Abstract
Wilms tumor (WT) is the most common childhood kidney cancer worldwide and poses a cancer health disparity to black children of sub‐Saharan African ancestry. Although overall survival from WT at 5 years exceeds 90% in developed countries, this pediatric cancer is alarmingly lethal in sub‐Saharan Africa and specifically in Kenya (36% survival at 2 years). Although multiple barriers to adequate WT therapy contribute to this dismal outcome, we hypothesized that a uniquely aggressive and treatment‐resistant biology compromises survival further. To explore the biologic composition of Kenyan WT (KWT), we completed a next generation sequencing analysis targeting 10 WT‐associated genes and evaluated whole‐genome copy number variation. The study cohort was comprised of 44 KWT patients and their specimens. Fourteen children are confirmed dead at 2 years and 11 remain lost to follow‐up despite multiple tracing attempts. TP53 was mutated most commonly in 11 KWT specimens (25%), CTNNB1 in 10 (23%), MYCN in 8 (18%), AMER1 in 5 (11%), WT1 and TOP2A in 4 (9%), and IGF2 in 3 (7%). Loss of heterozygosity (LOH) at 17p, which covers TP53, was detected in 18% of specimens examined. Copy number gain at 1q, a poor prognostic indicator of WT biology in developed countries, was detected in 32% of KWT analyzed, and 89% of these children are deceased. Similarly, LOH at 11q was detected in 32% of KWT, and 80% of these patients are deceased. From this genomic analysis, KWT biology appears uniquely aggressive and treatment‐resistant. © 2015 The Authors. Genes, Chromosomes & Cancer Published by Wiley Periodicals, Inc.
INTRODUCTION
Wilms tumor (WT) is the most common childhood kidney cancer worldwide and arises disparately and most prevalently among children of black sub‐Saharan African ancestry, regardless of original nationality, country of immigration, or subsequent generation (Stiller and Parkin, 1990; Breslow et al., 1993, 1994). Although survival from WT in developed countries now exceeds 90% at 5 years, dismal outcomes are experienced in low‐income nations of sub‐Saharan Africa. For example, our recent efforts to establish a comprehensive WT Registry and Tissue Repository in Kenya have shown that overall survival at 2 years remains alarmingly and unacceptably low at 36% in this resource‐challenged country (Abdallah and Macharia, 2001; Axt et al., 2013; Libes et al., 2014b). While a lack of standardized treatment protocols, an inconsistent availability of chemotherapeutics, and frequent care abandonment contribute significantly to this poor outcome from WT in Kenya, we have had reason, based on consistent clinical observations of its lethal behavior, to postulate that a unique and potentially more aggressive biology imparts a major obstacle to treatment efficacy (Murphy et al., 2012a; Libes et al., 2014a).
WT is a genetically heterogeneous disease arising in the context of several classical mutations that, depending on the stage of kidney organogenesis and the respective sequence in which each occurs, determine its histology and biology (Gadd et al., 2012; Scott et al., 2012). The combined frequency of three genetic alterations fundamental to Wilms tumorigenesis, specifically WT1, CTNNB1, and WTX (i.e., AMER1 or FAM123B), has been estimated to occur in roughly one‐third of WT, whereas aberrant expression of IGF2 has been shown to occur in 70% of WT specimens (Huff, 2011; Gadd et al., 2012). Furthermore, WT maintenance and disease progression are associated with the altered expression of multiple other genes, such as TP53, MYCN, CITED1, SIX2, TOP2A, and CRABP2 (Lovvorn et al., 2007; Schaub et al., 2007; Williams et al., 2011, 2015; Murphy et al., 2012b, 2014 2014; Libes et al., 2014a; Pierce et al., 2014). Specifically, mutations in TP53 and accumulation of its protein product, TP53, are a common finding in unfavorable histology (UH) WT and a notorious marker of treatment resistance (Lahoti et al., 1996; Sredni et al., 2001; Natrajan et al., 2007; Maschietto et al., 2014).
Within developed countries of North America and Europe, recent advances in WT therapy and outcome have evolved to modify the intensity of treatment algorithms according to specific biological properties. Specifically, combined loss of heterozygosity (LOH) at 1p and 16q in favorable histology (FH) WT has been associated with treatment resistant disease and portends a poor outcome, albeit only occurring in ∼5% of FHWT cases (Grundy et al., 1994, 2005; Dome et al, 2014). An even more recent prognostic marker of poor outcome is copy number gain (CNG) at 1q in FHWT specimens, which too has been associated with adverse biologic behavior (Hing et al., 2001; Natrajan et al., 2006; Perotti et al., 2012; Gratias et al., 2013). The presence of these biologic variables, specifically LOH of 1p and 16q, has been incorporated into the current Children's Oncology Group (COG) therapy paradigm to warrant a more intensive drug regimen up front for FHWT (Dome et al., 2014). Loss of genetic material at 4q, 11q, and 14q has also emerged as features of UHWT and poor prognosis (Wittmann et al., 2007; Williams et al., 2011). However, the frequency and prognostic consequence of these genetic and chromosomal alterations in WT among patients residing in the resource‐constrained nation of Kenya have not been previously characterized and may serve as a biologic road map for other sub‐Saharan African countries.
Building on our recent proteomic efforts to clarify the molecular basis for the persistently poor survival from WT in Kenya, we hypothesized that specimens from children in this disadvantaged country would harbor genetic signatures of biologically aggressive and treatment resistant disease.
MATERIALS AND METHODS
Kenyan Wilms Tumor Patients
To study the molecular composition of and survival from WT in Kenya, we established a comprehensive patient registry, consecutively enrolling children who were treated at four collaborating hospitals beginning January 1, 2008 (Axt et al., 2013; Libes et al., 2014b). Concomitantly, we established a Kenyan WT tissue repository to archive corresponding specimens for biological study (Libes et al., 2014a). Through December 2014, 263 Kenyan WT patients have been registered into this database. Available tissue blocks (formalin fixed and paraffin embedded) of registered patients were shipped bi‐annually to Vanderbilt University for molecular analysis; specimens from 146 Kenyan WT (KWT) patients could be located within the study time frame.
Histologic Analysis
Because resources to archive WT specimens consistently and in a timely manner are limited in Kenya, and because treatment regimens are not currently standardized there, we performed upfront a thorough histologic analysis of all shipped tissue blocks to verify diagnosis and to assure the highest tissue quality for genomic analysis. Briefly, 5 μm sections were obtained from each tissue block and stained with hematoxylin and eosin (H&E). A fellowship‐trained pediatric pathologist (HC) was blinded to all clinical and research data before histologic review of each tissue section. Specimens were reviewed on two separate occasions to determine pathologic diagnosis, histology (i.e., using COG criteria and the presence of diffuse anaplasia to define unfavorable histology), integrity of fixation, and tissue viability (Faria et al., 1996). Due to many WT patients receiving neoadjuvant chemotherapy in Kenya as a principal cause of tissue necrosis, we identified 44 different patient specimens as being of sufficient integrity to perform these genetic studies; the remaining 102 specimens showed predominant tissue necrosis either from treatment effect or delayed fixation that precluded reliable molecular analysis and therefore were excluded. Ten KWT specimens had adjacent kidney available for control germ line analysis, but only five tumor and kidney blocks could be paired, given current archiving methodologies and tumor specimen necrosis.
