Abstract
Alphaviruses have a nucleocapsid core composed of the RNA genome surrounded by an icosahedral lattice of capsid protein. An insertion after position 186 in the capsid protein produced a strongly temperature-sensitive growth phenotype. Even when the structural proteins were synthesized at the permissive temperature (28°C), subsequent incubation of the cells at the non-permissive temperature (37°C) dramatically decreased mutant capsid protein stability and particle assembly. Electron microscopy confirmed the presence of cytoplasmic nucleocapsids in mutant-infected cells cultured at the permissive temperature, but these nucleocapsids were not stable to sucrose gradient separation. In contrast, nucleocapsids isolated from mutant virus particles had similar stability to that of wildtype virus. Our data support a model in which cytoplasmic nucleocapsids go through a maturation step during packaging into virus particles. The insertion site lies in the interface between capsid proteins in the assembled nucleocapsid, suggesting the region where such a stabilizing transition occurs.
Keywords: alphavirus, nucleocapsid, virus assembly, virus budding, packaging, temperature-sensitive mutant
INTRODUCTION
Alphaviruses comprise a genus of small, enveloped plus-sense RNA viruses containing ∼40 members (Kuhn, 2013; Strauss and Strauss, 1994). Alphaviruses such as Chikungunya virus, Venezuelan equine encephalitis virus (VEEV) and Eastern equine encephalitis virus cause serious human diseases including arthritis and encephalitis (Griffin, 2013; Schwartz and Albert, 2010). There are currently no antiviral therapies for human alphavirus infections, and an approved human vaccine is only available for VEEV (Griffin, 2013; Paessler and Weaver, 2009). The alphaviruses Sindbis virus (SINV) and Semliki Forest virus (SFV) are less pathogenic for humans and have been intensively studied to characterize the entry, biogenesis, structure and assembly of members of this genus, and to identify potential targets for antiviral therapies.
Alphavirus particles contain a nucleocapsid (NC) core consisting of the RNA genome packaged with the viral capsid protein (reviewed in Kuhn, 2013). This central core is surrounded by the virus lipid bilayer containing the E1 and E2 transmembrane glycoproteins (Li et al., 2010; Voss et al., 2010). Alphaviruses infect cells by receptor-mediated endocytosis and low pH-triggered membrane fusion, which releases the NC into the cytoplasm (Kielian, 2014). Following release, the NC is disassembled and the RNA is translated to generate the viral RNA replication complex (Kuhn, 2013). During virus biogenesis the structural proteins are translated as a polyprotein from a subgenomic RNA, with the capsid protein located at the N-terminus of the polyprotein, followed by the E2 precursor protein (termed PE2/p62) and the E1 membrane fusion protein. The capsid protein is a chymotrypsin-like autoprotease that folds co-translationally and cleaves itself from the polyprotein (Aliperti and Schlesinger, 1978; Hahn et al., 1985; Nicola et al., 1999). The membrane proteins PE2 and E1 are then co-translationally translocated into the endoplasmic reticulum, form a heterodimer, and are transported through the secretory pathway to the plasma membrane. During transport PE2 is processed into the mature E2 protein by furin (deCurtis and Simons, 1988; Zhang et al., 2003). Virus particles bud from the plasma membrane.
Alphaviruses have a roughly spherical structure that is highly organized and contains 240 copies each of the capsid, E2 and E1 proteins (Cheng et al., 1995; Kuhn, 2013; Paredes et al., 1993; Tang et al., 2011; Zhang et al., 2011). The capsid protein forms a T=4 icosahedral lattice in which the N-terminal basic region (∼residues 1–113) associates with the viral RNA in the interior, and the protease domain (residues 114–264) forms 12 pentameric and 30 hexameric capsomers on the outside of the capsid shell (Choi et al., 1997; Choi et al., 1991; Tang et al., 2011; Zhang et al., 2011; Zhang et al., 2002). The E2/E1 membrane protein dimers are arranged in a T=4 icosahedral lattice composed of trimeric spikes. The cytoplasmic tail of each E2 protein interacts with a hydrophobic pocket on the C-terminal domain of the capsid protein. This 1:1 interaction of E2 and capsid is specifically required for virus budding (Jose et al., 2012; Lee et al., 1996; Zhao et al., 1994).
Cytoplasmic NC are assembled in alphavirus-infected cells and are detectable by electron microscopy at ∼6h post-infection (Brown et al., 1972; Kuhn, 2013). Biochemical and morphological studies indicate that the structure of cytoplasmic NC differs from that of the NC in released virus particles (Coombs et al., 1984; Lamb et al., 2010), suggesting that the NC organization changes through its association with the envelope protein lattice during budding (Jose et al., 2012; Pletnev et al., 2001). However, the structure of the cytoplasmic NC is sensitive to purification (Mukhopadhyay et al., 2002), complicating its interpretation. In addition, only a fraction of the cytoplasmic capsid protein actually buds into virus. Thus, it is not clear if NC undergoes a maturational step, and if so, how such maturation might take place.
Here we characterized the unique phenotype of SINV and SFV capsid mutants with insertions at the subunit interface of capsid pentamers and hexamers in the viral nucleocapsid. The mutants showed strongly temperature-sensitive growth, but even at the permissive temperature the insertion destabilized the cytoplasmic nucleocapsids to sucrose gradient sedimentation. In contrast, nucleocapsids released from budded mutant virus particles were stable to gradient sedimentation. Thus, the capsid insertion mutants revealed a biochemical difference between nucleocapsids in the infected cell cytoplasm and those in virus particles. Our data also suggest that the transition from the cytoplasmic NC state to that in virus particles involves the interface between capsid proteins.
RESULTS
Characterization of SINV capsid 186-TC insertion mutant
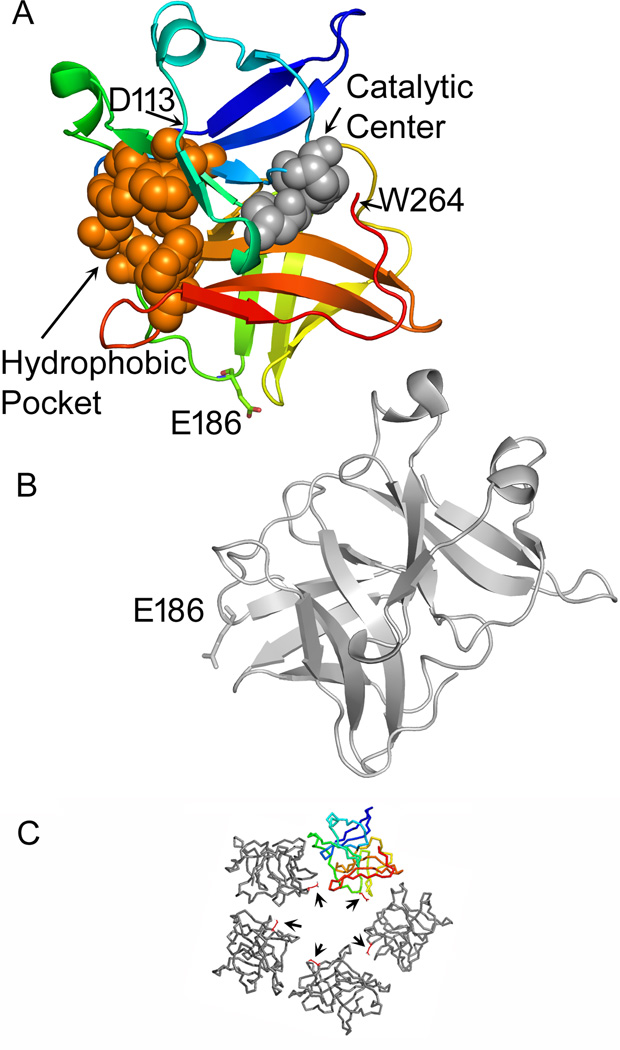
In earlier work we introduced tetracysteine (TC) tags into various positions on the SINV capsid protein (Zheng and Kielian, 2013). The tag was well-tolerated at several positions just N-terminal to the protease domain, allowing for live cell imaging studies. Insertions at several positions within the protease domain decreased virus growth by 1–7 logs. Unique among these mutants, insertion of the tag after capsid residue E186 in the protease domain produced strongly temperature-sensitive (ts) growth. This mutant was termed 186-TC. Examination of the structure of the SINV capsid protein showed that E186 is located at the tip of a flexible loop just before the A2 β-strand in the capsid protease domain (Fig. 1) (Choi et al., 1991). In addition, based on cryo-EM reconstruction of SINV and fitting studies (Tang et al., 2011; Zhang et al., 2002), E186 lies within the capsid-capsid interface of pentamers and hexamers on the surface of the virus nucleocapsid (Fig. 1).
