Abstract
Activation-induced cytidine deaminase (AID) mediates cytosine deamination and underlies two central processes in antibody diversification: somatic hypermutation and class-switch recombination. AID deamination is not exclusive to immunoglobulin loci; it can instigate DNA lesions in non-immunoglobulin genes and thus, stringent checks are in place to constrain and restrict its activity. Recent findings have provided new insights into the mechanisms that target AID activity to specific genomic regions, revealing an involvement for non-coding RNAs associated with polymerase pausing and with enhancer transcription as well as genomic architecture. We review these findings and integrate them into a model for multi-level regulation of AID expression and targeting in immunoglobulin and non-immunoglobulin loci. Within this framework we discuss gaps in understanding, and outline important areas of further research.
Keywords: AID, SHM, CSR, Immunoglobulin loci
AID AND ANTIBODY MATURATION
Millions of antigens, some innocuous others infectious, challenge the mammalian immune system daily. Adaptive immune responses to these challenges depend on B and T lymphocytes, which express specialized cell-surface receptors. Membrane-bound antigen receptors on B-lymphocytes recognize specific, pathogen-derived determinants. The B cell repertoire contains a tremendous diversity of antibody molecules, exceeding 109 specificities, first achieved during B cell development via V(D)J recombination. This process assembles unique combinations of exons encoding the amino-terminal variable regions of immunoglobulin heavy (IgH) and light chains (IgL) from component variable (V), diversity (D), and joining (J) segments [1-3]. Recombination yields mature but antigen-inexperienced naïve IgM+ B cells, which exit the bone marrow to secondary lymphoid organs such as the spleen and lymph nodes. Although V(D)J recombination generates a diverse primary repertoire of antibodies, immunoglobulin (Ig) genes undergo two additional DNA alteration events to enhance the specificity and functionality of antibodies: somatic hypermutation (SHM) and class-switch recombination (CSR). Together, these processes generate antigen-specific, high-affinity, switched B cells that secrete antibodies with unique effector functions [4-6].
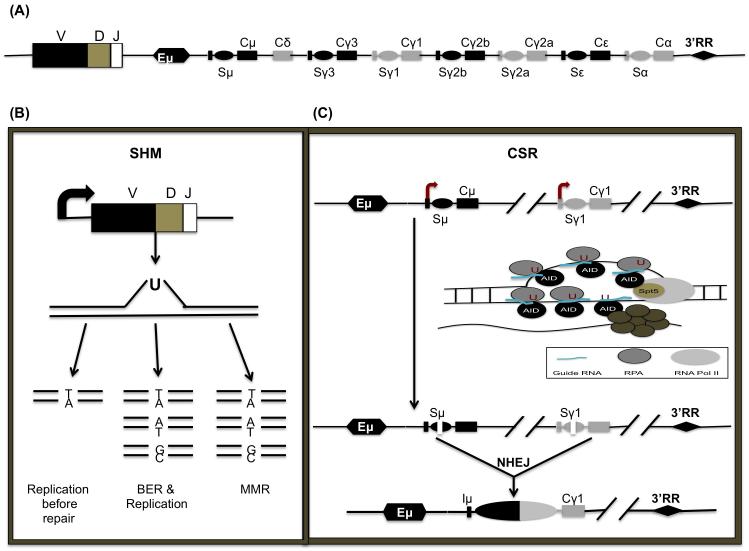
Both SHM and CSR require the enzyme activation-induced cytidine deaminase (AID). In addition to immunity enhancing antibody-diversification processes, AID promotes cancer-causing chromosomal translocations within both Ig and non-Ig loci [7, 8]. AID deaminates cytosine (C) in DNA and converts it to uracil (U), resulting in U:G mismatch lesions. These DNA lesions are then converted into point mutations during SHM and into DNA double-stranded breaks (DSBs) during CSR or aberrant chromosomal translocations. AID shows selectivity and deaminates single-stranded DNA (ssDNA) or supercoiled DNA that creates single-stranded patches of DNA [9, 10], but does not deaminate double-stranded DNA (dsDNA) or DNA:RNA hybrids [11, 12]. During SHM, point mutations and sometimes insertions and deletions, are introduced at a very high rate into the recombined variable region exons encoding IgH and IgL, which in some instances will generate B cells with increased antigen affinity. SHM requires transcription and occurs primarily, but not exclusively, at RGYW “hot-spot” motifs present in variable region exons (where R = purine base, Y = pyrimidine base, and W = A or T nucleotide). It has been proposed that AID-target ssDNA is generated as transcription bubbles or due to supercoilicity during transcription at the immunoglobulin locus [6, 13] (Figure 1B).
Figure 1.
Targeting of AID to the immunoglobulin locus (Ig): (A) Structure of the Ig heavy-chain locus after V(D)J recombination. The variable region V(D)J is followed by a series of constant regions, each specifying a different antibody isotype. Each constant region gene is comprised of a transcription unit with a cytokine-inducible promoter (P), an intervening (I)-exon, S region, and CH exons. The C regions are preceded by corresponding switch (S) repetitive sequences. Distal enhancers are present at the 3′ end of the IgH locus. (B) During SHM, mutations are introduced into the rearranged variable region genes. SHM begins with transcription through the variable region. ssDNA is created. If replication begins prior to the replacement of the U, a transition mutation (Ts) will appear at the original G:C to T:A. If BER removes the U prior to replication, Ts or Tv (transversion) mutations will replace the G:C. Finally, if the U is removed by the MMR pathway, both Ts and Tv mutations will occur at G:C and A:T base pairs. (C) CSR is initiated by transcription through the S regions, which leads to the formation of stable R-loops. RNA polII stalls at various regions due to the formation of secondary DNA structures like R loops. Following stalling, AID interacts with its cofactor, Spt5 and guide RNA’s which target AID to complementary S region DNA based on sequence information provided by the guide RNAs. The ssDNA is stabilized by the ssDNA binding protein complex RPA. Binding of the exosome degrades or displaces the nascent transcript to generate ssDNA, an AID substrate on the template strand.
