Abstract
The plant hormone jasmonate (JA) plays crucial roles in regulating plant responses to herbivorous insects and microbial pathogens and is an important regulator of plant growth and development1–7. Key mediators of JA signaling include MYC transcription factors, which are repressed by JAZ transcriptional repressors at the resting state. In the presence of active JA, JAZ proteins function as JA co-receptors by forming a hormone-dependent complex with COI1, the F-box subunit of an SCF-type ubiquitin E3 ligase8–11. The hormone-dependent formation of the COI1–JAZ co-receptor complex leads to ubiquitination and proteasome-dependent degradation of JAZ repressors and release of MYC proteins from transcriptional repression3,10,12. The mechanism by which JAZ proteins repress MYC transcription factors and how JAZ proteins switch between the repressor function in the absence of hormone and the co-receptor function in the presence of hormone remain enigmatic. Here we show that Arabidopsis MYC3 undergoes pronounced conformational changes when bound to the conserved Jas motif of the JAZ9 repressor. The Jas motif, previously shown to bind to hormone as a partially unwound helix, forms a complete α-helix that displaces the N-terminal helix of MYC3 and becomes an integral part of the MYC N-terminal fold. In this position, the Jas helix competitively inhibits MYC3 interaction with the MED25 subunit of the transcriptional Mediator complex. Our study elucidates a novel molecular switch mechanism that governs the repression and activation of a major plant hormone pathway.
To understand the structural basis of the interactions between MYC transcription factors and JAZ repressors, we first used yeast two-hybrid assays to determine the JAZ-binding regions within MYC2, MYC3, and MYC4. A conserved ~200 amino acid (aa 55-259, aa 44-234 and aa 55-253 in MYC2, MYC3 and MYC4, respectively) region within the N-termini of all three proteins that encompasses the previously defined JAZ-interacting domain (JID)13,14 and the transcription activation domain (TAD)13,15 was sufficient to interact with JAZ9 (Extended Data Fig. 1a, 2a). Similarly, we identified a 17 amino acid region within the Jas motif of JAZ9 (polyA-Jas) that is required and sufficient to interact with MYC3 (Extended Data Fig. 1b). Interestingly, this Jas motif shares the same segment of JAZ proteins that interacts with COI116, but is four amino acids shorter at the N-terminus (Extended Data Fig. 1c). We confirmed these results using AlphaScreen luminescence proximity assays with His6-tagged MYC proteins and biotinylated JAZ8, JAZ9, and JAZ12 peptides (Extended Data Fig. 1d, 2b).
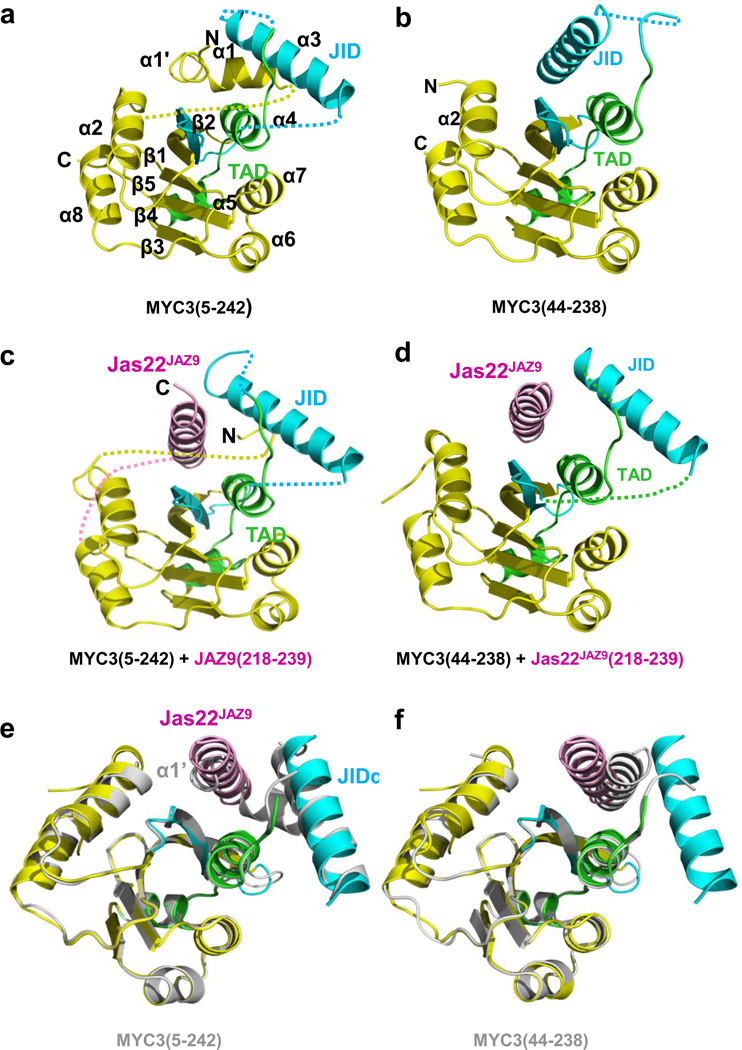
Based on our mapping results, we generated fifteen MYC2/3/4 N-terminal truncated proteins of various lengths (Extended Data Fig. 1d, 2b). MYC3(44-238) and MYC3(5-242) yielded high quality crystals that diffracted X-rays to 2.2 Å and 2.1 Å resolution, respectively (Extended Data Table 1). We solved the structure of selenomethionine-modified MYC3(44-238) by the Se-SAD phasing method and the structure of MYC3(5-242) by molecular replacement using the structure of MYC3(44-238) as search model (Fig. 1a, b and Extended Data Fig. 3). The proteins formed a helix-sheet-helix sandwich fold, in which eight α-helices are wrapped around a central five-stranded antiparallel β-sheet (Fig. 1a). Remarkably, while a hallmark of acidic TAD is that they are unstructured when not bound to a target in the transcriptional machinery17–19, the MYC3 TAD is well resolved and forms a loop-helix-loop-helix motif that packs against the JID with the N-terminal TAD helix and against β-strands 3–5 with the C-terminal TAD helix (Fig. 1a, b and Extended Data Fig 3). To our knowledge, this is the first example in which a non-complexed acidic TAD has a well resolved structure. The JID consists of the top (β2) strand of the β-sheet, the long α3-helix and two unresolved linkers (Fig. 1a, b and Extended Data Fig 3a). In MYC3(5-242), the JID forms together with the α4-helix of the TAD a groove. The N-terminal MYC helix (α1) is connected by a sharp ~90° kink to a loop that adopts a partial, stretched-out helical conformation (α1’, amino acids 6-16) that occupies the groove formed by the JID and TAD to cap the central β-sheet (Fig. 1a and Extended Data Fig. 3a). In N-terminally truncated MYC3 [MYC3(44-238), which lacks α1’+α1], the JID rearranges to adopt a position similar to that of α1’ in MYC3(5-242) to substitute for α1’ to cap the β-sheet in the fold (Fig. 1b). We performed hydrogen deuterium exchange (HDX) experiments to detect the surface accessibility and structural dynamics of MYC3(5-242) in solution (Extended Data Fig. 4). While the central β-sheet has a very stable structure and is well protected from deuterium exchange, the α1/ α1’ helix region has a very high deuterium exchange rate, suggesting that it has a very dynamic structure and forms only transiently in solution. This is consistent with the high B-factor values of the α1/ α1’ helix in the MYC3(5-242) crystal structure (Extended Data Fig. 5). While peptides corresponding to the JID helix were not resolved in HDX experiments, the JID helix also has a high B-factor (Extended Data Fig. 5), indicating that its position is dynamic as well. The MYC3(5-242) and MYC3(44-238) apo crystal structures therefore likely represent structure snapshots of two or more alternative MYC3 conformations in solutions.
Figure 1. Structures of MYC3 N-terminus in apo and Jas peptide-bound states.
a, Apo MYC3(5-242). b, Apo MYC3(44-238). c, MYC3(5-242) bound to the 22 amino acid JasJAZ9 motif peptide. Blue: JID, green: TAD, pink: Jas peptide. Dotted lines indicate the unresolved linkers that flank α1’/α1 and the JID helices. d, MYC3(44-238) bound to the 22 amino acid JasJAZ9 motif peptide. e and f, Overlay of the MYC3(44-238)–JAZ9 complex with apo MYC3(5-242) (e) and apo MYC3(44-238) (f). The complex structure is shown in color overlaid on the apo structures in grey. Binding of the JAZ9 Jas22 helix (pink) displaces the α1’ helix to become an integral part of the fold.
To crystallize a complex between MYC and the Jas motif, we synthesized a set of nine JAZ8, JAZ9, and JAZ12 Jas peptides of different lengths and complexed them with the above-mentioned set of the MYC N-terminal proteins. After extensive trials, we succeeded in obtaining crystals for MYC3(44-238) in complex with a 22-aa Jas peptide (S218-M239) from JAZ9 (Jas22JAZ9). However, no crystals for MYC3(5-242)-Jas peptide complexes could be obtained, suggesting that Jas complexes with α1’/α1 helix-containing MYC3 is less stable and/or conformationally more dynamic. To test this hypothesis, we generated a covalent fusion between MYC3(5-242) and Jas22JAZ9 separated by a 12 amino acid flexible linker to increase the stability of the MYC3(5-242) and Jas22JAZ9 complex and reduce conformational flexibility. This fusion protein formed high quality crystals and allowed us to solve the structure of the MYC3(5-242)–Jas22JAZ9 complex (Fig. 1c) at a resolution of 2.4 Å (Extended Data Table 1). The most striking aspect of the MYC3(5-242)–Jas22JAZ9 complex is that the Jas peptide formed a single, continuous helix that displaced the dynamic α1’/α1 helix in apo MYC3(5-242) (Supplementary Video 1). Correspondingly, the JID helix rearranged its conformation and the displaced α1’/α1 helix became almost completely disordered, suggesting the increase in disorder as the likely reason for the recalcitrance of this complex to crystallize. The Jas helix adopted a position in the groove that is almost superimposable with that of α1’ (Fig. 1e). In this position the Jas helix is nestled between the α4-helix of the TAD and the strand and helix of the JID to make extensive interactions with both the JID and the TAD and to become an integral part of the structural fold. In addition, we also determined the structure of MYC3(44-238), lacking the α1’/α1 helix, in complex with Jas22JAZ9 peptide at a resolution of 1.95 Å (Extended Data Table 1). In this structure, the Jas peptide also adopted a helical conformation and binds to the JID/TAD groove in the same way as seen in the MYC3(5-242)+Jas22JAZ9 fusion complex (Fig. 1d, 1f). Both α1’/α1 and the JID helix have long, unresolved linkers that appear to provide flexibility for their displacements/rearrangements by the Jas helix. Together, our analyses of the MYC3 apo and MYC3–JAZ complex structures indicate that occupancy of the groove (either by α1’ or the JID helix in apo MYC3, or by the Jas helix in the MYC–JAZ complex) is critical for formation of the overall MYC N-terminal fold.
