Abstract
Nucleoplasmin (Npm) is an abundant histone chaperone in vertebrate oocytes and embryos. During embryogenesis, regulation of Npm histone binding is critical for its function in storing and releasing maternal histones to establish and maintain the zygotic epigenome. Here we demonstrate that Xenopus laevis Npm post-translational modifications (PTMs) specific to the oocyte and egg promote either histone deposition or sequestration, respectively. Mass spectrometry and Npm phosphomimetic mutations used in chromatin assembly assays identified hyperphosphorylation on the N-terminal tail as a critical regulator for sequestration. C-terminal tail phosphorylation and PRMT5-catalyzed arginine methylation enhance nucleosome assembly by promoting histone interaction with the second acidic tract of Npm. Electron microscopy reconstructions of Npm and TTLL4 activity towards the C-terminal tail demonstrate that oocyte- and egg-specific PTMs cause Npm conformational changes. Our results reveal that PTMs regulate Npm chaperoning activity by modulating Npm conformation and Npm-histone interaction leading to histone sequestration in the egg.
INTRODUCTION
During early Xenopus laevis embryogenesis, rapid and synchronous cell division occurs in the absence of transcription. Activation of the zygotic genome is concomitant with the mid-blastula transition (MBT) (Almouzni and Wolffe, 1995; Newport and Dasso, 1989). This transcriptional quiescence necessitates that the cells survive solely on the maternally stored proteins and mRNAs, including histones (Sun et al., 2014). Regulation of the switch from storage to deposition of histones is critical for maintaining the pool of stored histones and simultaneously supporting rapid genome replication. The regulation between histone binding and release is therefore essential for establishing and maintaining the zygotic epigenome.
Nucleoplasmin (Npm; encoded by the npm2a and npm2b alloallelic genes) is a histone chaperone for histones H2A-H2B and is highly expressed in the oocyte and through the early stages of embryogenesis (Bouleau et al., 2014; Litvin and King, 1988). Its high concentration led to the hypothesis that Npm stores histones H2A-H2B in the egg (Finn et al., 2012; Keck and Pemberton, 2013). Npm is one of three Npm family members found in vertebrates (Finn et al., 2012). Npm forms a stable homopentamer comprised of individual 22 kDa subunits, and its hydrophobic core domain (amino acids 16–120) is responsible for pentamerization and extreme heat stability (Dutta et al., 2001), while the N- and C-termini are disordered (Bañuelos et al., 2003; Dutta et al., 2001). Npm contains three acidic tracts: A1, A2, and A3. The C-terminal intrinsically disordered domain contains a bipartite nuclear localization sequence, A2 and A3, and the extreme C-terminus containing positive amino acids (Dutta et al., 2001; Prado et al., 2004). Previous biochemical and electron microscope analyses revealed that the core is sufficient to bind histones, but the tail also engages in histone binding (Arnan et al., 2003; Ramos et al., 2014; Ramos et al., 2010; Taneva et al., 2009). The functional significance of the tail binding is unknown. Npm is extensively post-translationally modified (PTM).
Npm is phosphorylated during oogenesis and hyperphosphorylated upon progesterone-induced meiosis II (Banuelos et al., 2007; Cotten et al., 1986; Leno et al., 1996; Sealy et al., 1986; Tamada et al., 2006; Taneva et al., 2008). This hyperphosphorylation is critical for sperm DNA decondensation and protamine removal (Banuelos et al., 2007; Leno et al., 1996). Npm with Ser to Asp phosphomimetic mutations on predicted, but not known, phosphorylation sites showed an increase in affinity for histones H2A-H2B (Taneva et al., 2009). We previously showed that PRMT5 methylates Npm on its C-terminus (Wilczek et al., 2011). Glutamylation, an isopeptide addition of a glutamic acid to the γ-carboxyl of a primary chain glutamate residue occurs on the Npm-family member Nucleophosmin (Npm1) (van Dijk et al., 2008). Glutamylation is also found on histone chaperone Nap1 (Regnard et al., 2000) and was originally identified in tubulin (Edde et al., 1990; Janke et al., 2008), where it was shown to recruit binding partners (Sirajuddin et al., 2014).
An earlier comparison of histone deposition on plasmid DNA by oocyte Npm (oNpm) and egg Npm (eNpm) demonstrated specific Npm nucleosome assembly in the egg (Cotten et al., 1986; Sealy et al., 1986). This observation contrasted starkly with the hypothesis that Npm stores histones and suggested that Npm PTMs may regulate histone storage.
Here, we show that Npm is developmentally modified to regulate its function in histone storage and release. We present high-resolution mass spectrometry analysis revealing Npm arginine methylation and glutamylation on the C-terminal flexible tail and phosphorylation on both N- and C-terminal tails. Npm purified from the egg sequestered histones both from DNA and from another histone chaperone Nap1. Through the use of phosphomimetic mutations and PRMT5 methyltransferase treatment of Npm, we show that N- and C-terminal PTMs promote sequestration and deposition, respectively. Our TTLL4 glutamyltransferase treatment and electron microscope reconstruction of rNpm, oNpm, and eNpm demonstrates substantial PTM-dependent conformational changes and our biochemical analyses show that regulated acidic patch accessibility is a significant contributor to histone deposition activity. Our results support a model in which developmentally-regulated Npm PTMs alter its conformation and its histone deposition activity to balance the requirements of maternal histone storage and rapid zygotic genome duplication.
RESULTS
Nucleoplasmin is modified in a developmentally regulated fashion
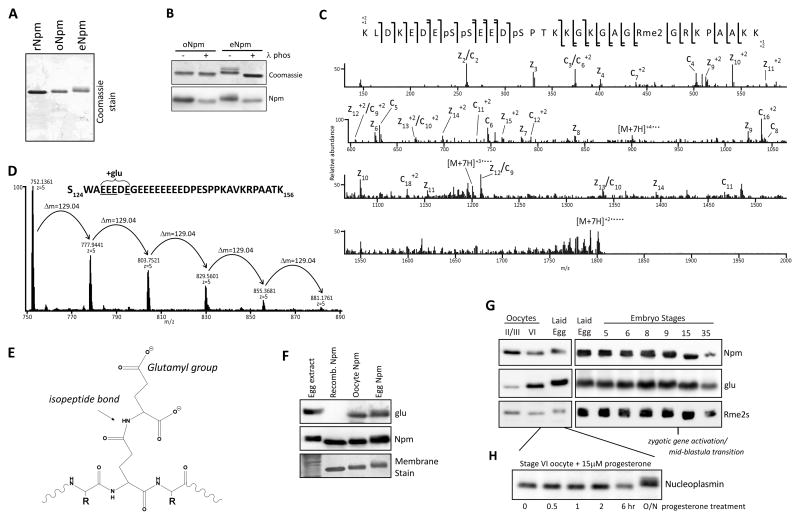
To test developmental Npm expression we isolated total protein from Xenopus laevis embryos and adult tissues. Immunoblots of these total lysates confirmed that Npm is only present in the early embryos and the ovaries and not in any other differentiated tissues (Figure S1A). We observed a distinct Npm mobility shift in lysates starting at the laid egg and lasting until Stage 9, concomitant with the MBT and the onset of zygotic gene expression (Figure S1B)(Cotten et al., 1986). We purified intact, non-proteolyzed rNpm pentamer by enrichment for full-length protein on MonoS column and isolated Npm from Xenopus laevis surgically extracted oocytes and laid eggs (Figure 1A and Figure S1C). Both purified oNpm and eNpm showed a mobility shift compared to rNpm that was relieved by λ-phosphatase treatment (Figure 1B). These results confirmed that oNpm is phosphorylated and eNpm is hyperphosphorylated (Cotten et al., 1986).
Figure 1. Npm PTM identification and characterization.
A. Purified rNpm, oNpm, and eNpm. oNpm and eNpm both show mobility shift compared to rNpm. B. λ-phosphatase treatment of purified oNpm and eNpm. The collapsed gel retardation confirms the shift is due to phosphorylation. C. Low resolution ETD MS/MS spectrum of the [M+7H]7+ charge state of K165-K195 peptide of oNpm. Complete localization of all PTMs is apparent from the sequence coverage map (top). D. Averaged full MS spectrum of Npm S124-K156 from oocyte displaying the distribution of polyglutamylation within the region of E127-E131. Δm=129.04 Da indicates an addition of a single glutamyl group. E. Chemical structure of a glutamylated peptide on a backbone glutamate reveals the increased and wider distribution of negative charge. F. Immunoblot of egg extract and purified rNpm, oNpm, and eNpm confirms glutamylation on oNpm and eNpm in vivo. G. Immunoblot of oocyte, laid egg, and embryo lysates probing for glutamylation and arginine symmetric dimethylation. H. Hyperphosphorylation of Npm occurred after overnight incubation of oocytes in progesterone to promote maturation and GVBD. Also See Figure S1.