Figure 1. Location of E186 in the SINV capsid-capsid interface.
The fitting of the SINV capsid protease domain into the viral nucleocapsid structure is described in reference (Zhang et al., 2002) and (PDB: 1LD4). A capsid pentamer from that structure is shown in panel C, with E186 shown as a red stick structure (arrows). The capsid protein shown in color and the gray protein located clockwise to it in the pentamer in C are displayed in the larger cartoon structures in A and B respectively, with E186 indicated on each. The colored subunit (A) is displayed in blue (starting at the N-terminal D113) to red (C-terminal W264). The residues that form the catalytic triad (H141, D163, and S215) are shown as space filling structures in grey. The residues that form the hydrophobic pocket into which the E2 cytoplasmic tail inserts (Lee et al., 1996; Skoging et al., 1996) are shown as space filling structures in orange. Figure was generated using PyMOL (DeLano, 2002).
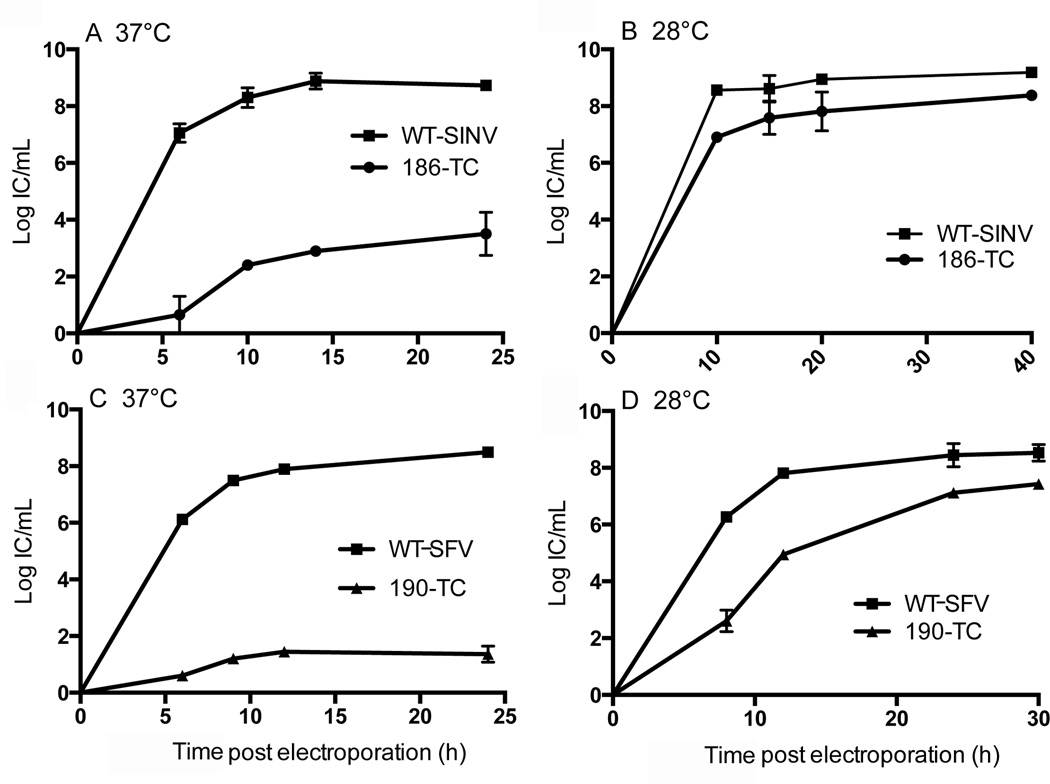
To quantitatively assess the temperature-sensitive phenotype, BHK-21 cells were electroporated with WT or 186-TC viral RNA and the kinetics of progeny virus production at 37°C and 28°C were determined. When the electroporated cells were incubated at 37°C, the titers of the 186-TC mutant were ∼4–6 logs lower than those of WT SINV at all time points (Fig. 2A). However, when the cells were incubated at 28°C efficient production of 186-TC was observed at 10 h post-electroporation and by 40 h the titer of 186-TC was less than 1 log lower than that of WT SINV (Fig. 2B). Thus growth of the 186-TC mutant was strongly temperature-sensitive. Mutant virus stocks prepared by 30h growth at 28°C were shown to maintain the ts phenotype when tested on fresh cells, and thus we did not observe significant generation of revertants within the time span of our experiments.
Figure 2. Growth curves of virus mutants.
BHK-21 cells were electroporated with WT or mutant viral RNAs and incubated at 37°C (A, C) or 28°C (B, D) for the indicated times. Mutants contained the TC motif inserted after SINV residue 186 (186-TC, panels A and B) or after the analogous SFV residue 190 (190-TC, panels C and D). At each time point, the culture medium was collected and the progeny virus titered by infectious center assay on BHK-21 cells. Data shown are the average of two independent experiments with the range indicated by the bars.
Growth properties of SINV capsid 186 insertion mutants
To test whether the temperature-sensitive phenotype of 186-TC is dependent on the specific sequence introduced, we generated SINV mutants with a variety of insertions at the same position. Substitution of all of the cysteine residues in the TC motif with serine (186-TSer) or replacement of the 12 residue TC motif with a 13 residue motif of a different sequence (186-LAP) resulted in strongly temperature-sensitive virus growth in each case (Table 1). To determine the effect of the length of the insertion, we tested insertions that contained only a half or a quarter of the TC sequence (1/2 TC, 1/4TC). Both of these mutants had a temperature-sensitive growth phenotype, but growth at the non-permissive temperature progressively improved as the size of the insertion was reduced. Together our data demonstrate that the temperature-sensitive phenotype was independent of specific residues or amino acid sequence and that smaller insertions produced a milder phenotype.
Table 1.
Growth properties of SINV capsid E186 insertion mutants
| Insertion at E186a | final titer at 37 °C vs. WTb | final titer at 28 °C vs. WTc |
|---|---|---|
| TC [FLNCCPGCCMEP] | 4–6 logs lower | 0.5–1 log lower |
| TSer [FLNSSPGSSMEP] | 3–4 logs lower | 0.5–1 log lower |
| ½ TC [FLNCCP] | 3–4 logs lower | 0.5–1 log lower |
| ¼ TC [FLN] | 2–3 logs lower | 0.5–1 log lower |
| LAP d [GFEIDKVWYDLDA] | 6–7 logs lower | 2–3 logs lower |
The indicated motifs were inserted into the SINV capsid protein after residue E186. Amino acid sequence of each motif is given in brackets.
BHK cells were electroporated with WT or SINV mutant RNAs, incubated for 24 h at 37°C, and the progeny virus titer determined by infectious center assay. Results represent the average of two independent experiments, with the log of the WT SINV titer averaging 8.5.
BHK cells were electroporated with WT or SINV mutant RNAs, incubated for 30 h at 28°C, and the progeny virus titer determined by infectious center assay. Results represent the average of two independent experiments, with the log of the WT SINV titer averaging 8.6.
LAP is a peptide motif developed for live cell fluorescent labeling (Uttamapinant et al., 2010).
Specific infectivity of 186-TC at 37°C versus 28°C
To address whether the entry of 186-TC is temperature-sensitive, the specific infectivities of 186-TC and WT SINV were compared at 37°C and 28°C. [35S]-labeled viruses were prepared by growth at 28°C, and then incubated with BHK cells at 37°C or 28°C to allow endocytic entry and virus fusion. Further infection was then blocked with 20mM NH4Cl and primary infection was quantitated after incubation at either temperature, and normalized to virus radioactivity (Table 2). The specific infectivity of the 186-TC mutant was slightly less than that of WT SINV at 37°C (∼31% of WT), and was also slightly lower than the WT at 28°C (∼ 62% of WT). Thus, the specific infectivity suggested that entry was not responsible for the 4–6 log reduction in mutant virus titer at 37°C vs. 28°C. This experiment also suggested that, once assembled, the mutant virus particles have similar stability to 37 °C incubation as the WT virus.
Table 2.