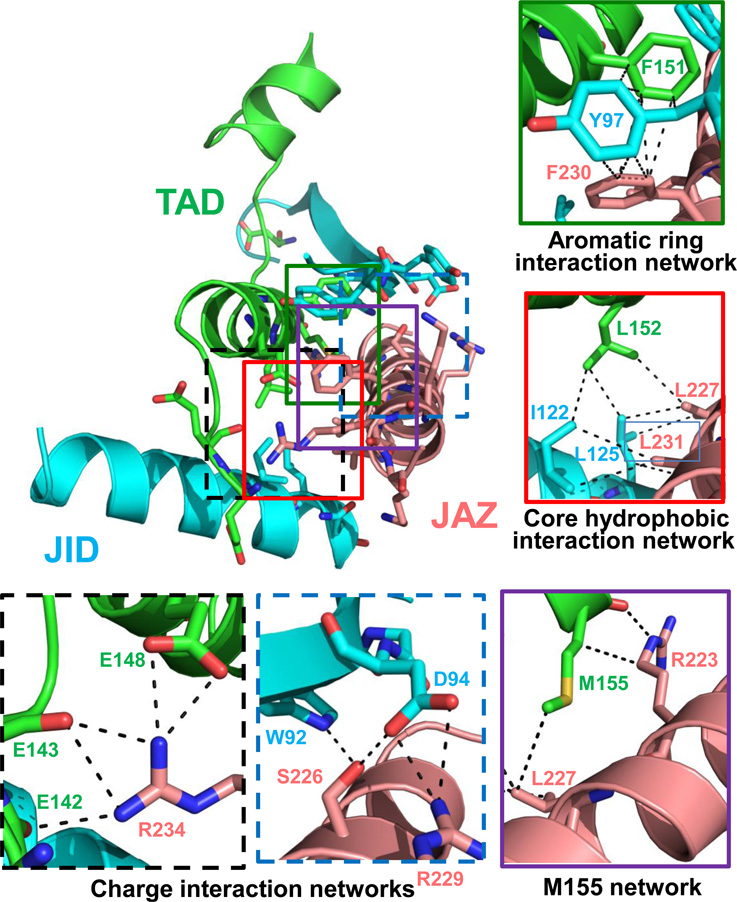
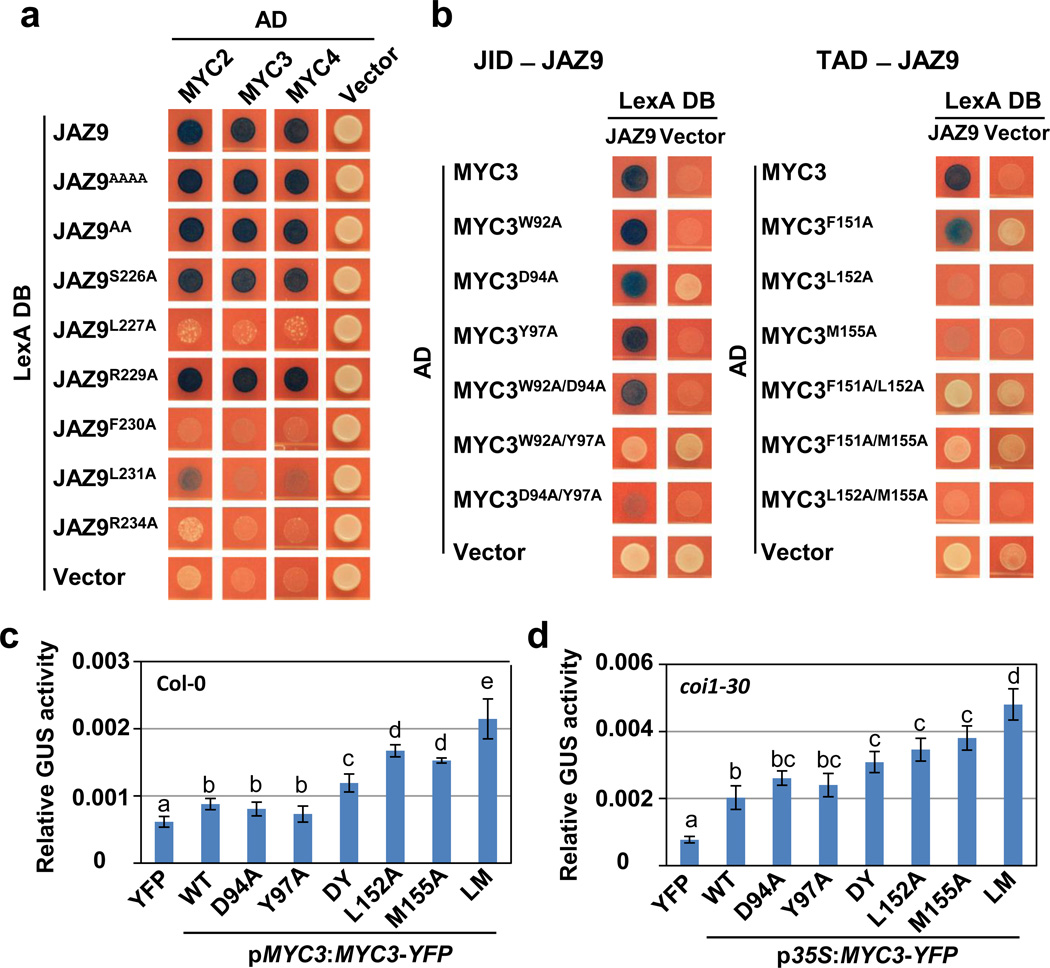
In the JAZ9–MYC3 complex, the JAZ peptide forms five main interaction networks with the TAD–JID surface (Fig. 2 and Extended Data Table 2): (i) R234 of JAZ9 forms salt bridges and hydrogen bonds with three glutamate residues (E142, E143, and E148) of the TAD, (ii) R229 and S226 form charge and hydrogen bond interactions with JID D94 and W92, (iii) L227 and L231 form a core hydrophobic network with TAD L152 and JID I122 and L125, and (iv) F230 interacts with JID Y97 and TAD F151 in an aromatic ring network. In addition, R223 and L227 have both hydrogen bond and/or hydrophobic interactions with TAD M155. In contrast, the first five amino acids (SVPQA) of the Jas motif that are critical for the co-receptor function of JAZ proteins (i.e., hormone-dependent binding to COI116) made no critical interactions with MYC3, consistent with our Y2H and AlphaScreen data (Extended Data Fig. 1b, d, 2b). Consistent with the structural data, mutational analysis showed that key interface residues of Jas, JID and TAD have important roles in MYC–JAZ interactions (Fig. 3a, b and Extended Data Fig. 6). In addition, the structure also provides an explanation for the partial in vivo relief of MYC3 repression by the MYC3D94N mutation observed previously20, as MYC3D94N lost interaction with a subset of JAZs, including JAZ3, JAZ4 and JAZ9 (Extended Data Fig. 6).
Figure 2. Jas peptide forms extensive interactions with the JID–TAD surface in MYC3.
JID–TAD–Jas structure with important interacting residues shown in stick presentation. Details of key interaction networks are shown as solid boxes (hydrophobic interaction networks) or dashed boxes (charge interaction networks). For clarity, not all interacting residues are shown in the detail boxes, compared to the non-boxed overview figure.
Next, we transfected the MYC-responsive pJAZ2::GUS reporter21 together with wildtype and mutant MYC3 expression plasmids into Arabidopsis protoplasts. As shown in Fig. 3c, mutant MYC3 proteins that were defective in interaction with multiple JAZ proteins (Extended Data Fig. 6) were partially relieved in repression (i.e., increased reporter gene activity). Moreover, the extent at which mutations compromised MYC3 interactions with JAZ proteins correlated with the increase in reporter gene activity and the magnitude of changes in reporter gene activity could be further accentuated by expressing MYC3 mutant proteins from the strong cauliflower mosaic virus 35S promoter in coi1-30 mutant protoplasts, in which all JAZ repressors are presumably stabilized (Fig. 3d). Together, these data validate the MYC3–JAZ9 complex structure and provide strong evidence that amino acid interactions identified in the MYC3–JAZ9 complex structure are important for MYC3 repression in planta.
Figure 3. Mutational analysis of the JAZ9–MYC3 interaction.
a, Yeast two-hybrid analysis of the interaction between JAZ9 mutant proteins and wild type MYC2, MYC3, and MYC4 proteins. JAZ9AAAA contains S218A, V219A, P220A and Q221A mutations and JAZ9AA contain R223A and K224A mutations. The experiment was repeated three times with same results. b, Yeast two-hybrid analysis between wild type JAZ9 protein and MYC3 proteins with mutations in the JID (left panel) or TAD (right panel). The experiment was repeated three times with same results. c and d, Alanine replacements of JAZ9-interacting amino acids of MYC3 increase MYC3 target gene expression in wildtype Col-0 and coi1-30 mutant plants, respectively. Arabidopsis protoplasts from wild-type (c) or coi1-30 mutant (d) were transfected with a MYC3-responsive JAZ2::GUS reporter together with YFP alone or MYC3-YFP constructs under control of the native MYC3 (c) or cauliflower mosaic virus 35S (d) promoter as indicated. A 35S::LUC reporter construct was co-transfected as control. GUS activities were normalized to the luciferase activity. Data shown are means of four independent transfections (n=4 biological replicates, error bars, s.d.). Different letters above the columns indicate the significant differences from each other (p<0.05) in pJAZ2:Gus reporter activities from transient expression of indicated MYC3 variants, as determined by Tukey–Kramer multiple comparison analysis. DY: the D94A/Y97A double mutant; LM: the L152A/M155A double mutant. The experiment was repeated three times with similar results.
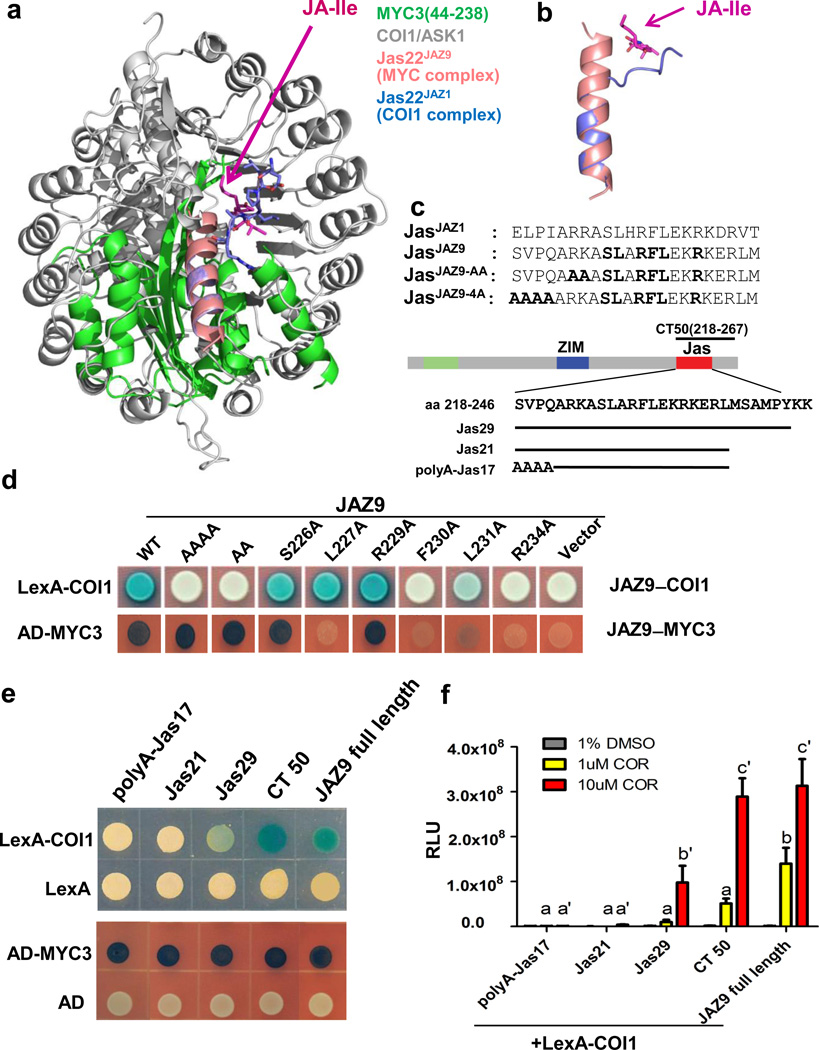
The Jas motif is required for its repressor function through interaction with MYC but also for its co-receptor function through interaction with COI122. While the JasJAZ9 peptide in the MYC3 complex formed a continuous helix (Fig. 1c, d), representing the rest state of JAZ, the JasJAZ1 peptide in the previously determined COI1–JA-Ile–Jas co-receptor structure (PDB: 3OGL) adopted a bipartite conformation with an N-terminal part stretched to form a distinct loop region followed by a shorter C-terminal helix16, as illustrated by the structural alignment in Fig. 4a, b and Supplementary Video 2. In addition to the JasJAZ9–MYC3 complex, we solved the structure of the JasJAZ1–MYC3 complex. As shown in the structure alignment in Extended Data Fig. 7, the Jas helices of JAZ9 and JAZ1 overlap very well, confirming the Jas conformational change between MYC-bound (resting stage) and COI1-bound (hormone-activated stage) is likely common in MYC interaction with different JAZ transcriptional repressors.