We identified the precise PTMs on purified oNpm and eNpm by mass spectrometry. Treatment of Npm with chymotrypsin yielded peptides of appropriate size and hydrophobicity for reversed-phase HPLC separations and mass spectrometric analyses. We obtained complete sequence coverage of the entire protein, including what we determined to be the three most heavily modified regions: the N-terminus, the C-terminus, and within the second acidic patch (Figure S2A). Npm2a and npm2b gene products have 11 modest amino acid alterations and a four amino acid deletion in the second acidic patch (Figure S2B). We observed that peptides from the shorter gene product, Npm2B (196 amino acids), were between 5 and 7 times more abundant than Npm2A. All subsequent mass spectrometry probed peptides from Npm2B (hereafter referred to as Npm).
High resolution MS1 analysis of the major C-terminal tryptic peptide revealed a distribution species related to one another by multiples of 79.9663 Da and 14.01565 Da, indicative of variable phosphorylation and methylation, respectively (Figure 1C). Electron-transfer dissociation (ETD) MS/MS sequencing revealed S172, S173 and S177 as the phosphorylated residues and arginine 187 as the dimethylated residue.
N-terminal phosphorylation increased between the two biological states, from a majority unphosphorylated in the oocyte sample to 50% doubly phosphorylated and 25% triply phosphorylated in the egg sample (Figure S2C,D). Sequencing of the N-terminal peptides by both ETD and CAD MS/MS revealed that S15 was never phosphorylated, and that the majority of the phosphorylations were distributed over S5, T7 and S8. The major forms of the C-terminal region of Npm from oocyte and egg contained two phosphorylations (S172 and S173), and three phosphorylations (S172, S173 and S177), respectively. Serine 144 was nearly 100% hyperphosphorylated in the egg. S144 and S177 are well conserved between Xenopus and human Npm (Figure S2E). Phosphorylation on threonine 96 on the peptide V91-F101 was <1% in both the oocyte sample and the egg sample and did not have significant change in abundance between the two samples. We also observed full N-terminal acetylation. In both oocyte and egg Npm, R187 dimethylation was roughly twice the abundance of monomethylation with approximately 5% unmodified.
A pattern of multiple 129.04 Da additions observed on the oocyte derived peptide S124-K156 indicated glutamylation (Figure 1D,E). The major form of this peptide was roughly 60% monoglutamylated relative to the unmodified form, and contained up to five total glutamylations in decreasing relative abundance. ETD MS/MS analysis localized these modifications to the glutamate residues within the region A126EEEDEG132. To further confirm the presence of glutamylation, we probed egg extract, rNpm, oNpm and eNpm by immunoblot using antibodies raised against glutamylated β-tubulin fragments (Spano and Frankfurter, 2010). Two of these antibodies, TTSG1 and TT-βIII, recognized oNpm and eNpm but not rNpm, confirming the glutamylation of Npm in vivo (TT-βIII was used in most experiments and labeled as “glu,” Figure 1F, S2F).
We observed simultaneous glutamylation and S144 phosphorylation in oocytes and eggs (Figure S2G). Immunoblot analysis on embryo lysates of different stages confirmed Npm phosphorylation, glutamylation and arginine methylation (Figure 1G). Arginine methylation occurred early in oogenesis at stages II/III, whereas glutamylation was enriched later in stage VI oocytes (Figure 1G). Embryos probed for Npm modifications showed the persistence of arginine methylation and glutamylation past the MBT. We also confirmed that hyperphosphorylation, detected as gel retardation, occurs upon germinal vesicle breakdown (GVBD) after progesterone treatment (Figure 1H).
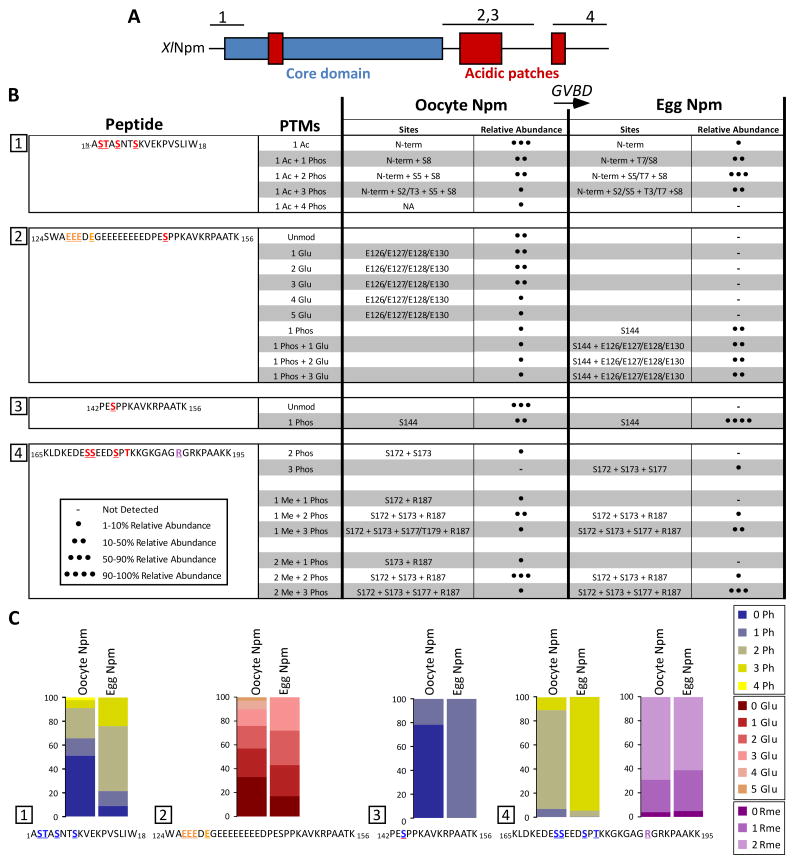
We summarized the modifications found on Npm with their relative abundances in heat maps in Figure 2, demonstrating a marked increase in the total abundance and density of phosphorylation and glutamylation upon GVBD and egg laying.
Figure 2. Npm modifications dynamically change upon oocyte maturation to the egg as determined by mass spectrometry.
A. Npm domains (hydrophobic core in blue, acidic patches in red) and regions analyzed by mass spectrometry (numbered above). B. A summary of the PTMs found on the four major peptides analyzed by mass spectrometry in both the oocyte and egg forms of Npm are shown. The peptide sequence is shown on the left column. Each row of the table shows the PTMs, the individual sites and their relative abundances (- not detected; ● 1–10% relative abundance; ●● 11–50% relative abundance; ●●● 51–90% relative abundance; ●●●● 91–100% relative abundance). C. The relative abundances of PTMs on each of the four peptides are shown in heatmaps to demonstrate the change upon GVBD from the oocyte to the egg. Also See Figure S2.