Next Generation Sequencing Analysis
To explore the genetic and chromosomal alterations in KWT, genomic DNA was isolated from all 44 WT and 10 adjacent kidney specimens using the QIAamp DNA FFPE Tissue Kit according to the manufacturer's protocol (Qiagen, Valencia, CA). Briefly, four paraffin sections at 10 μm each were acquired from the highest quality tissue block of each KWT patient (Vanderbilt Translational Pathology Shared Resource). After removal of wax in xylene, tissue sections were digested and genomic DNA was isolated and purified. To evaluate the presence of mutations in 10 WT‐associated genes (WT1, CTNNB1, AMER1 [i.e., WTX], IGF2, TP53, MYC‐N, CITED1, SIX2, CRABP2, and TOP2A), genomic DNA was analyzed using next generation sequencing (NGS) technology for single nucleotide variations, insertions, and deletions in these targeted loci. Briefly, multiplex amplicon sequencing libraries were prepared using an amplicon gene primer panel that targeted coding regions of these 10 genes. Input DNA was quantified using the high‐sensitivity dsDNA assay on the Qubit fluorometer and normalized to 4 ng/μl. The multiplex PCR was performed in eight reactions per sample using a custom Qiagen GeneRead DNA‐seq Panel following manufacturer's protocol without deviation (Qiagen).
Data Quality Control and Analysis
Variant calling was performed using the standard Genome Analysis Toolkit Haplotype Caller pipeline (GATK version 3.1‐1, http://www.broadinstitute.org/gatk/) (McKenna et al., 2010). Single nucleotide variant (SNV) mutation calls were made using the following threshold filters: (1) each candidate mutation had to pass GATK Variant Quality Score Recalibration filtering, (2) DP (depth) filtering was greater than 10, (3) Genotype Quality was greater than 30, (4) SNV was not observed in the 10 adjacent kidneys, and (5) the allele frequency in the 1,000 Genomes Project was lower than 0.2%. For small insertions and deletions (indels), we further manually inspected, using “samtools tview,” the alignment and removed calls close to the end of aligned reads. Additionally, we used MuTect software to call SNV mutations further to detect low allele fraction (AF) mutations (Cibulskis et al., 2013). Given the lower number of adjacent kidney specimens that could be located, we combined these ten germ line controls as a single sample and ran the MuTect analysis of individual tumors against this combined sample. We selected mutation calls with AF greater than 0.1 for further analysis. Mutation calls are reported for those SNVs occurring only in the WT specimens, and not concomitantly in the adjacent kidneys, and are annotated using ANNOVAR (version 2014jul14) (Wang et al., 2010). Only non‐synonymous SNV mutations with predicted deleteriousness in one of the algorithms implemented in ANNOVAR, and indel mutations in exonic regions, are called as potentially functionally significant. In aggregate, this strict approach to mutation calls yielded the greatest possible confidence, given the constrained resources.
Copy Number Variation and Loss of Heterozygosity Analysis
To evaluate copy number variations (CNV) and LOH at genomic regions that associate with adverse behavior of WT, we contracted with Affymetrix (Santa Clara, CA), which has a unique platform to analyze whole‐genome DNA isolated from FFPE specimens (Malek et al., 2011; Wang et al., 2012). Genomic DNA was available from 34 of these KWT specimens for this analysis and was shipped to Affymetrix to perform the OncoScanTM FFPE Assay Kit, as described (Singh et al., 2015). Data were compared against two Affymetrix controls, and quality control metrics were applied according to manufacturer standards. Nexus Express for OncoScanTM 3.0 (BioDiscovery, Hawthorne, CA) was used to generate all data figures and to analyze statistical significance, as described (Wang et al., 2012). Significance (P < 0.05) of chromosomal changes between group comparisons (e.g., dead versus alive and unfavorable versus favorable histology) is shown with a horizontal bar (blue is copy gain, red is copy loss, and yellow is LOH) in the row designated “Significant.” Furthermore, this OncoScan array interrogates, using molecular inversion probes, 74 somatic mutations in 9 genes, including KRAS (Singh et al., 2015).
RESULTS
Kenyan Wilms Tumor Patients
For this cohort of 44 KWT patients, 11 children were and remain lost to follow‐up (LTFU) after various intervals of adjuvant treatment following tumor resection, and their outcomes could not be accurately estimated despite exhaustive tracing efforts. Among those patients for whom vital status could be accurately determined through the medical record and multiple tracing calls (n = 33), 14 children are confirmed deceased. This cohort of 44 KWT included 8 specimens that showed diffuse unfavorable histology (UH; 18%), and 5 of these children are deceased (63%). Among the 36 patients having favorable histology (FH), 9 are deceased (25%). A total of 19 children (43%) received variable neoadjuvant therapy before resection, but the precise extent (i.e., specific drugs and cumulative dosing) could not be determined reliably from review of existing medical records.
Next Generation Sequencing Analysis
Among this study sample of 44 KWT specimens, potentially deleterious mutations were detected in all 10 target genes sequenced but at a variable frequency (Table 1). In descending order of occurrence, TP53 was mutated most commonly in 11 KWT specimens (25%), CTNNB1 in 10 KWT specimens (23%), MYCN in 8 (18%), AMER1 (i.e., WTX) in 5 (11%), WT1 and TOP2A in 4 each (9%), IGF2 in 3 (7%), and CITED1, SIX2, and CRABP2 in 1 KWT specimen each (2%). Multiple of these mutations are previously reported “hot spots” in WT arising in patients from other regions of the world, whereas certain mutations are unreported in the COSMIC database and may be novel and unique to this Kenyan cohort (Table 1). Concomitant mutations in CTNNB1 were detected in two of the four KWT specimens having a WT1 mutation (Maiti et al., 2000; Gadd et al., 2012).
Table 1.