Figure 4. Distinct conformations of the Jas helix in the COI1–JAZ coreceptor complex vs. the JAZ–MYC complex.
a, Structure overlay of the MYC3–JasJAZ9 complex with the COI1/Ask–JA-Ile–JasJAZ1 complex (PDB: 3OGL). b, Close up of the Jas motif overlay from the MYC3–JasJAZ9 complex (pink) and from the COI1/Ask–JA-Ile–JasJAZ1 complex (blue). c, Alignment of the Jas motifs from JAZ1 and JAZ9. Mutationally analyzed conserved single residues and N-terminal amino acid stretches in JAZ9 are highlighted in bold. The position and sequence of JAZ9 fragments are indicated below the simplified JAZ9 diagram. d, Yeast two-hybrid analysis of the interaction between JAZ9 and either COI1 (top) or MYC3 (bottom). The experiment was repeated three times with same results. e, Yeast two-hybrid analysis of the interaction between JAZ9 C-terminal fragments and either COI1 (top) or MYC3 (bottom). One µM coronatine was used in yeast two-hybrid assays for the COI1-JAZ interaction. The experiment was repeated three times with similar results. f, Quantitative yeast two-hybrid analysis of the interaction between JAZ9 truncations and COI1, with β-galactosidase reporter gene activity determined by Beta-Glo assay (n=3 biological replicates, error bars, s.d.). Different letters above the columns indicate significant differences from each other (p<0.05) in COI1 interaction with indicated JAZ9 fragments at a given test concentration of COR (i.e., 1 µM or 10 µM), as determined by two way ANOVA with Bonferroni posttest. The experiment was repeated three times with similar results.
In the JasJAZ1–COI1 complex, the loop region of the JasJAZ1 helix is formed by the five moderately conserved N-terminal amino acids of the Jas motif (Extended Data Fig. 1c) that directly interact with the JA-Ile hormone (Fig. 4a, b), and is required for Jas–JA-Ile–COI1 co-receptor complex formation16. When we mutated the corresponding N-terminal amino acids of JAZ9 to alanine (JAZ9-4A and JAZ9-AA; Fig. 4c), JAZ9 lost interaction with COI1 in yeast two-hybrid assays, but not with MYC3 (Fig. 4d), consistent with the MYC3 complex structure. Mutations in the middle of the Jas motif (S226A-R234A) affected binding to both MYC3 and COI1, albeit to different degrees. In addition, residues that are C-terminal to the Jas motif enhance JAZ9 interaction with COI1 in yeast two-hybrid assays, but are not critical for its interaction with MYC3 (Fig. 4e, f), which is consistent with a previous study of JAZ2, JAZ3, and JAZ10 interactions with COI1 and MYC223. Together, these results indicate that COI1 and MYC3 potentially compete for binding to the central part of the Jas motif, but that COI1 makes additional critical interactions with JAZ9 outside of the MYC3-interacting region, including the previously unrecognized hormone-dependent unwinding of the N-terminal helix of the Jas motif (Supplementary Video 2). These additional interactions may allow COI1 to drive JAZ ubiquitination and dissociation of the extensive JAZ–MYC interaction upon JA-Ile stimulation.
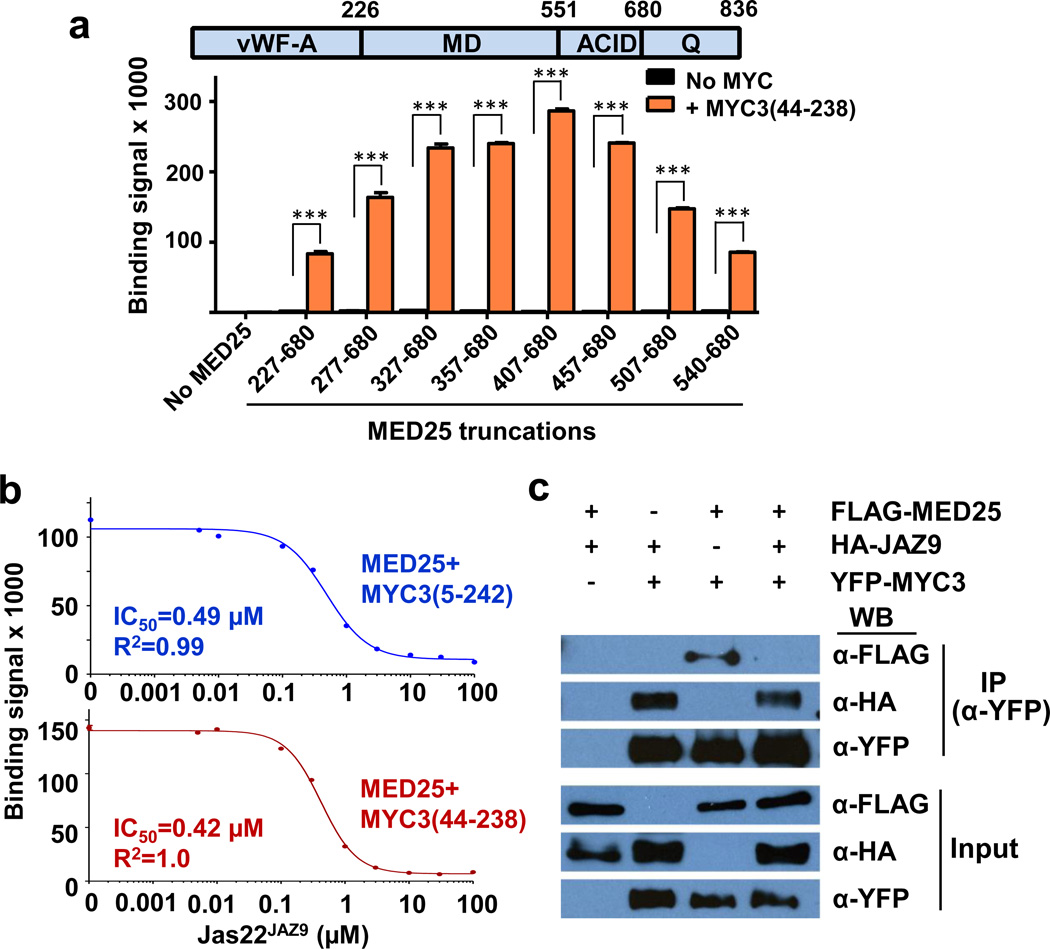
MED25 is a subunit of the Mediator complex that recruits RNA polymerase II to the promoters of JA responsive genes24 and is required for various JA responses13,15,25, including Arabidopsis susceptibility to Pseudomonas syringae bacterial infection (Extended Data Fig. 7c) and JA-induced inhibition of Arabidopsis root growth (Extended Data Fig. 7d). We found that MYC3(44-238) also directly binds MED25 and that a fragment (aa540-680) encompassing the MED25 Activator Interaction Domain (ACID) is sufficient to bind to MYC3 (Fig. 5a), analogous to what has previously been reported for MYC213. Since the MYC3 TAD makes critical interactions with JAZ repressors and is required for MYC3–JAZ9 complex formation (Fig. 2, Fig. 3b, and Extended Data Fig. 6), we explored the intriguing possibility that MYC3 binding of JAZ9 and MED25 are mutually exclusive. To test this prediction in a defined system, we performed AlphaScreen interaction assays between MED25(407-680) and both MYC3(44-238) and MYC3(5-242) in the presence of increasing amounts of untagged Jas22JAZ9 peptide. As shown in Fig. 5b, the JAZ peptide competitively inhibited the MYC–MED25 interaction with an IC50 of ~420 nM [MYC3(44-238)] and ~490 nM [MYC3(5-242)]. We further tested competition in planta by transiently expressing combinations of tagged MED25, JAZ9, and MYC3 in Nicotiana tabacum leaves. As shown in Fig. 5c, co-immunoprecipitation of MED25 with MYC3 was strongly reduced upon co-expression of JAZ9. Together, these results demonstrate that the Jas motif of JAZ proteins and the ACID domain of MED25 likely bind to a shared MYC3 surface, and that JAZ repressors can compete the MYC3 interaction with MED25 (and possibly other coactivators) in vitro and in planta.
Figure 5. Jas motif peptide competes the MYC3–MED25 interaction.
a, MYC3 interacts with the ACID domain of MED25. Note that that sequences N-terminal to ACID contribute to binding and/or stability. AlphaScreen interaction assay between biotinylated MYC3(44-238) and His6Sumo-MED25 fragments (n=3 technical replicates, error bars, s.d.). ***indicate significant differences (p<0.01) compared to the no-MYC control by Student's t-test. vWF-A: von Wildebrandt Factor A domain (responsible for Mediator binding), MD: Middle Domain, ACID: Activator Interaction Domain, Q: Q-rich C-terminal region. The experiment was repeated three times with similar results. b, JasJAZ9 peptide competes the interaction between MED25 and MYC3. AlphaScreen competition assay (n=3 technical replicates, error bars, s.d.). Jas22JAZ9: untagged JAZ9(218-239). MED25: His6Sumo-MED25(407-680). MYC3(5-242): biotin-MYC3(5-242). MYC3(44-238): biotin-MYC3(44-238). The experiment was repeated three times with similar results. An enlarged Fig. 5b with visible error bars is shown in Supplementary Fig. 1 and associated original data in Supplementary Table 1. c, Interference of the MYC3–MED25 interaction by JAZ9 in planta. FLAG-MED25 (+), HA-JAZ9 (+) and YFP-MYC3 (+) and respective vector controls carrying FLAG, HA or YFP tags (−) under control of the CaMV 35S promoter were transiently expressed in Nicotiana tabacum leaves. Protein extracts were immunoprecipitated (IP) with an anti-YFP antibody and analyzed by western blot (WB) with HA, FLAG, or YFP antibodies. The experiment was repeated three times with similar results. The original blots from which the images were cropped are shown in Supplementary Fig. 2.
