SUMMARY
Invading viral DNA can be recognized by the host cytosolic DNA sensor, cyclic GMP-AMP (cGAMP) synthase (cGAS), resulting in production of the second messenger cGAMP, which directs the adaptor protein STING to stimulate production of type I interferons (IFNs). Although several DNA viruses are sensed by cGAS, viral strategies targeting cGAS are virtually unknown. We report here that Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF52, an abundant gammaherpesvirus-specific tegument protein, subverts cytosolic DNA sensing by directly inhibiting cGAS enzymatic activity through a mechanism involving both cGAS- and DNA-binding. Moreover, ORF52 homologues in other gammaherpesviruses also inhibit cGAS activity and similarly bind cGAS and DNA, suggesting conserved inhibitory mechanisms. Furthermore, KSHV infection evokes cGAS-dependent responses that can limit the infection, and an ORF52-null mutant exhibits increased cGAS signaling. Our findings reveal a mechanism through which gammaherpesviruses antagonize host cGAS DNA sensing.
INTRODUCTION
Cytosolic DNA derived from microbial pathogens represents a potent pathogen-associated molecular pattern (PAMP) that triggers the host innate immune responses by stimulating production of type I interferons (IFNs). The cyclic guanosine monophosphate–adenosine monophosphate (cGAMP) synthase (cGAS) has recently been identified as the principal sensor of cytosolic DNA (Cai et al., 2014; Sun et al., 2013; Wu et al., 2013). Binding of DNA to cGAS activates its enzymatic activity, producing cGAMP from ATP and GTP (Cai et al., 2014; Civril et al., 2013; Gao et al., 2013b; Kranzusch et al., 2013; Li et al., 2013a; Zhang et al., 2014). As a second messenger, cGAMP binds to and activates the stimulator of interferon genes (STING) in infected cells, as well as neighboring cells, through cell-cell junctions (Ablasser et al., 2013a; Ablasser et al., 2013b; Gao et al., 2013c). Active STING then activates TANK-binding kinase 1 (TBK1) to phosphorylate and activate interferon regulatory factor 3 (IRF3), ultimately leading to expression of type I IFNs (Barber, 2014; Tanaka and Chen, 2012). DNA viruses, including herpes simplex virus 1 (HSV-1), vaccinia virus, and adenovirus, as well as retroviruses, such as HIV-1, have been shown to be sensed by cGAS (Dai et al., 2014; Gao et al., 2013a; Lam et al., 2014; Li et al., 2013b). Because activation of cGAS elicits a potent antiviral response (Li et al., 2013b; Schoggins et al., 2014), viruses must possess mechanisms to subvert the cGAS-cGAMP signaling pathway to establish successful infection. To date, no such mechanisms have been described.
Kaposi’s sarcoma-associated herpesvirus (KSHV) is the causative agent of Kaposi’s sarcoma (KS), primary effusion lymphoma, and a subset of multicentric Castleman’s disease (Cesarman et al., 1995; Chang et al., 1994; Ganem, 2007; Soulier et al., 1995). Like other herpesviruses, KSHV exhibits two alternative life cycles: latent and lytic. KSHV primarily establishes latency, during which only a handful of genes are expressed and no progeny are produced. Lytic replication constitutes expression of the full complement of viral genes in a temporal cascade, ultimately resulting in the production of progeny virions (Ganem, 2007). A low level of spontaneous lytic reactivation occurs in the lesions of KSHV-associated diseases, and is believed to be required for viral persistence and pathogenesis (Ganem, 2010). Although the capsid-enclosed herpesviral DNA is believed to be delivered into the nucleus, where herpesviruses replicate their genomes, viral DNA could leak into the cytosol and subsequently be sensed by cGAS (Horan et al., 2013; Paludan et al., 2011). It is thus possible that KSHV infection could elicit cGAS-dependent responses and that the virus possesses a mechanism(s) to subvert cGAS-cGAMP signaling in order to evade the innate immune response. However, no viral strategies that target cGAS have been described. We report here that KSHV ORF52, a gammaherpesvirus-specific tegument protein, inhibited cGAS enzymatic activity via a mechanism involving its binding to DNA and cGAS. Furthermore, ORF52 homologues in other gammaherpesviruses also inhibited cGAS. Moreover, we found that KSHV primary infection elicits cGAS- and STING- dependent responses that can be partially mitigated by ORF52. Our results reveal KSHV ORF52 as an inhibitor of cGAS, and we propose to name it KSHV inhibitor of cGAS, KicGAS.
RESULTS
KSHV ORF52 inhibits cGAS DNA-sensing signaling
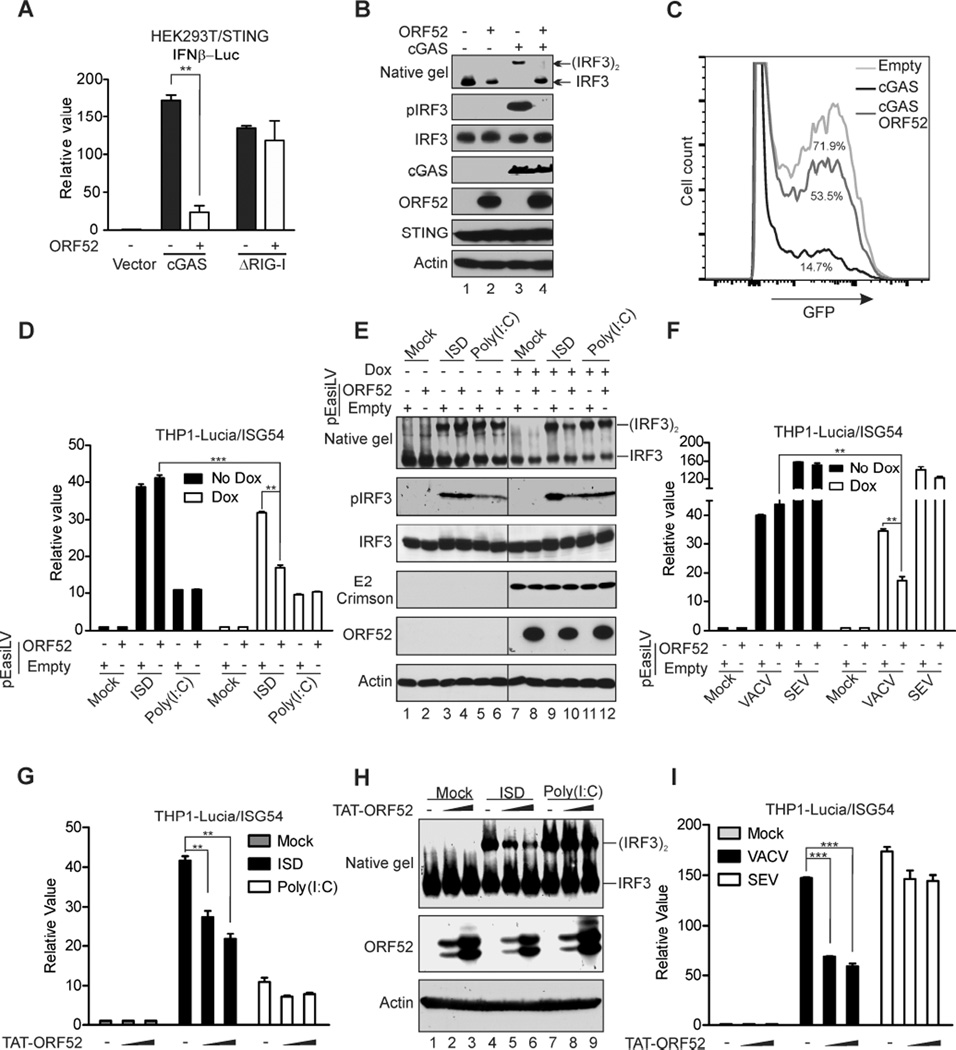
We reasoned that a potential cGAS inhibitor would be a virion component, localize to the cytoplasm, and interact with DNA and/or cGAS. Systematic analysis of all KSHV proteins for inhibition of cGAS-dependent IFNβ production revealed 8 viral protein candidates as cGAS signaling antagonists (Figure S1A). Among them, ORF52 was the only protein confirmed to bind to DNA (Figure S1, B and C). ORF52 was previously shown to be an abundant virion protein (Zhu et al., 2005) and localize exclusively to the cytoplasm (Sander et al., 2008), making it a prime candidate for an inhibitor of cGAS. To determine whether ORF52 has an effect on the cGAS signaling pathway, we co-expressed cGAS with IFNβ luciferase reporter in the presence or absence of ORF52 in HEK293T cells stably expressing STING (HEK293T/STING). Transfection of cGAS increased IFNβ promoter-driven luciferase activity by more than 150-fold, but co-expression with ORF52 reduced cGAS-induced luciferase activity by more than 80% (Figure 1A). The inhibition appeared to be specific to cGAS, because ORF52 had little effect on luciferase activity induced by the cytosolic RNA sensor RIG-I (Figure 1A). In agreement with the inhibition of luciferase reporter activity, cGAS-mediated dimerization and phosphorylation of IRF3 were inhibited by ORF52 (Figure 1B) in a dose-dependent manner (Figure S2A). Furthermore, while HEK293T/STING cells expressing cGAS displayed a strong antiviral effect to vesicular stomatitis virus (VSV-GFP) infection (14.7% infection), co-expression with ORF52 partially relieved this effect (53.5% infection), suggesting that ORF52 specifically inhibits cGAS-dependent antiviral responses (Figure 1C).