Alterations in 10 Target Genes Among Kenyan Wilms Tumors
| Chromosome | REF | Alteration | Location | Function | Gene | Amino acid change | Tumor | Cosmic68 | esp‐6500si | SIFT | Polyphen 2_HVAR | LRT | Mutation taster | Mutation/assessor | FATHMM | Radial SVM | LR | Cadd gt10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | G | A | exonic | nonsynonym | CRABP2 | NM_001878:exon2:c.C178T:p.R60C | KWT‐39 | COSM98340 | N/A | B | B | D | D | M | T | T | T | 15.34 |
| 2 | G | A | exonic | nonsynonym | SIX2 | NM_016932:exon1:c.C29T:p.T10M | KWT‐10 | N/A | N/A | D | P | U | D | M | D | D | D | 25.5 |
| 2 | C | T | exonic | nonsynonym | MYCN | NM_005378:exon2:c.C131T:p.P44L | KWT‐22, 38 | COSM35624 | N/A | D | D | N | D | M | T | T | T | 27.3 |
| 2 | C | T | exonic | nonsynonym | MYCN | NM_005378:exon2:c.C220T:p.P74S | KWT‐21, 30 | N/A | N/A | P | P | N | D | L | T | T | T | N/A |
| 2 | C | T | exonic | nonsynonym | MYCN | NM_005378:exon2:c.C173T:p.T58M | KWT‐26 | N/A | N/A | D | D | N | D | M | T | T | T | 23.1 |
| 2 | G | A | exonic | nonsynonym | MYCN | NM_005378:exon2:c.G191A:p.S64N | KWT‐29 | N/A | N/A | D | D | D | D | M | T | T | T | 21 |
| 2 | A | C | exonic | nonsynonym | MYCN | NM_005378:exon3:c.A1111C:p.S371R | KWT‐43, 44 | N/A | N/A | B | B | N | D | M | T | T | T | 16.62 |
| 3 | A | G | exonic | nonsynonym | CTNNB1 | NM_001098209:exon3:c.A121G:p.T41A | KWT‐16 | COSM5664 | N/A | P | P | D | D | M | T | T | T | 23.7 |
| 3 | T | C | exonic | nonsynonym | CTNNB1 | NM_001098209:exon3:c.T133C:p.S45P | KWT‐5, 27 | COSM5663 | N/A | D | P | D | D | M | T | T | T | 23.6 |
| 3 | C | T | exonic | nonsynonym | CTNNB1 | NM_001098209:exon3:c.C134T:p.S45F | KWT‐17 | COSM5667 | N/A | D | D | D | D | M | T | T | T | 24.8 |
| 3 | G | A | exonic | nonsynonym | CTNNB1 | NM_001098209:exon4:c.G439A:p.E147K | KWT‐30 | N/A | N/A | P | P | D | D | M | T | T | T | 23.8 |
| 3 | A | T | exonic | nonsynonym | CTNNB1 | NM_001098209:exon6:c.A801T:p.E267D | KWT‐13, 39 | N/A | N/A | B | B | D | D | N | T | T | T | 10.11 |
| 3 | G | A | exonic | nonsynonym | CTNNB1 | NM_001098209:exon7:c.G955A:p.G319S | KWT‐4, 26 | N/A | N/A | B | B | D | D | L | T | T | T | 20.6 |
| 3 | C | G | exonic | nonsynonym | CTNNB1 | NM_001098209:exon7:c.C1006G:p.L336V | KWT‐9 | N/A | N/A | D | P | D | D | M | T | T | T | 22.8 |
| 11 | G | A | exonic | nonsynonym | WT1 | NM_000378:exon1:c.C34T:p.P12S | KWT‐9a | N/A | N/A | B | B | N/A | D | N | T | T | T | 18.07 |
| 11 | C | T | exonic | nonsynonym | WT1 | NM_000378:exon1:c.G37A:p.A13T | KWT‐25 | N/A | N/A | P | B | N/A | D | N | T | T | T | 21 |
| 11 | GC | G | exonic | frameshift | WT1 | NM_000378:exon1:c.520delG:p.A174fs | KWT‐17a | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 11 | G | T | exonic | nonsynonym | WT1 | NM_000378:exon2:c.C719A:p.P240H | KWT‐29 | N/A | N/A | P | P | U | D | L | D | D | D | 28.6 |
| 11 | G | A | exonic | nonsynonym | IGF2 | NM_001042376:exon3:c.C319T:p.R107W | KWT‐8 | N/A | N/A | D | P | U | N | L | D | D | D | 14.03 |
| 11 | G | A | exonic | nonsynonym | IGF2 | NM_000612:exon4:c.C317T:p.P106L | KWT‐30 | N/A | N/A | D | P | U | D | M | D | D | D | 16.73 |
| 11 | G | A | exonic | nonsynonym | IGF2 | NM_000612:exon4:c.C515T:p.P172L | KWT‐3 | N/A | N/A | B | B | U | N | N | D | T | T | 11.4 |
| 17 | G | T | exonic | nonsynonym | TP53 | NM_001126115:exon1:c.C56A:p.P19H | KWT‐33 | COSM259150; 259151; 259152; 259149; 11476 | N/A | D | D | D | D | M | D | D | D | 16.15 |
| 17 | C | T | exonic | nonsynonym | TP53 | NM_001126115:exon1:c.G128A:p.R43H | KWT‐12 | COSM10648; 99022; 1640851; 99024; 99023; 99914 | N/A | D | D | D | A | M | D | D | D | 31 |
| 17 | G | A | exonic | nonsynonym | TP53 | NM_001126118:exon3:c.C22T:p.P8S | KWT‐18 | N/A | 0.0052 | B | B | N | N | L | D | D | D | N/A |
| 17 | G | C | exonic | nonsynonym | TP53 | NM_001126118:exon3:c.C56G:p.P19R | KWT‐19 | N/A | 0.0004 | D | B | N | N | N | D | D | D | N/A |
| 17 | C | T | exonic | nonsynonym | TP53 | NM_001126115:exon3:c.G337A:p.G113S | KWT‐10 | COSM1640833; 121037; 121035; 6932; 121036 | 0.0001 | D | D | D | D | M | D | D | D | 33 |
| 17 | G | A | exonic | nonsynonym | TP53 | NM_001126115:exon7:c.C734T:p.T245I | KWT‐3, 18 | N/A | N/A | B | B | N | D | M | D | D | D | 14.92 |
| 17 | G | A | exonic | nonsynonym | TP53 | NM_001126115:exon7:c.C737T:p.S246F | KWT‐13 | N/A | N/A | D | P | N | D | M | D | D | D | 22 |
| 17 | G | A | exonic | nonsynonym | TP53 | NM_001126115:exon7:c.C739T:p.R247C | KWT‐8, 40 | N/A | N/A | B | B | N | D | L | D | D | D | 12.79 |
| 17 | C | T | exonic | nonsynonym | TP53 | NM_001126115:exon7:c.G740A:p.R247H | KWT‐13, 23 | COSM44189 | N/A | B | B | N | N | L | D | D | D | 13.21 |
| 17 | C | G | exonic | nonsynonym | TP53 | NM_001126115:exon7:c.G740C:p.R247P | KWT‐1, 13, 23 | N/A | N/A | B | B | N | D | L | D | D | D | 12.75 |
| 17 | G | A | exonic | nonsynonym | TP53 | NM_001126115:exon7:c.C742T:p.H248Y | KWT‐23 | N/A | N/A | P | B | N | D | M | D | D | D | 13.43 |
| 17 | C | T | exonic | nonsynonym | TOP2A | NM_001067:exon6:c.G497A:p.G166E | KWT‐1 | N/A | N/A | D | D | D | D | H | D | D | D | 28.2 |
| 17 | C | T | exonic | nonsynonym | TOP2A | NM_001067:exon6:c.G521A:p.S174N | KWT‐1 | N/A | N/A | D | D | D | D | H | T | D | D | 32 |
| 17 | G | A | exonic | nonsynonym | TOP2A | NM_001067:exon9:c.C989T:p.A330V | KWT‐1 | N/A | N/A | B | B | N | D | N | T | T | T | N/A |
| 17 | C | T | exonic | nonsynonym | TOP2A | NM_001067:exon12:c.G1444A:p.G482R | KWT‐30 | N/A | N/A | D | D | D | D | H | T | T | T | 33 |
| 17 | C | T | exonic | nonsynonym | TOP2A | NM_001067:exon19:c.G2279A:p.G760D | KWT‐27 | N/A | N/A | D | D | D | D | H | T | D | D | 32 |
| 17 | A | C | exonic | nonsynonym | TOP2A | NM_001067:exon21:c.T2518G:p.W840G | KWT‐41 | N/A | N/A | D | D | D | D | M | T | T | T | 19.42 |
| 17 | A | G | exonic | nonsynonym | TOP2A | NM_001067:exon21:c.T2525C:p.I842T | KWT‐30 | N/A | N/A | B | B | D | D | L | T | T | T | 14.54 |
| 17 | C | T | exonic | nonsynonym | TOP2A | NM_001067:exon25:c.G3214A:p.E1072K | KWT‐1 | N/A | N/A | P | P | D | D | M | T | T | T | 33 |
| X | G | C | exonic | stopgain | AMER1 | NM_152424:exon2:c.C125G:p.S42X | KWT‐40 | N/A | N/A | N/A | N/A | N | A | N/A | N/A | N/A | N/A | 21.7 |
| X | G | A | exonic | stopgain | AMER1 | NM_152424:exon2:c.C1072T:p.R358X | KWT‐1 | COSM 193868, 22960 | N/A | N/A | N/A | D | A | N/A | N/A | N/A | N/A | 38 |
| X | C | A | exonic | nonsynonym | AMER1 | NM_152424:exon2:c.G2014T:p.G672W | KWT‐44 | N/A | N/A | D | D | N | D | L | T | T | T | N/A |
| X | C | T | exonic | nonsynonym | AMER1 | NM_152424:exon2:c.G2440A:p.V814M | KWT‐30 (F) | N/A | N/A | D | P | N/A | D | N | T | T | T | 14.9 |
| X | G | A | exonic | nonsynonym | AMER1 | NM_152424:exon2:c.C2510T:p.S837F | KWT‐14 | N/A | N/A | D | D | N/A | D | N | T | T | T | 16.45 |
| X | T | G | exonic | nonsynonym | AMER1 | NM_152424:exon2:c.A2543C:p.K848T | KWT‐14 | N/A | N/A | D | D | N/A | D | N | T | T | T | 12.42 |
| X | G | A | exonic | nonsynonym | AMER1 | NM_152424:exon2:c.C2545T:p.H849Y | KWT‐14 | N/A | N/A | P | B | N/A | D | N | T | T | T | N/A |
| X | C | T | exonic | nonsynonym | CITED1 | NM_001144886:exon3:c.G124A:p.V42M | KWT‐30 (F) | N/A | N/A | D | P | N | N | L | T | T | T | 18.79 |
nonsynonym = nonsynonymous alteration
Wilms tumor specimens having combined mutations in WT1 and CTNNB1.
