Abstract
The goal of the present study was to systematically identify endogenous microRNAs in endothelial cells that regulate mRNAs encoded by genes relevant to hypertension. Small RNA deep sequencing was performed in cultured human microvascular endothelial cells. Of the 50 most abundant microRNAs identified, 30 had predicted target mRNAs encoded by genes with known involvement in hypertension or blood pressure regulation. The cells were transfected with anti-miR oligonucleotides to inhibit each of the 30 microRNAs and the mRNA abundance of predicted targets was examined. Of 95 microRNA-target pairs examined, the target mRNAs were significantly up-regulated in 35 pairs and paradoxically down-regulated in 8 pairs. The result indicated significant suppression of the abundance of mRNA encoded by ADM by endogenous miR-181a-5p, ATP2B1 by the miR-27 family, FURIN by miR-125a-5p, FGF5 by the let-7 family, GOSR2 by miR-27a-3p, JAG1 by miR-21-5p, SH2B3 by miR-30a-5p, miR-98, miR-181a-5p, and the miR-125 family, TBX3 by the miR-92 family, ADRA1B by miR-22-3p, ADRA2A by miR-30a-5p and miR-30e-5p, ADRA2B by miR-30e-5p, ADRB1 by the let-7 family and miR-98, EDNRB by the miR-92 family, and NOX4 by the miR-92 family, miR-100-5p, and miR-99b-5p (n=3–9, P<0.05 vs. scrambled anti-miR). Treatment with anti-miR-21 decreased blood pressure in mice fed a 4% NaCl diet. Inhibition of the microRNAs targeting NOX4 mRNA increased H2O2 release from endothelial cells. The findings indicate widespread, tonic control of mRNAs encoded by genes relevant to blood pressure regulation by endothelial microRNAs and provide a novel and uniquely informative basis for studying the role of microRNAs in hypertension.
Keywords: Endothelial Cells, microRNA, hypertension, blood pressure, reactive oxygen species
Introduction
MicroRNAs (miRNAs) are small (~18–22 nts) endogenous RNA molecules that act as powerful and versatile regulators of gene expression. They generally act to suppress the expression of mRNAs with a complementary 3′ untranslated region, through translational repression or mRNA degradation. Approximately 2,000 microRNAs have been identified in human1 and predicted to regulate thousands of mRNAs.
Several miRNAs have been shown to regulate gene expression or biological processes relevant to blood pressure regulation or the development of hypertension2–6 and it is conceivable that many more miRNAs may be involved. It is, however, challenging to identify these miRNA-target relationships given the promiscuity of miRNA targeting and the uncertainty of target prediction.7 A single miRNA is capable of regulating several mRNA targets and multiple miRNAs could target the same mRNA.
In this study, we aimed to identify miRNAs that would regulate mRNAs encoded by genes in human endothelial cells related to vascular function and blood pressure regulation. Through knockdown of endogenous miRNAs that were 1) highly expressed in human vascular endothelial cells, and 2) predicted to target mRNA sequences of hypertension-related genes, we have identified 35 miRNA-target pairs in which the endothelial microRNAs tonically reduce the abundance of the target mRNAs that are relevant to blood pressure regulation or hypertension.
Methods
See Online Supplemental Information for detailed and additional Methods.
RNA analysis
RNA was extracted from human dermal microvascular endothelial cells (HMVEC-D)8 as previously described9 or using the Aurum Total RNA 96 Kit (BioRad). Small RNA deep sequencing was performed and data analyzed as previously described.10 qPCR analysis was perfomed using SYBR green chemistry.9
Inhibition of endogenous miRNA
HMVEC-D or mice were treated with locked nucleic acid (LNA)-modified anti-miRs (Exiqon).4, 11
Chronic blood pressure measurement
Blood pressure was measured using radiotelemetry in chronically instrumented male C57BL6/J mice (Jackson Lab).
H2O2 release
H2O2 release was measured by Amplex Red assay (Life Technologies).
Statistics
A Student’s t-test was used to make comparisons between mRNA expression levels in anti-miR and scrambled oligonucleotides treated cells. Blood pressure data was analyzed using the Holm-Sidak method for pairwise multiple comparisons. Data are expressed as means ± SE.
Results
The strategy for selecting miRNAs and hypertension-related target genes is described in Supplemental Figure S1A. We first identified microRNAs expressed in human endothelial cells by small RNA deep sequencing analysis of cultured human dermal microvascular endothelial cells (HMVEC-D). A complete list of identified miRNAs can be found in Supplemental Table S1. We detected 212 known human miRNAs in both replicates.
