Abstract
In striated muscle, the protein troponin complex turns contraction on and off in a calcium-dependent manner. The calcium-sensing component of the complex is troponin C, which is expressed from the TNNC1 gene in both cardiac muscle and slow-twitch skeletal muscle (identical transcript in both tissues) and the TNNC2 gene in fast-twitch skeletal muscle. Cardiac troponin C (cTnC) is made up of two globular EF-hand domains connected by a flexible linker. The structural C-domain (cCTnC) contains two high affinity calcium-binding sites that are always occupied by Ca2+ or Mg2+ under physiologic conditions, stabilizing an open conformation that remains anchored to the rest of the troponin complex. In contrast, the regulatory N-domain (cNTnC) contains a single low affinity site that is largely unoccupied at resting calcium concentrations. During muscle activation, calcium binding to cNTnC favors an open conformation that binds to the switch region of troponin I, removing adjacent inhibitory regions of troponin I from actin and allowing muscle contraction to proceed. Regulation of the calcium binding affinity of cNTnC is physiologically important, because it directly impacts the calcium sensitivity of muscle contraction. Calcium sensitivity can be modified by drugs that stabilize the open form of cNTnC, post-translational modifications like phosphorylation of troponin I, or downstream thin filament protein interactions that impact the availability of the troponin I switch region. Recently, mutations in cTnC have been associated with hypertrophic or dilated cardiomyopathy. A detailed understanding of how calcium sensitivity is regulated through the troponin complex is necessary for explaining how mutations perturb its function to promote cardiomyopathy and how post-translational modifications in the thin filament affect heart function and heart failure. Troponin modulating drugs are being developed for the treatment of cardiomyopathies and heart failure.
Keywords: Hypertrophic cardiomyopathy, dilated cardiomyopathy, heart failure, calcium sensitizer, levosimendan, sarcomere modulators
Biological context of troponin C
Muscle contraction is produced by the sliding of actin thin filaments against myosin thick filaments. In cardiac and skeletal muscle, thin and thick filaments are organized into highly polarized contractile units known as sarcomeres, which are linked in tandem and bundled together to form myofilaments. This arrangement gives these tissues their characteristic “striated” appearance.
The calcium-dependent contraction of striated muscle is controlled by the thin filament through the action of tropomyosin and the troponin complex, first discovered in 19642,3. The thin filament has a periodicity of about 38.5 nm1, comprising fourteen actin monomers arranged like a twisted “double string of beads”. Tropomyosin is a homodimeric parallel coiled-coil that polymerizes in a head-to-tail manner along the entire length of the thin filament (except at the Z-disc) 4, with each tropomyosin homodimer associated with one troponin complex and seven actin monomers. In the absence of troponin, tropomyosin lies along a positively charged groove 5,6, though it is able to shift or rotate considerably along the filament axis. At resting Ca2+ concentrations, the troponin complex anchors tropomyosin into a “blocked” position that sterically hinders the approach of myosin towards actin. An increase in free calcium concentration results in a conformational change within the troponin complex that releases tropomyosin into a “closed” position that allows weak actin-myosin interaction7. Strong binding of myosin to actin further shifts tropomyosin to the “open” position.
The troponin complex is made up of three components8–10: the calcium binding subunit, troponin C (TnC); the inhibitory subunit, troponin I (TnI); and an elongated protein, troponin T (TnT), that binds both TnC and TnI and anchors the entire complex to tropomyosin. For each of the subunits, homologous genes encode isoforms specific to the different types of striated muscle: cardiac, slow-twitch (type I) skeletal, and fast-twitch (type II) skeletal. The TNNC1 gene (3p21.1) is expressed in both slow skeletal and cardiac tissue (we will refer to it as cardiac troponin C, cTnC), whereas the TNNC2 gene (20q12-q13.11) encodes fast skeletal troponin C (hereafter referred to as sTnC)11–15. During development, both cTnC and sTnC are expressed in embryonic skeletal muscle, but expression of the cTnC gene is subsequently turned off during the transition to fast-twitch muscle16,17,18.
The Ca2+-dependent regulation of striated muscle contraction by troponin has been a fascinating area for biochemical and biophysical study since its discovery more than 50 years ago. TnC expresses well in E. coli and is the only soluble globular protein of the sarcomeric thin filament. Moreover, its role within the thin filament has been extensively studied, using well-established methods to reconstitute actin, tropomyosin, and troponin into functional filaments10,19. This review will focus on cardiac troponin C, highlighting its structure and function within the troponin complex. In recent times, the cardiac troponin complex has become important for understanding the pathogenesis, diagnosis, and treatment of cardiac diseases, especially as a target for the design of cardiac drugs. Underscoring its critical function, cTnC is highly conserved (96.8%– 99.4% = 1–6 sequence differences) across 61 known TnC sequences that have been cloned from 41 vertebrate and invertebrate species to date20.
Structure of cardiac troponin C
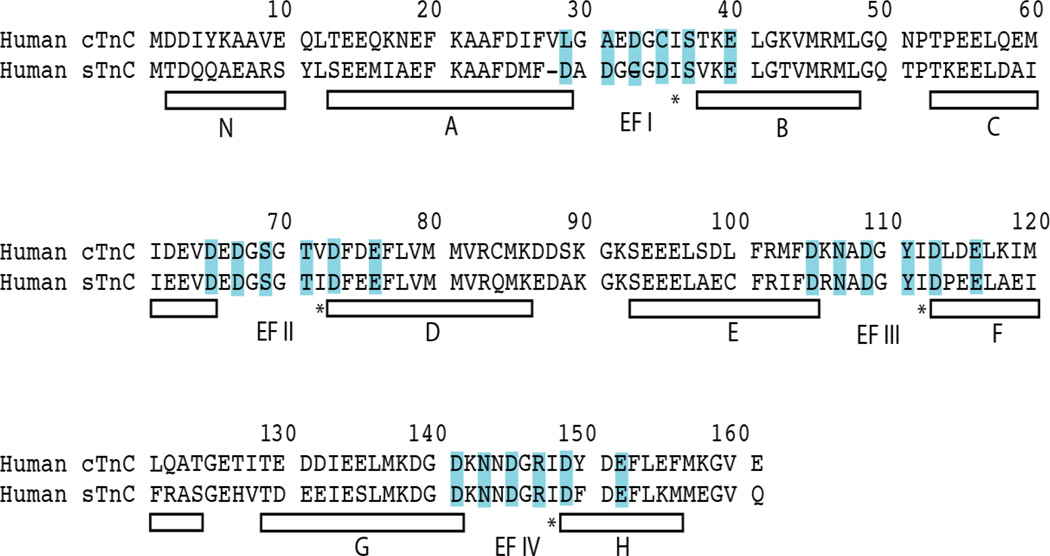
Troponin C is an 18-kDa member of the EF-hand Ca2+-binding protein family, first described in the X-ray crystal structure of parvalbumin in 197321. The family derives its name from a characteristic helix-loop-helix motif, in which six residues contribute oxygen ligands to define an octahedral Ca2+-binding site: 1(X), 3(Y), 5(Z), 7(−Y), 9(−X), and 12(−Z). Most of the ligands are polar amino acid sidechains, whereas the residue in the –Y position contributes a backbone carbonyl oxygen, and the sidechain in the –X position often indirectly coordinates Ca2+ via a bridging water molecule. The –Z position is almost always glutamate, which provides the only bidentate ligand, a carboxyl group, which changes the coordination geometry from octahedral to pentagonal bipyramidal. Both sTnC and cTnC comprise four EF-hand helix-loop-helix motifs as potential Ca2+-binding sites (I-IV), except that site I in cTnC is inactive due to an insertion (V28) and two key Ca2+-binding amino acid substitutions (D29L and D31A) (see Figure 1).
Figure 1.
Amino acid sequence comparison of human cTnC and sTnC. Helices are denoted by white bars below the sequences. In cTnC, residues 87–92 comprise the inter-domain helix. Calcium-coordinating EF-hand positions 1, 3, 5, 7, 9, and 12 are highlighted in cyan. Note, however, that EF-hand I in cTnC is defunct. Position 8 of each EF-hand is marked with an asterisk, denoting the hydrogen bonding central β-sheet-forming residue in each EF-hand.
The first three-dimensional structures of fast skeletal TnC were solved by X-ray crystallography in 1985, full-length turkey sTnC22 and full-length chicken sTnC23. sTnC is organized into two domains, each containing two Ca2+-binding EF-hands. In both structures, the two Ca2+-binding sites of the N-terminal domain (sNTnC) were unoccupied, while two Ca2+ ions were bound to the C-terminal domain (sCTnC) (Figure 2A). Comparison of the two homologous N- and C-domains showed that sNTnC was in a closed state, while sCTnC was in an open state, leading to the suggestion that Ca2+ binding to sNTnC would lead to a structural transition24, causing helices B+C to rotate away from helices N+A+D and exposing a large hydrophobic patch. A short anti-parallel β-sheet formed between EF-hands I and II (centered at position 8, see Figure 1) acts as a hinge for these sub-domain movements. This closed-to-open transition was confirmed in 1995 with the NMR solution structure of sTnC in the fully Ca2+-saturated state 25,26.
Figure 2.
Structures of sTnC and cTnC: A. sTnC•2Ca2+ (X-ray, pdb 5TNC); B. sTnC•4Ca2+ (X-ray, pdb 2TN4); C. cTnC•3Ca2+ (NMR, pdb 1AJ4). Ca2+ ions are shown as spheres.
Subsequent X-ray structures of sNTnC•2Ca2+ and sTnC•4Ca2+ have been reported27,28. The X-ray structures provide precise details of the metal coordination sites not obtainable from solution NMR methods. The main difference between the X-ray and solution structures of sTnC is found in the central linker connecting the N- and C-domains. The linker forms a rigid α-helix in the crystal forms, but it is unstructured and flexible in solution, highlighting a key caveat in the interpretation of X-ray crystal structures, that crystal packing contacts can induce structure in otherwise flexible regions. There is extensive solution NMR data on isolated TnC to suggest that the two domains can adopt a wide range of orientations: 15N relaxation measurements showing that the linker region is very dynamic and that the two domains tumble independently29, and paramagnetic relaxation enhancement (PRE) studies measuring the range of distances between domains30.
The NMR solution structure of cTnC•3Ca2+ was determined in 199731. Like sTnC, cTnC also adopts a dumbbell shape with a flexible linker (Figure 2B). As expected the structural C-domain of cTnC binds two Ca2+ ions and adopts an open conformation. A comparison of the regulatory domain of cTnC (cNTnC) in both the apo and Ca2+-bound states31,32 revealed that cNTnC remains strikingly closed in both the absence and presence of calcium, unlike sNTnC. This is likely a consequence of a defunct site I. In sNTnC, a site I-disrupting mutation E41A similarly abolished the link between Ca2+-binding and transition to the open state33.
Structure and function of cardiac troponin C within the troponin complex
Binding of Ca2+ to the regulatory NTnC domain is the key event that links cytoplasmic calcium influx to muscle contraction. Ca2+ binding allows NTnC to interact with the switch region of troponin I, removing the adjacent inhibitory region of TnI from its binding site on actin. It is primarily the inhibitory region of TnI34, and to a lesser degree, its C-terminal tail35,36, that anchors the troponin-tropomyosin complex to the blocked position that prevents actin-myosin interaction and shuts off muscle contraction in the absence of Ca2+.
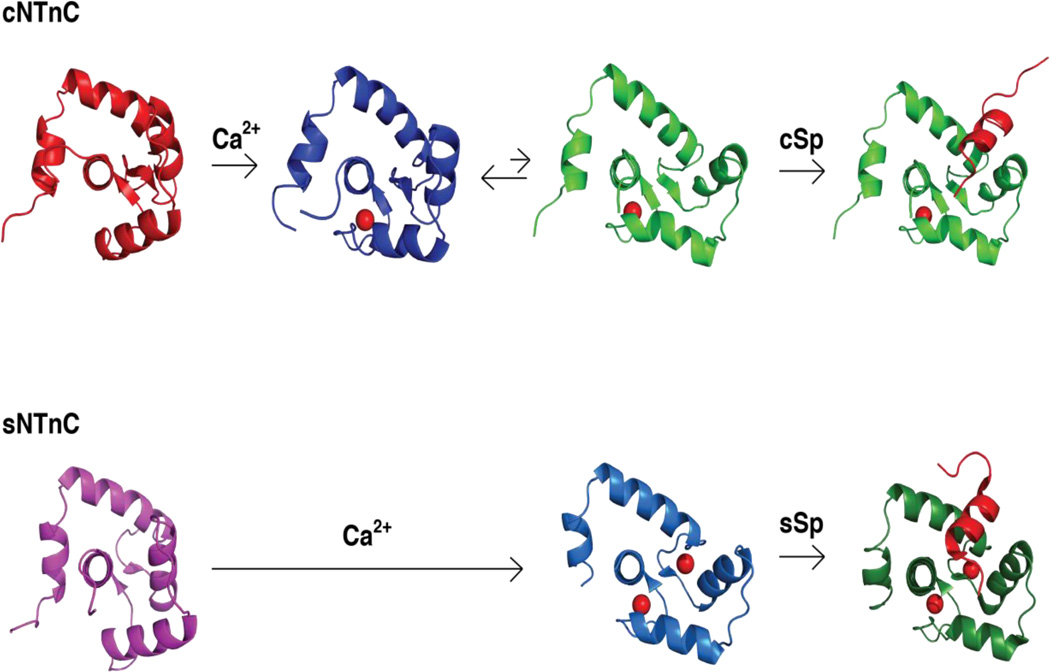
The closed-to-open transition observed in sNTnC suggests a mechanism for Ca2+-mediated regulation, whereby Ca2+ binding is coupled to the opening of a “sticky” hydrophobic surface. This is the central paradigm for calmodulin, the ubiquitous Ca2+-sensing protein that binds and regulates a plethora of targets via its versatile hydrophobic surfaces37. Indeed, sNTnC binds to the switch region of fast skeletal TnI via the corresponding hydrophobic surface exposed by Ca2+ binding38. Despite its significant homology to sNTnC, it was initially uncertain whether cNTnC would bind its target in a similar mode, since its Ca2+-bound form is predominantly closed. Nevertheless, the solution NMR structure of cNTnC•Ca2+•cTnI 147–163 revealed an open state for cNTnC39. The bound cTnI147–163 switch peptide adopts an α-helical conformation spanning residues 149–159, which wedges into the AB helical interface and inserts Ile148 and Met153 into the hydrophobic pocket to stabilize the open conformation of cNTnC (Figure 3).
