Abstract
By introducing a capping step and controlling reaction parameters, the assembly of metallic nanoparticle aggregates can be achieved using a small molecule crosslinker. Aggregates can be assembled from particles of varied size and composition and the size of the aggregates can be systematically adjusted. Following cell uptake of 60 nm aggregates, the aggregates are stable and non-toxic to macrophage cells up to 55mM Au.
Keywords: Gold nanoparticles, nanoaggregates, nanoclusters, biocompatible, covalent assembly
Early work on metallic nanoparticle aggregates made use of materials resulting from uncontrolled aggregation of citrate stabilized gold particles induced either by mixing with organic solvents[1] or salts[2]. These preparations enabled the discovery of many promising properties of metallic nanoparticle aggregates, such as their enhancement of raman spectroscopy[3], but these aggregates were poorly suited for biological applications due to the irreproducible nature of the random assembly. Small molecule crosslinkers would be ideal for controlling the assembly of metallic nanoparticles, as such crosslinkers are relatively straightforward to synthesize and this offers the opportunity to systematically control crosslinker properties. However, it has proven extremely difficult to use small molecule crosslinkers for nanoparticle aggregation, likely due to the challenge of controlling relative rates of initiation, propagation and termination. This is probably the result of the similarity between species responsible for initiation (a single particle coated in the small molecule crosslinker) and propogation (a cluster of particles coated in the small molecule crosslinker) and the lack of a true termination step. As a result, most efforts to use small molecule crosslinkers have resulted in the preparation of unstable aggregates that eventually undergo uncontrolled agglomeration. Here we show that small molecule crosslinkers can be used for the controlled assembly of metallic nanoparticles by adjusting the concentration of starting nanoparticles or the concentration of the crosslinker and introducing a termination step by capping reactive surfaces on aggregates.
In the absence of a small molecule crosslinker alternative, a number of different techniques for controlling the assembly of nanoaggregates have been developed.[4] Most previous controlled assemblies of metallic nanoparticle aggregates for biological applications have relied on macromolecule crosslinkers,[5] most commonly DNA.[6] Aggregates prepared in this manner have been used for ex vivo diagnostics[7] and proof-of-principle studies on the potential of aggregates for drug delivery[8] or for improved biodistirbution via accelerated clearance from the liver relative to solid nanoparticles of similar size.[9] However, DNA is an expensive crosslinker and, more importantly, is a bioactive molecule that can be cleaved by nucleases in the blood,[10] and/or induce an immune response[11].
Recently, gold nanoparticle aggregates have been assembled by simply mixing particles with block copolymers that induce assembly.[12, 13] These structures have shown promise for biological applications, but limited control over the aggregate assembly has been described in either case. In addition, as is suggested by the heterogeneous structures observed when some of these aggregates were internalized by cells,[12] non-covalently assembled aggregates may be susceptible to reorganization or other alterations in vivo. In a related example, magnetic iron oxide nanoparticles and CdSe quantum dots were assembled in surfactant to produce core-shell aggregates with the iron oxide particles serving as the core and the quantum dots serving as the shell.[14] While control over aggregate size was demonstrated for these aggregates, they required encapusaltion within a silica shell to take advantage of their dual magnetic and fluorescent properties for biological appliations. The use of small molecule crosslinkers for assembly would ideally combine the advantages of all of these approaches, without the various disadvantages, and allow for the direct synthesis of stable, biologically compatible aggregates in a controlled fashion.
There are limited examples of covalent assembly of metallic nanoparticle aggregates using small molecule crosslinkers, but they have been ill suited for biological applications. The Grzybowski[15] group used a photoswitchable small molecule crosslinker to assemble aggregates. The ability to switch the conformation of the hydrophobic dithiol crosslinker was used to control the relative rates of initiation and propagation, resulting in aggregates with relatively uniform morphology. However, these aggregates were hydrophobic and described as being very “sticky”, rendering them incompatible with biological applications. Intriguingly, the Brust[16] group reported the assembly of gold nanoparticle aggregates in toluene using a small dithiol alkane, where the assembly was highly sensitive to the concentration of the crosslinker. The hydrophobicity of these aggregates again makes them incompatible with biological applications, but this work suggested that it is possible to control the assembly of metallic nanoparticle aggregates using small molecule crosslinkers.
Here we demonstrate that the controlled assembly of biocompatible metallic nanoparticle aggregates can be realized by: using a crosslinker miscible in water, controlling the concentration of starting nanoparticles or crosslinker and capping reactive thiols present on the aggregates’ surfaces after initial assembly. The aggregates can be assembled from starting nanoparticles of varied size and composition and the size of the resulting aggregates can be tuned. The aggregates prepared using this method are relatively homogenous in size and morphology. The gold aggregates are biocompatible and show no toxicity when incubated with macrophage cells up to 55 mM Au. Finally, the aggregates are highly stable and appear unchanged after uptake by cells. It is expected that this straightforward and inexpensive assembly of highly stable nanoparticle aggregates will expand the biological applications of this class of materials. Furthermore, this method for preparing aggregates is highly modular as the crosslinker, the building block nanoparticles and the exterior coating can all be independently varied and the use of alternative crosslinkers and capping agents will enable applications in diverse material applications.