CSR preferentially occurs in germinal centers, microanatomical structures within secondary lymphoid organs where mature B cells encounter antigen presented on helper T cells [14]. The mouse IgH locus is comprised of eight constant region (CH) exons, with Cμ most proximal to the variable region segments. Newly generated naïve B cells initially express IgM encoded by Cμ exons. Upon CSR, the default Cμ exchanges for an alternative set of downstream CH exons. During this process, B cells switch from expressing IgM to producing different classes of antibodies (e.g., IgG, IgE, or IgA), which are encoded by unique CH genes (e.g., Cγ, Cε, and Cα) (Figure 1A). CSR occurs between highly repetitive and evolutionarily conserved sequences termed switch (S) regions that precede each set of CH exons. Transcription through S regions appears to promote formation of DNA:RNA hybrid structures such as R-loops. These structures create ssDNA substrates for AID-mediated cytidine deamination followed by repair of DNA double-strand breaks by components of the base-excision repair (BER) and mismatch repair (MMR) pathways. End-joining of DSBs between two S regions via nonhomologous end-joining (NHEJ) excises the intervening DNA sequence. This process juxtaposes the rearranged V(D)J segment with a new set of constant region exons, generating different classes of antibodies that retain their original specificity but acquire new effector functions (Figure 1C).
The discovery of AID by subtractive cDNA hybridization from the mouse CH12F3 B lymphoma cells and its essential role in SHM and CSR has propelled efforts to characterize its function for over a decade [15-17]. Despite considerable progress in our understanding of AID activity, how AID selects its genomic targets remains unclear. Earlier models suggesting specific and exclusive AID recruitment to the Ig locus have been revised in light of several findings demonstrating the broad nature of AID targeting. Still, mutation rates at the Ig locus are orders of magnitude greater than at non-Ig loci [18, 19]. This bias, which appears even in mice over-expressing AID, suggests that AID is targeted by tightly regulated mechanisms [20]. Recent findings point to a role for genomic architecture and non-coding RNA in AID targeting, involving mechanisms that include switch RNA, divergent transcription, polymerase pausing and the action of the RNA exosome complex at certain loci, notably at enhancers [21-24]. We review these findings here, integrating these new insights into earlier findings on AID targeting. From this discussion we present a framework summarizing the current understanding of the multi-level mechanisms that regulate AID expression and targeting in Ig and non-Ig loci.
REGULATION OF AID
While the primary and physiological role of AID is to drive antibody diversification by introducing DNA lesions at the Ig loci, AID also poses a threat to genomic integrity. Mistargeted AID activity is known to be a major underlying cause of oncogenic translocations, which are hallmarks of many B cell malignancies [7, 8]. Ectopic expression of AID leads to mutation even in non-B cells [25, 26], therefore, regulation of AID expression and function is important for the development of an efficient immune system and maintenance of genomic integrity (Figure 2).
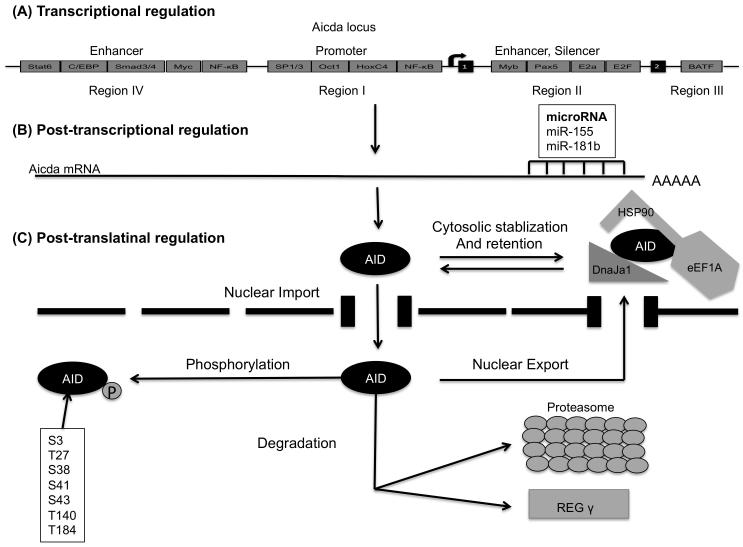
Figure 2.
AID regulation: (A) AID transcription is controlled through the binding of transcription factors to four distinct DNA regions (regions I to IV) of the AID gene locus. (B) After transcription, the Aicda mRNA can be negatively regulated by microRNAs, including miR-155 and miR-181b that target the 3′ UTR of Aicda mRNA. (C) To restrict the number of AID molecules that can target the Ig or non-Ig loci, Hsp90 stabilizes cytoplasmic AID whereas ubiquitylation or REGγ degrades nuclear AID. AID enters the nucleus passively through nuclear pores or actively through its putative nuclear localization signal. AID also actively exports the nucleus by binding to exportin1. Phosphorylation of AID affects its activity. Several Other phosphorylation sites have been identified, but S38 is the best characterized [104].