In the past decade, despite the identification of analogous hormone perception and transcriptional gene regulation that underpins several major hormone signal transduction pathways in plants26, no crystal structures of the transcriptional repressor-transcription factor complexes have been solved. The crystal structure of the MYC-JAZ complex reported here therefore provides the first structural insight into the mechanism of transcriptional repression in plant hormone signaling. Our structural, biochemical, and in planta analyses suggest that JAZ repressors employ a novel dual repression mechanism, which involves not only epigenetic modifications of the target gene chromatin structure through TPL co-repressors, as demonstrated previously27, but also direct inhibition of MYC binding to MED25 (and possibly other co-activators) as an integral part of a mechanism of preventing transcriptional activation of JA response genes (Extended Data Fig. 8). In addition, we discovered distinct JAZ conformations in the MYC–JAZ resting complex vs. the JAZ–COI1 hormone-activated complex16, providing the first structural insight into the switch mechanism between transcriptional repression and hormone-dependent transcriptional activation in a major plant hormone signalling pathway.
Methods
Protein preparation
Wild type MYC3(44-238) was expressed as a fusion protein with a cleavable N-terminal His6Sumo tag from a modified pSUMO (LifeSensors) expression vector. BL21 (DE3) cells transformed with the expression plasmid were grown in LB broth at 16 °C to an OD600 of ~1.0 and induced with 0.1 mM IPTG for 16 h. Cells were harvested, resuspended in 100 ml extract buffer (20 mM Tris, pH 8.0, 200 mM NaCl, and 10% glycerol) per six liters of cells, and passed three times through a French Press with pressure set at 1,000 Pa. The lysate was centrifuged at 18,500 rpm in a Sorvall SS34 rotor for 30 min, and the supernatant was loaded on a 50 ml Nickel HP column. The column was washed with 600 ml of 10% buffer B (20 mM Tris, pH 8.0, 200 mM NaCl, 500 mM imidazole, and 10% glycerol) and eluted with 200 ml of 50% buffer B, followed by 100 ml of 100% buffer B. The eluted His6Sumo-MYC3(44-238) was dialyzed against extract buffer and cleaved overnight with SUMO protease at a protease/protein ratio of 1:1000 at 4°C. The cleaved His6Sumo tag was removed by passing through a 5 ml Nickel HP column, and the protein was further purified by chromatography though a HiLoad 26/60 Superdex 200 gel filtration column in 25 mM Tris, pH 8.0, 200 mM ammonium acetate, 1 mM dithiothreitol and 1 mM EDTA. To prepare the protein-ligand complex, we mixed Jas22JAZ1, Jas22JAZ8, Jas22JAZ9 or Jas22JAZ12 peptides with purified MYC3(44-238) proteins at a 1.5:1 molar ratio.
The expression and purification of MYC3(5-242) followed the same method as for MYC3(44-238) described above. To prepare the protein-ligand complex, we mixed Jas22JAZ1, Jas22JAZ8, Jas22JAZ9 or Jas22JAZ12 peptides with purified MYC3(5-242) proteins at a 1.5:1 molar ratio. To prepare MYC3(44-238) selenomethionyl (Se-Met) protein for phase determination, we followed the same methods as described previously28. Purification of Se-Met MYC3(44-238) proteins followed the same protocol as for MYC3(44-238) native protein except that the procedure was performed more quickly to avoid protein oxidization. The MYC3(5-242)-Jas22JAZ9 complex was constructed as a fusion protein with His6Sumo-MYC3(5-242) at the N-terminus and Jas22JAZ9 at the C-terminus, separated by a flexible GSAGSAGSAGSA (4xGSA) linker [His6Sumo-MYC3(5-242)-4xGSA-Jas22JAZ9]. The expression and purification of the fusion protein followed the same methods as for MYC3(44-238).
Small-scale purification of His6Sumo-tagged MYC2/3/4 protein fragments (including JID and TAD domain) for binding studies with Jas peptides followed the same methods as for MYC3(44-238), except that the His6Sumo tag was not removed. Small-scale purification of His6Sumo-tagged MED25 protein fragments (including the ACID domain) for binding studies with biotinylated MYC3(44-238) followed the same methods as for MYC3(44-238), except that the His6Sumo tag was not removed. To express and purify biotinylated MYC3(44-238) and MYC3(5-242) protein for binding studies (Fig. 5a) and JAZ Jas competition assays (Fig. 5b), we followed the methods described previously29.
Crystallization
The apo-MYC3(5-242) crystals were grown at 20 °C in sitting drops containing 0.2 µl of purified MYC3(5-242) protein at a concentration of 10 mg/ml and 0.2 µl of well solution containing 0.2 M magnesium chloride, 0.1 M Tris, pH 8.5, 30% (w/v) polyethylene glycol 4,000 for 3 days. The Se-Met MYC3 (44-238) crystals were grown at 20 °C in sitting drops containing 0.2 µl of the purified protein at a concentration of 15 mg/ml and 0.2 µl of well solution containing 0.2 M sodium chloride, 0.1 M Bis-Tris, pH 5.5, and 25% (w/v) polyethylene glycol 3,350. Crystals of about 100 µm in length appeared in 3 days. The MYC3(44-238)–Jas22JAZ9 complex crystals were grown at 20 °C in sitting drops containing 0.2 µl of the purified complex proteins at a concentration of 15 mg/ml and 0.2 µl of well solution containing 0.2 M magnesium chloride, 0.1 M Tris, pH 8.5, and 30% (w/v) polyethylene glycol 4,000 for 3 days. The MYC3(44-238)–Jas22JAZ1 complex crystals were grown at 20 °C in sitting drops containing 0.2 µl of the purified complex proteins at a concentration of 15 mg/ml and 0.2 µl of well solution containing 3.5 M sodium formate. Crystals of about 80 µm in length appeared in 2 days. The MYC3(5-242)-Jas22JAZ9 fusion protein crystals were grown at 20 °C in sitting drops containing 0.2 µl of the purified fusion proteins at a concentration of 15 mg/ml and 0.2 µl of well solution containing 0.2 M magnesium nitrate, 20% (w/v) polyethylene glycol 3,350. Crystals of about 100 µm in length appeared in 3 days. All crystals were serially transferred to the well solution with 20% (v/v) ethylene glycol before flash freezing in liquid nitrogen.
Data collection and structure determination
Data collections were performed at sector 21-ID (LS-CAT) beam lines of the APS synchrotron using single native MYC3(5-242) crystals, MYC3(44-238)-Jas22JAZ9 complex crystals, and Se-Met substituted MYC3(44-238) crystals at different wavelengths. All diffraction data were processed using XDS30, and merged using Aimless of the CCP4 suite31. Initial phasing was tried by using the single wavelength anomalous dispersion (SAD) method based on anomalous diffraction of sulfur atoms as previously described32. S-SAD phasing using a combined data set of 11 native MYC3(5-242) crystals collected at 1.77 Å was not successful, probably due to the non-isomorphism of the individual crystals. To solve the phase problem, the Se-Met substituted MYC3(44-238) crystals were prepared and a single data set was collected at a peak wavelength of 0.9787 Å (Extended Data Table 1). Se-SAD phasing was performed by using the Phenix Autosol program. Five out of six selenium atoms were found with an FOM of 0.41. The Phenix autobuild program generated an initial model of 286 residues with an Rwork/Rfree of 0.35/0.40. Further model improvements were performed using Coot33 and refined using the Refmac program in CCP434 to a final model with an R factor of 0.21 and an Rfree factor of 0.26. The MYC3(5-242) apo structure, MYC3(5-242)–Jas22JAZ9, MYC3(44-238)–Jas22JAZ9 and MYC3(44-238)–Jas22JAZ1 complex structures were solved by using the molecular replacement program Phaser35 with the Se-Met MYC3(44-238) structure as a search model. The model for the α1’/ α1 helices for the MYC3(5-242) structure, for the Jas22JAZ9 peptide for the MYC3(44-238)–Jas22JAZ9 and MYC3(5-242)–Jas22JAZ9 complex structures and for the Jas22JAZ1 peptide for the MYC3(44-238)–Jas22JAZ1 complex structure were built based on the electron density maps using Coot.
AlphaScreen luminescence proximity assays
In vitro interactions between MYC3 and Jas peptides or MED25 fragments were assessed by luminescence–proximity AlphaScreen (Perkin Elmer) technology as described previously29,36,37. Reactions contained 50 nM His6Sumo-MYC3 protein bound to nickel-acceptor beads and 50 nM synthesized biotinylated Jas peptides bound to streptavidin donor beads (Extended Data Fig. 1d and 2b) or 50 nM His6Sumo-MED25-ACID protein bound to nickel-acceptor beads and 50 nM biotin--MYC3(44-238) bound to streptavidin donor beads (Fig. 5a). The results were based on an average of three experiments with standard errors typically less than 10% of the measurement.
For the competition assay (Fig. 5b), non-biotinylated Jas22JAZ9 peptide was added into the reaction at concentrations of 0, 5, 10, 100, 300, 1,000, 3,000, 10,000, 30,000 and 100,000 nM. The results were based on an average of three experiments with stand errors typically less than 10% of the measurement.
Mutagenesis
Site-directed mutagenesis was carried out using the QuickChange Method (Agilent). Mutations for all plasmid constructs were confirmed by sequencing.
Yeast two-hybrid (Y2H) assays
Most of the constructs for Y2H assays used in this study were described previously22,38,39. The coding sequences of full length MYC2, MYC3 and MYC4, and MYC N-terminal and C-terminal fragments were PCR-amplified and cloned into pGilda (Clontech). PCR-based deletions were conducted following the manufacturer’s protocol (Stratagene). Detailed protocols for Y2H assays were described previously38.
Plant materials and growth conditions
Arabidopsis plants used in this study were previously described39 or were ordered from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Arabidopsis seeds were stratified for 3 days at 4 °C before planting. The soil-grown plants were placed in a controlled growth chamber at 23 °C with a 12-h-day (80 µmol s−1m−2 cool-white fluorescent light)/12-h-night cycle.
Transient expression in tobacco leaves and Arabidopsis protoplasts
For transient expression in tobacco leaves, coding sequences of JAZ9, MED25 and MYC3 were cloned into pJYP003, pJYP011 and pJYP018 (Yao and He, unpublished), respectively, to create p35S:3xHA-JAZ9, p35S:3xFLAG-MED25 and p35S:YFP-MYC3 fusion constructs, which were transfected as previously described39. Protein extracts were immunoprecipitated (IP) with an anti-YFP antibody and analyzed by western blot (WB) with HA, FLAG, or YFP antibodies as previously described39. For transient expression in Arabidopsis mesophyll protoplasts, MYC3 (no stop codon) with or without its promoter (the 2-kb sequence upstream of the start codon) were PCR-amplified and cloned into pENTR-D/TOPO vector (Invitrogen) to create entry clones. Then, the MYC3 or pMYC3:MYC3 inserts were introduced into pSAT4A-DEST-Venus or pBR-DEST-Venus (Yao and He, unpublished) to create the p35S:MYC3-YFP or pMYC3-MYC3-YFP constructs. The JAZ2 promoter (the 2-kb sequence upstream of its start codon) was cloned into pBR-Gus (Yao and He, unpublished) to create pJAZ2:GUS reporter constructs. The transient expression assays using pBS-35S-Luc as transfection control followed a published protocol40. A 35S::LUC reporter construct was co-transfected as control. GUS activities were normalized to the luciferase activity.