Figure 1. Inhibition of cGAS DNA-sensing signaling by KSHV ORF52.
(A) KSHV ORF52 inhibits cGAS-induced IFNβ promoter activity. HEK293T–STING cells were transfected with IFNβ luciferase reporter and expression plasmids as indicated, and luciferase activity was assayed 24 h after transfection. The relative luciferase activity was expressed as arbitrary units by normalizing firefly luciferase activity to Renilla luciferase activity. (B) KSHV ORF52 inhibits cGAS-induced IRF3 dimerization and phosphorylation. HEK293T-STING cells were transfected with expression plasmids as indicated. Twenty-four h after transfection, cell lysates were analyzed for IRF3 dimerization by native gel electrophoresis. IRF3 phosphorylation and expression levels of the transfected genes were monitored by immunoblotting with antibodies. (C) KSHV ORF52 inhibits cGAS-induced antiviral response. HEK293T-STING cells transfected with specified plasmids were infected with vesicular stomatitis virus (VSV-GFP). The percentage of GFP-positive cells, as analyzed by FACS, is shown for each condition. (D–F) ORF52 inhibits cGAS DNA sensing signaling in THP-1 cells. THP1 Lucia™ ISG cells harboring pEasiLV-ORF52 or pEasiLV-empty were untreated or induced with 2 µg/ml doxycycline for 60 h then transfected with ISD45 or Poly(I:C) (2µg/ml) (D–E), or infected with VACV or SEV (F). The activity of secreted Lucia luciferase was determined, or cell lysates were harvested then analyzed for IRF3 dimerization and phosphorylation (E). (G–I) TAT-ORF52 protein inhibits cGAS DNA-sensing signaling in THP-1 cells. THP1 Lucia™ ISG cells were incubated with 1.25 µM or 2.5 µM TAT-ORF52 protein for 3 h, then transfected with ISD45 or Poly(I: C) (2µg/ml) (G and H) or infected with VACV or SEV (I). Analyses of luciferase activity or IRF3 dimerization were performed as in (D–F). **p<0.01 and ***p<0.001; Student’s t-test. See also Figures S1 and S2.
The human monocyte cell line THP-1 expresses most cytosolic DNA sensors and has been used extensively for their study (Sun et al., 2013; Wu et al., 2013; Zhang et al., 2011). We made use of the reporter cell line THP1 Lucia™ ISG (Invivogen), which expresses luciferase from a gene under the control of an IRF3-inducible ISG54 promoter, as a surrogate assay to measure induction of the innate immune response by nucleic acid sensors. We transduced these cells with a lentiviral vector expressing ORF52 from a doxycycline-inducible promoter or with control empty vector (pEasiLV). We found that expression of ORF52 reduced luciferase activity induced by interferon stimulatory DNA 45-mer (ISD45), but not by RNA Poly(I:C) (Figure 1D). ORF52 expression also reduced the dimerization and phosphorylation of IRF3, confirming its specific inhibition of DNA- but not RNA-induced activation of IRF3 (Figure 1E). Similarly, ORF52 also reduced luciferase activity induced by the DNA virus vaccinia (VACV), but not by the RNA virus Sendai (SEV) (Figure 1F), in a dose-dependent manner (Figure S2B). Furthermore, purified ORF52 protein directly transfected after fusion with TAT cell-penetrating peptide (Figures S2, C-E) also inhibited DNA- but not RNA-dependent responses, further confirming our previous results (Figures 1, G-I). Together, these data suggest that KSHV ORF52 selectively inhibits cGAS DNA-sensing signaling.
ORF52 inhibits cGAS enzymatic activity
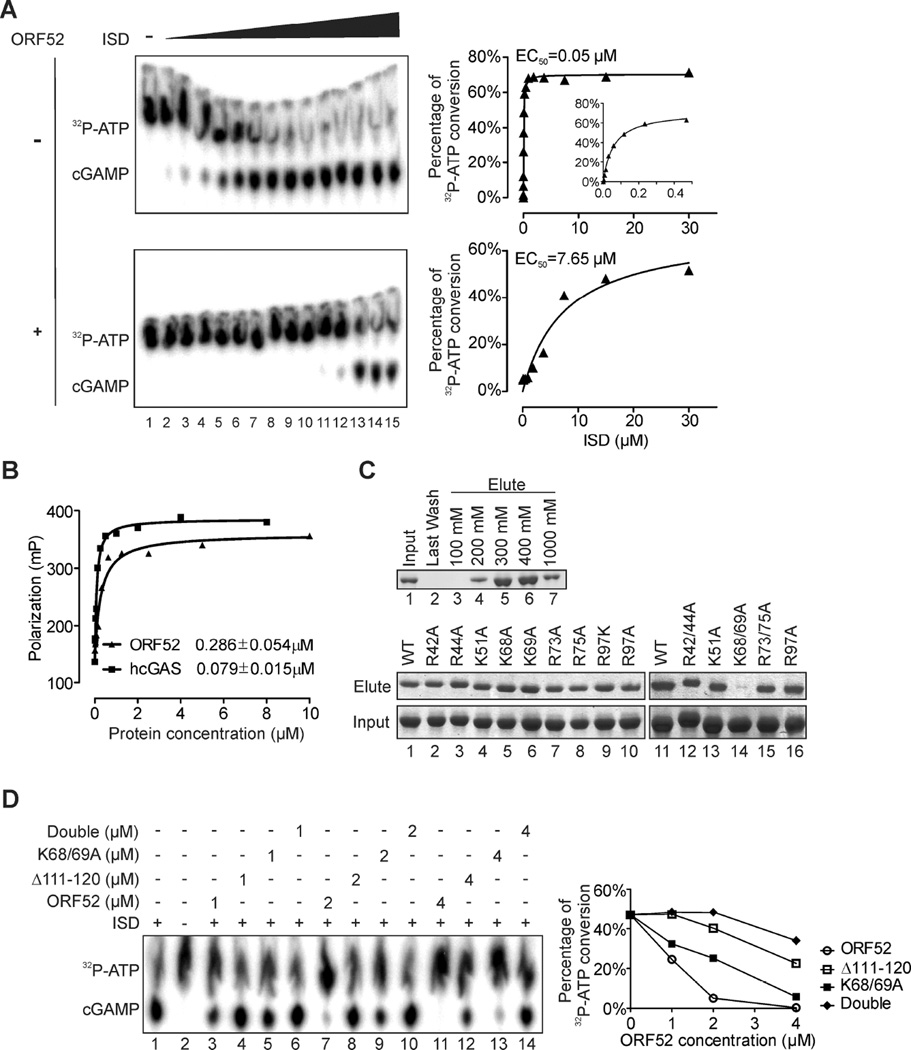
Although ORF52 inhibited DNA- and cGAS-triggered dimerization and phosphorylation of IRF3, it had no apparent effect on cGAMP-induced activation of IRF3 (Figures 2, A and B), suggesting that ORF52 inhibits cGAS directly. Because cGAS catalyzes synthesis of cGAMP upon binding to DNA, we examined the effect of ORF52 protein on cGAS enzymatic activity. We expressed human cGAS (hcGAS) and ORF52 in bacteria and purified them to homogeneity (Figure 2C). The purified hcGAS catalyzed the production of cGAMP only when ISD45 was included in the reaction. Approximately 70% of [α-32P]-ATP was converted to cGAMP in 2 hours under our assay conditions (Figure 2D). Inclusion of ORF52 in the reaction reduced production of cGAMP in a dose-dependent manner, from 40% at an ORF52:cGAS molar ratio of 1:1 to complete inhibition at a ratio of 4:1 (Figure 2, D and E). We found that ORF52 could also inhibit the enzymatic activity of mouse cGAS (mcGAS) in a dose-dependent manner (Figure 2F). These results indicate that ORF52 directly inhibits cGAS enzymatic activity, resulting in decreased cGAMP synthesis.