Nucleoplasmin post-translational modifications alters histone binding and chaperoning activity
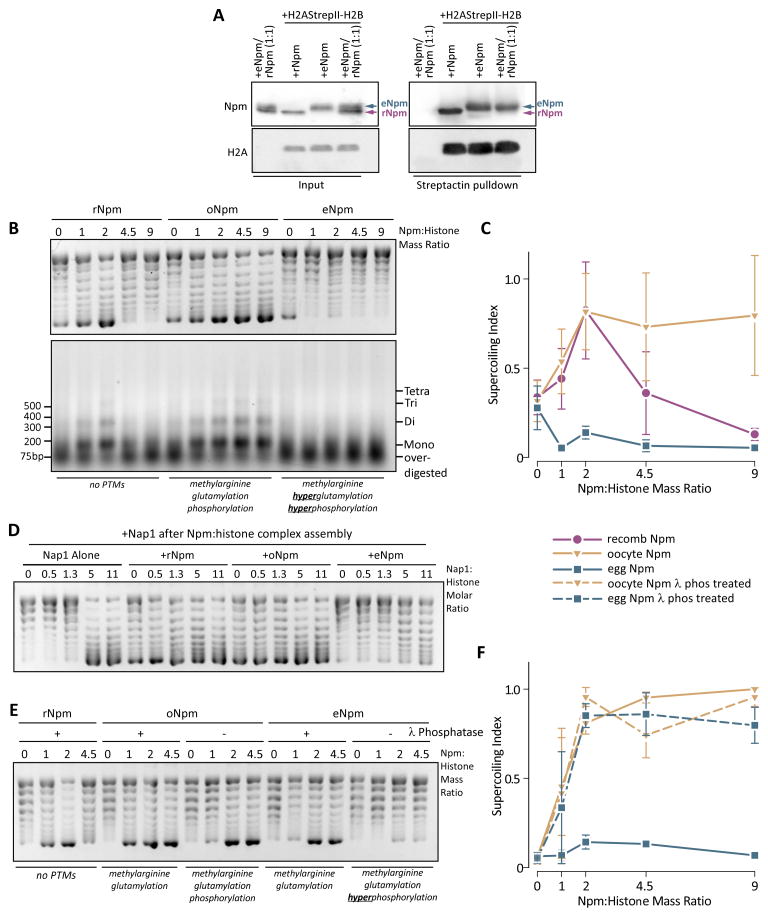
We first determined if the Npm PTMs caused a difference in histone binding. We incubated C-terminally StrepII (SII)-tagged H2A(SII)/H2B dimers with an equimolar quantity of rNpm, eNpm, or a 1:1 mixture of rNpm/eNpm and then isolated interacting complexes on Streptactin resin. Both rNpm and eNpm were co-precipitated with the tagged histone dimers, but eNpm outcompeted rNpm for histone binding in the mixed sample (Figure 3A). We also performed the same experiment using oNpm. Both oNpm and rNpm were co-precipitated with tagged histones even in a 1:1 molar mixture of oNpm and rNpm (Figure S3A). We concluded that eNpm has a higher affinity for histone dimers than rNpm or oNpm.
Figure 3. PTMs alter the in vitro histone binding and deposition of oNpm and eNpm.
A. Competitive co-precipitation assay of rNpm and eNpm. StrepII tagged H2A-H2B dimer complexed with rNpm, eNpm or a 1:1 molar mixture of rNpm and eNpm was precipitated with streptactin resin and blotted for Npm and H2A. eNpm migrates slower on the gel allowing separation from rNpm. B. Chromatin assembly assay using rNpm, oNpm, and eNpm. Concentrations of Npm used for each sample are indicated as a mass ratio to histones. Plasmid supercoiling (top) and MNase digestion of the same reaction (bottom) are shown. C. The quantification of histone deposition in (B). Histone deposition is expressed as a ratio of supercoiled band (bottom-most band) to total DNA (“supercoiling index”). The values represent the normalized mean of three replicates ± s.d. D. Chromatin assembly assay with a mixture of Npm and increasing amounts of XlNap1. The numbers over the lane represent Nap1:histone molar ratio. Npm was kept at 5:1 molar ratio to histones (equivalent to 2:1 mass ratio). E. Chromatin assembly assay with λ-phosphatase treated (+) or mock treated (-) Npm. F. quantification of (E), shown as in (C). The legend for (C) and (F) are shown between them. Also See Figure S3.
To test Npm histone chaperoning activity, we measured chromatin assembly in the presence of ISWI, an ATP-dependent chromatin remodeler, and hyperacetylated core histones purified from HeLa cells (Figure S3B). Using this system, we observed nearly 100% of plasmid supercoiled with recombinant X. laevis Nap1. Micrococcal nuclease (MNase) treatment of the product yielded bands corresponding to poly-nucleosomes in an ISWI dependent manner (Figure S3C), confirming our appropriate experimental design (Fyodorov and Kadonaga, 2003). Furthermore, we observed poor supercoiling and high background with Npm in the absence of ISWI (Figure S3D) further demonstrating the necessity of ISWI in the assay.
We first tested rNpm, containing no PTMs and oNpm and eNpm with PTMs in the chromatin assembly assay. DNA and core histone concentrations were kept constant while each species of Npm was titrated relative to core histone mass. We observed significant plasmid super-coiling for rNpm and oNpm (Figure 3B top panel). MNase treatment of the supercoiled product yielded bands corresponding to polynucleosomes, confirming bona fide nucleosome assembly by rNpm and oNpm (Figure 3B bottom panel). However, the histone deposition by rNpm was reduced above a 2:1 mass ratio. oNpm exhibited constant histone deposition above the 2:1 ratio up to 15:1, the highest ratio tested (Figure 3B,D, Figure S3E). The MNase treatment of rNpm and oNpm assembled nucleosomes also exhibited a band of overdigested DNA that was absent from Nap1 assembled nucleosomes (Figure 3B, S3C bottom panel). This difference in histone deposition between Npm and Nap1 suggests a relatively low efficiency of histone deposition for rNpm and oNpm in vitro, as previously reported (Sealy et al., 1986).
In contrast, eNpm did not deposit histones in our chromatin assembly assay (Figure 3B,C). To test the possibility that eNpm may have an altered threshold concentration for histone deposition, we performed chromatin assembly up to 15:1 mass ratio but were still unable to observe histone deposition (Figure S3E). Npm binds chromatin in the egg and DNA in vitro (Lu et al., 2012; Okuwaki et al., 2012). To exclude the possibility that the DNA binding prevented eNpm histone deposition, we performed a native gel shift assay using Npm and a linear DNA fragment. We confirmed that rNpm, but not wildtype or phosphatase-treated oNpm or eNpm, weakly bound DNA demonstrating that eNpm histone sequestration was not due to DNA binding (Figure S3F). To further test the ability of Npm to sequester histones, we titrated Nap1 into Npm preincubated with histones and observed that eNpm, but not rNpm or oNpm, prevented histone deposition by Nap1 (Figure 3D). This result indicated that eNpm specifically sequesters histones from DNA and other chaperones.
Since we identified hyperphosphorylation as the main difference between oNpm and eNpm, we tested the role of Npm phosphorylation in chromatin assembly. Dephosphorylation of eNpm with λ-phosphatase restored robust histone deposition indicating that hyperphosphorylation is critical for eNpm histone sequestration (Figure 3E,F), including at higher chaperone mass ratios (Figure S3G). We concluded that phosphorylation was essential for histone sequestration, while glutamylation and/or arginine methylation were responsible for enhanced deposition at higher mass ratios.
Post-translational modifications alter the conformation of Nucleoplasmin
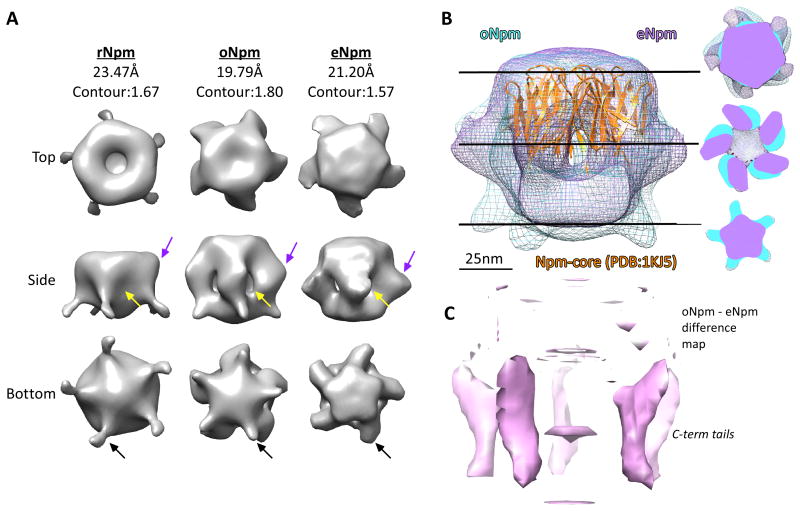
Our mass spectrometry identified PTMs solely on the N- and C-terminal flexible tails of Npm. We hypothesized that PTMs may alter the conformation of Npm in these intrinsically disordered regions and therefore performed electron microscopy reconstruction of each Npm species (Figure 4A, S4). The Npm pentamer core crystal structure fit well into a globular domain found in all three reconstructions (Figure 4B, PDB:1K5J shown in orange). Comparison of rNpm (rNpm) with oNpm and eNpm reconstructions showed several regions of altered density (Figure 4B, C). In rNpm we observed density on the lateral face of Npm immediately adjacent to the core when displayed at a contour covering 100% mass. This density was absent in oNpm and eNpm (Figure 4A, yellow arrows) forming a hollow space instead. On the lateral face of Npm core we observed protruded density on oNpm and eNpm (Figure 4B purple arrows). This protrusion was larger in eNpm and rotated around the pentamer axis relative to oNpm (Figure 4B, right hand panel). Another striking difference among the Npm species was a tailed structure protruding from the distal face progressively disappearing as more PTMs accumulate from rNpm to oNpm and to eNpm (Figure 4A, black arrows), also shown in the difference map between eNpm and oNpm (Figure 4C).