Root growth inhibition assay
Arabidopsis wild-type (Col-0) and med25 (pft1-2; SALK_129555)25 seedlings were used for the root growth inhibition assay. Seeds were surface-sterilized, stratified at 4°C, and germinated on ½ strength MS agar plates containing 1 µM, 3 µM or 10 µM MeJA or 0.1% DMSO (control). Plates were placed vertically in a growth chamber (16 h light/8 h dark light cycle, 100 µE s−1m−2 light intensity) for 10 days before pictures were taken, and root lengths were measured with ImageJ software (http://rsbweb.nih.gov/ij/).
Bacterial infection
Pst DC3000 infection assays in Arabidopsis plants were performed as described previously41. Briefly, 5-week old Arabidopsis plants were dip-inoculated with Pst DC3000 bacterial suspension (1×108 cfu/ml, 0.025% Silwet-L77). Bacterial growth was determined 3 days after inoculation.
Hydrogen-deuterium exchange (HDX) mass spectrometry
HDX of MYC3(5-242) in 20 mM Tris pH 8.0, 200 mM ammonium acetate, 1 mM EDTA, and 7% glycerol was performed at 4 °C using an automated system described previously42. Briefly, protein was incubated in a D2O buffer for a range of exchange times from 10 s to 1 h before quenching the deuterium exchange reaction with an acidic quench solution (pH 2.4) containing of 3 M Urea and 1% TFA. All mixing and digestions were carried out on a LEAP Technologies Twin HTS PAL liquid handling robot housed inside a temperature controlled fridge. Protein digestion was performed in-line with chromatography using an immobilized pepsin column. Mass spectra were acquired on a Q Exactive hybrid quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific). Percent deuterium exchange values for peptide isotopic envelopes at each time point were calculated and processed using the Workbench Software43.
The unit of measure represented as a single value is percent deuterium incorporation (% D), which is determined by initially calculating the intensity weighted average (centroid) of all spectral data within defined m/z limits. The % D is then determined by comparing the result to defined minimum (0%) and maximum m/z values (100%) for each peptide. The minimum and maximum m/z values are determined using experimentally observed undeuterated and fully deuterated controls.
The data representing each peptide are reduced to single value in the following manner: For each sample, the three individual time-point replicate % D (done in triplicate) at each time point are averaged. The mean of these values is then presented as a single value, representing the overall change in deuterium incorporation for the sample. The first number in brackets is the representation of the propagation of error for the sample, which is determined by a root means squared approach using the standard deviations from each individual time point. The second number in brackets is the charge state of the detected peptide.
Extended Data Fig. 4a shows cumulative peptides fragments that were detected in MS/MS. Shorter fragments (4–10 residues) provide higher resolution information than longer peptides. Therefore, they supersede longer fragments (> 10 residues) and were used to manually overlay onto the atomic structure as in the case of Fig. 2b. No subtraction was employed. The peptide set used for structural overlay contained the shortest fragments from the complete data set (all peptides).
Data analysis, statistics and experimental repeats
The specific statistical method used, the sample size and the results of statistical analyses are described in the relevant figure legends. Sample size was determined based on experimental trials and in consideration of previous publications on similar experiments to allow for confident statistical analyses. All reported results were reproduced in at least three independent experiments.
Extended Data
Extended Data Figure 1. Mapping of the JAZ9–MYC3 interface.
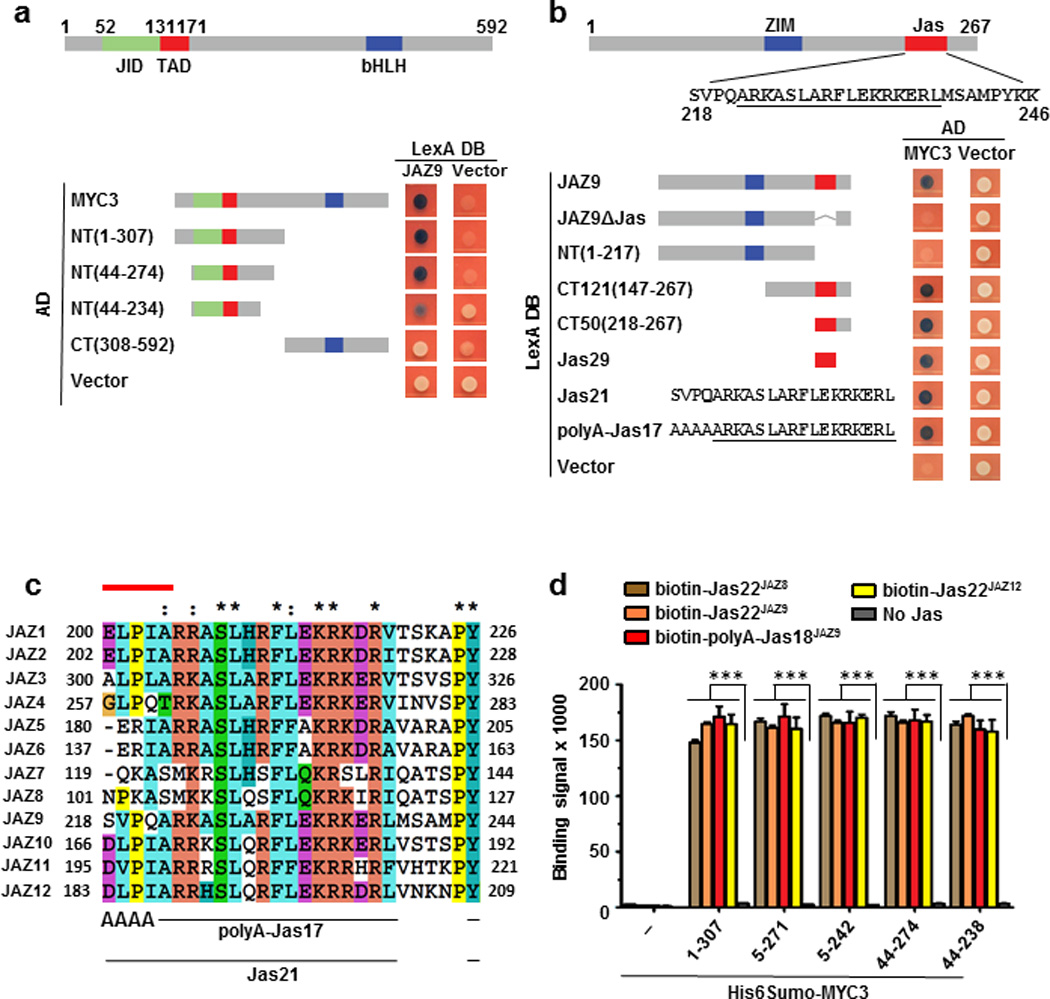
a and b, Yeast two-hybrid analysis of the interaction between LexA-JAZ9 and BD42(AD)-MYC3 constructs. Simplified diagrams of MYC3 (a) and JAZ9 (b) proteins are shown on top. Blue yeast colonies indicate a positive interaction between two proteins. The experiment was repeated three times with same results. c, Sequence alignment of the Jas motif of the 12 A. thaliana JAZ proteins. The N-terminal five amino acids that are unwound in the crystal structure of the COI1-ligand-JAZ co-receptor complex23 are indicated by a red line on top of JasJAZ1. Asterisks denote amino acids conserved in all of the sequences and colons denote similar amino acids. d, Interaction between purified His6Sumo-MYC3 N-terminal proteins and biotinylated Jas motif peptides by AlphaScreen luminescence proximity assay (n=3 technical replicates, error bars, s.d.). *** indicate significant differences (p<0.01) comparing to the no-Jas control by Student's t-test. The experiment was repeated three times with similar results.
Extended Data Figure 2. Interactions of MYC2 and MYC4 with Jas motifs of JAZ8, JAZ9, and JAZ12.
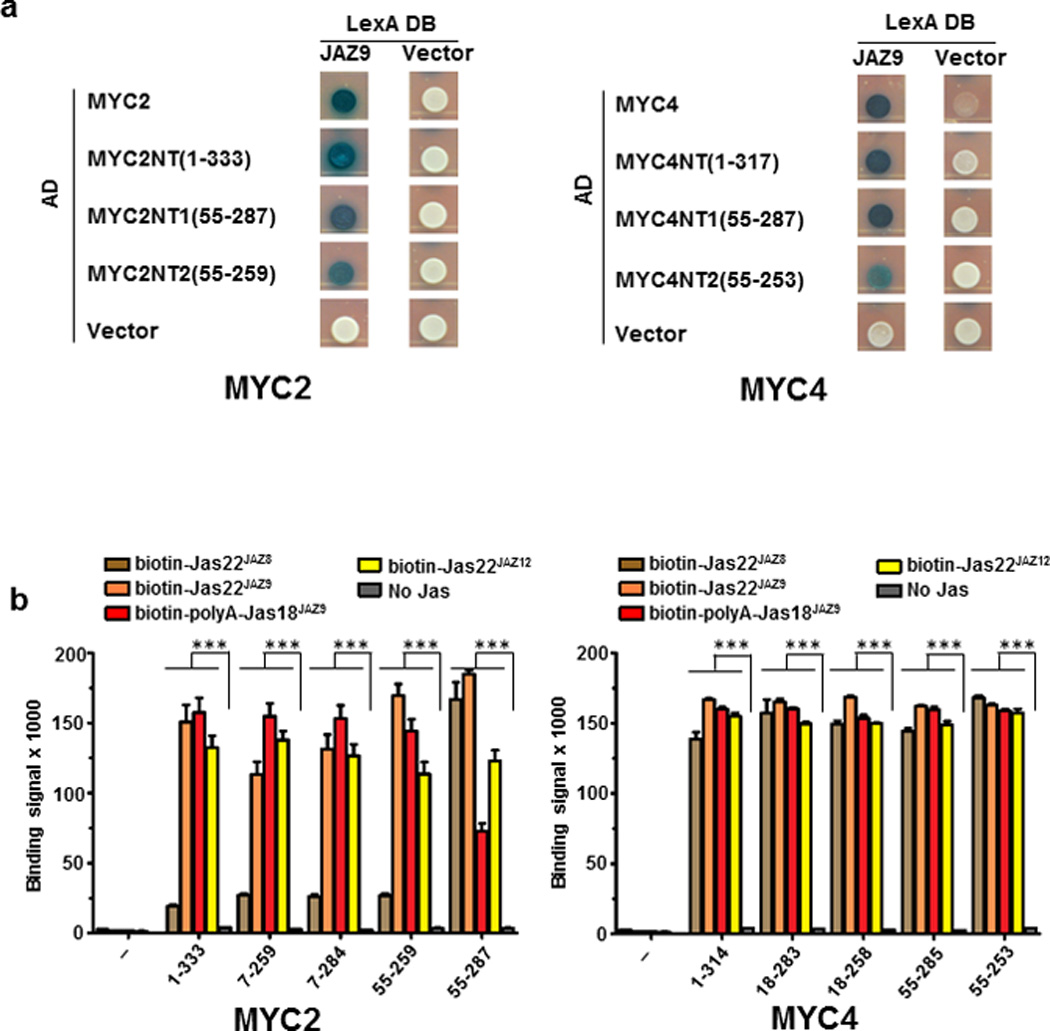
a, Yeast two-hybrid assays between MYC N-terminal proteins and full length JAZ9. The experiment was repeated three times with same results. b, AlphaScreen assay between His6Sumo-tagged MYC N-terminal proteins and biotinylated JAZ peptides (n=3 technical replicates, error bars, s.d.). ***indicate significant differences (p<0.01) comparing to no-Jas control by Student's t-test. The experiment was repeated three times with similar results.
