Abstract
Suberin, a major constituent of the potato periderm, is known to promote the production of thaxtomins, the key virulence factors of the common scab-causing agent Streptomyces scabiei. In the present study, we speculated that suberin affected the production of glycosyl hydrolases, such as cellulases, by S. scabiei, and demonstrated that suberin promoted glycosyl hydrolase activity when added to cellulose-, xylan-, or lichenin-containing media. Furthermore, secretome analyses revealed that the addition of suberin to a cellulose-containing medium increased the production of glycosyl hydrolases. For example, the production of 13 out of the 14 cellulases produced by S. scabiei in cellulose-containing medium was stimulated by the presence of suberin. In most cases, the transcription of the corresponding cellulase-encoding genes was also markedly increased when the bacterium was grown in the presence of suberin and cellulose. The level of a subtilase-like protease inhibitor was markedly decreased by the presence of suberin. We proposed a model for the onset of S. scabiei virulence mechanisms by both cellulose and suberin, the main degradation product of cellulose that acts as an inducer of thaxtomin biosynthetic genes, and suberin promoting the biosynthesis of secondary metabolites including thaxtomins.
Keywords: cellulase, glycosyl hydrolases, proteomics, Streptomyces scabiei, thaxtomins
Streptomyces species are Gram-positive bacteria that belong to the phylum Actinobacteria and exhibit complex morphological development. After germination, Streptomyces spores produce germ tubes that grow by tip extension and branch to form a vegetative mycelium. This mycelium differentiates by forming a second aerial mycelium, which eventually septates to form spores. Although most Streptomyces species are saprophytic soil inhabitants, some, such as Streptomyces scabiei, are plant pathogens (35). S. scabiei is one of the main agents of potato common scab. This plant pathogen induces symptoms ranging from corky patches to deep-pitted lesions on the surface of tubers, which reduce the value of affected potatoes (20). The pathogenicity of S. scabiei depends on the production of thaxtomins (17, 19), secondary metabolites that inhibit the biosynthesis and deposition of cellulose in plant cells (47).
The transcription of S. scabiei thaxtomin biosynthetic genes (txt genes) is induced in the presence of plant-derived materials such as suberin and cellobiose (31). The role of cellobiose in triggering the production of thaxtomins has been elucidated. Cellobiose binds the AraC/XylS transcriptional regulator, TxtR, thereby allowing the expression of txt genes (23). Furthermore, cellobiose has been shown to inhibit the attachment of the cellulose-utilization repressor CebR to binding sites of the txt gene cluster (14). On the other hand, the effects of suberin on txt gene expression remain unclear. Lerat et al.(31, 32) proposed that suberin activated the differentiation and production of secondary metabolites including thaxtomins in several Streptomyces species.
Suberin is a recalcitrant plant polymer composed of a polyaromatic domain covalently linked to a polyaliphatic domain (5). The polyaromatic domain, a lignin-like structure, is embedded in the primary cell wall and largely consists of the hydroxycinnamic acid, monolignol (4). The polyaliphatic domain is composed of polyesters of long-chain fatty acids and glycerol, which are located between the primary cell wall and the plasma membrane. Its chemical structure is related to cutin, a polymer covering the aerial parts of plants (39). Suberin has been suggested to play a protective role against common scab disease. A previous study reported a relationship between the phenolic acid composition of suberin and cultivar resistance to common scab (49), while Khatri et al.(26) observed the higher suberization of the potato periderm in a common scab-tolerant cultivar than in a susceptible one.
As soil-dwelling organisms, Streptomyces species feed on plant polymers. This genus is known for its ability to produce a wide variety of extra-cellular enzymes involved in the degradation of cellulose, hemicellulose, and recalcitrant plant polymers such as lignin (40, 51). S. scabiei has the ability to colonize the potato periderm. The outer layer of the periderm, the phellem, consists of suberized cells (26). Evidence to support S. scabiei at least partially degrading suberin is increasing. This bacterium was previously shown to grow on suberin as a source of carbon (29) and produced esterases in its presence (1). Furthermore, the S. scabiei genome encodes potential suberinase genes (29). One of these genes, sub1 is known to be specifically induced in the presence of suberin and cutin (29). A secretome analysis of S. scabiei cultures grown in the presence of suberin has also been conducted in order to identify the enzymes potentially involved in the degradation of suberin (30). Several enzymes predicted to play a role in lipid metabolism were identified as candidate enzymes involved in the degradation of suberin. However, these enzymes accounted for a small fraction of the S. scabiei secretome, whereas glycosyl hydrolases represented the most abundant functional group of proteins in the supernatant of suberin-containing medium (30). Several cellulases have been identified among the glycosyl hydrolases; however, most S. scabiei strains exhibit no or poor growth on cellulose (13).
The effects of suberin on the production of glycosyl hydrolases and expression of cellulase-encoding genes were examined in the present study. Furthermore, the secretomes of S. scabiei grown in media containing either cellulose or both cellulose and suberin were compared. A model of the onset of S. scabiei virulence mechanisms by both cellulose and suberin was proposed.
Materials and Methods
Bacterial strain and growth conditions
Streptomyces scabiei strain EF-35 was isolated in Canada from a common scab lesion on a potato tuber (12). A bacterial inoculum was prepared by inoculating approximately 108 spores in 25 mL of YME (4 g L−1 of glucose, 4 g L−1 of yeast extract, and 10 g L−1 of malt extract). The culture was incubated with shaking (250 rpm) for 48 h at 30°C, after which the bacterial cells were recovered by centrifugation (3,500×g) for 10 min. The bacterial inoculum was obtained by resuspending the pellet in 5 volumes of saline (NaCl 0.85%). The inoculum (200 μL) was transferred to 50 mL of a control medium (CM) composed of 0.5 g L−1 l-asparagine, 0.5 g L−1 K2HPO4, 0.2 g L−1 MgSO4·7H2O, 10 mg L−1 FeSO4·7H2O, and 0.05% (w/v) casein hydrolysate (Sigma, St-Louis, MO, USA) supplemented or not with 0.1% (w/v) suberin and/or 0.5% (w/v) of an additional carbon source: insoluble microcrystalline cellulose Avicel (EMD Millipore, Billerica, MA, USA), xylan (oat spelt, Sigma), or lichenin (also known as lichenan) (Megazyme, Wicklow, Ireland). Cultures were incubated with shaking (250 rpm) for 5 d at 30°C.
Suberin used in this study was extracted from potato tubers, as described in Kolattukudy and Agrawal (28). The amount of residual glucose in the purified polymer was determined according to Blakeney et al.(6). The alditol acetate derivatives of glucose, obtained by the hydrolysis, reduction, and acetylation of the suberin preparation, were quantified by gas chromatography on a Varian 3800 GC equipped with a flame-ionization detector (FID) and CP-Sil 88 column (48). The experiment was carried out with five replicates.
