual threshold deteriorations about 5dB HL in average at all frequencies. Recent audiogram at 29 years old shows severe to profound hearing loss across most frequencies (Figure 3B, upper panel). There is no obvious tinnitus.
Figure 3.
Pedigree (A) and Audiograms (B) of probands of Family 5879, Family 8989.
Hearing loss of Patient 8989-III:4 (Figure 3A lower panel, male/26 years old) started at 16 years, presenting with hearing loss of the high-frequencies which was followed by an increase of the low- and mid-frequency threshold values with advancing age (Figure 3B, lower panel).
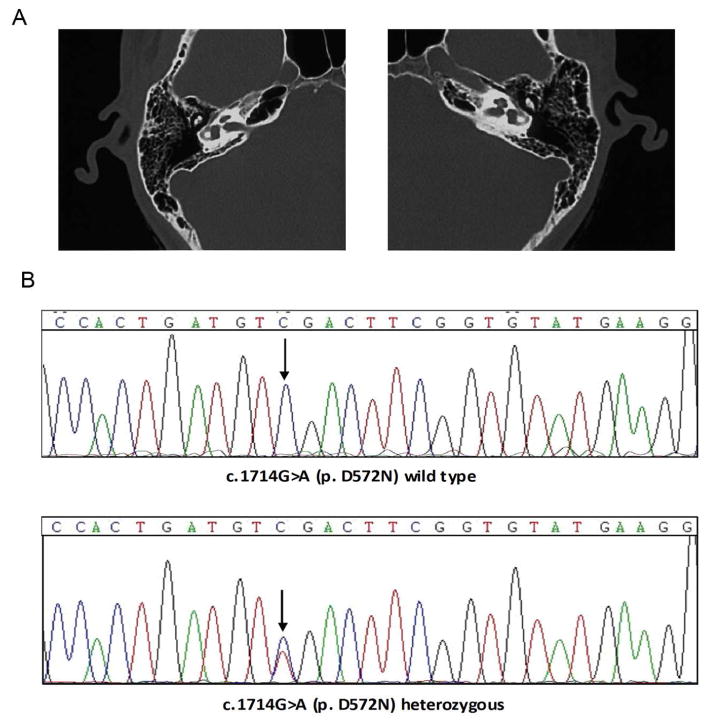
Tandem gait and Romberg tests did not reveal any symptoms of vestibular dysfunction in all family members. CT scans of the proband ruled out inner-ear malformations (Figure 2A). All affected members of three families had oral communication capacities with the help of lip reading. The general physical examinations for all recruited family members were completely normal. Affected individuals did not have delays in gross motor development, which is consistent with the phenotype reported for DFNA36. Individual V47 of Family 5315 underwent unilateral cochlear implantation, and the electrodes were completely inserted into the right cochlea. Residual hearing was preserved well and speech was improved obviously after surgery.
Figure 2. Temporal bone CT and mutational analysis of TMC1 in family 5315.
(A) Temporal bone CT of V47 shows no structural abnormality. (B) Electropherograms show the p. D572N mutation in TMC1. Both variants co-segregated with the phenotype and were absent in 308 ethnicity-matched controls.
Target deaf gene capture and massively paralleled sequencing
We sequenced all coding exons and about 100 bp of flanking intronic sequence of 82 deaf genes in three affected samples and three unaffected members of Family 5315. A missense mutation was detected in a heterozygous state and was caused by a G-to-A transition at nucleotide 1714 in exon 15 of TMC1, resulting in the replacement of aspartic acid, a polar/small/charged amino acid, with an uncharged polar amino acid, asparagine, in the intracellular region of TMC1.
Mutation analysis
By direct sequencing and digestion reactions, all participating members (affected and unaffected) in Family 5315, 308 ethnic-matched negative samples and 67 Chinese ADNSHL cohorts were genotyped to identify the mutation. In Family 5315, we found that the TMC1 heterozygous c.1714G>A (p.D572N) mutation co-segregated with all affected family members (Figure 2B). The penetrance of hearing loss in all family members over 10 years old was 100% based on the data from all investigated individuals. The mutation was absent from 308 negative samples (Figure 2C).
We also confirmed one 7-year-old male (Individual V45) with normal hearing in this family carrying the c.1714G>A mutation. Based on this, we were able to predict his hearing trend and provide his family with hearing protective protocols and therapeutic strategies.