Figure 3.
Comparison of the Ca2+- and cTnI switch peptide-induced structural changes in cNTnC and sNTnC. The pdb code for the structures are cNTnC•apo (1SPY), cNTnC•Ca2+ (1AP4), cNTnC•Ca2+•cSp (1MXL), sNTnC•apo (1TNP), sNTnC•2Ca2+ (1AVS), sNTnC•2Ca2+•sSp (1YTZ). Ca2+ ions are shown as spheres. Cardiac switch peptide (cSp) and fast skeletal switch peptide (sSp) are shown in red in the far right complexes.
Most of the structural work on cTnC has focused on its place in the cardiac troponin complex. However, cTnC is also the isoform for troponin C in slow-twitch skeletal muscle, forming a complex with slow skeletal TnI (ssTnI) and slow skeletal TnT (ssTnT). In fact, ssTnI is also present in the fetal heart, where it is postulated to allow the troponin complex to function in a more acidic environment40. The Ca2+ affinity of cTnC is reduced under acidic conditions through direct competition between H+ and Ca2+ ions. However, replacement of cTnI in cardiac muscle with ssTnI restores calcium sensitivity at low pH41. This acid-resistance was found to depend on a single amino acid substitution: the replacement of Ala162 in cTnI with the corresponding His130 in ssTnI42. The positively charged imidazole ring of His130 forms a salt bridge with Glu19 on the surface of cNTnC, stabilizing the cNTnC-ssTnI complex and restoring Ca2+-binding affinity under acidic conditions43. In the complex, His130 has a pKa of ~6.744, becoming more positively charged as the intracellular pH drops below 7. Recent NMR studies of cNTnC•Ca2+ bound to fsTnI115-131, as well as cNTnC•Ca2+ bound to A162H–cTnI147-163, have confirmed the importance of this electrostatic interaction45. The interaction is also present in the X-ray structure of the fast skeletal core troponin complex46, suggesting that it contributes to pH insensitivity in fast skeletal muscle as well. Some have advocated cardiac gene therapy to introduce the TnI A162H mutation in patients with severe cardiac ischemia47, since the Ca2+ sensitivity of the ischemic heart is reduced by cytosolic acidosis 40.
While the regulatory NTnC domain is generally accepted to play the crucial role of turning muscle contraction on and off in response to calcium, the structural CTnC domain keeps troponin C firmly anchored to the rest of the troponin complex throughout the cardiac cycle. The X-ray crystal structure of the cardiac troponin complex (Figure 4), cTnC•3Ca2+•cTnI33-209•cTnT183-28848, is indispensible for understanding the spatial relationships between troponin C, I, and T. (Note that the numbering of cTnI and cTnT is inconsistent throughout the literature. For cTnI, discrepancies arise because the N-terminal methionine residue is removed and replaced by an acetyl group, and it is variably included in the numbering. In this review, we will not include the initiator methionine so the first residue is Ala1. For cTnT, alternatively spliced forms result in varying lengths. We will use the numbering for the 288-residue version of human cTnT used in the crystal structure48.) The overall core domain structure is dominated by α-helical elements, most notably the IT arm (consisting of cCTnC, cTnI41-135 and cTnT225-276). cTnI41-135 forms two long α helices, cTnI41-79 and cTnI88-135. cTnI41-60 binds to the hydrophobic cleft of cCTnC, and then residues 62–80 continue the long helix well beyond cCTnC. Residues 80–87 form a loop that interacts with cTnT, and then cTnI88–135 proceeds back towards the opposite face of cCTnC while forming a coiled coil with cTnT225-276. cTnT225-276 is positioned between cCTnC and cTnI88–135, with cTnT259-270 binding extensively to the Ca2+-bound EF-hands III and IV of cTnC.
Figure 4.
X-ray structure of cardiac troponin core complex, cTnC•3Ca2+•cTnI34–161•cTnT221–276 (pdb 1J1E). cTnC is colored in green and the 3 bound Ca2+ are represented by magenta spheres, cTnI is colored in blue, and cTnT is colored in red. Regions N-terminal and C-terminal to cTnT221–276 are known to anchor the entire complex to tropomyosin and have been drawn in as squiggles to show that their structures are unknown. cTnI136–145 and cTnI162–209 are known to interact with actin to maintain the blocked state of the thin filament. These regions are shown as blue squiggles to signify that they are unstructured in the calcium-activated state, but acquire an as-of-yet undetermined structure bound to actin in the blocked state.
Limited proteolysis of troponin T (rabbit fast skeletal) yields two fragments, TnT1 and TnT249,50, with TnT2 featured in the troponin X-ray crystal structure. Binding studies of TnT2 have shown that both51,52 the region N-terminal53 and C-terminal54 to the IT arm (cTnT225-276) contribute to the interaction with tropomyosin, suggesting that the IT arm is rigidly fixed to tropomyosin. The C-terminus of cTnT, cTnT275-288, is also immediately adjacent to the cTnI128–147 inhibitory region, and both are essential for maintaining the blocked state of actin-tropomyosin55. On the other hand, one group has reported that the region corresponding to cTnT182-215 interacts with actin and contributes to the activation of actomyosin ATPase56. Thus, the IT arm and regions immediately adjacent to it are key for binding the troponin complex to tropomyosin as well as for stabilizing the inactive blocked state or the active open state of troponin-tropomyosin on actin.
Work performed prior to the publication of the X-ray structure of the cardiac troponin complex in 2003 suggested a possible interaction between the cTnI128–147 inhibitory region and the structural cCTnC domain. An NMR structure of cCTnC•2Ca2+•cTnI128-14757 shows residues L134-K139 of cTnI forming a helix upon interaction with the E and H helices of cCTnC•2Ca2+. However, since this arrangement of cTnI128-147 on cCTnC would be displaced by cTnI41-60, and cTnI88–135 is positioned on the opposite side of cCTnC as part of the IT arm (Figure 4), the observed cCTnC-cTnI128–147 interaction is unlikely to occur within the context of intact troponin complex. This example illustrates how binding studies on simplified systems need to be interpreted with caution.
Although N- and C-domains of troponin C are connected by a flexible linker, there are additional intermolecular interactions within the whole troponin complex that can fix the relative domain orientations of cTnC. The domain positions have recently been described using Förster resonance energy transfer (FRET) in reconstituted cardiac troponin complex 58 as well as with fluorescence polarization in intact thin filament59.
In cardiac TnC, the orientation of cNTnC relative to cCTnC is fixed by the direct interaction of the N-terminal 31-amino acid cardiac-specific region of cTnI with cNTnC 60–62. This region is critical to the regulation of calcium sensitivity in the cardiac sarcomere. Phosphorylation of cTnI in response to β-adrenergic sympathetic stimulation was first identified in 197663. β-adrenergic stimulation leads to production of cyclic AMP and activation of protein kinase A, which phosphorylates cTnI Ser22 and Ser23 (in addition to many other targets) 65,66. Other kinases have been found to phosphorylate this site as well including PKC, PKD1, and PKG67, showing Ser22/Ser23 to be an important regulatory node for integrating multiple signaling pathways. Phosphorylation at this site has been consistently shown to decrease the calcium sensitivity of the cardiac sarcomere64.
Recently, we have used solution NMR spectroscopy to study cTnI1-73 in complex with cTnC •3Ca2+68. In this complex, cTnI39-60 binds to cCTnC, stabilizing an α helix in cTnI41-67 and a type VIII turn in cTnI38-41 that brings cTnI19-37 into close proximity with the negatively charged surface of cNTnC (opposite the hydrophobic surface that binds the switch peptide, cTnI148-158). The interaction with cTnI19-37 is electrostatic in nature, which allows cTnI1–37 to maintain a disordered state even while in complex with cNTnC. In contrast to a previous model69 for the cNTnC-cTnI1–37 complex, the interaction does not induce secondary structure in cTnI1–37, it does not favor the open state of cNTnC, and it does not directly affect the Ca2+-binding affinity of cNTnC. However, it does fix the positioning of cNTnC relative to the rest of the troponin complex. We propose that this domain orientation is optimal for cNTnC binding to the cTnI148-158 switch region, and this is how cTnI19-37 indirectly increases the Ca2+ affinity of cNTnC within the context of the cardiac thin filament. Binding of cTnI148-158 stabilizes the Ca2+-bound open conformation of cNTnC, thus increasing its Ca2+ binding affinity. The cNTnC domain is released by protein kinase A (PKA) phosphorylation of cTnI Ser22 and Ser23, which disrupts the electrostatic interactions between cTnI19-37 and cNTnC.
The domain positioning of cNTnC relative to cCTnC is stabilized by the electrostatic interaction between cNTnC and cTnI19-37, but it is fully defined by additional weak interactions: a contact between Ala7 in the N helix of cNTnC and Ala42/Ser43 of the cTnI41-80 helix bound to cCTnC, as well as an interaction between the negatively charged C-terminal residue Glu161 of cCTnC and positively charged residues in the cTnC interdomain linker. These interactions are present in the X-ray crystal structure of the cardiac troponin complex48, and they bring together the N helix of cNTnC, the interdomain linker, and the C-terminus of cCTnC in the presence of unphosphorylated cTnI19-43 to define the interdomain orientation of cTnC.
Energetics of binding and conformational transitions
The calcium binding affinity of cNTnC must be exquisitely tuned to the cytosolic Ca2+ fluctuations within the cardiomyocyte. The cardiomyocyte free cytosolic Ca2+ concentration during diastole is about 100 nM, reaching a peak of about 1 µM during systole70. The half-activating concentration of free Ca2+ in cardiac myofilaments is typically 1–4 µM71,72.
The calcium binding affinity of the regulatory cNTnC domain is determined by the relative stability of the Ca2+-bound state versus the apo state. In Ca2+-binding EF-hand proteins that undergo a closed-to-open conformational transition, Ca2+ binding is driven by electrostatic interactions as well as entropically favorable release of water molecules73. This is offset by the exposure of a large hydrophobic patch upon transition to the open state. In cNTnC, the fact that EF-hand I is defunct means that there is no positive cooperativity of Ca2+ binding (at least for the isolated cNTnC domain), and the transition to the open state is less favorable, so that the closed state predominates in the absence of cTnI switch peptide. The structure of cNTnC in the closed conformation was recently highlighted by an X-ray structure of cNTnC in complex with Cd2+, providing for the first time a high resolution model of EF-hand II metal ion coordination in the closed state. Most of the Ca2+-coordinating interactions observed in the open state48 (see Figure 1) were intact in the closed state, except that D67 (the 3Y position of EF-hand II) and S69 (position 5Z) do not participate in the coordination of Cd2+ 74. This partial coordination may account for the increased rate of Ca2+ release in the closed state when compared to the cTnI switch peptide-stabilized open state (see below).
cNTnC is predominantly in the closed state when it is bound to Ca2+. However, it is now apparent that Ca2+-bound cNTnC exists in a dynamic equilibrium between closed and open forms (Figure 3). The opening involves movement of the BC helices as a unit away from the NAD helices unit. MD simulations have detected a concerted tilting motion of helices B and C away from helix A, corresponding to the opening motion of cNTnC•Ca2+ 75. NMR amide proton relaxation dispersion experiments reveal that many residues in the hinge regions of cNTnC experience a conformational exchange process with a time scale of ~30 µs 76,77. It was estimated that the open state is populated at about 20%78. This value is consistent with recent solution NMR paramagnetic relaxation enhancement (PRE) data from four spin-labeled monocysteine constructs of isolated cTnC, suggesting that the open state is populated ~27% in the calcium-saturated cTnC79,79, compared with ~0% in the apo state. Together these data support a model in which Ca2+ binding creates a dynamic equilibrium between the ‘closed’ and ‘open’ structural states to prime cNTnC for subsequent binding of the cTnI148-158 switch region.
Hydrophobic binding of cTnI148-158 to the large hydrophobic pocket of cNTnC stabilizes its open state, increasing its Ca2+ binding affinity. Using fluorescent IAANS labeling at Cys35 and Cys84, a Ca2+ binding KD of about 2 µM was determined for cNTnC in the context of isolated cTnC80. When cTnC was combined with cTnI and cTnT to make full troponin complex, KD of cNTnC became 0.3 µM, indicating about one order of magnitude tighter Ca2+ binding. This is due to a slowing of Ca2+ release, from >1000 s−1 from isolated cTnC at 15°C81 to 122 s−1 in the presence of cTnI and 33–42 s−1 in the presence of cTnI and cTnT82.
