Abstract
There are very few studies about K-ras mutations in colorectal cancer (CRC) from developing countries such as Iran. It is therefore essential to conduct studies to learn about the molecular signature of such tumors, allowing the determination of an appropriate management plan. In the present study, we aimed to determine the frequency and types of K-ras mutations among patients with CRC in Iran.
Formalin-fixed paraffin-embedded specimens of 100 cases of CRC were collected from hospitals affiliated with Shiraz University of Medical Sciences (June 2011 to June 2013). All of the H&E slides were examined and proper slide with a minimum of necrosis and maximum of well-preserved tumor cells (at least 70% tumor in each slide) were selected. Recurrent, metastatic, and post chemotherapy cases were excluded from the study. Mutation of codons 12 and 13 of K-ras gene by PCR was performed, followed by direct sequencing by Sanger method.
From 100 eligible cases (55 male and 45 females with mean age of 59 years), 32% had mutant K-ras gene; the most common substitution was 12G>C followed by 12G>A and 13G>A, respectively.
It is found that K-ras mutation rate, among the selected population of the southern province of Iran, was as high as 32% (codon 12: 71.8% and in codon 13: 25% and one in both codons: 3.1%).
Keywords: K-ras, Colorectal cancer, Frequency, Iran
Introduction
Colorectal cancer is one of the most frequent cancers worldwide and about half a hundred thousand people die annually from colorectal cancer (CRC).1
CRC is a major cause of morbidity and mortality in western developed countries; however, this cancer is decreasing in these countries. However, according to the recent studies, the incidence of CRC in Asian countries is increasing by 2 to 4 folds.2 Recently, the management of metastatic colorectal cancer (mCRC) has considerably improved due to a number of novel drugs, including targeted agents like bevacizumab, cetuximab, and panitumumab.3 In recent years, more studies have been focused on the role of K-ras mutation in the growth and histopathology of the tumor, clinical outcomes, and management choice of large bowel cancer.1,3,4
Epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that involve in cell development. With activation of the EGFR, the receptor causes activation of its kinase action. Afterward, the downstream signal cascade is started, which comprises activation of K-ras GTPase and Erk/Map kinase ensuing in cell proliferation. EGFR expression is noticed in up to 80% of CRC. Tumors with a high level of EGFR expression usually have unfortunate prognosis.5
Naturally turned on K-ras not barely uphold tumor beginning, but also tumor growth, survival, succession, local assault, metastasis, vascular density, and even immune response.1 In addition, mutated K-ras promotes angiogenesis and, in turn, helps tumor succession. Antiangiogenic remedy with bevacizumab has established to be effectual in K-ras mutant CRC.2 It is also revealed that expression of activated K-ras ease tumor invasion and metastasis.2
This proto-oncogene is regularly mutated (30-50% in different surveys) in CRC. Roughly 90% of the activating mutations, that are influential solitary amino acid replacement in the GTPase pocket that guide to a block of the activity of K-ras p21 protein, are recognized in codons 12 (GGT) and 13 (GGC) of exon 1 and almost 5% in codon 61 (CAA) situated in exon 2. The most regularly found kinds of mutations are G>A and G>T transitions.4 K-ras testing is said to have a vital improvement in the treatment of CRCs, especially after metastasis for anti-EGFR therapy.1,3
There are very few studies about the frequency of K-ras mutation from the Middle East and Iran; consequently, in the present study, we have analyzed K-ras mutation rate and spectrum in largest referral hospitals of southern Iran to determine the exact frequency of this mutation in CRCs.
Materials and Methods
From June 2011 to June 2013, formalin-fixed paraffin-embedded specimens of all patients with CRC that underwent surgical removal of the whole tumor were collected from hospitals affiliated to Shiraz University of Medical Sciences (Namazi and Faghihi Hospitals). All of the H&E slides were examined and proper slide with a minimum of necrosis and maximum of well-preserved tumor cells (at least 70% viable tumor cells)5 were selected. Recurrent, metastatic and post chemotherapy cases were excluded from the study, because we intended to select pure cases with no manipulation in order to have the exact and true incidence of this mutation in our region.