By screening this mutation in 67 affected members from ADNSHL families, we found two probands from Family 5879 and Family 8989 carrying the c.1714G>A mutation who had bilateral hearing loss present with a sloping audiogram at the age of onset, similar to the audiogram of affected members in Family 5315. The mutation prevalence in 67 ADNSHL families with sloping audiogram is 4.4%. The pedigree of Family 5879 and Family 8989 are shown in Figure 3A and the audiograms of the probands are shown in Figure 3B.
The p. D572N mutation identified in Family 5315 was implicated in high-frequency hearing loss. This mutation changes a negatively charged residue aspartic acid to a neutral asparagine at 572, possibly resulting in a loss of hydrogen bonds. None of these mutations in TMC1 were detected in unaffected members of families, nor were they found in DNA from 308 unrelated Chinese controls.
Haplotype analysis
To determine whether the p.D572N mutation of Families 5315, 5879 and 8989 derived from a common founder, we analyzed their linked haplotypes of nine STR markers in three probands and one negative control of Family 5315. All of the tested STR markers had no linked haplotype between the three families.
DISCUSSION
Recent advances in DNA enrichment and NGS have allowed for mutation screening in a large number of genes simultaneously, which is cost effective compared to Sanger sequencing when the number of genes is large [Diaz-Horta et al., 2012].
Gene mutations associated with late-onset progressive disorders are most likely to result in less drastic changes in protein structure and function. The progressive nature of these diseases could then be explained by a gradual increase in the ratio of damaged to normal proteins or changes in protein levels [Petersen 2002]. Pan et al. have identified that dominant mutation p.M412K in Tmc1 affects pore-forming of the transduction channel, which likely have secondary effects on hair cell function and survival. Change in protein levels reduces single-channel current level and calcium permeability and is probably superfluous [Pan et al., 2013].
Previous studies have shown that TMC1 is specifically required for hair cell mechanoelectrical transduction as functionally redundant stereocilia components and is likely to be subunit of the channel itself. Change in protein levels reduces single-channel current level and calcium permeability and is probably superfluous [Kawashima et al., 2011; Pan et al., 2013].
The predicted structure of TMC1 is similar to that of the α-subunit of voltage-dependent K+ channels, which has six α-helical TM segments and intracellular N and C termini [Hanlon et al., 2002]. It was predicted that TMC1 might be an ion channel or transporter which mediated K+ homeostasis in the inner ear [Keresztes et al., 2003]. The first four TM domains of the K+ channel α-subunit act as voltage sensors for activation gating [Li-Smerin et al., 2000], whereas the intervening segment between TM5 and TM6 appears to confer channel selectivity [Hanlon et al., 2002]. One conserved TMC1 sequence variant in this study (c.1714G>A (p.D572N)) lies within a large cytoplasmic loop between TM4 and TM5 that includes the TMC domain and a potential pore-forming loop [Labay et al., 2010]. Although these amino acid residues were non-conserved when human TMC1 was aligned with eight proteins from other species, alignment with other human and murine TMC proteins showed that these residues are conserved in members of the TMC subfamily A, TMC1, TMC2 and TMC3[Keresztes et al., 2003; Kurima et al., 2003].
In this study, we identified the first Chinese family (Family 5315) with ADNSHL caused by the c.1714G>A (p.D572N) mutation of TMC1 through targeted deaf gene capture and NGS. Furthermore, we identified additional two families (Family 5879 and 8989) segregating the same mutation from additional 67 Chinese ADNSHL families with sloping audiogram. TMC1 c.1714G>A (p.D572N) mutation prevalence is 4.4% and absent in 308 ethnicity-matched, unrelated controls.
Our study identified TMC1 c.1714G>A (p.D572N) as the ADNSHL-associated gene in multiple Chinese deaf families for the first time.
CONCLUSIONS
In summary, we describe the clinical and genetic characteristics of three sporadic Chinese families with postlingual ADNSHL caused by TMC1 c.1714G>A. For the first time, TMC1 c.1714G>A mutation has been reported in Chinese population and has an important impact on clinical patient management, genetic counseling, molecular diagnosis, and development of advanced therapeutic strategies.
