Abstract
In human and wildlife populations, the natural microbiota plays an important role in health maintenance and the prevention of emerging infectious diseases. In amphibians, infectious diseases have been closely associated with population decline and extinction worldwide. Skin symbiont communities have been suggested as one of the factors driving the different susceptibilities of amphibians to diseases. The activity of the skin microbiota of amphibians against fungal pathogens, such as Batrachochytrium dendrobatidis, has been examined extensively, whereas its protective role towards the cutaneous infectious diseases caused by Amphibiocystidium parasites has not yet been elucidated in detail. In the present study, we investigated, for the first time, the cutaneous microbiota of the Italian stream frog (Rana italica) and characterized the microbial assemblages of frogs uninfected and infected by Amphibiocystidium using the Illumina next-generation sequencing of 16S rRNA gene fragments. A total of 629 different OTUs belonging to 16 different phyla were detected. Bacterial populations shared by all individuals represented only one fifth of all OTUs and were dominated by a small number of OTUs. Statistical analyses based on Bray-Curtis distances showed that uninfected and infected specimens had distinct cutaneous bacterial community structures. Phylotypes belonging to the genera Janthinobacterium, Pseudomonas, and Flavobacterium were more abundant, and sometimes almost exclusively present, in uninfected than in infected specimens. These bacterial populations, known to exhibit antifungal activity in amphibians, may also play a role in protection against cutaneous infectious diseases caused by Amphibiocystidium parasites.
Keywords: amphibian, skin microbiota, Illumina sequencing, Amphibiocystidium, Rana italica
It is commonly accepted that plants and animals harbor assemblages of microbes and that such symbiotic microorganisms play crucial roles for their hosts (4, 40). The importance of natural microbiota in health maintenance and the prevention of diseases has been demonstrated in several systems (7, 15, 18, 21, 28, 52). Interactions between the host, symbiotic microbes, and pathogens have been shown to affect disease outcomes (28), and this represents a matter of special concern because emerging infectious diseases are increasingly threatening human and wildlife populations (32, 33).
The skin is the soft outer covering of vertebrates that interfaces with the environment. It provides the first line of defense from external factors, such as pathogens and harmful substances, prevents excessive water loss, and guarantees insulation and sensation. In amphibians, skin is a particularly complex organ as it plays key roles in respiration, osmoregulation, thermoregulation, pigmentation, chemical communication, and pathogen defense (10). Amphibian skin, owing to the presence of a glycoprotein-rich mucous layer, harbors a rich microbial community and, thus, represents a complex ecosystem shaped by interactions with the environment and host factors that influence colonization (48). Amphibians synthesize species-specific oligosaccharides that, together with components of the innate and adaptive immune systems, such as lysozyme, antimicrobial peptides, and mucosal antibodies, mediate the colonization process by specific microbial populations from the host’s environment (48).
Amphibians are faced with decline globally and emerging infectious diseases have been closely associated with this decline and, ultimately, extinction (5). Susceptibility to diseases may vary among individuals, populations, or species of amphibians, and their well-developed skin symbiotic communities have been suggested as one of the driving factors for these differences (4, 33, 41). The activity of amphibian cutaneous bacteria against the skin pathogen Batrachochytrium dendrobatidis (Bd) has attracted a lot of attention (2, 28–30). This fungus causes chytridiomycosis, an emerging infectious disease thought to be responsible for amphibian decline and extinction worldwide (22). On the other hand, some Bd-naive populations of amphibians have the ability to persist and co-exist with Bd, and population survival has been suggested to depend on the proportion of individuals with anti-Bd bacteria (24, 36, 56). Many factors have been suggested to play a role in amphibian disease resistance including host genetics, immunology, skin peptides, and environmental factors (48). Furthermore, bacteria isolated from amphibian skin are known to produce anti-fungal molecules that inhibit the growth of Bd (28), and have even been shown to reduce amphibian mortality under controlled experimental conditions (30).
Besides chytridiomycosis, amphibians are potentially threatened by other cutaneous diseases, among which include infections by parasites of the genus Amphibiocystidium in the class Mesomycetozoea, a group of microorganisms at the boundary between animals and fungi (42, 51). Such infections are macroscopically detectable by the presence of swellings in the skin on any part of the body (6, 26, 47) and may be associated with local declines in frog populations (17). In 2007, an infection by parasites of the genus Amphibiocystidium was detected for the first time in a population of the Italian stream frog (Rana italica) in Central Italy and is still occurring (49). R. italica is a stream-breeding anuran endemic of the Peninsular Italy, mainly occurring in forested areas associated with shady streams with a rocky substrate along the Apennine chain and adjacent hilly ranges (11), and is regarded as a species in need of strict protection by the European Union (Annex IV in the Council Directive 92/43/EEC).
A more detailed characterization of the microbial community structure and dynamics is needed in order to gain a better understanding of the relationship between the composition of the skin microbiota and health status. As in other environments, culture-based approaches are able to capture only a small fraction of the bacterial diversity present on amphibian skin (39). In this respect, next-generation sequencing technologies are advantageous because they provide a thorough description of microbial communities, including uncultivable members, due to high-throughput 16S rRNA gene tag-deep sequencing (25). Although this approach has been increasingly applied in recent studies to investigate skin-associated microbial communities in amphibians (3, 23, 35, 38, 41, 53), to the best of our knowledge, it has never been used to directly address bacterial populations with a potential defensive role against a cutaneous infectious disease, such as the infection by parasites of the genus Amphibiocystidium. Members of this genus, as well as other mesomycetozoeans, are uncultivable (42) and, thus, it is not possible to perform in vitro experiments in order to explore the inhibitory activity of cutaneous bacteria against Amphibiocystidium. The aim of the present study was to obtain information on the composition of the skin microbiota in the Italian stream frog (R. italica) using the Illumina next-generation sequencing of 16S rRNA gene fragments, and also to investigate differences in microbial assemblages between frogs infected and uninfected by Amphibiocystidium in order to infer the potential role of skin-associated bacteria in the defense against this parasite.