The Aicda gene codes for AID and is located on chromosome 6 and 12 in mice and humans, respectively. Early studies identified members of the E-protein family as the key regulators of CSR [27]. Specifically, it was demonstrated that E-protein levels increase in activated B-lineage cells and that interfering with E-protein expression blocked CSR. Subsequently, Sayegh and colleagues showed that members of the E-protein family directly activated AID expression to induce CSR [28]. More recent studies have demonstrated that four distinct and conserved DNA regions (regions I to IV) regulate the expression of AID in activated B-lineage cells (Fig.2). Region I functions as a TATA-less promoter containing the binding sites for HoxC4/Oct and NF-kB/Sp1/Sp3 [29]. This region is responsible for estrogen-mediated activation and progesterone-mediated repression of AID [30]. Silencer elements in region II bind the repressor proteins E2f and c-Myb to counter the activity of nearby transcriptional activators in a fashion unrestricted to B cells [31]. This region also includes binding sites for B-cell-specific transcription factors Pax5 and E-protein family members, linking E-proteins and cis-regulatory elements into a common framework [29]. Region 3, which has a BATF-binding site, is required to sustain physiological levels of AID expression [32]. Enhancer-containing region 4 binds to factors whose expression is up regulated during B cell activation, including NF-κB, STAT6, and SMAD3/4. c-Myc is also reported to bind this region and regulate AID expression [29, 31]. Recently, using ChIA-PET, Kieffer-Kwon et al. identified three additional enhancers tethered at a long distance (up to 50 kb) to the AID gene promoter [33]. An additional layer of regulation, microRNAs miR-155 and miR-181b were shown to regulate Aidca abundance by specifically binding to the conserved target sites on the 3′ UTR of Aicda mRNA [34, 35] (Figure 2B). Interestingly, mice with a mutation in the putative miR-155 binding site in the Aicda gene had higher AID mRNA and protein amounts resulting in Myc-Igh translocations [36]. How all these various factors act in concert to induce AID expression remains to be determined, but it seems clear that E-protein activity plays a critical part in this process (Figure 2A).
As AID is a mutator protein, its presence in the nucleus is tightly regulated by active nuclear import, cytoplasmic retention, and efficient nuclear export. The majority of AID is sequestered to the cytoplasm. Nuclear entry of AID depends on importin-3 and a conformational nuclear localization signal (NLS) at the C-terminus generated upon protein folding [37]. Ubiquitin-dependent and independent pathways target the vast majority of nuclear AID to proteasomes for degradation [38, 39] (Figure 2C).
Several phosphorylation sites of AID affect CSR, SHM and translocations without perturbing AID stability or deamination potential. These sites include serine-3 (S3), threonine-140 (T140), and serine-38 [40]. Phosphorylation of AID at serine-38 has been extensively characterized. Phosphorylation of serine-38, mediated by protein kinase A (PKA), is critical for AID to interact with AP endonuclease, which actively generates a high density of breaks required for CSR [41] (Figure 2C).
AID TARGETING
S regions and R loops
S regions are 1–12 kb repetitive sequences enriched with AID “hot-spot” 5′-RGYW-3′ motifs. Mammalian S regions are G-rich on the non-template strand and capable of forming secondary structures like R-loops. Evidence suggesting the role of S regions in CSR came from studies where deletion of Sμ impaired CSR to all isotypes while deletion of Sγ1 specifically impaired switching to IgG1[42]. S regions are also hyper accessible, compared to CH regions as evident by the epigenetic marks. Both donor and acceptor S regions are specifically enriched for acetylation and methylation marks generally associated with open chromatin, for example, H3K9/K14ac, H3K27ac, H4K8ac, and H3K4me3 [43-45]. The S regions undergo high levels of transcription linked with their ability to serve as recombination targets. This transcription is thought to promote the formation of R-loops by hybridizing the template strand of the switch region with the nascent RNA, ultimately leading to the looping out of the non-template strand as ssDNA (Figure 1C). Early studies in in vitro systems demonstrated that a transcribed, synthetic S region DNA fragment in its physiological orientation supported AID deamination and CSR in B cells whereas the reverse orientation, unable to form R-loops, supported neither AID-mediated deamination nor CSR [46, 47]. Later studies using bisulfite modification and sequencing confirmed the involvement of R-loops in AID targeting and the importance of the Sγ3 and Sγ2b regions in promoting CSR [48]. However, other studies in UNG−/−Msh2−/− mice did not show any preference for or unique mutational features in R-loops. Thus, it has been suggested that although R-loops enhance CSR, they may not be necessary for AID targeting [49, 50]. This observation is also consistent with AID targeting to non-R loop regions.
Trans-acting factors and AID targeting
E-proteins are among the best characterized factors that may promote AID targeting [51-53]. The first report implicating E2A in AID targeting overexpressed the E2A isoform E47 in the DT40 B cell line, which resulted in enhanced gene conversion; conversely, deletion of E2A in DT40 cells resulted in defective SHM [54, 55]. Two copies of the E2A recognition motif was sufficient to enhance SHM of an artificial hypermutable insert within an Igk transgene without affecting the expression of the transgene [53]. Later, it was shown that an artificial GFP-expressing transgene containing just three E2A binding motifs in the context of both Igk enhancers acquired a spectrum of mutations [52]. Furthermore, it was noted that non-Ig genes mistargeted by AID tend to be linked to cis-regulatory elements that contain E-box sites. Surprisingly, conditional E2A ablation in mature B cells and germinal center B cells in transgenic mice did not show defects in CSR or SHM, although germinal center development was affected [56]. Given that forced expression of Id-proteins, E-protein antagonists, completely blocked CSR, we suggest that multiple E-proteins control CSR and SHM.
Transcription and Targeting of AID to DNA
Sequencing of rearranged Ig loci encoding antibodies to the hapten phosphocholine showed that mutations were only present within V regions, and no mutations were detected upstream of the promoters or in downstream constant regions [57, 58]. Finer sequencing analysis revealed that mutations were infrequent in the early transcribed region, downstream of C exons and 5′ of the promoter, but instead were mainly concentrated in a 1–1.5 kb region from the 3′ region of the rearranged V(D)J promoter [59]. In 1996, Peters and Storb proposed a connection between transcription and SHM. They found that the normally unmutated Ck exon in an Igκ transgene would mutate when the Vk promoter was placed just 5′ from the Ck exon [60]. This study strongly supported the notion that transcription regulates SHM targeting. Peters and Storb also hypothesized that a mutator, now known to be AID, associated with the transcription initiation apparatus to induce the mutations they observed.