RESULTS
Aggregate Synthesis and Scope of Nanoparticle Building Blocks
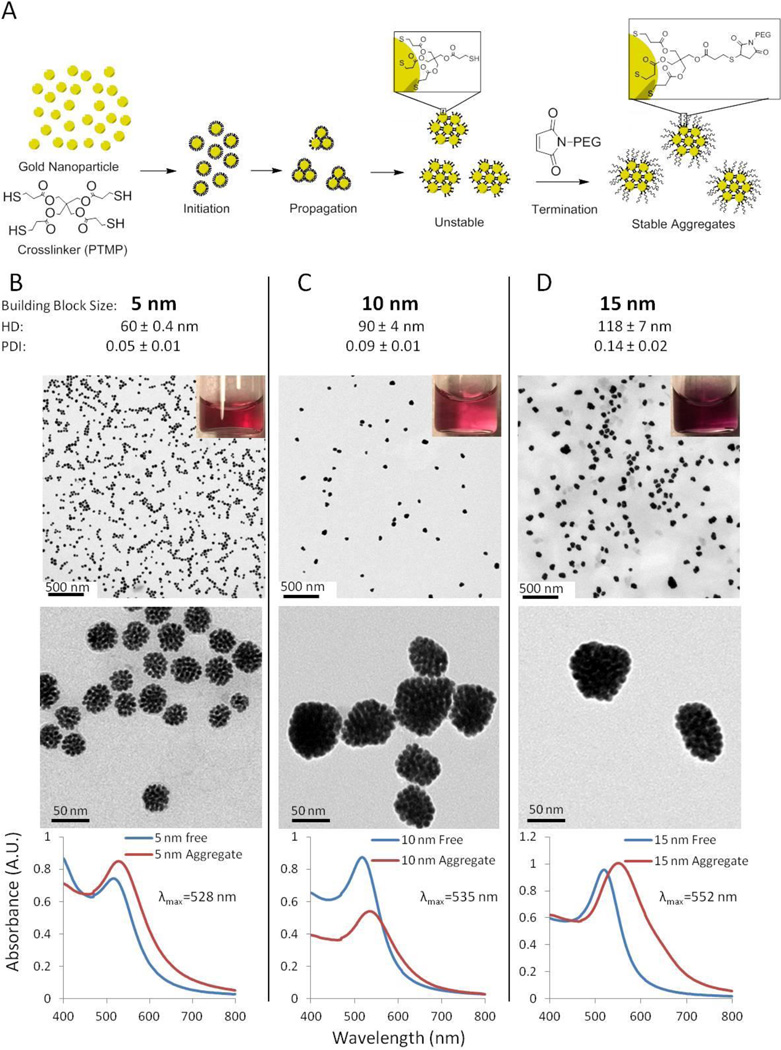
Due to its recent use in assembling gold nanoparticle oligomers,[17] we chose pentaerythritol tetrakis (3-mercaptopropionate) (PTMP) as our water miscible crosslinker and evaluated if it could be used to assemble 5 nm gold nanoparticles into aggregates. Order of addition, mechanism of mixing (ie. stirring, shaking or rocking), time, temperature and solvent were evaluated. It was found that aggregates could be prepared by adding an aqueous solution of gold nanoparticles to a solution of PTMP and PEG in a mixture of ethanol and water, followed by shaking at room temperature for 2 hours on a tabletop shaker at top speed and then letting the reaction stand on the benchtop for 24 hours. However, these aggregates were “sticky”, just as the Grzybowski lab described for their aggregates assembled only from a dithiol crosslinker.[15] As a result, the aggregates were very difficult to handle as they would stick to most surfaces they came in contact with, such as vial walls or tubing. We hypothesized that the “stickiness” resulted from free thiols on the surface of the aggregates, so we capped them by adding a solution of PEG2000-maleimide and shaking for another 2 hrs (Figure 1A). The aggregates could then be collected by centrifugation, washed and purified by filtration through a 0.2 µm membrane. The aggregates had a zeta potential of −23.2 mV and were easily handled. Dynamic light scattering (DLS) measurements found that the resulting aggregates had a hydrodynamic diameter (HD) of 60 nm and a very low polydispersity index (PDI) of 0.05. The low polydispersity is a measure of the relative uniformity between the aggregates formed, which is suggestive of growth occurring via monomer-cluster growth mechanism. This is discussed more fully in the next section. Transmission electron microscopy (TEM) confirmed that the aggregates were relatively homogeneous in size and morphology (Figure 1B). The size of the aggregates by TEM (40 nm) correlates well with that observed by DLS (60 nm). As is generally observed for nanoparticles, the hydrodynamic diameter is greater than the size of the gold core seen in TEM images. This size difference is the result of the hydrodynamic diameter measuring the presence of organic ligands on the surface and the solvating layer surrounding the particle aggregates, while only the metallic particles themselves are visualized in TEM. Thermogravimetric analysis (TGA) indicated that the aggregates were composed of 11% by weight crosslinker, 14% by weight PEG-maleimide and 75% gold (Supplementary Figure 1). In order to evaluate how generalizable the protocol was, in addition to the 5 nm gold nanoparticles, aggregates were also prepared from 10 and 15 nm gold nanoparticles (Figure 1C,D) and silver and platinum nanoparticles (Supplementary Figure 2).