(F) designates a female patient for the X‐chromosome genes, AMER1 and CITED1, only.
Score (dbtype)
SIFT (sift)
PolyPhen 2 HDIV (pp2_hdiv)
PolyPhen 2 HVar (pp2_hvar)
LRT (lrt)
MutationTaster (mt)
MutationAssessor (ma)
FATHMM (fathmm)
MetaSVM (metasvm)
MetaLR (metalr)
CADD
Categorical Prediction
D: Deleterious (sift<=0.05); T: tolerated (sift>0.05)
D: Probably damaging (>=0.909), P: possibly damaging (0.447<=pp2_hdiv<=0.909); B: benign (pp2_hdiv<=0.446)
D: Deleterious; N: Neutral; U: Unknown
A: (“disease‐causing‐automatic”); “D” (“disease‐causing”); “N” (“polymorphism”); “P” (“polymorphism‐automatic”)
H: high; M: medium; L: low; N: neutral. H/M means functional and L/N means non‐functional
D: Deleterious; T: Tolerated
D: Deleterious; T: Tolerated
D: Deleterious; T: Tolerated
Combined Annotation Dependent Depletion
Interestingly, three of these targeted genes showed multiple mutations within a given specimen (Table 1). Specifically, TP53 was mutated thrice each in KWT‐13 and −23 and twice in KWT‐18. AMER1 was mutated thrice in KWT‐14, while TOP2A was mutated at four separate positions in KWT‐1, and twice in KWT‐30.
Copy Number Variation and Loss of Heterozygosity Analysis
In the sub‐group of 34 KWT available for whole genome copy number analysis, chromosomal instability was detected readily and in a pattern associated with poor prognosis in developed countries (Fig. 1). Specifically, CNG at 1q, an emerging feature of adverse WT biology (Hing et al., 2001; Gratias et al., 2013), was detected in 11 (32%) of the KWTs analyzed, a frequency similar to other regions of the world; of these children, 8 are confirmed deceased, and only 1 is confirmed alive at 2 years (2 patients remain LTFU; Table 2; Fig. 2). CNG at 1q was significantly associated with death among this KWT cohort (Fig. 2). One unexpected finding from these studies was the frequent occurrence of LOH at region 16p11.2‐11.1, which is a locus rich in TP53 target genes (Table 2) (Hurst et al., 2012). Taken together with the frequency of mutations in TP53 and of CNL and LOH at 17p13.1 (i.e., the TP53 locus) observed in this cohort, it appears that loss of TP53 activity is common in and important to KWT biology (Tables 1 and 2; Fig. 2).
Figure 1.
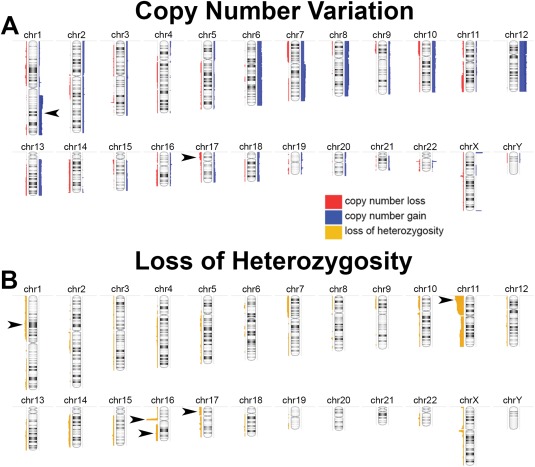
A: Whole genome view of copy number gain (blue) and loss (red) across 34 Kenyan WTs. Arrowheads denote gain at 1q and loss at 17p. B: Whole genome view of LOH (yellow) across the same KWT specimens. Arrowheads denote regions of interest to WT biology. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table 2.
Histology, Mutations, Outcome, and Copy Number Variations Among Kenyan Wilms Tumor Cohort
| Tumor | Histology | Genes altered | Outcome | LOH 16p11.2‐11.1 (multiple TP53 target genes) | LOH 17p13.1 (TP53) | LOH 11p15.5 | LOH 11p13 (WT1) | CNG 1q | LOH 1p | LOH 16q | LOH 11q |
|---|---|---|---|---|---|---|---|---|---|---|---|
| KWT1 | Favorable | TP53, TOP2A, AMER1 | LTFU | Yes | Yes | ||||||
| KWT2 | Favorable | Alive | Yes | ||||||||
| KWT3 | Favorable | IGF2, TP53 | Dead | Yes | Yes | Yes | |||||
| KWT4 | Favorable | CTNNB1 | Dead | Yes (deletion) | Yes | Yes | Yes | Yes | |||
| KWT5a | Favorable | CTNNB1 | LTFU | Yes | Yes | Yes | |||||
| KWT6 | Favorable | LTFU | |||||||||
| KWT7 | Favorable | Alive | Yes | Yes | Yes | Yes | |||||
| KWT8 | Favorableb | IGF2, TP53 | LTFU | ||||||||
| KWT9 | Unfavorable | CTNNB1, WT1 | Relapse, Alive | Yes | Yes | Yes | |||||
| KWT10a | Unfavorable | SIX2, TP53 | Dead | Yes | Yes | Yes | Yes | Yes | Yes | ||
| KWT11a | Favorablea | Alive | Yes | ||||||||
| KWT12a | Unfavorable | TP53 | Dead | Yes | Yes | Yes | Yes | Yes | Yes | ||
| KWT13a | Favorableb | CTNNB1, TP53 | Alive | Yes | |||||||
| KWT14 | Favorable | AMER1 | Alive | Yes | Yes | ||||||
| KWT15a | Favorable | Dead | Yes | Yes | Yes | Yes | |||||
| KWT16 | Favorable | CTNNB1 | Dead | Yes | Yes | ||||||
| KWT17a | Favorable | CTNNB1, WT1 | Alive | Yes | Yes | Yes | |||||
| KWT18a | Unfavorable | TP53 | Alive | Yes | Yes | Yes | Yes | Yes | Yes | ||
| KWT19a | Favorable | TP53 | LTFU | Yes | Yes | Yes | Yes | Yes | |||
| KWT20 | Favorable | Alive | Yes | ||||||||
| KWT21 | Favorable | MYCN | LTFU | Yes | |||||||
| KWT22a | Favorable | MYCN | LTFU | Yes | Yes | ||||||
| KWT23 | Favorableb | TP53 | Alive | ||||||||
| KWT24 | Favorableb | Alive | |||||||||
| KWT25 | Favorableb | WT1 | Alive | ||||||||
| KWT26 | Unfavorable | MYCN, CTNNB1 | Alive | ||||||||
| KWT27 | Favorableb | CTNNB1, TOP2A | Alive | Yes | Yes | Yes | |||||
| KWT28 | Favorable | LTFU | |||||||||
| KWT29 | Favorableb | MYCN, WT1 | LTFU | ||||||||
| KWT30 | Favorable | MYCN, CTNNB1, IGF2, TOP2A, AMER1, CITED1 | Alive | Yes | Yes | Yes | |||||
| KWT31 | Favorable | Alive | |||||||||
| KWT32 | Favorableb | Dead | Yes | Yes | Yes | ||||||
| KWT33a | Favorable | TP53 | Dead | Yes (deletion) | Yes | ||||||
| KWT34a | Favorableb | Relapse, Alive | Yes | Yes | Yes | ||||||
| KWT35a | Favorable | Alive | Yes | ||||||||
| KWT36 | Favorable | LTFU | |||||||||
| KWT37a | Favorableb | Alive | Yes | Yes | Yes | Yes | |||||
| KWT38a | Favorable | MYCN | LTFU | ||||||||
| KWT39a | Favorable | CRABP2, CTNNB1 | Dead | Yes | Yes | Yes | |||||
| KWT40a | Favorable | TP53, AMER1 | Dead | Yes | Yes | Yes | Yes | ||||
| KWT41 | Favorableb | TOP2A | Dead | Yes | Yes | Yes | Yes | ||||
| KWT42a | Unfavorable | Dead | Yes | Yes (deletion) | Yes | Yes | Yes | ||||
| KWT43 | Unfavorable | MYCN | Dead | Yes | Yes | ||||||
| KWT44a | Unfavorable | MYCN, AMER1 | Dead | Yes | Yes | Yes | Yes | Yes | |||
| Total (n = 34) | UH = 8 (18%) | 27 (79%) | 6 (18%) | 22 (65%) | 18 (53%) | 11 (32%) | 3 (9%) | 5 (15%) | 11 (32%) |
Designates those patients who received some dose of neoadjuvant chemotherapy.