K-ras mutation in codons 12 and 13 was evaluated in formalin-fixed paraffin-embedded (FFPE) tissue by PCR and DNA sequencing by the Sanger method as described by Nagasaka et al.6
DNA was extracted from FFPE tissue by using DNPTm kit after deparaffinization. PCR was performed according to the published method by Nagasaka et al.6 Primers used are shown below:
| PCR primers: | ||
| GCTGAAAATGACTG | Forward | 113 bp |
| AATATAAACTTGT | ||
| TTGTTGGATCATATTCGTCCAC | Reverse | |
| Sequencing primer:1 | ||
| TGGATCATATTCGTCCACAA | Reverse |
Results
Hundred CRC cases were analyzed during the study period. The age range was 21-87 (59.08±15.55) years. Of the 100 eligible cases, 55 were male and 45 females. Mutation in codon 12 was found in 23 cases (71.8% of mutant cases) and in codon 13, there were 8 cases (25% of mutant cases). figures 1 and 2 show the PCR and DNA sequencing of selected cases.
Figure 1.
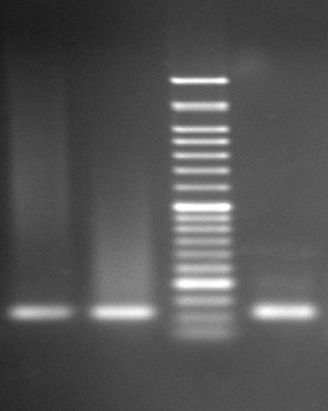
The picture shows K-ras gel electrophoresis 1. First Left: Positive control, K-ras codon 13 positive G12C/+ (GGT→TGT), 2. Second left: Cell line control, K-ras codon 13 positive G13N/+ (GGC→AAC), 3. First right: Patient sample.
Figure 2.

The figure shows DNA sequencing of mutation in codon 13 (with 2G to C substitution GGC→AAC).
Normal codon 12 (Glycine: GGT) and codon 13 (Alaninie: GGC) were found in 68% of the 100 cases of CRC. One case showed mutation in both codons (3.1% of mutant cases). In this case, mutation in both codons 12 and 13 has been detected. In this case there were substitutions of Alanine instead of Glycine in codon 12 (GCT instead of GGT) and Aspartic acid instead of Alanine in codon 13 (GAT instead of GGT).
table 1 shows the different types of detected mutations in this group of Iranian patients along with their respective amino acids. As the table shows, the most common mutation in our patients was in codon 12, with substitution of Alanine instead of Glycine (GCT instead of GGT), which was present in 12% of the cases. The second most common substitution was in codon 12, with substitution of Aspartic acid instead of Alanine (GAT instead of GGT) in 9% of the cases. In codon 13, the most common substitution was in Aspartic acid instead of Glycine in 6% of the cases (GAC instead of GGC).
Table 1.
Frequency of different types of K-ras mutations in the current study
| Type of substitution | Frequency (%) | Amino acid |
|---|---|---|
| Normal | ||
| (12GGT) | 68 | 12 Glycine (Gly/G) |
| (13GGC) | 13 Alanine (Ala/A) | |
| 12GCT | 12 | Alanine (Ala/A) |
| 12GAT | 9 | Aspartic acid (Asp/D) |
| 12AGT | 1 | Serine (Ser/S) |
| 12TGT | 1 | Cysteine (Cys/C) |
| 12GCT,13GAC | 1 | Ala, Asp |
| 13GAC | 6 | Asp |
| 13AGC | 1 | Ser |
| 13CGC | 1 | Arginine (Arg/R) |
| Total | 100 | - |
Discussion
CRC is a major cause of morbidity and mortality in western developed countries. Such rates are largely lower in developing countries of Asia and Africa, where studies on the prevalence and raw data about the disease are very blurred and not definite.1
Therefore, the present study was designed in order to determine the frequency of codons 12 and 13 mutations in K-ras gene among a population of Iranian patients with CRC. This is the first report from southern Iran and the fourth report from the whole country.7-10
Similar to other studies from around the world, our experience was that K-ras codon 13 mutations would be less frequent than those of codon 12. The frequency of mutations involving Kras codon 13 was detected in 8% of patients with colorectal cancer (in comparison to 24% of codon 12). Moreover, we observed that 75% of these mutations were GGC to GAC (Gly>Asp), which is similar to the bulk of existing literature.2-4 In other studies from different geographic parts of Iran (excluding the south) the frequency of K-ras mutation was from 12.5% to 37.4%.8-10
In a study performed by Bishehsari et al.,8 tumor samples from 182 Iranian colorectal cancer patients (170 sporadic cases and 12 HNPCC cases) were screened for K-ras mutations at codons 12, 13 and 61 by sequencing analysis. K-ras mutations were observed in 68/182 (37.4%) cases of CRC. Mutational analysis in the present study, however, showed that up to 32 % of patients had mutations in K-ras gene either involving codon 12 or 13. A possible source of the lower frequency of K-ras mutations in our study may be that sequencing of codon 61 was not performed resulting in a comparatively reduced reported frequency of mutations. In Bishehsari’s study, the frequency of mutation was HNPCC associated sporadic MSI-H and sporadic microsatellite-stable (MSS) tumors. But, the G13D substitution was more frequent in HNPCC (3/4, 75%) and sporadic MSI-H (7/11, 63.6%) tumors compared with sporadic MSS tumors (11/53, 20.4%) (P-value<0.01). Most common substitutions were 12G→A(GGT>GAT) (Gly→Asp). It is concluded that while the frequency of K-ras mutations could be similar with respect to these two populations, the mutational spectrum could be differentially influenced by genetic and environmental factors.