Extracellular protein quantification and concentration determination
Extracellular proteins were recovered by centrifuging the bacterial cultures (3,500×g) for 15 min at 4°C. The protein concentrations of the S. scabiei culture supernatants were determined according to Bradford (7) using bovine serum albumin as a standard. In proteomic assays, supernatants were concentrated to a final volume of 500 μL using Amicon Ultra-15 Centrifugal Filters-3K, and the proteins were resuspended in 5 volumes of 100% pre-chilled acetone and kept for 3 h at −20°C. Proteins were recovered by centrifugation (14,000×g, 20 min, 4°C) and the protein pellets were air-dried and resuspended in 80 μL of a buffer composed of 8 M urea, 2% (w/v) CHAPS, 2% (v/v) IPG buffer pH 4–7 (GE Healthcare, Buckinghamshire, UK), 18.15 mM DTT, and 0.002% bromophenol blue stock solution in 50 mM Tris-base. A final centrifugation (14,000×g) was carried out for 5 min at 4°C to remove insoluble material and the protein solutions were immediately subjected to a proteomic analysis.
Isolation of genomic DNA
Bacterial biomass was estimated by cellular DNA quantification. Genomic DNA was extracted according to the procedure of Kieser et al.(27). Genomic DNA was resuspended in 500 μL of a buffer containing 10 mM Tris-HCl and 1 mM disodium EDTA (pH 8.0) (TE buffer) and further purified. Two microliters of RNase (10 mg mL−1) was added to the DNA solution followed by an incubation for 1 h at 37°C. Chloroform (250 μL) was added to the DNA solution and mixed by inversion. The mixture was centrifuged for 10 min at 3,500×g. Two hundred and fifty microliters of 3 M sodium acetate, pH 5.2 and 1 mL of 2-propanol were mixed into the upper phase and the solution was centrifuged for 10 min at 3,500×g. The pellet was rinsed with 70% ethanol and resuspended in 400 μL of TE buffer. DNA was then quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s specifications.
Enzymatic assays
Cellulase, xylanase, and licheninase activities associated with S. scabiei culture supernatants were determined according to Lever (33) as further described in Komeil et al.(30).
Proteomics analysis
Extracellular proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% [w/v] SDS-PAGE) according to Komeil et al.(30). Horizontal slices were cut across the SDS-PAGE gel and used to perform a proteomic analysis. In-gel protein digestion and mass spectrometry were carried out at the Proteomics Platform of the Eastern Quebec Genomics Center (Quebec City, QC, Canada) using a quadrupole time-of-flight mass spectrometer Qq-TOF (Sciex, Concord, ON, Canada) coupled to HPLC for the separation of peptides used with a nanospray source. This system allowed for the resolution of more than 30,000 FWHM with an accuracy of <2 ppm and an acquisition rate of 50 MS/MS s−1.
All MS/MS spectra were analyzed for peptide identification using Mascot (Matrix Science, London, UK; version 2.2.0) to search the S. scabiei strain 87.22 Uniref100 database based on trypsin digestion, a fragment ion mass tolerance of 0.50 Da, and a parent ion tolerance of 2.0 Da. The search results were uploaded to the scaffold software program and a filter was set with a 99% minimum protein ID probability with a minimum number of two unique peptides in the protein, in which the cut-offs for peptide thresholds were 90%. Protein functions were reanalyzed using various databases such as GenBank, Pfam protein-domain/family, COG (50), and CAZy (34) as well as PRIAM (10) and KEGG resources (25). Phobius (24), SignalP 4.1 (43), SecretomeP (2), TatP (3), and Tatfind 1.4 (45) analyses were used to predict protein cellular localizations. A normalized spectral abundance factor (NSAF) was calculated for each protein that met the filtering criteria by dividing the proportion between the number of spectral counts (SpC) of a protein in culture medium and its molecular weight (MW) by the sum of SpC/MW of all proteins meeting the filtering criteria and found in the same medium (41).
Gene expression
The effects of carbon sources on gene expression were determined as follows. Five milliliters of each culture medium was sampled after 5 d of growth. One milliliter of stop solution (ethanol/acidic phenol, 95:5, [v/v]) was added to each sample to prevent RNA degradation. Bacterial cells were recovered by a 10-min centrifugation at 3,500×g and stored at −80°C until RNA extraction. Total RNA was extracted from cells with the NucleoSpin RNA II kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions. Cell lysis was improved by a sonication step (2×30 s) before phenol/chloroform extraction. An additional DNA digestion step was performed after elution with the Turbo DNA-free kit (Ambion, Austin, TX, USA). cDNA was synthesized by reverse transcription using 2 μg of total RNA with the First-Strand cDNA Synthesis kit (GE Healthcare) using 72% G+C-rich random hexamers. Quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) of gene transcripts was performed using Mx3000P (Agilent Technologies, Santa Clara, CA, USA) with the SYBR Green PCR master mix and JumpStart Taq DNA Polymerase (Sigma). PCR cycles were 95°C for 5 min, followed by 35 cycles at 95°C for 15 s and 60°C for 45 s. The primers used in this study are listed in Table 1. Gene gyrA was used as a reference for the internal control of relative quantification. Relative gene expression was determined using the comparative CT method (44).
Table 1.