Materials and Methods
Sample collection
R. italica specimens were collected in April 2010 from a population in the Nestore Valley (Central Italy). The capture and handling of R. italica specimens were conducted according to the permission granted by the Italian Ministry of Environment (issue no. DPN-2009-0011282). Frogs were captured by hand using fresh latex gloves each time. Before swabbing, each frog was rinsed twice with sterile distilled water in order to remove transient bacteria not associated with the skin (37). In each specimen, sterile cotton-tipped swabs were drawn across the skin of the lateral, ventral, and dorsal surfaces of the body, thighs, and feet ten times for each site. Each swab was recovered into a sterile vial and stored at −20°C for later analyses. Frogs infected by parasites of the genus Amphibiocystidium were first identified by the presence of gross swellings on the skin and then confirmed by a histopathological examination following a previously reported method (45, 46). Six specimens, three uninfected (indicated as R151S, R155S, and R156S) and three infected (indicated as R150M, R152M, and R154M), were selected for a next generation sequencing analysis of the skin microbiota. All chosen frogs were females, with the only exception of R154M, which was a male. Specimen weights ranged from 6.2 to 9.4 g (average 8.2 g), while snout-vent lengths ranged from 40.25 to 45.09 mm (average 42.07 mm).
Metagenomic DNA extraction and 16S rRNA gene fragment sequencing
Metagenomic DNA was extracted from each swab using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA). Swabs were loaded directly into the bead tubes provided and the remaining extraction procedure was performed as previously reported (20).
The V5-V6 hypervariable regions of the 16S rRNA gene were amplified in 50-μL volume reactions using the 5× HOT FIREPol Blend Master Mix (Solis BioDyne, Tartu, Estonia) with 1 μM each of the primers 783F and 1027R (25). A 6-bp barcode was also included at the 5′ end of the 783F primer to allow sample pooling and subsequent sequence sorting. Cycling conditions included initial denaturation at 94°C for 5 min followed by 29 cycles of 94°C for 50 s, 47°C for 30 s, and 72°C for 30 s and a final extension at 72°C for 5 min. The amplified products were purified with the Wizard SV PCR purification kit (Promega Corporation, Madison, WI, USA) and DNA quantity and purity were spectrophotometrically evaluated by NanoDrop (Thermo Scientific, USA).
Purified amplicons with different barcodes were pooled in 100-μL samples with a DNA concentration of 40 ng μL–1. Multiplexed sequencing of all the pooled samples was performed on a single Illumina Hiseq 1000 lane, using a paired-end 2×100 base-pair protocol and 4.0 sequencing chemistry. Cluster extraction and base-calling processing analyses were performed using the Illumina CASAVA Analysis software, version 1.8. Illumina Hiseq 1000 sequencing was carried out at BMR Genomics, Padua, Italy. The entire sequence dataset is available in the European Nucleotide Archive of the EMBL database with study accession number PRJEB7174.
Each sequence was assigned to its original sample according to its barcode. A quality cut-off was then applied in order to remove sequences i) that did not contain the barcode, ii) with an average base quality value (Q) lower than 30. The barcode was removed from sequences before further processing. The reverse read of each paired-end sequence was reverse-complemented and merged with the corresponding forward read, inserting 10 Ns in between (12). The number of reads recovered from each sample ranged from 612,786 to 1,016,572, while 10,000 reads per sample were randomly selected to normalize further analyses on the bacterial community structure (25).
The clustering of operational taxonomic units (OTUs) was carried out on the forward reads that contained the entire V6 region using the UPARSE-OTU algorithm (19). The minimum identity between each OTU member sequence and the representative sequence (i.e. the sequence that showed the minimum distance to all other sequences in the OTU) was set to 97%. OTUs that comprised only one read were removed. The taxonomic classification of the OTUs was carried out with the RDP classifier (54) using the representative sequence for each OTU.
Irrespective of their OTU assignment, all the filtered and merged reads were also given a taxonomic attribution using the stand-alone version of RDP Bayesian Classifier, applying 50% of confidence as suggested for sequences shorter than 200 bp (12).
Statistical analysis
A cluster analysis of OTUs was carried out using the heatmap function in the R statistical environment. Heatmap is a color-shaded matrix that is useful for a quick overview or exploratory analysis of data, in which color intensity indicates the relative value of the occurrence of the OTU (a light color indicates a low value, while a dark color indicates a high value). A hierarchical plot was produced using an average linkage cluster analysis based on the Bray-Curtis dissimilarity matrix.
An analysis of similarity (ANOSIM) (13) was performed for comparisons of similarity between infected and uninfected groups. ANOSIM is a non-parametric, permutation-based test for significant differences in compositions or variations between groups, and returns a test statistic, R, with a P-value for significance. The R statistic typically ranges between 0 and 1, with a higher value indicating a greater degree of distinction among groups. ANOSIM was calculated using the Vegan package for the R software (44). In order to visualize similarities among samples, a non-metric Multidimensional Scaling (nMDS) analysis was employed using the Bray-Curtis dissimilarity matrix. The goal of nMDS is to reduce information from multiple variables into two dimensions using rank orders so that they can be plotted and interpreted in the phase space. All statistical analyses and graphical representations were carried out using the R framework and ggplot2 package (44, 55).
Results
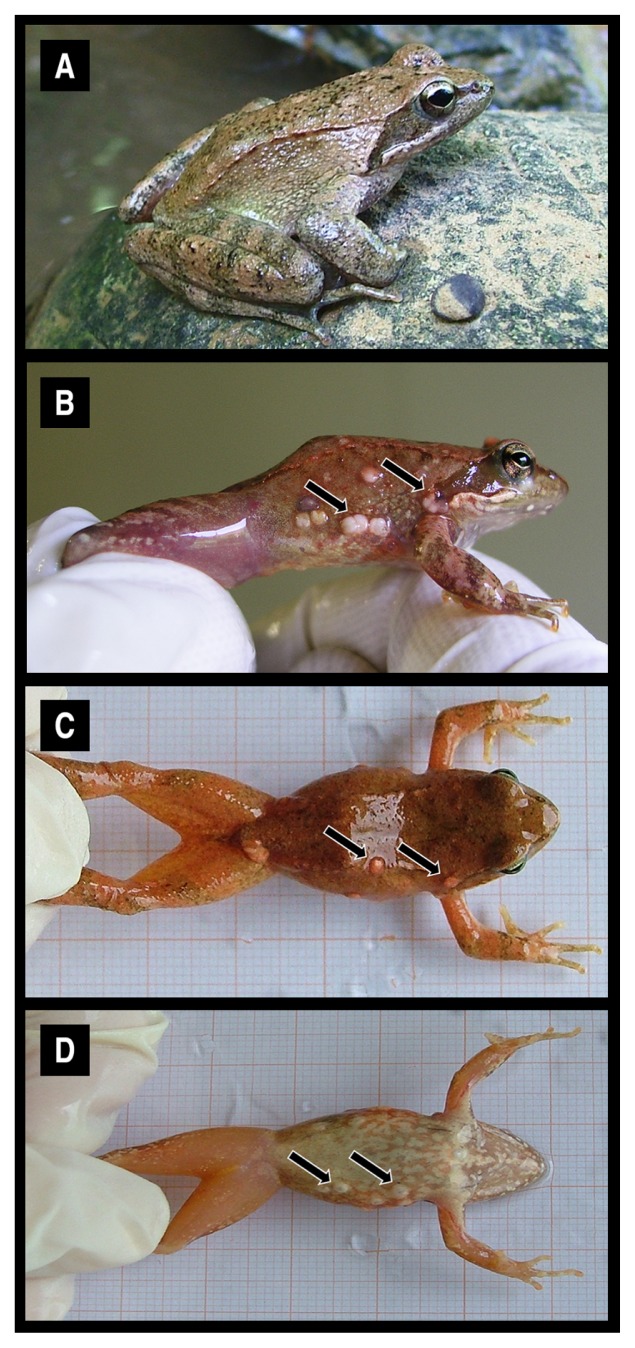
The R. italica specimens collected were divided into two groups: uninfected and infected by parasites of the genus Amphibiocystidium. Infected frogs exhibited gross swellings distributed on the dorsal and ventral skin of the head, torso, and limbs (Fig. 1). Further histopathological examinations indicated that the swellings appeared as spherical, ovoid, and/ or U-shape cysts filled with walled endospores. Three representative specimens of each group were selected in order to characterize the skin microbiota.