We compiled a list of protein-coding genes that included 1) 29 genes found to be associated with hypertension in human genome-wide association studies (GWAS),12, 13 and 2) 28 genes known to be functionally relevant to vascular endothelial cells and the regulation of blood pressure or development of hypertension. Most of the GWAS genes did not have known functional roles in endothelial cells or blood pressure regulation. The second group of genes included genes involved in cortisol processing, sympathetic signaling, vasoconstriction, endothelin signaling and oxidative stress response. These genes are shown in Supplemental Table S2.
We then searched miRNA target prediction databases for mRNAs predicted to be targeted by the 50 most abundant miRNAs detected in HMVEC-D and compared those with the 57 hypertension-related genes shown in Supplemental Table S2. We found 30 of the 50 most abundant miRNAs in HMVEC-D were predicted to target at least one hypertension-related gene, forming 99 potential miRNA-target pairs. The 99 potential miRNA-target pairs are shown in Table 1.
Table 1.
Abundant miRNAs in HMVEC-D and their predicted targets relevant to hypertension.
| Rank by abundance | miRbase v18 ID | Predicted target mRNAs relevant to hypertension |
|---|---|---|
| 1 | hsa-miR-10b-5p | TBX5 |
| 2 | hsa-miR-22-3p | ADRA1B |
| 3 | hsa-miR-10a-5p | TBX5 |
| 4 | hsa-let-7a-5p | TBX5, ADRB1, EDN1, FGF5 |
| 5 | hsa-miR-21-5p | JAG1 |
| 6 | hsa-miR-100-5p | NOX4 |
| 7 | hsa-miR-181a-5p |
SH2B3, ADM, ATP2B1 SH2B3, ADRB1, Catalase, ADRA1D, PLEKHA7, EDNRA, ADRA2A, |
| 8 | hsa-miR-30a-5p | ADRA2B, ATP2B1 |
| 9 | hsa-let-7f-5p | TBX5, ADRB1, EDN1, FGF5 |
| 10 | hsa-miR-26a-5p | JAG1, ADM |
| 11 | hsa-miR-92a-3p | TBX3, EDNRB, NOX4, ADRB1, ADM |
| 12 | hsa-miR-27b-3p | EDNRA, GOSR2, ATP2B1 |
| 13 | hsa-miR-125a-5p | FURIN, SH2B3 |
| 14 | hsa-miR-99b-5p | NOX4 |
| 15 | hsa-let-7i-5p | TBX5, ADRB1, EDN1, FGF5 |
| 16 | hsa-let-7e-5p | TBX5, ADRB1, EDN1, FGF5 |
| 17 | hsa-miR-92b-3p | ADM, TBX3, EDNRB, NOX4, ADRB1 |
| 18 | hsa-miR-186-5p | NOX2, JAG1 |
| 19 | hsa-let-7b-5p | TBX5, ADRB1, EDN1, FGF5 |
| 20 | hsa-miR-98 |
EDN1, TBX5, ADRB1, SH2B3 CAT, ADRA1D, PLEKHA7, EDNRA, ADRA2A, ADRA2B, ATP2B1, |
| 21 | hsa-miR-30d-5p | SH2B3, ADRB1 |
| 22 | hsa-miR-27a-3p | GOSR2, EDNRA, ATP2B1 |
| 23 | hsa-miR-181b-5p | ADM, SH2B3, ATP2B1 |
| 24 | hsa-miR-125b-5p | FURIN, SH2B3 |
| 25 | hsa-miR-16-5p | FURIN |
| 26 | hsa-let-7g-5p | TBX5, ADRB1, EDN1, FGF5 |
| 27 | hsa-miR-103a-3p |
FURIN CAT, ADRA1D, PLEKHA7, EDNRA, ADRA2A, ADRA2B, ATP2B1, |
| 28 | hsa-miR-30e-5p | SH2B3, ADRB1 |
| 29 | hsa-miR-25-3p | ADRB1, ADM, TBX3 |
| 30 | hsa-let-7c | TBX5, ADRB1, EDN1 |
To examine whether the predicted targets of the potential miRNA-target pairs were under tonic control by the endogenous miRNAs in HMVEC-D, we utilized locked nucleic acid (LNA)-modified anti-miRs to inhibit each of the 30 miRNAs individually. One would expect the mRNA abundance of a predicted target gene to increase following the anti-miR treatment if the predicted target mRNA is indeed suppressed by the endogenous microRNA via mRNA degradation.