Figure 2. ORF52 inhibits cGAS enzymatic activity.
(A–B) ORF52 affects the cGAS-DNA-sensing signaling pathway upstream of cGAMP. THP1 Lucia™ ISG cells harboring pEasiLV-ORF52 or pEasiLV-empty were induced with 2 µg/ml doxycycline for 60 h, then transfected with ISD45 or cGAMP (2µg/ml). IRF3 dimerization was analyzed (A) or the secreted Lucia activity was determined (B) as in Figure. 1. (C) Coomassie blue staining of purified hcGAS and ORF52 proteins. (D–E) ORF52 inhibits hcGAS enzymatic activity. Purified hcGAS protein (1 µM) was incubated with ISD45 (1 µM), [α-32P]-ATP, ATP and GTP in reaction buffer containing different amount of ORF52 protein (0, 1, 2, or 4 µM). The reactions were stopped at the indicated time by boiling for 5 minutes. Synthesis of cGAMP was analyzed by thin-layer chromatography (TLC) (D), and the efficiency of cGAMP production was plotted against time (E). (F) ORF52 inhibits mouse cGAS activity. Mouse cGAS (mcGAS; 1 µM) was used for the enzyme assay and TLC as described in (D). ***p<0.001; Student’s t-test.
DNA binding is required for ORF52 to inhibit cGAS activity
Because cGAS activity depends on DNA, we performed assays with increasing concentration of ISD (up to a DNA:cGAS molar ratio of 30:1) in the presence or absence of ORF52 (ORF52:cGAS of 4:1). At low-intermediate concentrations of DNA, cGAS activity was completely inhibited by ORF52 (Figure 3A). However, at high concentrations of DNA (DNA:cGAS molar ratio of >4:1), the inhibition of cGAS activity by ORF52 could be partially overcome. To further quantify the effect of ORF52 on cGAS activity, we plotted the multiple turnover reaction rate at a given cGAS concentration against the concentration of DNA in order to obtain the EC50 value of DNA (the concentration of DNA at which cGAS reaches half its maximum activity). The EC50 was significantly greater when 4 µM ORF52 was present (7.65 µM) than when it was absent (0.05 µM) (Figure 3A), suggesting that ORF52 may sequester DNA from cGAS.
Figure 3. ORF52 binding to DNA is required for its inhibition of cGAS activity.
(A) ORF52 affects the kinetics of cGAS-dependent cGAMP production. In vitro enzyme assay was performed as in Figure 2D with 2-fold serial dilutions of ISD45 (starting from 30 µM, from right to left) in presence or absence of ORF52 (4 µM) for 2 h. cGAMP production was detected by TLC and the initial reaction velocity was plotted against substrate concentration together with least-square fit to the Michaelis-Menten equation. (B) Fluorescence polarization analysis of hcGAS or ORF52 binding to DNA. FAM-labeled ISD45 was incubated with different amounts of ORF52 or hcGAS protein, and the Kd values were obtained from least-square fit to a hyperbolic binding isotherm. (C) Mutagenesis of ORF52 reveals that K68/69 are critical for its binding to DNA. GST-ORF52 protein was pulled down by dsDNA cellulose beads, and then eluted with the indicated salt concentration (upper panel). The GST-ORF52 charge-deficient mutants were pulled down by dsDNA cellulose beads, eluted with 300 mM NaCl, separated by SDS-PAGE, and visualized by Coomassie staining. (D) ORF52 mutants deficient in cGAS/DNA-binding display reduced inhibition of cGAS activity. In vitro enzyme assay was performed in the presence or absence of the indicated amount of ORF52 wild-type or mutant proteins, and the percentage of cGAMP production was plotted for each protein concentration (right panel). See also Figure S3.
However, cGAS apparently binds to DNA more efficiently than ORF52, as revealed by fluorescence polarization DNA binding assay (Figure 3B), raising the question of whether ORF52 inhibits cGAS activity solely through DNA binding. To address the extent to which DNA sequestration contributes to cGAS inhibition, we screened ORF52 mutants deficient in DNA binding, using a DNA binding assay with dsDNA-coupled cellulose beads (Figure 3C upper panel). We found that one ORF52 mutant (K68/69A) exhibited substantially reduced DNA binding ability (Figure 3C lower panel, lane 14). We then examined inhibition of cGAS by ORF52-K68/69A and found that loss of DNA binding compromised inhibition, supporting the role of DNA binding by ORF52 in cGAS inhibition (Figure 3D).
ORF52 interacts with cGAS
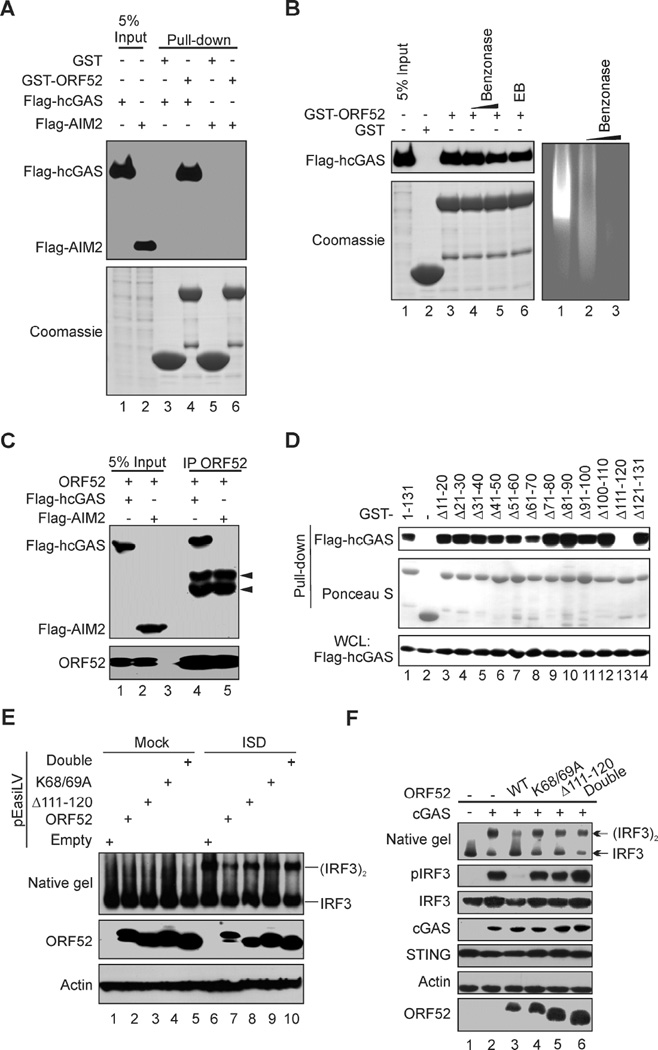
Although ORF52 inhibited DNA-induced cGAS-cGAMP signaling, it had little effect on DNA-induced inflammasome responses mediated by another cytosolic DNA sensor, AIM2 (Figure S3A and B) (Burckstummer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009). The specific inhibition of cGAS by ORF52 prompted us to investigate the possibility of an interaction between the two proteins. Indeed, we found that GST-ORF52 pulled down Flag-tagged hcGAS efficiently, but not Flag-AIM2 under the same conditions (Figure 4A). The interaction between ORF52 and cGAS was evidently not dependent on DNA or RNA, because treatment with Benzonase®, an enzyme that digests both DNA and RNA, or inclusion of ethidium bromide (EB) had little effect on it (Figure 4B). Importantly, ORF52 and cGAS also interact with each other in cells, as revealed by their co-immunoprecipitation (Figure 4C) and subcellular co-localization (Figure S3C). To further study their interactions, we mapped the interaction domains using a series of internal 10-aa deletion mutants of GST-ORF52 in GST pull-down assays and found that loss of aa111–120 in ORF52 (ORF52 Δ111–120) abolished its binding to cGAS (Figure 4D), which was confirmed by co-immunoprecipitation assays (Figure S3D). This deletion also compromised the inhibition of cGAS by ORF52 (Figure 3D). The K68/69A and Δ111–120 double mutant further reduced inhibition of cGAS (Figure 3D), supporting the idea that ORF52 blocks cGAS activity in part through their interaction. When expressed ectopically in THP-1 cells, ORF52 inhibited DNA-induced dimerization and phosphorylation of IRF3, but ORF52 mutants deficient in cGAS- or DNA-binding exhibited reduced inhibition (Figure 4E). Similar results were obtained in HEK293T/STING cells (Figure 4F), and in THP-1 cells transfected with TAT-ORF52 WT or mutant protein (Figures. S3, E-G). These results corroborate our in vitro assays, suggesting that optimal inhibition of cGAS activity requires both the cGAS-binding and DNA-binding properties of ORF52.