Extended Data Figure 3. Arrangement of secondary structure elements in MYC3(5-242).
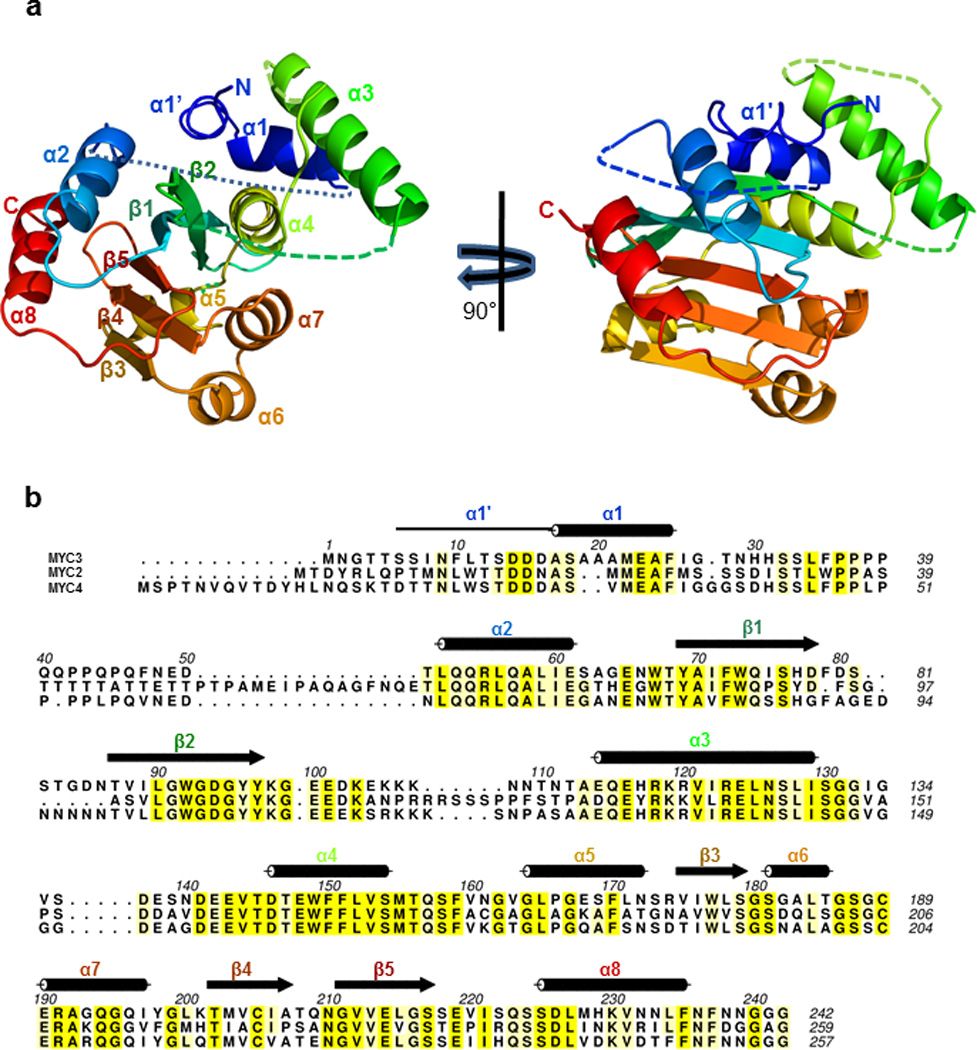
a, Rainbow color scheme of MYC3(5-242) in two orientations, from blue (N-terminus) to red (C-terminus). b, Secondary structure elements overlaid on the sequence alignment of MYC2, MYC3, and MYC4 N-terminal proteins. Note that α1’ (solid line) is a loop with partial helix character and is connected to α1 by a 90° kink.
Extended Data Figure 4. Surface accessibility and structural dynamics of MYC3(5-242) revealed by hydrogen deuterium exchange mass spectrometry (HDX).
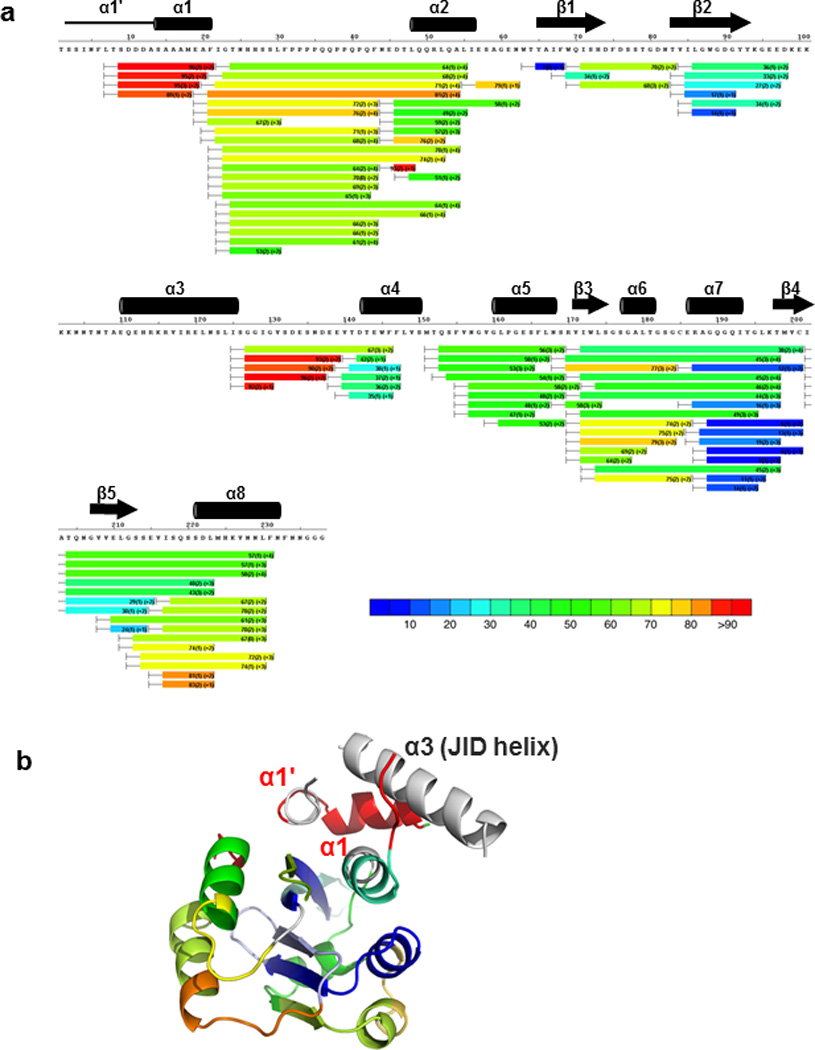
a, HDX heat map of MYC3(5-242). The color bar indicates % deuterium exchange. Three experimental repeats were performed for each HDX time point. b, HDX heat map overlaid onto the MYC3(5-242) apo structure. Peptides corresponding to the JID helix were not resolved (no HDX information, grey color), preventing a definitive assessment of the dynamics of the JID helix in solution.
Extended Data Figure 5. B-factor presentations of the four crystal structures.
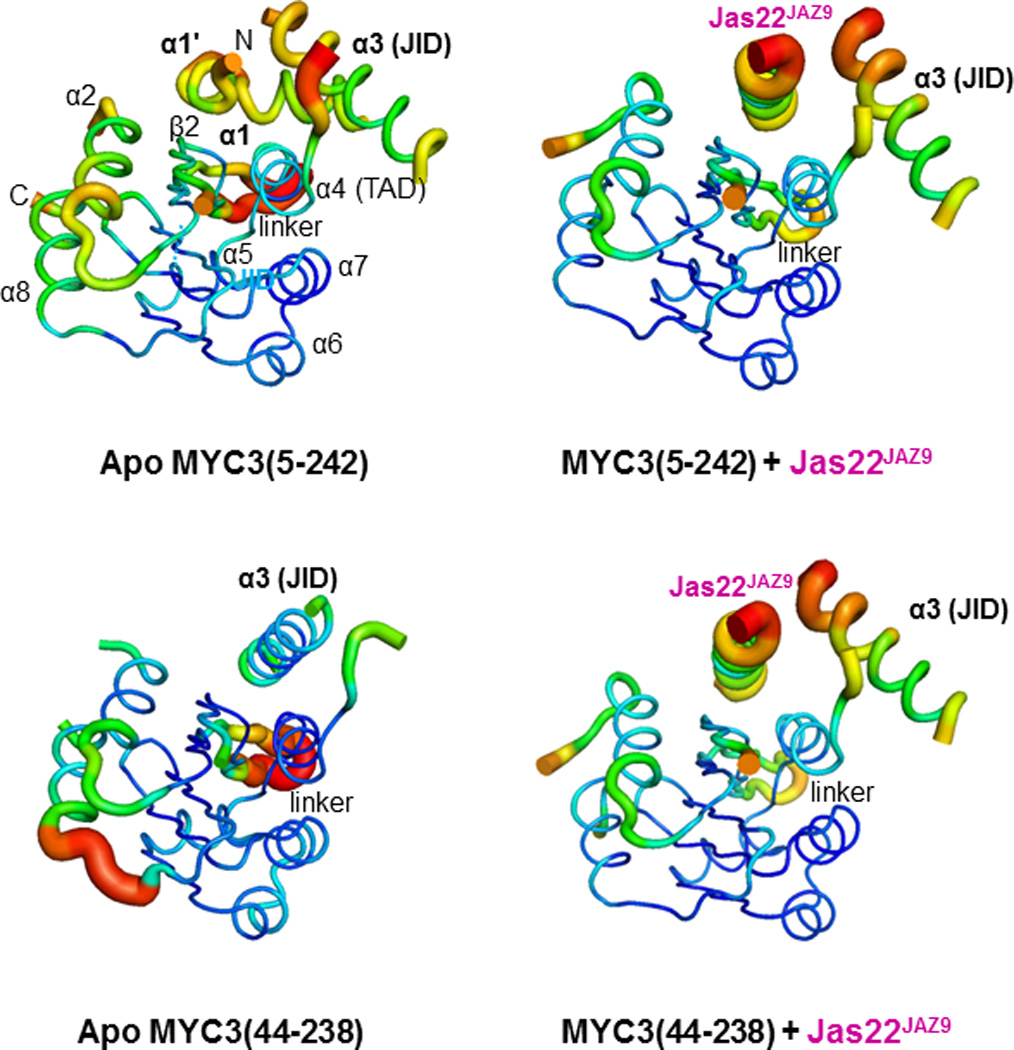
The B-factor indicates the dynamic mobilities of different resolved parts within the structure. The thicker the lines and the warmer the color, the higher is the mobility. Other than two linker regions (linker), the three helices that can occupy the JID helix have the highest B-factors in all four structures. The difficulties in crystallizing the MYC3(5-242)–Jas22JAZ9 complex are therefore likely due to its high conformational flexibility due to the presence of all three dynamic helices as well as the unfolding of the α1’/α1 helix. Covalent fusion to MYC3(5-242) likely stabilizes the conformational flexibility of Jas22JAZ9 and the complex. Note that the presence of the α1’/α1 helix does not interfere with the ability of MYC3 to bind the JAZ peptide (compare MYC3(5-242) and MYC3(44-238) in Fig. 5b).