Prior to initiating the large-scale anti-miR experiment we tested the efficacy of the anti-miR treatment in endothelial cells in the 96-well format. HMVEC-D were transfected with 10 nM LNA anti-miR-29a using Lipofectamine 2000. Effective knockdown of miR-29a was confirmed (Supplemental Figure S2).
The large-scale anti-miR study was performed as outlined in Supplemental Figure S1B. Briefly, LNA anti-miRs targeting the 30 most abundant miRNAs predicted to target mRNAs of interest from Table 1 were transfected individually into HMVEC-D in triplicate (using 90 wells of a 96-well plate) with control scrambled LNA anti-miR transfected into the remaining 6 wells. This was performed in a total of three 96-well plates. RNA was then extracted using Aurum kit prior to qRT-PCR analysis of target mRNAs of interest (see Table 1). Abundance of mRNAs in response to anti-miR treatment was expressed as % change relative to cells treated with scrambled anti-miR for each qPCR run.
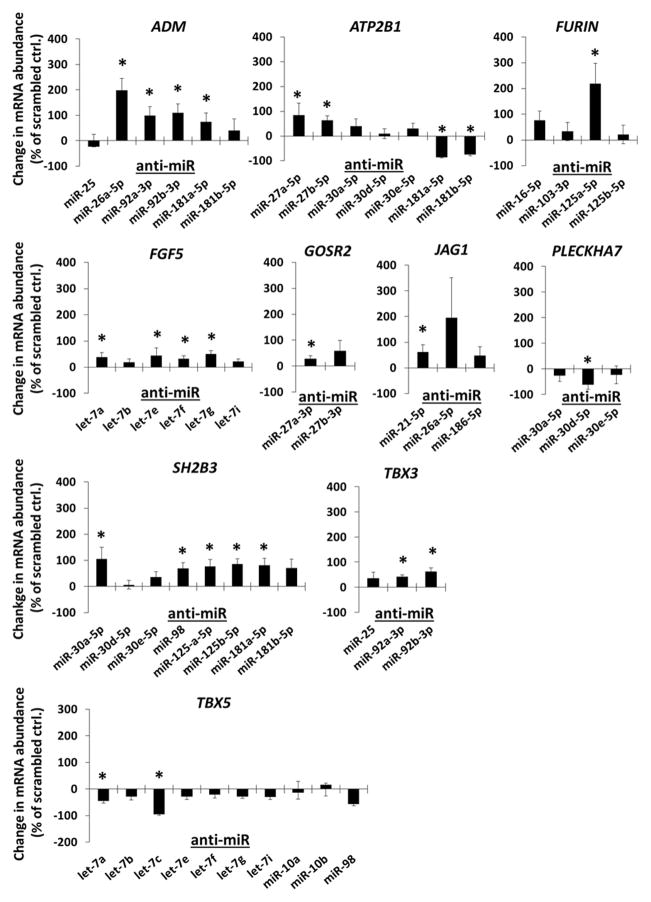
The effect of the anti-miR treatment on the abundance of mRNA encoded by hypertension risk genes identified by GWAS is shown in Figure 1. Based on significant increases of mRNA abundance following anti-miR treatment, our data indicate that the abundance of mRNA encoded by the following hypertension GWAS genes were suppressed by endogenous miRNAs in endothelial cells predicted to target them: adrenomedullin (ADM) by miR-26a-5p, miR-92a-3p, miR-92b-3p, miR-181a-5p and miR-181b-5p, plasma membrane calcium-transporting ATPase 1 (ATP2B1) by miR-27a-5p, miR-27b-5p, miR-30a-5p, miR-181a-5p and miR-181b-5p, FURIN by miR-125a-5p, fibroblast growth factor 5 (FGF5) by numerous members of the let-7 family of miRNAs, including let-7a, let-7e, let-7f, and let-7g, Golgi SNAP receptor complex member 2 (GOSR2) by miR-27a, jagged 1 (JAG1) by miR-21-5p, SH2B adapter protein 3 (SH2B3) by miR-30a-5p, miR-98, miR-125a-5p, miR-125b-5p and miR-181a-5pm, and T-box 3 (TBX3) by miR-92a-3p and miR-92b-3p.
Figure 1.