Solution NMR spectroscopy was also used to assess the binding of Ca2+ and cTnI147–163 to cNTnC at 30°C83. KD of ~20 µM and ~150 µM were determined for Ca2+ and cTnI147–163 peptide binding, respectively. The Ca2+ release rate was estimated to be 5000 s−1 in the absence of cTnI. The off rate of cTnI147-163 was about 5000 s−1, suggesting a rapid equilibrium between bound and free states.
A subsequent fluorescence study suggested that IAANS labeling at Cys35 and Cys84 altered the Ca2+-binding properties of cNTnC, so a monocysteine C35S/T53C/C84S mutant was put forward84. This mutant was used to study the Ca2+ binding characteristics of cTnC in the context of cardiac troponin complex (cTn) versus slow skeletal troponin complex (ssTn), as well as in the presence of actin, tropomyosin, and myosin S1 subfragment. In the isolated cardiac complex, a Ca2+ KD of 0.65 µM was measured, compared with 0.25 µM in the slow skeletal complex. The higher Ca2+ affinity of the ssTn complex can be expected from the enhanced affinity of the ssTnI switch region for cTnC. Upon addition of thin filament (actin and tropomyosin), the Ca2+ KD of cTn increased to 5 µM, explainable by the binding of cTnI128-147 to actin, making the cTnI148–158 switch region less available for binding to cNTnC. Addition of myosin S1 fragment (to troponin complex + actin + tropomyosin) changed the Ca2+ KD of cTn to 0.78 µM, likely because the formation of actomyosin rigor complexes shifts the thin filament to an open state that releases cTnI128–147 from actin. These studies demonstrate how the Ca2+ binding affinity of cNTnC is exquisitely dependent on the availability of cTnI148–158 switch region for binding. cTnI148–158 is tethered to actin via adjacent segments34 when the thin filament is in the blocked state, but released and available for binding cNTnC in the thin filament closed or open states. (Note that cNTnC itself has closed and open states that are distinct from the thin filament states, for which “closed” refers to the positioning of tropomyosin when cNTnC is bound to calcium, and the “open state” refers to a further shift of tropomyosin when myosin forms strong cross-bridges to actin.)
There are no known post-translational modifications of cTnC that modulate its calcium affinity. However, there are numerous phosphorylation sites on cTnI85 that impact the calcium affinity of cTnC, the most important of which is Ser22/23 in humans. We propose that many of these phosphorylation sites, including Ser22/23, impact cNTnC calcium binding by modulating the availability of cTnI148–158 switch peptide for binding (the interaction between the unphosphorylated cardiac-specific N-terminal extension and cNTnC orients it optimally for binding cTnI148–158, thus increasing the effective concentration of cTnI148–158). Moreover, modifications to TnT, tropomyosin, actin, or myosin can also impact the blocked-closed-open equilibrium of the thin filament to impact cTnI148–158 availability and calcium sensitivity.
In contrast to cNTnC, cCTnC binds Ca2+ with very high affinity. In isolated cTnC, the Ca2+ binding KD is about 40 nM86, decreasing to about 3 nM in intact troponin complex80. The very high affinity of CTnC is a consequence of the instability of its apo state. A closed-to-open transition does not occur in skeletal or cardiac CTnC, which appears to form a molten globule in the absence of Ca2+ 87,88. Ca2+ binding induces the formation of well defined tertiary structure, demonstrating that the sequence of CTnC is optimized for the open conformation, but not the closed. cTnI39–60 binds to Ca2+-saturated cTnC with very high affinity83,89. An affinity of 3 nM was measured for cTnI38–57 via surface plasmon resonance and ELISA90. The affinity for intact cTnI in the presence of cTnT is likely even stronger, given the additional contacts formed within the IT arm.
Mg2+ is chemically similar to Ca2+ and able to compete for the same binding sites in many EF-hand-containing proteins91. A classic equilibrium dialysis study of Ca2+- and Mg2+-binding in fast skeletal TnC showed that Mg2+ competes for the high affinity Ca2+ binding sites in the structural CTnC domain, whereas the lower affinity sites in the regulatory NTnC are Ca2+-specific92. (The same study also identified two non-competitive Mg2+ sites.) More recent fluorescence studies, however, have demonstrated that Mg2+ is also able to compete with Ca2+ in the NTnC domain, reducing the apparent calcium affinity by about a factor of 2 at a Mg2+ concentration of 1–3 mM93. Thus, the presence of Mg2+ in the muscle cytoplasm would have a calcium desensitizing effect, since Mg2+ itself does not trigger muscle contraction. Compared to Ca2+, Mg2+ binding tends to favor the closed conformation91,93, so cNTnC cannot bind cTnI switch peptide (in the open conformation) in the absence of Ca2+. In contrast, cCTnC is able to adopt an open conformation and bind cTnI33-80 when loaded with Mg2+ instead of Ca2+, as demonstrated by an NMR structure 94,95.
Hypertrophic cardiomyopathy-associated mutations
Hypertrophic cardiomyopathy (HCM) is a common genetic disease, having an estimated prevalence of ~1:50096,97. Although there are some rare infiltrative causes, HCM is generally a disease of the sarcomere, with a sarcomeric mutation identifiable in about half of affected patients. Mutations in the myosin heavy chain β and cardiac myosin binding protein-C are most common, followed by mutations in cardiac troponin I and troponin T98,99. One challenge in identifying definitive mutations is that many mutations have incomplete penetrance. As the name suggests, the most striking feature of HCM is abnormal hypertrophy of the ventricles, most classically prominent in the interventricular septum. The hypertrophy can be so severe that it causes a left ventricular outflow tract obstruction. The hypertrophied ventricles are also prone to ventricular arrhythmias that can cause sudden death. One challenge in the treatment of HCM is identifying patients at greatest risk of sudden death who would benefit from placement of an implantable cardioverter defibrillator. Other common complications include atrial fibrillation, diastolic dysfunction, myocardial ischemia, and mitral regurgitation100. To date, 7 mutations associated with HCM (A8V, L29Q, A31S, C84Y, Q122AfsX30, E134D, D145E) and 7 mutations associated with dilated cardiomyopathy (DCM) (Y5H, Q50R, E59D/D75Y, M103I, D145E, I148V, G159D) in cTnC have been reported. A recent review101 has summarized the biochemical characterization of these mutations, but we will briefly survey them here.
The first publication of a TNNC1 mutation associated with cardiomyopathy was in 2001102. An L29Q mutation was identified in a 60-year-old male HCM patient with 15 mm thickness of the septal and posterior left ventricular wall (normal is <11 mm and 13 mm is the minimum in adults required for HCM diagnosis). Ca2+ sensitivity of the L29Q–cTnC mutant has been reported to be decreased103, increased104,105, or unchanged106. These discrepancies have been suggested to be at least partly due to the different model systems used in these studies, although the same range of divergent results have also been reported for isolated protein components. Given the wide range of experimental findings in the absence of further supportive genetic evidence, it is possible that L29Q represents an incidental polymorphism.
An important study describing HCM mutations in troponin C was published in 2008, in which 1025 HCM patients were screened for mutations in TNNC1, and all positive hits were further screened at fifteen other known HCM susceptibility loci107. Four patients were identified with missense mutations in TNNC1 and no other locus, and these had left ventricular maximum wall widths ranging from 19 to 26 mm. The same authors later identified another mutation, A31S, in a five-year-old boy with a history of ventricular fibrillation and a mean ventricular wall thickness of 20 mm 108. None of these patients had affected family members with the same mutation. The five mutations, A8V, A31S, C84Y, E134D, and D145E were expressed recombinantly in troponin C and exchanged into porcine muscle fibers. All of the mutants except E134D were associated with increased calcium sensitivity109–112, so E134D may be a clinically insignificant variant. The observed increases in calcium sensitivity are consistent with what is observed in HCM-associated mutations involving other thin filament proteins 99. It seems plausible that increased calcium sensitivity of the thin filament leads to increased muscle activation and exuberant muscle hypertrophy.
HCM-associated mutations in cNTnC highlight the potential mechanisms by which the calcium affinity of cNTnC can be increased. The A8V mutation was found to increase calcium sensitivity of reconstituted thin filaments, particularly when cTnI was unphosphorylated 113. Ala8 is part of the N helix, which plays the key role in defining the domain orientation of cNTnC relative to the rest of the troponin complex, so it is possible that the A8V mutation stabilizes the positioning of the cNTnC domain. Ala31 is located in the loop region of EF hand I, so the A31S mutation may favor the conformation of this loop when cNTnC is in the open form. Cys84 contacts the switch region of cTnI in the open conformation and the BC loop in the closed conformation, so the C84Y mutation may either stabilize the open complex bound to cTnI or destabilize the closed conformation of cNTnC. In summary, we propose that the A8V, A31S, and C84Y increase the Ca2+-binding affinity of cNTnC by favoring the active form of cNTnC, either by stabilizing the activating orientation of the cNTnC domain, shifting the conformational equilibrium from the closed to open state, or directly enhancing binding to the cTnI switch peptide.
HCM-associated mutations identified in the cCTnC domain have far more destabilizing effects than those identified in the cNTnC domain. Asp145 is Ca2+-coordinating residue 5(Z) of EF-hand IV, and the D145E mutation was found to drastically reduce the Ca2+ binding affinity of cCTnC112,114,. Another HCM-associated mutation with a profound effect on the C-domain is Q122AfsX30, in which a frameshift mutation obliterates the last half of cCTnC and replaces it with 30 mistranslated residues115. This mutation was identified in a patient with histologically confirmed HCM after sudden death at the age of 19. The mutation was also identified in other family members with HCM. How these drastic mutations in the structural cCTnC domain contribute to HCM is currently unknown.
Dilated cardiomyopathy-associated mutations
Familial dilated cardiomyopathy (DCM) causes thin dilated ventricles associated with a decreased left ventricular ejection fraction. Compared with HCM, there is a wider variety of mutations, both sarcomeric and non-sarcomeric, that give rise to familial DCM, though DCM is more rare, with an estimated prevalence of ~1:5000 99,117.
G159D was the first DCM mutation reported118. It co-segregated with five affected family members, including the proband, who developed heart failure at the age of 21 and received a heart transplant two years later. A subsequent study identified a sixth family member who developed severe systolic heart failure requiring a heart transplant at the age of 3119. Thus, the genetic evidence supporting G159D as a causative mutation in DCM is the most extensive of any TNNC1 mutation. G159D-cTnC was shown to have similar Ca2+-binding affinity to wildtype-cTnC in isolation, but a reduced Ca2+ affinity was observed in intact muscle fiber experiments 120–123. Further studies demonstrated that G159D blunted the effect of cTnI phosphorylation at Ser22 and Ser23 124–126. Gly159 is located near the C-terminus of cTnC, where we postulate that the G159D mutation disrupts the delicate positioning of cNTnC relative to cCTnC.
The double mutant E59D/D75Y was identified in one patient 127. The same study also found that rat cardiomyocytes transfected with D75Y mutation, but not E59D, had greatly reduced contractility. Mutant D75Y-cTnC appears to have reduced Ca2+-binding affinity when tested in isolation, but the reduction in affinity appears more pronounced when it is incorporated into intact troponin complex or reconstituted thin filaments123. Asp75 is located position 11 of EF-hand II, though it is not directly involved in coordinating Ca2+. It is possible that the D75Y mutation favors the closed conformation of EF-hand II.
The Q50R mutation was identified in a 16-month-old with dilated cardiomyopathy, whose mother also had peripartum cardiomyopathy. The maternal grandmother and two of her sisters had a history of dilated cardiomyopathy as well, and the mutation co-segregated with disease in at least four family members across three generations128. Thus, the genetic evidence supporting Q50R as a DCM-causative mutation is strong, though no biochemical studies have been published to date. Q50 is located in the BC loop between EF-hands I and II, in a position where the Q50R mutation may produce an electrostatic repulsion with R147 of cTnI, disrupting binding of the switch region, cTnI148–158.
The remaining DCM-associated mutations in TNNC1 were identified in a study129 that examined a cohort of 312 patients with DCM. A total of 14 genes were sequenced, identifying mutations in up to 27% of the patients. There were four DCM-associated TNNC1 mutations identified: Y5H, M103I, D145E, and I148V. Y5H was detected in one patient, who developed heart failure at two weeks of age and later received a heart transplant at age of 15. Neither parent had DCM or the Y5H mutation. The father and the proband both had a mutation identified in MYH7, the beta myosin heavy chain, so it is possible that Y5H was only an incidentally observed mutation. All of the TNNC1 mutations identified in the study yielded small changes in calcium sensitivity, but Y5H had the largest Ca2+ desensitizing effect and also reduced the effect of PKA phosphorylation109. Tyr5 is located in helix N of cNTnC, where it packs against Arg83 of helix D. It is possible that the Y5H mutation creates an electrostatic repulsion that destabilizes helix N and disrupts positioning of the cNTnC domain. The M103I mutation was detected in a patient and a sister both with DCM 129. However, this family had two additional sisters without this mutation, but who had cardiac conduction abnormalities and/or syncope. Met103 is located in helix E of EF-hand III and binds to cTnI57-64. It is interesting to note that there is an isoleucine at the homologous position in skeletal TnC, so the M103I mutation would not be expected to cause a major perturbation. I148V was identified in a single patient with DCM129. Ile148 is located in the EF-hand IV loop region and is part of the small β-sheet at the base of the hydrophobic pocket. Other EF-hand proteins contain a valine at this site, so I148V is a relatively conservative substitution. Thus, M103I and I148V mutations would be expected to have only a minor impact on cCTnC structure and function, so it is uncertain whether these are truly clinically significant. In contrast, the D145E mutation, also independently identified in an unrelated HCM study (see section on HCM above)107, drastically reduces the calcium affinity of cCTnC. It is possible that this mutation primarily causes HCM, but HCM can progress towards a DCM phenotype in later stages.