Specific infectivity of 186-TC SINV and 190-TC SFV mutants
| Infectivity at 37°C (IC/cpma) | Infectivity at 28°C(IC/cpma) | |||
| SINV | 186-TC SINV | WT SINV | 186-TC SINV | WT SINV |
| 1.2 ± 0.1 | 3.9 ± 0.2 | 51 ± 16 | 82 ± 4 | |
| SFV | 190-TC SFV | WT SFV | 190-TC SFV | WT SFV |
| 1.6 ± 0.4 | 3.4 ± 0.7 | 38 ± 19 | 34 ± 7 | |
Purified [35S]-labeled WT SINV, WT SFV, 186-TC SINV or 190-TC SFV virus stocks were allowed to infect BHK cells for 1–1.5 h at 37°C or 3.5–4 h at 28°C. Further infection was then blocked by addition of 20 mM NH4Cl, and the infected cells were quantitated after incubation at 37°C for 7 h or at 28°C for 16h. Data represent the average and range from two independent experiments, and are expressed as the ratio of infectious centers to virus radioactivity. The somewhat higher infectivity observed at 28 °C reflects the more efficient detection of primary infection with longer time of culture.
Surface expression of the envelope proteins is reduced in mutant-infected cells at 37°C
To characterize the production of mutant virus proteins at the non-permissive temperature, we tested monoclonal antibodies (mAbs) to the SINV envelope proteins. Our available SINV E1 or E2 mAbs failed to recognize all forms of the E1/E2 proteins in infected cells (unpublished data). We therefore tested whether insertions in the Semliki Forest virus (SFV) capsid produced the same phenotype as in SINV. TC and TSer insertions were introduced after the SFV capsid E190 residue (SFV numbering of analogous position). Both SFV 190-TC and 190-TSer showed strongly temperature-sensitive growth (Fig. 2C, 2D and data not shown). The specific infectivity of the 190-TC mutant was slightly less than that of WT SFV at 37°C (∼47% of WT), and approximately equivalent to that of the WT at 28°C (Table 2). Thus, the effect of the insertion was conserved between SINV and SFV, and we therefore used SFV 190-TC and/or SINV 186-TC interchangeably in subsequent experiments.
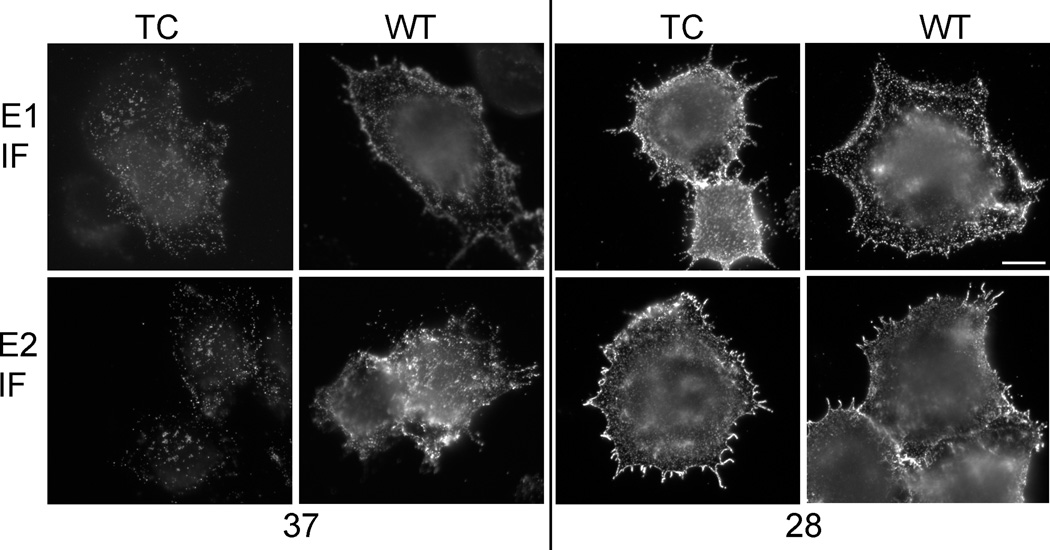
Cells were electroporated with SFV 190-TC RNA and incubated at 37 or 28°C, and surface expression of the E1 and E2 proteins was tested by immunostaining with mAbs against the E1 or E2 proteins on ice (Fig. 3). WT- and mutant-infected cells cultured at 28°C showed abundant surface expression of both the E1 and E2 envelope proteins. The mean fluorescence intensity of E2 and E1 immunostaining was comparable between the WT and mutant-infected cells (data not shown). However, mutant-infected cells cultured at 37°C showed decreased surface expression of both the E2 and E1 proteins (Fig. 3). The mean fluorescence intensity of E2 staining was 14 ± 4 (mean ± SD, N=20 in each case) for TC compared to 53 ± 20 for WT, and the mean intensity of E1 staining was 18 ± 6 for TC and 42 ± 15 for WT. Decreased intracellular staining for the mutant envelope proteins was observed when the cells were permeabilized. Under these conditions the mean intensity for E2 was 27 ± 9 for TC and 55 ± 20 for WT, and for E1 was 35 ± 15 for TC and 47 ± 25 for WT. Pulse labeling studies of cells equivalently electroporated with WT or mutant viral RNA showed that capsid protein levels were decreased in lysates of mutant-infected cells, particularly at the non-permissive temperature. The ratio of mutant capsid/WT capsid was 42% ±11% in cells infected at 28°C, while at 37 the ratio was 20%±5% (mean and STDEV, n=3). These results suggested that the auto-protease cleavage of the mutant capsid protein was decreased, resulting in an overall decrease in expression of the structural proteins, which was most severe at the non-permissive temperature.
Figure 3. The plasma membrane expression of WT and mutant E1 and E2 proteins.
BHK-21 cells were electroporated with WT SFV or 190-TC mutant viral RNA and incubated at 37°C for 7 h or 28°C for 15h. Cells were then incubated with mAbs against the E1 or E2 protein (Kielian et al., 1990) on ice, fixed with 3% paraformaldehyde, and stained with secondary antibody. Images shown are representative examples of two independent experiments. The scale bar represents 10 µm.
Temperature-sensitive assembly of SFV 190-TC
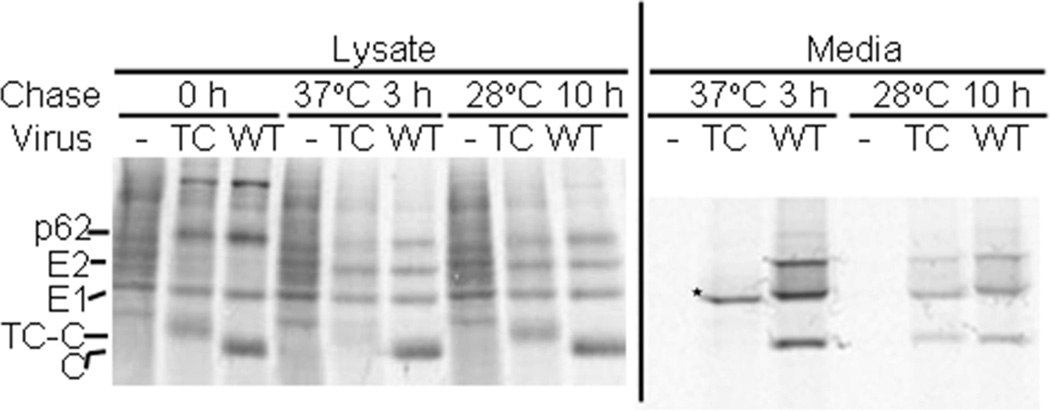
We then tested whether the assembly of SFV 190-TC virus particles was defective at 37°C. Cells were infected via electroporation with WT SFV or the 190-TC mutant RNA, pulse-labeled at the permissive temperature of 28 °C to permit efficient protein production, and then chased at either 37 °C or 28 °C. At each chase time the cells were lysed and virus particles released into the chase media were captured by immunoprecipitation. Aliquots of the cell lysates were directly analyzed by SDS-PAGE (Fig. 4, left panel). Both WT- and 190-TC-infected cells showed efficient expression of the p62, E1 and capsid proteins at the 0h time point, and maturation of p62 to E2 during chase at either 37 or 28°C. The E1 and E2 proteins were readily detected after chase at either temperature, indicating that E1 and E2 in the 190-TC-infected cells were stable at 37°C after synthesis at 28°C. However, capsid protein in the 190-TC-infected cells rapidly disappeared during the chase at 37°C but not at 28°C, suggesting that the capsid protein or nucleocapsid produced at 28°C was unstable when incubated at the non-permissive temperature. Selective loss of capsid protein at 37°C was also observed in cells infected with SINV 186-TC (data not shown).