Effect of anti-miR transfections in HMVEC-D on the abundance of mRNAs encoded by hypertension risk genes identified by human GWAS. HMVEC-D were transfected with the indicated LNA anti-miR or scrambled LNA anti-miR (10 nM) for 48 hours. mRNA abundance for predicted targets genes was measured by real-time RT-PCR and expressed as % change relative to cells treated with scrambled anti-miR on the same qPCR plate. ADM, adrenomedullin; ATP2B1, plasma membrane calcium-transporting ATPase 1; FGF5, fibroblast growth factor 5; GOSR2, Golgi SNAP receptor complex member 2; JAG1, jagged 1; PLECKHA7, pleckstrin homology domain containing, family A member 7; SH2B3, SH2B adapter protein 3; TBX3, T-box 3; TBX5, T-Box 5. N=3–9, *, P<0.05 vs. scrambled anti-miR.
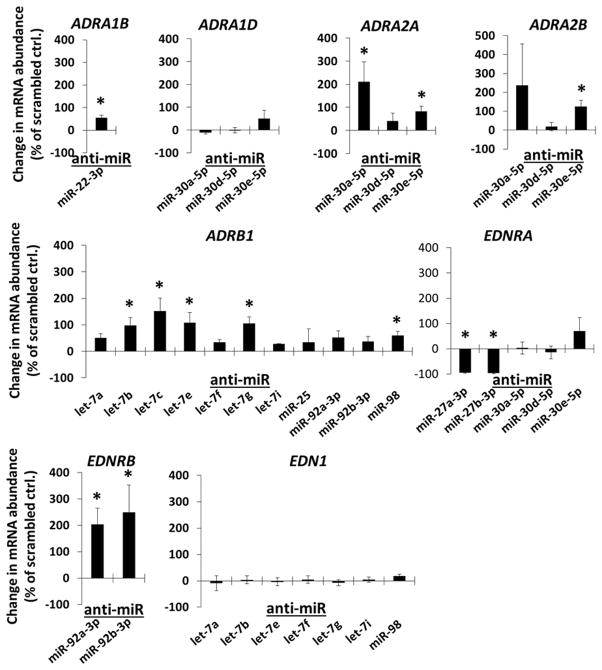
The effects of anti-miR treatment on mRNAs encoded by selected genes involved with sympathetic and endothelial signaling are shown in Figure 2. Adrenergic receptor A1b (ADRA1B) mRNA was suppressed by endogenous miR-22-3p in endothelial cells, adrenergic receptor A2A (ADRA2A) by miR-30a-5p, adrenergic receptor A2B (ADRA2B) by miR-30e-5p, adrenergic receptor B1 (ADRB1) by let-7b, let-7c, let-7e and let-7g, and endothelin receptor B (EDNRB) by miR-92a-3p and miR-92b-3p.
Figure 2.
Effect of anti-miR transfections in HMVEC-D on the abundance of mRNAs encoded by endothelial signaling genes relevant to hypertension. HMVEC-D were transfected with the indicated LNA anti-miR or scrambled LNA anti-miR (10 nM) for 48 hours. mRNA abundance of predicted targets was measured by real-time RT-PCR and expressed as % change relative to cells treated with scrambled anti-miR on the same qPCR plate. ADRA1B, adrenergic receptor A1B; ADRA1D, adrenergic receptor AID; ADRA2A, adrenergic receptor A2A; ADRA2B, adrenergic receptor A2B; ADRB1, adrenergic receptor B1; EDNRA, endothelin receptor A; EDNRB, endothelin receptor B; EDN1, endothelin 1. N=5–9, *, P<0.05 vs. scrambled anti-miR.
We also evaluated the potential for highly abundant miRNAs to target reactive oxygen species (ROS) and oxidative stress-related genes in HMVEC-Ds. We found that NADPH oxidase 4 (NOX4) mRNA was suppressed by endogenous miR-92a-3p, miR-92b-3p, miR-99b-5p and miR-100-5p, while catalase (CAT) mRNA and NADPH oxidase 2 (NOX2) mRNA were not significantly suppressed by the miRNAs studied (Figure 3).
Figure 3.