Figure 1. Synthesis of gold nanoparticle aggregates using a small molecule crosslinker.
(A) Unstable aggregates are synthesized by combining gold nanoparticles and a crosslinker (PTMP) in the presence of PEG. It is hypothesized that the nanoparticles are initially coated with PTMP before aggregating in small clusters. The aggregates are stabilized by capping free thiols with a PEG-maleimide. Aggregates were assembled from (B) 5, (C) 10 and (D) 15 nm spherical gold nanoparticles. HD and PDI are presented as the mean +/− standard error of the mean from measurements of 12, 3 and 4 separate preparations of the aggregates from 5 nm, 10 nm and 15 nm starting particles, respectively. Representative TEM images show the homogeneity of the aggregates and their structure at increasing magnification. UV-Vis spectroscopy indicates the characteristic red shift of maximal absorbance upon aggregation.
Mechanism of formation and control of aggregate size
The relatively homogenous spherical morphology observed suggested that the aggregates formed from a monomer-cluster growth mechanism, as this morphology is characteristic of this growth mechanism.[18] The Grzybowski lab performed measurements and modeling that also supported this mechanism for the growth of their “sticky” aggregates.[19] In this mechanism, there is the initial formation of small clusters which then grow by interacting with and binding to individual particles until all of the free nanoparticles are consumed. As a result, assuming the rate of aggregation is significantly faster than the rate of initiation, it should be possible to control the final size of the assembled aggregates by increasing the free particles/initial clusters ratio. This could be achieved either by increasing the number of free particles or by decreasing the number of initial clusters. We hypothesized that increasing the concentration of gold nanoparticle building blocks would serve to increase the free particles remaining after initial cluster formation and that increasing the concentration of PTMP would decrease the number of initial clusters because any free site for reaction on a given particle would be more likely to react with free PTMP in solution than PTMP on another particle.
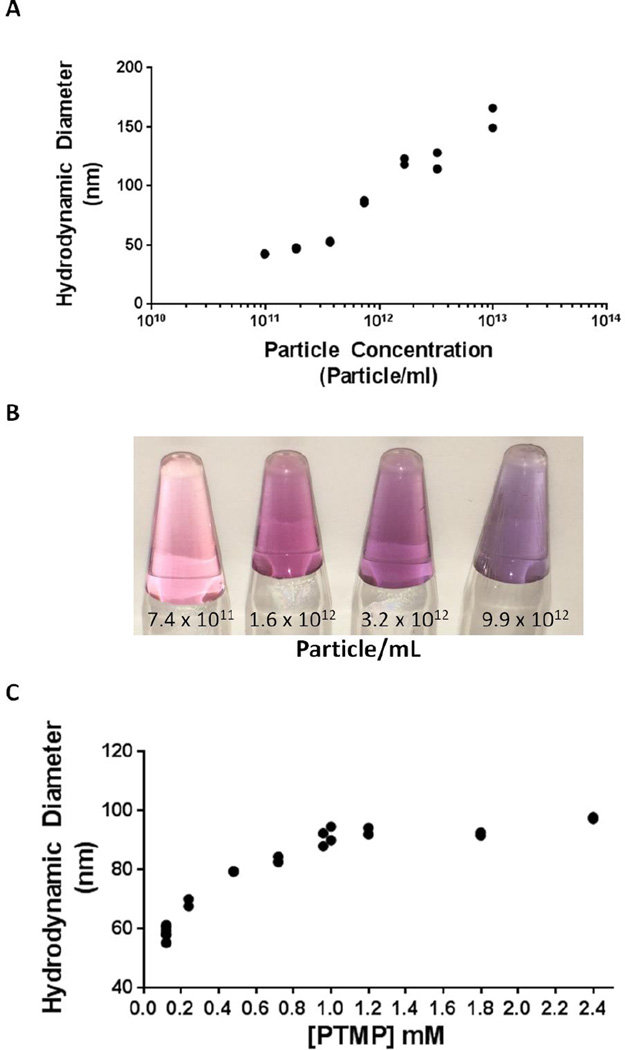
Indeed, we found that the size of the aggregates produced depended linearly on the concentration of nanoparticles added to the reaction. While the concentration of PTMP and the reaction volume were held constant, increasing the concentration of 10 nm nanoparticles from 9.8 × 1010 to 9.9 × 1013 particles/mL resulted in aggregates linearly increasing in HD from 40 to 160 nm. (Figure 2, Supplementary Figure 3 for TEM). Alternatively, holding the concentration of nanoparticles and the reaction volume constant, increasing the concentration of PTMP from 0.12 mM to 2.4 mM resulted in a logarithmic increase in aggregate HD from 60 to 100 nm. Interestingly, if the concentration of PTMP is too low, either no assembly or uncontrolled agglomeration is observed (Figure S4). This might explain why there are so few previous reports of nanoparticle assembly using small molecule crosslinkers, as the phenomenon only occurs for specific ranges of [nanoparticles] and [crosslinker]. For all of our experiments, assembly was only observed if the value of (# of thiols)/nm2 was at least 93 (Figure S5). We speculate that adjusting the concentration of starting nanoparticles and crosslinker is a general strategy to assemble nanoparticles using small molecule crosslinkers.