Figure 4. Interaction between ORF52 and cGAS underlies the specific inhibition of cGAS-mediated signaling.
(A) ORF52 binds to cGAS but not AIM2. Purified GST-ORF52 or GST proteins were used to pull down lysates of HEK293T cells transiently expressing Flag-hcGAS or Flag-hAIM2. The interaction was detected by western blot analysis using anti-FLAG antibody. (B) The interaction between ORF52 and hcGAS is not dependent on DNA. HEK293T cell lysate expressing Flag-hcGAS was untreated, or treated with Benzonase® (0.1 U/µl or 1 U/µl) or EB, for 2 h on ice, followed by pull-down assay with GST-ORF52 or GST protein. The interacted protein was detected with anti-FLAG antibody (left panel). The lysate DNA was separated by agarose gel electrophoresis and stained with EB (right panel). (C) ORF52 interacts with cGAS in vivo. Cells were transfected as indicated and lysates were immunoprecipitated with anti-ORF52 antibody, followed by western blot analysis with anti-FLAG and anti-ORF52 antibodies. Arrows indicate heavy chains of IgG. (D) Mapping the region of ORF52 required for its interaction with hcGAS. Serial 10-aa deletion mutants of GST-ORF52 were used to pull down lysates of HEK293T cells transiently expressing Flag-hcGAS. Interaction was detected by western blot analysis with anti-FLAG antibody, and comparable protein level was confirmed by Ponceau S. (E) THP1 Lucia™ ISG cells harboring ORF52-pEasiLV (WT or mutants) were mock treated or induced with doxycycline for 60 h, followed by mock transfection or transfection with ISD45. IRF3 dimerization was analyzed by native gel electrophoresis. (F) HEK293T/STING cells were transfected with expression plasmids of cGAS and ORF52 WT/mutants as indicated. Twenty-four h after transfection, cell lysates were analyzed for IRF3 dimerization by native gel electrophoresis. IRF3 phosphorylation and expression levels of the transfected genes were analyzed by western blot. See also Figures S3 and S4.
Interestingly, the cGAS-binding mutant, ORF52 Δ111–120, also shows weakened DNA binding activity similar to that of K68/69A (Kd of 2.23 vs 2.21 µM) (Figure S4A), making it difficult to distinguish the contributions of protein-protein and protein-DNA interactions from the loss of cGAS inhibition. We therefore mapped regions of cGAS involved in its interaction with ORF52. Our results showed that regions between aa1–160 and aa212–382 were involved in binding to ORF52 (Figure S4B). Consistently, we found that ORF52 inhibited the activity of the full-length cGAS more efficiently than the truncated form (aa161-522) (Figures S4C–E).
ORF52 homologues also inhibit cGAS
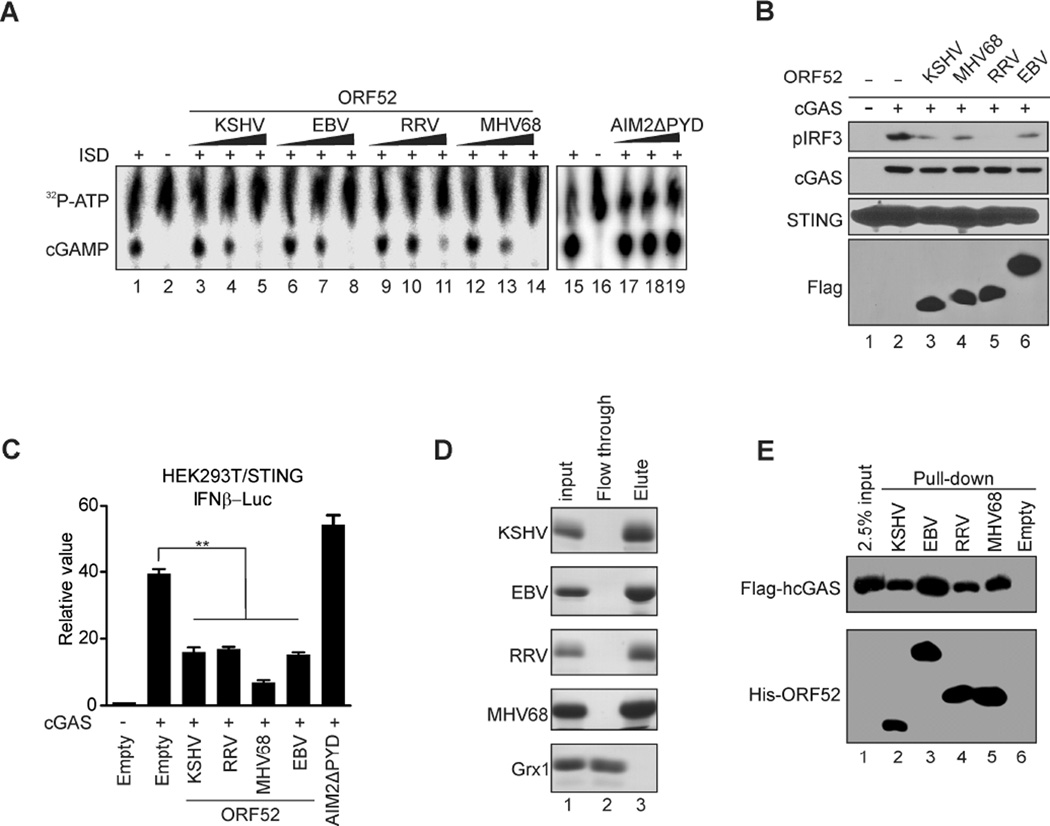
ORF52 is a gammaherpesvirus specific tegument protein (Bortz et al., 2003; Johannsen et al., 2004; O'Connor and Kedes, 2006; Zhu et al., 2005) and shares a high degree of conservation among them (Figure S5A). To investigate whether ORF52 homologues from other gammaherpesviruses also inhibit cGAS activity, we expressed them in bacteria and purified them to homogeneity (Figure S5B). Our in vitro enzymatic assay showed that these ORF52 homologues also inhibited cGAS activity (Figure 5A), and that they inhibited cGAS-dependent activation of IRF3 in cells (Figures 5, B and C). Moreover, these homologues all bound to DNA (Figure 5D and Figure S5C) and to cGAS (Figure 5E and Figure S5D) comparably, suggesting an evolutionarily-conserved mechanism for the inhibition of cGAS within gammaherpesviruses.
Figure 5. Homologues of ORF52 in other gammaherpesviruses also inhibit cGAS activity.
(A) ORF52 homologs but not AIM2 DNA binding domain (AIM2ΔPYD) can inhibit hcGAS activity. The enzyme assay and TLC were performed as in Figure 2D, but with ORF52 homologs or AIM2ΔPYD. (B–C) ORF52 homologs inhibit the cGAS-induced IFN response. Experiments were performed as described in Figure 1A and 1B, but with ORF52 homologs. (D) ORF52 homologs bind to dsDNA. dsDNA cellulose beads were used to pull down His-tagged ORF52 protein and glutaredoxin 1 (GRX1) proteins, then the beads were boiled in 2 × Laemmli loading buffer and the eluted proteins were analyzed by SDS-PAGE and Coomassie staining. (E) ORF52 homologs all bind to hcGAS. His-ORF52 proteins were used to pull down lysates of HEK293T cells expressing Flag-hcGAS, and the input/eluates were probed with anti-FLAG or anti-His antibody. See also Figure S5.