Figure 4. Assembly properties of SFV 190-TCat 37 °C vs. 28°C.
BHK-21 cells were electroporated with WT SFV or 190-TC mutant viral RNA, incubated for 15 h at 28°C, pulse-labeled with [35S]-methionine/cysteine for 1 h at 28°C, and chased for 0 h or 3 h at 37 °C or 10 h at 28 °C. At the end of each chase period the media were collected and the cells were lysed. Left panel: Aliquots of the cell lysates were analyzed directly by SDS-PAGE. The positions of the envelope proteins p62, E2 and E1 and of the 190-TC-capsid (TC-C) and WT capsid (C) proteins are indicated. Note that TC-C migrates slower than C, and that there is a non-specific protein that migrates slightly above the E1 protein. Right panel: Samples of the chase media were immunoprecipitated with a polyclonal antibody against the E1 and E2 glycoproteins in the absence of detergent to recover intact virus particles (Liao and Kielian, 2006). The proteolytically truncated form of E1 E1s (Lu et al., 2001) is labeled with an asterisk. Shown is a representative example of two independent experiments.
Analysis of the chase media showed that both WT and 190-TC viruses were produced at 28°C, with all of the structural proteins clearly detected (Fig. 4, right panel). However, at 37°C the mutant-infected cells produced no detectable virus. Instead, only a faster-migrating form of E1 termed E1s was detected, as indicated by the asterisk. E1s is a proteolytically truncated form of E1 previously shown to be produced in cells in which virus assembly is defective (Lu et al., 2001; Zhao and Garoff, 1992). A comparable defect in virus particle production at 37°C was observed in pulse-chase studies of SINV 186-TC-infected cells (data not shown). Thus, the TC-capsid proteins did not support virus assembly at the non-permissive temperature.
Electron microscopy of mutant-infected cells
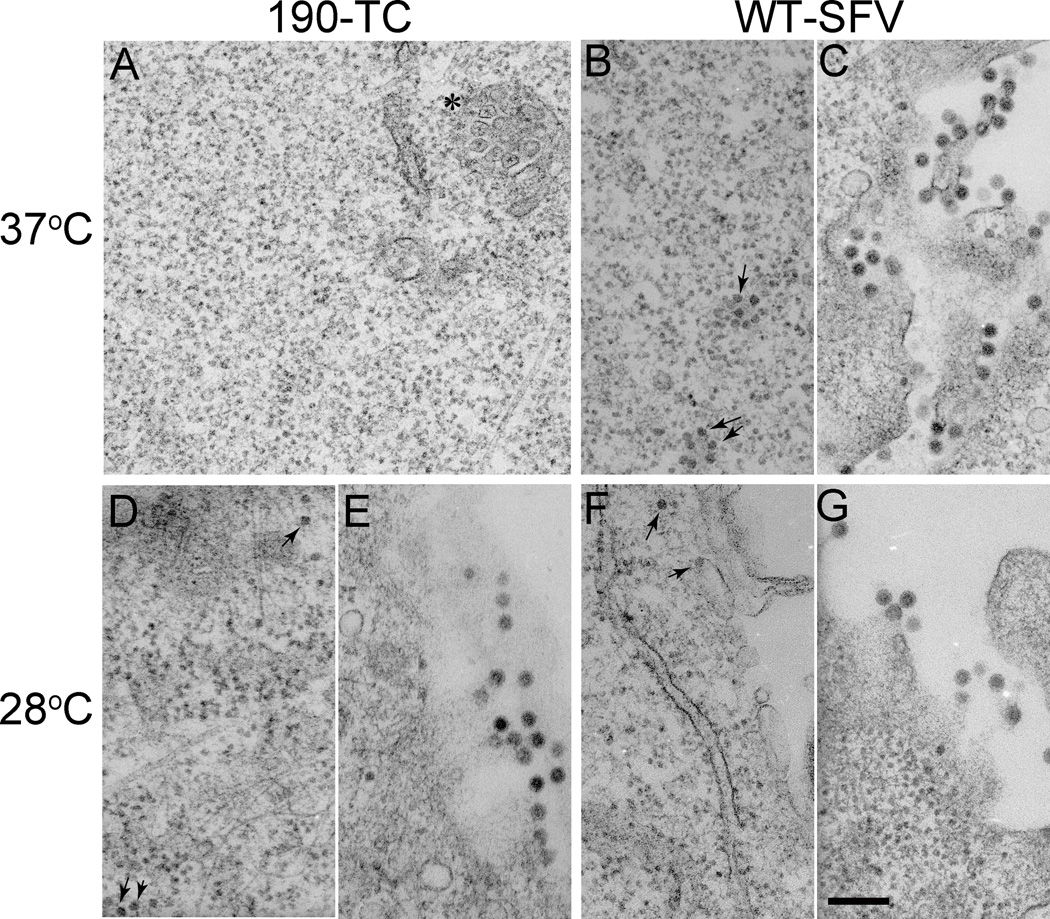
We next wished to evaluate the effect of the TC motif on the assembly and stability of cytoplasmic nucleocapsids. Cells were infected via electroporation with WT- or 190-TC- virus RNA, cultured at 37°C or 28°C and examined by electron microscopy (EM) (Fig. 5). The characteristic membranous replication structures termed cytopathic vacuole I (CPVI) (Kuhn, 2013; Salonen et al., 2005) were used as a marker to identify virus-infected cells (Fig. 5A, indicated by the asterisk). Although CPVI were readily detected in 190-TC infected cells at 37°C (Fig. 5A), no cytoplasmic nucleocapsids or virus particles were observed in any of the infected cells. In contrast, both nucleocapsids and virus particles were apparent in WT-infected cells (Fig. 5B and 5C). The lack of cytoplasmic nucleocapsids presumably is due to the observed reduction in 190-TC capsid protein generation and capsid protein stability at 37°C (Fig. 4), but may also reflect effects of the insertion on nucleocapsid assembly.
Figure 5. Electron microscopy of WT SFV or 190-TC mutant-infected cells.
BHK-21 cells were electroporated with WT SFV (B, C, F, G) or 190-TC mutant (A, D, E) viral RNA and incubated at 37°C for 2 h. Cells were then incubated at 37°C for 10 h (A, B, C) or 28°C for 18 h (D, E, F, G), and processed for electron microscopy. Panels C, E, and G show representative examples of the morphology of budding virus particles. Panel B, D, and F show representative examples of cytoplasmic nucleocapsids (indicated by arrows). Panel A is a representative image of 190-TC infected cells at 37°C with a cytopathic vacuole I structure indicated by the asterisk. All images were acquired at a magnification of 20,000 X, with the scale bar representing 200 nm.
EM analysis of cells cultured at 28°C showed abundant cytoplasmic nucleocapsids in both WT and 190-TC infected cells, with comparable morphology and diameters of ∼40nm (Fig. 5D vs. 5F, indicated by arrows). Virus particles were observed budding from the plasma membrane of both WT and 190-TC infected cells, with similar particle morphologies, diameters of ∼70 nm, and central dense nucleocapsid cores (Fig. 5E vs. 5G). Thus, at 28°C the 190-TC infected cells produced both cytoplasmic nucleocapsids and virus particles.