Figure 2. Aggregate Size Can Be Controlled by Particle Concentration or Crosslinker Concentration.
(A) For 10 nm starting material particles aggregated with 0.59 mM PTMP, the hydrodynamic diameter increases from 40 nm up to 160 nm with increasing concentration of particles. (B) The increase in size is reflected in a shift in color from pink to purple of solutions of aggregates in water. (C) For 5 nm starting material particles aggregated at a concentration of 5 × 1013, the hydrodynamic diameter increases from 60 nm up to 100 nm as PTMP concentration increases from 0.12 to 2.4 mM. Each condition tested is at least n ≥ 2, all values are included on the graph, though in some cases the two replicates overlap almost completely.
Biocompatibility of Aggregates
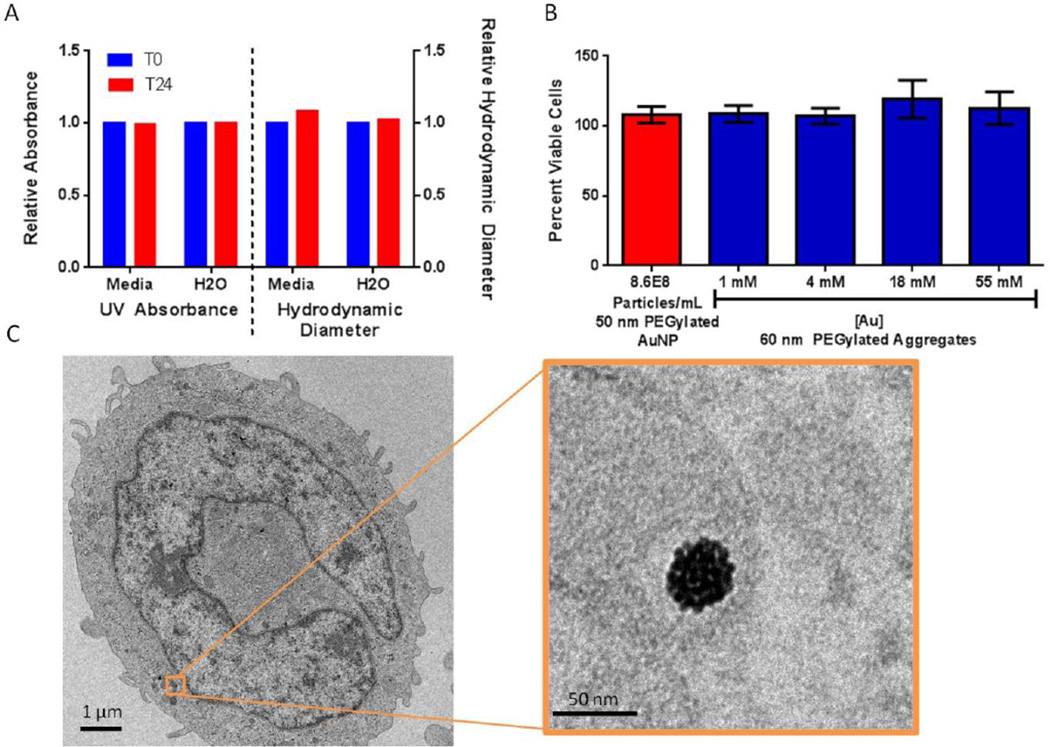
The PEGylated aggregates were readily dispersed in water, dimethyl sulfoxide (DMSO) and dimethyl formamide (DMF). Interestingly, similar to PEG itself, the aggregates could also be easily dispersed in chloroform which may facilitate materials applications where an organic solvent is desired. The PEG-Maleimide-capped aggregates were stable in water on the benchtop for at least 1 year with no change in HD (60.4 nm at t=0 and 60.0 nm after 1 year) and little change in PDI (0.05 at t=0 and 0.07 after 1 year) (Figure S6). The PEG-Maleimide-capped aggregates were stable in complete cell media with small changes in HD (60.4 nm at t=0 and 65.4 nm at t= 24hr), and PDI (0.01 at t=0 and 0.13 at t=24 hr) and no change in the absorbance of the aggrgates in complete media after 24 hours (Figure 3A). We treated macrophage cells (THP-1) for 24 hours with escalating doses (1mM Au up to 55 mM Au) of 60 nm PEG-maleimide-capped aggregates assembled from 5 nm particles. Macrophage cells were selected for initial testing as they readily uptake nanoparticles and as a result, all intravenously administered nanoparticles interact with macrophages.[20] The cells treated with escalating doses of PEG-maleimide-capped gold aggregates showed no change in cell metabolism compared to cells receiving no treatment (Figure 3B). Furthermore, TEM imaging showed that THP-1 cells treated with 60 nm aggregates (18 mM Au) showed no change in morphology after 24 hours and the aggregates were found in vesicles within the cells (Figure 3C, Figure S8). Remarkably, the aggregates showed no change in morphology when entrapped within cell vesicles. This is in sharp contrast to previous work with noncovalently assembled aggregates.[21] Thus, the aggregates appear to be biocompatible with reasonable stability in aqueous salt solutions, no cytotoxic behavior at the concentrations tested and the ability to be taken up into cells without significant perturbation. This retention of structure after internalization may be advantageous for processes that depend on nanoparticle organization, such as laser induced nanobubbles[22] and surface enhanced Raman spectroscopy[3].