KSHV can elicit a cGAS DNA sensing response that is counteracted by ORF52
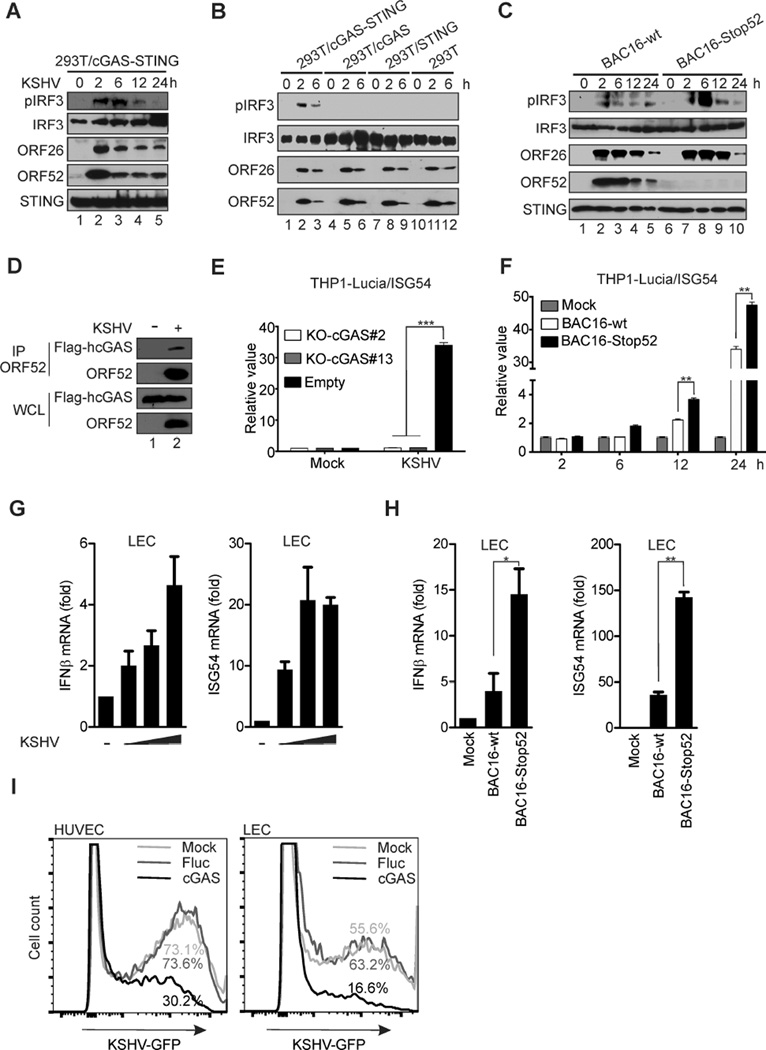
KSHV infection triggered the innate immune response in HEK293T cells reconstituted with cGAS and STING, as evidenced by increased pIRF3 signal (Figure 6A). Phosphorylation of IRF3 was most prominent at early times (2 and 6 h) but decreased at later times (12 and 24 h) post-infection (hpi). The phosphorylation was apparently dependent on the cGAS-STING DNA-sensing pathway, because the signal was not detected unless both cGAS and STING were expressed (Figure 6B). Our results are consistent with increasing evidence that herpesviral DNA can be sensed by cGAS (Sun et al., 2013; Li et al., 2013b; Yang et al., 2015). To determine the effect of ORF52 on sensing of KSHV by cGAS, we infected HEK293T/cGAS-STING cells with KSHV wild-type (BAC16-wt) or an ORF52-null mutant virus (BAC16-Stop52). As shown in Figure 6C, the ORF52-null virus induced a higher level of pIRF3 at 6 and 12 hpi, indicating a role of ORF52 in inhibiting the innate immune response to KSHV primary infection. The ORF52 protein delivered into cells from incoming virions apparently interacted with cGAS (Figure 6D), suggesting that ORF52 may interfere with cGAS during KSHV primary infection. When THP1 Lucia™ ISG cells were infected with KSHV, an increase of luciferase activity was observed, but this increase was abolished if cGAS was knocked out by CRISPR/Cas9-mediated genome editing (Figure 6E and Figure S6), providing evidence that KSHV is sensed by a cGAS-dependent pathway. Importantly, there was an additional increase of luciferase activity in cells infected with ORF52-null virus (Figure 6F), confirming a role of ORF52 in inhibition of cGAS-dependent responses during KSHV infection. We next used human primary lymphatic endothelial cells (LECs), because they are believed to be the target cells of KSHV and are highly permissive to KSHV infection. KSHV infection of LECs caused a noticeable increase in IFNβ and ISG54 mRNA levels at 6 hpi, (Figure 6G). In agreement with a role of ORF52 in inhibition of cGAS-DNA sensing, a more dramatic increase of IFNβ and ISG54 mRNA levels was observed in LECs infected with BAC16-Stop52 virus (Figure 6H). To determine whether activation of cGAS inhibits KSHV infection, we adopted a strategy that has been used by Dr. Charles Rice’s group to systematically examine the antiviral effect of hundreds of ISGs (Schoggins et al., 2011; Schoggins et al., 2014). Among those ISGs, cGAS was found to exert an antiviral effect on a variety of DNA and RNA viruses, including MHV68. We transduced LECs with either cGAS-expressing or control luciferase-expressing lentivirus, then infected them with GFP-labeled KSHV virions. As shown in Figure 6I, we found that ectopic expression of cGAS suppressed the permissiveness of primary endothelial cells to KSHV infection, indicating that cGAS exerts a potent antiviral effect on KSHV. Together, these experiments demonstrate that KSHV triggers a cGAS-dependent immune response during primary infection, and that ORF52 plays a critical role in inhibiting this response.
Figure 6. KSHV ORF52 antagonizes cGAS-dependent induction of the innate immune response to primary infection.
(A) HEK293T/cGAS-STING cells were infected by KSHV. At the indicated time after infection, cell lysates were analyzed by western blotting with the indicated antibodies. (B) KSHV primary infection and analysis of the indicated cell lines was performed as in (A). (C) HEK293T/cGAS-STING cells were infected by KSHV BAC16-wt or BAC16-Stop52 mutant. At the indicated time after infection, cell lysates were analyzed by western blotting with the indicated antibodies. (D) Flag-cGAS/293T cells were infected with KSHV at 50 genome copies/cell for 30 min, then cell lysates were prepared and immunoprecipitated with anti-ORF52 4H4 antibody. The immunocomplexes were analyzed by western blot with indicated antibodies. (E) cGAS was knocked out by CRISPR/Cas9-mediated genome editing in THP1 Lucia™ ISG cells. The knockout cell line and control were infected by KSHV, and the activity of secreted Lucia luciferase was assessed at 24 hpi. (F) THP-1 Lucia™ ISG cells were infected by KSHV BAC16-wt or BAC16-Stop52 mutant. Luciferase activity was measured as in (E). (G) LECs were mock treated or infected by KSHV at 25, 50, or 100 genome copies/cell. RNAs were isolated from cells and reverse transcribed. The levels of IFNβ and ISG54 mRNA were measured by qRT-PCR at 6 hpi. (H) LECs were infected by KSHV BAC16-wt or BAC16-Stop52 at 50 copies/cell. The levels of IFNβ and ISG54 mRNA were measured as in (G). (I) HUVECs or LECs were transduced with lentiviruses as indicated. At 24 h after transduction, they were infected with BAC16-wt, followed by fixation and FACS analyses of the GFP-positive cells the next day. *p<0.05 and **p<0.01; Student’s t-test. See also Figure S6.
DISCUSSION
The genomic DNA of herpesviruses accounts for about 10%–20% of virion mass (Heine et al., 1974; Nealon et al., 2001), constituting a significant PAMP within the incoming virions. Although herpesviruses deliver and replicate their genome in the nucleus, the incoming DNA could be recognized by cGAS (Horan et al., 2013; Orzalli and Knipe, 2014; Paludan et al., 2011; Sun et al., 2013; Yang et al., 2015). Stimulation of the cGAS-cGAMP DNA-sensing pathway results in activation of IRF3 and production of IFNs not only in infected cells, but also in neighboring cells, causing repression of viral replication, elimination of virus-infected cells by apoptosis, and subsequent activation of adaptive immune responses. Thus, it is intuitive that herpesviruses should possess mechanisms to prevent the sensing of their DNA. This would be especially true for KSHV, because periodic lytic replication and de novo infection occur naturally and are required for long-term persistence and pathogenesis in its hosts. Indeed, a screening of KSHV ORFs revealed that eight proteins substantially inhibited cGAS-STING-dependent DNA-sensing signaling. Notably, five of these proteins, ORF36, ORF45, ORF52, ORF55, and ORF64, have been shown or predicted to be tegument proteins (Bortz et al., 2003; Johannsen et al., 2004; O'Connor and Kedes, 2006; Zhu et al., 2005). Although these viral proteins could be involved in other aspects of immune evasion, the presence of multiple antagonists of the cGAS-STING pathway attests to the importance of preventing recognition of virion-contained PAMPs, including viral DNA. This also supports our previous observation that KSHV infection induced only a mild antiviral effect in human primary fibroblast cells, because the KSHV virion contains potent antagonists of host innate antiviral immune responses (Zhu et al., 2010).