Mutant cytoplasmic nucleocapsids are unstable
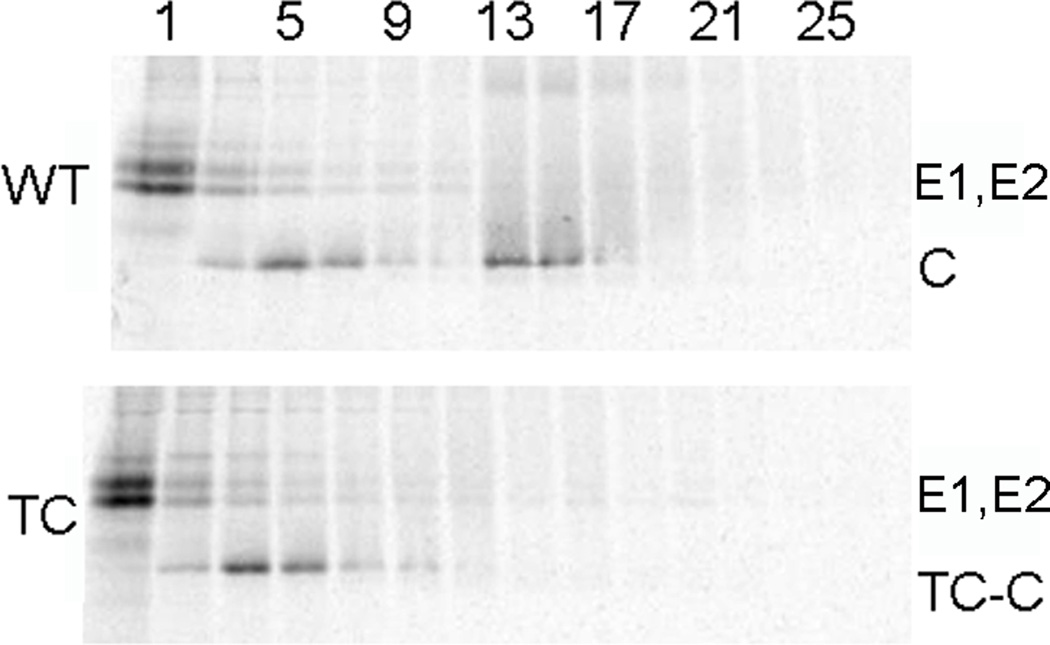
We then analyzed the cytoplasmic nucleocapsids from WT-SINV and TC-186 SINV mutant-infected cells by sucrose gradient sedimentation. Cells were infected with either virus, radiolabeled with [35S] at 28°C, lysed in NP-40-containing lysis buffer, and separated on sucrose gradients, and the fractions analyzed by SDS-PAGE and fluorography (Fig. 6). As previously observed in the pulse-chase experiments (Fig. 4), the WT and mutant structural proteins were comparably expressed at 28°C. The E1 and E2 envelope proteins remained at the top of the gradient (fraction 1) while the ribosome-bound capsid protein was detected with a peak at fraction 5, as previously observed (Ulmanen et al., 1976). As expected, the WT nucleocapsid sedimented with a peak at fractions 13–15 (Ulmanen et al., 1976). Surprisingly, however, the TC mutant capsid protein was not recovered in fractions 13–15, even though our EM results indicated that the mutant formed cytoplasmic nucleocapsids. Cell homogenates prepared and gradient analyzed in the absence of detergent showed a similar cytoplasmic nucleocapsid peak for WT infected cells but no comparable peak for mutant infected cells (data not shown). Gradient sedimentation analysis of SFV 190-TC infected cells also failed to detect cytoplasmic nucleocapsids (data not shown). Thus the mutant cytoplasmic nucleocapsids were unstable to gradient analysis.
Figure 6. Gradient stability of 186-TC SINV cytoplasmic nucleocapsids.
BHK-21 cells were infected with WT or 186-TC mutant SINV and labeled for 14 h with [35S]-methionine/cysteine at 28°C. Cells were then solubilized in lysis buffer, layered onto sucrose gradients, and separated by sedimentation. Aliquots of the fractions were analyzed by SDS-PAGE, with fraction 1 representing the top of the gradient. The positions of the E1, E2 and capsid (C or TC-C) proteins are indicated. Shown is a representative example of three independent experiments.
Mutant nucleocapsids show increased stability after packaging into virus particles
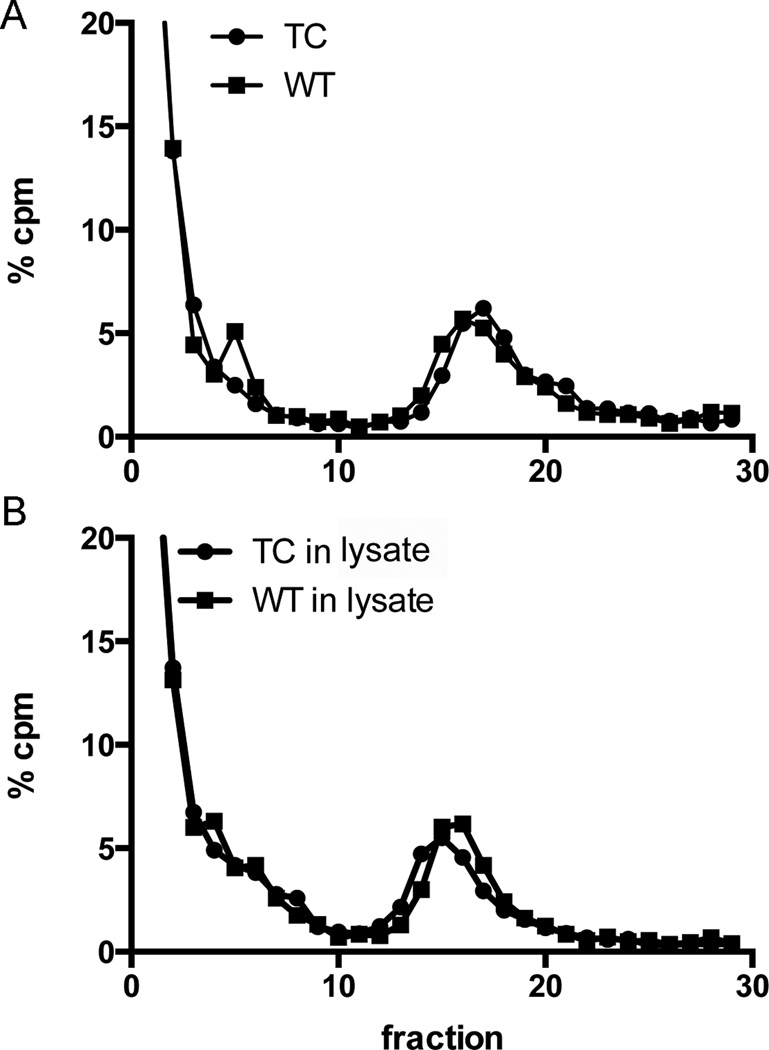
We then asked if the mutant nucleocapsids in virus particles also showed reduced stability. [35S]-labeled 186-TC SINV or WT SINV virus particles were produced, purified on sucrose gradients, solubilized with the same lysis buffer as that used to collect cytoplasmic nucleocapsids, and analyzed by sucrose gradient sedimentation. A clear nucleocapsid peak was detected in both the TC mutant and WT virus lysates samples (Fig. 7A).
Figure 7. Gradient stability of 186-TC SINV viral nucleocapsids.
Purified [35S]labeled WT or 186-TC mutant SINV viruses were solubilized in (A) lysis buffer alone or (B) lysates prepared from 186-TC mutant-infected BHK cells that were cultured at 28°C as in Fig. 6 but without radiolabeling. Solubilized virus samples were layered onto sucrose gradients and separated by sedimentation. Fractions were quantitated by scintillation counting, and the percentage of total radioactivity in each fraction was determined. Shown is a representative example of two independent experiments.
We tested the possibility that the cytoplasmic nucleocapsids were being destabilized by factors present in the lysates of mutant-infected cells. Purified labeled viruses were mixed with unlabeled lysates from mutant-infected cells cultured at 28°C as in Fig. 6, and the mixtures analyzed by sucrose gradient sedimentation. Both mutant and WT SINV samples gave similar nucleocapsid peaks (Fig. 7B), without significant decrease vs. the samples analyzed directly from virus (Fig. 7A). Similar results were obtained if the samples were incubated with lysates from WT-infected cells (data not shown). In addition, we also tested the thermostability of virus nucleocapsids when incubated in lysis buffer alone at either 37°C for 2 h or 28°C for 4 h. The recovery and position of the nucleocapsid peak was unchanged in both the WT and mutant samples (data not shown). Together, our results indicated that once the mutant nucleocapsid was packaged into virus particles, it remained stable to sucrose gradient sedimentation even if incubated at the non-permissive temperature or in cell lysates.