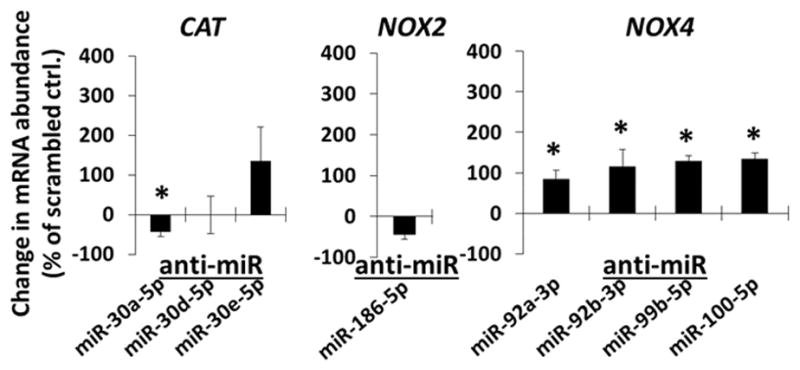
Effect of anti-miR transfections in HMVEC-D on the abundance of mRNAs encoded by reactive oxygen species-related genes. HMVEC-D were transfected with the indicated LNA anti-miR or scrambled LNA anti-miR (10 nM) for 48 hours. mRNA abundance for predicted targets genes was measured by real-time RT-PCR and expressed as % change relative to cells treated with scrambled anti-miR on the same qPCR plate. CAT, catalase; NOX2, NADPH oxidase 2; NOX4, NADPH oxidase 4. N=3–6, *, P<0.05 vs. scrambled anti-miR.
Several predicted target mRNAs were paradoxically down-regulated following anti-miR treatment (Figures 1, 2, 3), suggesting unconventional or indirect effects of the miRNAs. Four of the 99 pairs in Table 1 were not experimentally tested because of limited material.
In total, our data indicates that, of the 95 predicted miRNA-target pairs tested, suppression of the endogenous miRNAs in HMVEC-D resulted in up-regulation of target mRNAs encoded by hypertension-related target genes in 35 pairs (Figure 4).
Figure 4.
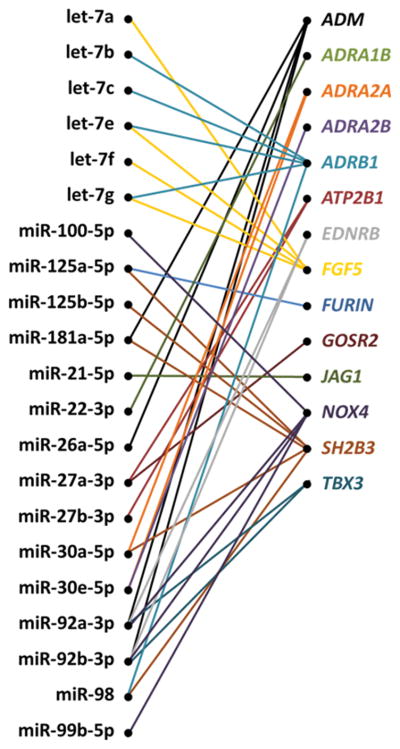
Summary of miRNA-target pairs supported by the anti-miR transfection experiment. The mRNA abundance for the predicted targets was significantly increased in HMVEC-D following inhibition of the indicated endogenous miRNA. See Figures 1–3 for full names of the targets.
To evaluate the functional relevance of miRNAs identified in the screening portion of the study in hypertension related processes we performed additional studies using in vivo and in vitro models. We tested whether endogenous miR-21-5p could affect blood pressure of C57BL/6J mice. Intraperitoneal administration of LNA anti-miR-21 induced a transient increase of blood pressure in mice fed a 0.4% NaCl diet compared to mice receiving an LNA scrambled anti-miR (Figure 5). A second injection of anti-miR was given nine days later. Blood pressure did not change on the day after the second injection when the mice were still on the 0.4% NaCl diet but decreased significantly after the mice were switched to a 4% NaCl diet and remained decreased to the end of the experiment (Figure 5). Heart rate increased transiently.
Figure 5.
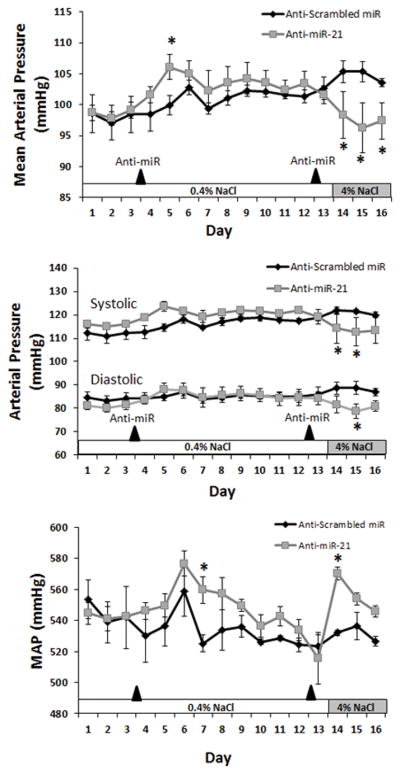
Effect of anti-miR-21 treatment on arterial blood pressures in mice. Blood pressure and heart rate were measured in C57BL/6J mice on AIN-76A diet containing 0.4% NaCl or 4% NaCl and receiving intraperitoneal injections of either scrambled LNA anti-miR or anti-miR-21 (10 mg/kg body weight) on days 3 and 12 as indicated. Blood pressure and heart rate were averaged over each 24 hour period (day) throughout the 16 day protocol. N=4/group, * P<0.05 vs scrambled anti-miR.