Fig. 1.
Specimens of Italian stream frogs (Rana italica) uninfected (panel A) and infected by a cutaneous parasite of the genus Amphibiocystidium (panels B, C, D). Arrows indicate the swellings commonly found on the lateral (B), dorsal (C) and ventral (D) skin of infected individuals.
The total number of microbial OTUs found across the two groups of specimens and calculated applying a threshold of 97% sequence similarity was 717, which was reduced to 629 after removing OTUs with only one sequence read. A total of 127 OTUs were shared by all six specimens. Of these, OTU_001, OTU_002, and OTU_003, identified as belonging to the families Comamonadaceae, Moraxellaceae, and Pseudomonadaceae, respectively, showed abundances ranging from 16.03% to 20.49% and together represented more than 53% of all the shared sequences. Considering OTU_004, OTU_005, and OTU_006, with abundances ranging from 4.31% to 4.74% and belonging to the families Methylophilaceae, Flavobacteriaceae, and Oxalobacteraceae, respectively, these six OTUs represented more than 66% of the common bacterial community.
The OTU richness found across samples, which ranged from 293 to 410, and the other diversity indexes are shown in Table 1. The bacterial diversity indexes were similar in specimens belonging to each group (i.e., uninfected and infected frogs), with uninfected frogs showing a generally higher bacterial diversity than that of infected frogs. The only exception was specimen R154M, an infected frog featuring diversity indexes closer to those obtained in uninfected specimens.
Table 1.
Bacterial alpha diversity of individual frogs based on the OTU distribution
| Specimena | S | H | D | J |
|---|---|---|---|---|
| R151S (U) | 410 | 3.8 | 0.93 | 0.63 |
| R155S (U) | 394 | 3.4 | 0.90 | 0.57 |
| R156S (U) | 375 | 3.3 | 0.87 | 0.56 |
| R150M (I) | 293 | 2.7 | 0.79 | 0.47 |
| R152M (I) | 355 | 2.6 | 0.78 | 0.45 |
| R154M (I) | 376 | 3.5 | 0.92 | 0.59 |
Specimens uninfected and infected by Amphibiocystidium are indicated as U and I, respectively.
S, OTU richness; H, Shannon index; D, Simpson index; J, Pielou index.
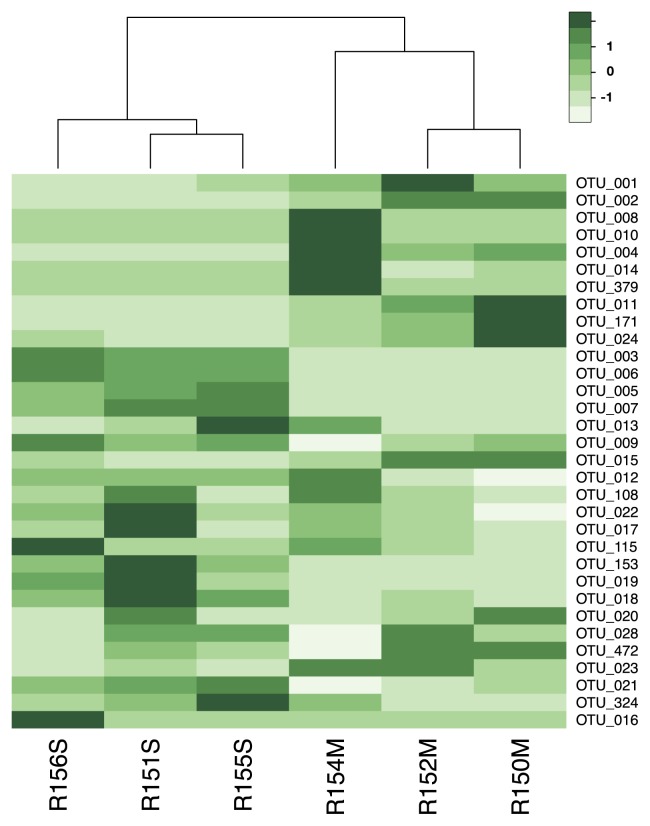
The community structures of the skin microbiota were shown in heatmaps reflecting relative OTU abundances across the six specimens (Fig. 2, a heatmap including only OTUs with a minimum abundance of 200 reads, and Fig. S1, heatmaps with OTU abundance cut-offs ranging from 5 to 600 reads). At all the considered OTU abundance cut-offs, the cluster analysis based on Bray-Curtis distances partitioned the six samples into two main groups, which corresponded to the bacterial communities of uninfected and infected frogs. Furthermore, the distances among uninfected frogs appeared to be shorter than those among infected frogs.
Fig. 2.
Heatmap showing the relative abundances of the OTUs across specimens uninfected (R151S, R155S, R156S) and infected (R150M, R152M, R154M) by Amphibiocystidium and clusters based on Bray-Curtis distances. The OTU abundance cut-off was set to a minimum of 200 reads. Darker green indicates higher values of abundance.