In another study of Iranian sporadic colorectal cancer patients, Shemirani et al.9 investigated 48 patients for K-ras codons 12 and 13 mutations using PCR sequencing. They found that 12.5% of carcinoma specimens exhibited K-ras mutation, including 10.4% in codon 12 (Gly12Asp) and 2.1% in codon 13 (Gly13Cys). All of the K-ras amino acid change in codon 12 was Glycine to aspartic acid (G12A). The frequency of mutations reported by Shemirani is considerably lower comparing to our study (32%). However, it should be mentioned that relatively small number of individuals studied by Shemirani might diminish the impact of their results. Such diversity in the results could be explained by considering the geographical and environmental differences between northern and southern Iranian populations.
Compared with the current study, tables 2 and 3 show the reported frequency of different K-ras mutation from different geographic areas of the world and Iran. As previously mentioned, we observed that 75% of K-ras mutations were GCC to GTC, which is in contrast to the bulk of existing literature where GGC>GAC (Gly>Asp) is the predominant type. Studies have shown that GGC>GAC (Gly>Asp) substitution in codon 13 is associated with poor prognostic outcomes, including reduced survival rate, less stable cancer and disease relapse. However, further specific studies in this field are required to address the prognostic significance of codon 13 mutation subtypes among Iranian CRC patients.
Table 2.
Comparison of different studies about K-ras gene status in CRC all around the world
| Study | Year | Method | Sample size | Mutation prevalence (%) | Most common substitution |
|---|---|---|---|---|---|
| Iraq2 | 2012 | PCRReverse hybridization to oligospecific probes | 50 | 48 | 12G>T |
| Codon 12, 13 | |||||
| Saudi Arabia11 | 2011 | PCRsequencing | 46 | 32 | 12G>A |
| Codon 12, 13 | |||||
| Turkey12 | 2013 | Codon 12, 13 | 145 | 37.9 | - |
| Jordan13 | 2012 | RTPCRbased assay Sanger sequencing | 100 | 44 | 12G>A |
| Codon 12, 13 | |||||
| India14 | 2009 | PCRsequencing | 53 | 22.64 | 12G>A |
| Codon 12, 13 | |||||
| Japan15 | 2000 | PCRsequencing | 18 | 50 | 12G>T |
| Only Codon 12 | |||||
| Thailand16 | 2013 | PCRsequencing | 200 | 23 | 12G>A |
| codon 12, 13 and 61 | |||||
| China17 | 2010 | PCRsequencing | 101 | 32.7 | 12G>A |
| codon 12, 13, 61 | |||||
| Korea18 | 2010 | PCRsequencing | 92 | 28.3 | 12G>A |
| Codon 12, 13 | |||||
| Italy19 | 2011 | PCRsequencing (fluorescencebased) | 478 | 30 | 12G>A |
| Codon12, 13, 61 | |||||
| UK20 | 2013 | PCRsequencing | 69 | 49.3 (46.4) | 12G>A |
| Codon12, 13, 61 | |||||
| Germany21 | 2009 | PCRsequencing | 1018 | 39.3 | 12G>A |
| Codon 12, 13 | |||||
| Slovenia22 | 2010 | RTPCR | 302 | 45.5 | 12G>A |
| Codon 12, 13 | |||||
| USA23 | 2013 | PCRsequencing | 171 | 31.6 | 12G>T |
| Codon 12, 13 | |||||
| Australia24 | 2013 | PCRsequencing | 776 | 28 | - |
| Codon 12, 13 | |||||
| Tunisia25 | 2012 | PCR sequencing | 52 | 23.07 | - |
| Codon 12, 13 | |||||
| Moroco26 | 2010 | PCRmelting and direct sequencing | 62 | 29 | - |
| Codon 12, 13 | |||||
| Current study | 2013 | PCRsequencing | 100 | 32 | 12G>C |
Table 3.