Primers used in this study
| Gene assignation | Predicted function of the corresponding protein | Primer sets (5′ to 3′) |
|---|---|---|
| SCAB_24291 | Gyrase A (gyrA) | For: GGACATCCAGACGCAGTACA Rev: CTCGGTGTTGAGCTTCTCCT |
| SCAB_5981 | Cellulase B precursor CelB | For: TGTTCAACGGGTGCCATTAC Rev: ACATAGCCGTACGAGATGCT |
| SCAB_16431 | Cellulase CelA1 | For: CATGAACCAGGCGCAGATA Rev: CCATGTAGACCCAGGTGTTG |
| SCAB_17001 | Cellulase | For: TCGTCCAGCTGGTGATCTA Rev: GTCGATGTACTGCGTCTTGT |
| SCAB_17011 | Cellulase | For: GAGGCGTACAGCTACCTCCTGTG Rev: GTAGAAGGAGTTGGTCGGCTGGTC |
| SCAB_17021 | Cellulase | For: GACACCTACACCTGGAAGAAC Rev: CTTCTCCTTGCGGTTGAAGA |
| SCAB_36371 | Xylanase/Cellulase | For: CTGAGAAGCCCGGGAAATC Rev: CACCCGTCACACACATGAA |
| SCAB_37051 | Cellulase/Xylanase | For: ATTCTCCGGAAGCACATCAC Rev: TCCTCGAAGACCTCGTTCA |
| SCAB_51081 | Cellulase | For: GGCATCAACTGGTTCGGTTTCGAG Rev: TGTTGTAGCCCAGCGACTTCATCT |
| SCAB_90081 | Cellulase B precursor | For: TACAACGGCTGCCACTAC Rev: CTCGCATAGCTGTAGGAGATAC |
| SCAB_90091 | Cellulase | For: AGGACAGCAGATACATCGAGGAGT Rev: TCAAGAAGATCGGCAACTGTGTCG |
| SCAB_90101 | Cellulase | For: TCGAGTTGGTCGCTGAAATG Rev: AAGCGTGCCGTTGTAGTT |
Results
Effects of the presence of suberin in polysaccharide-containing culture media on Streptomyces scabiei growth, extracellular protein production, and enzymatic activities
The amount of glucan remaining embedded in the suberin preparation added to the culture medium was determined by quantifying glucose concentrations. The average concentration of residual glucose was 103±8 μg mg purified suberin−1. The amount of glucose from suberin thus represented approximately 2% of the sugar present in control medium (CM) supplemented with cellulose (CM+C), xylan (CM+X), or lichenin (CM+L). The effects of adding suberin to the three latter media on S. scabiei growth, extracellular protein production, and enzymatic activity were then determined. The addition of suberin had different effects on S. scabiei growth depending on the polysaccharide that was available as the main carbon source. Suberin did not affect growth in xylan-, improved growth in cellulose-, and inhibited growth in lichenin-containing medium (Table 2). While suberin appeared to be a better growth substrate than cellulose for S. scabiei, this was not the case for either xylan or lichenin. Biomass, estimated by DNA quantification, was approximately 8 to 10-fold higher in the presence of these substrates than in CM+S (Table 2).
Table 2.
Comparison of Streptomyces scabiei growth, extracellular protein concentrations, and enzymatic activities between suberin-containing control medium (100%) and control medium supplemented with a polysaccharide in the presence or absence of suberin
| Mediuma | Relative growth (%)b,c | Relative protein concentration (%)b,d | Enzymatic activity testedb | Enzymatic activity (%)b,e |
|---|---|---|---|---|
| CM+X | 793 | 20 | Xylanolytic activity | 41 |
| CM+X+S | 747 | 20 | Xylanolytic activity | 126 |
| CM+L | 965 | 10 | Licheninase activity | 92 |
| CM+L+S | 472 | 21 | Licheninase activity | 387 |
| CM+C | 36 | 79 | Cellulase activity | 39 |
| CM+C+S | 105 | 80 | Cellulase activity | 170 |
Control medium supplemented with xylan (CM+X), xylan and suberin (CM+X+S), lichenin (CM+L), lichenin and suberin (CM+L+S), microcrystalline cellulose (CM+C), or microcrystalline cellulose and suberin (CM+C+S).
Data are the mean of three experiments.
Growth was estimated by cellular DNA quantification.
Protein concentration was estimated in μg μg of DNA−1.
Enzymatic activity was estimated in U μg of DNA−1.
The addition of suberin did not modify the levels of extracellular proteins produced per μg of bacterial DNA in CM supplemented with cellulose, xylan, or lichenin. However, there were approximately 5 to 10-fold more extracellular proteins in control medium supplemented with suberin (CM+S, 141.7±11.7 μg μg of DNA−1) than in CM supplemented with xylan (CM+X, 28.2±2.6 μg μg of DNA−1) or lichenin (CM+L, 14.8±1.8 μg μg of DNA−1). No significant differences were observed in the amounts of extracellular proteins associated with CM+S and CM supplemented with cellulose (Table 2).
A 3- to 4-fold increase in cellulase, xylanase, or licheninase activity was observed when suberin was added to CM+C, CM+X, and CM+L media. Licheninase activity was similar in CM+L (17.4±0.8 U μg of DNA−1) and CM+S (19.0±4.3 U μg of DNA−1), whereas cellulase and xylanase activities were higher in CM+S (1.8±0.5 U μg of DNA−1, 35.1±8.2 U μg of DNA−1, respectively) than in CM+C (0.7±0.6 U μg of DNA−1) or CM+X (14.5±1.1 U μg of DNA−1), respectively.
Effects of suberin on the Streptomyces scabiei secretome in cellulose-containing medium
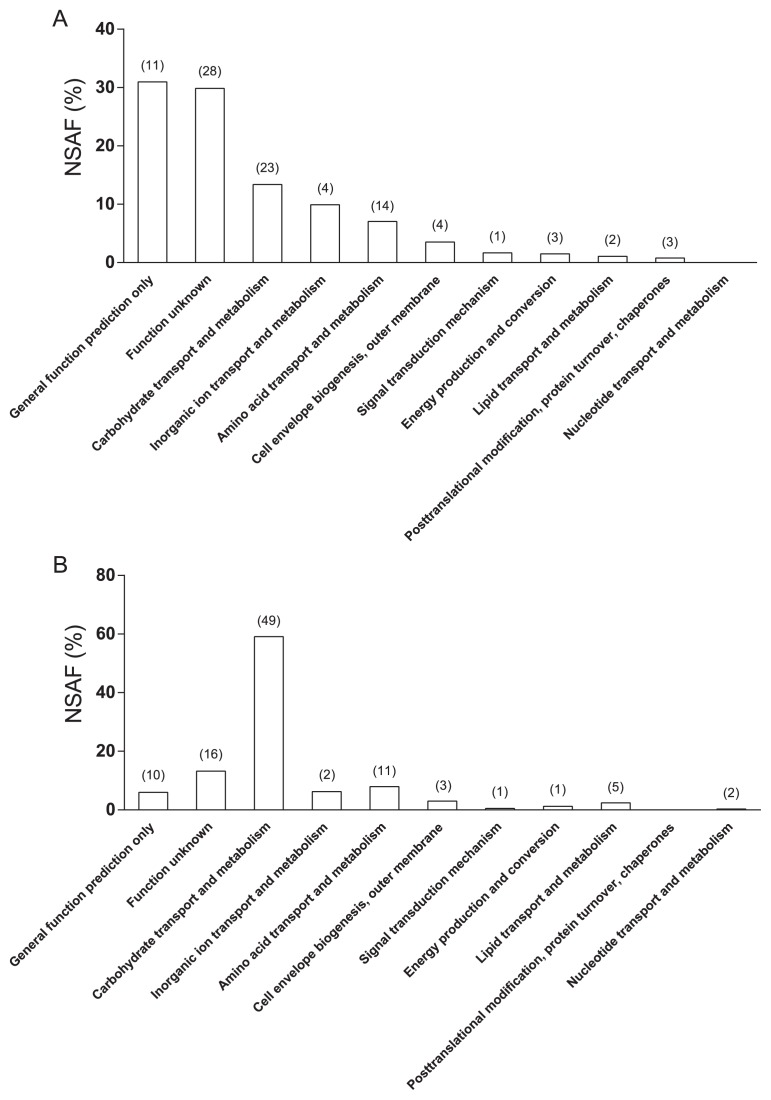
The addition of suberin to a cellulose containing-medium only slightly affected the diversity of the secretome. A total of 115 and 125 proteins met the filtering criteria in CM+C and in the same medium supplemented with suberin (CM+C+S), respectively (Table S1). A total of 22 and 25 proteins were predicted to have an intracellular localization in CM+C and CM+C+S, respectively. The other predicted extracellular proteins were divided into 11 functional groups. The distribution of the secreted proteins within the functional groups differed according to the growth medium. Fig. 1 shows a normalized spectral abundance factor (NSAF) and the number of proteins associated with each functional group. In CM+C, proteins associated with the “general function predicted only” group had a NSAF of 31% and included the most abundant protein in the secretome (SCAB_8801), a subtilase-like protease inhibitor with an NSAF of 26.02% (Fig. 1A and Table S1). Proteins of unknown function included 28 proteins with a NSAF of 30%. The functional group “carbohydrate transport and metabolism” was composed of 23 proteins and had a NSAF of 13% (Fig. 1A). Of these, 25 (NSAF=14.18%) were putative glycosyl hydrolases (GH) belonging to seven GH families (Table 3). The most abundant GH were the putative cellulase C9ZD50 (NSAF of 1.86%) and a putative xylanase A (C9ZE95, NSAF of 1.84%). The GH group included nine putative cellulases with a combined NSAF of 3.9% (Table 3).
