Summary
The mechanistic target of rapamycin complex 1 (mTORC1) senses diverse signals to regulate cell growth and metabolism. It has become increasingly clear that mTORC1 activity is regulated in time and space inside the cell, but direct interrogation of such spatiotemporal regulation is challenging. Here we describe a genetically encoded mTORC1 activity reporter (TORCAR) that exhibits a change in FRET in response to phosphorylation by mTORC1. Co-imaging mTORC1 activity and calcium dynamics revealed that a growth factor-induced calcium transient contributes to mTORC1 activity. Dynamic activity maps generated using subcellularly targeted TORCAR uncovered mTORC1 activity not only in cytosol and at the lysosome but also in nucleus and at the plasma membrane. Furthermore, a wide distribution of activities was observed upon growth factor stimulation, whereas leucine ester, an amino acid surrogate, induces more compartmentalized activities at the lysosome and in nucleus. Thus, mTORC1 activities are spatiotemporally regulated in a signal-specific manner.
INTRODUCTION
The mechanistic target of rapamycin (mTOR) is a highly conserved serine/threonine protein kinase. mTOR is the key catalytic component of two structurally and functionally distinct complexes named mTORC1 (mTOR Complex 1) and mTORC2 (mTOR Complex 2). The rapamycin-sensitive mTORC1 is composed of three essential components: the kinase mTOR, the signature subunit Raptor (regulatory associated protein of mTOR), and the mLST8 subunit (mammalian lethal with sec13 protein 8) that is also present in the rapamycin-insensitive mTORC2. As a critical signaling hub, mTORC1 senses diverse signals, such as growth factors (GFs), amino acids (AAs), stress, oxygen, and changes in energy levels. When activated, mTORC1 promotes protein and lipid synthesis and inhibits autophagy to control cell growth and cell metabolism. Due to its pivotal role in a large number of cellular processes, aberrant regulation of mTORC1 has been shown to be associated with human cancers, cardiovascular disease, and metabolic disorders such as type 2 diabetes.
The activation of mTORC1 is exquisitely regulated in response to distinct upstream cues. GF activates mTORC1 via the PI3K/Akt pathway and the Erk/RSK pathway at the level of the Tuberous Sclerosis Complex 2 (TSC2) (Dibble and Manning, 2013). Within the TSC complex (TSC1/TSC2/TBC1D7), TSC2 functions as a gatekeeper for mTOR activity by acting as a GTPase-activating protein (GAP) towards Rheb GTPase, thus promoting GTP hydrolysis and inhibiting Rheb. GTP-bound Rheb is an essential and direct upstream activator of mTORC1 (Dibble and Manning, 2013). Akt and Erk, as well as RSK downstream of Erk, phosphorylate the TSC2 and thereby allow Rheb to activate mTORC1. Unlike growth factors, amino acids communicate to mTORC1 through the Rag GTPase complex and the Ragulator complex that anchors the Rags to the lysosome (Bar-Peled et al., 2012; Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008). In response to amino acids, the Rag GTPase complex is activated and subsequently recruits mTORC1 to the lysosome, where the Rheb GTPase is also present. Therefore, the close proximity of mTORC1 and Rheb at the lysosome allows the immediate activation of the complex, assuming Rheb is loaded with GTP and active. When amino acids are absent, the Rag GAP complex (GATOR1) inactivates the Rag GTPases and leads to the dissociation of mTORC1 from the lysosomal surface (Bar-Peled et al., 2013).
Given that both GF and AA signals specifically impinge on mTORC1, an intriguing question is how this signaling specificity is achieved inside the cell. Spatial compartmentalization may be a key mechanism. Indeed, mTOR has been shown to be localized to several distinct subcellular compartments, including the lysosome, mitochondria, plasma membrane, endoplasmic reticulum and nucleus, although in some cases the evidence is disputable (Betz and Hall, 2013). At the lysosome, the best characterized site of mTORC1 activation, Menon et al have recently shown that growth factor signaling pathways directly activate mTORC1, independent of amino acids signaling, by promoting the release of the inhibitory TSC complex from lysosomes (Menon et al., 2014). It has also been reported in a parallel study that Rag GTPase recruits TSC2 to the lysosomal surface upon amino acid removal to inactivate mTORC1 (Demetriades et al., 2014). Therefore, both the activation and inactivation of mTORC1 may be intricately regulated in a signal- and location-specific manner. However, aside from the lysosome, the activity of mTORC1 at various other cellular locations has not been well characterized.
To shed light on compartmentalized mTORC1 signaling, we generated and characterized a genetically encoded mTORC1 activity reporter (TORCAR) to enable the characterization of mTORC1 signaling dynamics in single living cells. By co-imaging TORCAR along with a calcium probe, we uncovered that a transient intracellular calcium increase induced by PDGF contributes to mTORC1 activity. To reveal the spatial regulation of mTORC1, TORCAR was then targeted to distinct subcellular locations, including lysosome, the plasma membrane, and the nucleus. Surprisingly, we detected GF-stimulated mTORC1 activity at all the examined locations, including the nuclear site in dispute, whereas leucine ester-induced mTORC1 activity only occurs at the lysosome and in the nucleus, indicating that amino acid sensing and signaling to mTOR are rather local.
RESULTS
Development of mTORC1 activity reporters
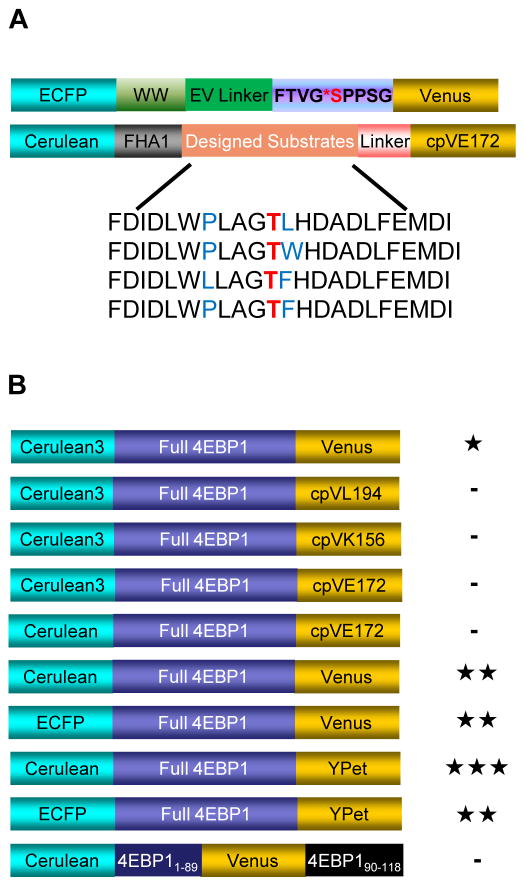
To monitor mTORC1 activity in living cells, we designed a series of biosensor candidate constructs based on different strategies for generating a FRET (Fluorescence resonance energy transfer) -based kinase activity biosensor (Ni et al., 2006; Zhou et al., 2012). In general, such biosensors consist of a kinase activity-dependent molecular switch sandwiched between a FRET-capable fluorescent protein (FP) pair. Our first strategy utilized an engineered molecular switch flanked with a cyan FP (CFP)/yellow FP (YFP) FRET pair. An engineered molecular switch is constructed by coupling a sensing domain, which usually is a peptide substrate that can be recognized and phosphorylated by a kinase of interest, to a domain that specifically binds to phosphoamino acids (Phosphoamino acid-binding domain, PAABD). When phosphorylated, the substrate falls into the binding pocket of the PAABD, and this engineered conformational change in the molecular switch can be read out as a change in FRET. Thus, by analogy to the previously engineered PKA activity reporter (AKAR) (Zhang et al., 2001), PKC activity reporter (CKAR) (Violin et al., 2003), and AMPK activity reporter (AMPKAR) (Tsou et al., 2011), we designed several candidate constructs based on the mTORC1 substrate consensus motifs identified by a study using positional peptide screening (Hsu et al., 2011). The designed mTORC1 substrate sequences were further modified to accommodate the binding preference of FHA1, a phosphothreonine binding domain, to prompt a phosphorylation-dependent conformational change (Figure 1A). Similarly, a candidate biosensor using a fragment of Ulk1 containing the mTORC1-specific phosphorylation site Ser757 (Kim et al., 2011) and the phosphoserine/phosphothreonine binding WW domain was also constructed (Figure 1A). However, when tested in living cells, these candidate biosensors did not show any responses when mTORC1 activity was stimulated.