We also tested whether the miRNAs we found to target NOX4 mRNA would affect H2O2 levels. The release of H2O2 from HMVEC-D was significantly increased after the cells were transfected with anti-miRs for miR-92a-3p, miR-92b-3p, miR-99-5p, and miR-100-5p when compared to cells transfected with scrambled anti-miR controls (Figure 6).
Figure 6.
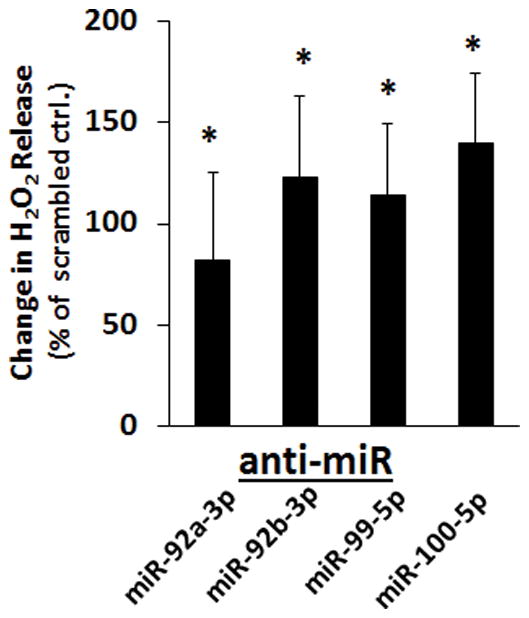
H2O2 release from HMVEC-D following transfection of anti-miRs that increased NOX4 mRNA abundance (from Figure 3). H202 release was measured with an Amplex Red assay over a 5.5 hour period initiated 48 hours after the HMVEC-D were transfected with scrambled anti-miR (N=12), anti-miR-92a-3p, anti-miR-92b-3p, anti-miR-99-5p, and anti-miR-100-5p (N=6/group). H2O2 release is expressed as % change relative to cells treated with scrambled anti-miR on the same assay plate. *P <0.05 vs. scrambled anti-miR.
Discussion
Only a few miRNAs and miRNA-target pairs have been shown to be involved in hypertension or physiological processes related to blood pressure regulation. Expression of the miR-143/145 gene cluster has been shown to be important in facilitating smooth muscle contractility.2 miR-29 plays an important role in regulating extracellular matrix in the kidneys in a rat model of salt-induced hypertension and renal injury.3 miR-192 targets Na+/K+-ATPase beta 1 and is involved in regulating urine volume in mice fed a high-salt diet.4 miR-181a and miR-663 were found to target renin, apoptosis-inducing factor mitochondrion-associated 1 (AIFM1) and apolipoprotein E (APOE) mRNAs.5, 14 11β-hydroxysteroid dehydrogenase type 2 may also be targeted by several miRNAs.6
The current study has substantially expanded this list by identifying 35 miRNA-target pairs in which mRNA encoded by hypertension-related genes was suppressed by endogenous miRNAs in human vascular endothelial cells. The findings indicate widespread, tonic control of gene expression relevant to blood pressure regulation by endothelial microRNAs. The vast majority of the 35 miRNA-target pairs identified in the current study have not been tested in previous studies. These findings provide an extensive, novel, and uniquely informative basis upon which numerous studies can be developed to investigate the role of microRNAs in hypertension and the mechanisms involved.
Of the 35 miRNA-target pairs we identified, 20 pairs involve mRNA encoded by hypertension risk genes nominated by human GWAS. Fibroblast growth factor signaling has been shown to be important for maintaining let-7 levels in endothelial cells.15 FGF5 has been shown to stimulate angiogenesis of human umbilical vein endothelial cells.16 The let-7 miRNA family is predicted to target mRNAs encoded by numerous angiogenic gene in addition to FGF5.17 Adrenomedullin is a proangiogenic and vasodilatory substance which acts to reduce blood pressure. Several proteins, including adrenomedullin (Adm) and endothelin-1 (ET-1), are known substrates of Furin in the endothelium.18 The functional relevance to hypertension or endothelial cells remains unknown for the other GWAS gene we analyzed. We found anti-miR-21 treatment reduced mean arterial pressure in mice fed a 4% NaCl diet, suggesting that endogenous miR-21 may affect blood pressure response to salt intake. It remains to be determined whether this effect of miR-21 is mediated by the targeting of JAG1 mRNA identified in the present study and what underlies the transient increase of blood pressure in mice on the 0.4% NaCl diet.