The ANOSIM analysis revealed a slight difference between the two groups (R=0.74; P=0.091). The nMDS analysis revealed that frogs uninfected by Amphibiocystidium had a more distinct bacterial composition than that of the infected frogs (Stress = 0.062) (Fig. S2).
In order to gain further insights into the identity and distribution of the bacterial populations in the skin of uninfected and infected frogs, the filtered and merged sequences obtained from each sample, irrespective of their OTU assignment, were given a taxonomic attribution as described in the Methods section. Excluding phylotypes represented by only one sequence and/or present in only one specimen, we detected a total of 16 different phyla. Of these, the dominant phyla, namely, those with a relative average abundance higher than 1%, were Proteobacteria (73.75%), Bacteroidetes (11.49%), Actinobacteria (4.78%), Firmicutes (2.24%), and Cyanobacteria (1.39%), which showed similar average abundances in uninfected and infected frogs, with the only exception of Bacteroidetes, which was higher in the former (14.86%) than in the latter (8.12%).
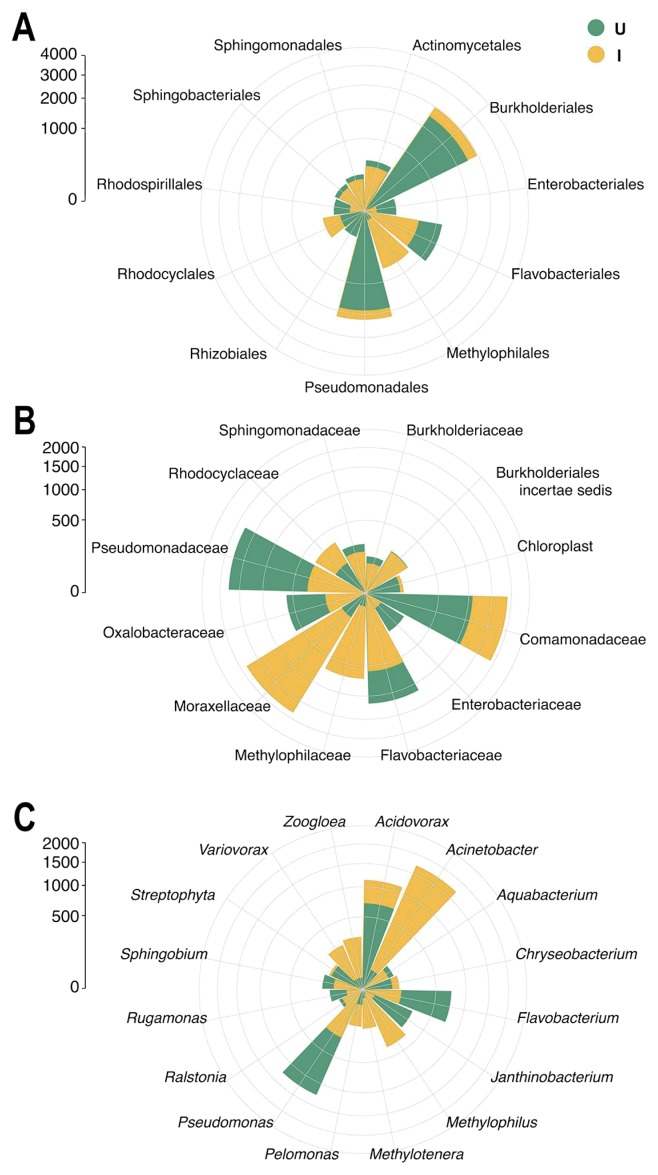
Among the most abundant orders shown in Fig. 3A, Burkholderiales, Pseudomonadales, and Actinomycetales presented average abundances that were similar in the skin of uninfected and infected frogs (37.81% and 39.49%, 28.15%, and 29.58%, and 7.40% and 5.06%, respectively). In contrast, Flavobacteriales were more abundant in uninfected specimens than in infected specimens (17.58% and 7.72%, respectively), while Methylophilales was abundant in infected frogs only (9.17%) (Fig. 3A).
Fig. 3.
Polar plots showing the average abundances of the most frequent bacterial phylotypes, at the taxonomic levels of order (A), family (B), and genus (C), found on the skin of specimens uninfected (U, in green) and infected (I, in yellow) by Amphibiocystidium.
The bacterial families Pseudomonadaceae, Flavobacteriaceae, and Oxalobacteraceae were more abundant in the skin of uninfected frogs than in that of infected frogs (31.15% and 5.06%, 20.24% and 8.94%, 10.41% and 2.46%, respectively) (Fig. 3B). Comamonadaceae, despite being more prevalent in the infected specimens than in the uninfected specimens (29.23% and 18.98%, respectively), were abundant in both groups (Fig. 3B). Furthermore, bacterial populations belonging to the families Moraxellaceae and Methylophilaceae were among the most abundant in the infected group of frogs only, in which they showed average abundances of 28.66% and 10.72%, respectively (Fig. 3B).
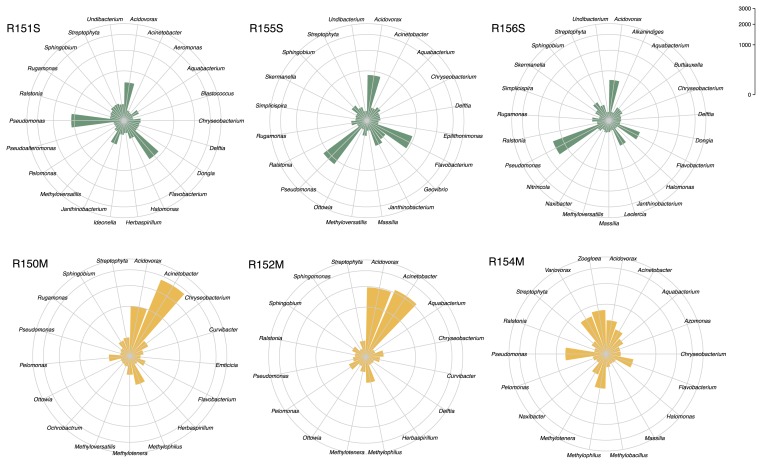
The average abundances of the most frequent bacterial genera in the two groups of specimens are shown in Fig. 3C, while the polar plots in Fig. 4 show the most abundant bacterial genera found in each frog specimen. Bacteria belonging to the genus Acidovorax showed similar average abundances in uninfected and infected frogs (19.01% and 23.22%, respectively), even though the abundance values were more similar among specimens in the uninfected group (ranging from 16.49% to 21.34%) than those in the infected group (ranging from 10.50% to 37.79%). The genera Pseudomonas and Flavobacterium showed higher average abundances in uninfected than in infected frogs (33.82% and 5.26%, 20.17% and 2.99%, respectively), and these bacteria within the latter group were abundant in specimen R154M only. Furthermore, the genus Janthinobacterium was among the most abundant in uninfected specimens only, with an average of 7.57%. In contrast, bacteria belonging to the genera Acinetobacter, Methylophilus, and Methylotenera were abundant in the infected frogs only, with averages of 35.37%, 7.54%, and 2.95% respectively. Two genera, namely, Zoogloea and Variovorax, were only abundant in the infected specimen R154M (17.80% and 14.94%, respectively).