Figure 1.
Domain structures of TORCAR candidate constructs. (A) Domain structures of candidate constructs based on a generalized modular design. The top construct used a fragment of Ulk1 as the substrate motif and the WW domain as the PAABD, and the bottom constructs used the consensus substrate motifs from peptide positioning screening studies. (B) Domain structures of candidate constructs based on putative conformational change in full-length or fragments of 4EBP1 upon phosphorylation. -, <5% response; ★, 5–10% response; ★★, 10–15% response; ★★★, >15% response. See also Figure S1.
As an alternative strategy, any naturally existing conformationally responsive element can be used as the switch when constructing a biosensor (Calleja et al., 2007). To test this strategy, we designed biosensors using full-length or truncated fragments of 4EBP1 (eIF4E binding protein 1) sandwiched between CFP and YFP (Figure 1B and Figure S1A). The phosphorylation of 4EBP1 is thought to induce a conformational change that leads to its release from eIF4E, thereby relieving its repression of translation (Fletcher and Wagner, 1998; Gosselin et al., 2010). In addition, another well characterized mTORC1 substrate (Burnett et al., 1998), p70 S6K, was tested in a similar format (Figure S1B). Among these constructs, the one containing full-length 4EBP1 sandwiched between Cerulean and Venus was shown to be responsive (Figure 1B). Serum- and amino acid- starved (hereafter referred to as double-starved, indicated as “DS”, and the starvation condition used if not otherwise indicated) NIH3T3 fibroblasts expressing this construct showed a consistent increase in the cyan-over-yellow (C/Y) emission ratio in response to PDGF stimulation, reflecting a decrease in energy transfer efficiency due to the putative phosphorylation-induced conformational change in 4EBP1. To improve the dynamic range of the biosensor, we tested various combinations of fluorescent proteins as FRET pairs (Figure 1B). The biosensor with Cerulean/YPet as the FRET pair showed the largest increase in emission ratio (C/Y) in response to PDGF stimulation.
Characterization of mTORC1 activity reporter (TORCAR)
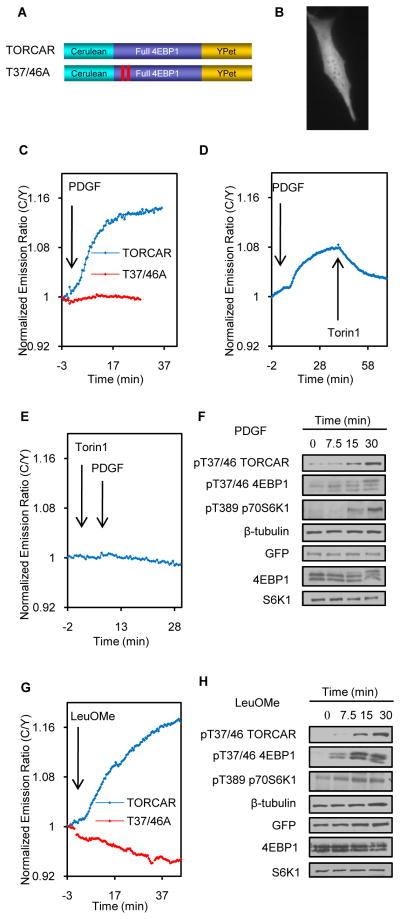
NIH3T3 cells expressing the biosensor containing full-length 4EBP1 flanked by Cerulean and YPet (Figure 2A) exhibited uniform cyan and yellow fluorescence throughout the cell, suggesting the reporter is distributed evenly in both the cytosol and nucleus (Figure 2B). PDGF addition induced a 13.3 ± 6.0% increase in emission ratio (C/Y) with a t1/2 of 12.2 ± 3.2min (Figure 2C, blue trace, n = 33). To examine whether the increase in the ratio (C/Y) is indicative of an increase in mTOR kinase activity, we added torin1, a potent mTOR inhibitor, following PDGF stimulation. Torin1 addition immediately reversed the reporter response (Figure 2D, n = 8). In addition, pretreating NIH3T3 cells with torin1 abolished the response to PDGF (Figure 2E, n = 6), suggesting that the reporter response reflects the mTOR kinase activity. Addition of torin1 to non-starved 3T3 cells expressing this reporter also led to a rapid decrease in emission ratio, indicating the presence of basal activity (n = 6, Figure S2A). We further examined whether the TORCAR response is specific to mTORC1. In these experiments, the addition of rapamycin, a selective inhibitor for mTORC1, following PDGF stimulation, reversed the PDGF-induced response (Figure S2B, n = 8). Pretreatment with rapamycin also blocked the PDGF-induced response (Figure S2C, n = 4). Taken together, these results indicate that this biosensor specifically detects mTORC1 activity in cells; therefore, we named it TORCAR (mTORC1 Activity Reporter).
Figure 2.
Characterization of TORCAR. (A) Domain structure of TORCAR and TORCAR (T37/46A). (B) Image of an NIH3T3 cell expressing TORCAR. Scale bar = 10 μm. (C) Representative time course of the TORCAR response to PDGF. Double starved NIH3T3 cells expressing TORCAR (blue trace, mean response ± SEM as 13.3 ± 6.0%, mean t1/2 ± SEM as 12.2 ± 3.3 min, n = 33) or TORCAR-T37/46A (red trace, n = 10) were stimulated with 50 ng/ml PDGF. (D) PDGF-induced TORCAR response was reversed upon addition of torin1 (1 μM) (n = 8). (E) Pretreatment with torin1 (200 nM) abolished the PDGF-induced TORCAR response (n = 6). (F) Immunoblots showing the time-course for the phosphorylation levels of TORCAR, endogenous 4EBP1 (p-T37/46), and p70 S6K1 (p-T389) (n = 3). Double starved NIH3T3 cells expressing TORCAR were stimulated with PDGF (50 ng/ml) for the indicated times. (G) Representative time course of the TORCAR FRET response to Leucine methyl ester. Double starved NIH3T3 cells expressing TORCAR (blue trace, n = 6) or TORCAR-T37/46A (red trace, n = 5) were stimulated with 7.5 mM LeuOMe. (H) Immunoblots showed the time-course for the phosphorylation levels of TORCAR, endogenous 4EBP1 (p-T37/46), and p70 S6K1 (p-T389) (n = 3). Double starved NIH3T3 cells expressing TORCAR were stimulated with LeuOMe (7.5 mM) for the indicated times. See also Figure S2.