Figure 4. Oocyte and egg PTMs alter Npm conformation.
A. Top, side, and bottom views of reconstructions of rNpm, oNpm, and eNpm three-dimensional structure from negatively stained electron micrographs of each protein. The density maps are displayed at contours covering 100% mass, as shown at the top. B. A side view reconstruction of oNpm (blue) and eNpm (purple) are modeled with the PDB:1KJ5 core domain structure fit (orange). The scale bar is shown in nm. Central slices of the reconstruction are shown on the right. C. A difference map between the oNpm and eNpm models is shown in pink illustrating the altered conformation and rotation of the C-terminal tails about the pentamer axis. Also See Figure S4.
Analysis of Nucleoplasmin domains required for histone deposition
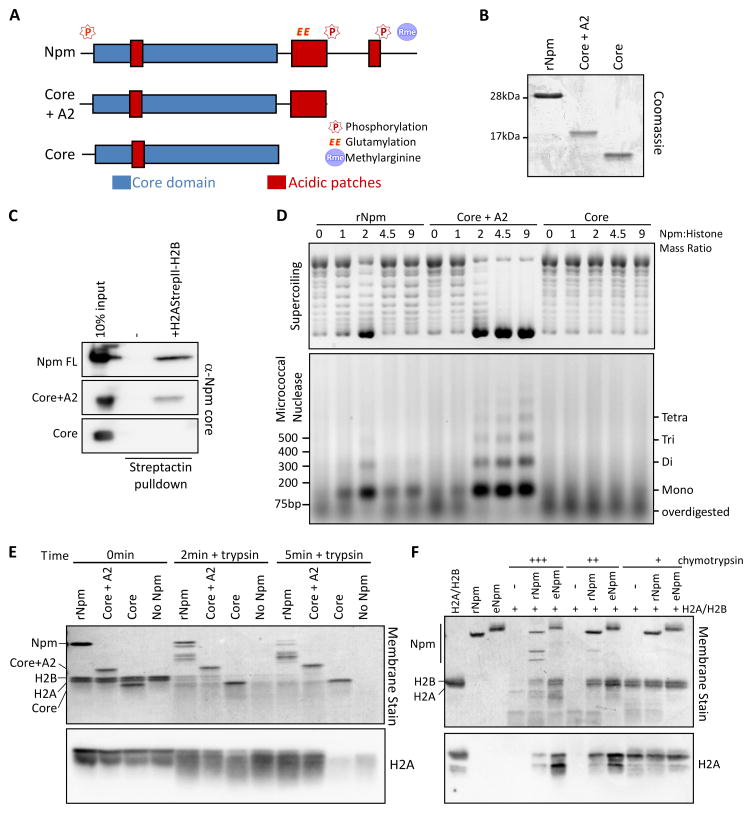
Since PTMs occurred adjacent to the A2 and A3 acidic patches on the C-terminal tail, we hypothesized that they may be regulating the interactions between the acidic tracts and the basic histones. To test this hypothesis, we first determined the domains of Npm necessary for histone deposition. We constructed a mutant truncating the region past A2 (Core+A2) and a mutant truncating the entire tail including A2 (Core; Figure 5A,B). Using StrepII-tagged H2A(SII)-H2B dimer co-precipitation and a polyclonal antibody raised against the core domain of Npm (Figure S5A), we confirmed that Core+A2, but not Core alone, binds histones (Figure 5C), consistent with the previously reported lower affinity of the core domain to histones (Taneva et al., 2009). We then performed chromatin assembly using these mutants and observed robust histone deposition by Core+A2, which showed almost 100% histone deposition and nucleosome assembly, and no histone deposition with Core (Figure 5D). This result indicated that A2 is critical for histone deposition while the extreme C-terminal tail suppressed rNpm histone deposition at high mass ratios.
Figure 5. Npm A2 binding to histone is critical for histone deposition.
A. Npm truncation mutants used in the domain analysis. The core domain is indicated in blue and acidic patches in red. PTM sites are noted above the schematic. B. Purified Npm truncation mutants used in the assay. C. Co-precipitation assay of Npm truncation mutants and histones. StrepII tagged H2A-H2B dimer complexed with full-length Npm, Core+A2, or Core mutants of Npm is precipitated with streptactin resin and blotted for Npm. D. Chromatin assembly assay using Npm truncation mutants. Plasmid supercoiling (top) and MNase digestion of the same reaction (bottom) are shown. E. Protease protection assay of full-length Npm, Core+A2 and Core mutants of Npm digested with trypsin. The Direct Blue 71 whole membrane stain and immunoblot for H2A are shown at 0, 2, and 5 min post-digestion. F. Protease protection assay using rNpm and eNpm with increasing amounts of chymotrypsin. The Direct Blue 71 whole membrane stain and immunoblot for H2A are shown. Also See Figure S5.
We further tested how A2 binds histones using a protease protection assay. In this assay the Npm-histone H2A-H2B complex was digested with a protease and the interaction domain was determined by a difference in digestion rate, monitored via immunoblot probing H2A at different time points. We employed trypsin and chymotrypsin, with unique digestion sites on Npm and H2A (Figure S5B,C) to completely monitor the interaction domain. Using the full-length and truncation mutants of Npm, we observed a significantly slower digestion rate of H2A with full-length and Core+A2 compared to Histone alone or Core mutant (Figure 5E). After 10 min of digestion, Core mutant and histone alone samples had almost complete H2A digestion while a significant amount still remained with full-length and Core+A2 (Figure S5D), consistent with A2 binding histones. We also performed protease protection assays comparing eNpm and oNpm to rNpm. Using trypsin, we observed higher H2A protection with oNpm and eNpm compared to rNpm (Figure S5E). We also probed the digested proteins for A2 glutamylation and confirmed that glutamylation, and therefore A2, still remained in the digestion products (Figure S5F). In parallel, we performed the protection assay with increasing concentrations of chymotrypsin, a protease with distinct cleavage sites, and confirmed that eNpm protected H2A better than rNpm did (Figure 5F). From these truncation analyses and protease protection assays, we concluded that A2 is critical for histone deposition and that oNpm and eNpm PTMs promote enhanced interaction surface for histones.
Specific function of Npm PTMs in regulation of histone deposition
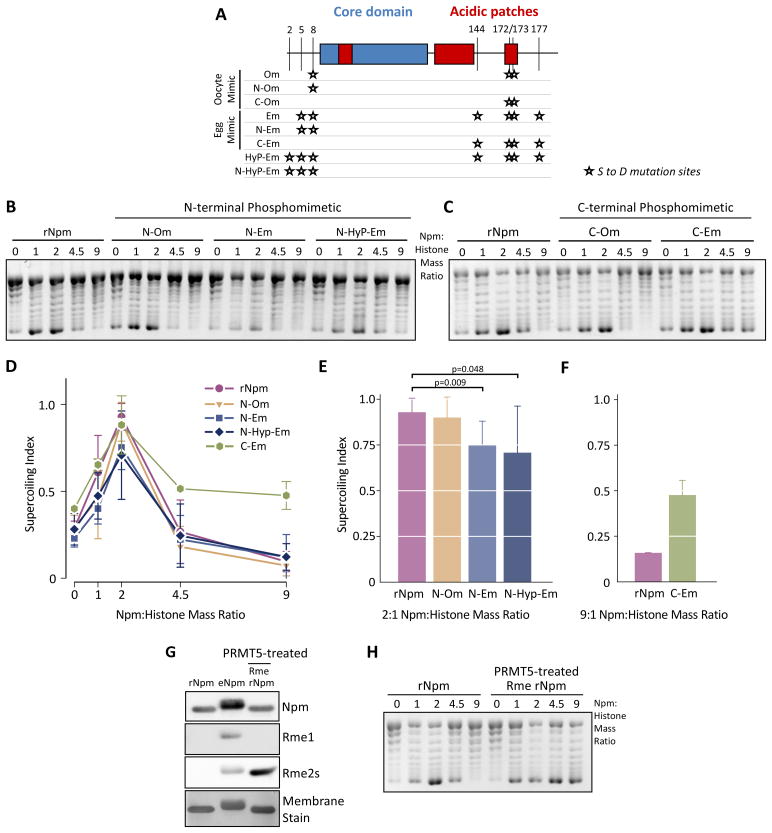
To determine how phosphorylation alters Npm chaperone activity, we made Ser-to-Asp phosphomimetic mutants at the identified phosphorylation sites (Figure 6A and S6A). We had observed that the N-terminal phosphorylation pattern was different from that on the C-terminal tail. In the C-terminal tail S144 and S177 were nearly 100% phosphorylated in the egg. Conversely, the N-terminal tails contained an ensemble of PTMs, with the majority of oNpm peptides containing zero to one phosphorylations and eNpm peptides containing two to three phosphorylations at 60% and 50% relative abundance, respectively (Figure 2C). Furthermore, we observed an N-terminal peptide containing three phosphorylations at 25% abundance in eNpm compared to less than 10% in oNpm (Figure 2C). This difference in phosphorylation patterns suggested that the N-terminal and C-terminal tail phosphorylation might distinctly regulate functions of Npm. Therefore, we made separate N- and C-terminal phosphomimetic mutants (Figure 6A).