Comparison of different studies about K-ras gene status in CRC in different parts of Iran
| Study | Year | Method | Sample size | Mutation prevalence (%) | Most common substitution |
|---|---|---|---|---|---|
| Shemirani et al.9 | 2011 | Codon 1213 | 48 CRC | 12.5 | 12G→A (GGT>GAT) |
| PCRsequencing | (Gly→Asp) | ||||
| Sobhani et al.10 | 2010 | Codon 1213 | 59 | 20.3 | 12G→A (GGT>GAT) |
| PCRsequencing | (Gly→Asp) | ||||
| Bishehsari et al.8 | 2006 | Codon12, 13, 61 | 182 | 37.4 | 12G→A (GGT>GAT) |
| PCRsequencing | (Gly→Asp) | ||||
| Current study | 2013 | PCRsequencing | 100 | 32 | 12G>C |
Yet, another important aspect of this study to be discussed is about multiple K-ras mutations. There was only one detected case (1%) with both codons 12 and 13 mutations. As revealed in a recent review by Macedo et al.,27 multiple mutations are not unusual in patients suffering from CRC. It is argued that the presence of a heterogeneous group of neoplastic cells inside the tumor along with increased genetic instability in cells that progressively acquire mutations are possible theories in explaining the concomitant detection of codons12 and 13 mutations. Macedo further reported that the frequency could range from 0.2% to 16.4%. Although the specificity and sensitivity of the method used in the present study are confirmed, it is proposed that in such cases testing the specimen using other methods along with analyzing several DNA samples taken from different parts of the tumor may shed more light. Considering data scarcity, regarding the true clinical impact of multiple K-ras mutations, this area remains to be further studied through practical experiences.
There ought to be a relatively large number of factors, which could be linked to the variations in the observed frequencies of K-ras mutations in different studies. Among these, environmental factors may be the most important parameter in the studies of neighboring countries, reporting similar results in contrast to those of western populations. However, detection of mutations using different methodologies provides a different range of sensitivity and specificity, which might serve as another reason for such diversity. Another yet important origin of variations in the literature regarding frequency of K-ras gene mutations are the number and type of the tested specific mutations. Environmental factor has a major impact on different populations with diverse lifestyles, dietary habits, and variable exposures to carcinogens.
Conclusion
Frequency of K-ras in our study was similar to some reports from other parts of the world. According to our results, in comparison with other parts of the world, there is marked variation in the reported frequency and type of K-ras mutations around the world. We recommend multicenter studies with large numbers of CRC to clarify the reason for these differences. It is worthy to note that the main point of this multicenter study should be using the same methodology across different centers.
Acknowledgment
This study was supported by the research project No-2012 at Shiraz University of Medical Sciences.
Conflicts of Interest: None declared.
References
- 1.Jiang Y, Kimchi ET, Staveley-O'Carroll KF, Cheng H, Ajani JA. Assessment of K-ras mutation:a step toward personalized medicine for patients with colorectal cancer. Cancer. 2009;115:3609–17. doi: 10.1002/cncr.24434. [DOI] [PubMed] [Google Scholar]
- 2.Al-Allawi NA, Ismaeel AT, Ahmed NY, Merza NS. The frequency and spectrum of K-ras mutations among Iraqi patients with sporadic colorectal carcinoma. Indian J Cancer. 2012;49:163–8. doi: 10.4103/0019-509x.98943. [DOI] [PubMed] [Google Scholar]
- 3.Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.can-06-0191. [DOI] [PubMed] [Google Scholar]
- 4.Palmirotta R, Savonarola A, Formica V, Ludovici G, Del Monte G, Roselli M, et al. A novel K-ras mutation in colorectal cancer. A case report and literature review. Anticancer Res. 2009;29:3369–74. [PubMed] [Google Scholar]
- 5.Ross JS. Clinical implementation of KRAS testing in metastatic colorectal carcinoma:the pathologist's perspective. Arch Pathol Lab Med. 2012;136:1298–307. doi: 10.5858/arpa.2011-0478-RA. [DOI] [PubMed] [Google Scholar]