We then asked whether the FRET ratio change is dependent on the phosphorylation of the reporter by mTORC1. 4EBP1 is known to be phosphorylated at multiple sites, including T37, T46, S65, T70, S83 and S112 (Fadden et al., 1997; Heesom et al., 1998). Phosphorylation at T37 and T46 was shown to be mTORC1-specific (Gingras et al., 1999; Thoreen et al., 2009). A mutant TORCAR containing T37A and T46A in the 4EBP region failed to respond to PDGF and torin1 addition (Figure 2C, red trace, n = 10). Consistent with these data, the time-course for PDGF-induced phosphorylation at T37/46 of 4EBP1 in TORCAR, as well as endogenous 4EBP1 (T37/46) and p70 S6K1 (T389), correlated with the emission ratio change (Figure 2F). We next examined whether TORCAR is able to detect mTORC1 activity in other cell types. Similar to the response in NIH3T3 cells, insulin induced an increase in the C/Y emission ratio in double-starved HeLa cells (Figure S2D, n = 4) and 3T3-L1 adipocytes (Figure S2E, n = 10) expressing TORCAR, suggesting that TORCAR also provides a selective FRET readout for mTORC1 in these cell types.
Amino acids, especially leucine, have been shown to activate mTORC1 (Dodd and Tee, 2011; Jewell and Guan, 2013). Indeed, TORCAR responded to a complete amino acid mixture with an emission ratio change of 11.2 ± 3.0% (n = 4, Figure S2F) and 4.2 ± 2.2% (n = 10, Figure S2G) in double starved 3T3 cells and amino acid-starved 3T3 cells, respectively. Compared with a total amino acid mixture, leucine appeared to be a weaker stimulus of mTORC1 activity, indicated by both the TORCAR response (Figure S2H and S2I) and the phosphorylation of mTORC1 substrates (Figure S2J). To further characterize TORCAR, we treated NIH3T3 cells with leucine O-methyl ester (LeuOMe), which bypasses amino acid transporters and diffuses freely across membranes, after which it is hydrolyzed to native leucine (Reeves, 1979; Zoncu et al., 2011). The addition of leucine ester also induced an increase in the C/Y emission ratio (Figure 2G, blue trace, n = 6) that was dependent on the presence of T37 and T46 (Figure 2G, red trace, n = 5). Parallel WB analysis confirmed that leucine ester induces the phosphorylation of T37/46 in both TORCAR and endogenous 4EBP1(T37/46) and p70 S6K1 (T389) (Figure 2H). Furthermore, stimulation with PDGF and amino acids showed additive effects on the TORCAR response (Figure S2K and S2L) and phosphorylation (Figure S2M). Consistent with previous studies (Roux et al., 2004; Tee et al., 2003), the phorbol ester PMA also induced a specific response (Figure S2N and S2O), presumably via the ERK/RSK pathway.
PDGF-induced calcium transient contributes to mTORC1 activity
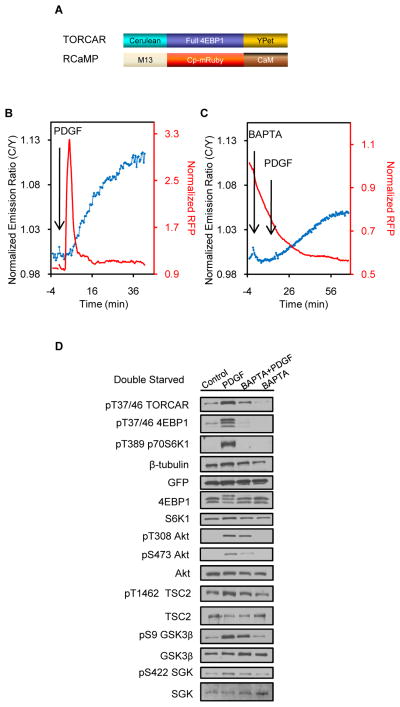
Previous studies indicated that the intracellular calcium plays a role in mTORC1 activation. For example, the leucine-induced activation of S6K1 has been shown to require the mobilization of intracellular Ca2+ in skeletal myoblasts, which is mediated by the SHP-2/PLC β4/IP3 pathway (Mercan et al., 2013). It has also been suggested that amino acids or insulin treatment in HeLa cells induced a rise in intracellular Ca2+, which triggered mTORC1 activation (Gulati et al., 2008). We therefore asked whether calcium contributes to GF-stimulated TORC1 activity. To test the involvement of calcium, we co-expressed a calcium probe, RCaMP, with TORCAR in NIH3T3 cells to simultaneously track calcium dynamics and mTORC1 activity changes. RCaMP consists of a red FP (RFP, cp-mRuby) flanked by calmodulin (CaM) and the Ca2+/CaM-binding peptide M13 (Figure 3A) (Akerboom et al., 2013). When Ca2+ binds to CaM in RCaMP, it induces a conformational change that leads to an increase in RFP intensity. Co-imaging TORCAR and RCaMP revealed a transient peak of intracellular calcium in NIH3T3 cells upon PDGF stimulation (Figure 3B, red trace), which occurs right before the onset of the TORCAR response (Figure 3B, blue trace,13.2 ± 6.9%, n = 25). Blocking the extracellular calcium using EGTA in calcium-free medium did not prevent the PDGF-induced calcium transient, suggesting that this calcium increase originates from intracellular store (Figure S3A, n = 4). To examine whether this PDGF-induced intracellular calcium increase contributes to mTORC1 activity, we treated cells with a cell-permeant calcium chelator, BAPTA-AM, before PDGF stimulation. BAPTA-AM treatment abolished the calcium increase and decreased the amplitude of the TORCAR response by approximately 40% (7.7 ± 2.9%, n = 18, p<0.0001), suggesting that calcium plays a significant role in mTORC1activity (Figure 3C). Consistent with this observation, WB analysis confirmed that the PDGF-induced phosphorylation of TORCAR at 4EBP T37/46 was reduced by BAPTA-AM (Figure 3D). In addition, BAPTA alone also caused a slight decrease in the TORCAR emission ratio (Figure 3C and S3B), as well as a slight reduction in the phosphorylation of TORCAR (Figure 3D and S3C), suggesting that both basal calcium and the PDGF-induced calcium spike may contribute to mTORC1 activity.
Figure 3.