During CSR, DNA mutations also occur in S region recombination junctions; but unlike SHM, where mutations are comprised primarily of nucleotide substitutions, the S regions show substitutions as well as deletions, inversions, and insertions. Analysis of S region mutations in mice deficient for DNA-repair proteins UNG and Msh2 revealed that mutations begin 150 bp after the TSS of the S region intronic promoter [50], suggesting that here too, the promoter defines the 5′ boundary of mutation. Other ex vivo studies in cultured splenic B cells revealed that bacterial lipopolysaccharide (LPS) induced germ-line transcription through Cγ2b and Cγ3, allowing CSR to IgG2b and IgG3, whereas LPS together with interleukin-4 induced Cγ1 and Cε transcription and CSR to IgG1 and IgE [61-63]. TGFβ was also found to stimulate Igα non-coding transcripts and subsequent CSR to IgA [64]. Alteration or deletion of the I-exon promoter was also found to affect CSR [14]. Altogether, these studies demonstrated that cytokine signaling activated germ line transcription associated with CSR.
It is now well established that the primary sequence of S regions, R-loops and transcription establish a permissive environment for AID activity in B cells. However, more regulatory steps appear to be involved. Studies of the distribution of Pol II across the Igh locus have helped to elucidate how transcription helps AID targeting. Using chromatin immunoprecipitation (ChIP), several groups observed an increase in Pol II occupancy at Sμ and Sγ3 regions in activated B cells, extending from the intronic promoter to the S region boundary [45, 65].
Early elongating Pol II moves through the promoter to the promoter proximal pause site as phosphorylated Pol II at serine 5 (ser5) on its C-terminal domain (CTD). Once Pol II escapes, productive elongation of transcription leads to the loss of p-ser5 and enrichment of p-ser2 at the CTD [66, 67]. The distribution of Pol II at the Ig locus was consistent with Pol II stalling and an elongation barrier in the S regions, and also tightly correlated with the observed distribution of mutations [60, 65]. This observation supports the model initially proposed by Peters and Storb that associated Pol II stalling with SHM [60]. Stalling has also been reported for non-immunoglobulin targets (c-myc, c-myb and Pim-1) of AID [68-71]. So it appears that Pol II stalling is important for AID targeting. An shRNA screen in the CH12F3 B cell lymphoma line provided the first direct evidence for a role of Pol II stalling in SHM and CSR. A Pol II elongation and stalling factor, Suppressor of Ty5 homolog (Spt5), was also found to be required for CSR [72]. Spt5 directly associated with AID, and its depletion reduced AID recruitment to Ig and non-Ig loci [72]. Moreover, ChIP-seq studies of AID showed that AID occupancy was associated with stalled Pol II in promoter-proximal regions [73, 74]. Later studies showed that several other transcription elongation factors like Pol II-associated factor 1 (Paf1) complex, histone chaperone suppressor of Ty6 (Spt6) and histone chaperone FACT complex could promote antibody diversification by regulating the association of AID with Pol II [75, 76]. Adaptor proteins 14-3-3 were also found to recruit AID to DNA through their interaction with RGYW sequences [77]. Finally, recent studies found that polypyrimidine-tract-binding protein-2 (PTBP2) promoted the targeting of AID to S regions by interacting with both sense and anti-sense S region transcripts [78]. A recent study found that G-quadruplexes formed by the intronic switch RNA recruit AID to the S region DNA. Splicing of primary switch region transcript resulted in an intronic lariat structure. Debranching enzyme (DBR1) converted the lariat to a linear exon-free switch transcript that guided AID to the complementary S region DNA by forming G-quadruplex or G-quadruplex-like structures [21]. Two hyper-IgM syndrome patients having the G133V mutation in the putative G-quadruplex RNA-binding domain of AID, increases the relevance of the switch RNA - AID interaction in CSR [79]. Altogether, these studies establish RNA as a cofactor in AID targeting to S regions. RNA-mediated recruitment of AID to DNA may enhance AID specificity for the Ig locus and have implications for the aberrant targeting of AID to non-Ig loci. In fact G-quadruplex structures were found in the first exon and intron of c-Myc corresponding to common AID-mediated breakpoints in c-Myc/IgH translocations. Recent studies (described in detail below) found that anti-sense and convergent transcription at super-enhancers facilitate mistargeting of AID to non-Ig locius; so there is possibility that RNA-mediated recruitment of AID is reason for the higher specificity and mutation rate observed at the immunoglobulin locus. But a detailed more careful study is required to reach any conclusion. Next generation sequencing based approach to find out the transcripts associated with AID can give us more clear picture about it but it requires better antibodies as well as better bioinformatics approaches to map repetitive sequences.
The RNA exosome complex and AID targeting
Non-template strands liberated during transcription are predictable AID substrates. However, how AID accesses DNA templates associated with ongoing nascent RNA transcription is less obvious. Both strands are similarly targeted by AID: Mice deficient for DNA-repair proteins UNG and Msh2 revealed no marked preference for targeting of the transcribed versus non-transcribed strand [50]. Anti-sense transcription across the IgH locus has been proposed to facilitate the interaction of AID with the template strand [80] but was not found to be essential for CSR [40, 81]. Intriguing, recent findings indicate that the RNA exosome complex is involved. In these studies it was shown that the RNA exosome promoted access of AID to the target DNA template strand by degrading associated transcripts. Furthermore, they found that the RNA exosome complex promotes AID deamination of both template and non-template strands of in vitro transcribed SHM substrates. They also found that the the RNA exosome complex associates with AID and accumulates on S regions in an AID-dependent manner in B cells activated for CSR. These studies suggest that the RNA exosome acts as a cofactor for AID targeting to both template and non-template strands [82].
