Introduction
The pleiotropic cytokine, interleukin-6 (IL-6), is produced by many cells and tissues, and plays a major role in normal physiology and inflammatory responses. It has been widely employed in research on population health and aging because circulating levels of IL-6 tend to rise in old age, with obesity, and following stressful life events (Fried et al., 1998; Friedman et al., 2005; Kiecolt-Glaser et al., 2003). It has also been associated with chronic stress and vulnerability to depression (Bob et al., 2010; Lutgendorf et al., 1999; Miller et al., 2002). Although blood levels of IL-6 are often quantified in isolation, its biological actions are determined by two distinct membrane bound glycoproteins expressed on the surface of target cells: 1) a classical transmembrane IL-6 receptor (mIL-6r), and 2) a signal-transducing non-ligand binding subunit, gp130, which is activated by a complex formed by IL-6 and the soluble form of the IL-6 receptor (sIL-6r) (Kallen, 2002; Peters et al., 1998). Further, the sIL-6r has also been shown to moderate central actions of IL-6 within the brain (Schöbitz et al., 1995). Therefore, to more completely understand variation in IL-6 synthesis and responses across individuals, it is important to also quantify the soluble receptor, which was done in the following study.
IL-6 and sIL-6r are coded by different genes and controlled by distinct mechanisms of expression (Crichton et al., 1996; Jones et al., 2001; Lust et al., 1992). In addition, sIL-6r may be produced either by alternative mRNA splicing or by proteolytic cleavage and shedding from the surface of cells (Jones et al., 2001; Müllberg et al., 1993). Thus, it is of significance to determine the extent and similarity of the genetic constraints on IL-6 and its soluble receptor. It is known that the magnitude of the IL-6 response in inflammatory conditions can be affected by different single nucleotide polymorphisms (SNPs) associated with the IL-6 gene (Bruunsgaard et al., 2004; Sen et al., 2011; Walston et al., 2007). Accordingly, an examination of IL-6 responses to a strong inflammatory stimulus yielded high heritability estimates (de Craen et al., 2005). However, this general conclusion is often, and inappropriately, overgeneralized to all aspects of IL-6 synthesis and release. In the absence of inflammatory stimuli, SNPs have a weak or no association with baseline levels of IL-6 in the blood stream (Bagli et al., 2003; Bennermo et al., 2004; Brull et al., 2001; Burzotta et al., 2001; Herbert et al., 2006; Lieb et al., 2004; Nauck et al., 2002; Sen et al., 2011; Shah et al., 2013; van Oijen et al., 2006; Wernstedt et al., 2004). On the other hand, SNPs affecting sIL-6r production appear to be more consistently associated with its levels in systemic circulation (Galicia et al., 2004; Rafiq et al., 2007; Sasayama et al., 2012; van Dongen et al., 2014).
Serum IL-6 and sIL-6r are both influenced by body adiposity, although IL-6 appears to be affected to a greater extent than the soluble receptor (Crichton et al., 1996; Mehra and Storfer-Isser, 2006; Mohamed-Ali et al., 1999; Pini et al., 2012). Accordingly, sIL-6r can serve as an independent biomarker for certain pathological conditions. It does not always react to the same physiological stimuli as IL-6; it is not always responsive to changes in IL-6 levels, and it does not play a role in all IL-6 pathways (Hurst et al., 2001; Kallen, 2002; Lamas et al., 2013; Mehra et al., 2006; Montero-Julian, 2001; Peters et al., 1998). Adipocytes, as well as the macrophages embedded in fat tissue, are known to be a major source of the IL-6 found in blood, especially in the non-inflammatory, healthy state (Coppack, 2007; Fried et al., 1998; Khaodhiar et al., 2004; Suganami and Ogawa, 2010; Weisberg et al., 2003; Wisse, 2004). The adipokine leptin also stimulates the release of IL-6 from leukocytes and macrophages (Agrawal et al., 2011; Behrendt et al., 2010; Kredel et al., 2013). Although these biological pathways often interact in a bidirectional and reciprocal manner, there is considerable evidence to suggest that adiposity exerts a far greater influence on inflammatory physiology (Miller, 2003; Welsh et al., 2010). Given the strong association between obesity and IL-6, it is likely that heritable factors influencing weight gain would also have a parallel effect on IL-6, a hypothesized linkage specifically tested in our analyses.
Likewise, the acute phase reactant, C-reactive protein (CRP) has been consistently associated with adiposity and inflammatory conditions, including cardiovascular disease (Carroll et al., 2009; Gupta et al., 2012; Saijo et al., 2004). Obesity can increase production of CRP by the liver as well as in adipose tissue (Anty et al., 2006; Calabro et al., 2005). Although previous genetic and heritability studies have demonstrated direct and independent effects on CRP levels(de Maat et al., 2004; Dehghan et al., 2011; Pankow et al., 2001; Wörns et al., 2006), there is evidence suggesting a causal relationship between the genetics of obesity and circulating CRP levels. SNPs associated with body mass index (BMI) can influence blood levels of CRP, while the converse has not been demonstrated (Holmes et al., 2014; Welsh et al., 2010). Thus, to further probe the unique nature and strength of the association between adiposity and IL-6, we also considered the relationship between adiposity and CRP. It is known that both CRP and IL-6 are involved in inflammatory response pathways, but CRP production appears to be more readily stimulated by IL-6, even within adipose tissue (Anty et al., 2006; Calabro et al., 2005; Heinrich et al., 1990; Volanakis, 2001). In addition, circulating levels of CRP and IL-6 are regulated independently by different genes (Shah et al., 2013). By analyzing these associations in identical and fraternal twins, we were able to directly compare and contrast the relative influence of adiposity on both CRP and IL-6, and to consider the reciprocal relationship between CRP and IL-6.
Previous twin studies have determined that adiposity, measured as BMI, is highly heritable (Hjelmborg et al., 2008; Schousboe et al., 2003; Segal et al., 2008). Associations between a genetic score, consisting of 14 SNPs related to BMI, and multiple cardiovascular and inflammatory traits including both IL-6 and CRP, were recently assessed (Holmes et al., 2014). However, it is still not known whether the heritable influence of obesity on IL-6 and its soluble receptor are coordinated, and whether CRP is affected in a similarly linked manner. We probed these relationships by comparing IL-6, sIL-6r and CRP levels in identical and fraternal adult twins, who also varied in adiposity and anthropometric concordance.
Twin studies take advantage of the different degree of genetic relatedness between monozygotic (MZ) and dizygotic (DZ) twins to estimate the relative contribution of genetic and environmental effects contributing to the phenotypic variance of a trait, as well as to the covariance between traits. Typical twin studies rely on the assumption that MZ twins share 100% and DZ twins share 50% of their genes, while both types of twins, regardless of zygosity, share a common rearing environment (Hall, 2003). In addition to the shared environment, it is also possible to discern effects of the non-shared environment, reflecting individual experiences not shared in common. Greater phenotypic similarity for MZ twins than found in DZ twins would be indicative of higher heritable contributions. On the other hand, when MZ and DZ twins present a similar phenotype, more variance attributable to life style and common environmental processes is assumed. Likewise, when monozygotic twins are discordant, it is usually attributed to unshared environmental influences (Boomsma et al., 2002; Christensen et al., 2001; Duffy et al., 1998; Hjelmborg et al., 2008).
We utilized bivariate ACE heritability models based on structural equation modeling (SEM) to estimate the proportion of IL-6, sIL-6r and CRP variation accounted for by genetic, shared and unshared environmental factors, as well as the heritable influences shared with adiposity. Our a priori hypothesis was that obesity would have a larger effect on IL-6 than on sIL-6r. Secondarily, the a posteriori hypothesis was tested: obesity would exert a,heritable influence on CRP, but one that was only moderately associated with the genetics of IL-6. We also compared IL-6 and sIL-6r intra-class correlations (ICCs) between co-twins in order to confirm the greater similarity between MZ co-twins than between DZ co-twins, indicative of heritable influences. To further test this hypothesis, we compared the ICC for MZ co-twins with that of genetically unrelated control participants matched to each MZ twin case. Assuming that environmental and lifestyle factors, BMI in particular, play a greater role in accounting for IL-6, we predicted that ICCs between the actual MZ co-twins would be no higher than for control adults who were closely matched on age, gender, BMI and a socio-economic index (SEI). By matching control subjects to the twin cases on these attributes, we simulated 4 of the major environmental and host factors known to influence IL-6 (Ershler and Keller, 2000; Friedman et al., 2005; Hjelmborg et al., 2008; Johnson and Krueger, 2005; O’Connor et al., 2007; Wisse, 2004). Hence, we could infer whether BMI and demographics (i.e., age, gender and SEI) accounted for the concordance between matched controls and co-twins. Given the likelihood of strong genetic constraints on sIL-6r, we anticipated small or negligible ICCs for sIL-6r with the unrelated, matched controls.
These analyses were possible because the recruitment strategy of a large survey of health and aging in the United States, Midlife Development in the United States (MIDUS), which included an over-sampling of twin siblings. It provided the unique opportunity to determine the heritability of circulating IL-6, sIL-6r and CRP, as well as the heritable contribution of obesity.
Methods
Participants
Participants were drawn from the MIDUS II Biomarker project, 2004–2009, a continuation of an earlier MIDUS 1 survey supported by the MacArthur Foundation in 1995–96. In addition to a representative probability sample, MIDUS 1 recruited a national sample of twin pairs, from which the current cohort was selected. Between 2003–2005, blood specimens were obtained, enabling the determination of cytokines and other biomarkers for each twin pair (Love et al., 2010). The twin sample was comprised of 73 monozygotic and 32 dizygotic same-sex twin pairs, as well as 37 matched controls. In addition, we took advantage of BMI and IL-6/sIL-6r data on 830 unrelated participants. The same dataset was utilized for the CRP analyses, with information available on 72 monozygotic twins, 31 dizygotic twins, and 826 unrelated participants.