Calcium plays a role in GF-induced mTORC1 activity. (A) Domain structures of TORCAR and RCaMP. (B) Double starved NIH3T3 cells co-expressing TORCAR and RCaMP responded to PDGF with a calcium transient and an increase in FRET ratio (C/Y) (mean response ± SEM as 13.2 ± 6.9%, n = 25). (C) Pretreatment with BAPTA-AM, an intracellular calcium chelator, antagonized the calcium peak and lowered the TORCAR response to PDGF (mean response ± SEM as 7.7 ± 2.9%, n = 18, p<0.0001 by two-tailed t test). (D) Immunoblots showed the phosphorylation levels of TORCAR, endogenous 4EBP1 (p-T37/46), p70 S6K1 (p-T389), phospho-Akt and total Akt upon different treatments (n = 3). Control: no drug added. PDGF: PDGF (50 ng/ml) was added at time 0 and cells were collected after 30 min. BAPTA+PDGF: BAPTA-AM (20 μM) was added at time 0 and PDGF (50 ng/ml) was added after 15 min; cells were collected 30 min after PDGF addition. BAPTA: BAPTA-AM (20 μM) was added and cells were collected after 15 min. See also Figure S3.
We further asked whether BAPTA-AM impairs PDGF-induced Akt activity. Previously we developed the Akt activity reporter (AktAR) to allow the real-time tracking of intracellular Akt activity (Gao and Zhang, 2008). A new version of AktAR with an enhanced dynamic range was generated then, by replacing the Cerulean of AktAR with Cerulean3, a brighter variant of CFP (Markwardt et al., 2011) (Figure S3D–F). Imaging with this improved Akt activity reporter, AktAR2, showed that pretreatment with BAPTA-AM reduced the PDGF-stimulated AktAR2 response (2.8 ± 2.9%, n = 8, Figure S3G), compared to the response without BAPTA-AM pretreatment (9 ± 6%, n = 10, p<0.01, Figure S3H), in these double-starved cells. Of note, amino acid starvation (Figure S3H and S3I) significantly lowered PDGF-induced Akt activity compared to control cells (Figure S3F), implying that amino acids deprivation suppressed PDGF-stimulated Akt activity (Figure S3J) (Tato et al., 2011). Furthermore, whereas the PDGF-induced phosphorylation of Akt T308 was not significantly altered, the phosphorylation of Akt S473, which is targeted by mTORC2, was attenuated by BAPTA-AM (Figure 3D). In addition, BAPTA pretreatment also lowered the phosphorylation of S422 SGK1, another mTORC2 target (Figure 3D), suggesting that mTORC2 activity is affected. These data suggest that both mTORC1 and mTORC2 activities are affected by calcium.
Lysosomal Akt activity during GF-induced mTORC1 activation
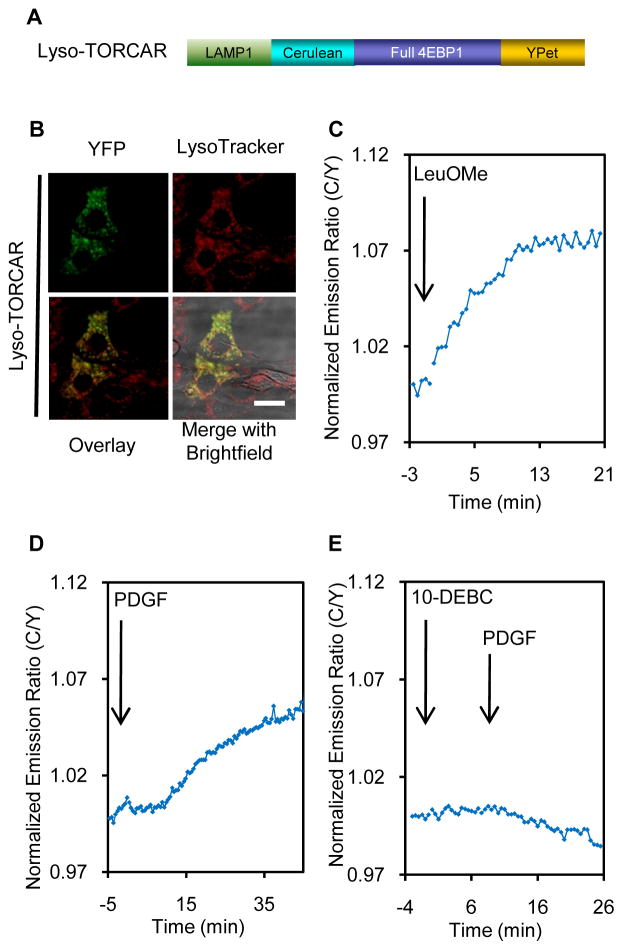
Lysosomal mTORC1activity has remained an important point of interest. In response to amino acids the Rag complex recruits mTOR from a poorly characterized cytoplasmic location to the lysosomal surface (Zoncu et al., 2011), suggesting lysosomes act as a signaling platform for mTORC1 regulation and function. To detect mTORC1 activity at the lysosome, we generated a lysosome-targeted TORCAR (Lyso-TORCAR) by attaching LAMP1 (lysosome associated membrane protein 1) to the N-terminus of TORCAR (Figure 4A). NIH3T3 cells expressing Lyso-TORCAR exhibited fluorescence in small punctate structures that can be co-stained with LysoTracker Red, a red lysosomal marker (Figure 4B).
Figure 4.
mTORC1 activity at the lysosomal membrane. (A) Domain structures of lysosome-targeted TORCAR (Lyso-TORCAR). (B) Image ofNIH3T3 cells expressing Lyso-TORCAR, co-stained with LysoTracker Red. Scale bar = 10 μm. (C) Lyso-TORCAR responded to LeuOMe (mean response ± SEM as 4.6 ± 2.5%, n = 5). (D) PDGF addition induced a Lyso-TORCAR response (mean response ± SEM as 6.8 ± 2.1%, n = 8). (E) Pretreating double-starved NIH3T3 cells with the Akt inhibitor, 10-DEBC (30 μM, 5 min), abolished the PDGF-induced Lyso-TORCAR response (n = 4). See also Figure S4.
We first tested the response of Lyso-TORCAR to leucine methyl ester, which diffuses freely across membranes and is hydrolyzed to native leucine within lysosomes. Consistent with the notion that leucine activates mTORC1 at the lysosome, a 4.6 ± 2.5% response was observed in double-starved NIH3T3 cells (Figure 4C, n = 5). The negative control, Lyso-TORCAR-TA (T37/46A) (Figure S4A) failed to respond to leucine ester, suggesting that the response is phosphorylation-dependent (Figure S4B, n = 4). Lyso-TORCAR also responded to a total amino acid mixture and leucine in double starved or amino acid starved 3T3 cells (Figure S4C–F).
In contrast to the mechanisms of activation by amino acids, which involves the Rag complex, growth factors activate mTORC1 largely via Akt/TSC/Rheb signaling (Laplante and Sabatini, 2009). Recently it has been shown that the TSC complex localizes to the lysosome (Demetriades et al., 2014; Dibble et al., 2012; Menon et al., 2014), where subpopulations of Rheb and mTOR are also present (Ohsaki et al., 2010; Sancak et al., 2010). We then asked whether Lyso-TORCAR could detect growth factor-induced mTORC1 activity at this location. As shown in Figure 4D, PDGF treatment indeed stimulated a 6.8 ± 2.1% response in double-starved NIH3T3 cells expressing Lyso-TORCAR (n = 8), indicating PDGF induced mTORC1 activity on the lysosomal outer membrane. The T-to-A mutation, Lyso-TORCAR-TA (T37/46A), abolished the response (Figure S4G, n = 3).