Fig. 4.
Polar plots showing the abundances of the most frequent bacterial genera, found on the skin of specimens uninfected (R151S, R155S, R156S) and infected (R150M, R152M, R154M) by Amphibiocystidium.
Discussion
Our understanding of the composition and role of the microbiota associated with plants and animals, including humans, is increasing due to the application of high-throughput next-generation sequencing technologies in microbial ecology. Microbial communities living on animal skin represent a particularly interesting scenario as they are continuously exposed to the influence of the external environment. Nevertheless, these studies have largely been limited to the human skin microbiome (39). Among wild animals, amphibians, due to the absence of fur or feathers, provide an excellent model system for studying skin-associated microbial communities, which are considered to mediate susceptibility to diseases because they provide the first line of defense against pathogens (28, 37, 56).
In the present study, we used the Illumina next-generation sequencing of 16S rRNA gene fragments to characterize the skin microbiota of the Italian stream frog (R. italica) and investigate its relationship with a cutaneous infectious disease caused by a mesomycetozoean parasite of the genus Amphibiocystidium. A similar approach has been used in recent studies to address different aspects of the amphibian skin microbiota, such as the bacterial community composition in co-habiting amphibian species (41), variabilities across species, sampling sites, and developmental stages (35), occurrence in the wild and stability through time in captivity (3, 38), and its relationship with environmental microbes (23, 53). Nevertheless, to the best of our knowledge, this is the first study on the skin microbiota of the Italian stream frog and, in particular, the first to show differences between infected and uninfected frogs with the aim of addressing bacterial populations with the potential to reduce susceptibility to a cutaneous disease in amphibians. Unlike the chytrid fungal skin pathogen Bd, it is not possible to culture Amphibiocystidium with the aim of carrying out growth inhibition assays (42); therefore, we hypothesized that the identification of protective bacterial taxa may be achieved by comparing the community composition in the skin of uninfected frogs with that of infected frogs. McKenzie and collaborators (41), who studied co-habiting amphibian species, found that one individual of the Western chorus frog (P. triseriata) was positive for Bd; however, due to the lack of other positive individuals, they were unable to draw any concrete conclusions regarding possible interactions between the infection and skin bacterial community. Previous studies demonstrated that the composition of the skin bacterial community was largely species-specific (41), that a strong developmental shift occurred in skin microbes following metamorphosis (35), and that pond sites represented a secondary influencing factor because, in a host species-specific manner, amphibian skin may select microbes that are generally present in low abundance in the environment (53). Therefore, in the present study, we only considered specimens belonging to the same species and same developmental stage sampled on the same day at the same site, with the aim of isolating the absence/presence of the cutaneous infectious disease from other factors structuring the skin microbiota.
In our survey, we found 629 different OTUs, which showed a per-sample abundance ranging from 293 to 410 and belonging to 16 different phyla. Using a similar approach, Costello et al.(14) demonstrated that the human skin microbiome was comprised of 18 different bacterial phyla, while McKenzie et al.(41) found between 10 and 18 unique skin bacterial phyla in amphibians, depending on the species. Therefore, the richness of the skin bacterial community that we discovered in the Italian stream frog was similar to that found in humans and other amphibian species. Our results also confirmed the ability, provided by the next-generation sequencing of 16S rRNA genes, to examine the skin bacterial community composition in more depth in respect to other culture-dependent or culture-independent approaches. Woodhams et al.(56) cultured 40 unique bacterial isolates belonging to only 3 different phyla from the skin of Rana muscosa, while Lauer et al.(37) found 19 different bacterial populations, representing only 4 different phyla, in eastern red-backed salamanders (Plethodon cinereus) using gradient gel electrophoresis 16S rRNA fingerprinting. We demonstrated that the bacterial community of all specimens was dominated by a limited number of phyla, namely Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes, and Cyanobacteria, which were largely the same taxa that were found to be dominant in similar studies (35, 41, 53). Furthermore, in our specimens, the shared bacterial community, namely, the bacterial populations present in the skin of all individuals, was composed of approximately one fifth of all OTUs and dominated by a small number of OTUs. These results are consistent with previous findings obtained using a next-generation sequencing approach, which showed that a few phylotypes dominated the cutaneous bacterial communities of several amphibian species (35, 38, 41, 53).
By comparing frogs uninfected and infected by Amphibiocystidium in the present study, a generally higher bacterial diversity was detected in the former. Our results also showed that the two groups had distinct bacterial community structures, as revealed by an nMDS analysis and cluster analysis based on Bray-Curtis distances, indicating that a relationship may exist between the bacterial community and infection occurrence. Furthermore, the shorter distances observed among the uninfected frogs than among the infected frogs indicated that the bacterial communities in the former were more homogenous than in the latter, suggesting the presence of a core microbiota with putative protective activity against the parasitic infection in uninfected frogs. The persistence of a Bd-naïve population of Rana muscosa was previously associated with a high proportion of individuals with anti-Bd bacteria (36). These findings suggested that protection, similar to herd immunity, may be based on a proportion of individuals needing to have anti-Bd bacteria in order to protect the population and allow co-existence with the pathogen. Since Amphibiocystidium infection was detected in the frog population inhabiting the same collection site of the present study in 2007 (unpublished data), a similar mechanism of protection may also occur against this amphibian skin pathogen. Thus, a closer examination of the composition of bacterial communities, and particularly the identification of bacterial phylotypes with a different distribution between infected and uninfected individuals, may provide an insight into bacterial populations with a protective role.
Most of the bacterial populations detected in the present study belonged to the phylum Proteobacteria, and, in particular, the orders Burkholderiales, Pseudomonadales, and Methylophilales. Even if Burkholderiales and Pseudomonadales showed similar abundances in the skin of uninfected and infected frogs at the order level, their distribution between the two groups varied at lower taxonomic ranks.