Figure 3.
A) Nanoaggregates are stable in complete cell media up to 24 hour at room temperature. B) Representative data demonstrating the percentage of viable cells for each experimental group normalized to THP-1 cells receiving no treatment. 50 nm pegylated particles were used as an additional control. C: Uptake of 60 nm nanoaggregates in a single cell, inset shows an aggregate in a vesicle within the cell. Scale Bar is 1 µm and 50 nm(inset).
CONCLUSION
The controlled assembly of biocompatible nanoparticle aggregates using small molecule crosslinker has been a long standing challenge, likely owing to difficulties in controlling rates of initiation, propagation and termination. Here we demonstrate that adjusting the [nanoparticles] and [crosslinker] allows for the preparation of relatively homogenous aggregates from metallic nanoparticles of varied composition and size, presumably by controlling the rates of initiation and propagation. Capping reactive thiols on the formed aggregates with PEG-maleimide provides a termination step and renders the aggregates stable and biocompatible. The size of the aggregates can be systematically adjusted. Since the coating of the aggregates is introduced by a simple capping using a maleimide, it should be possible to prepare aggregates with many functional coatings – such as antibodies, aptamers and fluorescent dyes. Moreover, linkers can be employed with different functional properties, such as pH dependent cleavage. The aggregates are stable in cell media, show no toxicity thus far and are unchanged following cell uptake. Future work will take advantage of the modular synthesis and biocompatibility to explore these aggregates as drug delivery vehicles and imaging agents. The availability of highly uniform aggregates may also make them useful for other materials applications.
Experimental Section
All materials were used as supplied. Citrate stabilized gold colloid suspensions (5 nm, 10 nm, and 15 nm) were purchased from Ted Pella. CellTiterGlo was purchased from Promega and Quant-IT PicoGreen Assay was purchased from Life Technologies. Phenol free-RPMI 1640 media was puerchased from Life Technologies. RPMI 1640 media and THP-1 cells were purchased from ATCC. 10× PBS was purchased from Corning. All other compounds were purchased from Sigma Aldrich. Pentaerythritol tetrakis (3-mercaptopropionate) was handled under Nitrogen at all times. NOTE: If PTMP is exposed to air and then used for the aggregation protocol, the size of the resulting aggregates cannot be accurately predicted.
Standard Aggregate Synthesis
A solution of polyethylene glycol (PEG2000) in diH2O (0.9 mg/mL) was prepared, for each reaction 178.5 µL was added to an eppendorf tube. A solution of pentaerythritol tetrakis (3-mercaptopropionate) (PTMP) in ethanol (0.4 mg/mL) was also prepared and for each reaction 116 µL was added to the water containing PEG2000. 500 µL of water containing 5 nm gold nanoparticles (5 × 1013 particles/mL) was added dropwise to the water-ethanol solution. The reaction solution was shaken for 2 hours on a tabletop shaker at top speed. After shaking, the reaction was left on the benchtop at room temperature for 24 hours.
The synthesis can also be performed without the initial addition of PEG and similar aggregates are formed (Figure S10)
Standard PEG-Maleimide Capping
A solution of PEG2000-Maleimide in diH2O (20 mg/mL) was prepared and for each reaction 430 µL was added to the reaction solution after it had stood for 24 hours at room temperature. The reaction was shaken at top speed on a table top shaker for 2 hours at room temperature. The particles were pelleted by centrifuging at 10,000g for 10 minutes. The supernatant was removed and the particle pellet was resuspended in 500 µL milliQ water. Particles were filtered using a 25 mm 0.2 µm polycarbonate track etch filter in a Swinnex housing. The filter was washed with 1.5 mL of milliQ water. The filtrate was pelleted via centrifugation (10,000g for 10 minutes) and resuspended in 500 µL of milliQ water.
Alternative Conditions for testing
[PTMP] Variation
The concentration of PTMP was varied from 200 nM to 2.4 mM while maintaining the total reaction volume and the amount of gold nanoparticles. Conditions were varied according to the table below. Final volume of ethanol was held constant at 116 µL. Volume of water containing PEG2000 was held constant at 178.5 µL (0.90 mg/mL).