DISCUSSION
Temperature-sensitive alphavirus capsid mutants
A variety of SINV and SFV ts mutants have been previously described and shown to have genetic defects at various locations in the non-structural and structural proteins (Burge and Pfefferkorn, 1966; Keränen and Kääriäinen, 1974; Strauss et al., 1976). Several of these ts mutants contain amino acid substitutions close to the capsid protease domain. At the non-permissive temperature these mutants are defective in autoprotease cleavage and thus in capsid protein production (Hahn et al., 1985). A SINV mutant with substitutions at capsid positions 180 and 183 shows reduced particle specific infectivity and mild reductions in growth at both low and elevated temperatures (Lee and Brown, 1994). Interestingly, this mutant can produce both T=4 and T=7 virus particles at low temperature, suggesting alterations in capsid-capsid interactions.
Here we describe a unique temperature-sensitive phenotype caused by TC insertion in the SINV and SFV capsid proteins. The mutants had 4–6 log differences in growth between the permissive and non-permissive temperatures. It is striking that the TC mutants had such a strong temperature-sensitive phenotype compared to that of the previously described point mutants at positions 180/183. This appears to reflect the size of the 12 residue TC insertion, as smaller insertions decreased the effect (Table 1). The insertion reduced capsid autocleavage particularly at 37°C, decreasing production of the capsid and envelope proteins. Although E186 is on the opposite side of the protease domain from the catalytic center (Fig. 1), our data suggest that an insertion at this position affected the overall folding of the capsid protein at the non-permissive temperature, and thus reduced the activity of the protease.
In addition to these effects on protein production, the TC insertion mutants were strongly impaired in virus assembly at 37°C even when the viral proteins were synthesized at 28°C. At least two possible explanations could account for this temperature-sensitive assembly. First, the mutant capsid proteins might not produce stable NC at 37°C, resulting in a block in the assembly of the complete virus particle. In support of this model, the mutant capsid protein synthesized at 28°C was degraded during a 37°C chase (Fig. 4), and the cytoplasmic NC observed in infected cells at 28°C were not stable to sucrose gradient sedimentation. The destabilization might be even more drastic at 37°C and thus prevent the formation of cytoplasmic NC. However, mutations in capsid that block the formation of cytoplasmic NC can still result in viral NC formation and budding driven by the icosahedral interactions of the envelope proteins (Forsell et al., 1996; Forsell et al., 2000; Skoging-Nyberg and Liljestrom, 2001), while we observed no virus production at 37°C. Another possibility is that the maturation step that transitions the cytoplasmic NC to the mature viral NC could be temperature-sensitive for the TC mutants. In this model, NC would be assembled during the 37°C chase period but would be unable to “switch” to the mature NC state, and this block would inhibit particle formation and lead to capsid degradation. Further studies on the ability of cytoplasmic NC to incorporate into virus particles and the importance of NC maturation will be required to evaluate these and other possible models.
Different states of nucleocapsids
Four types of alphavirus NCs have been characterized biochemically and/or morphologically. These species are: NC obtained from the cytoplasm of the infected cell, NC core-like particles (CLPs) assembled in vitro from recombinant capsid protein and nucleic acid (e.g., Tellinghuisen et al., 1999), NC isolated by detergent lysis of mature virus, and the NC present in mature, intact virus. Early biochemical analyses suggest that cytoplasmic nucleocapsids and the nucleocapsids isolated from viruses differ in their sedimentation properties and sensitivity to ribonuclease (Coombs et al., 1984). Comparisons of cytoplasmic NC and in vitro assembled CLPs show that they have similar biochemical properties, suggesting similar structures (Mukhopadhyay et al., 2002). However, both of these NC species are sensitive to damage by gradient purification, precluding high-resolution reconstruction by cryo-electron microscopy (Mukhopadhyay et al., 2002). Morphological studies indicate that the capsid pentamers in CLPs are rotated ∼10° counterclockwise with the density at the fivefold axes projecting outwards, as compared to the NC present in mature, intact virus (Mukhopadhyay et al., 2002). Cryo-EM studies of VEEV cytoplasmic NC and isolated NC from mature virus also indicate conformational differences between these NC states, as well as with that of NC in the mature intact virus (Lamb et al., 2010; Paredes et al., 2003). Variations among these results may reflect differences in the alphaviruses used in these studies and the cellular proteins present in the various NC preparations, as well the technical difficulties of the reconstructions (Mukhopadhyay et al., 2002). In spite of these issues, though, in aggregate the evidence supports a change in the NC structure produced during virus budding.
Our results with the SINV and SFV TC mutants support a maturational change between cytoplasmic NC and NC of mature virus particles. Capsid protein position SINV186/SFV190 is located at the intersubunit interface of capsid pentamers and hexamers (Choi et al., 1991; Tang et al., 2011) (Fig. 1). The position suggests that the TC insertion caused spatial hindrance to the subunit contacts of the cytoplasmic NC, making them unstable to gradient sedimentation. The interaction with the envelope protein lattice during budding would induce a rearrangement of the NC, changing these contacts and making the NC more stable to gradient purification. Our results thus suggest that the capsid interface is involved in the transition to the mature viral NC.
Possible roles of NC states
Changes in the properties of the NC during virus biogenesis could have important consequences for virus infection. The nascent NC is stable during virus assembly yet it must be able to disassemble during virus entry into new host cells. Several models have been suggested to explain this NC change (reviewed in Garoff et al., 2004; Jose et al., 2009; Singh and Helenius, 1992a). NC disassembly during entry involves capsid protein binding to ribosomes (Singh and Helenius, 1992b; Wengler and Gros, 1996; Wengler and Wengler, 1984), and it was proposed that the ribosomes in infected cells gradually become saturated with capsid protein, making them unable to disassemble progeny NC. However, a similar loss of NC disassembly activity occurs in replicon-expressing cells that don't produce capsid protein (Singh et al., 1997). In addition, studies of NC from infected cells vs. budded virus (albeit produced by detergent lysis) show no difference in their disassembly by ribosomes (Wengler and Gros, 1996). Another suggestion is that the incoming NC is destabilized by exposure to endosomal acidity, but to date there is no direct evidence to support this model. The interactions of the envelope proteins with the NC during budding might also prime the virus for uncoating (Jose et al., 2012; Pletnev et al., 2001), making viral NC more sensitive than the cytoplasmic NC to ribosomes, low pH and/or other factors. Our results add to the evidence that changes in the NC structure occur during alphavirus budding, but further studies are required to determine their functional significance during virus infection.
MATERIALS AND METHODS
Cells
BHK-21 cells were cultured in complete BHK medium (Dulbecco's modified Eagle's medium containing 5% fetal calf serum, 10% tryptose phosphate broth, 100 U penicillin/ml, and 100 µg streptomycin/ml) at 37°C.
Construction of SINV and SFV infectious clones
The WT SINV used in our studies was derived from the dsTE12Q infectious clone provided by Dr. Beth Levine (Hardwick and Levine, 2000). SINV 186-capsid insertions were generated by overlapping PCR using primers encoding the optimized TC sequence FLNCCPGCCMEP (Adams et al., 2002), the TSer sequence FLNSSPGSSMEP, the ½ TC sequence FLNCCP, the ¼ TC sequence FLN, or the LAP sequence GFEIDKVWYDLDA (Uttamapinant et al., 2010). The resultant DNA fragments containing the capsid insertions were subcloned into the dsTE12Q infectious clone using the HpaI/BclI sites (Zheng and Kielian, 2013). The LAP peptide was inserted for comparison because the sequence is completely different from TC motif and does not contain cysteine residues. The WT SFV used in this study was prepared from the pSP6-SFV4 infectious clone (Liljeström et al., 1991). Insertions after residue 190 of the SFV capsid protein were generated by overlapping PCR using primers encoding the TC or TSer sequence, and the fragment was subcloned into the pSP6-SFV4 infectious clone using the XbaI/NsiI sites.
DNA sequence analysis (Genewiz Inc., North Brunswick, NJ) of the entire SINV HpaI/BclI or SFV XbaI/NsiI PCR fragments was performed to confirm the presence of the desired mutations and absence of additional mutations. Two independent infectious clones of each virus mutant were tested to confirm the phenotype.
Production and titration of virus stocks
Viral RNAs were in vitro transcribed from the XhoI-linearized SINV infectious clone or the SpeI-linearized SFV infectious clone. RNAs were electroporated into BHK-21 cells to generate virus stocks (Liljeström et al., 1991). All stocks were prepared by incubation of electroporated cells for 2 h at 37°C, followed by a 30 h incubation at 28°C. In subsequent studies, although we have indicated the route of infection for individual experiments, we found that virus infection by RNA electroporation or by addition of virus stocks prepared at 28°C gave similar results.