Fig. 1.
Normalized spectral abundance factor (NSAF) within functional groups of Streptomyces scabiei strain EF-35 secreted proteins associated with control medium supplemented with microcrystalline cellulose (A) or both microcrystalline cellulose and suberin (B). Values in brackets represent the number of proteins found within functional groups.
Table 3.
Proteins involved in carbohydrate transport and metabolism were secreted into control medium supplemented with either microcrystalline cellulose or microcrystalline cellulose and suberin
| UniProt accession number | Corresponding gene in the S. scabiei 87.22 genome | Putative function | CAZy classification | Normalized spectral abundance factor (%) | |
|---|---|---|---|---|---|
|
| |||||
| CM+C | CM+C+S | ||||
| Cellulases | |||||
| C9YVP5 | SCAB_5981 | Cellulase B precursor CelB | CE1 | Da | 0.94 |
| C9Z0D5 | SCAB_8871 | Cellulase | CBM13 | 0.12 | 0.38 |
| C9ZD50 | SCAB_16431 | Cellulase CelA1 | GH6 | 1.86 | 3.06 |
| C9ZEP9 | SCAB_17001 | Cellulase | GH6, CBM2 | D | 1.51 |
| C9ZEQ0 | SCAB_17011 | Cellulase | GH48, CBM2 | 0.18 | 1.58 |
| C9ZEQ1 | SCAB_17021 | Cellulase | GH74, CBM2 | NDb | 1.47 |
| C9YUZ2 | SCAB_36371 | Xylanase/cellulase | GH10, CBM2 | 0.19 | 1.77 |
| C9YW88 | SCAB_37051 | Cellulase/xylanase | GH10 | 0.46 | 2.06 |
| C9YTK2 | SCAB_51081 | Cellulase | GH5, CBM2 | 0.22 | 0.75 |
| C9ZB17 | SCAB_77391 | Cellulose 1,4-β-cellobiosidase | NFc | ND | 0.22 |
| C9Z351 | SCAB_86311 | Cellulase | GH5 | ND | 0.30 |
| C9Z9L5 | SCAB_90081 | Cellulase B precursor | GH12, CBM2 | 0.30 | D |
| C9Z9L6 | SCAB_90091 | Cellulase | GH48, CBM2 | 0.34 | 2.38 |
| C9Z9L7 | SCAB_90101 | Cellulase | GH6, CBM2 | 0.19 | 0.48 |
| Other proteins involved in carbohydrate transport and metabolism | |||||
| C9ZBE6 | SCAB_0631 | α-l-fucosidase | GH29, CBM13 | ND | 0.13 |
| C9YYV2 | SCAB_3881 or SCAB_22931 | Arabinofuranosidase | GH62, CBM13 | ND | 0.68 |
| C9YUC5 | SCAB_4961 | Glucuronoarabinoxylan endo-1,4-β-xylanase | GH30 | ND | 0.49 |
| C9YUG2 | SCAB_5351 | ABC-type sugar transport system | NF | 1.27 | 0.71 |
| C9YVN3 | SCAB_5851 | Glycosyl hydrolase | CBM32 | ND | 0.17 |
| C9YVP9 | SCAB_6021 | Endo β-1,4-xylanase | GH10 | 0.25 | 1.37 |
| C9YYN8 | SCAB_7551 | Glycosyl hydrolase | NF | 0.62 | D |
| C9Z1T6 | SCAB_9291 or SCAB_91051 | Lactonase | NF | ND | 0.26 |
| C9Z1U5 | SCAB_9381 | Exo-α-sialidase | NF | ND | 0.53 |
| C9Z507 | SCAB_11431 | Glycosyl hydrolase | GH43 | 0.22 | 1.11 |
| C9Z878 | SCAB_13491 | Glucan endo-1,3-β-d-glucosidase | GH64 | ND | 0.13 |
| C9ZD59 | SCAB_16521 | Arabinofuranosidase | GH43, CBM42 | ND | 1.81 |
| C9ZD61 | SCAB_16551 | Mannosidase | GH26, CBM23 | ND | 0.11 |
| C9YT63 | SCAB_19561 | β-fructofuranosidase | GH43, CBM13 | ND | 0.23 |
| C9YUL1 | SCAB_19941 | Arabinofuranosidase | GH43, CBM42 | 0.18 | 0.36 |
| C9YVX8 | SCAB_21021 | Xylose ABC transporter substrate-binding protein | NF | 0.49 | ND |
| C9Z5F4 | SCAB_42381 | ABC transporter substrate-binding protein | NF | 0.26 | ND |
| C9Z5L1 | SCAB_42951 | Glucose/Sorbosone dehydrogenase | NF | 0.93 | 9.47 |
| C9Z737 | SCAB_43661 | Galactan endo-1,6-β-galactosidase | GH30, CBM13 | D | 0.49 |
| C9YY37 | SCAB_54441 | Enolase | NF | 0.18 | ND |
| C9Z2N2 | SCAB_57161 | Endo-β-1,6-galactanase | GH30 | ND | 0.21 |
| C9Z451 | SCAB_57751 | Cellobiose-binding transport system associated | NF | 1.46 | 1.72 |
| C9ZDW4 | SCAB_63891 | ABC-type xylose transport-system, periplasmic | NF | 1.05 | 0.19 |
| C9ZFW2 | SCAB_66021 | β-xylosidase | GH43, CBM13 | ND | 0.32 |