Twin Recruitment
Twin pairs were recruited by asking randomly selected correspondents from about 50,000 households across 48 states whether there were twin pairs in the immediate family. With their permission, twin pairs were referred to MIDUS II recruiters. The recruited twin pairs were related to original correspondents, reared together but living apart as adults, ranging from 25 to 74 years in age, residing in the continental U.S., English speakers, possessing a residential telephone number, and were mentally and physically capable of participating in interviews and questionnaires. They had to be healthy enough to travel to one of 3 Institutes for Clinical and Translational Research (ICTR) for the Biomarker project, where they spent the night before sample collection on the following morning.
Socio-Demographics, Zygosity, Clinical and Biological Measures
Zygosity was determined by self-report in MIDUS 1. Similarity of eye and hair colors, as well as the degree to which their identity was confused by others during childhood, were among the criteria for twin determination. This approach is more than 95% accurate when compared to blood tests (Nichols and Bilbro, 1966). Each sibling’s Socioeconomic Index (SEI) was derived using income, educational attainment and occupation categories from the 1990 Census classification (Hauser and Warren, 1997) and incorporated into MIDUS II, 2004–2006.
Clinical and biological measures were assessed for a total of 1255 participants in the Biomarker project who consented to the overnight stay, either in Madison, WI, Los Angeles, CA, or Washington DC. Participants arrived on Day 1 at one of the three sites where they were admitted to the hospital research unit. They completed a medical history and physical exam, as well as a self-administered questionnaire. Smoking can affect IL-6 and could interact with genetic factors that influence transcription and release of IL-6 (Bruunsgaard et al., 2004; Semlali et al., 2012; Zhou et al., 2014). Therefore, we also examined the concordance of the smoking history between MZ and DZ co-twins. Smoking history was assessed on the clinical questionnaire by asking: “Have you ever smoked cigarettes regularly?“. Fasted blood samples were obtained between 0500 and 0700, and sera frozen until analyzed. All sample collections and analyses were approved by the Health Sciences Institutional Review Board at the University of Wisconsin-Madison, as well as by the IRBs at UCLA and Georgetown University. All participants provided informed consent. Nursing staff followed standardized procedures detailed in a general “Manual of Procedures”, as well as specific “Guidelines for Collecting and Processing Biomarkers” in order to maintain consistency.
Cytokine Assessment
Serum IL-6 levels were determined for all 1255 biomarker project participants by high-sensitivity enzyme-linked immunosorbent assay (ELISA) (Quantikine, R&D Systems, Minneapolis, MN), with a lower sensitivity of detection at 0.16 pg/mL. All values were quantified in duplicate; any value over 10 pg/mL was re-run with sera diluted to fall on the standard curve. The laboratory intra-assay coefficient of variance (CV) was 4.1% and the inter-assay CV was 12.9% (generated by inclusion of a low and high IL-6 serum pool in each assay). Sandwich ELISA kits were also employed to quantify sIL-6r levels (Quantikine, R&D Systems). Sera were diluted 1:100 so values would fall on the standard reference curve from 7 to 2000 pg/mL. Thus, the effective assay range for sIL-6r was 0.7–200 ng/mL. The intra-assay and inter-assay CVs were 2.0% and 6.9%, respectively. Serum CRP levels were determined for all subjects via particle-enhanced immunonephelometric assay, and high values used as exclusion criteria.
Statistical Analysis
Nine twin pairs were excluded when a sibling had CRP levels indicative of sickness (above 10 mg/L), or they were discrepant on smoking status or chronic illness. In addition, because there were insufficient numbers of African-Americans (only 2 twin pairs), and there are known race differences in cytokine and CRP levels (Carroll et al., 2009; Coe et al., 2011; Crawford et al., 2006), both pairs were excluded. Opposite-sex, dizygotic twin pairs were also excluded from the analysis because there were too few pairs to include in the model. In order to achieve normal distributions, IL-6, sIL-6r, CRP and BMI were natural log-transformed before statistical calculations.
Linear regressions were employed to examine the effect of BMI on IL-6, sIL-6r, and CRP for singleton birth participants in the main Biomarker sample, after excluding the twin participants. Including the twins in these regression analyses would have violated assumption of independent observations. Similarly, these regression analyses were run on only the Caucasian participants of European descent, because of the known differences in African-Americans (Coe et al., 2011).
In order to estimate the additive genetic and environmental effects contributing to the IL-6, sIL-6r and CRP variance, including the covariance with factors affecting BMI, we fitted bivariate ACE models to these data by using Cholesky’s decomposition approaches in SEM (Karmakar et al., 2012). SEM allows for testing whether covariance matrices of hypothetical models fit the covariance matrix of the actual data. They further allow for distinguishing the submodels that fit the data most parsimoniously. Classical ACE models decompose the phenotypic variance into three categories, A (Additive Genetic Effects), C (Common Environmental Effects) and E (Unshared Environmental Effects plus residual/error variance). Bivariate ACE models estimate A, C and E effects for two phenotypic traits and, in addition, estimate the covariance between each effect across phenotypes. We defined a saturated ACE covariance model as well as submodels for AE covariance (negligible sharing of common environmental effects) and CE covariance (negligible sharing of additive genetic effects) which were tested against the data covariance matrices, providing the bases upon which one may identify the models that best fits the data. The Cholesky’s decomposition of phenotypic effects allowed us to parse out the additive genetic effects unique to IL-6, sIL-6r, or CRP from those shared with BMI by comparing the various covariance submodels, while allowing univariate variance components to vary freely. This same approach was used in order to test the genetic covariance shared between CRP and IL-6.
These biometric models were run in MX using a script adapted from file rawVC4a.mx, provided with the MX software (Neale et al., 2003; Posthuma and Boomsma, 2005). Because MZ and DZ twins share 100% and 50% of their independently segregating genes, respectively, SEM covariance coefficients for additive genetic effects were set to 1 for MZ twin pairs and to ½ for DZ pairs. Covariance coefficients for common environment effects were set to 1 and those attributed to unshared environment effects were set to 0. Age and Gender were included in Cholesky’s decomposition models as covariates in order to prevent indirect inflation of ICCs that could potentially confound heritability estimates (McGue and Bouchard, 1984). Saturated and nested covariance submodels (ACE, AE and CE) were fit to the data and submodels were tested against the saturated ACE models. Non-significant differences in Chi-square probabilities allowed us to discern nested submodels that provided fit of the data. The models that best described our data were identified on the basis of maximum likelihood estimates, -2LL, (−2*log-likelihood = −2*log(C − χ2 /2)), and relative fit indices, Akaike’s Information Criterion, AIC, (AIC = χ2 − 2*d.f.), and Bayesian Information Criterion, BIC (BIC= χ2 − d.f. * ln(n)). Lower AIC and BIC values indicate models that fit the observed data most parsimoniously.
Intra-class correlations for IL-6, sIL-6r, and smoking history were calculated for the MZ and DZ co-twins (Hawkins, 1989; Hotelling, 1953; Sedgwick, 2013). One-tailed Fisher’s r-to-z tests assessed whether IL-6 and sIL-6r ICCs for MZ twins were significantly greater than for DZ twins. To further evaluate the influence of obesity on IL-6 and sIL-6r, one-tailed Fisher’s r-to-z tests were used to compare the ICCs between MZ co-twins to the ICCs between 37 MZ twins and unrelated controls matched for age, gender, BMI and SEI. Smoking history was assessed using a two-tailed Fisher’s r-to-z test to examine the presence of possibly confounding differences between MZ and DZ ICCs. Except for the biometrical models, analyses were determined with SPSS 19.
Results
Twin Sample
The MIDUS Biomarker project has been shown previously to be comparable to the larger MIDUS participants for most socio-demographic, health status, and health behavior indicators, but not race (Love et al., 2010). The latter was not an issue for the current analyses because the 2 African-American twin pairs were excluded. Table 1 presents the socio-demographic and clinical information, and mean cytokine values for MZ and DZ twins and the unrelated controls who were matched with the cases, as well as for the remaining Biomarker participants from which the controls were selected.
Table 1.
Sample descriptives for primary variables used in the twin analyses
| MZ | DZ | Control | Larger Sample | |||||
|---|---|---|---|---|---|---|---|---|
| Age | 52.8 | (11.2) | 50.2 | (9.1) | 52.0 | (8.5) | 56.0 | (11.9) |
| BMI (Kg/m2) | 27.6 | (5.4) | 29.6 | (5.7) | 25.6 | (3.4) | 29.4 | (6.1) |
| IL-6 (pg/mL) | 2.1 | (1.7) | 2.7 | (2.1) | 2.5 | (1.7) | 2.9 | (3.0) |
| sIL-6r (pg/mL) | 37401 | (10582) | 33611 | (7826) | 34600 | (9407) | 36412 | (10351) |
| CRP | 2.10 | (2.22) | 2.54 | (2.26) | 2.12 | (1.87) | 3.02 | (4.78) |
| SEI | 44.6 | (13.9) | 44.8 | (15.8) | 44.2 | (10.7) | 43.0 | (14.1) |
| Ever Smoked | 36.0% | 39.0% | 43.3% | 46.5% | ||||
| Males | 62 | 16 | 20 | 394 | ||||
| Females | 84 | 48 | 17 | 436 | ||||
| Total Participants | 146 | 64 | 37 | 830 | ||||
Mean values and standard deviations, or counts for numbers of participants.
MZ, Monozygotic Twins; DZ, Dizygotic twins; the Larger Sample was comprised of all other subjects in Biomarker project, after excluding the twins and African-American participants.