Figure 6. N- and C-terminal PTMs distinctly regulate Npm histone deposition.
A. Schematic showing the Npm Ser-to-Asp phosphomimetic mutations we prepared. The mutation sites (★) are indicated by residue number (top) and the mutant protein name (left). B. Chromatin assembly assay using N-terminal phosphomimetic mutant Npm proteins. C. Chromatin assembly assay using C-terminal phosphomimetic mutant Npm proteins. D. Supercoiling index of (B) represented as in Figure 3D. The normalized mean of seven replicates ± s.d. is shown. E. Supercoiling index of (B) at 2:1 mass ratio is shown. p values (top) were calculated using Student’s t-test. F. The quantification of C-Em and rNpm at 9:1 mass ratio are shown. The result represents the normalized mean of two replicate ± s.d. G. rNpm, eNpm, and PRMT5-treated rNpm were immunoblotted for mono- (Rme1) and symmetric dimethylarginine (Rme2s) as shown. H. Chromatin assembly assay using rNpm and PRMT5-treated rNpm. Also See Figure S6.
To test the specific function of discrete Npm phosphorylations in histone chaperone function, we performed chromatin assembly assays using the N-terminal phosphomimetic mutations. All mutants exhibited rNpm-like histone deposition with a peak at 2:1 mass ratio, while deposition by the egg mimic (N-Em, 2 sites) and the hyperphosphorylated egg mimic (N-HyP-Em, 3 sites) was reduced at the 2:1 ratio (Figure 6B). This result strongly suggested that N-terminal phosphorylation is critical for eNpm sequestration of histones. Conversely, the C-Em 4 site C-terminal phosphomimetic mutant exhibited enhanced histone deposition above the 2:1 ratio (Figure 6C). Quantification of multiple replicates of these supercoiling assays confirmed the essential differences in histone deposition activity of N- and C-terminal phosphomimetic mutants (Figure 6D). Egg N-terminal phosphomimetic mutants had significant reductions in deposition at the 2:1 ratio when analyzed with a two-tailed t test (Figure 6E), while C-terminal phosphomimetic mutants enhanced histone deposition at the 9:1 ratio (Figure 6F).
Our chromatin assembly assay with λ-phosphatase treated Npm showed that glutamylation and/or arginine methylation is sufficient to block rNpm inhibition of histone deposition at higher mass ratios (Figure 3E). Therefore, we tested if in vitro methylation would cause a similar response. We treated rNpm with the PRMT5 arginine methyltransferase and confirmed that treated and repurified Npm was dimethylated (Figure 6G). As hypothesized, we observed enhanced chromatin assembly with the PRMT5-treated Npm (Figure 6H, 4.5 and 9 mass ratios). Npm arginine methylation may act synergistically with phosphorylation to alter its histone deposition activity. Therefore, we performed the chromatin assembly assay with PRMT5-treated Em to test if phosphorylation and methylation together caused histone sequestration. However, we observed similar histone deposition with arginine methylated Em (Figure S6B).
These results supported our hypothesis that N- and C-terminal phosphorylation distinctly regulate Npm histone deposition. C-terminal phosphorylation promotes enhanced histone deposition while N-terminal phosphorylation is critical for sequestration.
Regulated accessibility of the Npm A2 acidic patch
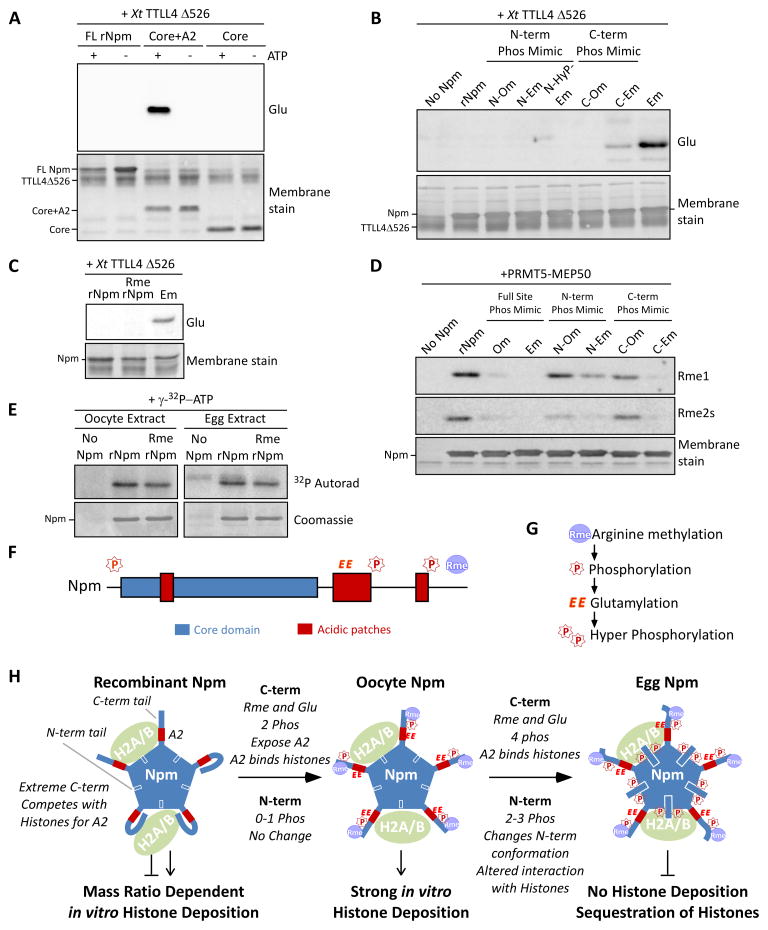
In the Npm domain analysis and protease protection assays we observed that PTMs promoted interaction between A2 and histones. Considering that the C-terminal tail past A2 is enriched for positive amino acids (Figure S2B), an intramolecular interaction may limit histone accessibility to the A2. To test this hypothesis, we measured the efficiency of glutamylation using recombinant Xenopus tropicalis Tubulin-Tyrosine-Ligase Like 4 catalytic domain (TTLL4Δ526) (Regnard et al., 2000; van Dijk et al., 2008). Since the endogenous glutamylation occurs on A2, we used this efficiency of glutamylation as a read-out of A2 exposure. Using full-length, Core+A2, and Core truncation mutants of Npm, we observed that Core+A2 but not the full length or Core mutant was efficiently glutamylated by TTLL4 (Figure 7A). This observation was consistent with our hypothesis that the extreme C-terminal region interacts with A2 and limits its accessibility to other proteins.
Figure 7. Nucleoplasmin phosphorylation, glutamylation, and methylation crosstalk reveals a developmental mechanism for regulated histone accessibility.
A. X. tropicalis TTLL4Δ526 glutamylation of Npm truncation mutants. Immunoblot for glutamylation (Glu) and Direct Blue 71 membrane stain for total protein are shown. B. Glutamylation assay using phosphomimetic mutants of Npm as in (A). C. Glutamylation assay using PRMT5-treated rNpm (“Rme rNpm”) and Em phosphomimetic mutant, as in (A). D. Arginine methylation assay using phosphomimetic mutants of Npm and XlPRMT5-MEP50. Immunoblot for mono- (Rme1) and symmetric dimethylarginine (Rme2s) and the membrane stain are shown. E. Kinase assay of rNpm and arginine methylated rNpm (Rme rNpm) in egg extract and oocyte extract supplemented with γ-32P-ATP; shown are autoradiogram (top) and Coomassie stained gel (bottom). The weak band appearing in the Npm negative lane in the egg extract is the endogenous Npm from the extract. F. Npm domains with identified sites of modification (“P”, phosphorylation; “EE”, glutamylation; “Rme”, arginine methylation) shown above. G. Our inferred biological order of events during oogenesis. H. A model summarizing our results. Shown are the N-terminal tail (blue box with white outline), A2 (red box), and C-terminal tail (thick blue line) with representative PTMs for each stage. Also See Figure S7.