The RNA exosome is a macromolecular complex containing 3′–5′ exoribonuclease properties required for the processing of structural RNAs and the degradation of improperly processed premRNA [83]. The exosome can only load onto RNA species associated with an accessible 3′ region, consistent with stalled Pol II sites that exhibit nascent RNA molecules with exposed 3′ ends due to Pol II backtracking [82, 84, 85]. This feature allows the exosome access to the AIDSpt5-Pol II complex present at stalled loci. Once the exosome accesses the loci, it generates ssDNA on the template strand by displacing and/or degrading the nascent RNA [82].
Since immunoglobulin locus is the primary target of AID mutagenic reactions so its activity is tightly regulated here. Remarkable advancement of this field now established that successful AID activity in Ig locus require initiation of transcription, pausing of the polymerase, RNA exosome mediated accessibility and complementary guide RNA associated AID mediated hypermutation and recombination. To increase its targeting specificity AID involves several complementary and multilayered regulatory mechanisms. But compared to relatively small size of this protein for which conformational domains are not clear, the number of proposed cofactors is surprisingly high and certainly require further study to determine the relative contributions of these co-factors.
AID TARGETING AT NON-IMMUNOGLOBULIN LOCI
While preferentially targeting V region exons and S region DNA, AID also acts across genomic regions that are not associated with the Ig loci. These off-target effects of AID have pathological consequences like the development of B-cell lymphomas [8, 19, 20]. Indeed aberrant AID activity induces aberrant genomic lesions as well as SHM-like activity at non-immunoglobulin loci [8, 19, 20]. Specifically, AID was found to be associated with mutations across the c-Myc, Pim1, Jund, Bcl2, Cd79b, RhoH and Pax5 loci, promoting the development of diffuse large B cell lymphoma [19, 20, 86-88]. Conspicuous among chromosomal translocations associated with AID are those that involve the c-Myc and IgH loci in Burkitt′s lymphoma [8]. Another AID target, Bcl6, is also mutated in B cell lymphomas, normal human tonsillar germinal center B cells and memory B cells [18, 89-91]. An increase in AID expression correlates with the transition from the chronic stage of chronic myeloid leukemia to fatal B lymphoid blast crisis [92]. Mutation analysis in diffuse large-cell lymphomas also revealed several AID-linked mutated non-Ig genes absent in normal B cells [93]. Recent study using improved replication protein A chromatin immunoprecipitation (RPA-ChIP) that detects asymmetric RPA recruitment with high precision 236 total targets in 53BP1−/−IgkAID B cells [94]. Non-Ig AID targets mimic the mutation profiles at V genes. Like Ig targets, the mutations at non-Ig targets were also focused on RGYW hotspots and localized in promoter-proximal regions [6, 19, 95-97]. Transcription was also found to be associated with non-Ig targets but transcription alone cannot explain why only some transcribed non-Ig genes are targeted by AID. However, it is important to note that the mutation rates on non-Ig sites are 10 to 100-fold lower than that at V genes [19]. This preferential targeting to Ig loci has been studied and debated for over 20 years. A recent study using the chicken B cell line DT40 found that the Ig enhancers strongly activated SH in neighboring genes [98]. Using their highly sensitive GFP-based reporter assay termed the DIVAC assay, authors demonstrated that chicken, mouse, and human Ig locus enhancers and enhancer-like elements were core diversification activator (DIVAC) sequences that worked together to target SH. They also found that these elements relied on a common set of well-characterized transcription factor binding motifs (three E-boxes and three other putative sites: CBF, C/EBP and PU.1). On the basis of these findings, they proposed two plausible models for how SH could be targeted to Ig genes: 1) DIVAC-bound factor(s), actively recruit AID; or, 2) DIVACs induce changes in the Pol II transcription initiation or elongation complex, making the transcribed DNA more accessible to AID.
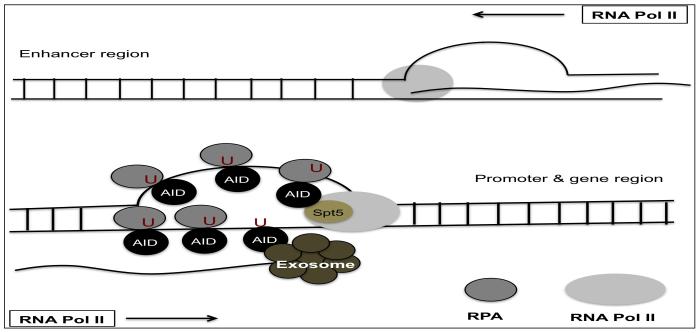
To find out how AID selects its targets in non-Ig region, Wang et al. compared AID-mediated translocations in two different cell types, B cells and mouse embryonic fibroblasts (MEFs). They found distinct sets of hotspot in each cell type concentrated in highly transcribed but stalled genes. They also found that transcription alone was insufficient to recruit AID activity. Instead, target genes in both fibroblasts and B cells were also enriched with chromatin modifications typical of active enhancers (H3K27Ac) and active transcription (H3K36me3). They proposed that the combination of both transcription and chromatin modification could direct the AID hotspot [99]. Another study using next generation sequencing, identified three transcription factor binding sites (E-box motifs, YY1 and C/EBP-β) common to 83 genes expressed in Ung−/− Msh2−/− mice that may work together to recruit AID [100]. Two other recent studies identified AID off-target DNA double-strand break (DSB) sites in the activated B cells genome and analyzed these data with an extensive array of epigenetic, nuclear architecture, and transcriptional data sets. They found that most of non-Ig targets of AID lie within active gene bodies and super-enhancers [22, 94]. These studies also revealed that the majority of AID-mediated lesions were found near TSS, which are topologically highly interconnected with multiple promoters and enhancers. AID-mediated DSBs correlated well with strong convergent transcription, in which normal sense transcription of the gene overlapped with super-enhancer-derived antisense enhancer RNA (eRNA) transcription. These studies also proposed that collision of two RNA polymerases moving in opposite directions could result in stalling, serving to recruit AID. Together, these properties are thought to create a nuclear microenvironment suitable for AID-mediated deamination (Figure 3). Additional regulatory mechanisms like requirements for specific transcription factors [98] and RNA processing by the exosome complex [23] also seems to play a role, as not all super-enhancers loci having convergent transcription are AID targets [94].