Effect of BMI on IL-6, sIL-6r and CRP
After adjusting the regression models for age and gender by including them as covariates, BMI accounted for 7.8% of the variance in IL-6 levels (β = 0.28, F[1,829] = 42.4, p < 0.001), and 18.0% of the variance in CRP levels (β = 0.43, F[1,826] = 70.0, p < 0.001), but predicted only 0.3% of the variance in sIL-6r (β = 0.07, F[1,829] = 3.2, p = 0.02). In keeping with our predictions, BMI exerted a stronger effect on IL-6 than on the soluble IL-6 receptor among unrelated MIDUS participants, as well as had a substantial effect on CRP.
Heritability Estimates
Non-significant chi-square difference tests indicated that CE and AE covariance submodels for BMI X IL-6, BMI X sIL-6r, BMI X CRP and CRP X IL-6 presented an equivalent fit for the data as did the saturated ACE covariance models. In addition, lower relative fit indices (AIC and BIC) identified the AE submodels as most parsimoniously fitting all three sets of BMI covariance data, and the CE submodel as a better fit for the CRP X IL-6 data (Table 3). In the three bivariate BMI models, additive genetic effects and unshared environmental effects comprised almost all of the influences acting independently on BMI, thus providing two available pathways for covariance with IL-6, sIL-6r, and CRP (Table 4). Covariance was notably high in accounting for the heritable associations between BMI and IL-6 (83.0%). The proportions presented in Table 4 were calculated based on the SEM path coefficient estimates.
Table 3.
Comparison of nested model statistics
| Covariance Models | −2LL | D.F. | AIC | BIC | Difference χ2 | p-value | |
|---|---|---|---|---|---|---|---|
| BMI × IL-6 | ACE | 39.22 | 405 | −770.77 | −922.81 | ||
| CE | 41.52 | 406 | −770.48 | −924.00 | 2.29 | 0.13 | |
| AE | 40.69 | 406 | −771.31 | −924.41 | 1.46 | 0.23 | |
| BMI × sIL-6r | ACE | −216.23 | 405 | −1026.23 | −1050.54 | ||
| CE | −215.63 | 406 | −1027.63 | −1052.57 | 0.60 | 0.44 | |
| AE | −215.99 | 406 | −1027.99 | −1052.75 | 0.25 | 0.62 | |
| BMI × CRP | ACE | 1396.80 | 401 | 594.80 | −232.80 | ||
| CE | 1398.10 | 402 | 594.10 | −234.47 | 1.31 | 0.25 | |
| AE | 1396.81 | 402 | 592.81 | −235.12 | 0.01 | 0.91 | |
| CRP × IL-6 | ACE | 1415.15 | 401 | 613.15 | −223.63 | ||
| CE | 1415.46 | 402 | 611.46 | −225.79 | 0.31 | 0.57 | |
| AE | 1415.48 | 402 | 611.47 | −225.78 | 0.33 | 0.57 | |
−2LL (−2 * Log-Likelihood), maximum likelihood estimate; AIC, Akaike’s Information Criterion; BIC, Bayesian Information Criterion; Lower -2LL, AIC and BIC indicate models that more parsimoniously fit the data; Non-significance in chi-square (χ2) values between saturated and nested models indicate equivalent fit of data; Bold highlighted font indicates the model that most parsimoniously fit the data, based on AIC and BIC.
Table 4.
Proportion of genetic and environmental effects for best-fit models
| Variance / Covariance Components |
Estimated Effects (95% C.I.) | |||||
|---|---|---|---|---|---|---|
| Additive genetic (A) | Common Environment (C) | Unshared Environment (E) |
||||
| BMI | 68.8% | (41.4–78.5) | 0.0% | 31.2% | (21.5–44.8) | |
| IL-6 | 26.1% | (12.5–51.8) | 7.6% | (0–7.6) | 66.3% | (48.2–84.3) |
| Covariance | 83.0% | 17.0% | ||||
| BMI | 68.2% | (21.1–78.9) | 1.3% | (1.1–44.7) | 30.5% | (21.1–44.0) |
| sIL-6r | 42.2% | (7.7–82.9) | 35.6% | (0–67.9) | 22.2% | (15.3–32.2) |
| Covariance | 33.7% | 66.3% | ||||
| BMI | 70.6% | (27.2–79.8) | 0.0% | 29.4% | (20.2–42.7) | |
| CRP | 18.2% | (1.2–50.0) | 13.7% | (0.0–36.6) | 68.1% | (50.0–88.2) |
| Covariance | 54.5% | 45.5% | ||||
| CRP | 15.2% | (0.0–31.7) | 14.7% | (2.8–44.2) | 70.1% | (52.8–90.0) |
| IL-6 | 0.0% | (0.0–35.0) | 37.8% | (6.4–53.0) | 62.2% | (45.4–79.9) |
| Covariance | 51.3% | 48.7% | ||||
Best-fit models were chosen on the basis of the AIC and BIC.
These effects are also portrayed in Figure 1 for IL-6 and in Figure 2 for its soluble receptor, which show the discrete effects parsed by specific sources of variation. These estimations indicated further that the suggestion of a genetic constraint on IL-6 levels was not attributable specifically to the genetics of IL-6, but rather was driven more indirectly by factors shared with the predisposition for BMI (Figure 1). Even when inflated by considering the high covariance with BMI (Table 4), the total genetic effect estimated for IL-6 was still small (averaging 26.2%). In contrast, the heritable influences on the soluble receptor for IL-6 were significantly stronger (averaging 42.2%). Although sIL-6r also showed some covariance with genetic factors associated with BMI (33.7%, Table 4), path coefficients indicated the effects attributable to BMI were minimal (Figure 2). In addition, a much larger portion of the total phenotypic variance was unique to sIL-6r (Figure 2). CRP also exhibited a high genetic covariance with BMI (54.5%, Table 4); however, not to the same extent as the IL-6 covariance (83.0%). Further, the additive genetic effects on CRP were almost evenly divided between those shared with BMI and factors unique to CRP (Figure 3). Moreover, our models did not reveal shared genetic effects between CRP and IL-6 (Figure 4). The covariance between CRP and IL-6 phenotype was split between shared and unshared environmental effects (Table 4).
Fig. 1.
Path coefficient estimates for bivariate BMI X IL-6 model, including the corresponding percent contributions of additive genetic effects, and common and unshared environmental effects affecting IL-6. The model also parses effects specific to IL-6 from those shared with BMI. The most parsimonious model indicated that BMI and IL-6 share both genetic and unshared environmental effects. However, model estimates further indicated that the additive genetic effect acting on IL-6 is mostly, shared with BMI.
Fig. 2.
The path coefficient estimates for the bivariate BMI X sIL-6r model also parsed the genetic effects specific to sIL-6r from those shared with BMI. This model similarly points to a shared covariance between genetic and unshared environmental effects. However, most of the additive genetic effects explaining the sIL-6r phenotype were independent of BMI, thus very distinct from the pattern of genetic constraints found for IL-6.
Fig. 3.
The most parsimonious path coefficient estimates for the bivariate BMI X CRP model also indicated shared covariance between genetic and unshared environmental effects. Unlike the bivariate model for IL-6, however, the phenotypic variance attributed to the additive genetic effects was split between those shared with BMI and some independent of BMI.
Fig. 4.
The path coefficient estimates for the bivariate CRP X IL-6 model that most parsimoniously fit the data indicated a lack of genetic covariance between CRP and IL-6, attributing the phenotypic covariance to common and unshared environmental effects.