This GF-induced lysosomal mTORC1 activity could result either from the translocation of mTORC1 activated elsewhere or from a subpool of lysosome-resident mTORC1 being activated locally via the Akt/TSC1/2/Rheb pathway. To test whether lysosomal Akt activity is contributing to local mTORC1 activity, we monitored Akt activity on the lysosomal membrane using a lysosomal targeted Akt activity reporter, Lyso-AktAR2 (Figure S4A). Lyso-AktAR2 was constructed by tagging LAMP1 to the N-terminus of the improved Akt activity reporter, AktAR2. When expressed, Lyso-AktAR2 exhibited punctate fluorescence in the cytosol that co-localized with LysoTracker Red (Figure S4H). PDGF stimulated a 6.0 ± 2.6% response from the lysosomal Akt reporter (Figure S4I, n = 4), whereas the T/A negative control of the lysosomal Akt reporter showed no response (Figure S4J, n = 3), suggesting that PDGF induced Akt activity on the lysosomal surface. Pretreatment with an Akt specific inhibitor, 10-DEBC at the concentration of 30μM, inhibited the lysosomal Akt activity induced by PDGF (Figure S4K, n = 3). Importantly, the PDGF-induced Lyso-TORCAR response was largely inhibited as well (Figure 4E, n = 4). Similar results were also observed when another Akt inhibitor, SH-5, was used (Figure S4L and S4M). Collectively, these results implied that growth factors efficiently and acutely induced mTORC1 activity increases at the lysosome and that these increases are, at least in part, mediated by lysosomal Akt activity. These data are consistent with the newly identified role of Akt in phosphorylating TSC2, leading to the dissociation of the TSC complex from the lysosome and the activation of mTORC1 at this location (Menon et al., 2014).
GF induced mTORC1 activity at the plasma membrane
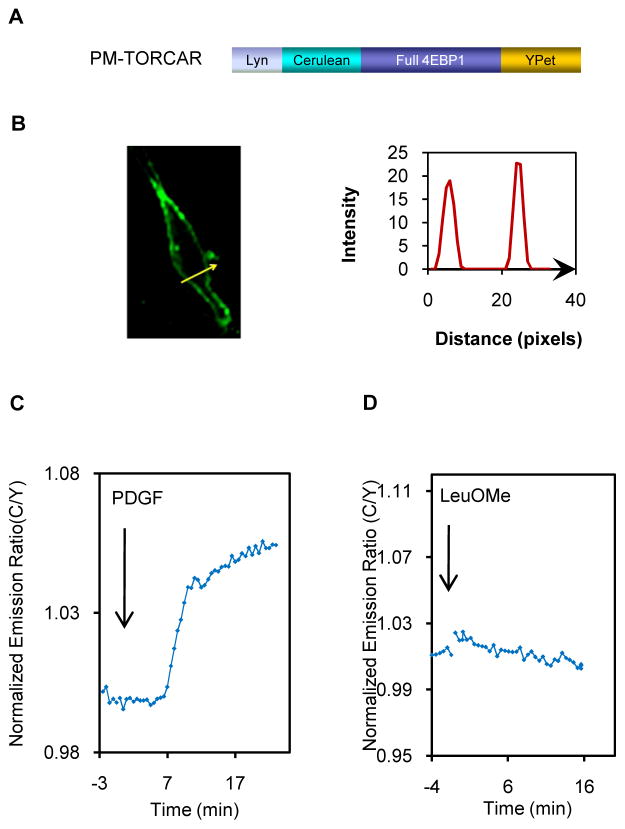
Upon GF stimulation, Akt is recruited to the plasma membrane through the specific binding of its PH domain to the accumulating PIP3 at the membrane, and this translocation event has been shown to be critical for Akt activation. Hence, we asked whether GF specifically stimulated mTORC1 activity at the plasma membrane. Previously, subcellular fractionation of endothelial cells revealed the presence of mTOR and Raptor in plasma membrane rafts (Partovian et al., 2008), which are cholesterol-rich, detergent-insoluble microdomains known to function as pivotal signaling platforms (Hanzal-Bayer and Hancock, 2007). Whereas TSC2 may be present at the plasma membrane (Wienecke et al., 1995), the plasma membrane localization of the proximal activator of mTORC1, Rheb, is unclear. Rheb has been suggested to localize throughout the endomembrane system but not at the plasma membrane, due to its lack of either palmitoylation sites or polybasic-rich sequences for plasma membrane targeting (Takahashi et al., 2005). On the other hand, an earlier study reported that Rheb is associated with the plasma membrane in NIH3T3 cells and enriched in the apparent membrane ruffle regions (Clark et al., 1997). We also found Rheb in membrane raft fractions from either plasma membrane or endomembrane (Figure S5A). To address whether the mTORC1 activity is present at the plasma membrane, we constructed TORCAR targeted to the plasma membrane (PM-TORCAR) by fusing the Lyn-targeting motif to the N-terminus of TORCAR (Gao and Zhang, 2008) (Figure 5A). When expressed in cells, PM-TORCAR is properly targeted to the plasma membrane, with the majority of fluorescence detected at the cell membrane(Figure 5B). Upon PDGF stimulation, PM-TORCAR showed a robust response (7.6 ± 3.3%, n = 30) (Figure 5C), whereas the T-to-A mutant of PM-TORCAR (Figure S5B) showed no response (Figure S5C, n = 3). Whereas it is still unclear if the TSC/Rheb pathway mediates the plasma membrane mTORC1 activity, it is noteworthy that another player, Ral GTPase, has been recently suggested to regulate the serum-induced translocation of mTORC1 to the plasma membrane (Martin et al., 2014), providing a possible mechanism underlying plasma membrane mTORC1 activity. Interestingly, Lyn-TORCAR showed no response to leucine methyl ester (Figure 5D, n = 3), indicating that the mTORC1activity detected at the plasma membrane is GF specific.