| C9ZFW3 | SCAB_66031 | Arabinofuranosidase | GH43, CBM42 | D | 1.09 |
| C9YYF0 | SCAB_70591 | Pectate lyase | PL9 | ND | 0.72 |
| C9Z2V1 | SCAB_72711 | Endo-1,4-β-xylanase | GH11 | D | 0.62 |
| C9Z2W0 | SCAB_72801 | Glycosyl hydrolase | NF | ND | 1.05 |
| C9Z4J7 | SCAB_74141 | α-N-furanosidase | GH51 | ND | 0.43 |
| C9Z623 | SCAB_74681 | Licheninase | NF | ND | 0.44 |
| C9ZAZ8 | SCAB_77201 | Glycosyl hydrolase | GH106 | ND | 2.33 |
| C9ZB22 | SCAB_77441 | α-arabinanase | GH93, CBM13 | 0.13 | 0.24 |
| C9ZCR4 | SCAB_78891 | Glycosyl hydrolase | GH30, CBM13 | ND | 0.17 |
| C9ZE74 | SCAB_79011 | Acetyl-xylan esterase | CE2 | ND | 0.80 |
| C9ZE94 | SCAB_79241 | Arabinofuranosidase | GH62, CBM13 | 0.26 | 1.16 |
| C9ZE95 | SCAB_79251 | Xylanase A | GH10, CBM13 | 1.84 | 10.25 |
| C9ZEC5 | SCAB_79561 | Glycosyl hydrolase | GHnc, CBM13 | ND | 0.30 |
| C9YU29 | SCAB_82021 | β-mannosidase | GH5, CBM2 | ND | 0.51 |
| C9Z1I5 | SCAB_85231 | Chitinase | GH19, CBM12 | 0.44 | 0.54 |
| C9Z804 | SCAB_89741 | Cellulose-binding protein | CBM33 | 0.74 | 1.05 |
D: detected. Peptides have not fulfilled the filtering criteria as described in the Materials and Methods section.
ND: not detected.
NF: no module has been found in the protein.
The addition of suberin to CM+C resulted in a marked reduction in the abundance of the subtilase-like protease inhibitor (NSAF of 1.1%) (Table S1). In contrast, suberin led to an increase in protein abundance in the “carbohydrate transport and metabolism” group (NSAF of 59%) (Fig. 1B). The number of proteins in this group was approximatively 2-fold lower in CM+C (23 proteins) than in CM+C+S (49 proteins) (Fig. 1). In CM+C+S, the two most abundant proteins with respective NSAF of 10.3% and 9.5% were a putative xylanase A (C9ZE95) and glucose/sorbosone dehydrogenase predicted to be involved in glucose metabolism (C9Z5L1) (Table S1). When suberin was added to CM+C, the diversity of GH increased (Table 3) and these GH were distributed among 19 GH families (Table 3). Thirteen cellulases produced in CM+C+S met the filtering criteria (Table 3). These 13 predicted cellulases were more abundant in CM+C+S than in CM+C. The only cellulase that was more abundant in CM+C than in CM+C+S was cellulase B (C9Z9L5), which accounted for NSAF of 0.30% in CM+C. This cellulase B was detected in CM+C+S, but did not pass the filtering criteria.
Effects of plant cell wall constituents on the expression of cellulase-encoding genes
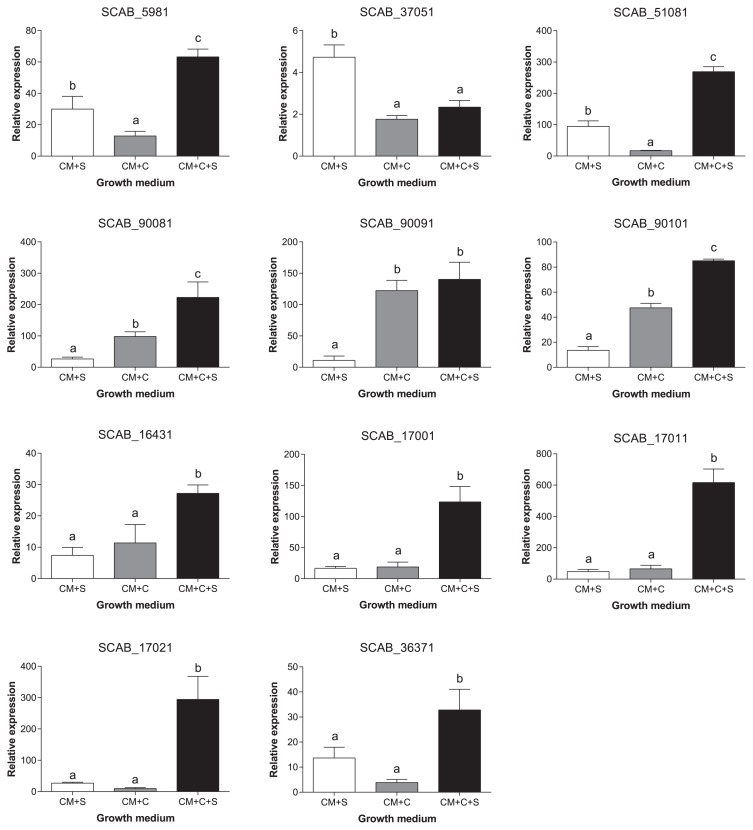
The expression of 11 genes encoding cellulases found in the S. scabiei secretome was monitored in different growth media. The addition of cellulose, suberin or both polymers to control medium (CM) induced the gene expression of all the cellulase-encoding genes tested. The expression levels of genes encoding cellulases SCAB_16431, SCAB_17001, SCAB_17011, SCAB_17021, and SCAB_36371 were similar in media containing either suberin (CM+S) or microcrystalline cellulose (CM+C). The expression of genes encoding cellulases (SCAB_90081, SCAB_90091 and SCAB_90101) was higher in control medium supplemented with cellulose than in CM+S. In contrast, the expression of the cellulase-encoding genes SCAB_5981, SCAB_37051, and SCAB_51081 was higher in CM+S than in CM+C. When suberin was added to microcrystalline cellulose-containing medium, cellulase-encoding gene expression increased significantly, from 1.2- to 32-fold, for all genes, except SCAB_90091 and SCAB_37051 (Fig. 2).
Fig. 2.