The genetic evidence behind some DCM-associated TNNC1 mutations is excellent. However, unlike thin filament HCM mutations, for which there is a consensus about increased myofilament calcium sensitivity, there is less agreement regarding the functional impact of DCM mutations. Although some studies document a decrease in calcium sensitivity, this has not been consistently demonstrated. One interesting observation in a number of DCM-associated mutations in the thin filament is an insensitivity to the effect of PKA-mediated phosphorylation of Ser22/Ser23 in cTnI130. Phosphorylation of cTnI is an important mechanism by which the calcium sensitivity of the cardiac sarcomere is regulated. Dephosphorylation of cTnI has been observed in heart failure of various etiologies, including DCM and ischemic cardiomyopathy131. A non-phosphorylatable R21C mutation in cTnI (it abolishes the PKA RRXS phosphorylation motif) was associated with hypertrophic cardiomyopathy and diastolic dysfunction132,133. It is possible that the insensitivity to cTnI phosphorylation seen in some DCM mutants is indicative of a structural derangement whereby positioning of the regulatory cNTnC domain by unphosphorylated cTnI is either unachievable or ineffective. In this case, the predominant defect would be a decrease in calcium sensitivity for which dephosphorylation of cTnI would be unable to compensate.
Drugs that bind the regulatory domain, cNTnC
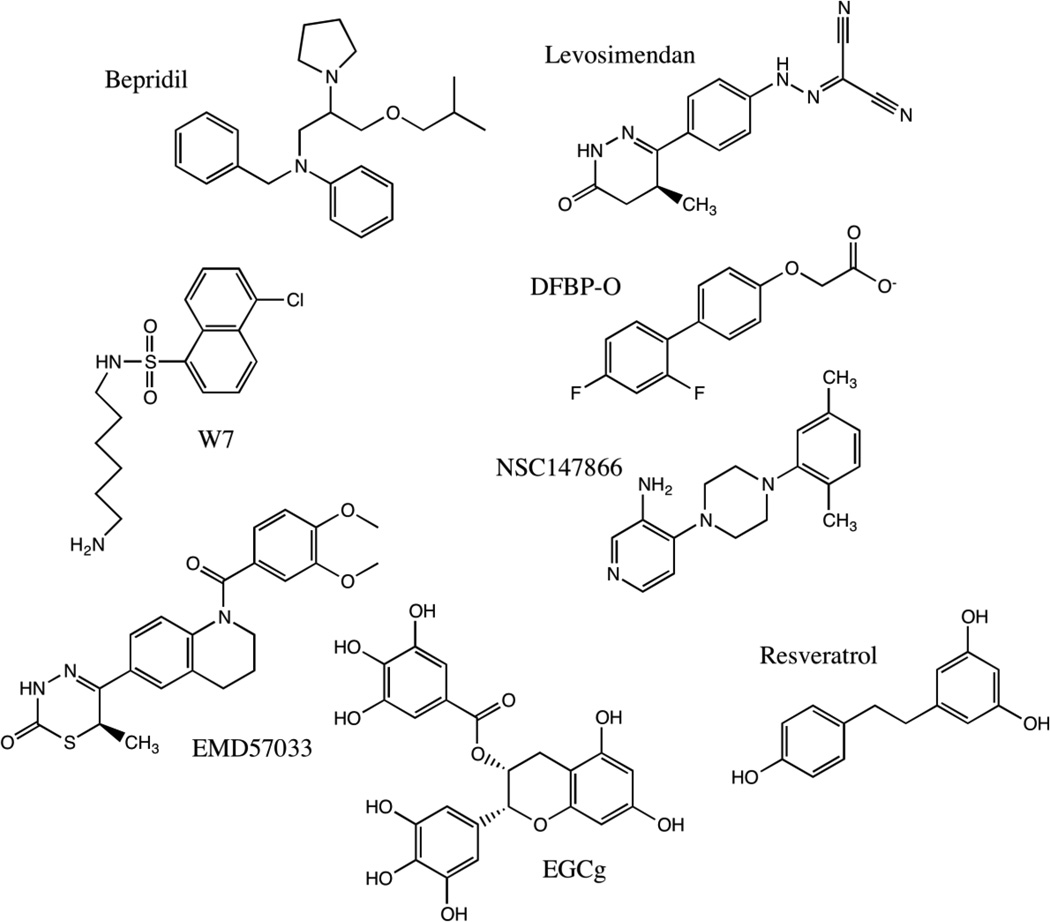
A number of small molecules have been found to bind to cardiac troponin C (see Figure 5). Of these, levosimendan is the most widely studied to date, licensed in some South American and European countries for use as a positive inotropic agent in acute decompensated systolic heart failure. Levosimendan has been shown to improve cardiac output and decrease symptoms of heart failure134. However, its use has been limited by associated cardiac arrhythmias and hypotension, and recent Phase III trials demonstrated no improvement in overall mortality135,136, though some meta-analyses suggest some mortality benefit137. Levosimendan has been suggested to bind to the regulatory domain of troponin C, cNTnC, but its exact mechanism of action has remained controversial138,139. One study failed to detect evidence of levosimendan binding to cTnC140, though small amounts of covalent adduct were detected after prolonged incubation, likely formed through levosimendan’s reactive malononitrile moiety (see Figure 5). It was later suggested that Cys84 is necessary for the covalent binding of levosimendan with cTnC 139,141, and this residue was changed to serine to produce a cysteineless mutant in some studies. Nevertheless, even with intact Cys84, no group has yet produced uniform stable cTnC•levosimendan complex for structural analysis. Moreover, the active metabolite of levosimendan, OR-1896142, does not possess the reactive malononitrile group and is unlikely to form a covalent conjugate. On the other hand, levosimendan does inhibit type 3 phosphodiesterase (PDE3) with nanomolar affinity142, and there are many other PDE3 inhibitors that act as positive inotropes and calcium sensitizers143, all of which also cause hypotension and arrhythmias as undesirable side effects144.
Figure 5.
Chemical structures of compounds known to bind to cardiac troponin C.
A levosimendan analogue (DFBP-O) was shown to bind to cNTnC-cTnI147–163 and increase the Ca2+ sensitivity of skinned cardiac trabeculae145. The structure of cNTnC•Ca2+•cTnI147–163•DFBP-O shows that DFBP-O binds to the interface between cNTnC and cTnI147–163 to enhance the binding of cTnI147–163 to cNTnC (see Figure 6). Using the structure of cNTnC•Ca2+•cTnI147–163•DFBP-O, a novel Ca2+ sensitizer 4-(4-(2,5-dimethylphenyl)-1-piperazinyl)-3-pyridinamine (NCI147866) was discovered using computational drug screening and verified by solution NMR 146. NCI147866 binds to both cNTnC •Ca2+ and the cNTnC•Ca2+•cTnI147-163 complex. Its presence increases the affinity of switch peptide to cNTnC by approximately a factor of two. Its affinity for cNTnC•Ca2+•cTnI147–163 is comparable to that of DFBP-O (KD 380 µM for both).
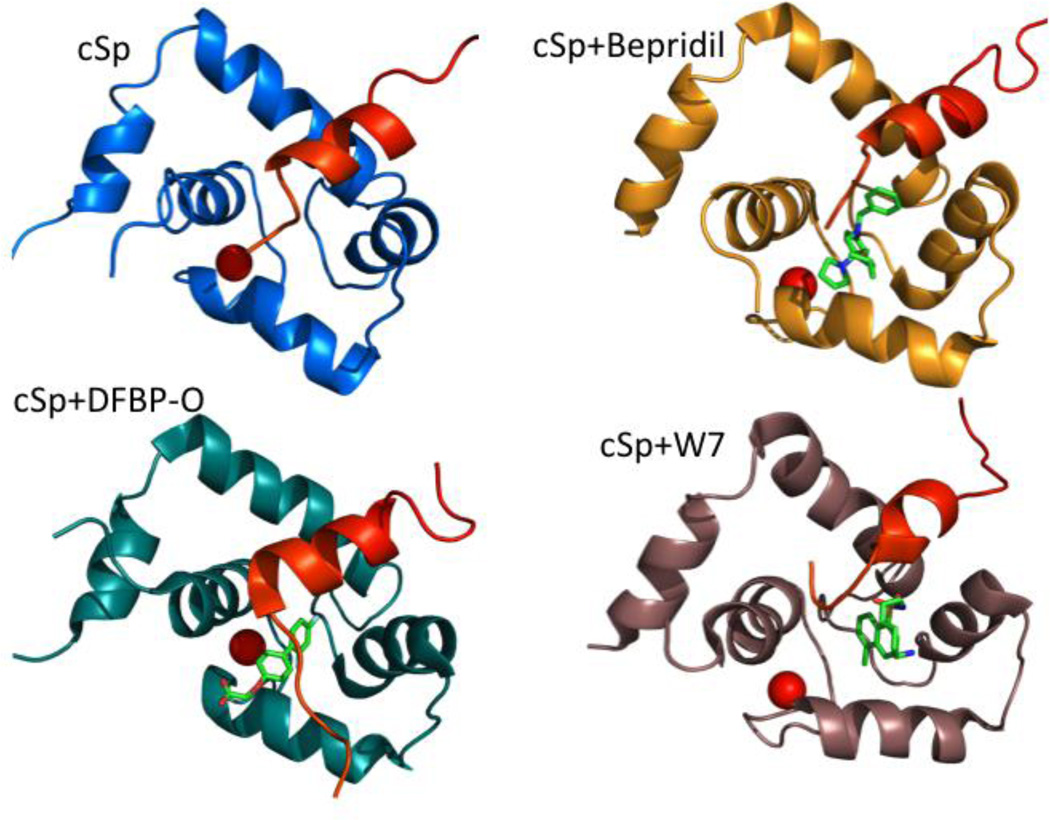
Figure 6.
Structures of cNTnC in complex with drugs and/or cTnI peptides: cNTnC•Ca2+•cSp (pdb 1MXL) is colored blue, cNTnC•Ca2+•cSp•bepridil (pdb 1LXF) is colored gold, cNTnC•Ca2+•cSp•DFBP-O (pdb 2L1R), cNTnC is colored teal, and cNTnC•Ca2+•cSp•W7 (pdb 2KRD) is colored brown. The cardiac switch peptide (cSp) is colored red, Ca2+ ions are shown as red spheres, and drugs are shown as sticks.
A CaM antagonist, W7 (N-(6-aminohexyl)-5-chloro-1-naphthalene sulfonamide) was found to inhibit the maximum ATPase activity and tension development in both skeletal and cardiac muscle fibers147,148. NMR titration studies have shown that W7 reduced the affinity of cTnI switch peptide for cNTnC from 154 µM to over 2000 uM149. An NMR structure of the ternary complex cNTnC•Ca2+•cTnI144–163•W7 (see Figure 6) revealed that W7 binding causes a considerable displacement of the cTnI switch peptide from its preferred binding site149. Thus, W7 acts as a troponin inhibitor and Ca2+ desensitizer by increasing the off rate of the cTnI switch peptide. Its mechanism of action stands in contrast to DFBP-O and NCI147866, which both act as troponin activators by enhancing the binding of cTnI switch peptide to cNTnC.
Bepridil is a Ca2+ channel blocker and a calmodulin (CaM) antagonist previously marketed as an anti-anginal agent. Bepridil increases the Ca2+ sensitivity of myofilaments, increasing actomyosin ATPase activity and force generation150. The X-ray structure of cTnC•3Ca2+•3bepridil provides detailed structural data on bepridil binding151. In this structure, three bepridil molecules stack together, but one molecule is bound to the hydrophobic pocket of cNTnC and another to the hydrophobic pocket of cCTnC, bringing the two domains together. The NMR structure of the cNTnC•Ca2+•cTnI147-163•bepridil ternary complex reveals a binding site for cTnI147-163 primarily located on the A/B interhelical interface and a binding site for bepridil in the hydrophobic core of cNTnC•Ca2+ 152. Bepridil seems to slightly weaken the binding of cTnI147-163 by displacing cTnI Ile148, but not to the same extent as W7.
In summary, cNTnC is an eminently “druggable” target with a large hydrophobic cavity already known to bind a number of compounds, albeit at low affinity. Moreover, cNTnC-binding compounds are able to act as troponin activators or inhibitors, depending on their effect on cTnI switch peptide binding. (It is preferable to refer to these compounds as troponin modulators rather than calcium sensitizers/desensitizers to distinguish their mechanism of action from other compounds that act through signaling pathways, like levosimendan.) Cardiomyopathy-associated mutations in troponin demonstrate that minute changes in troponin can have a dramatic effect on cardiac function and phenotype, so it is very possible that drugs targeting troponin will play a role in treating cardiomyopathies as well as heart failure in general. For fast skeletal troponin, a number of troponin activators developed by Cytokinetics are already at clinical and pre-clinical stages. We are currently working to develop the first high affinity cardiac troponin modulators.