The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/3BRfp3
Supplementary Material
Acknowledgments
This work was supported by grants from the Project of the National Natural Science Foundation of China (Grant Nos. 30801285, 81230020, 81200751, 81000415, 81371098, 81371096, 81400471, 81371352), grants from China Postdoctoral Science Foundation (2012M521878, 2013T60947), a grant from Military Twelfth Five-year Project of China (CWS115034), a grant from Minister of Science and Technology of China (2012BAI09B02), grants from the National Basic Research Program of China (973 Program) (2014CB541706, 2014CB541701) and a grant from Minister of Health of China (201202005). Work performed in the Lin lab was supported by a grant from NIH (R33 DC010476). We sincerely thank all the family members for their participation and cooperation in this study. We are also grateful to Dr. Guangchun Bai of Albany Medical College, New York State Department of Health for his valuable comments and suggestions.
Footnotes
Authors’ contributions
XG, SSH, PD, XL conceived of the study and participated in its design and draft the manuscript. YYY participated in the next generation sequencing. JCX, YBJ, YL participated in the data analysis. GJW, MYH, FY, DYK participated in the collection of clinical data and blood samples. All authors read and approved the final manuscript.
References
- Azaiez H, Booth KT, Bu F, Huygen P, Shibata SB, Shearer AE, Kolbe D, Meyer N, Black-Ziegelbein EA, Smith RJ. TBC1D24 mutation causes autosomal-dominant nonsyndromic hearing loss. Hum Mutat. 2014;35(7):819–823. doi: 10.1002/humu.22557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlouli A, Bonnet C, Abdi S, Bouaita A, Lelli A, Hardelin JP, Schietroma C, Rous Y, Louha M, Cheknane A, Lebdi H, Boudjelida K, Makrelouf M, Zenati A, Petit C. EPS8, encoding an actin-binding protein of cochlear hair cell stereocilia, is a new causal gene for autosomal recessive profound deafness. Orphanet J Rare Dis. 2014;9:55. doi: 10.1186/1750-1172-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, Rahile M, Lieberman S, Levy-Lahad E, Lee MK, Shomron N, King MC, Walsh T, Kanaan M, Avraham KB. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in Middle Eastern families. Genome Biol. 2011;12(9):R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoudi-Dehaghani E, Fallah MS, Tavakkoly-Bazzaz J, Bagherian H, Zeinali S. Allelic heterogeneity among Iranian DFNB7/11 families: report of a new Iranian deaf family with TMC1 mutation identified by next-generation sequencing. Acta Otolaryngol. 2014:1–5. doi: 10.3109/00016489.2014.969383. [DOI] [PubMed] [Google Scholar]
- Davoudi-Dehaghani E, Zeinali S, Mahdieh N, Shirkavand A, Bagherian H, Tabatabaiefar MA. A transversion mutation in non-coding exon 3 of the TMC1 gene in two ethnically related Iranian deaf families from different geographical regions; evidence for founder effect. Int J Pediatr Otorhinolaryngol. 2013;77(5):821–826. doi: 10.1016/j.ijporl.2013.02.021. [DOI] [PubMed] [Google Scholar]
- Diaz-Horta O, Duman D, Foster J, 2nd, Sirmaci A, Gonzalez M, Mahdieh N, Fotouhi N, Bonyadi M, Cengiz FB, Menendez I, Ulloa RH, Edwards YJ, Zuchner S, Blanton S, Tekin M. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One. 2012;7(11):e50628. doi: 10.1371/journal.pone.0050628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman TB, Griffith AJ. Human nonsyndromic sensorineural deafness. Annu Rev Genomics Hum Genet. 2003;4:341–402. doi: 10.1146/annurev.genom.4.070802.110347. [DOI] [PubMed] [Google Scholar]
- Gao X, Su Y, Guan LP, Yuan YY, Huang SS, Lu Y, Wang GJ, Han MY, Yu F, Song YS, Zhu QY, Wu J, Dai P. Novel compound heterozygous TMC1 mutations associated with autosomal recessive hearing loss in a Chinese family. PLoS One. 2013;8(5):e63026. doi: 10.1371/journal.pone.0063026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girotto G, Abdulhadi K, Buniello A, Vozzi D, Licastro D, d’Eustacchio A, Vuckovic D, Alkowari MK, Steel KP, Badii R, Gasparini P. Linkage study and exome sequencing identify a BDP1 mutation associated with hereditary hearing loss. PLoS One. 2013;8(12):e80323. doi: 10.1371/journal.pone.0080323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon MR, Wallace BA. Structure and function of voltage-dependent ion channel regulatory beta subunits. Biochemistry. 2002;41(9):2886–2894. doi: 10.1021/bi0119565. [DOI] [PubMed] [Google Scholar]