Represents nuclear unrest within favorable histology Wilms tumor.
LTFU: loss to follow up; LOH: loss of heterozygosity; CNG: copy number gain; UH: unfavorable histology.
Black filled cells denote specimens not analyzed for chromosomal aberrations.
Figure 2.
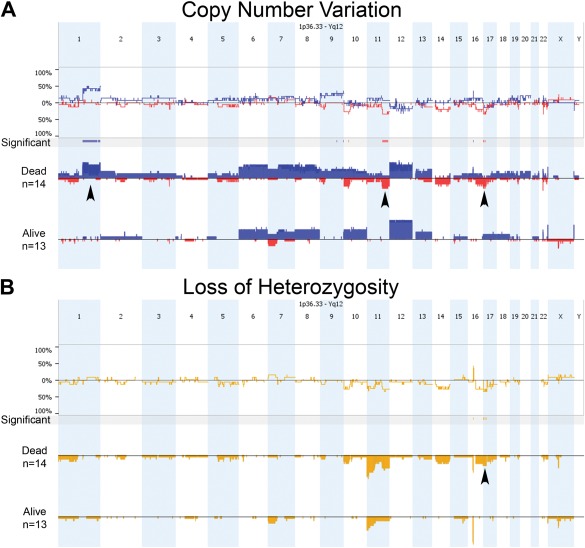
Comparison of copy number variation (A) and LOH (B) across the Kenyan WT genome between patients who died (n = 14) or survived until conclusion of the study (n = 13). A: Copy gain is denoted in blue and loss in red, and arrowheads highlight a statistically significant gain at 1q and loss at 11q among those who died. Other significant regions are noted. B: For LOH, only two regions were statistically different between outcome groups: 16p and 17p. The latter (arrowhead) covers the TP53 region. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
As expected, separating the KWT specimens according to histologic subtype revealed greater chromosomal instability among UH tumors, showing many significantly different regions for both CNV and LOH (Fig. 3). LOH at 1p and 16q are poor prognostic features of FHWT in developed countries, particularly when occurring together, and warrant more intensified therapy to reduce the risk for subsequent relapse (Grundy et al., 1994, 2005). Fortunately, this pair of allelic loss occurs in only 5% of FHWT patients in the developed world and was detected in only one of these KWT specimens, which showed UH, and that child is deceased from disease progression. CNL and LOH at 1p and 17p were more commonly associated with UH in this study (Fig. 3). Of further interest, copy number loss (CNL) at 11q was observed differentially in UH relative to FH KWT specimens and appears to be associated with death too. LOH at 11q was detected in 11 KWT specimens, and 80% of these patients are confirmed deceased (Table 2). Separated according to histology, LOH at 11q was present in 71% of UH KWT analyzed but in only 22% of FH KWT (Table 2).
Figure 3.
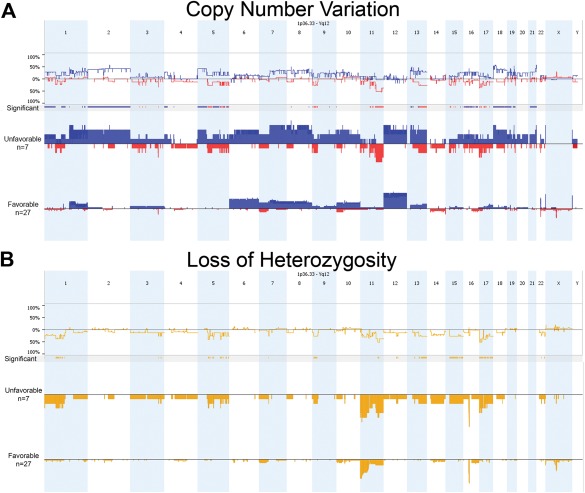
Comparison of copy number variation (A) and LOH (B) across the Kenyan WT genome between unfavorable (UH) and favorable (FH) histology specimens. As expected, UH specimens show greater variability relative to FH, as depicted by significant regions. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
KRAS Mutations
To explore the consistent gain at chromosome 12 observed in this KWT cohort and reported in other WT populations as well, we examined the OncoScan array data for somatic mutations in KRAS, which is the only of 9 genes included on this platform to be located on chromosome 12 and which has been shown in a transgenic WT model to drive disease progression (Clark et al., 2011; Yi et al., 2015). CNG at 12p12.1, the KRAS locus, was observed in 14 of the KWTs (47%), and 6 of these children (43%) are confirmed deceased. A total of six point mutations, which exceeded 2 standard deviations from the mean MutScore (Affymetrix OncoScanTM 3.0 platform), were detected in 11 KWT specimens; as an even stricter threshold of mutation confidence, five of these point mutations exceeded 3 standard deviations from the mean MutScore and were detected in three KWT specimens (Table 3).
Table 3.