Intra-Class Correlations
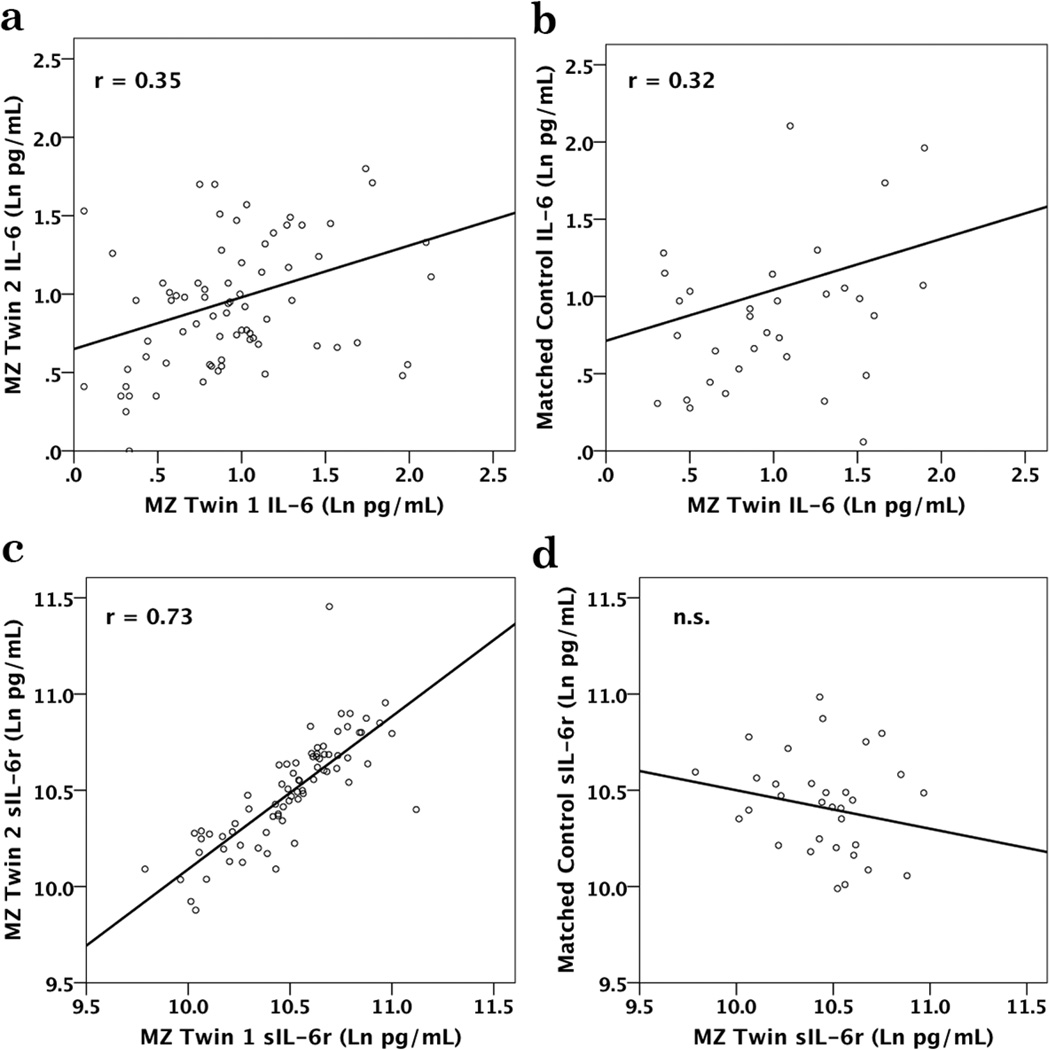
Stronger genetic constraints on sIL-6r were also clearly evident when comparing the trait ICCs between the twins and unrelated adult controls who had been matched on BMI, gender, age, and SEI (Table 2). This case/control analysis confirmed the importance of heritable pathways related to obesity, and indicated that the 4 matching criteria resulted in IL-6 values similar to the twins, whereas there was no evidence that these 4 attributes increased the likelihood of a correlation in sIL-6r between cases and controls. In addition, as shown in Table 2, the ICC for IL-6 between the MZ twins was not greater than for the DZ twins, demonstrating that the degree of relatedness did not influence IL-6 more than the concordance for obesity. In contrast, the sIL-6r levels were more similar for MZ twins, while DZ twins were divergent, supporting the greater heritability of sIL-6r. When a MZ participant was matched to an unrelated control by age, gender, BMI and SEI, the resulting ICC for IL-6 was nearly the same as for the MZ co-twin (Figures 5a & 5b). On the other hand, the sIL-6r of a twin was not significantly correlated with the value seen in the unrelated, matched control (Figures 5c & 5d). MZ and DZ twins were highly concordant for smoking history (r = 0.57, p < 0.001 and r = 0.41, p = 0.02, respectively). However, a history of smoking or abstinence did not significantly affect either the IL-6 or sIL-6r levels in this cohort.
Table 2.
Intra-class correlations and statistical difference between coefficients demonstrating MZ and DZ twins had similar values for IL-6, but differed for sIL-6r
| Intra-Class Correlation Coefficients and 95% C.I. | Fisher's r-to-z, z-values | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MZ | DZ | MC | MZ vs. DZ | MZ vs. MC | ||||||
| IL-6 | 0.35 (.17 – .50) | P<0.001 | 0.32 (−.02 –.60) | P=0.03 | 0.36 (.02 –.63) | P=0.02 | 0.15 | n.s. | 0.05 | n.s. |
| sIL-6r | 0.73 (.62 –.80) | p<0.001 | 0.48 (.17 –.70) | P=0.002 | −0.20 (−.51 –.16) | n.s. | 1.84 | p=.03 | 5.41 | p<0.001 |
MZ, Monozygotic Twins; DZ, Dizygotic Twins; MC, Matched Controls
Fig. 5.
Intra-class correlations are shown for IL-6 between MZ co-twins and between MZ twins and unrelated, matched controls, as well as for the sIL-6r associations. The correlation for IL-6 for MZ co-twins (A) was nearly identical to the correlation between MZ twins and unrelated controls matched on age, gender, BMI and SEI (B). Levels of sIL-6r in MZ co-twins were highly correlated (C), while sIL-6r values were not significantly correlated between MZ cases and their unrelated, matched controls (D).
Discussion
By taking advantage of the fact that monozygotic twins are genetically more similar than dizygotic twins, our analyses demonstrated that the genetic constraints on IL-6 and its soluble receptor are clearly distinct. In addition, the association between BMI and IL-6 was more evident than for sIL-6r. This differential relationship with adiposity affected the heritability estimates. By fitting bivariate ACE models, we calculated variance and covariance components, parsing out genetic and environmental effects specific to IL-6, sIL-6r, and CRP from those linked to the heritability of BMI. The approach more clearly revealed the degree of covariance in the additive genetic effects shared by IL-6 and BMI. Optimized path coefficients did not expose unique effects specific to the genetic control of IL-6, but rather an overlap of the heritable processes influencing both IL-6 and BMI in adults. These analyses also extend our understanding of the differential influence exerted by BMI on CRP. While the regression analyses did show that BMI affected serum CRP levels to a greater extent than IL-6, the heritability models indicated that the effects of BMI on IL-6 were largely due to shared genetics. In contrast, the effect of BMI on CRP was split between shared genetics and environmental influences. The genetic effects acting on CRP were evenly distributed between those shared with BMI and those unique to CRP genetics, whereas the additive genetic effects influencing IL-6 phenotype appeared to be more exclusively tied to BMI. In spite of the close relationship between CRP and IL-6, our heritability estimates point to environmental factors as the main source of covariance. Hence, in the unstimulated state without infection or chronic disease, the genetics underlying body adiposity appears to influence IL-6 and CRP levels in the blood through independent pathways.
In order to further validate our conclusions on the differential constraints regulating IL-6 and sIL-6r, we conducted a case/control analysis, with each twin matched to an unrelated individual on the basis of gender, age, BMI, and education. Given the strong influence these 4 variables have on IL-6, the ICCs attained for the matched controls were of the same magnitude as for the actual co-twin siblings. In contrast, the sIL-6r values were markedly discordant, confirming that the genetic constraint on the soluble receptor was less influenced by life style and other environmental factors.
Our heritability estimates for IL-6 in blood do contrast with some of the commonly held assumptions derived from other approaches. For example, it has been reported previously that both CRP and IL-6 levels are similar in twins, a finding that will emerge when the influence of adiposity is not taken into account, nor statistically considered as a contributing factor (Rooks et al., 2012; Wörns et al., 2006). In addition, there is a substantial literature reporting that allele polymorphisms affect IL-6 release, but that effect is most apparent in the context of inflammatory disorders, or when cells are activated in vitro by a proinflammatory stimulant (Bennermo et al., 2004; Brull et al., 2001; Burzotta et al., 2001; Shah et al., 2013). IL-6 gene-related polymorphisms include the SNPs rs1800795 and rs1800796 (Chatzikyriakidou et al., 2013; Chen et al., 2012; Vaughn et al., 2013). Although these polymorphisms do affect inflammatory responses in patients (Bruunsgaard et al., 2004; Sen et al., 2011; Walston et al., 2007), they do not appear to have a strong influence on basal IL-6 in the blood of a healthy individual. In addition, these SNPs do not have a strong effect on IL-6 transcription at baseline or even a large influence when the cells are activated by proinflammatory stimulants (Smith et al., 2012). That may help to explain why studies of the association between the -176C/G SNP and cardiovascular disease have been inconsistent (Brull et al., 2001; Burzotta et al., 2001; Lieb et al., 2004; Nauck et al., 2002). Similar concerns have also been raised about the predictive power of IL-6 related SNPs in meta-analyses of the literature on inflammatory disease (Dai et al., 2012; Di Bona et al., 2009; Lee et al., 2012; Nikolopoulos et al., 2008; Yang et al., 2012).
In contrast, several polymorphisms that affect the production and shedding of the soluble IL-6 receptor, including rs2228145, rs2228146, rs2229238, rs4072391, rs4537545 and rs8192284, reliably account for variation in basal levels (Ferreira et al., 2013; Lamas et al., 2013; Marinou et al., 2010; Rafiq et al., 2007; Reich et al., 2007; Rodríguez-Rodríguez et al., 2011; Sasayama et al., 2012; Wang et al., 2005). These SNPs have already been linked to poor prognosis in clinical conditions, including chronic inflammation, metabolic and psychological disorders (Ferreira et al., 2013; Hamid et al., 2004; Huth et al., 2006; Lamas et al., 2013; Marinou et al., 2010; Sasayama et al., 2012; Stephens et al., 2012; Zhang et al., 2013). Genetic admixture mapping also indicates robust correlations of the sIL-6r variation in blood to ancestry (Reich et al., 2007). Although both IL-6 and its soluble receptor play important roles in inflammatory processes, the production of sIL-6r seems to be more tightly regulated, while IL-6 is more sensitive to life style, diet and other factors associated with adiposity.
IL-6 is secreted by many cells, including fibroblasts, hepatocytes, endothelial cells, and, especially, adipocytes, in addition to leukocytes (Hamzic et al., 2013; Lepiller et al., 2013; Saiki et al., 2013; Salman et al., 2013). The diversity of these tissue sources is key to understanding the heritability of IL-6, especially given the strong genetic and familial influences on adiposity (Schousboe et al., 2003; Segal et al., 2008). Although some previous twin studies considered the influence of adiposity on IL-6 heritability by adjusting IL-6 values by BMI or waist-to-hip ratios, they did not account for the shared genetic covariance (de Maat et al., 2004; Sas et al., 2012; Su et al., 2008). Therefore, those genetic estimates were likely inflated by the shared influence with BMI.