- Hilgert N, Alasti F, Dieltjens N, Pawlik B, Wollnik B, Uyguner O, Delmaghani S, Weil D, Petit C, Danis E, Yang T, Pandelia E, Petersen MB, Goossens D, Favero JD, Sanati MH, Smith RJ, Van Camp G. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet. 2008;74(3):223–232. doi: 10.1111/j.1399-0004.2008.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Monahan K, Kurima K, Li C, Friedman RA, Griffith AJ, Van Camp G. Amino acid 572 in TMC1: hot spot or critical functional residue for dominant mutations causing hearing impairment. J Hum Genet. 2009;54(3):188–190. doi: 10.1038/jhg.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz A, Kohrman DC, Naz S. A frameshift mutation in GRXCR2 causes recessively inherited hearing loss. Hum Mutat. 2014;35(5):618–624. doi: 10.1002/humu.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworek TJ, Richard EM, Ivanova AA, Giese AP, Choo DI, Khan SN, Riazuddin S, Kahn RA. An alteration in ELMOD3, an Arl2 GTPase-activating protein, is associated with hearing impairment in humans. PLoS Genet. 2013;9(9):e1003774. doi: 10.1371/journal.pgen.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Karaguzel A, Caylan R, Heister A, Cremers FP, Cremers CW, Brunner HG, de Brouwer AP, Kremer H. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum Mutat. 2005;26(6):591. doi: 10.1002/humu.9384. [DOI] [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y, Brown KE, Pohl E, Bicknell LS, Kayserili H, Li Y, Tuysuz B, Nurnberg G, Kiess W, Koegl M, Baessmann I, Buruk K, Toraman B, Kayipmaz S, Kul S, Ikbal M, Turner DJ, Taylor MS, Aerts J, Scott C, Milstein K, Dollfus H, Wieczorek D, Brunner HG, Hurles M, Jackson AP, Rauch A, Nurnberg P, Karaguzel A, Wollnik B. CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat Genet. 2011;43(1):23–26. doi: 10.1038/ng.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Geleoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, Griffith AJ. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest. 2011;121(12):4796–4809. doi: 10.1172/JCI60405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Heller S. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 2003;4(1):24. doi: 10.1186/1471-2164-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri S, Makishima T, Friedman TB, Griffith AJ. A novel mutation at the DFNA36 hearing loss locus reveals a critical function and potential genotype-phenotype correlation for amino acid-572 of TMC1. Clin Genet. 2007a;71(2):148–152. doi: 10.1111/j.1399-0004.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- Kitajiri SI, McNamara R, Makishima T, Husnain T, Zafar AU, Kittles RA, Ahmed ZM, Friedman TB, Riazuddin S, Griffith AJ. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet. 2007b;72(6):546–550. doi: 10.1111/j.1399-0004.2007.00895.x. [DOI] [PubMed] [Google Scholar]
- Krawitz PM, Schweiger MR, Rodelsperger C, Marcelis C, Kolsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, Isau M, Fischer A, Dahl A, Kerick M, Hecht J, Kohler S, Jager M, Grunhagen J, de Condor BJ, Doelken S, Brunner HG, Meinecke P, Passarge E, Thompson MD, Cole DE, Horn D, Roscioli T, Mundlos S, Robinson PN. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42(10):827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Kuhlenbaumer G, Hullmann J, Appenzeller S. Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat. 2011;32(2):144–151. doi: 10.1002/humu.21400. [DOI] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, Ghosh M, Menon PS, Deshmukh D, Oddoux C, Ostrer H, Khan S, Deininger PL, Hampton LL, Sullivan SL, Battey JF, Jr, Keats BJ, Wilcox ER, Friedman TB, Griffith AJ. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet. 2002;30(3):277–284. doi: 10.1038/ng842. [DOI] [PubMed] [Google Scholar]