We then performed the glutamylation assay using the phosphomimetic mutants of Npm to test our hypothesis that phosphorylation on the C-terminal tail increases the exposure of A2 by disrupting the interaction between A2 and the extreme C-terminal region of Npm. As predicted, the C-Em mutant was glutamylated by TTLL4 (Figure 7B). Surprisingly, our mutant carrying seven phosphomimetic mutations over the full length (Em) greatly increased glutamylation compared to the C-Em mutant, while the N-Em mutant alone showed no glutamylation. This result was consistent with our hypothesis that C-terminal phosphorylation exposes A2, most likely by limiting the interaction between the extreme C-terminal region and A2.
Our observation that the N-terminal phosphomimetics enhance glutamylation synergistically with the C-terminal phosphomimetic indicated that N-terminal phosphorylation may itself causes conformational change leading to histone sequestration. We performed a chromatin assembly assay with oocyte, egg, and hyperphosphorylated egg phosphomimetic mutations over the full length of Npm, but we were unable to observe full sequestration (Figure S6C).
Specific function of Npm PTMs in modification crosstalk
The phosphomimetic mutations clearly promoted glutamylation of Npm. To test if there was additional crosstalk between modifications, we measured TTLL4 activity towards arginine methylated rNpm, but did not observe any increase in glutamylation compared to unmethylated rNpm (Figure 7C). We also tested if the phosphomimetic mutations would increase arginine methylation by treating the phosphomimetic mutants of Npm with purified Xl PRMT5-MEP50 complex and measuring mono- and symmetric dimethylation via immunoblot. The Om and Em full phosphomimetic Npm proteins were very poor substrates for PRMT5-MEP50, with intermediate inhibition with fewer Ser-to-Asp mutations on both the N- and C-terminal domains (Figure 7D). N-terminal phosphomimetic mutations specifically reduced dimethylarginine products. This observation was consistent with our initial observation that arginine methylation preceded glutamylation during oogenesis (Figure 1G). We also tested if arginine methylation increased phosphorylation of rNpm in egg and oocyte extracts, but we did not observe any significant change in phosphorylation in either extract (Figure 7E).
These observations indicate that the Nucleoplasmin PTMs occur in the following order during oogenesis: 1) arginine methylation; 2) basal phosphorylation; 3) glutamylation; and then 4) hyperphosphorylation upon GVBD (Figure 7F,G).
DISCUSSION
A wave of protein phosphorylation occurs upon breakdown of the Xenopus laevis germinal vesicle in the oocyte and the resumption of meiosis, regulating many processes (Ferrell, 1999). Npm hyperphosphorylation was previously shown to be required for sperm decondensation (Bañuelos et al., 2003; Frehlick et al., 2006; Leno et al., 1996; Ramos et al., 2005; Tamada et al., 2006). Other studies showed that Npm phosphomimetic mutants altered its histone binding ability, although these studies were limited due to the unknown sites of endogenous phosphorylation (Taneva et al., 2009; Taneva et al., 2008).
Our findings are summarized in a model in Figure 7H. In rNpm, without any modifications, the extreme C-terminal region may interact with A2 and limit its histone accessibility. The competition between the extreme C-terminal basic region and histones for A2 likely results in the mass ratio dependent histone deposition pattern that we observed. PTMs acquired by Npm during oogenesis, including basal phosphorylation, arginine methylation, and glutamylation, likely promote stronger interaction between A2 and histones, in part due to phosphorylation disrupting the interaction between the extreme C-terminal region and A2. Upon egg laying, two to three phosphorylations accumulate on the N-terminal tail and two further phosphorylations occur on the C-terminal tail. The accumulating phosphorylation on the N-terminal tail likely causes further conformational change that is critical for the eNpm histone sequestration.
Npm PTMs have substantial crosstalk to regulate histone deposition and sequestration
Glutamylation is catalyzed by TTLL polyglutamylases (Janke et al., 2005). Glutamyltransferase activity exists within Xenopus oocyte extracts (van Dijk et al., 2007) and egg extracts (data not shown), consistent with our observed Npm glutamylation and our in vitro TTLL4 modification of Npm. We previously demonstrated that PRMT5-MEP50 is abundant in eggs and methylates Npm (Wilczek et al., 2011). Phosphorylation of Npm is catalyzed by CK2 (Taylor et al., 1987; Vancurova et al., 1995) and the cyclin-dependent kinase cdc2/cdk1 (Cotten et al., 1986). S144 and S177, sites of hyperphosphorylation upon egg laying, are in a consensus sequence for cdk kinase activity (SPxK) and are highly conserved across metazoan Nucleoplasmin proteins. S8, S172 and S173 phosphorylation on both oNpm and eNpm is likely catalyzed by CK2, with a kinase motif of [S/T]xx[E/D] (Pandey et al., 2007; Vancurova et al., 1995).
We observed that phosphorylations on S5, 8, 144, and 177 are the critical difference between Npm in the oocyte and egg. Since Npm hyperphosphorylation occurred concomitant with GVBD and lasted until the MBT, the developmental period with simultaneous requirement for storage and release of maternal histones, we hypothesized that these PTMs would influence Npm histone chaperoning activity. Consistent with our hypothesis, we demonstrated that C-terminal phosphorylations and arginine methylation promote enhanced histone deposition, compared to the stoichiometry-dependent deposition promoted by rNpm.
We also demonstrated that the hyperphosphorylation on eNpm causes sequestration of histones. This eNpm sequestration was surprising as it contrasted with the previous observation that Npm histone deposition in vitro is enhanced by the egg PTMs (Sealy et al., 1986). Our purification strategy for Npm differed from the previous study in a few key ways: 1) we employed immediate heat purification of low speed extract to eliminate cycloheximide and cytochalasin B that had suppressed ionization in the mass spectrometer, possibly resulting in distinct phosphorylation states; 2) More phosphorylation was associated with poorer heat stability, indicating that different temperatures during purification may result in different eNpm states (Taneva et al., 2008). Another mass spectrometric analysis of eNpm reported a majority of three to four phosphorylations on the N-terminus (Banuelos et al., 2007). We observed that N-terminal phosphomimetic mutations reduced histone deposition, suggesting that the N-terminal phosphorylation is crucial to eNpm histone sequestration further supporting that the discrepancy in eNpm histone deposition in the literature may be due to different phosphorylation states in the N-terminal tail. These observations together suggest that the N-terminal phosphorylation may function as a switch between histone storage and release, where double phosphorylation leads to sequestration and further phosphorylation promotes the release. This hypothesis is supported by observations that N-terminal phosphorylation promotes sperm DNA decondensation, the hallmark event of fertilization (Bañuelos et al., 2003). Further experiments with phosphoserine-containing Npm mutants, rather than Ser-to-Asp mutants, will be necessary to fully test this hypothesis. Interestingly, histone deposition activity was not correlated with a specific charge state on the Npm protein as might be predicted for a presumed electrostatically-mediated process (Figure S7). Therefore, our work demonstrated that individual PTMs have substantial effects on Npm histone chaperoning activity, either through specific interaction sites and/or via conformational change.
Conformational change, regulated acidic patch accessibility, and histone chaperone activity
We favor the hypothesis that PTM-dependent conformational change alters Npm histone binding and chaperoning activity. In support of this hypothesis, we demonstrated through EM reconstruction and biochemical assays that Npm PTMs cause successive conformational changes upon maturation of the oocyte to the egg. In particular, our evidence supports a model in which C-terminal tail phosphorylation exposes A2 and promotes the interaction between A2 and histones causing enhanced deposition. We also demonstrated that arginine methylation does not cause a similar conformational change despite its enhancement of rNpm deposition activity. These observations indicated that arginine methylation induced strong histone deposition through a mechanism distinct from the A2 exposure to histones as seen with phosphorylation. Considering that arginine methylation occurs past A3, we hypothesize that arginine methylation may directly increase the affinity of the C-terminal tail, containing A2 and A3, to histones.