KRAS Mutations in Kenyan Wilms Tumor
| No. of mutations > 2 standard deviations beyond mean MutScore | No. of mutations > 3 standard deviations beyond mean MutScore | KRAS mutation | Cosmic_ID |
|---|---|---|---|
| 1 | 1 | p.G12A:c.35G>C | COSM522 |
| 2 | 2 | p.G12C/S:c.34G>T/A | COSM517; 516 |
| 5 | 1 | p.G12D/V:c.35G>A/T | COSM520; 521 |
| 5 | 1 | p.G13D:c.38G>A | COSM532 |
| 3 | 1 | p.Q61H:c.183A>C | COSM554 |
| 1 | 0 | p.Q61H:c.183A>T | COSM555 |
| 11 KWT—32% | 3 KWT—9% |
DISCUSSION
WT poses a significant cancer health disparity to black children of sub‐Saharan African ancestry, not only because of its more common occurrence among black populations worldwide, but also because of its persistently high lethality in resource‐constrained nations on the African continent, such as Kenya. To identify both societal and biological risk factors that contribute to the persistently dismal 2‐year survival from WT of only 36%, we, as a five‐institution collaborative research team (1 American and 4 Kenyan hospitals), established a Kenyan Wilms Tumor Registry and Tissue Repository, initially registering patients treated in 2008 (Axt et al., 2013; Libes et al., 2014b). Indeed, many barriers to adequate WT therapy, its completion, and the long‐term follow‐up of survivors compromise optimal outcomes. Yet, we asked the fundamental question whether a population‐specific biology was also a deleterious contributing factor. As a complement to our recent study that evaluated differences in peptide profiles among North American and Kenyan WT specimens, we conducted the present and first‐ever investigation to characterize the genetic and chromosomal alterations in the latter population, as much has been published on this genomic topic in the developed world (Murphy et al., 2012a; Libes et al., 2014a). WT is a genetically heterogeneous disease, and specific patterns of recurring mutations comprise the theory as to its tumorigenesis, whereas chromosomal aberrations have been associated with poor prognosis. Moreover, developed countries now risk‐stratify FHWT patients according to the presence or absence of LOH at both 1p and 16q, which together guide upfront intensity of therapy (Dome et al., 2014). Emerging as another poor prognostic indicator of WT outcome is CNG at 1q (Gratias et al., 2013). So, to optimize therapy in a low resource environment such as Kenya, it is necessary to identify both societal and biological risk factors that form the basis for the poor outcome from WT experienced there.
The foremost observation from this genomic analysis of KWT reveals a pattern of genomic instability that indeed associates with adverse biological behavior and treatment resistance seen in developed regions of the world. Specifically, CNG at 1q was detected at a similar frequency as in North American specimens but was associated with a nearly uniform risk for death (Hing et al., 2001; Gratias et al., 2013). This observation suggests that CNG at 1q in a KWT indicates treatment resistant and potentially lethal disease, and will require more intensive therapy upfront and a greater effort to retain these high risk patients in therapy through its completion and close monitoring for subsequent relapse post therapy. Interestingly, combined LOH at 1p and 16q was not observed among 34 FHWT specimens analyzed in this Kenyan cohort, but this paired genomic event was detected in one UH specimen, and predictably that child died from disease. LOH at both 1p and 16q, which commonly accompanies CNG at 1q, occurs in ∼5% of FHWT specimens and significantly reduces 5‐year survival in developed countries, but its frequency and effect on survival in sub‐Saharan countries remains to be clarified (Grundy et al., 2005). Importantly, CNL and LOH at 11q also emerged from this cohort of KWT as a feature of UH and an ominous risk for death (Klamt et al., 1998; Wittmann et al., 2007).
Accumulation of the TP53 protein in WT specimens has been associated with UH and treatment resistance (Lahoti et al., 1996; Sredni et al., 2001; Natrajan et al, 2007; Maschietto et al, 2014). It has been further postulated that TP53 mutation in WT is a late occurrence in its disease sequence and progression (Natrajan et al., 2007). In this cohort of KWT, TP53 was the most frequently mutated gene we tested, found in 25% of the specimens, and LOH at 17p, which covers TP53, was detected in 18% of specimens examined. This frequency of alterations in TP53 (i.e., 32% total having either a potentially deleterious mutation or LOH at 17p) exceeds those reported in other WT studies (Scott et al., 2012). One related finding of this Kenyan study was the common occurrence (79%) of LOH at a region on 16p that harbors a number of TP53 target genes (Ng et al., 1999). Although 16p is a region that can be prone to copy number variability, its specific variance among the Kenyan population is currently unknown and therefore the presence of constitutional polymorphisms could not be distinguished.
Taken together, these observations suggest that loss of TP53 and its wild‐type protein product potentially contribute fundamentally to KWT biology, although its functional significance in this context has yet to be defined. In parallel epidemiologic studies of this patient registry, we have reported that Kenyan children present with WT at an age typical for this disease, as documented in other populations (i.e., between 3 and 4 years); as a result, delayed presentation at a later stage in disease progression is not solely explanatory of these observed alterations in TP53. Copy number gain of MYCN is another feature of treatment‐resistant WT (Schaub et al., 2007; Williams et al., 2011). A recent article describes a similar frequency of MYCN alterations (18.5%) in a cohort of European WT and reports the same P44L mutation that was detected in two of these KWT (Wegert et al., 2015). Finally, we and others have observed consistent gain of whole chromosome 12 in WT. Because we have reported previously on activation of KRAS, which resides at 12p12.1, as a mechanism that drives tumor dissemination in a mouse model, we queried what changes may be occurring with KRAS in these KWT (Clark et al., 2011). KRAS CNG was frequent in almost half of these specimens, and mutations were observed relatively commonly too, three of which were detected with high confidence at p.G12, the site that was engineered into the transgenic model. For comparison, in a parallel screen of 20 North American WT specimens, we detected the p.G12D mutation in one patient tumor (5% mutation rate at this locus), which represents a similar variation frequency (data unpublished at time of this writing). These observations of KRAS alterations suggest a potentially targetable mechanism that drives the disease progression of KWT.
The authors would like to acknowledge several limitations of this study that temper interpretation of the results, which center principally around the challenges of conducting molecular research on tissues acquired from resource‐constrained countries. Foremost, stratifying the clinical significance of specific mutations and chromosomal alterations on outcomes among KWT patients is minimized by: (1) the lack of a nationally standardized therapeutic regimen, (2) a high frequency of patients to abandon care, and (3) a substantial loss to follow‐up rate. As a result, it is difficult to define clearly what genetic aberrations align with favorable or poor prognosis and with treatment efficacy when many children are not completing therapy. For example, we have been unable to determine a precise incidence and time interval for relapse and any effect this adverse event has on overall survival, as salvage therapy is not standardized or widely available in Kenya. Kenyan parents often view relapse as a non‐survivable condition and may not seek additional treatment, particularly when on‐therapy toxicity is so high (Axt et al., 2013; Libes et al., 2014a). Nevertheless, through exhaustive tracing efforts, we have been able to determine a reasonably accurate overall survival at two years for this KWT cohort that allowed evaluation of whole‐genome CNV between those who died or were alive at conclusion of the study. A second study limitation concerns the integrity and consistency of methods employed to archive WT tissues in Kenya. It is unknown for what duration and at what temperature a specimen may sit in pathology before formalin fixation, which together limit experimental approaches to reveal precise biological markers. To overcome this question of study tissue integrity, we first performed a thorough quality assurance histologic analysis to select the KWT specimens showing greatest viability, which would yield the greatest confidence of having analyzed tumor and not inflammatory or apoptotic cells. Third, given that many WT patients in Kenya are pre‐treated with neoadjuvant therapy (43% in this cohort), it is possible that we may have selected unintentionally a more treatment‐resistant cohort of specimens, wishing to avoid sequencing of tissues having a large fraction of necrosis. Unfortunately, it was not possible as another control measure to determine the precise dosing of neoadjuvant therapy or the effect it had on tumor regression in this KWT cohort. As a result, the increased incidence of UH may be real or may be artificial as a consequence of this histologic subtype to resist treatment, thereby imparting a bias in the selection of viable tissues. Nevertheless, our chromosomal comparison between histology types and vital status remain reliable, as the tissue specimens again were controlled for quality (i.e., viability). Finally, for analysis as germ line controls, we could locate only 10 adjacent kidney specimens from which a WT arose, and only half of these could be matched definitively to tumor samples. As a result, our mutation calls rarely may include potential polymorphisms unique to the Kenyan population; however, by combining genomic data from all 10 adjacent kidney specimens and excluding any single nucleotide variation arising in this “control” pool, we should have preserved strict integrity for mutation calls.