Compounds that bind to the structural domain, cCTnC
As noted above, bepridil also binds to the structural domain of cardiac troponin C, cCTnC, being a promiscuous molecule that also interacts with calmodulin and calcium channels. However, bepridil does not bind to cCTnC in the presence of cTnI. The hydrophobic patch of cCTnC is capable of binding a number of other compounds well known for their promiscuity: EMD 57033, resveratrol, and green tea catechin EGCg (epigallocatechin gallate) (see Figure 5). Of these, resveratrol and EGCg have been highlighted recently as an example of “PAINS” (pan-assay interference compounds) 153. Thus, although these compounds have demonstrated activity as calcium sensitizers or desensitizers, it is uncertain whether binding to cCTnC or some other target accounts for their observed activity.
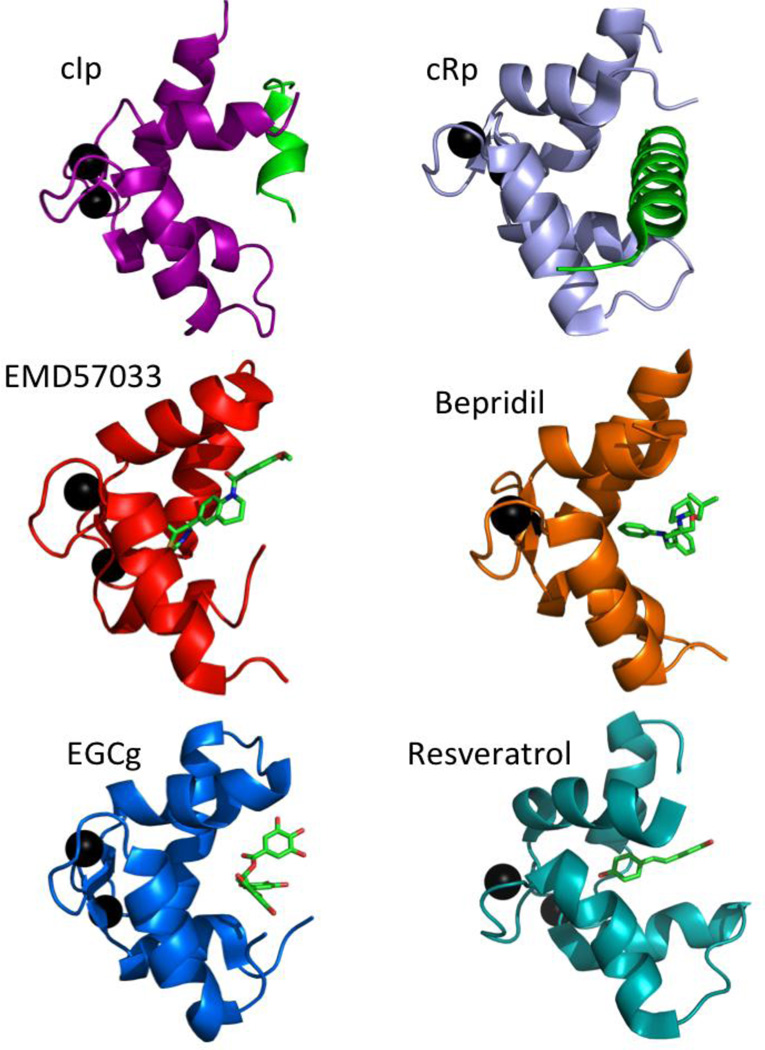
EMD 57033 is the (+)-enantiomer of EMD 53998. While the (−)-enantiomer, EMD 57439, is a PDE3 inhibitor, EMD 57033, enhances the Ca2+-sensitivity of both myofibrillar ATPase and force development in skinned muscle fibers154–156. EMD 57033 exerts a direct effect on actomyosin, increasing its ATPase activity. A recent study of EMD 57033 binding to myosin suggested that its binding site involved the SH3-like subdomain of the N-terminal domain 157. Direct binding of EMD 57033 to the C-domain of cTnC has been detected though tyrosine fluorescence 158 and solution NMR spectroscopy 140,159. In the high resolution structure of cCTnC•2Ca2+•EMD 5703389, the drug molecule is oriented such that the chiral group of EMD 57033 fits deep in the hydrophobic pocket and makes several key contacts with the protein (see Figure 7). EMD 57033 is completely displaced by the binding of cTnI34–71, but it is compatible with cTnI128–147 binding159. This suggests that EMD 57033 does not bind to cTnC in the context of intact troponin complex, but its interaction may become a factor if cCTnC becomes dissociated from the rest of the IT arm. In such a circumstance, EMD 57033 may even act as a calcium sensitizer by stabilizing the open conformation of cCTnC and priming it to bind cTnI128–147. However, there is currently no evidence that this ever occurs in vivo.
Figure 7.
Structures of cCTnC in complexes with drugs or cTnI peptides: cCTnC•2Ca2+•cIp (pdb 1OZS) is colored magenta, cCTnC•2Ca2+•cRp (pdb 1J1E) is colored slate, cCTnC•2Ca2+•EMD57033 (pdb 1IH0) is colored red, cCTnC•2Ca2+•bepridil (pdb 1DTL) is colored orange, cCTnC•2Ca2+•EGCg (pdb 2KDH) is colored blue, and cCTnC•2Ca2+•resveratrol (pdb 2L98) is colored cyan. Ca2+ ions are shown as black spheres, inhibitory peptide (cIp, cTnI128–147) and regulatory peptide(cRp, cTnI34–71) are shown in green, and drugs are shown as sticks.
The polyphenolic compounds EGCg160 and resveratrol161 were also found to bind to cCTnC, but not cNTnC. While EGCg is found in green tea, resveratrol is found in red wine. The NMR solution structures of cCTnC•2Ca2+•EGCg160 and cCTnC•2Ca2+•resveratrol161 show that these compounds bind to a similar pocket as EMD 57033 (see Figure 7). EGCg binding is more shallow than EMD 57033 binding, and unlike EMD 57033, EGCg can still bind cCTnC in the presence cTnI34–71, although the affinity is very weak, with a KD of about 1.8 mM. An examination of the cCTnC complexes presented in Figure 7 suggests that EGCg and cTnI128–147 bind more superficially to cCTnC than cTnI34–71 and the other drugs.
Summary
Over the past 40 years, significant progress has been made toward understanding the structure and function of cardiac troponin C. cTnC plays a critical role in the heart, coupling its electrical pacing system to its mechanical apparatus through the binding of Ca2+ ions. Calcium plays a dual role in cTnC, stabilizing the structural C-domain (via two high affinity binding sites), as well as activating the regulatory N-domain (via a single low affinity binding site), the key calcium sensor of cardiac contraction. cNTnC is a dynamic molecule, allowing for effective regulation of cardiac calcium sensitivity. First of all, cNTnC undergoes a rapid closed-to-open equilibrium, with the open state being the biologically active form that binds the troponin I switch peptide. Troponin modulating drugs can act by binding the open state, with troponin activators enhancing the affinity for the switch peptide and troponin inhibitors blocking this interaction. The cNTnC domain as a whole is dynamic, being connected to cCTnC and the rest of the troponin complex by a flexible linker. Stabilizing or disrupting the orientation of cNTnC via weak electrostatic interactions is an important regulatory mechanism readily impacted by phosphorylation of troponin I. Finally, the availability of the troponin I switch peptide as the thin filament alternates between blocked, closed, and open states, is an important downstream modulator of cNTnC function that can be impacted by modifications in myosin, actin, tropomyosin, and the rest of the troponin complex. All of these dynamic processes (the closed-to-open equilibrium, domain positioning, and switch peptide availability) are readily impacted by cardiomyopathy-causing mutations. Understanding the dynamic structural biology of cardiac troponin C within the thin filament will impact not only the treatment of the heritable cardiomyopathies, but also the treatment of heart failure of any etiology.
Highlights.
-
-
Cardiac troponin C (TNNC1) is the sarcomeric calcium sensor in cardiac and slow skeletal muscle
-
-
The C-domain has two high affinity Ca2+ binding sites and is anchored to troponin I and T
-
-
Ca2+ binding to the N-domain shifts muscle from relaxed to contracted states
-
-
Mutations in TNNC1 are associated with hypertrophic or dilated cardiomyopathy
-
-
Troponin modulators bind the open conformation of the N-domain and have therapeutic potential
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Cardiac Gene Wiki Review series – a series resulting from a collaboration between the journal GENE, the Gene Wiki Initiative, and the BD2K initiative. The Cardiac Gene Wiki Initiative is supported by National Institutes of Health (GM089820 and GM114833). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors would like to thank Brian Sykes for his mentorship and for valuable discussions. PMH is supported as a co-investigator by grant-in-aid G-14-0005884 from the Heart and Stroke Foundation of Canada. PMH is also supported by startup funds from the University of Alberta and an endowment from the June Hwang Professional Corporation.
The corresponding Gene Wiki entry for this review can be found here: http://en.wikipedia.org/wiki/Troponin_C_type_1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Yang S, Barbu-Tudoran L, Orzechowski M, et al. Three-dimensional organization of troponin on cardiac muscle thin filaments in the relaxed state. Biophys J. 2014;106(4):855–864. doi: 10.1016/j.bpj.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebashi S, Ebashi F. A new protein factor promoting contraction of actomyosin. Nature. 1964;203:645–646. doi: 10.1038/203645a0. [DOI] [PubMed] [Google Scholar]
- 3.Ebashi S, Ebashi F, Kodama A. Troponin as the ca++-receptive protein in the contractile system. J Biochem. 1967;62(1):137–138. doi: 10.1093/oxfordjournals.jbchem.a128628. [DOI] [PubMed] [Google Scholar]
- 4.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: How structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 5.Hitchcock-DeGregori SE. Tropomyosin: Function follows structure. Adv Exp Med Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- 6.von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519(7541):114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narita A, Yasunaga T, Ishikawa T, Mayanagi K, Wakabayashi T. Ca(2+)-induced switching of troponin and tropomyosin on actin filaments as revealed by electron cryo-microscopy. J Mol Biol. 2001;308(2):241–261. doi: 10.1006/jmbi.2001.4598. [DOI] [PubMed] [Google Scholar]
- 8.Schaub MC, Perry SV. The relaxing protein system of striated muscle. resolution of the troponin complex into inhibitory and calcium ion-sensitizing factors and their relationship to tropomyosin. Biochem J. 1969;115(5):993–1004. doi: 10.1042/bj1150993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebashi S. Separation of troponin into its three components. J Biochem. 1972;72(3):787–790. doi: 10.1093/oxfordjournals.jbchem.a129961. [DOI] [PubMed] [Google Scholar]
- 10.Greaser ML, Gergely J. Purification and properties of the components from troponin. J Biol Chem. 1973;248(6):2125–2133. [PubMed] [Google Scholar]
- 11.Gahlmann R, Wade R, Gunning P, Kedes L. Differential expression of slow and fast skeletal muscle troponin C. slow skeletal muscle troponin C is expressed in human fibroblasts. J Mol Biol. 1988;201(2):379–391. doi: 10.1016/0022-2836(88)90145-3. [DOI] [PubMed] [Google Scholar]
- 12.Schreier T, Kedes L, Gahlmann R. Cloning, structural analysis, and expression of the human slow twitch skeletal muscle/cardiac troponin C gene. J Biol Chem. 1990;265(34):21247–21253. [PubMed] [Google Scholar]
- 13.Gahlmann R, Kedes L. Cloning, structural analysis, and expression of the human fast twitch skeletal muscle troponin C gene. J Biol Chem. 1990;265(21):12520–12528. [PubMed] [Google Scholar]
- 14.Townsend PJ, Yacoub MH, Barton PJ. Assignment of the human cardiac/slow skeletal muscle troponin C gene (TNNC1) between D3S3118 and GCT4B10 on the short arm of chromosome 3 by somatic cell hybrid analysis. Ann Hum Genet. 1997;61(Pt 4):375–377. doi: 10.1046/j.1469-1809.1997.6140375.x. [DOI] [PubMed] [Google Scholar]
- 15.Townsend PJ, Yacoub MH, Barton PJ. Assignment of the human fast skeletal muscle troponin C gene (TNNC2) between D20S721 and GCT10F11 on chromosome 20 by somatic cell hybrid analysis. Ann Hum Genet. 1997;61(Pt 5):457–459. doi: 10.1046/j.1469-1809.1997.6150457.x. [DOI] [PubMed] [Google Scholar]
- 16.Dhoot GK, Perry SV. Distribution of polymorphic forms of troponin components and tropomyosin in skeletal muscle. Nature. 1979;278(5706):714–718. doi: 10.1038/278714a0. [DOI] [PubMed] [Google Scholar]
- 17.Toyota N, Shimada Y. Differentiation of troponin in cardiac and skeletal muscles in chicken embryos as studied by immunofluorescence microscopy. J Cell Biol. 1981;91(2 Pt 1):497–504. doi: 10.1083/jcb.91.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhoot GK, Frearson N, Perry SV. Polymorphic forms of troponin T and troponin C and their localization in striated muscle cell types. Exp Cell Res. 1979;122(2):339–350. doi: 10.1016/0014-4827(79)90310-0. [DOI] [PubMed] [Google Scholar]
- 19.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction I. biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246(15):4866–4871. [PubMed] [Google Scholar]
- 20.Gillis TE, Marshall CR, Tibbits GF. Functional and evolutionary relationships of troponin C. Physiol Genomics. 2007;32(1):16–27. doi: 10.1152/physiolgenomics.00197.2007. [DOI] [PubMed] [Google Scholar]
- 21.Kretsinger RH, Nockolds CE. Carp muscle calcium-binding protein II. structure determination and general description. J Biol Chem. 1973;248(9):3313–3326. [PubMed] [Google Scholar]
- 22.Herzberg O, James MN. Structure of the calcium regulatory muscle protein troponin-C at 2.8 A resolution. Nature. 1985;313(6004):653–659. doi: 10.1038/313653a0. [DOI] [PubMed] [Google Scholar]
- 23.Sundaralingam M, Bergstrom R, Strasburg G, et al. Molecular structure of troponin C from chicken skeletal muscle at 3-angstrom resolution. Science. 1985;227(4689):945–948. doi: 10.1126/science.3969570. [DOI] [PubMed] [Google Scholar]
- 24.Herzberg O, Moult J, James MN. A model for the Ca2+-induced conformational transition of troponin C. A trigger for muscle contraction. J Biol Chem. 1986;261(6):2638–2644. [PubMed] [Google Scholar]