Figure 3.
AID targeting to non-immunoglobulin targets: In gene bodies with interconnected transcriptional regulatory elements and high transcriptional activity, convergent transcription is directed by polymerases that proceed in both directions generating sense and anti-sense transcripts. Collision of these RNA polymerases moving in opposite directions stalls the polymerases. Stalled polymerases recruit AID with the help of factors Spt5, RPA, and the RNA exosome complex, exposing single-stranded DNA substrates to AID.
A recent study linked the divergent antisense transcription upstream of transcription start sites to the genome stability and targeting of AID [23]. Pefanis and colleagues used a mouse model (Exosc3COIN/+ and Exosc3COIN/COIN mice) in which the essential subunit of the RNA exosome complex Exosc3 can be conditionally deleted. They identified RNA exosome substrate non-coding RNAs (ncRNA) including TSS RNAs (xTSS-RNA) that were transcribed divergently from cognate coding gene transcripts. xTSS-RNA were highly expressed at genes targeted by AID-mediated mutation or chromosomal translocations in B cells. The RNA exosome was revealed to regulate ncRNA-recruited AID to single-strand DNA-forming sites of antisense and divergent transcription in the B-cell genome. In a follow-up study, using mouse ablated for the cellular RNA degradation machinery the authors showed that genes or canonical enhancers near super-enhancers expressed high levels of RNA exosome-regulated anti-sense RNAs around their TSSs or within gene bodies (x-asRNAs). These x-asRNAs appeared to regulate the interaction between super-enhancers and their target promoters/genes. This feature may provide a mode of long-range chromatin regulation [24]. Pefanis et al. also proposed that divergent transcription generates positive DNA supercoiling ahead of RNA polymerase complexes thus negatively supercoiling the intervening DNA that is divergently transcribing RNA polymerases. This configuration would favor R-loop formation, transcriptional stalling and RNA exosome recruitment [101]. These studies support a model in which RNA exosome-mediated RNA processing events recruit AID to arrested noncoding transcription complexes resulting in mutations or breaks [101].
Since divergently transcribed TSS at promoters and intragenic enhancers can create “convergent/overlapping” transcription that leads to head-to-head collisions between oncoming RNA polymerases, as converging RNA polymerases cannot bypass one another and result into RNA exosome-coupled premature transcription termination. Alternatively, positive supercoiling of the DNA ahead of each RNA polymerase would create an area of extensive positive supercoiling lying between the two RNA polymerases that will impede transcription elongation and may lead to transcriptional stalling prior to relaxation of the DNA by DNA gyrase. This extended transcriptional stalling could ultimately lead to RNA exosome-mediated premature transcription termination and AID recruitment.
CONCLUDING REMARKS
The discovery of AID in 1999 marked a significant advance in the field of B cell biology. Highly focused molecular analyses of SHM and CSR have revealed how simple deamination diversifies the antibody response. Based on these studies, we now have a preliminary working model of AID targeting in the Ig locus. According to this model, AID targeting to the Ig locus occurs in three steps: 1) Ig enhancers are required to initiate transcription 2) pausing of the polymerase involving Spt5, enables hypermutation and recombination by complementary guide RNA associated AID and 3) the RNA degrading exosome complex displaces nascent S transcripts thereby rendering both DNA strands accessible to deamination (Figure 1C). AID targeting specificity is maintained by several complementary and multilayered regulatory mechanisms. Like targeting of AID by interaction with RNA polymerase II in a co-transcriptional step whereas binding to switch RNA molecules in a post-transcriptional step is two distinct mechanisms. But the number of proposed co-factors is very high and certainly requires more in depth study to determine relative contributions of these co-factors. AID is also targeted to a small subset of non-Ig genes (AID “off-target” genes), which provokes recurrent mutations or DSBs. High levels of transcriptional activity, interconnected transcriptional regulatory elements, and convergent transcription are thought to provide the micro environment necessary for AID off-targets. It was proposed that opposing polymerases stall by either collision or extensive positive supercoiling, creating an environment favorable for AID targeting. These intriguing findings deserve further scrutiny. Not all gene bodies, positioned in close genomic proximity to super-enhancers with sites of convergent transcription are targeted by AID. It seems possible that these off-target effects are caused by distinct folding patterns of the chromatin fiber.
Despite this progress, many questions remain. One important question to investigate is how AID assembles its cofactors on its target at the Ig and non-Ig locus to deaminate DNA. This study will also distinguish the AID binding with its very specific mutating targets. Even with the knowledge of several co factors for AID targeting, there is still no coherent model that clearly explains why under physiological conditions AID activity is so exclusively specified for the Ig loci. Another prominent question is how double-stranded breaks separated by large genomic distances during the cleavage process find each other in space and time, since unlike VDJ joining, switch regions ligate only after double-stranded breaks are generated. Previous studies have suggested that geometric confinement determines the frequencies of such encounters [102]. Indeed recent studies predict that the first-passage times for genomic encounters are determined in large-part by geometric confinement. This analysis also predicts that double-stranded breaks generated in switch regions will find each other within seconds to minutes [103]. It will be important to validate these predictions using in vivo imaging approaches to track the motion of double-stranded breaks generated during CSR.