Figure 5.
mTORC1 activity at the plasma membrane. (A) Domain structures of plasma membrane-targeted TORCAR. (B) Confocal image of an NIH3T3 cell expressing PM-TORCAR. Line scans showed the intensity of PM-TORCAR across the plasma membrane (n = 3). Scale bar = 10 μm. (C) PM-TORCAR responded to PDGF (mean response ± SEM as 7.6 ± 3.3%, n = 30). (D) PM-TORCAR showed no response to leucine ester (n = 3). See also Figure S5 and Movie S1.
mTORC1activity is present in the nucleus
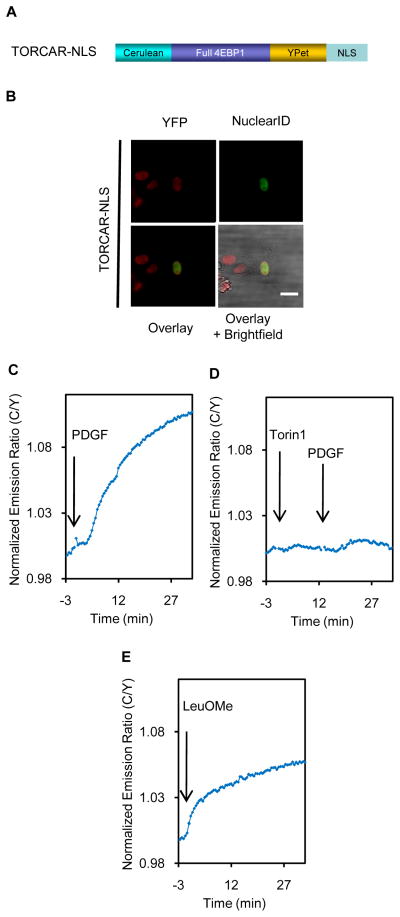
Many reports have suggested the presence of mTOR and Raptor (mTORC1), as well as the downstream effector S6K in the nucleus (Betz and Hall, 2013). In addition, the functional relevance of the nuclear mTORC1 has also been indicated (Workman et al., 2014). However, it is unclear whether functional endogenous mTORC1 exists in the nucleus. In fact, mTORC1 is considered to be non-nuclear and the major function of mTORC1 signaling is largely considered to be the result of mTOR kinase activity towards cytoplasmic substrates (Betz and Hall, 2013; Rosner and Hengstschlager, 2012). To probe the nuclear mTORC1 activity, we constructed a nuclear-targeted TORCAR (TORCAR-NLS) by tagging a nuclear localization sequence (NLS) to the C-terminus of TORCAR (Ananthanarayanan et al., 2005) (Figure 6A). TORCAR-NLS is localized exclusively in the nucleus when expressed in NIH3T3 cells, as indicated by co-staining with Nuclear-ID Red (Figure 6B). PDGF induced a 10 ± 4% response with a t1/2 of 13.3 ± 2.6min (n = 11), suggesting that there is a pool of mTORC1activity in nucleus (Figure 6C). Pretreatment with torin1 abolished the PDGF-induced response, indicating that the response is mTOR-specific (Figure 6D, n = 4). Curiously, leucine ester induced a similar response to that of PDGF (Figure 6E, 8.2 ± 1.8%, n = 7). These data suggested that both growth factor and amino acid signaling can induce mTORC1 activity in the nucleus, with kinetics on par with the cytosolic accumulation of mTORC1 activity.
Figure 6.
Nuclear mTORC1 activity. (A) Domain structures of nuclear-targeted TORCAR (TORCAR-NLS). (B) Image of anNIH3T3 cell expressing TORCAR-NLS. Scale bar = 10 μm. (C) TORCAR-NLS responded to PDGF with a mean response ± SEM as 10.2 ± 4.1% and mean t1/2 ± SEM as 13.3 ± 2.6 min (n = 11). (D) Pretreating cells with torin1 (1μM, 10 min) abolished the PDGF-induced TORCAR-NLS response (n = 4). (E) LeuOMe induced a TORCAR-NLS response (mean response ± SEM as 8.2 ± 1.8%, n = 7). See also Movie S2.
DISCUSSION
Distinct upstream signals impinge on mTORC1, which controls cell growth and metabolism. A critical question is how this complex senses these different inputs and transduces the specific signals to precisely control cell growth. The lysosomal activation of mTORC1 highlighted the signal- and location-specific regulation of mTORC1 and prompted us to examine the spatial regulation of mTORC1 activity. To achieve this goal, we needed a molecular tool that allowed for sensitive detection of the activity of the intact complex at different subcellular locations in live cells. We therefore developed a first-generation mTORC1 activity reporter, TORCAR. When expressed in cells, TORCAR responded to different stimuli, such as growth factor, total amino acid mixture, leucine, and PMA, suggesting that TORCAR can be used as a molecular tool to monitor endogenous mTORC1 activity. TORCAR offers several unique advantages. TORCAR can be genetically targeted to any specific subcellular location and is not restricted by the localization of endogenous substrates, thus allowing a more comprehensive mapping of mTORC1 activity. Genetic targeting also helps achieve high spatial resolution and specific detection of mTORC1 activities in subcellular regions, some of which may not be easily distinguished by standard microscopy (Mehta and Zhang, 2011), for example, signaling domains such as membrane microdomains and multiprotein complexes. In addition, TORCAR provides high temporal resolution in detecting the activity. In the co-imaging experiments, we showed that a PDGF-induced calcium spike immediately precedes the onset of TORC1 activity; this kind of temporal information is difficult to obtain using conventional approaches such as immunostaining. With a ratiometric readout of TORCAR, variations in illumination and cell thickness, etc, can also be cancelled out to provide more quantitative data, which allows us to compare the activities stimulated by different signals. Last but not least, TORCAR allows dynamic information to be obtained from the contexts of living cells, which avoids potential artifacts introduced by cell fixation. In summary, TORCAR complements the current approaches based on phosphorylation-specific antibodies and should be of great value to the field. Future efforts will further enhance TORCAR for example by improving its dynamic range, as shown in the case of other kinase activity reporters (Depry et al., 2010; Gao and Zhang, 2008).
There is a longstanding interest in investigating the link between calcium signaling and mTORC1 activation (Altamirano et al., 2009; Ghislat et al., 2012; Gulati et al., 2008; Mercan et al., 2013). Many possible links have been suggested, yet the exact role of calcium in mTORC1 activation remains to be elucidated. Downstream of calcium, several potential mTORC1 regulators and mechanisms have been proposed, including Ca2+/CaM, PI(3)P/hVps34, PA/PLD, PI(3,5)P2/PIKFYVE, and PI3K-C2α. For example, it has been suggested in HeLa cells that amino acids induced an intracellular calcium increase, which activates calmodulin and in turn promotes its binding to hVps34, a class III PI3K. The binding of Ca2+/CaM to hVps34 enhances the production of PI-3-phosphate, which is required for mTORC1 activation (Gulati et al., 2008).
It has also been indicated that calcium may regulate the activity of phospholipase D (PLD), thereby providing positive inputs to mTOR (Fang et al., 2001; Razmara et al., 2013). By co-imaging TORCAR with a calcium probe, we demonstrated that in double-starved NIH3T3 cells, PDGF induced a transient calcium spike that, most likely originates from intracellular calcium stores and partially contributes to the mTORC1 activity. Upstream of mTORC1, Akt activity appeared to be largely dependent on calcium as well. We show that intracellular calcium chelation attenuated the phosphorylation of Akt S473, but had no effect on the phosphorylation of Akt T308, which is consistent with previous findings (Farrer et al., 2010; Razmara et al., 2013). It has been shown that the S473 phosphorylation is less critical for regulating Akt activity towards TSC2 (Jacinto et al., 2006), which may explain the reduced effect of chelation of intracellular calcium onmTORC1 activity (40% reduction) compared with the effect on Akt activity (70% reduction).