| Final [PTMP] | PTMP in Ethanol Added |
Additional Ethanol Added |
5 nm AuNP in Water (µL) |
| 2.4 mM | 116 µL of a 8 mg/mL solution | - | 500 |
| 1.8 mM | 87.3 µL of a 8 mg/mL solution | 28.7 µL | 500 |
| 1.2 mM | 116 µL of a 4 mg/mL solution | - | 500 |
| 1.0 mM | 97 µL of a 4 mg/mL solution | 29 µL | 500 |
| 0.72 mM | 74.3 µL of a 4 mg/mL solution | 41.7 µL | 500 |
| 0.48 mM | 46.5 µL of a 4 mg/mL solution | 69.5 µL | 500 |
| 0.24 mM | 23.3 µL of a 4 mg/mL solution | 92.7 µL | 500 |
| 0.12 mM | 11.6 µL of a 4 mg/mL solution | 104.4 µL | 500 |
| 200 nM | 19.4 µL of a 4 µg/mL solution | 96.6 µL | 500 |
[Particle] Variation
10 nm gold nanoparticles were concentrated by centrifuging 24 mL of 10 nm stock gold nanoparticles (5.7e12 particles/mL) at 21,150 g for 30 minutes. The supernatant was removed from the pellet and the pellet was resuspended in 600 µL of diH2O resulting in a concentration of 4.5 × 1013 particles/ mL. Dilutions were made of the concentrated 10 nm particles. UV Absorbance was measured at 520 nm and particle concentration was determined using the extinction coefficient (9.55E7 M−1cm−1)[23]. Water containing PEG2000 (0.27 mg/mL, 87.8 µL) was mixed with 26.5 µL of ethanol containing PTMP (4 mg/mL), then 250 µL of water containing 10 nm gold nanoparticles at varying concentrations was added dropwise. The reaction solution was placed on a tabletop shaker for 2 hours at top speed. After shaking, the reaction was left on the benchtop at room temperature for 24 hours. The aggregates were capped with 98.1 µL of PEG-maleimide solution (20 mg/mL in water).
Particle Size
Nanoparticles were concentrated to equivalent total surface area from Ted Pella stock solutions via centrifugation (21,150g for 30 minutes) according to the table below. Surface area values were taken from the website of BBI solutions.[24] The aggregates were then synthesized according the standard aggregate synthesis above.
| Diameter of particles |
Initial Surface Area (cm2/mL)* |
Desired Surface Area (cm2/mL) |
Initial Volume for Centrifugation |
Volume Removed After Centrifugation |
| 5 nm | 39.3 | 39.3 | 500 µL | 0 µL |
| 10 nm | 17.9 | 39.3 | 1098 µL | 598 µL |
| 15 nm | 9.9 | 39.3 | 1985 µL | 1485 µL |
Particle Characterization
Dynamic Light Scattering and Zeta potential measurements were performed on the Zeta Pals instrument (Brookhaven Instruments Corporation). UV absorbance was measured on Ultrospec 3000pro (GE Lifesciences). In order to visualize the aggregates, 2 µL of aggregates solution were dried onto a formvar stabilized 200 mesh copper carbon grid purchased from TED Pella. TEM images were taken using FEI Tecnai 12 Twin. Extinction coefficients for aggregates synthesized from 5, 10 and 15 nm particles (Supplementary Figure 7) were determined by generating a standard curve between the UV absorbance at 530 nm and the concentration of gold determined via ICPMS. UV measurements were determined on Ultrospec 3000pro (GE Lifesciences). These samples were prepared for ICPMS by digesting them in 1 mL BDH Aristar ® Plus Nitric Acid (70%). The samples were diluted up to 10 mL with 2% Nitric Acid. Gold concentration was determined by ICPMS Analysis on Agilent 7500 Series using a concentric nebulizer, Scott type spray chamber, and torch with a fixed quartz injector torch. A CX interface was used. Plasma power was 1500 Watt. A standard curve was made using serial dilution of a 1ppm solution of gold standard solution (Spex CertiPrep). Data was analyzed quantitatively in a spreadsheet program. Extinction coefficients were used to determine the dose of Au added to the cells for viability assays and imaging.
For TGA analysis, 30 mL of capped or uncapped gold aggregates were prepared according to the standard synthesis but without the initial addition of PEG2000. The aggregates were concentrated by centrifugation and resuspended in water. Uncapped aggregates were difficult to resuspend and often stuck as a pellet. The pellet was able to be dislodged by pippetting and was then placed into the sample holder to dry. Particles were dried in the platinum sample holder under vacuum at 105°C. The sample was loaded into the sample pan of the TGA (Q50, TA Instruments) and the sample was heated to 200°C and held for 30 minutes to completely dry the sample and then heated up to 800°C at a rate of 20°C /minute. The starting material weight was the weight at the end of the drying period and the final gold weight was the weight at 800°C.
Cell Growth and Viability Experiments
THP-1 cells from ATCC (TIB-202) were cultured in complete RPMI 1640 ATCC formulation Media (10% FBS, 1% Penicillin and streptomycin, and 0.05 mM β-mercaptoethanol). THP-1 cells were grown in suspension. For each experiment 50,000 cells were added to each well and then treated with 5 ng/mL Phorbol 12-myristate 13-acetate (PMA, Sigma Aldrich) for 48 hours in order to differentiate the human monocytes THP-1 cells into macrophages[25]. After 48 hours, the supernatant was removed, the cells were washed with PBS, and 195 µL fresh media was added. Escalating doses of gold nanoaggregates determined by UV absorbance in 5 µL were added to each well for 24 hours at 37°C. Final concentration of gold ranged from 1 mM Au to 55 mM Au this concentration was calculated from the extinction coefficient for 60 nm aggregates assembled from 5 nm particles (14.39 MAu−1cm−1) (Figure S9). 50 nm PEGylated particles were added as a control, optical density of these aggregates (1.110 at 520 nm) was between 1 and 4 mM Au doses for the aggregates. Particle concentration is reported for the 50 nm particles based on extinction coefficient (1.935E10 Mparticles−1cm−1).[23] After a 24-hour treatment with particles, the media was removed and replaced with 100µL Cell Lysis Buffer (20mM Tris, 2mM EDTA, 150mM NaCl, 0.5% Triton X-100, pH 7.4). Cells were frozen to ensure complete cell lysis. ATP concentration at the time of lysis was measured using the CellTiter-Glo® Assay. ATP concentration is correlated with metabolic activity in cells. In the CellTiter-Glo® Assay, the CellTiter-Glo® substrate is converted into a luminescent substrate which is proportional to the amount of ATP in the cell lysate[26]. In order to normalize to cell number the amount of double stranded DNA in the cell lysate was measured by the fluorescence of PicoGreen® reagent. PicoGreen reagent fluoresces upon binding to double stranded DNA. Experimental conditions were normalized to the no treatment control.