HIGHLIGHTS.
-
❖
Regulation of AID expression and targeting ensures antibody diversification and genomic stability
-
❖
E proteins, B-lineage and transcription factors associated with B cell activation regulate AID expression
-
❖
Antisense transcription targets AID to Ig and non-Ig loci, and is regulated by the RNA exosome complex
-
❖
Intronic switch RNA targets AID to S regions for class switch recombination
ACKNOWLEDGEMENTS
The authors thank Masatoshi Aida for critical reading of the manuscript. The research in the Murre laboratory is supported by the NIH (RO1 AI109599-25).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 3.Bossen C, et al. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 6.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 7.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robbiani DF, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsa JY, et al. Negative supercoiling creates single-stranded patches of DNA that are substrates for AID-mediated mutagenesis. PLoS Genet. 2012;8:e1002518. doi: 10.1371/journal.pgen.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 12.Larijani M, Martin A. The biochemistry of activation-induced deaminase and its physiological functions. Semin Immunol. 2012;24:255–263. doi: 10.1016/j.smim.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 14.Stavnezer J, et al. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muramatsu M, et al. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 16.Muramatsu M, et al. Discovery of activation-induced cytidine deaminase, the engraver of antibody memory. Adv Immunol. 2007;94:1–36. doi: 10.1016/S0065-2776(06)94001-2. [DOI] [PubMed] [Google Scholar]
- 17.Revy P, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 18.Pasqualucci L, et al. BCL-6 mutations in normal germinal center B cells: evidence of somatic hypermutation acting outside Ig loci. Proc Natl Acad Sci U S A. 1998;95:11816–11821. doi: 10.1073/pnas.95.20.11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu M, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 20.Robbiani DF, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S, et al. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 2015;161:762–773. doi: 10.1016/j.cell.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng FL, et al. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pefanis E, et al. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 2014;514:389–393. doi: 10.1038/nature13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pefanis E, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell. 2015;161:774–789. doi: 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshikawa K, et al. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 26.Martin A, Scharff MD. Somatic hypermutation of the AID transgene in B and non-B cells. Proc Natl Acad Sci U S A. 2002;99:12304–12308. doi: 10.1073/pnas.192442899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quong MW, et al. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 1999;18:6307–6318. doi: 10.1093/emboj/18.22.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayegh CE, et al. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 29.Vuong BQ, Chaudhuri J. Combinatorial mechanisms regulating AID-dependent DNA deamination: interacting proteins and post-translational modifications. Semin Immunol. 2012;24:264–272. doi: 10.1016/j.smim.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mai T, et al. Estrogen receptors bind to and activate the HOXC4/HoxC4 promoter to potentiate HoxC4-mediated activation-induced cytosine deaminase induction, immunoglobulin class switch DNA recombination, and somatic hypermutation. J Biol Chem. 2010;285:37797–37810. doi: 10.1074/jbc.M110.169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran TH, et al. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat Immunol. 2010;11:148–154. doi: 10.1038/ni.1829. [DOI] [PubMed] [Google Scholar]
- 32.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Yebenes VG, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patenaude AM, et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 38.Uchimura Y, et al. REG-gamma associates with and modulates the abundance of nuclear activation-induced deaminase. J Exp Med. 2011;208:2385–2391. doi: 10.1084/jem.20110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aoufouchi S, et al. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaidyanathan B, et al. AIDing Chromatin and Transcription-Coupled Orchestration of Immunoglobulin Class-Switch Recombination. Front Immunol. 2014;5:120. doi: 10.3389/fimmu.2014.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuong BQ, et al. A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat Immunol. 2013;14:1183–1189. doi: 10.1038/ni.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khamlichi AA, et al. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat. Blood. 2004;103:3828–3836. doi: 10.1182/blood-2003-10-3470. [DOI] [PubMed] [Google Scholar]
- 43.Li G, et al. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 2013;5:702–714. doi: 10.1016/j.celrep.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeevan-Raj BP, et al. Epigenetic tethering of AID to the donor switch region during immunoglobulin class switch recombination. J Exp Med. 2011;208:1649–1660. doi: 10.1084/jem.20110118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L, et al. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 47.Shinkura R, et al. The influence of transcriptional orientation on endogenous switch region function. Nat Immunol. 2003;4:435–441. doi: 10.1038/ni918. [DOI] [PubMed] [Google Scholar]
- 48.Yu K, et al. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 49.Ronai D, et al. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue K, et al. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J Exp Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Staudt LM, Lenardo MJ. Immunoglobulin gene transcription. Annu Rev Immunol. 1991;9:373–398. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka A, et al. Attracting AID to targets of somatic hypermutation. J Exp Med. 2010;207:405–415. doi: 10.1084/jem.20090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michael N, et al. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 54.Conlon TM, Meyer KB. The chicken Ig light chain 3′-enhancer is essential for gene expression and regulates gene conversion via the transcription factor E2A. Eur J Immunol. 2006;36:139–148. doi: 10.1002/eji.200535219. [DOI] [PubMed] [Google Scholar]
- 55.Schoetz U, et al. E2A expression stimulates Ig hypermutation. J Immunol. 2006;177:395–400. doi: 10.4049/jimmunol.177.1.395. [DOI] [PubMed] [Google Scholar]
- 56.Kwon K, et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 57.Crews S, et al. A single VH gene segment encodes the immune response to phosphorylcholine: somatic mutation is correlated with the class of the antibody. Cell. 1981;25:59–66. doi: 10.1016/0092-8674(81)90231-2. [DOI] [PubMed] [Google Scholar]