We demonstrated that ORF52 bound to DNA and to cGAS, inhibited cGAS-STING-dependent activation of IRF3 in cells, and, most importantly, that ORF52 directly inhibited cGAS enzymatic activity. Although ORF52 binds to DNA, no known DNA binding domain can be recognized. Unlike most DNA-binding proteins that are usually localized in the nucleus, ORF52 is found exclusively in the cytoplasm (Figure S3C and Sander et al., 2008), a feature that might play an important role in the inhibition of cGAS. Although the DNA-binding capacity of ORF52 is required for inhibition of cGAS, it is apparently insufficient because OFR52 binds to DNA with less affinity than cGAS. Importantly, we found ORF52 also bound to cGAS but not to another cytosolic DNA sensor, AIM2. This difference explains why ORF52 inhibits cGAS-dependent activation of IRF3 but not AIM2-dependent inflammasome responses. The fact the AIM2 DNA binding domain cannot inhibit cGAS either in vitro or in vivo reaffirms that simple sequestration of DNA appears not to be an effective mechanism of inhibiting cGAS. Interestingly, ORF52 binds to both the catalytic domains aa212–382 and aa1–160, which appears to be a regulatory domain of cGAS (Sun et al., 2013). Unfortunately, because the DNA- and cGAS-binding properties of ORF52 are seemingly inseparable, the extent to which they contribute to the inhibition of cGAS is unknown. Nevertheless, our data suggest both DNA-binding and cGAS-binding are required for inhibition of cGAS by ORF52. Further structural studies of ORF52 interaction with cGAS and/or DNA will be crucial for delineation of the underlying molecular mechanisms.
Although ORF52 efficiently inhibits cGAS activity, ORF52-null mutant viruses only resulted in a moderate increase in IRF3-depedent responses. This is not surprising because KSHV is known to encode multiple genes of immune modulatory functions, and because our initial screening found at least eight ORFs that interfered with the cGAS-STING signaling pathway. Some of these have been shown to subvert innate immunity by means of diverse mechanisms. But, unlike ORF52 that targets cGAS directly, these KSHV proteins appear to interfere with downstream signaling components of cGAS, including IRFs. For example, K9 (vIRF-1), ORF36, and ORF45 all interfere with activation of IRFs (Gao et al., 1997; Hwang et al., 2009; Liang et al., 2012; Lin et al., 2001; Sathish and Yuan, 2011; Zhu et al., 2002); ORF64, a viral deubiquitinase (DUB), interferes with ubiquitination of the key components of innate immune signaling pathways (Inn et al., 2011; Wang et al., 2013). Because many viral immune-modulatory proteins are known to interfere with different aspects of immune responses, we expect that further studies will reveal additional roles and mechanisms by which these KSHV proteins subvert the host antiviral immune responses. The rich presence of antagonists of pattern recognition receptors (PRR) provides an ideal mechanism to suppress activation of sensing of virion-contained PAMPs, thus blocking the up-regulation of the next wave of ISGs.
Although ORF52 is known as an abundant tegument protein in virions (Bortz et al., 2007; Johannsen et al., 2004; O'Connor and Kedes, 2006; Zhu et al., 2005), its precise function has remained elusive (Anderson et al., 2014; Bortz et al., 2007; Wang et al., 2012). It is conserved among gammaherpesviruses but not in alpha or beta herpesviruses. Significantly, we found that ORF52 homologues also bound to cGAS and DNA and inhibited cGAS activity directly, suggesting that inhibition of cGAS by virion-contained ORF52 is a conserved mechanism for gammaherpesviruses.
Viral inhibitors of RNA sensing have been extensively studied, and several viral strategies to avoid dsRNA-mediated activation of RNA sensors have been characterized (Gack, 2014; Yoneyama et al., 2015; Zinzula and Tramontano, 2013). We show here that ORF52 is a viral inhibitor of the principal cytosolic DNA sensor, cGAS. We propose to name it KSHV inhibitor of cGAS (KicGAS). A growing number of viruses and bacteria have been shown to be sensed by cGAS (Dai et al., 2014; Gao et al., 2013a; Lam et al., 2014; Li et al., 2013b Collins et al., 2015; Hansen et al., 2014; Storek et al., 2015; Wassermann et al., 2015; Watson et al., 2015). Among hundreds of ISGs evaluated by Dr. Charles Rice’s group, cGAS is one of the few that exhibit potent antiviral effects on diverse groups of both DNA and RNA viruses (Schoggins et al., 2011; Schoggins et al., 2014). Viruses other than gammaherpesviruses are conceivably expected to encode cGAS antagonists. A recent study has revealed that the HSV-1 VP22 core domain has limited structural and sequence homology to MHV68 ORF52 (Hew et al., 2015). Because VP22 is also an abundant tegument protein (Spear and Roizman, 1972), it would be interesting to examine whether VP22 can inhibit cGAS, and how other large DNA viruses, such as alpha and beta herpesviruses and poxviruses, overcome cGAS-dependent DNA sensing. Collectively, our results have provided insights into the study of viral strategies to antagonize cGAS DNA-sensing signaling. The sensing of self and foreign DNA is delicately regulated (Ablasser et al., 2014; Konno et al., 2013; Liang et al., 2014), and the dysregulation of DNA sensing is associated with several human autoimmune diseases, such as systemic lupus erythematosus and Aicardi-Goutières syndrome (Ahn and Barber, 2014). Therefore, further delineation of the mechanisms by which KicGAS inhibits cGAS will aid in our understanding of this pathway, and may facilitate the development of therapeutics to treat or prevent diseases in which this pathway is dysregulated.
EXPERIMENTAL PROCEDURES
Cell culture and transfection
HEK293T and HeLa cells were cultured under 5% CO2 at 37 °C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics. iSLK-puro cells carrying BAC16 wild type or mutants were cultured in DMEM containing 10% FBS, 50 µg/ml G418, 1 µg/ml puromycin and 500 µg/ml hygromycin (Brulois et al., 2012; Myoung and Ganem, 2011). Conditions for induction of lytic reactivation in these cells were described previously (Fu et al., 2015). THP-1 and THP1 Lucia™ ISG cells (InvivoGen) were cultured in RPMI containing 10% FBS, and antibiotics. LECs and HUVECs were cultured in EBM-2 media supplemented with the EGM-2 MV bullet in collagen type I-coated dishes. Transient transfections were performed with Lipofectamine® 2000 according to the manufacturer’s protocol, or with polyethylenimine (PEI), or calcium chloride using standard protocols.
Luciferase reporter assays
HEK293T/STING cells seeded on 24-well plates were transiently transfected with 100 ng of the luciferase reporter plasmid together with a total of 300 ng of various expression plasmids and/or empty vector controls using Effectene Transfection Reagent (Qiagen). As an internal control, 10 ng of pRL-TK (Renilla luciferase) was transfected simultaneously. Luciferase assays were then performed at 24 h post-transfection according to the Promega Dual-Luciferase® Reporter Assay System protocol. The relative luciferase activity was expressed as arbitrary units by normalizing firefly to Renilla luciferase activity.
Induction of immune responses by nucleic acid transfection and virus infection
HEK293T or SLK cells were mock treated or transfected with ISD or Poly(I:C) (2 µg/ml). THP1-Lucia™ ISG cells carrying ORF52-pEasiLV or Empty-pEasiLV were untreated or treated with doxycycline (2 µg/ml) for 60 h, then seeded to new wells and either mock treated or transfected with ISD/Poly(I:C) (2 µg/ml) or cGAMP (2 µg/ml; Figure. 2A–B). Six hours post-transfection, cell lysates were harvested and analyzed by native PAGE (to detect IRF3 dimerization) or western blot. Alternatively, the media was collected at 16 h post-transfection and the activity of secreted Lucia luciferase was determined by addition of QUANTI-Luc™ (Invivogen) and analysis with a luminometer. To detect the immune response to virus infection, HEK293T cells were mock infected or infected with VACV, or SEV. THP1-Lucia cells carrying ORF52-pEasiLV or Empty-pEasiLV were mock treated or treated with doxycycline, then infected with VACV or SEV. Analysis of IRF3 activation and luciferase assay were performed as described above.
Protein purification
TAT-ORF52 protein purification was performed according to Dr. Dowdy’s group (Becker-Hapak and Dowdy, 2003). Further details, including methods used to purify His-tagged cGAS and ORF52, can be found in the supplemental information.
TAT-ORF52 protein transduction
THP1-Lucia cells were incubated with TAT-ORF52 proteins (1.25 or 2.5 µM) in serum-free media. After 3 h, cells were washed with complete media (RPMI + 10% FBS), then either transfected with ISD/Poly(I:C) or infected with VACV/SEV virus as described above. Delivery of protein into cells was confirmed by immunofluorescence staining using anti-ORF52 antibody.
GST pull-down assay
GST-ORF52 or mutant proteins were bound to glutathione agarose beads, and incubated for 3 h with lysates from HEK293T cells transiently expressing Flag-cGAS or Flag-AIM2. The beads were washed three times each with whole-cell lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM sodium orthovanadate [Na3VO4], 40 mM-glycerophosphate, 1mM sodium fluoride, 10% glycerol, 5 mM EDTA, 5 µg/ml of aprotinin, 5 µg/ml of leupeptin, 5 mM benzamidine, and 1 mM PMSF) and 1× PBS, then mixed with an equal volume of 2 × Laemmli loading buffer, and boiled for 10 minutes. The input/eluates were resolved by SDS-PAGE and analyzed by Coomassie staining and/or western blot.