- Kurima K, Yang Y, Sorber K, Griffith AJ. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 2003;82(3):300–308. doi: 10.1016/s0888-7543(03)00154-x. [DOI] [PubMed] [Google Scholar]
- Labay V, Weichert RM, Makishima T, Griffith AJ. Topology of transmembrane channel-like gene 1 protein. Biochemistry. 2010;49(39):8592–8598. doi: 10.1021/bi1004377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Smerin Y, Hackos DH, Swartz KJ. alpha-helical structural elements within the voltage-sensing domains of a K(+) channel. J Gen Physiol. 2000;115(1):33–50. doi: 10.1085/jgp.115.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makishima T, Kurima K, Brewer CC, Griffith AJ. Early onset and rapid progression of dominant nonsyndromic DFNA36 hearing loss. Otol Neurotol. 2004;25(5):714–719. doi: 10.1097/00129492-200409000-00011. [DOI] [PubMed] [Google Scholar]
- Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J Physiol. 2006;574(Pt 3):677–698. doi: 10.1113/jphysiol.2005.095661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Meyer CG, Gasmelseed NM, Mergani A, Magzoub MM, Muntau B, Thye T, Horstmann RD. Novel TMC1 structural and splice variants associated with congenital nonsyndromic deafness in a Sudanese pedigree. Hum Mutat. 2005;25(1):100. doi: 10.1002/humu.9302. [DOI] [PubMed] [Google Scholar]
- Murphy BC, Scriver CR, Singh SM. CpG methylation accounts for a recurrent mutation (c.1222C>T) in the human PAH gene. Hum Mutat. 2006;27(9):975. doi: 10.1002/humu.9447. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, Fennell T, Banks E, Ambrogio L, Cibulskis K, Kernytsky A, Gonzalez E, Rudzicz N, Engert JC, DePristo MA, Daly MJ, Cohen JC, Hobbs HH, Altshuler D, Schonfeld G, Gabriel SB, Yue P, Kathiresan S. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363(23):2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SK, Baek JI, Weigand KM, Venselaar H, Swarts HG, Park SH, Hashim Raza M, Jung DJ, Choi SY, Lee SH, Friedrich T, Vriend G, Koenderink JB, Kim UK, Lee KY. A missense variant of the ATP1A2 gene is associated with a novel phenotype of progressive sensorineural hearing loss associated with migraine. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Geleoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, Holt JR. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron. 2013;79(3):504–515. doi: 10.1016/j.neuron.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen MB. Non-syndromic autosomal-dominant deafness. Clin Genet. 2002;62(1):1–13. doi: 10.1034/j.1399-0004.2002.620101.x. [DOI] [PubMed] [Google Scholar]
- Pierce SB, Walsh T, Chisholm KM, Lee MK, Thornton AM, Fiumara A, Opitz JM, Levy-Lahad E, Klevit RE, King MC. Mutations in the DBP-deficiency protein HSD17B4 cause ovarian dysgenesis, hearing loss, and ataxia of Perrault Syndrome. Am J Hum Genet. 2010;87(2):282–288. doi: 10.1016/j.ajhg.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, Escaramis G, Jares P, Bea S, Gonzalez-Diaz M, Bassaganyas L, Baumann T, Juan M, Lopez-Guerra M, Colomer D, Tubio JM, Lopez C, Navarro A, Tornador C, Aymerich M, Rozman M, Hernandez JM, Puente DA, Freije JM, Velasco G, Gutierrez-Fernandez A, Costa D, Carrio A, Guijarro S, Enjuanes A, Hernandez L, Yague J, Nicolas P, Romeo-Casabona CM, Himmelbauer H, Castillo E, Dohm JC, de Sanjose S, Piris MA, de Alava E, San Miguel J, Royo R, Gelpi JL, Torrents D, Orozco M, Pisano DG, Valencia A, Guigo R, Bayes M, Heath S, Gut M, Klatt P, Marshall J, Raine K, Stebbings LA, Futreal PA, Stratton MR, Campbell PJ, Gut I, Lopez-Guillermo A, Estivill X, Montserrat E, Lopez-Otin C, Campo E. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, Ahmed ZM, Riazuddin S, Khan SN, Friedman TB. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet. 2010;86(3):378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RL, Lee K, Azeem Z, Antonellis PJ, Pollock LM, Khan S, Irfanullah, Andrade-Elizondo PB, Chiu I, Adams MD, Basit S, Smith JD, Nickerson DA, McDermott BM, Jr, Ahmad W, Leal SM. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am J Hum Genet. 2013;93(1):132–140. doi: 10.1016/j.ajhg.