Relative expression levels (±SD) of eleven targeted cellulases found in the secretome of Streptomyces scabiei EF-35 grown in control medium (CM) in the presence of suberin (CM+S, white bars), microcrystalline cellulose (CM+C, gray bars), or microcrystalline cellulose supplemented with suberin (CM+C+S, black bars). Data were normalized with the gyrA gene used as an internal control. Data shown are representative of three replicates. Data with the same letter are not significantly different (P<0.05, LSD test).
Discussion
A recent proteomic study (30) showed that a wide variety of extracellular glycosyl hydrolases were produced by the phytopathogen S. scabiei in the presence of suberin. Therefore, we speculated that suberin affects the extracellular enzyme production of S. scabiei when added to polysaccharides such as β-glucans (cellulose or lichenin) or xylan. The addition of suberin to glucan- or xylan-containing medium did not modify the amount of secreted proteins per μg of bacterial DNA. However, the quantity of extracellular proteins was markedly higher in suberin- and cellulose-containing media than in xylan- or lichenin-containing media. Therefore, the amount of extracellular proteins was lower in media supporting good growth (lichenin- or xylan-containing media) than in CM+S and cellulose-containing media. This upregulation in the responses of genes encoding enzymes that hydrolyze plant polysaccharides to carbon-starving conditions has been demonstrated previously (11, 53). As recalcitrant growth substrates, both cellulose and suberin may, thus, trigger the secretion of extracellular enzymes (18, 36, 37, 42). The poor ability of S. scabiei to grow in the presence of cellulose was unexpected considering the presence of several predicted cellulase- and β-glucanase-encoding genes in its genome. However, cellobiose, the main degradation product of cellulose, has been shown to be a poor inducer for endoglucanases in at least some streptomycetes (15), including S. scabiei. Although cellobiose uptake and catabolism have both been demonstrated in S. scabiei cells (31), S. scabiei cellulase activity in the supernatant of CM supplemented with cellobiose was only 0.05±0.02 U μg of DNA−1 (data not shown), but reached 0.7±0.6 U μg of DNA−1 in CM+C (see above in the Results section). The poor inducing ability of cellobiose may also explain why some streptomycetes showed higher cellulase activity on complex organic materials such as straw than on purified cellulose (15, 46, 54, 55).
The present study demonstrated that suberin was an as good as, or even a better inducer of glucanase and xylanase activities than polysaccharides. Furthermore, the addition of suberin to glucan- or xylan-containing medium increased glucanase or xylanase activity, respectively. This higher enzymatic activity may not have been due to the additional supply of polysaccharides provided by sugar residues attached to suberin because (i) the amount of glucose embedded in this lipidic polymer represented only 2% of the glucose contained in the medium, and (ii) the suberin-containing medium supernatant from the S. scabiei culture was unable to release reducing sugars from the suberin used in this study (data not shown).
Therefore, secretomes of S. scabiei cultivated in CM+C and CM+C+S were compared in order to determine if the supply of suberin induced specific glucanases or caused the general overproduction of glucanases. A proteomic analysis revealed that most GH that associated with the CM+C supernatant were predicted to cleave β-1,4 links (cellulases, xylanase, and chitinase), while several other types of GH were only found in CM+C+S medium (including pectate lyase, β-1,6 galactanase, α-N-furanosidase, α-arabinase, α-fucosidase, endo-1,3 β-d-glucosidase, and acetyl-xylan esterase). As with other soil streptomycetes, the S. scabiei genome encodes for a multiplicity of carbohydrate catabolic proteins, especially proteins involved in the degradation of plant-derived materials. The expression of genes coding for plant cell wall-degrading enzymes is sometimes induced by molecules that bear no structural relation to the substrate (21). For example, the production of cellulases in the absence of cellulose or its degradative products has previously been reported in Streptomyces albaduncus (18).
All cellulases found in CM+C were previously identified in S. scabiei cultures grown in the presence of suberin (30), and the amount of cellulases produced was generally higher when both polymers were present (this study). The expression of cellulase-encoding genes was also generally higher when bacteria were exposed to both substrates than to cellulose or suberin only. The presence of both cellulose and suberin had an additive or even synergistic effect on the transcription of most cellulase-encoding genes, suggesting that different environmental signals are necessary for ensuring maximal cellulase-encoding gene expression.
The stimulatory effects of suberin on cellulase-encoding gene transcription may have been due to its chemical composition. Chellapandi and Jani (9) showed that surfactants enhanced the production of endoglucanases in some Streptomyces isolates. Esterases that have been shown to be produced by S. scabiei in the presence of suberin (30, 38) presumably release long chain fatty acids with hydroxyl or epoxy moieties, which may have surfactant properties (37). Furthermore, previous studies reported that phenolic compounds, including those encountered in the aromatic fraction of suberin, regulated the production of cellulolytic enzymes in some Streptomyces strains (15), as well as in fungi (16, 52).
The stimulating effects of suberin on cellulase-encoding gene transcription may also have resulted from its ability to promote differentiation and secondary metabolism in Streptomyces species (32). Cellulase activity is low during primary metabolism in some Streptomyces species, but reaches a maximal level during secondary metabolism (9). However, Lerat et al.(31) demonstrated that cellobiose, the main degradation product of cellulose, blocked differentiation in S. scabiei, which exhibits a bald phenotype when grown in the presence of this disaccharide. The molecular mechanism by which cellobiose inhibits differentiation and secondary metabolism in different Streptomyces species currently remains unknown (31); however, the results of the present study suggest that the subtilase-like protease inhibitor encoded by SCAB_8801, the levels of which were approximately 20-fold higher in CM+C than in CM+C+S (Table S1), was involved in the phenomenon. The role of subtilase protease inhibitors in holding off differentiation has been documented in several Streptomyces species (8). A reduction in the amount of the subtilase-like protease inhibitor when suberin was added to a cellulose-containing medium is consistent with suberin promoting differentiation, even in the presence of cellobiose (31).