Cell Uptake Experiments
3 million THP-1 cells in a T75 flask were differentiated with 5 ng/mL PMA-media solution for 48 hours. The PMA media was removed and the cells washed with PBS. 9.5 mL of fresh media were added to the cells. 250 µL of gold nanoaggregates (70.3mM, determined by UV absorbance at 530) in water were added to cells and fresh media. Thus, the final concentration of aggregates in the flask was 18mM Au. After 24 hours at 37°C, the media containing excess aggregates was removed. The cells were washed with 5 mL of 1× PBS. Cells were removed from the flask using 0.25% Trypsin for 5 minutes at 37°C. The trypsination was terminated by the addition of fresh media. The cell-media mixture was centrifuged at 130g for 5 minutes to pellet the cells. The cell pellet was washed with 10 mL of 1× PBS and centrifuged to pellet. The cell pellet was then resuspended in 1 mL of EM grade 2% glutaraldehyde in 0.1 M Cacodylate buffer at pH 7.4. The fixed cells were stored overnight at 4°C. The next day the cells were embedded in 10% gelatin. After the gelatin became firm, the gelatin was minced and fixed in 2% glutaraldehyle in 0.1 M Cacodylate buffer at pH 7.4 for 30 min at room temperature. The cell pellets were washed three times with 0.1M Cacodylate buffer, pH7.2, post-fixed with 1% OsO4 in 0.1M Cacodylate buffer for 30 min and washed three times with 0.1M Cacodylate buffer. The samples were then dehydrated through 60%, 70%, 80%, 95% ethanol, 100% absolute ethanol (twice), propylene oxide (twice), and were left in propylene oxide/Eponate (1:1) overnight at room temperature. The vials were sealed. The next day the vials were left open for 2–3 hours to evaporate the propylene oxide. The samples were infiltrated with 100% Eponate and polymerized at ~64°C for 48 hours. Ultra-thin sections (~70 nm thick) were cut using a Leica Ultra cut UCT ultramicrotome with a diamond knife and picked up on 200 mesh copper EM grids. Grids were stained with 2% uranyl acetate for 10 minutes followed by Reynold’s lead citrate staining for 1 minute. Electron microscopy was done on an FEI Tecnai 12 transmission electron microscope equipped with a Gatan Ultrascan 2K CCD camera. The TEM was operated at 120 KeV.
Cell Media Stability
100 µL Nanoparticle aggregates (60 nm) were added to 900 µL phenol free RPMI 1640 media(10% FBS, 1% Penicillin and streptomycin, and 0.05 mM β-mercaptoethanol). UV absorbance was measured every 30 minutes for the first 6 hours and every hour for the next 18 hours. At 22 and 24 hrs, aggregates were removed from the cuvette, spun down (45 minutes at 21,150g). Aggregates were washed with 1 mL of DiH2O and pelleted a second time (10 minutes at 10,000g). Aggregates were suspened in 1 mL fresh DiH2O. Hydrodynamic Diameter was measured and recorded as a percentage of original size.
Salt Stability Experiments
Nanomaterials were aliquoted into plastic cuvettes. 50 µL 10× PBS (Potassium chloride 2 g/L, Monopotassium phosphate 2.4 g/L, Sodium chloride 80g/L, Disodium phosphate 14.4 g/L, Tris Ultrapure 24.2 g/L) was added to 450 µL aggregates. Each cuvette was examined after an hour for changes in absorbance and hydrodynamic diameter.
Supplementary Material
Acknowledgements
We gratefully acknowledge: Rachael Mooney for guidance with the CellTiter-Glo/PicoGreen assay, Marcia Miller, Zhuo Li and Ricardo Zerda for assistance with sample prep and TEM imaging, Warren Chan for editorial advice, ICP-MS instrumentation under the supervision of Nathan Dalleska at the Environmental Analysis Center at the California Institute of Technology. Studies were supported by generous funding from STOP Cancer and ThinkCure!. Julia Longmate was supported by The Rose Hills Foundation as part of the Eugene and Ruth Roberts Summer Academy. Research reported in this publication included work performed in the Electron Microscopy Core supported by the National Cancer Institute of the National Institutes of Health under award number P30CA33572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author. TGA data and analysis, TEM imaging of aggregates formed from platinum and silver nanoparticles, TEM imaging of aggregates of different size synthesized by varying starting particle concentration, demonstration of unsuitable [PTMP] and determination of extinction coefficients for the aggregates are provided in the SI.