Within Burkholderiales, bacteria of the family Oxalobacteraceae were more abundant in uninfected specimens. Bacteria belonging to the genus Janthinobacterium were almost exclusively present in uninfected specimens. Janthinobacterium species are among the most extensively studied bacteria with disease-protective roles in amphibians and have been cultured from the skin of various species. Janthinobacterium isolates, obtained from H. scutatum, showed antifungal activity in vitro against Mariannaea elegans and Rhizomucor variabilis (37), and a strain of J. lividum, isolated from the skin of Plethodon cinereus, was found to produce antifungal metabolites at concentrations that inhibited the growth of Bd (9). In vivo studies demonstrated that the addition of J. lividum to the skin of Rana muscosa prevented morbidity and mortality caused by such a pathogen (29) and also that these effects were, at least in the red-backed salamander (P. cinereus), related to increases in the concentration of the antifungal metabolite violacein on the skin (1). Using Illumina technology Loudon et al.(38) identified J. lividum in a large proportion of red-backed salamanders (P. cinereus) both in the wild and captivity. We also showed that another family of Burkholderiales, namely, the Comamonadaceae, and particularly those of the genus Acidovorax, despite being abundant in both groups, prevailed within infected specimens. Members of the family Comamonadaceae were previously shown to be abundant in the skin of amphibians, including frogs, salamanders, and newts (35, 38, 41, 43), and strains of Acidovorax sp. were isolated from the skin of Rana cascadae (50) and Hemidactylium scutatum (37).
Within the order Pseudomonadales, bacteria belonging to the family Pseudomonadaceae, particularly those of the genus Pseudomonas, were more abundant in uninfected than infected specimens. Using next-generation sequencing, Pseudomonadaceae were found to be among the most abundant phylotypes in frog and salamander species (38, 41). Furthermore, Kueneman et al.(35) showed that the occurrence of Pseudomonas was high on R. cascadae tadpoles and in the water of the lake from which these specimens were captured. Similar findings were reported by Walke et al.(53) for juvenile bullfrogs (Rana catesbeiana) and adult red-spotted newts (Notophthalmus viridescens), as well as the pond substrate inhabited by these individuals. Several Pseudomonas species are known to provide protection against pathogenic bacteria and fungi in many plant and animal hosts, and are used as probiotics in agriculture and aquaculture (16, 27, 31). In amphibians, a large number of culture-dependent studies have shown that many pseudomonads may be isolated from the skin of different species and produce antimicrobials capable of inhibiting bacterial and fungal pathogens, including Bd (28, 36, 37, 56). Within endangered mountain yellow-legged frogs (Rana sierrae and Rana muscosa), several Pseudomonas species, together with other bacteria capable of inhibiting Bd growth in vitro, were found to occur more frequently on individuals belonging to populations that co-existed with the pathogen and survived the infection than on those in populations that were extirpated by Bd infection (36, 56). Harris et al.(30) demonstrated that adding Pseudomonas reactans to salamanders (P. cinereus) infected with Bd ameliorated the effects of chytridiomycosis. We found that, within the Pseudomonadales, in contrast to that observed for Pseudomonadaceae, bacteria of the family Moraxellaceae, particularly those of the genus Acinetobacter, were among the most abundant in the infected group of frogs only. Despite Acinetobacter species showing anti-Bd activity in toads of the genus Atelopus (24), these bacteria have also been associated with diseased coral (Oculina patagonica) and stress-exposed fish (Salvelinus fontinalis) (8, 34). Thus, the occurrence of Acinetobacter species in R. italica may also be related to stressful conditions, such as the occurrence of Amphibiocystidium infection.
We found phylotypes of the order Methylophilales almost exclusively on the infected specimens. This order was largely represented by two genera of the family Methylophilaceae, namely Methylophilus and Methylotenera, bacteria commonly found in typical amphibian environments, such as mud as well as river and pond water. Methylotenera species were previously detected on Western chorus frogs (41), American bullfrogs (35), and red-spotted newts (53). Nevertheless, to the best of our knowledge, our results on R. italica are the first to suggest that these two bacterial genera may be related to stress conditions such as a pathogen infection.
Within the phylum Proteobacteria, frog R154M showed, besides those shared with other specimens, further dominant taxa uncharacteristic of its own bacterial community, such as the genera Zoogloea and Variovorax. Since R154M was the only male frog examined in the present study, we cannot exclude the possibility that its atypical bacterial community may have depended on gender, as previously reported (37).
The second most abundant phylum that we encountered in our survey was represented by Bacteroidetes, which was disproportionate in the two groups of specimens, being more abundant in uninfected frogs. This was largely attributed to the very high abundance of phylotypes belonging to the genus Flavobacterium (order Flavobacteriales, family Flavobacteriaceae) in the uninfected specimens. Bacteria belonging to Flavobacteriaceae were isolated (43, 50) and identified by next-generation sequencing (3, 35, 41) in the skin of frogs and other amphibians. Furthermore, Lauer et al.(37) detected Flavobacterium species among the most frequently occurring bacteria, isolated from the skin of salamanders, with antifungal activities, while Lam et al.(36) isolated a Flavobacterium strain showing anti-Bd activity in vitro from the skin of Rana muscosa. These findings, together with our result showing the abundance of Flavobacteria in uninfected specimens of R. italica, point toward the possible protective role of these bacteria against cutaneous diseases in amphibians.
Conclusion
We herein provided, for the first time, evidence for the cutaneous microbiota composition of R. italica, which appeared to be dominated by a few OTUs, similar to other amphibian species. By comparing uninfected and infected individuals, we observed a relationship between the structure of the skin bacterial community and occurrence of a cutaneous infectious disease caused by a mesomycetozoean of the genus Amphibiocystidium. We demonstrated that the abundances of phylotypes identified as Pseudomonas, Flavobacterium, and Janthinobacterium were higher in uninfected specimens than in infected specimens. Since members of these genera are known to exhibit antifungal activity in amphibians and taking into account the impossibility of culturing Amphibiocystidium to perform growth inhibition assays, these results led us to hypothesize that these bacteria may play a role in protecting R. italica against the Amphibiocystidium infection.