- 25.Slupsky CM, Sykes BD. NMR solution structure of calcium-saturated skeletal muscle troponin C. Biochemistry. 1995;34(49):15953–15964. doi: 10.1021/bi00049a010. [DOI] [PubMed] [Google Scholar]
- 26.Gagne SM, Tsuda S, Li MX, Smillie LB, Sykes BD. Structures of the troponin C regulatory domains in the apo and calcium-saturated states. Nat Struct Biol. 1995;2(9):784–789. doi: 10.1038/nsb0995-784. [DOI] [PubMed] [Google Scholar]
- 27.Strynadka NC, Cherney M, Sielecki AR, Li MX, Smillie LB, James MN. Structural details of a calcium-induced molecular switch: X-ray crystallographic analysis of the calcium-saturated N-terminal domain of troponin C at 1.75 A resolution. J Mol Biol. 1997;273(1):238–255. doi: 10.1006/jmbi.1997.1257. [DOI] [PubMed] [Google Scholar]
- 28.Houdusse A, Love ML, Dominguez R, Grabarek Z, Cohen C. Structures of four Ca2+-bound troponin C at 2.0 A resolution: Further insights into the Ca2+-switch in the calmodulin superfamily. Structure. 1997;5(12):1695–1711. doi: 10.1016/s0969-2126(97)00315-8. [DOI] [PubMed] [Google Scholar]
- 29.Gaponenko V, Abusamhadneh E, Abbott MB, et al. Effects of troponin I phosphorylation on conformational exchange in the regulatory domain of cardiac troponin C. J Biol Chem. 1999;274(24):16681–16684. doi: 10.1074/jbc.274.24.16681. [DOI] [PubMed] [Google Scholar]
- 30.Cordina NM, Liew CK, Gell DA, Fajer PG, Mackay JP, Brown LJ. Interdomain orientation of cardiac troponin C characterized by paramagnetic relaxation enhancement NMR reveals a compact state. Protein Sci. 2012;21(9):1376–1387. doi: 10.1002/pro.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sia SK, Li MX, Spyracopoulos L, et al. Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain. J Biol Chem. 1997;272(29):18216–18221. doi: 10.1074/jbc.272.29.18216. [DOI] [PubMed] [Google Scholar]
- 32.Spyracopoulos L, Li MX, Sia SK, et al. Calcium-induced structural transition in the regulatory domain of human cardiac troponin C. Biochemistry. 1997;36(40):12138–12146. doi: 10.1021/bi971223d. [DOI] [PubMed] [Google Scholar]
- 33.Service RF. Flexing muscle with just one amino acid. Science. 1996;271(5245):31. doi: 10.1126/science.271.5245.31. [DOI] [PubMed] [Google Scholar]
- 34.Tripet B, Van Eyk JE, Hodges RS. Mapping of a second actin-tropomyosin and a second troponin C binding site within the C terminus of troponin I, and their importance in the Ca2+- dependent regulation of muscle contraction. J Mol Biol. 1997;271(5):728–750. doi: 10.1006/jmbi.1997.1200. [DOI] [PubMed] [Google Scholar]
- 35.Ramos CH. Mapping subdomains in the C-terminal region of troponin I involved in its binding to troponin C and to thin filament. J Biol Chem. 1999;274(26):18189–18195. doi: 10.1074/jbc.274.26.18189. [DOI] [PubMed] [Google Scholar]
- 36.Rarick HM, Tu XH, Solaro RJ, Martin AF. The C terminus of cardiac troponin I is essential for full inhibitory activity and Ca2+ sensitivity of rat myofibrils. J Biol Chem. 1997;272(43):26887–26892. doi: 10.1074/jbc.272.43.26887. [DOI] [PubMed] [Google Scholar]
- 37.Ikura M, Ames JB. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: Two ways to promote multifunctionality. Proc Natl Acad Sci U S A. 2006;103(5):1159–1164. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercier P, Ferguson RE, Irving M, Corrie JE, Trentham DR, Sykes BD. NMR structure of a bifunctional rhodamine labeled N-domain of troponin C complexed with the regulatory "switch" peptide from troponin I: Implications for in situ fluorescence studies in muscle fibers. Biochemistry. 2003;42(15):4333–4348. doi: 10.1021/bi027041n. [DOI] [PubMed] [Google Scholar]
- 39.Li MX, Spyracopoulos L, Sykes BD. Binding of cardiac troponin-I147-163 induces a structural opening in human cardiac troponin-C. Biochemistry. 1999;38(26):8289–8298. doi: 10.1021/bi9901679. [DOI] [PubMed] [Google Scholar]
- 40.Metzger JM, Westfall MV. Covalent and noncovalent modification of thin filament action: The essential role of troponin in cardiac muscle regulation. Circ Res. 2004;94(2):146–158. doi: 10.1161/01.RES.0000110083.17024.60. [DOI] [PubMed] [Google Scholar]
- 41.Westfall MV, Metzger JM. Single amino acid substitutions define isoform-specific effects of troponin I on myofilament Ca2+ and pH sensitivity. J Mol Cell Cardiol. 2007;43(2):107–118. doi: 10.1016/j.yjmcc.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dargis R, Pearlstone JR, Barrette-Ng I, Edwards H, Smillie LB. Single mutation (A162H) in human cardiac troponin I corrects acid pH sensitivity of Ca2+-regulated actomyosin S1 ATPase. J Biol Chem. 2002;277(38):34662–34665. doi: 10.1074/jbc.C200419200. [DOI] [PubMed] [Google Scholar]
- 43.Pineda-Sanabria SE, Robertson IM, Li MX, Sykes BD. Interaction between the regulatory domain of cardiac troponin C and the acidosis-resistant cardiac troponin I A162H. Cardiovasc Res. 2013;97(3):481–489. doi: 10.1093/cvr/cvs348. [DOI] [PubMed] [Google Scholar]
- 44.Robertson IM, Holmes PC, Li MX, Pineda-Sanabria SE, Baryshnikova OK, Sykes BD. Elucidation of isoform-dependent pH sensitivity of troponin i by NMR spectroscopy. J Biol Chem. 2012;287(7):4996–5007. doi: 10.1074/jbc.M111.301499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robertson IM, Pineda-Sanabria SE, Holmes PC, Sykes BD. Conformation of the critical pH sensitive region of troponin depends upon a single residue in troponin I. Arch Biochem Biophys. 2014;552–553:40–49. doi: 10.1016/j.abb.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Vinogradova MV, Stone DB, Malanina GG, et al. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci U S A. 2005;102(14):5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson BR, Metzger JM. Cell biology of sarcomeric protein engineering: Disease modeling and therapeutic potential. Anat Rec (Hoboken) 2014;297(9):1663–1669. doi: 10.1002/ar.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the ca(2+)-saturated form. Nature. 2003;424(6944):35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 49.Jackson P, Amphlett GW, Perry SV. The primary structure of troponin T and the interaction with tropomyosin. Biochem J. 1975;151(1):85–97. doi: 10.1042/bj1510085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearlstone JR, Carpenter MR, Smillie LB. Primary structure of rabbit skeletal muscle troponin-T. sequence determination of four cyanogen bromide fragments, CB4, CB6, and CB7. J Biol Chem. 1977;252(3):978–982. [PubMed] [Google Scholar]
- 51.Pearlstone JR, Smillie LB. Identification of a second binding region on rabbit skeletal troponin-T for alpha-tropomyosin. FEBS Lett. 1981;128(1):119–122. doi: 10.1016/0014-5793(81)81095-2. [DOI] [PubMed] [Google Scholar]
- 52.Pearlstone JR, Smillie LB. The interaction of rabbit skeletal muscle troponin-T fragments with troponin-I. Can J Biochem Cell Biol. 1985;63(3):212–218. doi: 10.1139/o85-030. [DOI] [PubMed] [Google Scholar]
- 53.Jin JP, Chong SM. Localization of the two tropomyosin-binding sites of troponin T. Arch Biochem Biophys. 2010;500(2):144–150. doi: 10.1016/j.abb.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanokura M, Tawada Y, Ono A, Ohtsuki I. Chymotryptic subfragments of troponin T from rabbit skeletal muscle. interaction with tropomyosin, troponin I and troponin C. J Biochem. 1983;93(2):331–337. doi: 10.1093/oxfordjournals.jbchem.a134185. [DOI] [PubMed] [Google Scholar]
- 55.Franklin AJ, Baxley T, Kobayashi T, Chalovich JM. The C-terminus of troponin T is essential for maintaining the inactive state of regulated actin. Biophys J. 2012;102(11):2536–2544. doi: 10.1016/j.bpj.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oliveira DM, Nakaie CR, Sousa AD, Farah CS, Reinach FC. Mapping the domain of troponin T responsible for the activation of actomyosin ATPase activity. identification of residues involved in binding to actin. J Biol Chem. 2000;275(36):27513–27519. doi: 10.1074/jbc.M002735200. [DOI] [PubMed] [Google Scholar]
- 57.Lindhout DA, Sykes BD. Structure and dynamics of the C-domain of human cardiac troponin C in complex with the inhibitory region of human cardiac troponin I. J Biol Chem. 2003;278(29):27024–27034. doi: 10.1074/jbc.M302497200. [DOI] [PubMed] [Google Scholar]
- 58.Jayasundar JJ, Xing J, Robinson JM, Cheung HC, Dong WJ. Molecular dynamics simulations of the cardiac troponin complex performed with FRET distances as restraints. PLoS One. 2014;9(2):e87135. doi: 10.1371/journal.pone.0087135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sevrieva I, Knowles AC, Kampourakis T, Sun YB. Regulatory domain of troponin moves dynamically during activation of cardiac muscle. J Mol Cell Cardiol. 2014;75:181–187. doi: 10.1016/j.yjmcc.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Finley N, Abbott MB, Abusamhadneh E, et al. NMR analysis of cardiac troponin C-troponin I complexes: Effects of phosphorylation. FEBS Lett. 1999;453(1–2):107–112. doi: 10.1016/s0014-5793(99)00693-6. [DOI] [PubMed] [Google Scholar]
- 61.Abbott MB, Dong WJ, Dvoretsky A, et al. Modulation of cardiac troponin C-cardiac troponin I regulatory interactions by the amino-terminus of cardiac troponin I. Biochemistry. 2001;40(20):5992–6001. doi: 10.1021/bi0100642. [DOI] [PubMed] [Google Scholar]
- 62.Ward DG, Brewer SM, Cornes MP, Trayer IP. A cross-linking study of the N-terminal extension of human cardiac troponin I. Biochemistry. 2003;42(34):10324–10332. doi: 10.1021/bi034495r. [DOI] [PubMed] [Google Scholar]
- 63.Solaro RJ, Moir AJ, Perry SV. Phosphorylation of troponin I and the inotropic effect of adrenaline in the perfused rabbit heart. Nature. 1976;262(5569):615–617. doi: 10.1038/262615a0. [DOI] [PubMed] [Google Scholar]
- 64.Wattanapermpool J, Guo X, Solaro R. The unique amino-terminal peptide of cardiac troponin-i regulates myofibrillar activity only when it is phosphorylated. J Mol Cell Cardiol. 1995;27(7):1383–1391. doi: 10.1006/jmcc.1995.0131. [DOI] [PubMed] [Google Scholar]
- 65.Solaro RJ, van der Velden J. Why does troponin I have so many phosphorylation sites? fact and fancy. J Mol Cell Cardiol. 2010;48(5):810–816. doi: 10.1016/j.yjmcc.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang J, Guy MJ, Norman HS, et al. Top-down quantitative proteomics identified phosphorylation of cardiac troponin I as a candidate biomarker for chronic heart failure. J Proteome Res. 2011;10(9):4054–4065. doi: 10.1021/pr200258m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Solaro RJ, Henze M, Kobayashi T. Integration of troponin I phosphorylation with cardiac regulatory networks. Circulation Research. 2013;112(2):355–366. doi: 10.1161/CIRCRESAHA.112.268672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hwang PM, Cai F, Pineda-Sanabria SE, Corson DC, Sykes BD. The cardiac-specific N-terminal region of troponin I positions the regulatory domain of troponin C. Proc Natl Acad Sci U S A. 2014;111(40):14412–14417. doi: 10.1073/pnas.1410775111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howarth JW, Meller J, Solaro RJ, Trewhella J, Rosevear PR. Phosphorylation-dependent conformational transition of the cardiac specific N-extension of troponin I in cardiac troponin. J Mol Biol. 2007;373(3):706–722. doi: 10.1016/j.jmb.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 70.Bers DM. Calcium fluxes involved in control of cardiac myocyte contraction. Circ Res. 2000;87(4):275–281. doi: 10.1161/01.res.87.4.275. [DOI] [PubMed] [Google Scholar]
- 71.Wijnker PJ, Sequeira V, Foster DB, et al. Length-dependent activation is modulated by cardiac troponin I bisphosphorylation at Ser23 and Ser24 but not by Thr143 phosphorylation. Am J Physiol Heart Circ Physiol. 2014;306(8):H1171–H1181. doi: 10.1152/ajpheart.00580.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mullins PD, Bondarenko VE. A mathematical model of the mouse ventricular myocyte contraction. PLoS One. 2013;8(5):e63141. doi: 10.1371/journal.pone.0063141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gifford JL, Walsh MP, Vogel HJ. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J. 2007;405(2):199–221. doi: 10.1042/BJ20070255. [DOI] [PubMed] [Google Scholar]