- 58.Kim S, et al. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981;27:573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- 59.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 60.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 61.Lutzker S, et al. Mitogen- and IL-4-regulated expression of germ-line Ig gamma 2b transcripts: evidence for directed heavy chain class switching. Cell. 1988;53:177–184. doi: 10.1016/0092-8674(88)90379-0. [DOI] [PubMed] [Google Scholar]
- 62.Severinson E, et al. Induction of germ-line immunoglobulin heavy chain transcripts by mitogens and interleukins prior to switch recombination. Eur J Immunol. 1990;20:1079–1084. doi: 10.1002/eji.1830200520. [DOI] [PubMed] [Google Scholar]
- 63.Snapper CM, Paul WE. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. doi: 10.1126/science.3107127. [DOI] [PubMed] [Google Scholar]
- 64.Shockett P, Stavnezer J. Effect of cytokines on switching to IgA and alpha germline transcripts in the B lymphoma I.29 mu. Transforming growth factor-beta activates transcription of the unrearranged C alpha gene. J Immunol. 1991;147:4374–4383. [PubMed] [Google Scholar]
- 65.Rajagopal D, et al. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 67.Fabrega C, et al. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol Cell. 2003;11:1549–1561. doi: 10.1016/s1097-2765(03)00187-4. [DOI] [PubMed] [Google Scholar]
- 68.Bentley DL, Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986;321:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 69.Rohwer F, et al. The effect of IL-2 treatment on transcriptional attenuation in proto-oncogenes pim-1 and c-myb in human thymic blast cells. J Immunol. 1996;157:643–649. [PubMed] [Google Scholar]
- 70.Raschke EE, et al. Transcriptional regulation of the Ig kappa gene by promoter-proximal pausing of RNA polymerase II. J Immunol. 1999;163:4375–4382. [PubMed] [Google Scholar]
- 71.Schneider EE, et al. Regulation of c-myc and immunoglobulin kappa gene transcription by promoter-proximal pausing of RNA polymerase II. Curr Top Microbiol Immunol. 1999;246:225–231. doi: 10.1007/978-3-642-60162-0_28. [DOI] [PubMed] [Google Scholar]
- 72.Pavri R, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 74.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willmann KL, et al. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209:2099–2111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Begum NA, et al. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. J Biol Chem. 2012;287:32415–32429. doi: 10.1074/jbc.M112.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Z, et al. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat Struct Mol Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nowak U, et al. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nat Immunol. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahdaviani SA, et al. Novel mutation of the activation-induced cytidine deaminase gene in a Tajik family: special review on hyper-immunoglobulin M syndrome. Expert Rev Clin Immunol. 2012;8:539–546. doi: 10.1586/eci.12.46. [DOI] [PubMed] [Google Scholar]
- 80.Perlot T, et al. Antisense transcripts from immunoglobulin heavy-chain locus V(D)J and switch regions. Proc Natl Acad Sci U S A. 2008;105:3843–3848. doi: 10.1073/pnas.0712291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haddad D, et al. Replacement of Imu-Cmu intron by NeoR gene alters Imu germ-line expression but has no effect on V(D)J recombination. Mol Immunol. 2010;47:961–971. doi: 10.1016/j.molimm.2009.11.024. [DOI] [PubMed] [Google Scholar]
- 82.Basu U, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–363. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.(!!! INVALID CITATION !!! (Houseley et al., 2006, Lykke-Andersen et al., 2009))
- 84.Erie DA. The many conformational states of RNA polymerase elongation complexes and their roles in the regulation of transcription. Biochim Biophys Acta. 2002;1577:224–239. doi: 10.1016/s0167-4781(02)00454-2. [DOI] [PubMed] [Google Scholar]
- 85.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 86.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 87.Gordon MS, et al. Somatic hypermutation of the B cell receptor genes B29 (Igbeta, CD79b) and mb1 (Igalpha, CD79a) Proc Natl Acad Sci U S A. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muschen M, et al. Somatic mutation of the CD95 gene in human B cells as a side-effect of the germinal center reaction. J Exp Med. 2000;192:1833–1840. doi: 10.1084/jem.192.12.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shen HM, et al. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- 90.Gaidano G, et al. Frequent mutation of the 5′ noncoding region of the BCL-6 gene in acquired immunodeficiency syndrome-related non-Hodgkin’s lymphomas. Blood. 1997;89:3755–3762. [PubMed] [Google Scholar]
- 91.Migliazza A, et al. Frequent somatic hypermutation of the 5′ noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci U S A. 1995;92:12520–12524. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Klemm L, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lohr JG, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qian J, et al. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barreto VM, et al. Activation-induced deaminase: controversies and open questions. Trends Immunol. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 96.Storb U, et al. Somatic hypermutation of immunoglobulin and non immunoglobulin genes. Philos Trans R Soc Lond B Biol Sci. 2001;356:13–19. doi: 10.1098/rstb.2000.0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- 98.Buerstedde JM, et al. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 2014;12:e1001831. doi: 10.1371/journal.pbio.1001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Q, et al. Epigenetic targeting of activation-induced cytidine deaminase. Proc Natl Acad Sci U S A. 2014;111:18667–18672. doi: 10.1073/pnas.1420575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duke JL, et al. Multiple transcription factor binding sites predict AID targeting in non-Ig genes. J Immunol. 2013;190:3878–3888. doi: 10.4049/jimmunol.1202547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pefanis E, Basu U. RNA Exosome Regulates AID DNA Mutator Activity in the B Cell Genome. Adv Immunol. 2015;127:257–308. doi: 10.1016/bs.ai.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gostissa M, et al. IgH class switching exploits a general property of two DNA breaks to be joined in cis over long chromosomal distances. Proc Natl Acad Sci U S A. 2014;111:2644–2649. doi: 10.1073/pnas.1324176111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lucas JS, et al. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell. 2014;158:339–352. doi: 10.1016/j.cell.2014.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]