Endothelin-1 (EDN1) has been previously shown to be regulated by miR-125a/b in endothelial cells.19 EDN1 mRNA was not regulated by any of the miRNAs we tested based on our anti-miR experiment. NADPH oxidase 4 (NOX4) is an oxidoreductase that catalyzes the conversion of oxygen to different reactive oxygen species and could affect endothelial cell biology and blood pressure.20–22 We found that endogenous miR-92a-3p, miR-92b-3p, miR-99-5p and miR-100-5p reduced NOX4 mRNA abundance in HMVEC-D and reduced H2O2 release. We examined several other genes involved in the regulation of ROS (Supplemental Table S2). Their mRNA transcripts were either not predicted targets of abundant miRNAs in HMVEC-D, or their predicted regulation by abundant miRNAs was not supported by results of the anti-miR experiment.
The current study took a new approach to identify functionally important miRNAs by nominating nearly 100 miRNA-target pairs potentially relevant to hypertension and testing each of them in HMVEC-D. We used anti-miR treatments, which allowed us to assess the effect of endogenous miRNAs at their native levels of abundance and how partial, physiologically relevant reductions of specific endogenous miRNAs might lead to changes in the abundance of hypertension-related genes. Anti-miR was used to inhibit 30 miRNAs individually, making our study one of the largest studies using miRNA inhibition.
The study is not, however, a comprehensive evaluation of all miRNA-mRNA interactions that would be relevant to hypertension. Instead, we developed a largely unbiased strategy to identify some of the most promising candidate pairs and tested them. We focused on abundant miRNAs. It is possible that the less abundant miRNAs also play an important role in the regulation of hypertension-related genes. The absence or low-abundance of certain miRNAs might be important in allowing certain hypertension-related genes to be expressed, and upregulation of these miRNAs might inhibit the expression of hypertension-related genes. We used mRNA abundance as an index of miRNA effect.23 Several studies have shown that miRNA-target interactions can also involve translational repression, leaving mRNA levels unaffected.
Perspectives
The findings of the present study substantially increased the number of experimentally supported miRNA-target pairs with relevance to hypertension from a few to several dozen. The findings indicate widespread, tonic control of mRNAs encoded by hypertension-related genes by endothelial microRNAs and provide a novel and uniquely informative basis for studying the role of microRNAs in hypertension. It would also be valuable to apply the approach we developed to study hypertension in future studies. It would also be valuable to apply the approach we developed to study additional cell types important in hypertension. Finally, findings of the current study can be used as the basis for developing future studies to thoroughly investigate the functional role in hypertension of the miRNA-target pairs identified.
Supplementary Material
Novelty and Significance.
What is new?
Identification of 35 miRNA-target pairs in which mRNAs encoded by hypertension-related genes are suppressed by the miRNAs endogenously in human microvascular endothelial cells.
30 miRNAs were subjected to anti-miR inhibition, one of the largest numbers of miRNAs specifically manipulated in a single study.
The findings increased the number of experimentally supported miRNA-target pairs with relevance to hypertension from a few to several dozen.
What is relevant?
The findings indicate widespread, tonic control of mRNAs encoded by hypertension-related genes by endothelial microRNAs and provide a novel and uniquely informative basis for studying the role of microRNAs in hypertension.
Summary
We analyzed miRNA abundance in cultured human microvascular endothelial cells, identified and experimentally tested 95 potential miRNA-target pairs in which the target genes are relevant to hypertension, and demonstrated widespread, tonic control of mRNAs encoded by hypertension-related genes by endogenous endothelial miRNAs.
Acknowledgments
Sources of Funding:
This work was supported by US National Institutes of Health grants HL121233 and HL082798-6186 (ML).