Viral titers were measured by infectious center assays on BHK-21 cells. Cells were infected with serial dilutions of virus stocks for 1–1.5 h at 37°C or 3.5–4 h at 28°C. Media were then replaced with complete BHK media containing 20mM NH4Cl to prevent secondary infection, and incubated at 28°C overnight to allow viral protein expression. Infected cells were quantitated by immunofluorescence, using a polyclonal antiserum against the SFV E1 and E2 proteins (Kielian et al., 1990) or a mixture of monoclonal antibody (mAb) R2 and mAb R6 against the SINV E1 and E2 protein, respectively (Meyer et al., 1992). Hybridoma cells producing R2 and R6 were kindly provided by Dr. William Klimstra.
Surface expression of E1 and E2 proteins
To detect the expression of SFV glycoproteins on the surface of infected cells, electroporated BHK-21 cells were incubated at 37 °C for 7 h or for 15h at 28°C. The cells were then incubated for 30 min on ice with mAb against the E1 or E2 proteins (Kielian et al., 1990), washed three times on ice with PBS containing 0.2% BSA, and fixed with 3% paraformaldehyde on ice. Cells were then stained with AlexaFluor-dye conjugated secondary antibody (Life Technologies) and analyzed by immunofluorescence microscopy. Images were captured using identical settings, and micrographs were prepared using Adobe Photoshop CS4 (Adobe) using identical conditions. Quantitation was performed using the ImageJ program (Abramoff et al., 2004).
Virus assembly assay
Virus assembly was evaluated by pulse-chase analysis (Chanel-Vos and Kielian, 2004). BHK-21 cells were electroporated with WT or mutant viral RNAs, incubated at 37 °C for 2 h to promote cell adherence, and then incubated for 15h at 28°C. Cells were then pulse-labeled with 50 µCi/ml [35S]-methionine and cysteine (Express Labeling Mix; Perkin Elmer Life and Analytical Sciences) for 1 h at 28°C or for 30 min at 37°C, and chased in the absence of label for 3 h at 37°C or 10 h at 28°C. Aliquots of the chase media were immunoprecipitated with a polyclonal antibody against SFV E1/E2 or with SINV mAb R6 against the E2 protein. Immunoprecipitation of the media was performed in the absence of detergent to recover the intact virus particles (Liao and Kielian, 2006). The cells were lysed in the presence of 40µM N-ethylmaleimide to alkylate free cysteine residues in the TC tag. Aliquots of the cell lysates and the immunoprecipitated media samples were analyzed by SDS-PAGE and fluorography.
Electron microscopy of virus-infected cells
BHK-21 cells were electroporated with WT or mutant RNA, plated on 35-mm diameter dishes in complete BHK medium, incubated at 37°C for 2 h to promote cell adherence, and then incubated for 10 h at 37°C or 18 h at 28°C. Cells were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer for 30 min at room temperature. Samples were processed and stained by the Analytical Imaging Facility of the Albert Einstein College of Medicine as described (Zheng and Kielian, 2013), and examined on a JEOL 1200EX electron microscope at 80 kv. Images were recorded at a magnification of 20,000x and assembled with the software Adobe Photoshop CS.
Preparation of radiolabeled virus
To prepare [35S]-labeled SINV or SFV virus, BHK-21 cells were electroporated with viral RNAs, incubated at 37°C for 2 h, and transferred to 28°C for 6 h. Cells were labeled with 200 µCi/ml [35S]-methionine and cysteine at 28°C for 15 h. The labeling medium was collected, and the virus was pelleted and purified by banding on a Pfefferkorn gradient (Chanel-Vos and Kielian, 2004).
Gradient analysis of nucleocapsids
Samples containing virus nucleocapsids were prepared by solubilizing [35S]-labeled purified virus in lysis buffer (100 mM Tris, pH 7.4, 50 mM NaCl, 2 mM EDTA, 1% NP-40, 40 µM N-ethylmaleimide, 250U RNase inhibitor/mL and a protease inhibitor cocktail containing 1ug pepstatin/mL, 5 ug leupeptin/mL, 1mM phenylmethylsulfonyl fluoride, and 1% aprotinin) (Skoging-Nyberg and Liljestrom, 2001).
Lysates containing cytoplasmic nucleocapsids were prepared by solubilizing [35S]-labeled, virus-infected cells. Briefly, BHK-21 cells were infected with WT or 186-TC mutant SINV at a MOI of 1 for 9 h at 28°C. Cells were then labeled with 50 µCi/ml [35S]-methionine and cysteine at 28°C for 14 h. Cells were lysed in the lysis buffer described above, centrifuged for 5 min at 5000 rpm to remove nuclei, and incubated on ice with a final concentration of 25 mM EDTA for at least 20 min to dissociate polysomes (Skoging-Nyberg and Liljestrom, 2001).
Lysates of virus or cells were loaded onto 15–30% continuous sucrose gradients (wt/wt in 50mM Tris, pH 7.4, 100mM NaCl, 2mM EDTA, 1% NP-40). Gradients were centrifuged at 35,000 rpm for 2.5 h at 4°C using an SW41 rotor. 400 uL fractions were collected and virus protein radioactivity was quantitated by scintillation counting. Aliquots of the cell lysate gradients (alternate fractions) were analyzed by SDS-PAGE and fluorography.
Alternatively, cell homogenates were prepared in the absence of detergent by scraping cells off the plate in lysis buffer without 1% NP-40 and passing through a 20G needle 35 times. Homogenates were then centrifuged, EDTA-treated and analyzed as above, except that gradients were prepared without detergent.
HIGHLIGHTS.
We characterize an alphavirus capsid insertion mutation.
These capsid mutants are highly temperature sensitive for growth.
The insertion affects nucleocapsid stability.
Results suggest that the nucleocapsid is stabilized during virus budding.
ACKNOWLEDGMENTS
We thank all the members of our lab for their helpful discussions and experimental suggestions and Drs. Löic Etienne, Maria Guadalupe Martinez and Katie Stiles for critical reading of the manuscript. We thank Drs. Erik Snapp and Guadalupe Martinez for many helpful discussions and input on imaging, Youqing Xiang for her excellent technical assistance, and the staff of the Einstein Analytical Imaging Facility for their support.