TORCAR was successfully targeted to various subcellular locations, including the lysosome, plasma membrane and nucleus, where it detected multiple pools of mTORC1 activity. It appears that membrane compartments serve as specific signaling platforms for mTORC1 activation, as the direct and potent mTORC1 activator, Rheb, localizes to most endomembranes and anchors itself by farnesylation (Buerger et al., 2006; Takahashi et al., 2005). In many such locations, TSC1/2 is also present (Wienecke et al., 1995), implying that the Akt/TSC/Rheb axis may exist and spatially regulate mTORC1 signaling. Particularly at the lysosome, we detected Akt activity in response to PDGF, and blockade of this Akt activity significantly diminished lysosomal mTORC1 activity. This suggests that Akt, which either localized to the lysosomal surface or translocated there upon PDGF stimulation, is an indispensable signal to fully activate mTORC1.
Recently, Menon et al reported that the insulin-induced dissociation of the TSC complex from lysosomes requires Akt-mediated TSC2 phosphorylation (Menon et al., 2014). Our finding provides evidence that increased lysosomal Akt activity is critical for GF-induced lysosomal mTORC1 activity. It remains unclear whether GF signaling recruits Akt to the lysosome or a pool of lysosomal Akt is poised in situ to activate mTORC1.
We detected mTORC1 activity in the plasma membrane upon GF stimulation. Although there is still conflicting data on the presence of Rheb in the plasma membrane, a recent study by Martin et al suggested that Ral GTPase may also regulate mTORC1 signaling at the plasma membrane. This study showed that serum-induced mTOR recruitment to the plasma membrane is dependent on RalB GTPase, and that suppression of the RalB GTPase signaling at the plasma membrane reduced the serum-induced mTOR translocation to plasma membrane and mTOR-dependent phosphorylation of S6K. Although plasma membrane mTORC1 activity was not directly probed in this study, the importance of a plasma membrane pool of RalB in regulating mTOR membrane translocation and substrate phosphorylation may suggest an alternative model of mTORC1 activation, which could account for the mTORC1 activity in the plasma membrane observed in our study. Further studies would examine the mechanisms underlying the plasma membrane mTORC1 activity, whether the TSC complex dissociates from the plasma membrane upon the phosphorylation of TSC2 by Akt, as does lysosomal mTORC1 (Menon et al., 2014), or whether the Ral GAP/Ral pathway contributes more to the plasma membrane mTORC1 activity.
This study also identified the nucleus as a new location where both growth factor and amino acid stimulation can induce an increase in mTORC1 activity. Whether mTORC1 is functional and present in the nuclear compartment was a topic of debate. For instance, it has been suggested that there is little intact mTORC1 in the nucleus due to low affinity of nuclear raptor to mTOR than cytoplasmic raptor (Rosner and Hengstschlager, 2008). On the other hand, nuclear mTORC1 signaling has been suggested to play a role in some cellular processes, including promoting the transcription of metabolic genes for ribosome biogenesis, lipid formation, and mitochondrial function (Back and Kim, 2011; Workman et al., 2014). For instance, it has been shown that mTOR binds to TFIIIC, a transcription factor that binds to pol III promoters, leading to enhanced protein synthesis in response to nutrients and growth factors (Kantidakis et al., 2010). As we have shown, both GF and AA stimulation can lead to the accumulation of mTORC1 activity in the nucleus, highlighting the presence of functional mTORC1 in this location. However, the similar kinetics between nuclear TORCAR and diffusible TORCAR does not allow us to distinguish between the two models of nuclear mTORC1 activation – mTORC1 could either be activated in the nucleus as an independent pool, or it could be translocated to the nucleus after activation in the cytosol or another location.
Notably, whereas GF widely stimulates mTORC1 activity across different subcellular locations including the plasma membrane, lysosome, and nucleus, leucine ester-induced mTORC1 activity is more restricted to the lysosome and nucleus. This signal-specific pattern has important implications. First, it suggests that Akt, which is also widely distributed throughout the cell, may mediate the GF-sensitive pathway, whereas the AA-sensing machinery, which has been characterized for the lysosome but has not been explored at other locations, is restricted to specific subcellular compartments. Furthermore, it has been suggested the intracellular nutrients contributes to the basal activation of mTORC1, but further activation requires GF (Dibble and Manning, 2013; Hara et al., 1998). Our results suggested such basal activation may occur in a subset of compartments, whereas GF-stimulated further activation induces more global effects. Intriguingly, the lysosome and nucleus appear to be the sites where the two pathways converge, underscoring the importance of the mTORC1 signaling control within these two compartments. Further delineation of the molecular mechanisms by which mTORC1 is regulated by specific signals will advance our understanding of this signaling network and facilitate the identification of potential targets for treating human diseases that involve aberrant mTORC1 signaling.
EXPERIMENTAL PROCEDURES
Detailed experimental procedures are provided in the Extended Experimental Procedures.
Constructs
TORCAR was generated by sandwiching full-length 4EBP1 between a FRET pair, Cerulean and YPet. Details regarding subcellularly targeted TORCAR constructs are provided in the Extended Experimental Procedures. All constructs were verified by sequencing after subcloning into a modified version of the mammalian expression vector pcDNA3.
Live-Cell Imaging
Cells were washed once with modified HBSS and imaged in the dark at room temperature. Images were acquired on a Zeiss Axiovert 200M microscope with a cooled charge-coupled device camera, as previously described (Ananthanarayanan et al., 2005). Dual-emission ratio imaging was performed with a 420DF20 excitation filter, a 450DRLP dichroic mirror, and two emission filters, 475DF40 and 535DF25 for CFP and YFP, respectively. For RFP, a 568DF55 excitation filter, a 600DRLP dichroic mirror, and a 653DF95 emission filter were used. Exposure times were 50–500ms and images were taken every 30 s. Imaging data was analyzed with Metafluor 6.2 software (Universal Imaging, Downingtown, PA). Fluorescence images were background-corrected by deducting the background (regions with no cells) from the emission intensities of CFP or YFP. Regions of interest (ROIs) at the cell periphery representing the plasma membrane were used for analysis for PM-TORCAR. Traces were normalized by setting the emission ratio before addition of drugs as 1.
Statistical Analysis
Data are shown as means ± SEM. Means were compared by two-tailed t test.
Supplementary Material
Acknowledgments
We thank Nathanael S. Gray (Harvard University) for providing torin1, Gary C. H. Mo and Qiang Ni (Johns Hopkins University) for expert technical assistance. We also thank Carolyn Machamer and David Graham for discussions about the membrane fractionation experiments and Kirill Gorshkov and Sohum Mehta for careful reading of the manuscript. This work was supported by NIH grants R01 DK073368 (to J.Z.), DP1 CA174423 (to J.Z.) and DK084171 (to G.W.W.).