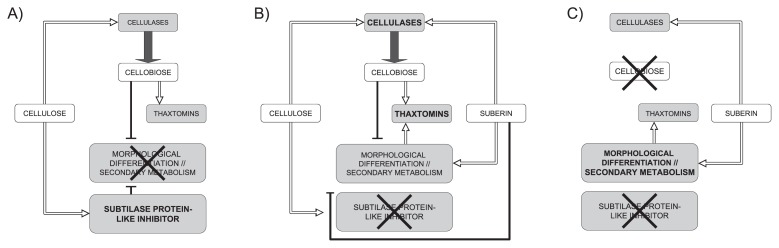
We propose the following model by which the constituents of the potato periderm promote the onset of S. scabiei virulence mechanisms (Fig. 3). The major periderm constituents, cellulose and suberin, both play a role in triggering the production of thaxtomins. A low amount of cellulases was produced in the presence of cellulose only (Fig. 3A), thereby allowing the release of cellobiose, the inducer of thaxtomin biosynthetic genes, which are the main S. scabiei virulence determinants (1, 22, 31). However, cellobiose has been shown to lock morphogenesis and secondary metabolism (31), possibly by affecting the production of a subtilase-like protease inhibitor (this work). Cellobiose, in maintaining S. scabies in primary metabolism, impaired the production of secondary metabolites such as thaxtomins and, consequently, only a small quantity of thaxtomins was produced. Cellulases were produced in the presence of suberin only (Fig. 3C), whereas cellobiose was not produced in the absence of accessible cellulose. Since inducers of thaxtomin biosynthetic genes are lacking, the biosynthesis of thaxtomins is low, relying only on signals promoting morphogenesis and secondary metabolism. In the presence of both suberin and cellulose (Fig. 3B), suberin may have a dual role in the onset of S. scabiei virulence. It may first stimulate the production of cellobiose, the transcriptional inducer of thaxtomin biosynthetic genes, by stimulating cellulase activity (this work) and the consequent release of cellobiose from cellulose. It may then inhibit the effects of cellobiose on secondary metabolism by acting as a signal molecule for morphogenesis, thereby promoting the production of secondary metabolites such as thaxtomins.
Fig. 3.
Model of the onset of Streptomyces scabiei virulence mechanisms by both cellulose and suberin. In the presence of cellulose only (A) or suberin only (C), the thaxtomin biosynthetic genes were only weakly expressed. In the first case, cellulases that cleave cellulose to release cellobiose were produced in low amounts. The liberated cellobiose acted as an inducer of thaxtomin biosynthetic genes. However, in the absence of an environmental signal that triggers secondary metabolism, cellobiose and/or cellulose locked S. scabiei in primary metabolism, possibly by allowing the production of a subtilase protease inhibitor and, thus, limiting the production of secondary metabolites such as thaxtomins,. In the presence of suberin only (C), secondary metabolism was promoted and cellulases were synthesized, whereas cellobiose, the inducer of thaxtomin biosynthetic genes was not produced due to the absence of cellulose. Thaxtomin biosynthetic genes were strongly expressed in the presence of both cellulose and suberin (B). Suberin and cellulose promoted cellulase activity and cellobiose released from cellulose induced thaxtomin biosynthetic genes. Suberin also triggered differentiation and secondary metabolism, thereby overcoming the actions of cellulose and cellobiose.
Supplementary Information
Acknowledgements
This work was supported by grants #018602 and #157930 from the Natural Sciences and Engineering Research Council of Canada (NSERC).
References
- 1.Beauséjour J, Goyer C, Vachon J, Beaulieu C. Production of thaxtomin A by Streptomyces scabies strains in plant extract containing media. Can J Microbiol. 1999;45:764–768. [Google Scholar]
- 2.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005;5:58. doi: 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bendtsen JD, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernards MA, Razem FA. The poly(phenolic) domain of potato suberin: A non-lignin cell wall bio-polymer. Phytochemistry. 2001;57:1115–1122. doi: 10.1016/s0031-9422(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 5.Bernards MA. Demystifying suberin. Can J Bot. 2002;80:227–240. [Google Scholar]
- 6.Blakeney AB, Harris PJ, Henry RJ, Stone BA. A simple and rapid preparation of alditol acetates for monosaccharide analysis. Carbohydr Res. 1983;113:291–300. [Google Scholar]
- 7.Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 8.Chater KF, Biró S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiol Rev. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- 9.Chellapandi P, Jani HM. Production of endoglucanase by the native strains of Streptomyces isolates in submerged fermentation. Braz J Microbiol. 2008;39:122–127. doi: 10.1590/S1517-838220080001000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claudel-Renard C, Chevale C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–6639. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- 12.Faucher E, Savard T, Beaulieu C. Characterization of actinomycetes isolated from common scab lesions on potato tubers. Can J Plant Pathol. 1992;14:197–202. [Google Scholar]
- 13.Faucher E, Paradis E, Goyer C, Hodge NC, Hogue R, Stall RE, Beaulieu C. Characterization of Streptomyces causing deep-pitted scab of potato in Quebec, Canada. Int J Syst Bacteriol. 1995;45:222–225. [Google Scholar]
- 14.Francis IM, Jourdan S, Fanara S, Loria R, Rigali S. The cellobiose sensor CebR is the gatekeeper of Streptomyces scabies pathogenicity. mBio. 2015;6:e02018–14. doi: 10.1128/mBio.02018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godden B, Legon T, Helvestein P, Pennickx M. Regulation of the production of hemicellulolytic and cellulolytic enzymes by a Streptomyces sp. growing on lignocellulose. J Gen Microbiol. 1989;135:285–292. doi: 10.1099/00221287-135-2-285. [DOI] [PubMed] [Google Scholar]
- 16.Golba B, Treutter D, Kollar A. Effects of apple (Malus×domestica Borkh.) phenolic compounds on proteins and cell wall-degrading enzymes of Venturia inaequalis. Trees-Struct Funct. 2012;26:131–139. [Google Scholar]
- 17.Goyer C, Vachon J, Beaulieu C. Pathogenicity of Streptomyces scabies mutants altered in thaxtomin A production. Phytopathology. 1998;88:442–445. doi: 10.1094/PHYTO.1998.88.5.442. [DOI] [PubMed] [Google Scholar]
- 18.Harchand RK, Singh S. Induction of cellulases in Streptomyces albaduncus by different substrates. Indian J Microbiol. 2001;41:45–49. [Google Scholar]
- 19.Healy FG, Wach M, Krasnoff SB, Gibson DM, Loria R. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol Microbiol. 2000;38:794–804. doi: 10.1046/j.1365-2958.2000.02170.x. [DOI] [PubMed] [Google Scholar]
- 20.Hill J, Lazarovits G. A mail survey of growers to estimate potato common scab prevalence and economic loss in Canada. Can J Plant Pathol. 2005;27:46–52. [Google Scholar]
- 21.Hodgson DA. Primary metabolism and its control in Streptomycetes: a most unusual group of bacteria. Adv Microb Physiol. 2000;42:47–238. doi: 10.1016/s0065-2911(00)42003-5. [DOI] [PubMed] [Google Scholar]