This work was supported by grants to M.K. from the National Institute of General Medicine (R01-GM057454) and by Cancer Center Core Support Grant NIH/NCI P30-CA13330. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medicine or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Aliperti G, Schlesinger MJ. Evidence for an autoprotease activity of sindbis virus capsid protein. Virol. 1978;90:366–369. doi: 10.1016/0042-6822(78)90321-5. [DOI] [PubMed] [Google Scholar]
- Brown DT, Waite MR, Pfefferkorn ER. Morphology and morphogenesis of Sindbis virus as seen with freeze-etching techniques. J.Virol. 1972;10:524–536. doi: 10.1128/jvi.10.3.524-536.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge BW, Pfefferkorn ER. Isolation and characterization of conditional-lethal mutants of Sindbis virus. Virol. 1966;30:204–213. doi: 10.1016/0042-6822(66)90096-1. [DOI] [PubMed] [Google Scholar]
- Chanel-Vos C, Kielian M. A conserved histidine in the ij loop of the Semliki Forest virus E1 protein plays an important role in membrane fusion. J.Virol. 2004;78:13543–13552. doi: 10.1128/JVI.78.24.13543-13552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng RH, Kuhn RJ, Olson NH, Rossman MG, Choi H-K, Smith TJ, Baker TS. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Lu GG, Lee S, Wengler G, Rossmann MG. Structure of Semliki Forest virus core protein. Proteins. 1997;27:345–359. doi: 10.1002/(sici)1097-0134(199703)27:3<345::aid-prot3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Choi HK, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- Coombs K, Brown B, Brown DT. Evidence for a change in capsid morphology during Sindbis virus envelopment. Virus Res. 1984;1:297–302. doi: 10.1016/0168-1702(84)90018-2. [DOI] [PubMed] [Google Scholar]
- deCurtis I, Simons K. Dissection of Semliki Forest virus glycoprotein delivery from the trans-Golgi network to the cell surface in permeabilized BHK cells. Proc.Natl.Acad.Sci.USA. 1988;85:8052–8056. doi: 10.1073/pnas.85.21.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL User's Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Forsell K, Griffiths G, Garoff H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996;15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H. Membrane proteins organize a symmetrical virus. EMBO J. 2000;19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H, Sjoberg M, Cheng RH. Budding of alphaviruses. Virus Res. 2004;106:103–116. doi: 10.1016/j.virusres.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Griffin DE. Alphaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Sixth ed. Philadelphia, PA: Lippincott, Williams and Wilkins; 2013. pp. 651–686. [Google Scholar]
- Hahn CS, Strauss EG, Strauss JH. Sequence analysis of three Sindbis virus mutants temperature-sensitive in the capsid protein autoprotease. Proc.Natl.Acad.Sci.USA. 1985;82:4648–4652. doi: 10.1073/pnas.82.14.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JM, Levine B. Sindbis virus vector system for functional analysis of apoptosis regulators. Meth.Enzym. 2000;322:492–508. doi: 10.1016/s0076-6879(00)22045-4. [DOI] [PubMed] [Google Scholar]
- Jose J, Przybyla L, Edwards TJ, Perera R, Burgner JW, Kuhn RJ., 2nd Interactions of the cytoplasmic domain of sindbis virus e2 with nucleocapsid cores promote alphavirus budding. J.Virol. 2012;86:2585–2599. doi: 10.1128/JVI.05860-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose J, Snyder JE, Kuhn RJ. A structural and functional perspective of alphavirus replication and assembly. Future Microbiol. 2009;4:837–856. doi: 10.2217/fmb.09.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keränen S, Kääriäinen L. Isolation and basic characterization of temperature-sensitivity mutants from Semliki Forest virus. Acta Pathol.Microbiol.Scand.Sect.B. 1974;82:810–820. doi: 10.1111/j.1699-0463.1974.tb02378.x. [DOI] [PubMed] [Google Scholar]
- Kielian M. Mechanisms of Virus Membrane Fusion Proteins. Annual Review of Virology. 2014;1:171–189. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- Kielian M, Jungerwirth S, Sayad KU, DeCandido S. Biosynthesis, maturation, and acid-activation of the Semliki Forest virus fusion protein. J.Virol. 1990;64:4614–4624. doi: 10.1128/jvi.64.10.4614-4624.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ. Togaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. Sixth ed. Philadelphia, PA: Lippincott, Williams and Wilkins; 2013. pp. 629–650. [Google Scholar]
- Lamb K, Lokesh GL, Sherman M, Watowich S. Structure of a Venezuelan equine encephalitis virus assembly intermediate isolated from infected cells. Virol. 2010;406:261–269. doi: 10.1016/j.virol.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Brown DT. Mutations in an exposed domain of Sindbis virus capsid protein result in the production of noninfectious virions and morphological variants. Virol. 1994;202:390–400. doi: 10.1006/viro.1994.1355. [DOI] [PubMed] [Google Scholar]
- Lee S, Owen KE, Choi H-K, Lee H, Lu G, Wengler G, Brown DT, Rossmann MG, Kuhn RJ. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Kielian M. Functions of the stem region of the Semliki Forest virus fusion protein during virus fusion and assembly. J.Virol. 2006;80:11362–11369. doi: 10.1128/JVI.01679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J.Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YE, Eng CH, Shome SG, Kielian M. In vivo generation and characterization of a soluble form of the Semliki Forest virus fusion protein. J.Virol. 2001;75:8329–8339. doi: 10.1128/JVI.75.17.8329-8339.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer WJ, Gidwitz S, Ayers VK, Schoepp RJ, Johnston RE. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J.Virol. 1992;66:3504–3513. doi: 10.1128/jvi.66.6.3504-3513.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Chipman PR, Hong EM, Kuhn RJ, Rossmann MG. In vitro-assembled alphavirus core-like particles maintain a structure similar to that of nucleocapsid cores in mature virus. J.Virol. 2002;76:11128–11132. doi: 10.1128/JVI.76.21.11128-11132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola AV, Chen W, Helenius A. Co-translational folding of an alphavirus capsid protein in the cytosol of living cells. Nature Cell Biol. 1999;1:341–345. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- Paessler S, Weaver SC. Vaccines for Venezuelan equine encephalitis. Vaccine. 2009;27(Suppl 4):D80–D85. doi: 10.1016/j.vaccine.2009.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ. Structure of isolated nucleocapsids from venezuelan equine encephalitis virus and implications for assembly and disassembly of enveloped virus. J.Virol. 2003;77:659–664. doi: 10.1128/JVI.77.1.659-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes AM, Brown DT, Rothnagel R, Chiu W, Schoepp RJ, Johnston RE, Prasad BVV. Three-dimensional structure of a membrane-containing virus. Proc.Natl.Acad.Sci.USA. 1993;90:9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletnev SV, Zhang W, Mukhopadhyay S, Fisher BR, Hernandez R, Brown DT, Baker TS, Rossmann MG, Kuhn RJ. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell. 2001;105:127–136. doi: 10.1016/s0092-8674(01)00302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Ahola T, Kaariainen L. Viral RNA replication in association with cellular membranes. Curr.Topics Microbiol.Immunol. 2005;285:139–173. doi: 10.1007/3-540-26764-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat.Rev.Micro. 2010;8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- Singh I, Helenius A. Nucleocapsid uncoating during entry of enveloped animal RNA viruses into cells. Semin.Virol. 1992a;3:511–518. [Google Scholar]
- Singh I, Helenius A. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J.Virol. 1992b;66:7049–7058. doi: 10.1128/jvi.66.12.7049-7058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh IR, Suomalainen M, Varadarajan S, Garoff H, Helenius A. Multiple mechanisms for the inhibition of entry and uncoating of superinfecting Semliki Forest virus. Virol. 1997;231:59–71. doi: 10.1006/viro.1997.8492. [DOI] [PubMed] [Google Scholar]
- Skoging U, Vihinen M, Nilsson L, Liljeström P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- Skoging-Nyberg U, Liljestrom P. M-X-I motif of semliki forest virus capsid protein affects nucleocapsid assembly. J.Virol. 2001;75:4625–4632. doi: 10.1128/JVI.75.10.4625-4632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss EG, Lenches EM, Strauss JH. Mutants of sindbis virus. I. Isolation and partial characterization of 89 new temperature-sensitive mutants. Virol. 1976;74:154–168. doi: 10.1016/0042-6822(76)90137-9. [DOI] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol.Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Jose J, Chipman P, Zhang W, Kuhn RJ, Baker TS. Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J Mol Biol. 2011;414:442–459. doi: 10.1016/j.jmb.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen TL, Hamburger AE, Fisher BR, Ostendorp R, Kuhn RJ. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli . J.Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I, Soderlund H, Kaariainen L. Semliki Forest virus capsid protein associates with the 60S ribosomal subunit in infected cells. J.Virol. 1976;20:203–210. doi: 10.1128/jvi.20.1.203-210.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamapinant C, White KA, Baruah H, Thompson S, Fernandez-Suarez M, Puthenveetil S, Ting AY. A fluorophore ligase for site-specific protein labeling inside living cells. Proc.Natl.Acad.Sci.USA. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- Wengler G, Gros C. Analyses of the role of structural changes in the regulation of uncoating and assembly of alphavirus cores. Virol. 1996;222:123–132. doi: 10.1006/viro.1996.0403. [DOI] [PubMed] [Google Scholar]
- Wengler G, Wengler GX. Identification of transfer of viral core protein to cellular ribosomes during the early stages of alphavirus infection. Virol. 1984;134:435–442. doi: 10.1016/0042-6822(84)90310-6. [DOI] [PubMed] [Google Scholar]
- Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W. 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 2011;30:3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Mukhopadhyay S, Pletnev SV, Baker TS, Kuhn RJ, Rossmann MG. Placement of the structural proteins in sindbis virus. J.Virol. 2002;76:11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Fugere M, Day R, Kielian M. Furin processing and proteolytic activation of Semliki Forest virus. J.Virol. 2003;77:2981–2989. doi: 10.1128/JVI.77.5.2981-2989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Garoff H. Role of cell surface spikes in alphavirus budding. J.Virol. 1992;66:7089–7095. doi: 10.1128/jvi.66.12.7089-7095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Lindqvist B, Garoff H, von Bonsdorff C-H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Kielian M. Imaging of the Alphavirus Capsid Protein during Virus Replication. J.Virol. 2013;87:9579–9589. doi: 10.1128/JVI.01299-13. [DOI] [PMC free article] [PubMed] [Google Scholar]