References
- 1.Lapotko D. Opt. Express. 2009;17:2538. doi: 10.1364/oe.17.002538. [DOI] [PubMed] [Google Scholar]
- 2.Schwartzberg AM, Grant CD, Wolcott A, Talley CE, Huser TR, Bogomolni R, Zhang JZ. The Journal of Physical Chemistry B. 2004;108:19191. [Google Scholar]; Albanese A, Chan WCW. ACS Nano. 2011;5:5478. doi: 10.1021/nn2007496. [DOI] [PubMed] [Google Scholar]
- 3.Hossain MK, Kitahama Y, Huang GG, Kaneko T, Ozaki Y. Appl. Phys. B. 2008;93:165. [Google Scholar]
- 4.Klinkova A, Choueiri RM, Kumacheva E. Chemical Society reviews. 2014;43:3976. doi: 10.1039/c3cs60341e. [DOI] [PubMed] [Google Scholar]
- 5.Giljohann DA, Seferos DS, Daniel WL, Massich MD, Patel PC, Mirkin CA. Angewandte Chemie International Edition. 2010;49:3280. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boal AK, Ilhan F, DeRouchey JE, Thurn-Albrecht T, Russell TP, Rotello VM. Nature. 2000;404:746. doi: 10.1038/35008037. [DOI] [PubMed] [Google Scholar]
- 6.Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Nature. 1996;382:609. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]; Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 7.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Science. 1997;277:1078. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 8.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Journal of the American Chemical Society. 2009;131:2072. doi: 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou LYT, Zagorovsky K, Chan WCW. Nat Nano. 2014;9:148. doi: 10.1038/nnano.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu X, Han MS, Mirkin CA. Angewandte Chemie. 2007;46:3468. doi: 10.1002/anie.200605249. [DOI] [PubMed] [Google Scholar]; Han G, Martin CT, Rotello VM. Chemical biology & drug design. 2006;67:78. doi: 10.1111/j.1747-0285.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- 11.Massich MD, Giljohann DA, Seferos DS, Ludlow LE, Horvath CM, Mirkin CA. Molecular Pharmaceutics. 2009;6:1934. doi: 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tam JM, Tam JO, Murthy A, Ingram DR, Ma LL, Travis K, Johnston KP, Sokolov KV. ACS Nano. 2010;4:2178. doi: 10.1021/nn9015746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Zaki A, Joh D, Cheng Z, De Barros ALB, Kao G, Dorsey J, Tsourkas A. ACS nano. 2013;8:104. doi: 10.1021/nn405701q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen O, Riedemann L, Etoc F, Herrmann H, Coppey M, Barch M, Farrar CT, Zhao J, Bruns OT, Wei H, Guo P, Cui J, Jensen R, Chen Y, Harris DK, Cordero JM, Wang Z, Jasanoff A, Fukumura D, Reimer R, Dahan M, Jain RK, Bawendi MG. Nature communications. 2014;5:5093. doi: 10.1038/ncomms6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klajn R, Bishop KJM, Fialkowski M, Paszewski M, Campbell CJ, Gray TP, Grzybowski BA. Science. 2007;316:261. doi: 10.1126/science.1139131. [DOI] [PubMed] [Google Scholar]
- 16.Hussain I, Zhang H, Brust M, Barauskas J, Cooper AI. Journal of colloid and interface science. 2010;350:368. doi: 10.1016/j.jcis.2010.06.016. [DOI] [PubMed] [Google Scholar]; Hussain I, Wang Z, Cooper AI, Brust M. Langmuir. 2006;22:2938. doi: 10.1021/la053126o. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Chen G, Yang M, Silber G, Xing S, Tan LH, Wang F, Feng Y, Liu X, Li S, Chen H. Nat Commun. 2010;1:87. doi: 10.1038/ncomms1089. [DOI] [PubMed] [Google Scholar]
- 18.Hiemenz PC, Rajagopalan R. Principles of colloid and surface chemistry. New York: Marcel Dekker; 1997. [Google Scholar]
- 19.Klajn R, Bishop KJ, Fialkowski M, Paszewski M, Campbell CJ, Gray TP, Grzybowski BA. Science. 2007;316:261. doi: 10.1126/science.1139131. [DOI] [PubMed] [Google Scholar]
- 20.Weissleder R, Nahrendorf M, Pittet MJ. Nat Mater. 2014;13:125. doi: 10.1038/nmat3780. [DOI] [PubMed] [Google Scholar]; Walkey CD, Olsen JB, Guo H, Emili A, Chan WC. J Am Chem Soc. 2012;134:2139. doi: 10.1021/ja2084338. [DOI] [PubMed] [Google Scholar]
- 21.Tam JM, Tam JO, Murthy A, Ingram DR, Ma LL, Travis K, Johnston KP, Sokolov KV. ACS nano. 2010;4:2178. doi: 10.1021/nn9015746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapotko D. Nanomedicine (Lond) 2009;4:813. doi: 10.2217/nnm.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Leng X, Wang M, Zhu Z, Dai Q. Ecotoxicol Environ Saf. 2011;74:1304. doi: 10.1016/j.ecoenv.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 24. [Google Scholar]
- 25.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Inflamm. res. 2007;56:45. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 26.Niles AL, Moravec RA, Riss TL. Current chemical genomics. 2009;3:33. doi: 10.2174/1875397300903010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.