- 22.Johnson EG, Joshi MV, Gibson DM, Loria R. Cellooligosaccharides released from host plants induce pathogenicity in scab-inducing Streptomyces species. Physiol Mol Plant Pathol. 2007;71:18–25. [Google Scholar]
- 23.Joshi MV, Bignell DRD, Johnson EG, Sparks JP, Gibson DM, Loria R. The AraC/XylS regulator TxtR modulates thaxtomin biosynthesis and virulence in Streptomyces scabies. Mol Microbiol. 2007;66:633–642. doi: 10.1111/j.1365-2958.2007.05942.x. [DOI] [PubMed] [Google Scholar]
- 24.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007;35:429–432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khatri BB, Tegg RS, Brown PH, Wilson CR. Temporal association of potato tuber development with susceptibility to common scab and Streptomyces scabiei-induced responses in the potato periderm. Plant Pathol. 2011;60:776–786. [Google Scholar]
- 27.Kieser T, Bibb MJ, Buttner MJ, Chatter KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Fondation; Norwich, UK: 2000. [Google Scholar]
- 28.Kolattukudy PE, Agrawal VP. Structure and composition of aliphatic constituents of potato tuber skin suberin. Lipids. 1974;9:682–691. [Google Scholar]
- 29.Komeil D, Simao-Beaunoir A-M, Beaulieu C. Detection of potential suberinase-encoding genes in Streptomyces scabiei strains and other actinobacteria. Can J Microbiol. 2013;59:294–303. doi: 10.1139/cjm-2012-0741. [DOI] [PubMed] [Google Scholar]
- 30.Komeil D, Padilla-Reynaud R, Lerat S, Simao-Beaunoir A-M, Beaulieu C. Comparative secretome analysis of Streptomyces scabiei during growth in the presence or absence of potato suberin. Proteome Sci. 2014;12:35. doi: 10.1186/1477-5956-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerat S, Simao-Beaunoir A-M, Wu R, Beaudoin N, Beaulieu C. Involvement of the plant polymer suberin and the disaccharide cellobiose in triggering thaxtomin A biosynthesis, a phytotoxin produced by the pathogenic agent Streptomyces scabies. Phytopathology. 2010;100:91–96. doi: 10.1094/PHYTO-100-1-0091. [DOI] [PubMed] [Google Scholar]
- 32.Lerat S, Forest M, Lauzier A, Grondin G, Lacelle S, Beaulieu C. Potato suberin induces differentiation and secondary metabolism in the genus Streptomyces. Microbes Environ. 2012;27:36–42. doi: 10.1264/jsme2.ME11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lever M. A new reaction for colorimetric determination of carbohydrates. Anal Biochem. 1972;47:273–279. doi: 10.1016/0003-2697(72)90301-6. [DOI] [PubMed] [Google Scholar]
- 34.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database CAZy in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loria R, Bukhalid RA, Fry BA, King RR. Plant pathogenicity in the genus Streptomyces. Plant Dis. 1997;81:836–846. doi: 10.1094/PDIS.1997.81.8.836. [DOI] [PubMed] [Google Scholar]
- 36.MacKenzie CR, Patel GB, Bilous D. Factors involved in hydrolysis of microcrystalline cellulose by Acetivibrio cellulolyticus. Appl Environ Microbiol. 1987;53:304–308. doi: 10.1128/aem.53.2.304-308.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martins I, Hartmann DO, Alves P, et al. Elucidating how the saprophytic fungus Aspergillus nidulans uses the plant polyester suberin as carbon source. BMC Genomics. 2014;15:613. doi: 10.1186/1471-2164-15-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McQueen DAR, Schottel JL. Purification and characterization of a novel extracellular esterase from pathogenic Streptomyces scabiei that is inducible by zinc. J Bacteriol. 1987;169:1967–1971. doi: 10.1128/jb.169.5.1967-1971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moire L, Schmutz A, Buchala A, Yan B, Stark RE, Ryser U. Glycerol is a suberin monomer. New experimental evidence for an old hypothesis. Plant Physiol. 1999;119:1137–1146. doi: 10.1104/pp.119.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moraïs S, Barak Y, Hadar Y, Wilson DB, Shoham Y, Lamed R, Bayer EA. Assembly of xylanases into designer cellulosomes promotes efficient hydrolysis of the xylan component of a natural recalcitrant cellulosic substrate. mBio. 2011;2:e00233–11. doi: 10.1128/mBio.00233-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, Van Sluyter SC, Haynes PA. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 42.Ofong AU, Pearce RB. Suberin degradation by Rosellinia desmazieresii. Eur J For Path. 1994;24:316–322. [Google Scholar]
- 43.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 44.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. Adaptation of protein secretion to extremely high salt concentrations by extensive use of the twin arginine translocation pathway. Mol Microbiol. 2002;5:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 46.Saritha M, Arora A, Surender S, Lata N. Streptomyces griseorubens mediated delignification of paddy straw for improved enzymatic saccharification yields. Bioresour Technol. 2013;135:12–17. doi: 10.1016/j.biortech.2012.11.040. [DOI] [PubMed] [Google Scholar]
- 47.Scheible W-R, Fry B, Kochevenko A, Schindelasch D, Zimmerli L, Somerville S, Loria R, Somerville CR. An Arabidopsis mutant resistant to thaxtomin A, a cellulose synthesis inhibitor from Streptomyces species. Plant Cell. 2003;15:1781–1794. doi: 10.1105/tpc.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao D, Dunlop WD, Lui EMK, Bernards MA. Immunostimulatory and anti-inflammatory polysaccharides from Tripterygium wilfordii: comparison with organic extracts. Pharm Biol. 2008;46:8–15. [Google Scholar]
- 49.Singhai PK, Sarma BK, Srivastava JS. Phenolic acid content in potato peel determines natural infection of common scab caused by Streptomyces spp. World J Microbiol Biotechnol. 2011;27:1559–1567. [Google Scholar]
- 50.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 51.Thierie J, Pennickx MJ. Modeling of competitive mutualistic relationships. Application to cellulose degradation by Streptomyces sp. strains. Curr Microbiol. 2007;55:507–511. doi: 10.1007/s00284-007-9022-7. [DOI] [PubMed] [Google Scholar]
- 52.Tsujiyama S-I, Sumida K, Ueno H. Influence of vanillin on the production of cellulytic and xylanolytic enzymes from a wood-rotting fungus, Corius versicolor. Mycoscience. 2001;41:527–532. [Google Scholar]
- 53.van Munster JM, Daly P, Delmas S, et al. The role of carbon starvation in the induction of enzymes that degrade plant-derived carbohydrates in Aspergillus niger. Fungal Genet Biol. 2014;72:34–47. doi: 10.1016/j.fgb.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vĕtrovský T, Steffen KT, Baldrian P. Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS One. 2014;9:e89108. doi: 10.1371/journal.pone.0089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeng J, Singh D, Laskar DD, Chen S. Degradation of native wheat straw lignin by Streptomyces viridosporus T7A. Int J Environ Sci Technol. 2013;10:165–174. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.