Twin analyses have a number of assumptions, including that there is a sharing of 100% of the genetic load by MZ twins versus 50% for DZ twins. It is also assumed that MZ and DZ siblings both share a similar influence of the common environments, and gene-environment interactions are not modeled in the standard twin model (i.e., ACE estimates are estimated as constant for the sample because potential moderator variables are not modeled). Some investigations have questioned the latter assumption based on the view that epigenetic modifications and individual perceptions may lead to a differential experience of even minor environmental events (Stenberg, 2012). Assuming a perfectly homogeneous and common environmental influence is probably not reasonable, so it will be important to verify our conclusions with a larger-scale gene-association study (Purcell, 2002; Tan et al., 2010). In addition, a substantial portion of the variance in IL-6 and CRP still remains to be accounted. Some variance may be attributed to reliability issues when relying on a single blood sample (Navarro et al., 2012). Determining the stability of IL-6 levels over time through multiple samples, would be of value for replicating and extending our heritability estimates. It should also be acknowledged that our participants were entirely American adults of European descent. IL-6 tends to be higher in certain races, including in those of African backgrounds, whereas it tends to be lower in some Asian populations (e.g., Japanese) (Coe et al., 2011). That was one reason why we opted not to include the small number of African-American twins in the current analyses. Similarly, body adiposity, IL-6 and CRP, as well as the relationships between adiposity and these inflammatory markers, vary not only by race, but also by gender (Carroll et al., 2009; Clifton, 2003; Coe et al., 2011). Hence, it is likely that estimates of shared genetic effects will vary by race and gender. Further validation for the driving effects of obesity on IL-6 and CRP would be achieved by controlled interventional studies targeting improvements in diet, exercise and weight reduction.
IL-6 affects not only immunity, but also cell growth, metabolism, and a number of critical brain functions. Further, cytokines are implicated in pain sensitization and sickness, including fever (Burton et al., 2011; May et al., 2009; Patel et al., 2012; Sasayama et al., 2012). Variation in IL-6 levels has been associated with psychological factors, including arousal, stress, and cognitive functioning. Thus, it makes sense that the secretion of IL-6 is dynamic and labile, while its modulatory receptor appears to be more conservatively constrained. This differential relationship is not seen for all cytokines, which often rely on the up- or down-regulation of the receptor to moderate the activity of the ligand (Krupp and Lane, 1981; Marchio et al., 1989; Seo et al., 2000). But it does make IL-6 an ideal biomarker for research on aging and health disparities across populations, because it is especially responsive to adiposity, a critical factor involved in the path to Type 2 diabetes and cardiovascular disease.
Gene association studies account for only 1–2% of basal IL-6 levels in circulation
Heritability models with identical and fraternal twins were used to estimate the genetic influences on IL-6 and the sIL-6r
Genetic influences on IL-6 were strongly affected by pathways associated with obesity
In contrast, genetic constraints on the soluble receptor were greater and independent of obesity
IL-6 is more responsive to lifestyle, while sIL-6r is under greater genetic control
Acknowledgements
This research was supported by a grant from the National Institute on Aging (P01 AG020166). The original MIDUS study was supported by the MacArthur Foundation Research Network on Successful Midlife Development. Specimen collection was facilitated by the General Clinical Research Centers program (M01-RR023942 [Georgetown], M01-RR00865 [UCLA]), and at UW from the Clinical and Translational Science Award (CTSA) program of the National Center for Research Resources (1UL1RR025011). The contributions of D. Brar and J. Wochos are gratefully acknowledged. WZA is a predoctoral fellow on a NIMH supported training program (5T32 MH018931).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
Amaral, WZ has no conflict of interest to declare
Krueger, RF does not have any conflicts of interest.
Ryff, CD does not have any conflicts of interest.
Coe, CL does not have any conflicts of interest.
Contributor Information
Robert F. Krueger, Email: krueg038@umn.edu.
Carol D. Ryff, Email: cryff@wisc.edu.
Christopher L. Coe, Email: ccoe@wisc.edu.
Reference
- Agrawal S, Gollapudi S, Su H, Gupta S. Leptin activates human B cells to secrete TNF-α, IL-6, and IL-10 via JAK2/STAT3 and p38MAPK/ERK1/2 signaling pathway. J. Clin. Immunol. 2011;31:472–478. doi: 10.1007/s10875-010-9507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anty R, Bekri S, Luciani N, Saint-Paul M-C, Dahman M, Iannelli A, Amor I, Ben, Staccini-Myx A, Huet P-M, Gugenheim J, Sadoul J-L, Le Marchand-Brustel Y, Tran A, Gual P. The inflammatory C-reactive protein is increased in both liver and adipose tissue in severely obese patients independently from metabolic syndrome, Type 2 diabetes, and NASH. Am. J. Gastroenterol. 2006;101:1824–1833. doi: 10.1111/j.1572-0241.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- Bagli M, Papassotiropoulos A, Hampel H, Becker K, Jessen F, Bürger K, Ptok U, Rao ML, Möller H-J, Maier W, Heun R. Polymorphisms of the gene encoding the inflammatory cytokine interleukin-6 determine the magnitude of the increase in soluble interleukin-6 receptor levels in Alzheimer’s disease. Results of a pilot study. Eur. Arch. Psychiatry Clin. Neurosci. 2003;253:44–48. doi: 10.1007/s00406-003-0405-x. [DOI] [PubMed] [Google Scholar]
- Behrendt P, Buchenauer T, Horn R, Brabant G, Jacobs R, Bode F, Stephan M, Nave H. Diet-induced obesity, exogenous leptin-, and MADB106 tumor cell challenge affect tissue leukocyte distribution and serum levels of cytokines in F344 rats. Endocrine. 2010;38:104–112. doi: 10.1007/s12020-010-9358-9. [DOI] [PubMed] [Google Scholar]
- Bennermo M, Held C, Stemme S, Ericsson C-G, Silveira A, Green F, Tornvall P. Genetic predisposition of the interleukin-6 response to inflammation: implications for a variety of major diseases? Clin. Chem. 2004;50:2136–2140. doi: 10.1373/clinchem.2004.037531. [DOI] [PubMed] [Google Scholar]
- Bob P, Raboch J, Maes M, Susta M, Pavlat J, Jasova D, Vevera J, Uhrova J, Benakova H, Zima T. Depression, traumatic stress and interleukin-6. J. Affect. Disord. 2010;120:231–234. doi: 10.1016/j.jad.2009.03.017. [DOI] [PubMed] [Google Scholar]
- Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat. Rev. Genet. 2002;3:872–882. doi: 10.1038/nrg932. [DOI] [PubMed] [Google Scholar]
- Brull DJ, Montgomery HE, Sanders J, Dhamrait S, Luong L, Rumley A, Lowe GDO, Humphries SE. Interleukin-6 gene-174G>C and-572G>C promoter polymorphisms are strong predictors of plasma interleukin-6 levels after coronary artery bypass surgery. Arterioscler. Thromb. Vasc. Biol. 2001;21:1458–1463. doi: 10.1161/hq0901.094280. [DOI] [PubMed] [Google Scholar]
- Bruunsgaard H, Christiansen L, Pedersen AN, Schroll M, Jørgensen T, Pedersen BK. The IL-6-174G>C polymorphism is associated with cardiovascular diseases and mortality in 80-year-old humans. Exp. Gerontol. 2004;39:255–261. doi: 10.1016/j.exger.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Burton MD, Sparkman NL, Johnson RW. Inhibition of interleukin-6 trans-signaling in the brain facilitates recovery from lipopolysaccharide-induced sickness behavior. J. Neuroinflamm. 2011;8:54. doi: 10.1186/1742-2094-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzotta F, Iacoviello L, Di Castelnuovo A, Glieca F, Luciani N, Zamparelli R, Schiavello R, Donati MB, Maseri A, Possati G, Andreotti F. Relation of the - 174 G/C polymorphism of interleukin-6 to interleukin-6 plasma levels and to length of hospitalization after surgical coronary revascularization. Am. J. Cardiol. 2001;88:1125–1128. doi: 10.1016/s0002-9149(01)02046-x. [DOI] [PubMed] [Google Scholar]
- Calabro P, Chang DW, Willerson JT, Yeh ETH. Release of C-reactive protein in response to inflammatory cytokines by human adipocytes: linking obesity to vascular inflammation. J. Am. Coll. Cardiol. 2005;46:1112–1113. doi: 10.1016/j.jacc.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Carroll JF, Fulda KG, Chiapa AL, Rodriquez M, Phelps DR, Cardarelli KM, Vishwanatha JK, Cardarelli R. Impact of race/ethnicity on the relationship between visceral fat and inflammatory biomarkers. Obesity (Silver Spring) 2009;17:1420–1427. doi: 10.1038/oby.2008.657. [DOI] [PubMed] [Google Scholar]