In order to better understand the protective role of skin microorganisms in R. italica, it will be necessary to carry out a similar approach in a larger amphibian population and explore further factors affecting the microbiota composition among uninfected and infected hosts, such as different time points, including seasonal cycles and different capture sites. Furthermore, since Kueneman et al.(35) demonstrated the occurrence of a marked shift in the skin microbial community following metamorphosis and considering that such a process involves changes in the immune system and in the skin structure that may lead to a greater risk of pathogen infections, it will also be necessary to analyze the microbiome of R. italica specimens in different life history stages in order to verify a possible relationship with Amphibiocystidium infection.
Supplementary Information
References
- 1.Becker MH, Brucker RM, Schwantes CR, Harris RN, Minbiole KP. The bacterially produced metabolite violacein is associated with survival of amphibians infected with a lethal fungus. Appl Environ Microbiol. 2009;75:6635–6638. doi: 10.1128/AEM.01294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker MH, Harris RN. Cutaneous bacteria of the red-back salamander prevent morbidity associated with a lethal disease. PLoS One. 2010;5:e10957. doi: 10.1371/journal.pone.0010957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker MH, Richards-Zawacki C, Gratwicke B, Belden LK. The effect of captivity on the cutaneous bacterial community of the critically endangered Panamanian golden frog (Atelopus zeteki) Biol Conserv. 2014;176:199–206. [Google Scholar]
- 4.Belden LK, Harris RN. Infectious diseases in wildlife: the community ecology context. Front Ecol Environ. 2007;5:533–539. [Google Scholar]
- 5.Blaustein AR, Gervasi SS, Johnson PTJ, Hoverman JT, Belden LK, Bradley PW, Xie GY. Ecophysiology meets conservation: understanding the role of disease in amphibian population declines. Phil Trans R Soc B. 2012;367:1688–1707. doi: 10.1098/rstb.2012.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borteiro C, Cruz JC, Kolenc F, et al. Dermocystid-chytrid coinfection in the neotropical frog Hypsiboas pulchellus (Anura: Hylidae) J Wild Dis. 2014;50:150–153. doi: 10.7589/2013-06-151. [DOI] [PubMed] [Google Scholar]
- 7.Bourne DG, Garren M, Work TM, Rosenberg E, Smith GW, Harvell CD. Microbial disease and the coral holobiont. Trends Microbiol. 2009;17:554–562. doi: 10.1016/j.tim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 8.Boutin S, Bernatchez L, Audet C, Derôme N. Network analysis highlights complex interactions between pathogen, host and commensal microbiota. Plos One. 2013;8:e84772. doi: 10.1371/journal.pone.0084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, Lam BA, Minbiole KPC. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the Salamander Plethodon cinereus. J Chem Ecol. 2008;34:1422–1429. doi: 10.1007/s10886-008-9555-7. [DOI] [PubMed] [Google Scholar]
- 10.Campbell CR, Voyles J, Cook DI, Dinudom A. Frog skin epithelium: electrolyte transport and chytridiomycosis. Int J Biochem Cell Biol. 2012;44:431–434. doi: 10.1016/j.biocel.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canestrelli D, Cimmaruta R, Nascetti G. Population genetic structure and diversity of the Apennine endemic stream frog, Rana italica—insights on the Pleistocene evolutionary history of the Italian peninsular biota. Mol Ecol. 2008;17:3856–3872. doi: 10.1111/j.1365-294X.2008.03870.x. [DOI] [PubMed] [Google Scholar]
- 12.Claesson MJ, Wang Q, O’Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O’Toole PW. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010;38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clarke KR, Warwick RM. A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar Ecol Prog Ser. 2001;216:265–278. [Google Scholar]
- 14.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danielsson J, Reva O, Meijer J. Protection of oilseed rape (Brassica napus) toward fungal pathogens by strains of plant-associated Bacillus amyloliquefaciens. Microb Ecol. 2007;54:134–140. doi: 10.1007/s00248-006-9181-2. [DOI] [PubMed] [Google Scholar]
- 16.Daskin JH, Alford RA. Context-dependent symbioses and their potential roles in wildlife diseases. Proc R Soc B. 2012;279:1457–1465. doi: 10.1098/rspb.2011.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Rosa I, Simoncelli F, Fagotti A, Pascolini R. The proximate cause of frog decline? Nature. 2007;447:E4–E5. doi: 10.1038/nature05941. [DOI] [PubMed] [Google Scholar]
- 18.Dillon RJ, Vennard CT, Buckling A, Charnley AK. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett. 2005;8:1291–1298. [Google Scholar]
- 19.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 20.Federici E, Pepi M, Esposito A, Scargetta S, Fidati L, Gasperini S, Cenci G, Altieri R. Two-phase olive mill waste composting: community dynamics and functional role of the resident microbiota. Bioresour Technol. 2011;102:10965–10972. doi: 10.1016/j.biortech.2011.09.062. [DOI] [PubMed] [Google Scholar]
- 21.Fierer N, Ferrenberg S, Flores GE, et al. From animalcules to an ecosystem: application of ecological concepts to the human microbiome. Annu Rev Ecol Evol Syst. 2012;43:137–155. [Google Scholar]
- 22.Fisher MC, Garner TWJ, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian Chytridiomycosis in space, time, and host. Ann Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- 23.Fitzpatrick BM, Allison AL. Similarity and differentiation between bacteria associated with skin of salamanders (Plethodon jordani) and free-living assemblages. FEMS Microbiol Ecol. 2014;88:482–494. doi: 10.1111/1574-6941.12314. [DOI] [PubMed] [Google Scholar]
- 24.Flechas SV, Sarmiento C, Cardenas ME, Medina EM, Restrepo S, Amezquita A. Surviving chytridiomycosis: differential anti-Batrachochytrium dendrobatidis activity in bacterial isolates from three lowland species of Atelopus. PLoS One. 2012;7:e44832. doi: 10.1371/journal.pone.0044832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzetti A, Tatangelo V, Gandolfi I, Bertolini V, Bestetti G, Diolaiuti G, D’Agata C, Mihalcea C, Smiraglia C, Ambrosini R. Bacterial community structure on two alpine debriscovered glaciers and biogeography of Polaromonas phylotypes. ISME J. 2013;7:1483–1492. doi: 10.1038/ismej.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.González-Hernández M, Denoël M, Duffus AJ, Garner TW, Cunnungham AA, Acevedo-Whitehouse K. Dermocystid infection and associated skin lesions in free-living palmate newts (Lissotriton helveticus) from southern France. Parasitol Int. 2010;59:344–350. doi: 10.1016/j.parint.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Haas D, Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 28.Harris RN, James TY, Lauer A, Simon MA, Patel A. Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of amphibian species. EcoHealth. 2006;3:53–56. [Google Scholar]