In summary, this targeted genomic and chromosomal analysis of KWT reveals a pattern of treatment‐resistance and late phases of the WT sequence despite a typical age at presentation for this disease globally. Mortality remains unacceptably high among this KWT cohort for multiple reasons, but an aggressive, treatment‐resistant biology may indeed contribute more to the dismal outcomes than previously anticipated. Standardization of WT care in Kenya will help to reduce overall mortality and will permit a better understanding of the clinical significance for the various molecular signatures, whether genomic or proteomic. Furthermore, simple and inexpensive immunohistochemical screening for TP53 as a marker of treatment resistant KWT could help to risk‐stratify patients in this low‐income nation. If resources and collaborations improve, a focused analysis for CNG at 1q and CNL at 11q could further guide the intensity of future treatment regimens in Kenya. Finally, the administration of drugs that target the β‐catenin or KRAS pathways may be of future benefit to treat these challenging KWT patients, assuming efficacy can be proven without violating the Declaration of Helsinki for research involving vulnerable populations.
ACKNOWLEDGMENTS
The authors would like to thank Travis Clark and Holli Dilks, formerly of the Vanderbilt TPSR, for their technical assistance to conduct the targeted next generation sequencing analysis of the KWT specimens.
The Kenyan Wilms Tumor Consortium: James N'Dungu, Oliver Oruko, Jessie Githanga, Fatmah Abdullah (Kenyatta National Hospital, University of Nairobi, Nairobi, Kenya); Joyce Musimbi, Festus Njuguna, Kirtika Patel (Moi University Teaching and Referral Hospital, Eldoret, Kenya); Gabriel Ellsworth, Michael Mwachiro, and Russell White (Tenwek Mission Hospital, Bomet, Kenya).
REFERENCES
- Abdallah FK, Macharia WM. 2001. Clinical presentation and treatment outcome in children with nephroblastoma in Kenya. East Afr Med J 78:S43–S47. [PubMed] [Google Scholar]
- Axt J, Abdallah F, Axt M, Githanga J, Hansen E, Lessan J, Li M, Musimbi J, Mwachiro M, Newton M, Ndung'u J, Njuguna F, Nzioka A, Oruko O, Patel K, Tenge R, Ukoli F, White R, O'Neill JA, Jr. , Lovvorn HN, III. 2013. Wilms tumor survival in Kenya. J Pediatr Surg 48:1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow N, Olshan A, Beckwith JB, Green DM. 1993. Epidemiology of Wilms tumor. Med Pediatr Oncol 21:172–181. [DOI] [PubMed] [Google Scholar]
- Breslow N, Olshan A, Beckwith JB, Moksness J, Feigl P, Green D. 1994. Ethnic variation in the incidence, diagnosis, prognosis, and follow‐up of children with Wilms' tumor. J Natl Cancer Inst 86:49–51. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PE, Polosukhina D, Love H, Correa H, Coffin C, Perlman EJ, de Caestecker M, Moses HL, Zent R. 2011. Beta‐catenin and K‐RAS synergize to form primitive renal epithelial tumors with features of epithelial Wilms' tumors. Am J Pathol 179:3045–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dome JS, Perlman EJ, Graf N. 2014. Risk stratification for wilms tumor: Current approach and future directions. Am Soc Clin Oncol Educ Book 2014:215–223. [DOI] [PubMed] [Google Scholar]
- Faria P, Beckwith JB, Mishra K, Zuppan C, Weeks DA, Breslow N, Green DM. 1996. Focal versus diffuse anaplasia in Wilms tumor–new definitions with prognostic significance: A report from the National Wilms Tumor Study Group. Am J Surg Pathol 20:909–920. [DOI] [PubMed] [Google Scholar]
- Gadd S, Huff V, Huang CC, Ruteshouser EC Dome JS, Grundy PE Breslow N, Jennings L, Green DM, Beckwith JB Perlman EJ. 2012. Clinically relevant subsets identified by gene expression patterns support a revised ontogenic model of Wilms tumor: A Children's Oncology Group Study. Neoplasia 14:742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratias EJ, Jennings LJ, Anderson JR, Dome JS, Grundy P, Perlman EJ. 2013. Gain of 1q is associated with inferior event‐free and overall survival in patients with favorable histology Wilms tumor: A report from the Children's Oncology Group. Cancer 119:3887–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy PE, Telzerow PE, Breslow N, Moksness J, Huff V, Paterson MC. 1994. Loss of heterozygosity for chromosomes 16q and 1p in Wilms' tumors predicts an adverse outcome. Cancer Res 54:2331–2333. [PubMed] [Google Scholar]
- Grundy PE, Breslow NE, Li S, Perlman E, Beckwith JB, Ritchey ML, Shamberger RC, Haase GM, D'Angio GJ, Donaldson M, Coppes MJ, Malogolowkin M, Shearer P, Thomas PR, Macklis R, Tomlinson G, Huff V, Green DM. 2005. Loss of heterozygosity for chromosomes 1p and 16q is an adverse prognostic factor in favorable‐histology Wilms tumor: A report from the National Wilms Tumor Study Group. J Clin Oncol 23:7312–7321. [DOI] [PubMed] [Google Scholar]
- Hing S, Lu YJ, Summersgill B, King‐Underwood L, Nicholson J, Grundy P, Grundy R, Gessler M, Shipley J, Pritchard‐Jones K. 2001. Gain of 1q is associated with adverse outcome in favorable histology Wilms' tumors. Am J Pathol 158:393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff V. 2011. Wilms' tumours: About tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 11:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst CD, Platt FM, Taylor CF, Knowles MA. 2012. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin Cancer Res 18:5865–5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klamt B, Schulze M, Thate C, Mares J, Goetz P, Kodet R, Scheulen W, Weirich A, Graf N, Gessler M. 1998. Allele loss in Wilms tumors of chromosome arms 11q, 16q, and 22q correlate with clinicopathological parameters. Genes Chromosomes Cancer 22:287–294. [DOI] [PubMed] [Google Scholar]
- Lahoti C, Thorner P, Malkin D, Yeger H. 1996. Immunohistochemical detection of p53 in Wilms' tumors correlates with unfavorable outcome. Am J Pathol 148:1577–1589. [PMC free article] [PubMed] [Google Scholar]
- Libes JM, Seeley EH, Li M, Axt JR, Pierce J, Correa H, Newton M, Hansen E, Judd A, McDonald H, Caprioli RM, Naranjo A, Huff V, O'Neill JA, Jr. , Lovvorn HN, III . 2014a. Race disparities in peptide profiles of North American and Kenyan Wilms tumor specimens. J Am Coll Surg 218:707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libes J, Oruko O, Abdallah F, Githanga J, Ndung'u J, Musimbi J, Njuguna F, Patel K, White J, Axt JR, O'Neill JA, Jr ., Shrubsole M, Li M, Lovvorn HN, III . 2014b. Risk factors for abandonment of Wilms tumor therapy in Kenya. Pediatr Blood Cancer 2014 Nov 8. doi: 10.1002/pbc.25312. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovvorn HN, Westrup J, Opperman S, Boyle S, Shi G, Anderson J, Perlman EJ, Perantoni AO, Wills M, de Caestecker M. 2007. CITED1 expression in Wilms' tumor and embryonic kidney. Neoplasia 9:589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti S, Alam R, Amos CI, Huff V. 2000. Frequent association of beta‐catenin and WT1 mutations in Wilms tumors. Cancer Res 60:6288–6292. [PubMed] [Google Scholar]