Footnotes
Disclosures:
None
References
- 1.Kozomara A, Griffiths-Jones S. MirBase: Annotating high confidence micrornas using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boettger T, Beetz N, Kostin S, Schneider J, Kruger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, Yeo NC, Liang M. Renal medullary micrornas in dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mladinov D, Liu Y, Mattson DL, Liang M. Micrornas contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets na+/k+-atpase beta1. Nucleic Acids Res. 2013;41:1273–1283. doi: 10.1093/nar/gks1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris BJ. Renin, genes, micrornas, and renal mechanisms involved in hypertension. Hypertension. 2015;65:956–962. doi: 10.1161/HYPERTENSIONAHA.114.04366. [DOI] [PubMed] [Google Scholar]
- 6.Rezaei M, Andrieu T, Neuenschwander S, Bruggmann R, Mordasini D, Frey FJ, Vogt B, Frey BM. Regulation of 11beta-hydroxysteroid dehydrogenase type 2 by microRNA. Hypertension. 2014;64:860–866. doi: 10.1161/HYPERTENSIONAHA.114.00002. [DOI] [PubMed] [Google Scholar]
- 7.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. Microrna-target pairs in the rat kidney identified by microrna microarray, proteomic, and bioinformatic analysis. Genome Res. 2008;18:404–411. doi: 10.1101/gr.6587008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang M, Pietrusz JL. Thiol-related genes in diabetic complications: A novel protective role for endogenous thioredoxin 2. Arterioscler Thromb Vasc Biol. 2007;27:77–83. doi: 10.1161/01.ATV.0000251006.54632.bb. [DOI] [PubMed] [Google Scholar]
- 9.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. Microrna-target pairs in human renal epithelial cells treated with transforming growth factor beta 1: A novel role of mir-382. Nucleic Acids Res. 2010;38:8338–8347. doi: 10.1093/nar/gkq718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriegel AJ, Liu Y, Liu P, Baker MA, Hodges MR, Hua X, Liang M. Characteristics of micrornas enriched in specific cell types and primary tissue types in solid organs. Physiol Genomics. 2013;45:1144–1156. doi: 10.1152/physiolgenomics.00090.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu X, Kriegel AJ, Liu Y, Usa K, Mladinov D, Liu H, Fang Y, Ding X, Liang M. Delayed ischemic preconditioning contributes to renal protection by upregulation of miR-21. Kidney Int. 2012;82:1167–1175. doi: 10.1038/ki.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehret GB. Genome-wide association studies: Contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12:17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Consortium for Blood Pressure Genome-Wide Association. Ehret GB, Munroe PB, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marques FZ, Campain AE, Tomaszewski M, Zukowska-Szczechowska E, Yang YH, Charchar FJ, Morris BJ. Gene expression profiling reveals renin mrna overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension. 2011;58:1093–1098. doi: 10.1161/HYPERTENSIONAHA.111.180729. [DOI] [PubMed] [Google Scholar]
- 15.Chen PY, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, Anderson DG, Kotelianski V, Wang F, Tellides G, Simons M. FGF regulates TGF-beta signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep. 2012;2:1684–1696. doi: 10.1016/j.celrep.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allerstorfer S, Sonvilla G, Fischer H, et al. FGF5 as an oncogenic factor in human glioblastoma multiforme: Autocrine and paracrine activities. Oncogene. 2008;27:4180–4190. doi: 10.1038/onc.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: An emerging next-generation cancer therapeutic. Curr Oncol. 2010;17:70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim W, Essalmani R, Szumska D, Creemers JW, Roebroek AJ, D’Orleans-Juste P, Bhattacharya S, Seidah NG, Prat A. Loss of endothelial furin leads to cardiac malformation and early postnatal death. Mol Cell Biol. 2012;32:3382–3391. doi: 10.1128/MCB.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D, Yang P, Xiong Q, Song X, Yang X, Liu L, Yuan W, Rui YC. MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertension. 2010;28:1646–1654. doi: 10.1097/HJH.0b013e32833a4922. [DOI] [PubMed] [Google Scholar]
- 20.Schroder K, Zhang M, Benkhoff S, Mieth A, Pliquett R, Kosowski J, Kruse C, Luedike P, Michaelis UR, Weissmann N, Dimmeler S, Shah AM, Brandes RP. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circulation Res. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. [DOI] [PubMed] [Google Scholar]
- 21.Ray R, Murdoch CE, Wang M, Santos CX, Zhang M, Alom-Ruiz S, Anilkumar N, Ouattara A, Cave AC, Walker SJ, Grieve DJ, Charles RL, Eaton P, Brewer AC, Shah AM. Endothelial Nox4 NADPH oxidase enhances vasodilatation and reduces blood pressure in vivo. Arterioscler Thromb Vasc Biol. 2011;31:1368–1376. doi: 10.1161/ATVBAHA.110.219238. [DOI] [PubMed] [Google Scholar]
- 22.Guo S, Chen X. The human nox4: Gene, structure, physiological function and pathological significance. J Drug Target. 2015:1–9. doi: 10.3109/1061186X.2015.1036276. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.