- 74.Zhang XL, Tibbits GF, Paetzel M. The structure of cardiac troponin C regulatory domain with bound Cd2+ reveals a closed conformation and unique ion coordination. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 5):722–734. doi: 10.1107/S0907444913001182. [DOI] [PubMed] [Google Scholar]
- 75.Lindert S, Kekenes-Huskey PM, Huber G, Pierce L, McCammon JA. Dynamics and calcium association to the N-terminal regulatory domain of human cardiac troponin C: A multiscale computational study. J Phys Chem B. 2012;116(29):8449–8459. doi: 10.1021/jp212173f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eichmuller C, Skrynnikov NR. A new amide proton R1rho experiment permits accurate characterization of microsecond time-scale conformational exchange. J Biomol NMR. 2005;32(4):281–293. doi: 10.1007/s10858-005-0658-y. [DOI] [PubMed] [Google Scholar]
- 77.Eichmuller C, Skrynnikov NR. Observation of microsecond time-scale protein dynamics in the presence of Ln3+ ions: Application to the N-terminal domain of cardiac troponin C. J Biomol NMR. 2007;37(2):79–95. doi: 10.1007/s10858-006-9105-y. [DOI] [PubMed] [Google Scholar]
- 78.McKay RT, Saltibus LF, Li MX, Sykes BD. Energetics of the induced structural change in a Ca2+ regulatory protein: Ca2+ and troponin I peptide binding to the E41A mutant of the N-domain of skeletal troponin C. Biochemistry. 2000;39(41):12731–12738. doi: 10.1021/bi001240u. [DOI] [PubMed] [Google Scholar]
- 79.Cordina NM, Liew CK, Gell DA, Fajer PG, Mackay JP, Brown LJ. Effects of calcium binding and the hypertrophic cardiomyopathy A8V mutation on the dynamic equilibrium between closed and open conformations of the regulatory N-domain of isolated cardiac troponin C. Biochemistry. 2013;52(11):1950–1962. doi: 10.1021/bi4000172. [DOI] [PubMed] [Google Scholar]
- 80.Johnson JD, Collins JH, Robertson SP, Potter JD. A fluorescent probe study of Ca2+ binding to the Ca2+-specific sites of cardiac troponin and troponin C. J Biol Chem. 1980;255(20):9635–9640. [PubMed] [Google Scholar]
- 81.Tikunova SB, Davis JP. Designing calcium-sensitizing mutations in the regulatory domain of cardiac troponin C. J Biol Chem. 2004;279(34):35341–35352. doi: 10.1074/jbc.M405413200. [DOI] [PubMed] [Google Scholar]
- 82.Gomes AV, Venkatraman G, Davis JP, et al. Cardiac troponin T isoforms affect the ca(2+) sensitivity of force development in the presence of slow skeletal troponin I: Insights into the role of troponin T isoforms in the fetal heart. J Biol Chem. 2004;279(48):49579–49587. doi: 10.1074/jbc.M407340200. [DOI] [PubMed] [Google Scholar]
- 83.Li MX, Saude EJ, Wang X, Pearlstone JR, Smillie LB, Sykes BD. Kinetic studies of calcium and cardiac troponin I peptide binding to human cardiac troponin C using NMR spectroscopy. Eur Biophys J. 2002;31(4):245–256. doi: 10.1007/s00249-002-0227-1. [DOI] [PubMed] [Google Scholar]
- 84.Davis JP, Norman C, Kobayashi T, Solaro RJ, Swartz DR, Tikunova SB. Effects of thin and thick filament proteins on calcium binding and exchange with cardiac troponin C. Biophys J. 2007;92(9):3195–3206. doi: 10.1529/biophysj.106.095406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang P, Kirk JA, Ji W, et al. Multiple reaction monitoring to identify site-specific troponin I phosphorylated residues in the failing human heart. Circulation. 2012;126(15):1828–1837. doi: 10.1161/CIRCULATIONAHA.112.096388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson JD, Potter JD. Detection of two classes of Ca2+ binding sites in troponin C with circular dichroism and tyrosine fluorescence. J Biol Chem. 1978;253(11):3775–3777. [PubMed] [Google Scholar]
- 87.Mercier P, Li MX, Sykes BD. Role of the structural domain of troponin C in muscle regulation: NMR studies of Ca2+ binding and subsequent interactions with regions 1–40 and 96–115 of troponin I. Biochemistry. 2000;39(11):2902–2911. doi: 10.1021/bi992579n. [DOI] [PubMed] [Google Scholar]
- 88.Brito RM, Krudy GA, Negele JC, Putkey JA, Rosevear PR. Calcium plays distinctive structural roles in the N- and C-terminal domains of cardiac troponin C. J Biol Chem. 1993;268(28):20966–20973. [PubMed] [Google Scholar]
- 89.Wang X, Li MX, Spyracopoulos L, et al. Structure of the C-domain of human cardiac troponin C in complex with the Ca2+ sensitizing drug EMD 57033. J Biol Chem. 2001;276(27):25456–25466. doi: 10.1074/jbc.M102418200. [DOI] [PubMed] [Google Scholar]
- 90.Ferrières G, Pugnière M, Mani J, et al. Systematic mapping of regions of human cardiac troponin I involved in binding to cardiac troponin C: N- and C-terminal low affinity contributing regions. FEBS Lett. 2000;479(3):99–105. doi: 10.1016/s0014-5793(00)01881-0. doi: http://dx.doi.org/10.1016/S0014-5793(00)01881-0. [DOI] [PubMed] [Google Scholar]
- 91.Grabarek Z. Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins. Biochim Biophys Acta. 2011;1813(5):913–921. doi: 10.1016/j.bbamcr.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Potter JD, Gergely J. The calcium and magnesium binding sites on troponin and their role in the regulation of myofibrillar adenosine triphosphatase. J Biol Chem. 1975;250(12):4628–4633. [PubMed] [Google Scholar]
- 93.Davis JP, Rall JA, Reiser PJ, Smillie LB, Tikunova SB. Engineering competitive magnesium binding into the first EF-hand of skeletal troponin C. J Biol Chem. 2002;277(51):49716–49726. doi: 10.1074/jbc.M208488200. [DOI] [PubMed] [Google Scholar]
- 94.Finley N, Dvoretsky A, Rosevear PR. Magnesium-calcium exchange in cardiac troponin C bound to cardiac troponin I. J Mol Cell Cardiol. 2000;32(8):1439–1446. doi: 10.1006/jmcc.2000.1174. [DOI] [PubMed] [Google Scholar]
- 95.Finley NL, Howarth JW, Rosevear PR. Structure of the Mg2+-loaded C-lobe of cardiac troponin C bound to the N-domain of cardiac troponin I: Comparison with the Ca2+-loaded structure. Biochemistry. 2004;43(36):11371–11379. doi: 10.1021/bi049672i. [DOI] [PubMed] [Google Scholar]
- 96.Zou Y, Song L, Wang Z, et al. Prevalence of idiopathic hypertrophic cardiomyopathy in china: A population-based echocardiographic analysis of 8080 adults. Am J Med. 2004;116(1):14–18. doi: 10.1016/j.amjmed.2003.05.009. [DOI] [PubMed] [Google Scholar]
- 97.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. echocardiographic analysis of 4111 subjects in the CARDIA study. coronary artery risk development in (young) adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 98.Chang AN, Parvatiyar MS, Potter JD. Troponin and cardiomyopathy. Biochem Biophys Res Commun. 2008;369(1):74–81. doi: 10.1016/j.bbrc.2007.12.081. [DOI] [PubMed] [Google Scholar]
- 99.Willott RH, Gomes AV, Chang AN, Parvatiyar MS, Pinto JR, Potter JD. Mutations in troponin that cause HCM, DCM AND RCM: What can we learn about thin filament function? J Mol Cell Cardiol. 2010;48(5):882–892. doi: 10.1016/j.yjmcc.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 100.Gersh BJ, Maron BJ, Bonow RO, et al. 2011 ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: A report of the american college of cardiology foundation/american heart association task force on practice guidelines. developed in collaboration with the american association for thoracic surgery, american society of echocardiography, american society of nuclear cardiology, heart failure society of america, heart rhythm society, society for cardiovascular angiography and interventions, and society of thoracic surgeons. J Am Coll Cardiol. 2011;58(25):e212–w260. doi: 10.1016/j.jacc.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 101.Kalyva A, Parthenakis FI, Marketou ME, Kontaraki JE, Vardas PE. Biochemical characterisation of troponin C mutations causing hypertrophic and dilated cardiomyopathies. J Muscle Res Cell Motil. 2014;35(2):161–178. doi: 10.1007/s10974-014-9382-0. [DOI] [PubMed] [Google Scholar]
- 102.Hoffmann B, Schmidt-Traub H, Perrot A, Osterziel KJ, Gessner R. First mutation in cardiac troponin C, L29Q, in a patient with hypertrophic cardiomyopathy. Hum Mutat. 2001;17(6):524. doi: 10.1002/humu.1143. [DOI] [PubMed] [Google Scholar]
- 103.Schmidtmann A, Lindow C, Villard S, et al. Cardiac troponin C-L29Q, related to hypertrophic cardiomyopathy, hinders the transduction of the protein kinase A dependent phosphorylation signal from cardiac troponin I to C. FEBS J. 2005;272(23):6087–6097. doi: 10.1111/j.1742-4658.2005.05001.x. [DOI] [PubMed] [Google Scholar]
- 104.Liang B, Chung F, Qu Y, et al. Familial hypertrophic cardiomyopathy-related cardiac troponin C mutation L29Q affects Ca2+ binding and myofilament contractility. Physiol Genomics. 2008;33(2):257–266. doi: 10.1152/physiolgenomics.00154.2007. [DOI] [PubMed] [Google Scholar]
- 105.Li AY, Stevens CM, Liang B, et al. Familial hypertrophic cardiomyopathy related cardiac troponin C L29Q mutation alters length-dependent activation and functional effects of phosphomimetic troponin I*. PLoS One. 2013;8(11):e79363. doi: 10.1371/journal.pone.0079363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dweck D, Hus N, Potter JD. Challenging current paradigms related to cardiomyopathies. are changes in the Ca2+ sensitivity of myofilaments containing cardiac troponin C mutations (G159D and L29Q) good predictors of the phenotypic outcomes? J Biol Chem. 2008;283(48):33119–33128. doi: 10.1074/jbc.M804070200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Landstrom AP, Parvatiyar MS, Pinto JR, et al. Molecular and functional characterization of novel hypertrophic cardiomyopathy susceptibility mutations in TNNC1-encoded troponin C. J Mol Cell Cardiol. 2008;45(2):281–288. doi: 10.1016/j.yjmcc.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Parvatiyar MS, Landstrom AP, Figueiredo-Freitas C, Potter JD, Ackerman MJ, Pinto JR. A mutation in TNNC1-encoded cardiac troponin C, TNNC1-A31S, predisposes to hypertrophic cardiomyopathy and ventricular fibrillation. J Biol Chem. 2012;287(38):31845–31855. doi: 10.1074/jbc.M112.377713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pinto JR, Siegfried JD, Parvatiyar MS, et al. Functional characterization of TNNC1 rare variants identified in dilated cardiomyopathy. J Biol Chem. 2011;286(39):34404–34412. doi: 10.1074/jbc.M111.267211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pinto JR, Reynaldo DP, Parvatiyar MS, et al. Strong cross-bridges potentiate the ca(2+) affinity changes produced by hypertrophic cardiomyopathy cardiac troponin C mutants in myofilaments: A fast kinetic approach. J Biol Chem. 2011;286(2):1005–1013. doi: 10.1074/jbc.M110.168583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Swindle N, Albury AN, Baroud B, Burney M, Tikunova SB. Molecular and functional consequences of mutations in the central helix of cardiac troponin C. Arch Biochem Biophys. 2014;548:46–53. doi: 10.1016/j.abb.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Swindle N, Tikunova SB. Hypertrophic cardiomyopathy-linked mutation D145E drastically alters calcium binding by the C-domain of cardiac troponin C. Biochemistry. 2010;49(23):4813–4820. doi: 10.1021/bi100400h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Albury AN, Swindle N, Swartz DR, Tikunova SB. Effect of hypertrophic cardiomyopathy-linked troponin C mutations on the response of reconstituted thin filaments to calcium upon troponin I phosphorylation. Biochemistry. 2012;51(17):3614–3621. doi: 10.1021/bi300187k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pinto JR, Parvatiyar MS, Jones MA, Liang J, Ackerman MJ, Potter JD. A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J Biol Chem. 2009;284(28):19090–19100. doi: 10.1074/jbc.M109.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chung WK, Kitner C, Maron BJ. Novel frameshift mutation in troponin C (TNNC1) associated with hypertrophic cardiomyopathy and sudden death. Cardiol Young. 2011;21(3):345–348. doi: 10.1017/S1047951110001927. [DOI] [PubMed] [Google Scholar]