- Chatzikyriakidou A, Voulgari PV, Lambropoulos A, Drosos AA. Genetics in rheumatoid arthritis beyond HLA genes: what meta-analyses have shown? Semin. Arthritis Rheum. 2013;43(1):29–38. doi: 10.1016/j.semarthrit.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Chen S, Chen T, Lai L, Chen J. Sequence variants of interleukin 6 (IL-6) are significantly associated with a decreased risk of late-onset Alzheimer’s disease. J. Neuroinflammation. 2012;9:21. doi: 10.1186/1742-2094-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Støvring H, McGue M. Do genetic factors contribute to the association between birth weight and blood pressure? J. Epidemiol. Community Health. 2001;55:583–587. doi: 10.1136/jech.55.8.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton PM. Diet and C-reactive protein. Curr. Atheroscler. Rep. 2003;5:431–436. doi: 10.1007/s11883-003-0032-z. [DOI] [PubMed] [Google Scholar]
- Coe CL, Love GD, Karasawa M, Kawakami N, Kitayama S, Markus HR, Tracy RP, Ryff CD. Population differences in proinflammatory biology: Japanese have healthier profiles than Americans. Brain Behav. Immun. 2011;25:494–502. doi: 10.1016/j.bbi.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppack SW. Pro-inflammatory cytokines and adipose tissue. Proc. Nutr. Soc. 2007;60:349–356. doi: 10.1079/pns2001110. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Yi Q, Smith JD, Shephard C, Wong M, Witrak L, Livingston RJ, Rieder MJ, Nickerson Da. Allelic spectrum of the natural variation in CRP. Hum. Genet. 2006;119:496–504. doi: 10.1007/s00439-006-0160-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton MB, Nichols JE, Zhao Y, Bulun SE, Simpson ER. Expression of transcripts of interleukin-6 and related cytokines by human breast tumors, breast cancer cells, and adipose stromal cells. Mol. Cell. Endocrinol. 1996;118:215–220. doi: 10.1016/0303-7207(96)03761-6. [DOI] [PubMed] [Google Scholar]
- Dai L, Liu D, Guo H, Wang Y, Bai Y. Association between polymorphism in the promoter region of Interleukin 6 (−174 G/C) and risk of Alzheimer’s disease: a meta-analysis. J. Neurol. 2012;259:414–419. doi: 10.1007/s00415-011-6164-0. [DOI] [PubMed] [Google Scholar]
- De Craen AJM, Posthuma D, Remarque EJ, van den Biggelaar AHJ, Westendorp RGJ, Boomsma DI. Heritability estimates of innate immunity: an extended twin study. Genes Immun. 2005;6:167–170. doi: 10.1038/sj.gene.6364162. [DOI] [PubMed] [Google Scholar]
- De Maat MPM, Bladbjerg EM, Hjelmborg JVB, Bathum L, Jespersen J, Christensen K. Genetic influence on inflammation variables in the elderly. Arterioscler. Thromb. Vasc. Biol. 2004;24:2168–2173. doi: 10.1161/01.ATV.0000143856.01669.e7. [DOI] [PubMed] [Google Scholar]
- Dehghan A, Dupuis J, Barbalic M, Bis JC, Eiriksdottir G, Lu C, et al. Metaanalysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–738. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bona D, Vasto S, Capurso C, Christiansen L, Deiana L, Franceschi C, Hurme M, Mocchegiani E, Rea M, Lio D, Candore G, Caruso C. Effect of interleukin-6 polymorphisms on human longevity: a systematic review and meta-analysis. Ageing Res. Rev. 2009;8:36–42. doi: 10.1016/j.arr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- Duffy DL, Mitchell CA, Martin NG. Genetic and environmental risk factors for asthma: a cotwin-control study. Am. J. Respir. Crit. Care Med. 1998;157:840–845. doi: 10.1164/ajrccm.157.3.9702070. [DOI] [PubMed] [Google Scholar]
- Ershler W, Keller E. Age-associated increased interleukin-6 gene expression, late-life diseases, and frailty. Annu. Rev. Med. 2000:245–270. doi: 10.1146/annurev.med.51.1.245. [DOI] [PubMed] [Google Scholar]
- Ferreira RC, Freitag DF, Cutler AJ, Howson JMM, Rainbow DB, Smyth DJ, Kaptoge S, Clarke P, Boreham C, Coulson RM, Pekalski ML, Chen W-M, Onengut-Gumuscu S, Rich SS, Butterworth AS, Malarstig A, Danesh J, Todd JA. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 2013;9:e1003444. doi: 10.1371/journal.pgen.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried SK, Bunkin DA, Greenberg AS. Omental and subcutaneous adipose tissues of obese subjects release interleukin-6: depot difference and regulation by glucocorticoid. J. Clin. Endocrinol. Metab. 1998;83:847–850. doi: 10.1210/jcem.83.3.4660. [DOI] [PubMed] [Google Scholar]
- Friedman EM, Hayney MS, Love GD, Urry HL, Rosenkranz Ma, Davidson RJ, Singer BH, Ryff CD. Social relationships, sleep quality, and interleukin-6 in aging women. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18757–18762. doi: 10.1073/pnas.0509281102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galicia JC, Tai H, Komatsu Y, Shimada Y, Akazawa K, Yoshie H. Polymorphisms in the IL-6 receptor (IL-6R) gene: strong evidence that serum levels of soluble IL-6R are genetically influenced. Genes Immun. 2004;5:513–516. doi: 10.1038/sj.gene.6364120. [DOI] [PubMed] [Google Scholar]
- Gupta NK, De Lemos JA, Ayers CR, Abdullah SM, McGuire DK, Khera A. The relationship between C-reactive protein and atherosclerosis differs on the basis of body mass index: The Dallas Heart study. J. Am. Coll. Cardiol. 2012;60:1148–1155. doi: 10.1016/j.jacc.2012.04.050. [DOI] [PubMed] [Google Scholar]
- Hall JG. Twinning. Lancet. 2003;362:735–743. doi: 10.1016/S0140-6736(03)14237-7. [DOI] [PubMed] [Google Scholar]
- Hamid YH, Urhammer SA, Jensen DP, Glümer C, Borch-Johnsen K, Jørgensen T, Hansen T, Pedersen O. Variation in the interleukin-6 receptor gene associates with type 2 diabetes in Danish whites. Diabetes. 2004;53:3342–3345. doi: 10.2337/diabetes.53.12.3342. [DOI] [PubMed] [Google Scholar]
- Hamzic N, Tang Y, Eskilsson A, Kugelberg U, Ruud J, Jönsson J-I, Blomqvist A, Nilsberth C. Interleukin-6 primarily produced by non-hematopoietic cells mediates the lipopolysaccharide-induced febrile response. Brain Behav. Immun. 2013;33:123–130. doi: 10.1016/j.bbi.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Hauser R, Warren J. Socioeconomic Indexes for occupations: a review, update, and critique. Sociol. Methodol. 1997;27:177–298. [Google Scholar]
- Hawkins D. Using U statistics to derive the asymptotic distribution of Fisher’s Z statistic. Am. Stat. 1989;43:235–237. [Google Scholar]
- Heinrich PC, Castell JV, Andus T. Interleukin-6 and the acute phase response. Biochem J. 1990;265:621–636. doi: 10.1042/bj2650621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert A, Liu C, Karamohamed S, Liu J. BMI Modifies Associations of IL-6 Genotypes with Insulin Resistance: The Framingham Study. Obesity. 2006;14:1454–1461. doi: 10.1038/oby.2006.165. [DOI] [PubMed] [Google Scholar]
- Hjelmborg JVB, Fagnani C, Silventoinen K, McGue M, Korkeila M, Christensen K, Rissanen A, Kaprio J. Genetic influences on growth traits of BMI: a longitudinal study of adult twins. Obesity. 2008;16:847–852. doi: 10.1038/oby.2007.135. [DOI] [PubMed] [Google Scholar]
- Holmes MV, Lange LA, Palmer T, Lanktree MB, North KE, Almoguera B, et al. Causal effects of body mass index on cardiometabolic traits and events: A Mendelian randomization analysis. Am. J. Hum. Genet. 2014;94:198–208. doi: 10.1016/j.ajhg.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. New light on the correlation coefficient and its transforms. J. RStat. Soc. Ser. B-Methodological. 1953;15:193–232. [Google Scholar]
- Hurst SM, Wilkinson TS, McLoughlin RM, Jones S, Horiuchi S, Yamamoto N, Rose-John S, Fuller GM, Topley N, Jones Sa. Il-6 and its soluble receptor orchestrate a temporal switch in the pattern of leukocyte recruitment seen during acute inflammation. Immunity. 2001;14:705–714. doi: 10.1016/s1074-7613(01)00151-0. [DOI] [PubMed] [Google Scholar]
- Huth C, Heid IM, Vollmert C, Gieger C, Grallert H, Wolford JK, et al. IL6 gene promoter polymorphisms and type 2 diabetes: joint analysis of individual participants’ data from 21 studies. Diabetes. 2006;55:2915–2921. doi: 10.2337/db06-0600. L, [DOI] [PubMed] [Google Scholar]
- Johnson W, Krueger RF. Genetic effects on physical health: lower at higher income levels. Behav. Genet. 2005;35:579–590. doi: 10.1007/s10519-005-3598-0. [DOI] [PubMed] [Google Scholar]
- Jones SA, Horiuchi S, Topley N, Yamamoto N, Fuller GM. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 2001;15:43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- Kallen K-J. The role of transsignalling via the agonistic soluble IL-6 receptor in human diseases. Biochim. Biophys. Acta. 2002;1592:323–343. doi: 10.1016/s0167-4889(02)00325-7. [DOI] [PubMed] [Google Scholar]
- Karmakar B, Malkin I, Kobyliansky E. Inheritance of dermatoglyphic traits in twins: univariate and bivariate variance decomposition analysis. Anthropol. Anzeiger. 2012;69:221–228. doi: 10.1127/0003-5548/2011/0134. [DOI] [PubMed] [Google Scholar]
- Khaodhiar L, Ling P-R, Blackburn GL, Bistrian BR. Serum Levels of Interleukin-6 and C-Reactive Protein Correlate With Body Mass Index Across the Broad Range of Obesity. J. Parenter. Enter. Nutr. 2004;28:410–415. doi: 10.1177/0148607104028006410. [DOI] [PubMed] [Google Scholar]