Extended Data Figure 6. Effects of mutations in MYC3 on the interactions between JAZ and MYC3 proteins in yeast two-hybrid assays.
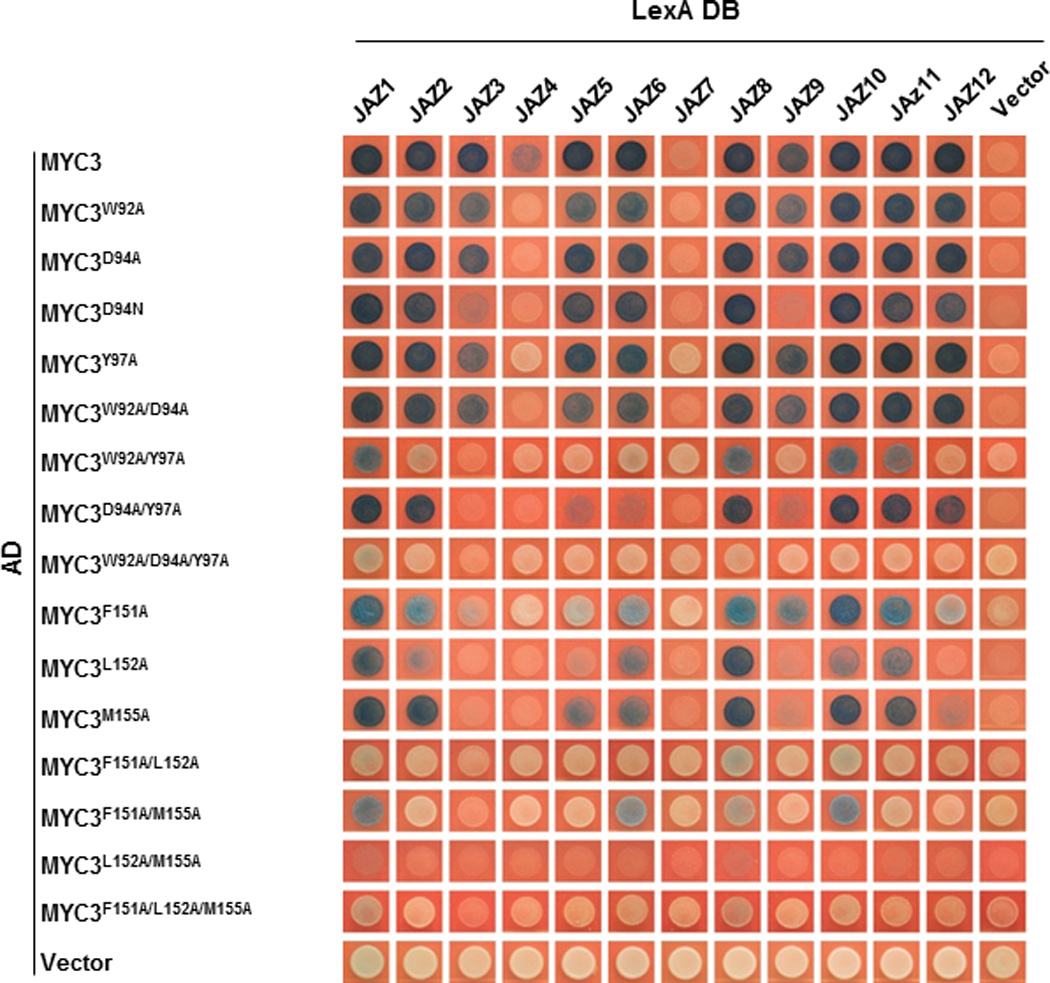
The development of blue yeast colonies indicates the positive interaction between two proteins. The experiment was repeated three times with same results.
Extended Data Figure 7. MYC3(44-238) in complex with Jas22JAZ1 and Jas22JAZ9 and phenotypes of the Arabidopsis med25 mutant.
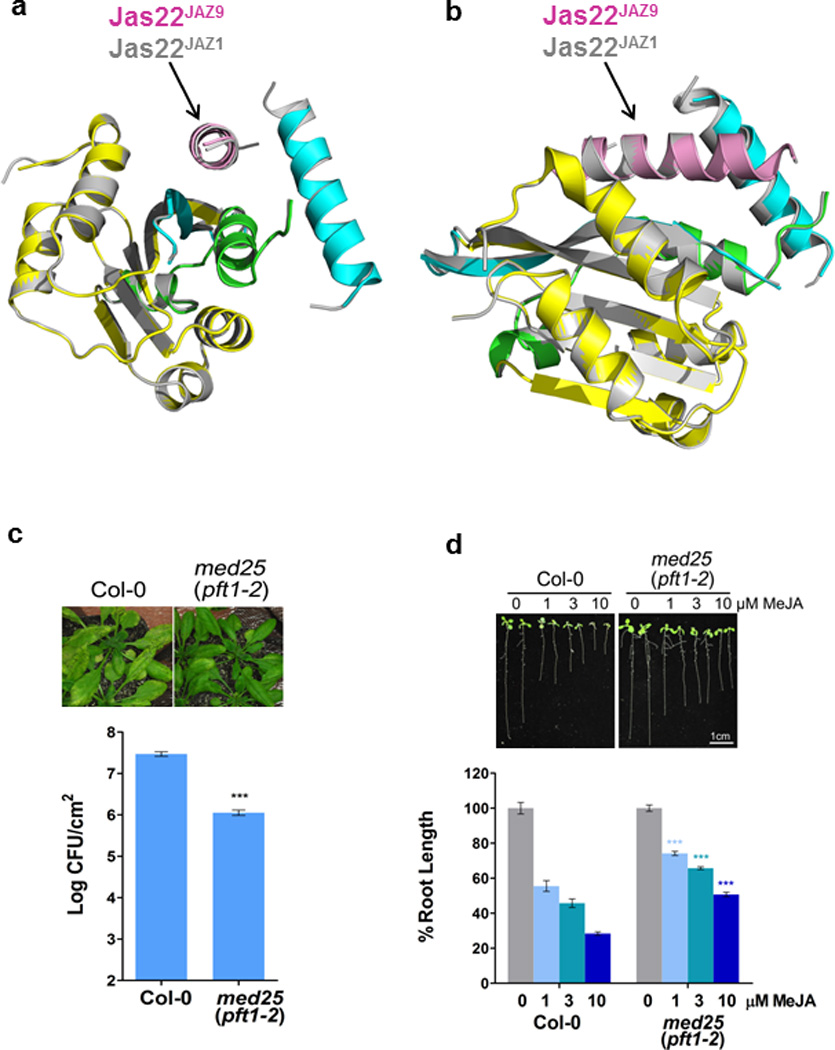
a and b, MYC3(44-238) in complex with Jas22JAZ1(aa 200-221; grey) overlaid with MYC3(44-238) in complex with Jas22JAZ9(aa 218-239; pink). c, Arabidopsis med25 mutant (pft1-2) plants are less susceptible to Pseudomonas syringae pv. tomato (Pst) DC3000 than Arabidopsis wild-type (Col-0) plants. Disease symptoms (chlorotic lesions; upper panel) and bacterial population (lower panel) of Arabidopsis wild-type (Col-0) and med25 mutant (pft1-2) plants 3 days after dip-inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 at 1×108 cfu/ml (n=4 biological replicates, error bars, s.e.m.). *** indicates a significant difference (p<0.001) in bacteria population between Col-0 and med25 mutant plants, as determined by two-tailed t test. The experiment was repeated 5 times with similar results and the images presented are representative of five repeats. d, med25 mutant plants are less sensitive to JA-induced root growth inhibition than wild-type (Col-0) plants. A representative picture (upper panel) and percentages of root growth inhibition (lower panel) of 10-day old wild-type (Col-0) and med25 mutant (pft1-2) Arabidopsis seedlings after treatment with 0.1% DMSO (control), 1 μM, 3 µM or 10 µM MeJA (n=15 biological replicates, error bars, s.e.m.). *** with different colors indicate the significant differences (p<0.001) between Col-0 and the med25 (pft1-2) mutant with the same concentration of MeJA treatment, as determined by two way ANOVA with Bonferroni posttest. The experiment was repeated four times with similar results and the images presented are representative of four repeats.
Extended Data Figure 8. A simplified diagram of the core components of the jasmonate signaling cascade.
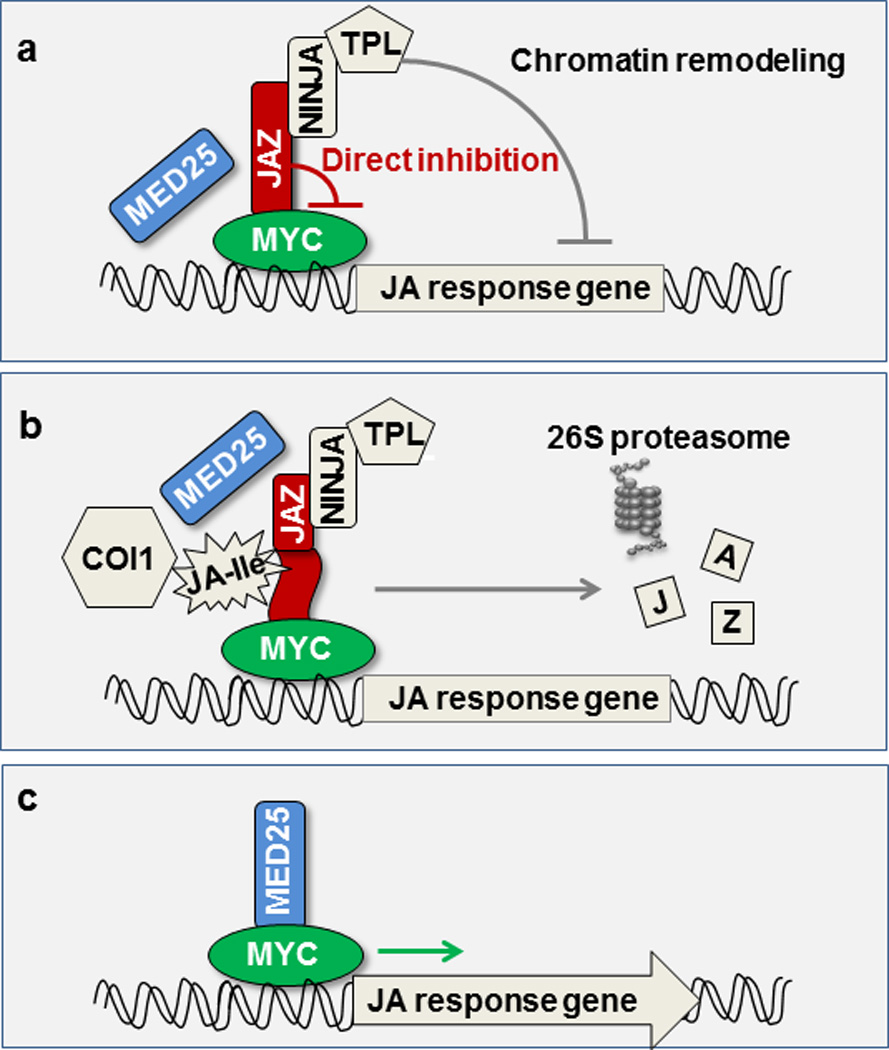
(a) In the resting stage, JA response gene expression is restrained by a family of JAZ transcriptional repressors. JAZ repressors bind and inhibit the MYC family of transcription factors through (i) direct inhibition and (ii) recruiting TOPLESS (TPL) co-repressors either directly or through the NINJA adaptor. TPL in turn recruits histone deacetylases/methyltransferases (not shown) to repress gene expression through chromatin remodeling. (b) In response to stress or developmental cues, plants synthesize JA-Ile, which serves as molecular glue to facilitate the formation of a co-receptor complex between JAZ and COI1. The formation of the COI1–JAZ co-receptor complex leads to ubiquitination and proteasome-dependent degradation of JAZ repressors. (C) JAZ-free MYCs interact with the MED25 subunit of the Mediator complex and recruit RNA polymerase II (not shown) to the promoters of JA responsive genes. Components examined in this study are colored.