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RL, Lee K, Giese AP, Ansar M, Amin-Ud-Din M, Rehn K, Wang X, Aziz A, Chiu I, Hussain Ali R, Smith JD, Shendure J, Bamshad M, Nickerson DA, Ahmed ZM, Ahmad W, Riazuddin S, Leal SM. Adenylate cyclase 1 (ADCY1) mutations cause recessive hearing impairment in humans and defects in hair cell function and hearing in zebrafish. Hum Mol Genet. 2014;23(12):3289–3298. doi: 10.1093/hmg/ddu042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos RL, Wajid M, Khan MN, McArthur N, Pham TL, Bhatti A, Lee K, Irshad S, Mir A, Yan K, Chahrour MH, Ansar M, Ahmad W, Leal SM. Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat. 2005;26(4):396. doi: 10.1002/humu.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen I, Helfmann S, Inagaki A, Predoehl F, Tabatabaiefar MA, Picher MM, Sommen M, Seco CZ, Oostrik J, Kremer H, Dheedene A, Claes C, Fransen E, Chaleshtori MH, Coucke P, Lee A, Moser T, Van Camp G. A mutation in CABP2, expressed in cochlear hair cells, causes autosomal-recessive hearing impairment. Am J Hum Genet. 2012;91(4):636–645. doi: 10.1016/j.ajhg.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, 2nd, Scherer S, Scheetz TE, Smith RJ. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Justice CM, Krishnan M, Wojciechowski R, Sung H, Cai J, Green T, Lewis D, Behneman D, Wilson AF, Bailey-Wilson JE. Old lessons learned anew: family-based methods for detecting genes responsible for quantitative and qualitative traits in the Genetic Analysis Workshop 17 mini-exome sequence data. BMC Proc. 2011a;5(Suppl 9):S83. doi: 10.1186/1753-6561-5-S9-S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE. Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet. 2011b;48(3):145–151. doi: 10.1136/jmg.2010.083568. [DOI] [PubMed] [Google Scholar]
- Tang WS, Qian D, Ahmad S, Mattox D, Todd NW, Harrison H, Huang S, Li Y, Wang Y, Li HW, Lin X. A low-cost exon capture method suitable for large-scale screening of genetic deafness by the massively-parallel sequencing approach. Genetic Testing & Mol Biomarker. 2011 doi: 10.1089/gtmb.2011.0187. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlili A, Rebeh IB, Aifa-Hmani M, Dhouib H, Moalla J, Tlili-Chouchene J, Said MB, Lahmar I, Benzina Z, Charfedine I, Driss N, Ghorbel A, Ayadi H, Masmoudi S. TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol Neurootol. 2008;13(4):213–218. doi: 10.1159/000115430. [DOI] [PubMed] [Google Scholar]
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, Hrabe De Angelis M, Avraham KB, Steel KP. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet. 2002;30(3):257–258. doi: 10.1038/ng848. [DOI] [PubMed] [Google Scholar]
- Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, Kanaan M. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87(1):90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G, Yao J, Wu B, Liu T, Wei Q, Liu C, Lu Y, Chen Z, Zheng H, Yang X, Cao X. Identification of OSBPL2 as a novel candidate gene for progressive nonsyndromic hearing loss by whole-exome sequencing. Genet Med. 2014 doi: 10.1038/gim.2014.90. [DOI] [PubMed] [Google Scholar]
- Yang T, Kahrizi K, Bazazzadeghan N, Meyer N, Najmabadi H, Smith RJ. A novel mutation adjacent to the Bth mouse mutation in the TMC1 gene makes this mouse an excellent model of human deafness at the DFNA36 locus. Clin Genet. 2010;77(4):395–398. doi: 10.1111/j.1399-0004.2009.01338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang D, Zong L, Zhao F, Guan L, Zhang P, Shi W, Lan L, Wang H, Li Q, Han B, Yang L, Jin X, Wang J, Wang Q. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PLoS One. 2014;9(5):e97064. doi: 10.1371/journal.pone.0097064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhao F, Zong L, Zhang P, Guan L, Zhang J, Wang D, Wang J, Chai W, Lan L, Li Q, Han B, Yang L, Jin X, Yang W, Hu X, Wang X, Li N, Li Y, Petit C, Wang HY, Wang Q. Exome sequencing and linkage analysis identified tenascin-C (TNC) as a novel causative gene in nonsyndromic hearing loss. PLoS One. 2013;8(7):e69549. doi: 10.1371/journal.pone.0069549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.