We also demonstrated that the N-terminal hyperphosphorylation causes further conformational change. Although the identification of the specific sites of conformational change or the interaction sites between Npm and histones leading to sequestration require further experiments, we note that the N-terminal tail is located near the A1 acidic patch in the crystal structure of Npm core domain (Hierro et al., 2001; Namboodiri et al., 2003; Platonova et al., 2011). It is possible that histone sequestration may result from further interaction between these regions and histones promoted by the N-terminal hyperphosphorylation. Furthermore, the N- and C-terminal tails are located side-by-side on the distal face of Npm core (Dutta et al., 2001). Although both tails are missing from the crystal structure, it is likely that the A2 on the C-terminal tail is immediately adjacent to the N-terminal tail. This close proximity suggests a synergistic function between the N-terminal phosphorylation and glutamylation on A2, consistent with our observation that the N- and C-terminal phosphomimetic mutations synergistically exposed A2 in our glutamylation assay. This synergistic nature and the lack of glutamylation may have contributed to our inability to recapitulate full histone sequestration.
Since N-terminal phosphomimetic mutations alone reduced histone deposition, the eNpm sequestration of histones may be independent of the C-terminal tail but dependent on the conformational change caused by N-terminal hyperphosphorylation. Consistent with this, our EM reconstruction of eNpm showed a conformational change on the lateral face of Npm core domain due to egg specific hyperphosphorylation. Our inability to recapitulate eNpm sequestration in full may also be due to the poor phosphomimetic nature of Ser-to-Asp mutations. We observed increased TTLL4-dependent glutamylation with the Ser-to-Asp mutations, but the overall efficiency of glutamylation was still poor. We estimated the glutamylation of Em mutant to be about 10% of glutamylation on Core+A2 mutant (data not shown), further indicating the poor phosphomimetic nature of Ser-to-Asp mutations.
Taken together, our observations demonstrate that the histone chaperoning activity of Npm is dynamically regulated through PTMs leading to sequestration in the egg and the N-terminus phosphorylation of Npm is the critical switch between storage and release.
EXPERIMENTAL PROCEDURES
See Supplemental Experimental Procedures for more detailed methods.
Xenopus extract preparation and histone purification
Xenopus egg and oocyte extracts, and histones were prepared as described (Banaszynski et al., 2010).
Npm protein preparation
Npm was purified from extracts as described (Sealy et al., 1986). Recombinant Npm, Npm truncations, and mutants were His-tagged, produced in E. coli and purified via Ni-affinity and MonoS cation exchange chromatography.
Electron Microscopy and Image Processing
Following optimization of the protein concentration, 3μl aliquots of specimen were applied to carbon-coated copper grids that had been glow discharged. The specimen was then stained on a drop of 2% Uranyl Acetate for 25 seconds. After removing the extra stain with filter paper, the grid was air-dried and examined under a transmission electron microscope. Micrographs were taken under a JEOL JEM-2100F electron microscope. A TVIPS TemCam-415 CCD camera recorded the specimens at a pixel size of 1.25A at specimen space. Image processing was completed using the Single Particle Reconstruction package: EMAN. Boxing of individual particles was done using a box-size of 85 px for each isolated Npm. Initial particle classification was done using multivariate statistical analysis. Initial models were all generated independently of one another. Two initial models were generated for each particle set: one with symmetry imposed, and one without. These were all subsequently refined at an angular step of fifteen to resolve low resolution details before refining at a lower angular step of seven to resolve higher resolution details. The initial model for eNpm required hand-selecting against misaligned particles in the initial particle stack. Refinements were carried out iteratively using reference-based statistical analysis from projections of the structures obtained in the previous iteration. Symmetry was released during early refinement in all three models in order to confirm convergence of refinement.
Chromatin assembly assays
Plasmid supercoiling and MNase digestions were all performed on plasmid pGIEO with hyperacetylated HeLa histones and purified chaperones as described (Fyodorov and Kadonaga, 2003).
Mass Spectrometry Analyses
Npm purified from Xenopus oocytes or laid eggs were reduced, alkylated, and digested with endoproteinase chymotrypsin (Schroeder et al., 2004). Self-packed precolumns and analytical columns were prepared as previously described (see Ficarro et al., 2009; Martin et al., 2000 and Supplemental). Aliquots of oocyte or egg Npm were bomb-loaded onto the precolumn, desalted with 0.1% acetic acid, and connected to the analytical column. Peptides were gradient-eluted into the mass spectrometer via reversed-phase HPLC at a flow rate of 60 nl/min (see supplemental for gradient conditions). Data-dependent MS/MS analyses were acquired on an LTQ-FT using either CAD or FETD (Earley et al., 2013). All data was searched using OMSSA and all spectra corresponding to Npm were manually interpreted.
Chemicals and Antibodies
Chemicals and reagents were obtained from Sigma (St. Louis, MO), RPI (Illinois) or Fisher Scientific (Pittsburgh, PA). We used the following antibodies in this study: a purified monoclonal Npm antibody (Dilworth et al., 1987), polyclonal rabbit antibody generated against full length recombinant Npm, anti-glutamylation TTbIII, SG I polyclonal antibodies (gift of Dr. Frankfurter, Spano and Frankfurter, 2010), and histone and methylarginine polyclonal antibodies (Wilczek et al., 2011).
Images and adjustment
Gel images were acquired using digital tools (Epson V700 scanner and GE LAS-4000) with 16 bit dynamic range. Complete images were levels adjusted as a whole to improve clarity without obscuring, eliminating, or misrepresenting any information present in the original.
Supplementary Material
Acknowledgments
We are grateful to S. Dilworth for the gift of a Nucleoplasmin monoclonal antibody, T. Frankfurter for the gift of anti-glutamylation antibodies, R. Heald for the gift of a Xenopus Nap1 clone, K. Ohsumi for Nap1 antibodies, and to D. Fyodorov and A. Emelyanov for ISWI pGIE0, TopoI constructs, and initial assistance and consultation with ISWI-dependent chromatin assembly assays. This work was supported by the National Institutes of Health [grant R01GM037537 to D.F.H., R01AI095382 to R.H.C., R01GM108646 to D.S., and training grant T32GM007491 to C.W.], by a UC Discovery Grant [to R.H.C.], and by startup funds from the Albert Einstein College of Medicine and the Alexander and Alexandrine Sinsheimer Foundation Scholar Award [to D.S.]. D.S. is supported by The American Cancer Society – Robbie Sue Mudd Kidney Cancer Research Scholar Grant [124891-RSG-13-396-01-DMC].
Footnotes
ACCESSION NUMBERS
Nucleoplasmin reconstructions were deposited in the EMDataBank under accession numbers EMD-2866 (egg Npm), EMD-2868 (oocyte Npm), and EMD-2869 (recombinant Npm).