- Kiecolt-Glaser JK, Preacher KJ, MacCallum RC, Atkinson C, Malarkey WB, Glaser R. Chronic stress and age-related increases in the proinflammatory cytokine IL-6. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9090–9095. doi: 10.1073/pnas.1531903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredel LI, Batra A, Stroh T, Kühl AA, Zeitz M, Erben U, Siegmund B. Adipokines from local fat cells shape the macrophage compartment of the creeping fat in Crohn’s Disease. Gut. 2013;62:852–862. doi: 10.1136/gutjnl-2011-301424. [DOI] [PubMed] [Google Scholar]
- Krupp M, Lane MD. On the mechanism of ligand-induced down-regulation of insulin receptor level in the liver cell. J. Biol. Chem. 1981;256:1689–1694. [PubMed] [Google Scholar]
- Lamas JR, Rodríguez-Rodríguez L, Tornero-Esteban P, Villafuertes E, Hoyas J, Abasolo L, Varadé J, Alvarez-Lafuente R, Urcelay E, Fernández-Gutiérrez B. Alternative splicing and proteolytic rupture contribute to the generation of soluble IL-6 receptors (sIL-6R) in rheumatoid arthritis. Cytokine. 2013;61:720–723. doi: 10.1016/j.cyto.2012.12.025. [DOI] [PubMed] [Google Scholar]
- Lee YH, Bae S-C, Choi SJ, Ji JD, Song GG. The association between interleukin- 6 polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm. Res. 2012;61:665–671. doi: 10.1007/s00011-012-0459-1. d. [DOI] [PubMed] [Google Scholar]
- Lepiller Q, Abbas W, Kumar A, Tripathy MK, Herbein G. HCMV activates the IL-6-JAK-STAT3 axis in HepG2 cells and primary human hepatocytes. PLoS One. 2013;8:e59591. doi: 10.1371/journal.pone.0059591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb W, Pavlik R, Erdmann J, Mayer B, Holmer SR, Fischer M, Baessler A, Hengstenberg C, Loewel H, Doering A, Riegger GA, Schunkert H. No association of interleukin-6 gene polymorphism (−174 G/C) with myocardial infarction or traditional cardiovascular risk factors. Int. J. Cardiol. 2004;97:205–212. doi: 10.1016/j.ijcard.2003.07.038. [DOI] [PubMed] [Google Scholar]
- Love GD, Seeman TE, Weinstein M, Ryff CD. Bioindicators in the MIDUS national study: protocol, measures, sample, and comparative context. J. Aging Health. 2010;22(8):1059–1080. doi: 10.1177/0898264310374355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lust JA, Donovan KA, Kline MP, Greipp PR, Kyle RA, Maihle NJ. Isolation of an mRNA encoding a soluble form of the human interleukin-6 receptor. Cytokine. 1992;4:96–100. doi: 10.1016/1043-4666(92)90043-q. [DOI] [PubMed] [Google Scholar]
- Lutgendorf SK, Garand L, Buckwalter KC, Reimer TT, Hong SY, Lubaroff DM. Life stress, mood disturbance, and elevated interleukin-6 in healthy older women. J. Gerontol. A. Biol. Sci. Med. Sci. 1999;54:M434–M439. doi: 10.1093/gerona/54.9.m434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchio A, Tiollais P, Dejean A. Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J. 1989;8:429–433. doi: 10.1002/j.1460-2075.1989.tb03394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinou I, Walters K, Winfield J, Bax DE, Wilson AG. A gain of function polymorphism in the interleukin 6 receptor influences RA susceptibility. Ann. Rheum. Dis. 2010;69:1191–1194. doi: 10.1136/ard.2008.100644. [DOI] [PubMed] [Google Scholar]
- May U, Schiffelholz T, Baier PC, Krueger JM, Rose-John S, Scheller J. IL-6-trans-signalling increases rapid-eye-movement sleep in rats. Eur. J. Pharmacol. 2009;613:141–145. doi: 10.1016/j.ejphar.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Bouchard TJ. Adjustment of twin data for the effects of age and sex. Behav. Genet. 1984;14:325–343. doi: 10.1007/BF01080045. [DOI] [PubMed] [Google Scholar]
- Mehra R, Storfer-Isser A, Kirchner HL, Johnson N, Jenny N, Tracy RP, Redline S. Soluble interleukin 6 receptor: A novel marker of moderate to severe sleep-related breathing disorder. Arch. Intern. Med. 2006;166:1725–1731. doi: 10.1001/archinte.166.16.1725. [DOI] [PubMed] [Google Scholar]
- Miller G. Pathways linking depression, adiposity, and inflammatory markers in healthy young adults. Brain. Behav. Immun. 2003;17:276–285. doi: 10.1016/s0889-1591(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Miller GE, Stetler CA, Carney RM, Freedland KE, Banks WA. Clinical depression and inflammatory risk markers for coronary heart disease. Am. J. Cardiol. 2002;90:1279–1283. doi: 10.1016/s0002-9149(02)02863-1. [DOI] [PubMed] [Google Scholar]
- Mohamed-Ali V, Goodrick S, Bulmer K, Holly JM, Yudkin JS, Coppack SW. Production of Soluble Tumor Necrosis Factor Receptors by Human Subcutaneous Adipose Tissue In Vivo. Am. J. Physiol. 1999;277:E971–E975. doi: 10.1152/ajpendo.1999.277.6.E971. [DOI] [PubMed] [Google Scholar]
- Montero-Julian FA. The soluble IL-6 receptors: serum levels and biological function. Cell. Mol. Biol. 2001;47:583–597. [PubMed] [Google Scholar]
- Müllberg J, Schooltink H, Stoyan T, Günther M, Graeve L, Buse G, Mackiewicz A, Heinrich PC, Rose-John S. The soluble interleukin-6 receptor is generated by shedding. Eur. J. Immunol. 1993;23:473–480. doi: 10.1002/eji.1830230226. [DOI] [PubMed] [Google Scholar]
- Nauck M, Winkelmann BR, Hoffmann MM, Böhm BO, Wieland H, März W. The interleukin-6 G(−174)C promoter polymorphism in the LURIC cohort: no association with plasma interleukin-6, coronary artery disease, and myocardial infarction. J. Mol. Med. 2002;80:507–513. doi: 10.1007/s00109-002-0354-2. [DOI] [PubMed] [Google Scholar]
- Navarro SL, Brasky TM, Schwarz Y, Song X, Wang CY, Kristal AR, Kratz M, White E, Lampe JW. Reliability of serum biomarkers of inflammation from repeated measures in healthy individuals. Cancer Epidemiol. Biomarkers Prev. 2012;21:1167–1170. doi: 10.1158/1055-9965.EPI-12-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale M, Boker S, Xie G, Maes H. VCU Box 900126. 6th edition. Richmond, VA: 2003. Mx: statistical modeling. [Google Scholar]
- Nichols RC, Bilbro J. The diagnosis of twin zygosity. Hum. Hered. 1966;16:265–275. doi: 10.1159/000151973. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos GK, Dimou NL, Hamodrakas SJ, Bagos PG. Cytokine gene polymorphisms in periodontal disease: a meta-analysis of 53 studies including 4178 cases and 4590 controls. J. Clin. Periodontol. 2008;35:754–767. doi: 10.1111/j.1600-051X.2008.01298.x. [DOI] [PubMed] [Google Scholar]
- O’Connor M-F, Motivala SJ, Valladares EM, Olmstead R, Irwin MR. Sex differences in monocyte expression of IL-6: role of autonomic mechanisms. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;293:R145–R151. doi: 10.1152/ajpregu.00752.2006. [DOI] [PubMed] [Google Scholar]
- Pankow JS, Folsom AR, Cushman M, Borecki IB, Hopkins PN, Eckfeldt JH, Tracy RP. Familial and genetic determinants of systemic markers of inflammation: The NHLBI family heart study. Atherosclerosis. 2001;154:681–689. doi: 10.1016/s0021-9150(00)00586-4. [DOI] [PubMed] [Google Scholar]
- Patel A, Zhu Y, Kuzhikandathil EV, Banks WA, Siegel A, Zalcman SS. Soluble interleukin-6 receptor induces motor stereotypies and co-localizes with gp130 in regions linked to cortico-striato-thalamo-cortical circuits. PLoS One. 2012;7:e41623. doi: 10.1371/journal.pone.0041623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M, Müller AM, Rose-John S. Interleukin-6 and soluble interleukin-6 receptor: direct stimulation of gp130 and hematopoiesis. Blood. 1998;92:3495–3504. [PubMed] [Google Scholar]
- Pini M, Rhodes DH, Castellanos KJ, Hall AR, Cabay RJ, Chennuri R, Grady EF, Fantuzzi G. Role of IL-6 in the resolution of pancreatitis in obese mice. J. Leukoc. Biol. 2012;91:957–966. doi: 10.1189/jlb.1211627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posthuma D, Boomsma DI. Mx scripts library: structural equation modeling scripts for twin and family data. Behav. Genet. 2005;35:499–505. doi: 10.1007/s10519-005-2791-5. [DOI] [PubMed] [Google Scholar]
- Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Res. 2002;5:554–571. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- Rafiq S, Frayling T, Murray A, Hurst A. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun. 2007;8:552–559. doi: 10.1038/sj.gene.6364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, et al. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am. J. Hum. Genet. 2007;80:716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Rodríguez L, Lamas JR, Varadé J, López-Romero P, Tornero-Esteban P, Abasolo L, et al. Plasma soluble IL-6 receptor concentration in rheumatoid arthritis: associations with the rs8192284 IL6R polymorphism and with disease activity. Rheumatol. Int. 2011;31:409–413. doi: 10.1007/s00296-010-1593-0. [DOI] [PubMed] [Google Scholar]
- Rooks C, Veledar E, Goldberg J, Bremner JD, Vaccarino V. Early trauma and inflammation: Role of familial factors in a study of twins. Psychosom. Med. 2012;74:146–152. doi: 10.1097/PSY.0b013e318240a7d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Kiyota N, Kawasaki Y, Miyazaki Y, Kashimura J, Fukuda M, Kishi R. Relationship between C-reactive protein and visceral adipose tissue in healthy Japanese subjects. Diabetes. Obes. Metab. 2004;6:249–258. doi: 10.1111/j.1462-8902.2003.0342.x. [DOI] [PubMed] [Google Scholar]