- 116.Sequeira V, Wijnker PJ, Nijenkamp LL, et al. Perturbed length-dependent activation in human hypertrophic cardiomyopathy with missense sarcomeric gene mutations. Circ Res. 2013;112(11):1491–1505. doi: 10.1161/CIRCRESAHA.111.300436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chang AN, Potter JD. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail Rev. 2005;10(3):225–235. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- 118.Mogensen J, Murphy RT, Shaw T, et al. Severe disease expression of cardiac troponin C and T mutations in patients with idiopathic dilated cardiomyopathy. J Am Coll Cardiol. 2004;44(10):2033–2040. doi: 10.1016/j.jacc.2004.08.027. [DOI] [PubMed] [Google Scholar]
- 119.Kaski JP, Burch M, Elliott PM. Mutations in the cardiac troponin C gene are a cause of idiopathic dilated cardiomyopathy in childhood. Cardiol Young. 2007;17(6):675–677. doi: 10.1017/S1047951107001291. [DOI] [PubMed] [Google Scholar]
- 120.Mirza M, Marston S, Willott R, et al. Dilated cardiomyopathy mutations in three thin filament regulatory proteins result in a common functional phenotype. J Biol Chem. 2005;280(31):28498–28506. doi: 10.1074/jbc.M412281200. [DOI] [PubMed] [Google Scholar]
- 121.Preston LC, Lipscomb S, Robinson P, et al. Functional effects of the DCM mutant Gly159Asp troponin C in skinned muscle fibres. Pflugers Arch. 2007;453(6):771–776. doi: 10.1007/s00424-006-0161-7. [DOI] [PubMed] [Google Scholar]
- 122.Dyer EC, Jacques AM, Hoskins AC, et al. Functional analysis of a unique troponin c mutation, GLY159ASP, that causes familial dilated cardiomyopathy, studied in explanted heart muscle. Circ Heart Fail. 2009;2(5):456–464. doi: 10.1161/CIRCHEARTFAILURE.108.818237. [DOI] [PubMed] [Google Scholar]
- 123.Dweck D, Reynaldo DP, Pinto JR, Potter JD. A dilated cardiomyopathy troponin C mutation lowers contractile force by reducing strong myosin-actin binding. J Biol Chem. 2010;285(23):17371–17379. doi: 10.1074/jbc.M109.064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Biesiadecki BJ, Kobayashi T, Walker JS, Solaro RJ, de Tombe PP. The troponin C G159D mutation blunts myofilament desensitization induced by troponin I Ser23/24 phosphorylation. Circ Res. 2007;100(10):1486–1493. doi: 10.1161/01.RES.0000267744.92677.7f. [DOI] [PubMed] [Google Scholar]
- 125.Preston LC, Ashley CC, Redwood CS. DCM troponin C mutant Gly159Asp blunts the response to troponin phosphorylation. Biochem Biophys Res Commun. 2007;360(1):27–32. doi: 10.1016/j.bbrc.2007.05.221. [DOI] [PubMed] [Google Scholar]
- 126.Dong WJ, Xing J, Ouyang Y, An J, Cheung HC. Structural kinetics of cardiac troponin C mutants linked to familial hypertrophic and dilated cardiomyopathy in troponin complexes. J Biol Chem. 2008;283(6):3424–3432. doi: 10.1074/jbc.M703822200. [DOI] [PubMed] [Google Scholar]
- 127.Lim CC, Yang H, Yang M, et al. A novel mutant cardiac troponin C disrupts molecular motions critical for calcium binding affinity and cardiomyocyte contractility. Biophys J. 2008;94(9):3577–3589. doi: 10.1529/biophysj.107.112896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Spaendonck-Zwarts KY, van Tintelen JP, van Veldhuisen DJ, et al. Peripartum cardiomyopathy as a part of familial dilated cardiomyopathy. Circulation. 2010;121(20):2169–2175. doi: 10.1161/CIRCULATIONAHA.109.929646. [DOI] [PubMed] [Google Scholar]
- 129.Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3(2):155–161. doi: 10.1161/CIRCGENETICS.109.912345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Memo M, Leung MC, Ward DG, et al. Familial dilated cardiomyopathy mutations uncouple troponin I phosphorylation from changes in myofibrillar ca(2)(+) sensitivity. Cardiovasc Res. 2013;99(1):65–73. doi: 10.1093/cvr/cvt071. [DOI] [PubMed] [Google Scholar]
- 131.Hamdani N, Paulus WJ, van Heerebeek L, et al. Distinct myocardial effects of beta-blocker therapy in heart failure with normal and reduced left ventricular ejection fraction. Eur Heart J. 2009;30(15):1863–1872. doi: 10.1093/eurheartj/ehp189. [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, Pinto JR, Solis RS, et al. Generation and functional characterization of knock-in mice harboring the cardiac troponin I-R21C mutation associated with hypertrophic cardiomyopathy. J Biol Chem. 2012;287(3):2156–2167. doi: 10.1074/jbc.M111.294306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gomes AV, Harada K, Potter JD. A mutation in the N-terminus of troponin I that is associated with hypertrophic cardiomyopathy affects the ca(2+)-sensitivity, phosphorylation kinetics and proteolytic susceptibility of troponin. J Mol Cell Cardiol. 2005;39(5):754–765. doi: 10.1016/j.yjmcc.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 134.Papp Z, Edes I, Fruhwald S, et al. Levosimendan: Molecular mechanisms and clinical implications: Consensus of experts on the mechanisms of action of levosimendan. Int J Cardiol. 2012;159(2):82–87. doi: 10.1016/j.ijcard.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 135.Mebazaa A, Nieminen MS, Packer M, et al. Levosimendan vs dobutamine for patients with acute decompensated heart failure: The SURVIVE randomized trial. JAMA. 2007;297(17):1883–1891. doi: 10.1001/jama.297.17.1883. [DOI] [PubMed] [Google Scholar]
- 136.Packer M, Colucci W, Fisher L, et al. Effect of levosimendan on the short-term clinical course of patients with acutely decompensated heart failure. JACC Heart Fail. 2013;1(2):103–111. doi: 10.1016/j.jchf.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 137.Landoni G, Biondi-Zoccai G, Greco M, et al. Effects of levosimendan on mortality and hospitalization. A meta-analysis of randomized controlled studies. Crit Care Med. 2012;40(2):634–646. doi: 10.1097/CCM.0b013e318232962a. [DOI] [PubMed] [Google Scholar]
- 138.Haikala H, Linden IB. Mechanisms of action of calcium-sensitizing drugs. J Cardiovasc Pharmacol. 1995;26(Suppl 1):S10–S19. [PubMed] [Google Scholar]
- 139.Pollesello P, Ovaska M, Kaivola J, et al. Binding of a new Ca2+ sensitizer, levosimendan, to recombinant human cardiac troponin C. A molecular modelling, fluorescence probe, and proton nuclear magnetic resonance study. J Biol Chem. 1994;269(46):28584–28590. [PubMed] [Google Scholar]
- 140.Kleerekoper Q, Putkey JA. Drug binding to cardiac troponin C. J Biol Chem. 1999;274(34):23932–23939. doi: 10.1074/jbc.274.34.23932. [DOI] [PubMed] [Google Scholar]
- 141.Sorsa T, Heikkinen S, Abbott MB, et al. Binding of levosimendan, a calcium sensitizer, to cardiac troponin C. J Biol Chem. 2001;276(12):9337–9343. doi: 10.1074/jbc.M007484200. [DOI] [PubMed] [Google Scholar]
- 142.Szilagyi S, Pollesello P, Levijoki J, et al. The effects of levosimendan and OR-1896 on isolated hearts, myocyte-sized preparations and phosphodiesterase enzymes of the guinea pig. Eur J Pharmacol. 2004;486(1):67–74. doi: 10.1016/j.ejphar.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 143.Endoh M. Cardiac Ca2+ signaling and Ca2+ sensitizers. Circ J. 2008;72(12):1915–1925. doi: 10.1253/circj.cj-08-0838. [DOI] [PubMed] [Google Scholar]
- 144.Amsallem E, Kasparian C, Haddour G, Boissel JP, Nony P. Phosphodiesterase III inhibitors for heart failure. Cochrane Database Syst Rev. 2005;(1):CD002230. doi: 10.1002/14651858.CD002230.pub2. (1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Robertson IM, Sun YB, Li MX, Sykes BD. A structural and functional perspective into the mechanism of Ca2+-sensitizers that target the cardiac troponin complex. J Mol Cell Cardiol. 2010;49(6):1031–1041. doi: 10.1016/j.yjmcc.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lindert S, Li MX, Sykes BD, McCammon JA. Computer-aided drug discovery approach finds calcium sensitizer of cardiac troponin. Chem Biol Drug Des. 2015;85(2):99–106. doi: 10.1111/cbdd.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Silver PJ, Pinto PB, Dachiw J. Modulation of vascular and cardiac contractile protein regulatory mechanisms by calmodulin inhibitors and related compounds. Biochem Pharmacol. 1986;35(15):2545–2551. doi: 10.1016/0006-2952(86)90052-3. [DOI] [PubMed] [Google Scholar]
- 148.Adhikari BB, Wang K. Interplay of troponin- and myosin-based pathways of calcium activation in skeletal and cardiac muscle: The use of W7 as an inhibitor of thin filament activation. Biophys J. 2004;86(1 Pt 1):359–370. doi: 10.1016/S0006-3495(04)74112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oleszczuk M, Robertson IM, Li MX, Sykes BD. Solution structure of the regulatory domain of human cardiac troponin C in complex with the switch region of cardiac troponin I and W7: The basis of W7 as an inhibitor of cardiac muscle contraction. J Mol Cell Cardiol. 2010;48(5):925–933. doi: 10.1016/j.yjmcc.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Solaro RJ, Bousquet P, Johnson JD. Stimulation of cardiac myofilament force, ATPase activity and troponin C ca++ binding by bepridil. J Pharmacol Exp Ther. 1986;238(2):502–507. [PubMed] [Google Scholar]
- 151.Li Y, Love ML, Putkey JA, Cohen C. Bepridil opens the regulatory N-terminal lobe of cardiac troponin C. Proc Natl Acad Sci U S A. 2000;97(10):5140–5145. doi: 10.1073/pnas.090098997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang X, Li MX, Sykes BD. Structure of the regulatory N-domain of human cardiac troponin C in complex with human cardiac troponin I147–163 and bepridil. J Biol Chem. 2002;277(34):31124–31133. doi: 10.1074/jbc.M203896200. [DOI] [PubMed] [Google Scholar]
- 153.Baell J, Walters MA. Chemistry: Chemical con artists foil drug discovery. Nature. 2014;513(7519):481–483. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 154.Ferroni C, Hano O, Ventura C, et al. A novel positive inotropic substance enhances contractility without increasing the Ca2+ transient in rat myocardium. J Mol Cell Cardiol. 1991;23(3):325–331. doi: 10.1016/0022-2828(91)90068-w. [DOI] [PubMed] [Google Scholar]
- 155.Solaro RJ, Gambassi G, Warshaw DM, et al. Stereoselective actions of thiadiazinones on canine cardiac myocytes and myofilaments. Circ Res. 1993;73(6):981–990. doi: 10.1161/01.res.73.6.981. [DOI] [PubMed] [Google Scholar]
- 156.Brixius K, Mehlhorn U, Bloch W, Schwinger RH. Different effect of the ca(2+) sensitizers EMD 57033 and CGP 48506 on cross-bridge cycling in human myocardium. J Pharmacol Exp Ther. 2000;295(3):1284–1290. [PubMed] [Google Scholar]
- 157.Radke MB, Taft MH, Stapel B, Hilfiker-Kleiner D, Preller M, Manstein DJ. Small molecule-mediated refolding and activation of myosin motor function. Elife. 2014;3:e01603. doi: 10.7554/eLife.01603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pan BS, Johnson RG., Jr Interaction of cardiotonic thiadiazinone derivatives with cardiac troponin C. J Biol Chem. 1996;271(2):817–823. [PubMed] [Google Scholar]
- 159.Li MX, Spyracopoulos L, Beier N, Putkey JA, Sykes BD. Interaction of cardiac troponin C with ca(2+) sensitizer EMD 57033 and cardiac troponin I inhibitory peptide. Biochemistry. 2000;39(30):8782–8790. doi: 10.1021/bi000473i. [DOI] [PubMed] [Google Scholar]
- 160.Robertson IM, Li MX, Sykes BD. Solution structure of human cardiac troponin C in complex with the green tea polyphenol, (−)-epigallocatechin 3-gallate. J Biol Chem. 2009;284(34):23012–23023. doi: 10.1074/jbc.M109.021352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Pineda-Sanabria SE, Robertson IM, Sykes BD. Structure of trans-resveratrol in complex with the cardiac regulatory protein troponin C. Biochemistry. 2011;50(8):1309–1320. doi: 10.1021/bi101985j. [DOI] [PMC free article] [PubMed] [Google Scholar]