AUTHOR CONTRIBUTIONS
TO conceived of and performed experiments, interpreted results, and wrote the manuscript. JJN performed mass spectrometry experiments, interpreted results, and wrote the manuscript. LX and BA performed electron microscopy reconstruction and interpreted results. CW performed Npm truncation and TTLL experiments. WW performed developmental analysis. SER performed protease protection assays. ESB purified and characterized CePRMT5 and XlPRMT5-MEP50. RHC directed and interpreted electron microscopy reconstruction. JS and DFH directed and interpreted mass spectrometry experiments. DS conceived of and interpreted experiments and wrote the manuscript. All authors read and approved the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnan C, Saperas N, Prieto C, Chiva M, Ausió J. Interaction of nucleoplasmin with core histones. J Biol Chem. 2003;278:31319–31324. doi: 10.1074/jbc.M305560200. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Allis CD, Shechter D. Analysis of histones and chromatin in Xenopus laevis egg and oocyte extracts. Methods. 2010;51:3–10. doi: 10.1016/j.ymeth.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bañuelos S, Hierro A, Arizmendi JM, Montoya G, Prado A, Muga A. Activation mechanism of the nuclear chaperone nucleoplasmin: role of the core domain. J Mol Biol. 2003;334:585–593. doi: 10.1016/j.jmb.2003.09.067. [DOI] [PubMed] [Google Scholar]
- Banuelos S, Omaetxebarria MJ, Ramos I, Larsen MR, Arregi I, Jensen ON, Arizmendi JM, Prado A, Muga A. Phosphorylation of both nucleoplasmin domains is required for activation of its chromatin decondensation activity. J Biol Chem. 2007;282:21213–21221. doi: 10.1074/jbc.M702842200. [DOI] [PubMed] [Google Scholar]
- Bouleau A, Desvignes T, Traverso JM, Nguyen T, Chesnel F, Fauvel C, Bobe J. Maternally inherited npm2 mRNA is crucial for egg developmental competence in zebrafish. Biol Reprod. 2014;91:43. doi: 10.1095/biolreprod.114.119925. [DOI] [PubMed] [Google Scholar]
- Cotten M, Sealy L, Chalkley R. Massive phosphorylation distinguishes Xenopus laevis nucleoplasmin isolated from oocytes or unfertilized eggs. Biochemistry. 1986;25:5063–5069. doi: 10.1021/bi00366a014. [DOI] [PubMed] [Google Scholar]
- Dilworth S, Black S, Laskey R. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- Dutta S, Akey IV, Dingwall C, Hartman KL, Laue T, Nolte RT, Head JF, Akey CW. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Molecular cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- Earley L, Anderson LC, Bai DL, Mullen C, Syka JE, English AM, Dunyach JJ, Stafford GC, Jr, Shabanowitz J, Hunt DF, et al. Front-end electron transfer dissociation: a new ionization source. Anal Chem. 2013;85:8385–8390. doi: 10.1021/ac401783f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Xenopus oocyte maturation: new lessons from a good egg. Bioessays. 1999;21:833–842. doi: 10.1002/(SICI)1521-1878(199910)21:10<833::AID-BIES5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Finn RM, Ellard K, Eirin-Lopez JM, Ausio J. Vertebrate nucleoplasmin and NASP: egg histone storage proteins with multiple. Faseb j. 2012;26:4788–4804. doi: 10.1096/fj.12-216663. [DOI] [PubMed] [Google Scholar]
- Frehlick L, Eirín-López J, Jeffery E, Hunt D, Ausió J. The characterization of amphibian nucleoplasmins yields new insight into their role in sperm chromatin remodeling. BMC Genomics. 2006;7:99. doi: 10.1186/1471-2164-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. In Methods in enzymology (United States) 2003:499–515. doi: 10.1016/S0076-6879(03)71037-4. [DOI] [PubMed] [Google Scholar]
- Hierro A, Arizmendi JM, De Las Rivas J, Urbaneja MA, Prado A, Muga A. Structural and functional properties of Escherichia coli-derived nucleoplasmin. A comparative study of recombinant and natural proteins. Eur J Biochem. 2001;268:1739–1748. [PubMed] [Google Scholar]
- Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? ‘Protein Modifications: beyond the usual suspects’ review series. EMBO reports. 2008;9:636–641. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava A, Strub J, Temurak N, van Dijk J, Boucher D, van Dorsselaer A. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- Keck KM, Pemberton LF. Histone chaperones link histone nuclear import and chromatin assembly. Biochim Biophys Acta. 2013;1819:277–289. [PubMed] [Google Scholar]
- Leno G, Mills A, Philpott A, Laskey R. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J Biol Chem. 1996;271:7253–7256. doi: 10.1074/jbc.271.13.7253. [DOI] [PubMed] [Google Scholar]
- Litvin J, King ML. Expression and segregation of nucleoplasmin during development in Xenopus. Development. 1988;102:9–21. doi: 10.1242/dev.102.1.9. [DOI] [PubMed] [Google Scholar]
- Lu Q, Lu Z, Liu Q, Guo L, Ren H, Fu J, Jiang Q, Clarke PR, Zhang C. Chromatin-bound NLS proteins recruit membrane vesicles and nucleoporins for nuclear envelope assembly via importin-alpha/beta. Cell Res. 2012;22:1562–1575. doi: 10.1038/cr.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namboodiri VM, Dutta S, Akey IV, Head JF, Akey CW. The crystal structure of Drosophila NLP-core provides insight into pentamer formation and histone binding. Structure. 2003;11:175–186. doi: 10.1016/s0969-2126(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Newport J, Dasso M. On the coupling between DNA replication and mitosis. J Cell Sci Suppl. 1989;12:149–160. doi: 10.1242/jcs.1989.supplement_12.13. [DOI] [PubMed] [Google Scholar]
- Okuwaki M, Sumi A, Hisaoka M, Saotome-Nakamura A, Akashi S, Nishimura Y, Nagata K. Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. Nucleic Acids Res. 2012;40:4861–4878. doi: 10.1093/nar/gks162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RA, Balamurugan P, Suresh M, Raghunath R, Sudhir Gopal T, Akhilesh A curated compendium of phosphorylation motifs. Nature biotechnology. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- Platonova O, Akey IV, Head JF, Akey CW. Crystal structure and function of human nucleoplasmin (npm2): a histone chaperone in oocytes and embryos. Biochemistry. 2011;50:8078–8089. doi: 10.1021/bi2006652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado A, Ramos I, Frehlick L, Muga A, Ausió J. Nucleoplasmin: a nuclear chaperone. Biochem Cell Biol. 2004;82:437–445. doi: 10.1139/o04-042. [DOI] [PubMed] [Google Scholar]
- Ramos I, Fernandez-Rivero N, Arranz R, Aloria K, Finn R, Arizmendi JM, Ausio J, Valpuesta JM, Muga A, Prado A. The intrinsically disordered distal face of nucleoplasmin recognizes distinct oligomerization states of histones. Nucleic Acids Res. 2014;42:1311–1325. doi: 10.1093/nar/gkt899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I, Martin-Benito J, Finn R, Bretana L, Aloria K, Arizmendi JM, Ausio J, Muga A, Valpuesta JM, Prado A. Nucleoplasmin binds histone H2A-H2B dimers through its distal face. J Biol Chem. 2010;285:33771–33778. doi: 10.1074/jbc.M110.150664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I, Prado A, Finn R, Muga A, Ausió J. Nucleoplasmin-mediated unfolding of chromatin involves the displacement of linker-associated chromatin proteins. Biochemistry. 2005;44:8274–8281. doi: 10.1021/bi050386w. [DOI] [PubMed] [Google Scholar]
- Regnard C, Desbruyeres E, Huet JC, Beauvallet C, Pernollet JC, Edde B. Polyglutamylation of nucleosome assembly proteins. J Biol Chem. 2000;275:15969–15976. doi: 10.1074/jbc.M000045200. [DOI] [PubMed] [Google Scholar]
- Schroeder MJ, Shabanowitz J, Schwartz JC, Hunt DF, Coon JJ. A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal Chem. 2004;76:3590–3598. doi: 10.1021/ac0497104. [DOI] [PubMed] [Google Scholar]
- Sealy L, Cotten M, Chalkley R. Xenopus nucleoplasmin: egg vs. oocyte. Biochemistry. 1986;25:3064–3072. doi: 10.1021/bi00358a049. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano AJ, Frankfurter A. Characterization of anti-beta-tubulin antibodies. In Methods Cell Biol (United States, 2010 Elsevier Inc) 2010:33–46. doi: 10.1016/S0091-679X(10)95003-6. [DOI] [PubMed] [Google Scholar]
- Sun L, Bertke MM, Champion MM, Zhu G, Huber PW, Dovichi NJ. Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development. Sci Rep. 2014;4:4365. doi: 10.1038/srep04365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada H, Van Thuan N, Reed P, Nelson D, Katoku-Kikyo N, Wudel J, Wakayama T, Kikyo N. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneva SG, Bañuelos S, Falces J, Arregi I, Muga A, Konarev PV, Svergun DI, Velázquez-Campoy A, Urbaneja MA. A mechanism for histone chaperoning activity of nucleoplasmin: thermodynamic and structural models. J Mol Biol. 2009;393:448–463. doi: 10.1016/j.jmb.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Taneva SG, Muñoz IG, Franco G, Falces J, Arregi I, Muga A, Montoya G, Urbaneja MA, Bañuelos S. Activation of nucleoplasmin, an oligomeric histone chaperone, challenges its stability. Biochemistry. 2008;47:13897–13906. doi: 10.1021/bi800975r. [DOI] [PubMed] [Google Scholar]
- Taylor A, Allende CC, Weinmann R, Allende JE. The phosphorylation of nucleoplasmin by casein kinase-2 is resistant to heparin. FEBS Lett. 1987;226:109–114. doi: 10.1016/0014-5793(87)80561-6. [DOI] [PubMed] [Google Scholar]
- van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283:3915–3922. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- van Dijk J, Rogowski K, Miro J, Lacroix B, Eddé B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Molecular cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Vancurova I, Paine T, Lou W, Paine P. Nucleoplasmin associates with and is phosphorylated by casein kinase II. Journal of Cell Science. 1995;108:779–787. doi: 10.1242/jcs.108.2.779. [DOI] [PubMed] [Google Scholar]
- Wilczek C, Chitta R, Woo E, Shabanowitz J, Chait BT, Hunt DF, Shechter D. Protein Arginine Methyltransferase Prmt5-Mep50 Methylates Histones H2A and H4 and the Histone Chaperone Nucleoplasmin in Xenopus laevis Eggs. Journal of Biological Chemistry. 2011;286:42221–42231. doi: 10.1074/jbc.M111.303677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.