Footnotes
AUTHOR CONTRIBUTIONS
X.Z., T.L.C., P.R.L., M.S., G.W.W., and J.Z. designed research; X.Z., T.L.C., P.R.L., and M.S. performed research; X.Z., T.L.C., P.R.L., M.S., G.W.W., and J.Z. analyzed data; X.Z. and J.Z. wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolo J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schuler C, Chen TW, Sarkisyan KS, Marvin JS, Bargmann CI, Kim DS, Kugler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER, Looger LL. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altamirano F, Oyarce C, Silva P, Toyos M, Wilson C, Lavandero S, Uhlen P, Estrada M. Testosterone induces cardiomyocyte hypertrophy through mammalian target of rapamycin complex 1 pathway. J Endocrinol. 2009;202:299–307. doi: 10.1677/JOE-09-0044. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan B, Ni Q, Zhang J. Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15081–15086. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back JH, Kim AL. The expanding relevance of nuclear mTOR in carcinogenesis. Cell Cycle. 2011;10:3849–3852. doi: 10.4161/cc.10.22.18329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 2012;150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja V, Alcor D, Laguerre M, Park J, Vojnovic B, Hemmings BA, Downward J, Parker PJ, Larijani B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. The Journal of biological chemistry. 1997;272:10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA. Regulation of TORC1 in Response to Amino Acid Starvation via Lysosomal Recruitment of TSC2. Cell. 2014;156:786–799. doi: 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depry C, Allen MD, Zhang J. Visualization of PKA activity in plasma membrane microdomains. Molecular bioSystems. 2010;7:52–58. doi: 10.1039/c0mb00079e. [DOI] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Molecular cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nature cell biology. 2013;15:555–564. doi: 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd KM, Tee AR. Leucine and mTORC1: a complex relationship. Am J Physiol Endocrinol Metab. 2011;302:E1329–1342. doi: 10.1152/ajpendo.00525.2011. [DOI] [PubMed] [Google Scholar]
- Fadden P, Haystead TA, Lawrence JC., Jr Identification of phosphorylation sites in the translational regulator, PHAS-I, that are controlled by insulin and rapamycin in rat adipocytes. The Journal of biological chemistry. 1997;272:10240–10247. doi: 10.1074/jbc.272.15.10240. [DOI] [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Farrer RG, Farrer JR, DeVries GH. Platelet-derived growth factor-BB activates calcium/calmodulin-dependent and -independent mechanisms that mediate Akt phosphorylation in the neurofibromin-deficient human Schwann cell line ST88–14. The Journal of biological chemistry. 2010;288:11066–11073. doi: 10.1074/jbc.M112.442244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher CM, Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang J. Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Biol Cell. 2008;19:4366–4373. doi: 10.1091/mbc.E08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislat G, Patron M, Rizzuto R, Knecht E. Withdrawal of essential amino acids increases autophagy by a pathway involving Ca2+/calmodulin-dependent kinase kinase-beta (CaMKK-beta) The Journal of biological chemistry. 2012;287:38625–38636. doi: 10.1074/jbc.M112.365767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes & development. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin P, Oulhen N, Jam M, Ronzca J, Cormier P, Czjzek M, Cosson B. The translational repressor 4E-BP called to order by eIF4E: new structural insights by SAXS. Nucleic Acids Res. 2010;39:3496–3503. doi: 10.1093/nar/gkq1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. The Journal of biological chemistry. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Heesom KJ, Avison MB, Diggle TA, Denton RM. Insulin-stimulated kinase from rat fat cells that phosphorylates initiation factor 4E-binding protein 1 on the rapamycin-insensitive site (serine-111) Biochem J. 1998;336 (Pt 1):39–48. doi: 10.1042/bj3360039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends in biochemical sciences. 2013;38:233–242. doi: 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature cell biology. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markwardt ML, Kremers GJ, Kraft CA, Ray K, Cranfill PJ, Wilson KA, Day RN, Wachter RM, Davidson MW, Rizzo MA. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TD, Chen XW, Kaplan RE, Saltiel AR, Walker CL, Reiner DJ, Der CJ. Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Molecular cell. 2014;53:209–220. doi: 10.1016/j.molcel.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Zhang J. Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annual review of biochemistry. 2011;80:375–401. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. Spatial Control of the TSC Complex Integrates Insulin and Nutrient Regulation of mTORC1 at the Lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercan F, Lee H, Kolli S, Bennett AM. Novel Role for SHP-2 in Nutrient-Responsive Control of S6 Kinase 1 Signaling. Mol Cell Biol. 2013;33:293–306. doi: 10.1128/MCB.01285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q, Titov DV, Zhang J. Analyzing protein kinase dynamics in living cells with FRET reporters. Methods. 2006;40:279–286. doi: 10.1016/j.ymeth.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Suzuki M, Shinohara Y, Fujimoto T. Lysosomal accumulation of mTOR is enhanced by rapamycin. Histochem Cell Biol. 2010;134:537–544. doi: 10.1007/s00418-010-0759-x. [DOI] [PubMed] [Google Scholar]
- Partovian C, Ju R, Zhuang ZW, Martin KA, Simons M. Syndecan-4 regulates subcellular localization of mTOR Complex2 and Akt activation in a PKCalpha-dependent manner in endothelial cells. Molecular cell. 2008;32:140–149. doi: 10.1016/j.molcel.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razmara M, Heldin CH, Lennartsson J. Platelet-derived growth factor-induced Akt phosphorylation requires mTOR/Rictor and phospholipase C-gamma1, whereas S6 phosphorylation depends on mTOR/Raptor and phospholipase D. Cell Commun Signal. 2013;11:3. doi: 10.1186/1478-811X-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves JP. Accumulation of amino acids by lysosomes incubated with amino acid methyl esters. The Journal of biological chemistry. 1979;254:8914–8921. [PubMed] [Google Scholar]
- Rosner M, Hengstschlager M. Cytoplasmic and nuclear distribution of the protein complexes mTORC1 and mTORC2: rapamycin triggers dephosphorylation and delocalization of the mTORC2 components rictor and sin1. Human molecular genetics. 2008;17:2934–2948. doi: 10.1093/hmg/ddn192. [DOI] [PubMed] [Google Scholar]
- Rosner M, Hengstschlager M. Detection of cytoplasmic and nuclear functions of mTOR by fractionation. Methods Mol Biol. 2012;821:105–124. doi: 10.1007/978-1-61779-430-8_8. [DOI] [PubMed] [Google Scholar]
- Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13489–13494. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, Yamanaka S. Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. The Journal of biological chemistry. 2005;280:32768–32774. doi: 10.1074/jbc.M506280200. [DOI] [PubMed] [Google Scholar]
- Tato I, Bartrons R, Ventura F, Rosa JL. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. The Journal of biological chemistry. 2011;286:6128–6142. doi: 10.1074/jbc.M110.166991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee AR, Anjum R, Blenis J. Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. The Journal of biological chemistry. 2003;278:37288–37296. doi: 10.1074/jbc.M303257200. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. The Journal of biological chemistry. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou P, Zheng B, Hsu CH, Sasaki AT, Cantley LC. A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab. 2011;13:476–486. doi: 10.1016/j.cmet.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienecke R, Konig A, DeClue JE. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. The Journal of biological chemistry. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- Workman JJ, Chen H, Laribee RN. Environmental signaling through the mechanistic target of rapamycin complex 1-mTORC1 goes nuclear. Cell Cycle. 2014;13 doi: 10.4161/cc.28112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ma Y, Taylor SS, Tsien RY. Genetically encoded reporters of protein kinase A activity reveal impact of substrate tethering. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Herbst-Robinson KJ, Zhang J. Visualizing dynamic activities of signaling enzymes using genetically encodable FRET-based biosensors from designs to applications. Methods Enzymol. 2012;504:317–340. doi: 10.1016/B978-0-12-391857-4.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.