In vitro enzyme assay
cGAS protein was incubated with ISD45, 100 µM ATP, 100 µM GTP, α32P-ATP (10 µCi), and the indicated amount of ORF52 protein in reaction buffer at 37 °C for the indicated time. The reaction was terminated by boiling for 5 minutes. One µL of each sample was applied to a PEI-Cellulose F thin-layer chromatography plate. Reaction products were resolved with 1 M (NH4)2SO4/1.5 M KH2PO4 (pH 3.8). Plates were dried at 80°C for 10 min, and radiolabeled products were detected by a Storm phosphorimager (GE Life Sciences). The signal was analyzed with ImageQuant and the data were fitted to a Michaelis-Menten-like equation: Y = Vmax*[ISD45]/( EC50 + [ISD45]). The EC50 value of DNA determined in this manner indicated the concentration of DNA at which cGAS reaches its half maximum activity. Detailed experimental procedures are shown in the Supplemental Information.
Double strand DNA cellulose pull-down assay
dsDNA cellulose (Sigma, D8515) was incubated with purified GST-ORF52 or mutant proteins in buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM sodium orthovanadate [Na3VO4], 1× protease inhibitor cocktail, and 1 mM PMSF) for 1h at 4°C, then washed 3 times with the same buffer. The protein bound beads were eluted step-wise with 10 mM Tris-HCl (pH 7.4) buffer containing different concentrations of NaCl (Figure 3C, upper panel),buffer containing 300 mM NaCl (Figure 3C, lower panel), or with 2 × Laemmli loading buffer (Figure. 5D). The input and eluted proteins were then applied to SDS-PAGE and stained by Coomassie blue.
Virus stock preparation and infection
For KSHV, virus stock preparation, quantification, and infection were performed as described previously (Fu et al., 2015). SEV was used at a final concentration of 50 HA units/ml. VACV was inactivated at 56°C for 30 minutes before usage and was used at an MOI of 10. For vesicular stomatitis virus infection, VSV-GFP was used at an MOI of 1. Cells were analyzed by FACS at 48 h post-infection. Further details can be found in the Supplemental Information
Quantitative real-time PCR analysis
LECs were mock treated or infected with KSHV for 6 h. The mRNA was extracted, reverse transcribed, and mRNA levels were determined by real time PCR. Expression levels were normalized to GAPDH expression and data are presented as fold induction over mock treatment control. More details can be found in the Supplemental Information.
CRISPR/Cas9-mediated genome editing
Potential guide RNAs (gRNAs) targeting the first exon of cGAS were analyzed using the CRISPR Design tool (crispr.mit.edu). Double-stranded oligos were cloned into the lentiCRISPRv1 vector and co-transfected with packaging plasmids into HEK293T cells. Lentiviral particles were collected and used to transduce THP1 Lucia™ ISG cells. #2 and #13 represent two single cell clones with efficient cGAS knockout. More details regarding generation and validation of these cell lines can be found in the Supplemental Information.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Kevin Brulois, Katherine Fitzgerald, Adolfo García-Sastre, David Gilbert, Young-Kwon Hong, Pingwei Li, Michael Malim, Jinjong Myoung, Charles Rice, Ren Sun, Hengli Tang, Scott Wong, Feng Zhang for reagents. We thank the Florida State University hybridoma facility for generating anti-ORF52 antibodies. We thank Dr. Lijun Sun for his helpful advice regarding the IRF3 dimerization assay. We also thank Ms. Jen Kennedy at the Florida State University for editorial assistance. We thank Drs. Betty Gaffney, Kenneth Roux, Beth Stroupe, Hengli Tang, and all members of the Zhu lab for helpful comments and discussion. This work was supported by NIH grants R01 DE016680 (to F. Z.), F31 CA183250 (to D. A.), R01 GM099604 and R01 GM66958 (to H. L.) and in part by the National Cancer Institute, NIH (contract number HHSN261200800001E).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: J.W., W.L., and F.Z. designed the experiments and analyzed the data. J.W., W.L., Y.S., D.A., F.B., J.G., T.H., S.M., and X. L. performed the experiments. J.W., W.L., D.A., H.L., and F.Z. wrote the paper. A.K., F.N., M.S., W.M., and D.W provided some reagents.
REFERENCES
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner K-P, Ludwig J, Hornung V. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013a;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 Deficiency Triggers Cell-Autonomous Immunity in a cGAS-Dependent Manner. J Immunol. 2014;192:5993–5997. doi: 10.4049/jimmunol.1400737. [DOI] [PubMed] [Google Scholar]
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013b;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Barber GN. Self-DNA, STING-dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol. 2014;31C:121–126. doi: 10.1016/j.coi.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Anderson MS, Loftus MS, Kedes DH. Maturation and Vesicle-Mediated Egress of Primate Gammaherpesvirus Rhesus Monkey Rhadinovirus Require Inner Tegument Protein ORF52. J Virol. 2014;88:9111–9128. doi: 10.1128/JVI.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35:88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- Becker-Hapak M, Dowdy SF. Protein transduction: generation of full-length transducible proteins using the TAT system. Curr Protoc Cell Biol. 2003;18:20.2.1–20.2.25. doi: 10.1002/0471143030.cb2002s18. [DOI] [PubMed] [Google Scholar]
- Bortz E, Wang L, Jia Q, Wu T-T, Whitelegge JP, Deng H, Zhou ZH, Sun R. Murine Gammaherpesvirus 68 ORF52 Encodes a Tegument Protein Required for Virion Morphogenesis in the Cytoplasm. J Virol. 2007;81:10137–10150. doi: 10.1128/JVI.01233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortz E, Whitelegge JP, Jia Q, Zhou ZH, Stewart JP, Wu T-T, Sun R. Identification of Proteins Associated with Murine Gammaherpesvirus 68 Virions. J Virol. 2003;77:13425–13432. doi: 10.1128/JVI.77.24.13425-13432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brulois KF, Chang H, Lee AS-Y, Ensser A, Wong L-Y, Toth Z, Lee SH, Lee H-R, Myoung J, Ganem D, et al. Construction and Manipulation of a New Kaposi's Sarcoma-Associated Herpesvirus Bacterial Artificial Chromosome Clone. J Virol. 2012;86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Cai X, Chiu YH, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's Sarcoma-Associated Herpesvirus-Like DNA Sequences in AIDS-Related Body-Cavity-Based Lymphomas. New Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner K-P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AC, Cai H, Li T, Franco LH, Li XD, Nair VR, Scharn CR, Stamm CE, Levine B, Chen ZJ, et al. Cyclic GMP-AMP Synthase Is an Innate Immune DNA Sensor for Mycobacterium tuberculosis. Cell Host & Microbe. 2015;17:820–828. doi: 10.1016/j.chom.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai P, Wang W, Cao H, Avogadri F, Dai L, Drexler I, Joyce JA, Li X-D, Chen Z, Merghoub T, et al. Modified Vaccinia Virus Ankara Triggers Type I IFN Production in Murine Conventional Dendritic Cells via a cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway. PLoS Pathog. 2014;10:e1003989. doi: 10.1371/journal.ppat.1003989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu J-W, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu B, Kuang E, Li W, Avey D, Li X, Turpin Z, Valdes A, Brulois K, Myoung J, Zhu F. Activation of p90 ribosomal S6 kinases by ORF45 of Kaposi's sarcoma-associated herpesvirus is critical for optimal production of infectious viruses. J Virol. 2015;89:195–207. doi: 10.1128/JVI.01937-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol. 2014;88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem D. Kaposi's sarcoma-associated herpesvirus. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2007. pp. 2847–2888. [Google Scholar]
- Ganem D. KSHV and the pathogenesis of Kaposi sarcoma: listening to human biology and medicine. J Clin Invest. 2010;120:939–949. doi: 10.1172/JCI40567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Wu J, Wu Y-T, Du F, Aroh C, Yan N, Sun L, Chen ZJ. Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science. 2013a;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney Barbara L, Zillinger T, Serganov Artem A, Liu Y, Jones Roger A, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013b;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov Artem A, Gaffney Barbara L, Shuman S, Jones Roger A, Deng L, et al. Structure-Function Analysis of STING Activation by c[G(2′,5′)pA(3′,5′)p] and Targeting by Antiviral DMXAA. Cell. 2013c;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Boshoff C, Jayachandra S, Weiss RA, Chang Y, Moore PS. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene. 1997;15:1979–1985. doi: 10.1038/sj.onc.1201571. [DOI] [PubMed] [Google Scholar]