- 29.Harris RN, Brucker RM, Walke JB, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009;3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- 30.Harris RN, Lauer A, Simon MA, Banning JL, Alford RA. Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis Aquat Org. 2009;83:11–16. doi: 10.3354/dao02004. [DOI] [PubMed] [Google Scholar]
- 31.Irianto A, Austin B. Probiotics in aquaculture. J Fish Dis. 2002;25:1–10. [Google Scholar]
- 32.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilpatrick AM, Briggs CJ, Daszak P. The ecology and impact of chytridiomycosis: an emerging disease of amphibians. Trends Ecol Evol. 2010;25:109–118. doi: 10.1016/j.tree.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Koren O, Rosenberg E. Bacteria associated with the bleached and cave coral Oculina patagonica. Microb Ecol. 2008;55:523–529. doi: 10.1007/s00248-007-9297-z. [DOI] [PubMed] [Google Scholar]
- 35.Kueneman JC, Parfrey LW, Woodhams DC, Archer HM, Knight R, McKenzie VJ. The amphibian skin-associated microbiome across species, space and life history stages. Mol Ecol. 2014;23:1238–1250. doi: 10.1111/mec.12510. [DOI] [PubMed] [Google Scholar]
- 36.Lam BA, Walke JB, Vredenburg VT, Harris RN. Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol Conserv. 2010;143:529–531. [Google Scholar]
- 37.Lauer A, Simon MA, Banning JL, Lam BA, Harris RN. Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. ISME J. 2008;2:145–157. doi: 10.1038/ismej.2007.110. [DOI] [PubMed] [Google Scholar]
- 38.Loudon AH, Woodhams DC, Parfrey LW, Archer H, Knight R, McKenzie V, Harris RN. Microbial community dynamics and effect of environmental microbial reservoirs on redbacked salamanders (Plethodon cinereus) ISME J. 2014;8:830–840. doi: 10.1038/ismej.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathieu A, Vogel TM, Simonet P. The future of skin metagenomics. Res Microb. 2014;165:69–76. doi: 10.1016/j.resmic.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 40.McFall-Mgai M, Hadfieldb MG, Bosch TCG, et al. Animals in a bacterial world, a new imperative for the life sciences. PNAS. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenzie JV, Bowers RM, Fierer N, Knight R, Lauber CL. Co-habiting amphibian species harbour unique skin bacterial communities in wild populations. ISME J. 2012;6:588–596. doi: 10.1038/ismej.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendoza L, Taylor JW, Ajello L. The class Mezomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu Rev Microbiol. 2002;56:315–344. doi: 10.1146/annurev.micro.56.012302.160950. [DOI] [PubMed] [Google Scholar]
- 43.Michaels CJ, Antwis RE, Preziosi RF. Impact of plant cover on fitness and behavioral traits of captive red-eyed tree frogs (Agalychnis callidryas) Plos One. 2014;9:e95207. doi: 10.1371/journal.pone.0095207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oksanen J, Blanchet FG, Kindt R, et al. Package Vegan: Community ecology package, version 2.0 10. 2013 http://CRAN.Rproject.org/package=vegan.
- 45.Pascolini R, Daszak P, Cunningham AA, Tei S, Vagnetti D, Bucci S, Fagotti A, Di Rosa I. Parasitism by Dermocystidium ranae in a population of Rana esculenta complex in Central Italy and description of Amphibiocystidium n. gen. Dis Aquat Org. 2003;56:65–74. doi: 10.3354/dao056065. [DOI] [PubMed] [Google Scholar]
- 46.Pereira CN, Di Rosa I, Fagotti A, Simoncelli F, Pascolini R, Mendoza L. The pathogen of frogs Amphibiocystidium ranae is a member of the Order Dermocystida in the Class Mesomycetozoea. J Clin Microbiol. 2005;43:192–198. doi: 10.1128/JCM.43.1.192-198.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raffel TR, Bommarito T, Barry DS, Witiak SM, Shackelton LA. Widespread infection of the eastern red-spotted newt (Notophthalmus viridescens) by a new species of Amphibiocystidium, a genus of fungus-like mesomycetozoean parasites not previously reported in North America. Parasitology. 2008;135:203–215. doi: 10.1017/S0031182007003708. [DOI] [PubMed] [Google Scholar]
- 48.Rollins-Smith LA, Ramsey JP, Pask JD, Reinert LK, Woodhams DC. Amphibian immune defenses against chytridiomycosis: impacts of changing environments. Integr Comp Biol. 2011;51:552–562. doi: 10.1093/icb/icr095. [DOI] [PubMed] [Google Scholar]
- 49.Rossi R, Barocco R, Paracucchi R, Di Rosa I. In: Carafa M, Di Francesco N, Di Tizio L, Pellegrini Mr, editors. Parasitism by Amphibiocystidium (Mesomycetozoea) in Rana italica Dubois, 1987 (Amphibia: Anura: Ranidae): preliminary data; Proceedings of the Ith Symposium of the Socìetas Herpetologica Italica, Section Abruzzo “Antonio Bellini”; Talea Edizioni, Atessa, CH. 2008. pp. 35–40. [Google Scholar]
- 50.Roth T, Foley J, Worth J, Piovia-Scott J, Pope K, Lawler S. Bacterial flora on Cascades frogs in the Klamath mountains of California. Comp Immunol Microbiol Infect Dis. 2013;36:591–598. doi: 10.1016/j.cimid.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Rowley JJL, Gleason FH, Andreou D, Marshall WL, Lilje O, Gozlan R. Impacts of mesomycetozoean parasites on amphibian and freshwater fish populations. Fun Biol Rev. 2013;27:100–111. [Google Scholar]
- 52.Shnit-Orland M, Sivan A, Kushmaro A. Antibacterial activity of Pseudoalteromonas in the coral holobiont. Microb Ecol. 2012;64:851–859. doi: 10.1007/s00248-012-0086-y. [DOI] [PubMed] [Google Scholar]
- 53.Walke JB, Becker MH, Loftus SC, House LL, Cormier G, Jensen RV, Belden LK. Amphibian skin may select for rare environmental microbes. ISME J. 2014;8:2207–2217. doi: 10.1038/ismej.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 56.Woodhams DC, Vredenburg VT, Simon MA, Billheimer D, Shakhtour B, Shyr Y, Briggs CJ, Rollins-Smith LA, Harris RN. Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biol Conserv. 2007;138:390–398. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.