- Malek JA, Mery E, Mahmoud YA, Al‐Azwani EK Roger L, Huang R, Jouve E, Lis R, Thiery JP, Querleu D Rafii A. 2011. Copy number variation analysis of matched ovarian primary tumors and peritoneal metastasis. PLoS One 6:e28561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschietto M, Williams RD, Chagtai T, Popov SD, Sebire NJ, Vujanic G, Perlman E, Anderson JR, Grundy P, Dome JS, Pritchard‐Jones K. 2014. TP53 mutational status is a potential marker for risk stratification in Wilms tumour with diffuse anaplasia. PLoS One 9:e109924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: A MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Res 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AJ, Axt JR, de Caestecker C, Pierce J, Correa H, Seeley EH, Caprioli RM, Newton MW, de Caestecker MP, Lovvorn HN, III. 2012a. Molecular characterization of Wilms' tumor from a resource‐constrained region of sub‐Saharan Africa. Int J Cancer 131:E983–E994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AJ, Pierce J, de Caestecker C, Taylor C, Anderson JR, Perantoni AO, de Caestecker MP, Lovvorn HN, III. 2012b. SIX2 and CITED1, markers of nephronic progenitor self‐renewal, remain active in primitive elements of Wilms' tumor. J Pediatr Surg 47:1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AJ, Pierce J, de Caestecker C, Ayers GD, Zhao A, Krebs JR, Saito‐Diaz VK, Lee E, Perantoni AO, de Caestecker MP Lovvorn HN, III . 2014. CITED1 confers stemness to Wilms tumor and enhances tumorigenic responses when enriched in the nucleus. Oncotarget 5:386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natrajan R, Williams RD Hing SN, Mackay A, Reis‐Filho JS, Fenwick K, Iravani M, Valgeirsson H, Grigoriadis A, Langford CF, Dovey O, Gregory SG, Weber BL, Ashworth A, Grundy PE, Pritchard‐Jones K Jones C. 2006. Array CGH profiling of favourable histology Wilms tumours reveals novel gains and losses associated with relapse. J Pathol 210:49–58. [DOI] [PubMed] [Google Scholar]
- Natrajan R, Little SE, Sodha N, Reis‐Filho JS, Mackay A, Fenwick K, Ashworth A, Perlman EJ, Dome JS, Grundy PE, Pritchard‐Jones K Jones C. 2007. Analysis by array CGH of genomic changes associated with the progression or relapse of Wilms' tumour. J Pathol 211:52–59. [DOI] [PubMed] [Google Scholar]
- Ng CC, Koyama K, Okamura S, Kondoh H, Takei Y, Nakamura Y. 1999. Isolation and characterization of a novel TP53‐inducible gene, TP53TG3. Genes Chromosomes Cancer 26:329–335. [PubMed] [Google Scholar]
- Perotti D, Spreafico F, Torri F, Gamba B, D'Adamo P, Pizzamiglio S, Terenziani M, Catania S, Collini P, Nantron M, Pession A, Bianchi M, Indolfi P, D'Angelo P, Fossati‐Bellani F, Verderio P, Macciardi F, Radice P. 2012. Genomic profiling by whole‐genome single nucleotide polymorphism arrays in Wilms tumor and association with relapse. Genes Chromosomes Cancer 51:644–653. [DOI] [PubMed] [Google Scholar]
- Pierce J, Murphy AJ, Panzer A, de Caestecker C, Ayers GD, Neblett D, Saito‐Diaz K, de Caestecker M, Lovvorn HN, III. 2014. SIX2 effects on Wilms tumor biology. Transl Oncol 7:800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub R, Burger A, Bausch D, Niggli FK, Schafer BW, Betts DR. 2007. Array comparative genomic hybridization reveals unbalanced gain of the MYCN region in Wilms tumors. Cancer Genet Cytogenet 172:61–65. [DOI] [PubMed] [Google Scholar]
- Scott RH, Murray A, Baskcomb L, Turnbull C, Loveday C, Al‐Saadi R, Williams R, Breatnach F, Gerrard M, Hale J, Kohler J, Lapunzina P, Levitt GA, Picton S, Pizer B, Ronghe MD, Traunecker H, Williams D, Kelsey A, Vujanic GM, Sebire NJ, Grundy P, Stiller CA, Pritchard‐Jones K, Douglas J, Rahman N. 2012. Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget 3:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Sahu DK, Goel M, Kant R, Gupta DK. 2015. Retrospective analysis of FFPE based Wilms' tumor samples through copy number and somatic mutation related Molecular Inversion Probe Based Array. Gene 565:295–308. [DOI] [PubMed] [Google Scholar]
- Sredni ST, de Camargo B, Lopes LF, Teixeira R, Simpson A. 2001. Immunohistochemical detection of p53 protein expression as a prognostic indicator in Wilms tumor. Med Pediatr Oncol 37:455–458. [DOI] [PubMed] [Google Scholar]
- Stiller CA, Parkin DM. 1990. International variations in the incidence of childhood renal tumours. Br J Cancer 62:1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: Functional annotation of genetic variants from high‐throughput sequencing data. Nucleic Acids Res 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cottman M, Schiffman JD. 2012. Molecular inversion probes: A novel microarray technology and its application in cancer research. Cancer Genet 205:341–355. [DOI] [PubMed] [Google Scholar]
- Wegert J, Ishaque N, Vardapour R, Georg C, Gu Z, Bieg M, Ziegler B, Bausenwein S, Nourkami N, Ludwig N, Keller A, Grimm C, Kneitz S, Williams RD, Chagtai T, Pritchard‐Jones K, van Sluis P, Volckmann R, Koster J, Versteeg R, Acha T, O'Sullivan MJ, Bode PK, Niggli F, Tytgat GA van Tinteren H, van den Heuvel‐Eibrink MM Meese E, Vokuhl C, Leuschner I, Graf N, Eils R, Pfister SM, Kool M Gessler M. 2015. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high‐risk blastemal type Wilms tumors. Cancer Cell 27:298–311. [DOI] [PubMed] [Google Scholar]
- Williams RD, Al‐Saadi R, Natrajan R, Mackay A, Chagtai T, Little S, Hing SN, Fenwick K, Ashworth A, Grundy P, Anderson JR, Dome JS, Perlman EJ, Jones C, Pritchard‐Jones K. 2011. Molecular profiling reveals frequent gain of MYCN and anaplasia‐specific loss of 4q and 14q in Wilms tumor. Genes Chromosomes Cancer 50:982–995. [DOI] [PubMed] [Google Scholar]
- Williams RD, Chagtai T, Alcaide‐German M, Apps J, Wegert J, Popov S, Vujanic G, van Tinteren H, van den Heuvel‐Eibrink MM, Kool M, de Kraker J, Gisselsson D, Graf N, Gessler M, Pritchard‐Jones K. 2015. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget 6:7232–7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann S, Zirn B, Alkassar M, Ambros P, Graf N, Gessler M. 2007. Loss of 11q and 16q in Wilms tumors is associated with anaplasia, tumor recurrence, and poor prognosis. Genes Chromosomes Cancer 46:163–170. [DOI] [PubMed] [Google Scholar]
- Yi Y, Polosukhina D, Love HD, Hembd A, Pickup M, Moses HL, Lovvorn HN, 3rd , Zent R, Clark PE. 2015. A murine model of K‐ras and beta‐catenin induced renal tumors express high levels of E2F1 and resemble human Wilms Tumor. J Urol 2015 Apr 28. doi: 10.1016/j.juro.2015.04.090. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]