- Hansen K, Prabakaran T, Laustsen A, Jørgensen SE, Rahbæk SH, Jensen SB, Nielsen R, Leber JH, Decker T, Horan KA, et al. Listeria monocytogenes induces IFNβ expression through an IFI16, cGAS and STING dependent pathway. The EMBO Journal. 2014;33:1654–1666. doi: 10.15252/embj.201488029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine JW, Honess RW, Cassai E, Roizman B. Proteins Specified by Herpes Simplex Virus XII. The Virion Polypeptides of Type 1 Strains. J Virol. 1974;14:640–651. doi: 10.1128/jvi.14.3.640-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hew K, Dahlroth SL, Pan LX, Cornvik T, Nordlund P. The core domain of VP22 from herpes simplex virus 1 reveals a surprising structural conservation in both the alpha and the gamma herpesvirus subfamilies. J Gen Virol. 2015 doi: 10.1099/vir.0.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan KA, Hansen K, Jakobsen MR, Holm CK, Soby S, Unterholzner L, Thompson M, West JA, Iversen MB, Rasmussen SB, et al. Proteasomal degradation of herpes simplex virus capsids in macrophages releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Kim KS, Flano E, Wu T-T, Tong LM, Park AN, Song MJ, Sanchez DJ, O'Connell RM, Cheng G, et al. Conserved Herpesviral Kinase Promotes Viral Persistence by Inhibiting the IRF-3-Mediated Type I Interferon Response. Cell Host & Microbe. 2009;5:166–178. doi: 10.1016/j.chom.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn K-S, Lee S-H, Rathbun JY, Wong L-Y, Toth Z, Machida K, Ou J-HJ, Jung JU. Inhibition of RIG-I-Mediated Signaling by Kaposi's Sarcoma-Associated Herpesvirus-Encoded Deubiquitinase ORF64. J Virol. 2011;85:10899–10904. doi: 10.1128/JVI.00690-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. Proteins of purified Epstein-Barr virus. Proc Natl Acad Sci USA. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN. Cyclic Dinucleotides Trigger ULK1 (ATG1) Phosphorylation of STING to Prevent Sustained Innate Immune Signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell Reports. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Stein S, Falck-Pedersen E. Adenovirus Detection by the cGAS/STING/TBK1 DNA Sensing Cascade. J Virol. 2014;88:974–981. doi: 10.1128/JVI.02702-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton Catherine T, Shelton Catherine L, Diao J, Zuo X, Kao CC, Herr Andrew B, Li P. Cyclic GMP-AMP Synthase Is Activated by Double-Stranded DNA-Induced Oligomerization. Immunity. 2013a;39:1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal Roles of cGAS-cGAMP Signaling in Antiviral Defense and Immune Adjuvant Effects. Science. 2013b;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Fu B, Wu F, Li X, Yuan Y, Zhu F. ORF45 of Kaposi's Sarcoma-Associated Herpesvirus Inhibits Phosphorylation of Interferon Regulatory Factor 7 by IKKε and TBK1 as an Alternative Substrate. J Virol. 2012;86:10162–10172. doi: 10.1128/JVI.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Seo Gil J, Choi Youn J, Kwak M-J, Ge J, Rodgers Mary A, Shi M, Leslie Benjamin J, Hopfner K-P, Ha T, et al. Crosstalk between the cGAS DNA Sensor and Beclin-1 Autophagy Protein Shapes Innate Antimicrobial Immune Responses. Cell Host & Microbe. 2014;15:228–238. doi: 10.1016/j.chom.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ, Jr, Barber GN, Hiscott J. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene. 2001;20:800–811. doi: 10.1038/sj.onc.1204163. [DOI] [PubMed] [Google Scholar]
- Myoung J, Ganem D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J Virol Methods. 2011;174:12–21. doi: 10.1016/j.jviromet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealon K, Newcomb WW, Pray TR, Craik CS, Brown JC, Kedes DH. Lytic Replication of Kaposi's Sarcoma-Associated Herpesvirus Results in the Formation of Multiple Capsid Species: Isolation and Molecular Characterization of A, B, and C Capsids from a Gammaherpesvirus. J Virol. 2001;75:2866–2878. doi: 10.1128/JVI.75.6.2866-2878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor CM, Kedes DH. Mass Spectrometric Analyses of Purified Rhesus Monkey Rhadinovirus Reveal 33 Virion-Associated Proteins. J Virol. 2006;80:1574–1583. doi: 10.1128/JVI.80.3.1574-1583.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Knipe DM. Cellular Sensing of Viral DNA and Viral Evasion Mechanisms. Annu Rev Microbiol. 2014;68:477–492. doi: 10.1146/annurev-micro-091313-103409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat Rev Immunol. 2011;11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander G, Konrad A, Thurau M, Wies E, Leubert R, Kremmer E, Dinkel H, Schulz T, Neipel F, Stürzl M. Intracellular Localization Map of Human Herpesvirus 8 Proteins. J Virol. 2008;82:1908–1922. doi: 10.1128/JVI.01716-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathish N, Yuan Y. Evasion and Subversion of Interferon-Mediated Antiviral Immunity by Kaposi’s Sarcoma-Associated Herpesvirus: an Overview. J Virol. 2011;85:10934–10944. doi: 10.1128/JVI.00687-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, Agay MF, Clauvel JP, Raphael M, Degos L, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- Spear PG, Roizman B. Proteins Specified by Herpes Simplex Virus V. Purification and Structural Proteins of the Herpesvirion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storek KM, Gertsvolf NA, Ohlson MB, Monack DM. cGAS and Ifi204 Cooperate To Produce Type I IFNs in Response to Francisella Infection. The Journal of Immunology. 2015 doi: 10.4049/jimmunol.1402764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING Specifies IRF3 Phosphorylation by TBK1 in the Cytosolic DNA Signaling Pathway. Science Signaling. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo H, Reyes N, Lee S, Bortz E, Guo F, Sun R, Tong L, Deng H. Distinct Domains in ORF52 Tegument Protein Mediate Essential Functions in Murine Gammaherpesvirus 68 Virion Tegumentation and Secondary Envelopment. J Virol. 2012;86:1348–1357. doi: 10.1128/JVI.05497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang K, Li J, Zheng C. Herpes Simplex Virus 1 Ubiquitin-Specific Protease UL36 Inhibits Beta Interferon Production by Deubiquitinating TRAF3. J Virol. 2013;87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassermann R, Gulen Muhammet F, Sala C, Perin Sonia G, Lou Y, Rybniker J, Schmid-Burgk Jonathan L, Schmidt T, Hornung V, Cole Stewart T, et al. Mycobacterium tuberculosis Differentially Activates cGAS- and Inflammasome-Dependent Intracellular Immune Responses through ESX-1. Cell Host & Microbe. 2015;17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- Watson Robert O, Bell Samantha L, MacDuff Donna A, Kimmey Jacqueline M, Diner Elie J, Olivas J, Vance Russell E, Stallings Christina L, Virgin Herbert W, Cox Jeffery S. The Cytosolic Sensor cGAS Detects Mycobacterium tuberculosis DNA to Induce Type I Interferons and Activate Autophagy. Cell Host & Microbe. 2015;17:811–819. doi: 10.1016/j.chom.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Wang J, Wu M, Li M, Wang Y, Huang X. Mesenchymal stem cells detect and defend against gammaherpesvirus infection via the cGAS-STING pathway. Sci Rep. 2015;5:7820. doi: 10.1038/srep07820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32C:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wu J, Du F, Xu H, Sun L, Chen Z, Brautigam Chad A, Zhang X, Chen Zhijian J. The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop. Cell Reports. 2014;6:421–430. doi: 10.1016/j.celrep.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu Y-J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Chong JM, Wu L, Yuan Y. Virion Proteins of Kaposi’s Sarcoma-Associated Herpesvirus. J Virol. 2005;79:800–811. doi: 10.1128/JVI.79.2.800-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A Kaposi’s sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci USA. 2002;99:5573–5578. doi: 10.1073/pnas.082420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu FX, Sathish N, Yuan Y. Antagonism of Host Antiviral Responses by Kaposi’s Sarcoma-Associated Herpesvirus Tegument Protein ORF45. PLoS ONE. 2010;5:e10573. doi: 10.1371/journal.pone.0010573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzula L, Tramontano E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: hide, mask, hit. Antiviral Res. 2013;100:615–635. doi: 10.1016/j.antiviral.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.