- Saiki R, Hayashi D, Ikuo Y, Nishimura K, Ishii I, Kobayashi K, Chiba K, Toida T, Kashiwagi K, Igarashi K. Acrolein stimulates the synthesis of IL-6 and C-reactive protein (CRP) in thrombosis model mice and cultured cells. J. Neurochem. 2013;127(5):652–659. doi: 10.1111/jnc.12336. [DOI] [PubMed] [Google Scholar]
- Salman BN, Vahabi S, Movaghar SE, Mahjour F. Proliferative and inductive effects of cyclosporin a on gingival fibroblast of child and adult. Dent. Res. J. (Isfahan) 2013;10:52–58. doi: 10.4103/1735-3327.111777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sas AA, Jamshidi Y, Zheng D, Wu T, Korf J, Alizadeh BZ, Spector TD, Snieder H. The age-dependency of genetic and environmental influences on serum cytokine levels: A twin study. Cytokine. 2012;60:108–113. doi: 10.1016/j.cyto.2012.04.047. [DOI] [PubMed] [Google Scholar]
- Sasayama D, Hori H, Teraishi T, Hattori K, Ota M, Matsuo J, Kawamoto Y, Kinoshita Y, Amano N, Kunugi H. Association of cognitive performance with interleukin-6 receptor Asp358Ala polymorphism in healthy adults. J. Neural Transm. 2012;119:313–318. doi: 10.1007/s00702-011-0709-3. [DOI] [PubMed] [Google Scholar]
- Schöbitz B, Pezeshki G, Pohl T. Soluble interleukin-6 (IL-6) receptor auguments central effects of IL-6 in vivo. FASEB J. 1995;6:659–664. doi: 10.1096/fasebj.9.8.7768358. [DOI] [PubMed] [Google Scholar]
- Schousboe K, Willemsen G, Kyvik KO, Mortensen J, Boomsma DI, Cornes BK, Davis CJ, Fagnani C, Hjelmborg J, Kaprio J, De Lange M, Luciano M, Martin NG, Pedersen N, Pietiläinen KH, Rissanen A, Saarni S, Sørensen TIA, Van Baal GCM, Harris JR. Sex differences in heritability of BMI: a comparative study of results from twin studies in eight countries. Twin Res. 2003;6:409–421. doi: 10.1375/136905203770326411. [DOI] [PubMed] [Google Scholar]
- Sedgwick P. Intraclass correlation coefficient. BMJ. 2013;346:f1816–f1816. [Google Scholar]
- Segal N, Feng R, McGuire S. Genetic and environmental contributions to body mass index: comparative analysis of monozygotic twins, dizygotic twins and same-age unrelated siblings. Int. J. Obes. 2008;33:37–41. doi: 10.1038/ijo.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlali A, Witoled C, Alanazi M, Rouabhia M. Whole cigarette smoke increased the expression of TLRs, HBDs, and proinflammory cytokines by human gingival epithelial cells through different signaling pathways. PLoS One. 2012;7:e52614. doi: 10.1371/journal.pone.0052614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Paine S, Chowdhury I. Impact of interleukin-6 promoter polymorphism and serum interleukin-6 level on the acute inflammation and neovascularization stages of patients with Eales’ Disease. Mol. Vis. 2011;17:2552–2563. [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Larsimont D, Ma Y, Laios I, Leclercq G. Regulation of estrogen receptor levels by ligand-induced release of compound(s) in MCF-7 cells. Mol. Cell. Endocrinol. 2000;164:19–29. doi: 10.1016/s0303-7207(00)00244-6. [DOI] [PubMed] [Google Scholar]
- Shah T, Zabaneh D, Gaunt T, Swerdlow DI, Shah S, Talmud PJ, Day IN, Whittaker J, Holmes MV, Sofat R, Humphries SE, Kivimaki M, Kumari M, Hingorani AD, Casas JP. Gene-centric analysis identifies variants associated with interleukin-6 levels and shared pathways with other inflammation markers. Circ. Cardiovasc. Genet. 2013;6:163–170. doi: 10.1161/CIRCGENETICS.112.964254. [DOI] [PubMed] [Google Scholar]
- Smith A, Zheng D, Palmen J, Pang D. Effects of genetic variation on chromatin structure and the transcriptional machinery: analysis of the IL6 gene locus. Genes Immun. 2012;13:583–586. doi: 10.1038/gene.2012.32. [DOI] [PubMed] [Google Scholar]
- Stenberg A. Interpreting estimates of heritability - A note on the twin decomposition. Econ. Hum. Biol. 2012;11:201–205. doi: 10.1016/j.ehb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Stephens OW, Zhang Q, Qu P, Zhou Y, Chavan S, Tian E, Williams DR, Epstein J, Barlogie B, Shaughnessy JD. An intermediate-risk multiple myeloma subgroup is defined by sIL-6r: levels synergistically increase with incidence of SNP rs2228145 and 1q21 amplification. Blood. 2012;119:503–512. doi: 10.1182/blood-2011-07-367052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Snieder H, Miller A, Ritchie J. Genetic and environmental influences on systemic markers of inflammation in middle-aged male twins. Atherosclerosis. 2008;200:213–220. doi: 10.1016/j.atherosclerosis.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganami T, Ogawa Y. Adipose tissue macrophages: their role in adipose tissue remodeling. J. Leukoc. Biol. 2010;88:33–39. doi: 10.1189/jlb.0210072. [DOI] [PubMed] [Google Scholar]
- Tan Q, Ohm Kyvik K, Kruse TA, Christensen K. Dissecting complex phenotypes using the genomics of twins. Funct. Integr. Genomics. 2010;10:321–327. doi: 10.1007/s10142-010-0160-9. d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen J, Jansen R, Smit D, Hottenga J-J, Mbarek H, Willemsen G, Kluft C, Penninx BWJ, Ferreira MA, Boomsma DI, de Geus EJC. The contribution of the functional IL6R polymorphism rs2228145, eQTLs and other genome-wide SNPs to the heritability of plasma sIL-6R levels. Behav. Genet. 2014;44(4):368–382. doi: 10.1007/s10519-014-9656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oijen M, Arp PP, de Jong FJ, Hofman A, Koudstaal PJ, Uitterlinden AG, Breteler MMB. Polymorphisms in the interleukin 6 and transforming growth factor beta1 gene and risk of dementia. The Rotterdam Study. Neurosci. Lett. 2006;402:113–117. doi: 10.1016/j.neulet.2006.03.050. [DOI] [PubMed] [Google Scholar]
- Vaughn C, Ochs-Balcom H, Nie J. No association between circulating levels and genetic variants of IL-6 and TNF-α and colon adenoma. Gastroenterol. Res. 2013;66:43–48. doi: 10.4021/gr529w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volanakis J. Human C-reactive protein: expression, structure, and function. Mol. Immunol. 2001;38:189–197. doi: 10.1016/s0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- Walston JD, Fallin MD, Cushman M, Lange L, Psaty B, Jenny N, Browner W, Tracy R, Durda P, Reiner A. IL-6 gene variation is associated with IL-6 and C-reactive protein levels but not cardiovascular outcomes in the Cardiovascular Health Study. Hum. Genet. 2007;122:485–494. doi: 10.1007/s00439-007-0428-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang Z, Chu W, Hale T, Cooper JJ, Elbein SC. Molecular screening and association analyses of the interleukin 6 receptor gene variants with type 2 diabetes, diabetic nephropathy, and insulin sensitivity. J. Clin. Endocrinol. Metab. 2005;90:1123–1129. doi: 10.1210/jc.2004-1606. [DOI] [PubMed] [Google Scholar]
- Weisberg SP, Mccann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW. Obesity is associated with macrophage accumulation. J. Clin. Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh P, Polisecki E, Robertson M, Jahn S, Buckley BM, De Craen AJM, Ford I, Jukema JW, Macfarlane PW, Packard CJ, Stott DJ, Westendorp RGJ, Shepherd J, Hingorani AD, Smith GD, Schaefer E, Sattar N. Unraveling the directional link between adiposity and inflammation: A bidirectional mendelian randomization approach. J. Clin. Endocrinol. Metab. 2010;95:93–99. doi: 10.1210/jc.2009-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernstedt I, Eriksson A-L, Berndtsson A, Hoffstedt J, Skrtic S, Hedner T, Hultén LM, Wiklund O, Ohlsson C, Jansson J-O. A common polymorphism in the interleukin-6 gene promoter is associated with overweight. Int. J. Obes. Relat. Metab. Disord. 2004;28:1272–1279. doi: 10.1038/sj.ijo.0802763. [DOI] [PubMed] [Google Scholar]
- Wisse BE. The inflammatory syndrome: the role of adipose tissue cytokines in metabolic disorders linked to obesity. J. Am. Soc. Nephrol. 2004;15:2792–2800. doi: 10.1097/01.ASN.0000141966.69934.21. [DOI] [PubMed] [Google Scholar]
- Wörns MA, Victor A, Galle PR, Höhler T. Genetic and environmental contributions to plasma C-reactive protein and interleukin-6 levels--a study in twins. Genes Immun. 2006;7:600–605. doi: 10.1038/sj.gene.6364330. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang F, Skrip L, Lei H, Wang Y, Hu D, Ding R. IL-6 gene polymorphisms and CAD risk: a meta-analysis. Mol. Biol. Rep. 2012;6:2589–2598. doi: 10.1007/s11033-012-2345-x. [DOI] [PubMed] [Google Scholar]
- Zhang H-Y, Feng L, Wu H, Xie X-D. The association of IL-6 and IL-6R gene polymorphisms with chronic periodontitis in a Chinese population. Oral Dis. 2013 doi: 10.1111/odi.12075. [DOI] [PubMed] [Google Scholar]
- Zhou X, An G, Chen J. Inhibitory effects of hydrogen sulphide on pulmonary fibrosis in smoking rats via attenuation of oxidative stress and inflammation. J. Cell. Mol. Med. 2014;XX:1–6. doi: 10.1111/jcmm.12254. [DOI] [PMC free article] [PubMed] [Google Scholar]