Extended Data Table 1.
X-ray data collection and refinement statistics for MYC3 structures.
| Native MYC3(5–242)# |
SeMet MYC3(44–238)# |
Native MYC3(44– 238)+JAZ9 complex# |
Native MYC3(44– 238)+JAZ1 complex# |
Native MYC3(5– 242)+JAZ9 complex# |
|
|---|---|---|---|---|---|
| PDB code | 4RRU | 4RQW | 4RS9 | 4YZ6 | 4YWC |
| Data collection | |||||
| Space group | P3221 | P212121 | P3221 | P3221 | C2221 |
| Cell dimensions | |||||
| a, b, c (Å) | 85.4, 85.4, 53.7 | 57.3, 76.6, 85.9 | 85.8, 85.8, 60.0 | 86.2, 86.2, 59.4 | 59.1, 110.6, 161.8 |
| a, b, g (°) | 90, 90, 120 | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Peak | |||||
| Wavelength | 1.0782 | 0.9787 | 0.9786 | 1.078 | 0.9787 |
| Resolution (Å) | 50–2.1 | 50–2.2 | 50–1.95 | 50–1.95 | 50–2.4 |
| Rsym or Rmerge | 0.054 (0.97)* | 0.050 (1.22)* | 0.057 (1.31)* | 0.049 (0.95)* | 0.139 (1.08)* |
| / / s/ | 18.2 (2.2)* | 20.6 (1.8)* | 23.7 (2.0)* | 29.7 (2.6)* | 10.3 (2.0)* |
| Completeness (%) | 100 (100)* | 99.9 (99.9)* | 100.0 (100.0)* | 100.0 (100.0)* | 99.9 (100.0)* |
| Redundancy | 7.3 (7.4)* | 8.2 (8.4)* | 12.0 (10.8)* | 12.2 (12.4)* | 8.2 (8.3)* |
| Refinement | |||||
| Resolution (Å) | 50–2.1 | 50–2.2 | 50–1.95 | 50–1.95 | 50–2.4 |
| No. reflections | 12780 | 18734 | 17922 | 17940 | 21103 |
| Rwork / Rfree | 0.234/0.277 | 0.214/0.263 | 0.200/0.234 | 0.184/0.217 | 0.239/0.295 |
| No. molecules per asymmetric unit |
1 | 2 | 1 | 1 | 2 |
| No. atoms | |||||
| Protein | 1447 | 2550 | 1338 | 1367 | 2729 |
| Ligand/peptide | 2 | 2 | 155 | 155 | 357 |
| Water | 71 | 57 | 137 | 160 | 100 |
| B-factors | |||||
| Protein | 60.0 | 62.5 | 46.9 | 40.3 | 58.8 |
| Ligand/peptide | 106.3 | 94.2 | 70.5 | 70.9 | 56.6 |
| Water | 57.4 | 65.4 | 58.3 | 55.9 | 56.8 |
| R.m.s deviations | |||||
| Bond lengths (Å) | 0.009 | 0.008 | 0.006 | 0.008 | 0.008 |
| Bond angles (°) | 1.34 | 1.27 | 1.01 | 1.09 | 1.24 |
The X-ray diffraction data were obtained from a single crystal.
Values in parentheses are for the highest-resolution shell.
Extended Data Table 2.
Main interacting residues between JAZ9 and MYC3
| JAZ9 aa |
Distance | Interaction | MYC3* aa |
|---|---|---|---|
| R223 | 3.1 Å | H-bond | M155 |
| 4.0 Å | VdW | M155 | |
| 4.0 Å | VdW | W92 | |
| S226 | 2.6 Å | H-bond | D94 |
| 2.8 Å | H-bond | W92 | |
| L227 | 3.8 Å | VdW | L125 |
| 4.1 Å | VdW | L125 | |
| 3.9 Å | VdW | L152 | |
| 4.2 Å | VdW | M155 | |
| R229 | 2.9 Å | Ionic | D94 |
| 3.4 Å | ionic | D94 | |
| F230 | 3.9 Å | VdW | Y97 |
| 4.5 Å | VdW | Y97 | |
| 4.1 Å | VdW | E148 | |
| 3.8 Å | VdW | F151 | |
| 3.9 Å | VdW | F151 | |
| L231 | 4.1 Å | VdW | L125 |
| 4.0 Å | VdW | N126 | |
| 4.5 Å | VdW | I122 | |
| 4.4 Å | VdW | I122 | |
| K233 | 3.2 Å | H-bond | Y97 |
| 3.4 Å | H-bond | Y96 | |
| R234 | 2.9 Å | H-bond | E143 |
| 3.3 Å | H-bond | E143 | |
| 2.9 Å | Ionic | E148 | |
| 3.0 Å | Ionic | E148 | |
| 3.0 Å | H-bond | E142 | |
| K235 | 3.0 Å | H-bond | N126 |
Green: TAD amino acid, cyan: JID amino acid, bold: residues whose mutation to alanine compromised the JAZ-MYC interaction in yeast two-hybrid and AlphaScreen assays. VdW: Van der Waals bond.
Supplementary Material
Acknowledgements
This research is supported by the Gordon and Betty Moore Foundation (GBMF3037, S.Y.H.), the China Scholarship Council (F.Z.), Van Andel Research Institute (H.E.X. and K.M.), the National Institutes of Health (R01 GM102545 to K.M. and R01AI060761 to S.Y.H.), and the Department of Energy (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science Grant DE–FG02–91ER20021 (S.Y.H.). We thank Stephanie Grant for administrative support and staff members of the Life Science Collaborative Access Team of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of APS was supported by the Office of Science of the US Department of Energy, under Contract No. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank Gregg Howe and Kyaw Aung for critical reading of this manuscript.
Footnotes
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under PDB ID codes 4RRU, 4RQW, 4RS9, 4YZ6 and 4YW (see Extended Data Table 1).
The authors declare no competing financial interests.
Contributions
H.E.X., K.M., M.Z, and S.Y.H. conceived the project and designed experiments, F.Z., J.Y., J.K., L.Z., V.Q.L., XFX, X.E.Z., J.C., J.B., P.R.G. performed and/or interpreted experiments. K.M., F.Z., J.Y. and S.Y.H. wrote the manuscript with support from all authors.
References
- 1.Browse J. Jasmonate: preventing the maize tassel from getting in touch with his feminine side. Sci Signal. 2009;2:pe9. doi: 10.1126/scisignal.259pe9. [DOI] [PubMed] [Google Scholar]
- 2.Wager A, Browse J. Social Network: JAZ Protein Interactions Expand Our Knowledge of Jasmonate Signaling. Front Plant Sci. 2012;3:41. doi: 10.3389/fpls.2012.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 4.Farmer EE, Almeras E, Krishnamurthy V. Jasmonates and related oxylipins in plant responses to pathogenesis and herbivory. Current opinion in plant biology. 2003;6:372–378. doi: 10.1016/s1369-5266(03)00045-1. [DOI] [PubMed] [Google Scholar]
- 5.Liechti R, Farmer EE. The jasmonate pathway. Science. 2002;296:1649–1650. doi: 10.1126/science.1071547. [DOI] [PubMed] [Google Scholar]
- 6.Pauwels L, Inze D, Goossens A. Jasmonate-inducible gene: What does it mean? Trends in plant science. 2009;14:87–91. doi: 10.1016/j.tplants.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonseca S, et al. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol. 2009;5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 9.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 11.Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- 12.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen R, et al. The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 2012;24:2898–2916. doi: 10.1105/tpc.112.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Calvo P, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cevik V, et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012;160:541–555. doi: 10.1104/pp.112.202697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheard LB, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melcher K. The strength of acidic activation domains correlates with their affinity for both transcriptional and non-transcriptional proteins. J. Mol. Biol. 2000;301:1097–1112. doi: 10.1006/jmbi.2000.4034. [DOI] [PubMed] [Google Scholar]
- 18.Sun X, Rikkerink EH, Jones WT, Uversky VN. Multifarious roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell. 2013;25:38–55. doi: 10.1105/tpc.112.106062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triezenberg SJ. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 20.Smolen GA, Pawlowski L, Wilensky SE, Bender J. Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics. 2002;161:1235–1246. doi: 10.1093/genetics/161.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa P, Browse J. The Arabidopsis JAZ2 promoter contains a G-Box and thymidine-rich module that are necessary and sufficient for jasmonate-dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 2012;53:330–343. doi: 10.1093/pcp/pcr178. [DOI] [PubMed] [Google Scholar]
- 22.Melotto M, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung HS, et al. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. Plant J. 2010;63:613–622. doi: 10.1111/j.1365-313X.2010.04265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazan K, Manners JM. MYC2: The Master in Action. Molecular Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 25.Kidd BN, et al. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell. 2009;21:2237–2252. doi: 10.1105/tpc.109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 27.Pauwels L, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 29.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Ke J, et al. Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat Struct Mol Biol. 2013;20:1377–1382. doi: 10.1038/nsmb.2710. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suino K, et al. The Nuclear Xenobiotic Receptor CAR; Structural Determinants of Constitutive Activation and Heterodimerization. Molecular Cell. 2004;16:893–905. doi: 10.1016/j.molcel.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 37.Xu HE, et al. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 38.Withers J, et al. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc Natl Acad Sci U S A. 2012;109:20148–20153. doi: 10.1073/pnas.1210054109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang DL, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 41.Yao J, Withers J, He SY. Pseudomonas syringae infection assays in Arabidopsis. Methods Mol Biol. 2013;1011:63–81. doi: 10.1007/978-1-62703-414-2_6. [DOI] [PubMed] [Google Scholar]
- 42.Chalmers MJ, et al. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 43.Pascal BD, et al. HDX workbench: software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23:1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.