- 6.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–94. doi: 10.1200/jco.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 7.Siddiqui AD, Piperdi B. KRAS mutation in colon cancer:a marker of resistance to EGFR-I therapy. Ann Surg Oncol. 2010;17:1168–76. doi: 10.1245/s10434-009-0811-z. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishehsari F, Mahdavinia M, Malekzadeh R, Verginelli F, Catalano T, Sotoudeh M, et al. Patterns of K-ras mutation in colorectal carcinomas from Iran and Italy (a Gruppo Oncologico dell'Italia Meridionale study):influence of microsatellite instability status and country of origin. Ann Oncol. 2006;17:vii91–6. doi: 10.1093/annonc/mdl959. [DOI] [PubMed] [Google Scholar]
- 9.Shemirani AI, Haghighi MM, Milanizadeh S, Taleghani MY, Fatemi SR, Damavand B, et al. The role of kras mutations and MSI status in diagnosis of colorectal cancer. Gastroenterol Hepatol Bed Bench. 2011;4:70–5. [ PMC Free Article] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobhani S, Ghaffarpour M, Mostakhdemin Hosseini Z, Kamali F, Nour Mohammadi Z, Houshmand M. The prevalence of common mutation frequency in K-ras codons 12, 13 in Iranian Colorectal Cancer patients. Genetics in the 3rd Millennium. 2010;8:2011–8. Persian. [Google Scholar]
- 11.Zekri J, Rizvi A, Al-Maghrabi J, bin Sadiq B. K-ras in colorectal cancer tumors from Saudi patients:Frequency, Clinco-pathological Association and Clinical Outcome. The Open Colorectal Cancer J. 2012;5:22–7. doi: 10.2174/1876820201205010022. [DOI] [Google Scholar]
- 12.Gunal A, Hui P, Kilic S, Xu R, Jain D, Mitchell K, et al. KRAS mutations are associated with specific morphologic features in colon cancer. J Clin Gastroenterol. 2013;47:509–14. doi: 10.1097/MCG.0b013e3182703030. [DOI] [PubMed] [Google Scholar]
- 13.Elbjeirami WM, Sughayer MA. KRAS mutations and subtyping in colorectal cancer in Jordanian patients. Oncol Lett. 2012;4:705–10. doi: 10.3892/ol.2012.785. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sameer AS, Chowdhri NA, Abdullah S, Shah ZA, Siddiqi MA. Mutation pattern of K-ras gene in colorectal cancer patients of Kashmir:a report. Indian J Cancer. 2009;46:219–25. doi: 10.4103/0019-509x.52956. [DOI] [PubMed] [Google Scholar]
- 15.Kusaka T, Fukui H, Sano Y, Ueda Y, Chiba T, Fujimori T. Analysis of K-ras codon 12 mutations and p53 overexpression in colorectal nodule-aggregating tumors. J Gastroenterol Hepatol. 2000;15:1151–7. doi: 10.1046/j.1440-1746.2000.02280.x. [DOI] [PubMed] [Google Scholar]
- 16.aChaiyapan W, Duangpakdee P, Boonpipattanapong T, Kanngern S, Sangkhathat S. Somatic mutations of K-ras and BRAF in Thai colorectal cancer and their prognostic value. Asian Pac J Cancer Prev. 2013;14:329–32. doi: 10.7314/APJCP.2013.14.1.329. [DOI] [PubMed] [Google Scholar]
- 17.Yunxia Z, Jun C, Guanshan Z, Yachao L, Xueke Z, Jin L. Mutations in epidermal growth factor receptor and KRAS in Chinese patients with colorectal cancer. BMC Med Genet. 2010;11:34–43. doi: 10.1186/1471-2350-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon MJ, Lee SE, Kang SY, Choi YL. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers:Comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–8. doi: 10.1016/j.prp.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Palomba G, Colombino M, Contu A, Massidda B, Baldino G, Pazzola A, et al. Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population:clues from Sardinia. J Transl Med. 2012;10:178. doi: 10.1186/1479-5876-10-178. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richman SD, Chambers P, Seymour MT, Daly C, Grant S, Hemmings G, et al. Intra-tumoral heterogeneity of KRAS and BRAF mutation status in patients with advanced colorectal cancer (aCRC) and cost-effectiveness of multiple sample testing. Anal Cell Pathol (Amst) 2011;34:61–6. doi: 10.3233/ACP-2011-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol Res Pract. 2009;205:858–62. doi: 10.1016/j.prp.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Licar A, Cerkovnik P, Ocvirk J, Novakovic S. KRAS mutations in Slovene patients with colorectal cancer:frequency, distribution and correlation with the response to treatment. Int J Oncol. 2010;36:1137–44. doi: 10.3892/ijo_00000596. [DOI] [PubMed] [Google Scholar]
- 23.Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–12. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 24.Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26:825–34. doi: 10.1038/modpathol.2012.240. [DOI] [PubMed] [Google Scholar]
- 25.Sammoud S, Khiari M, Semeh A, Amine L, Ines C, Amira A, et al. Relationship between expression of ras p21 oncoprotein and mutation status of the K-ras gene in sporadic colorectal cancer patients in Tunisia. Appl Immunohistochem Mol Morphol. 2012;20:146–52. doi: 10.1097/PAI.0b013e3182240de1. [DOI] [PubMed] [Google Scholar]
- 26.Bennani B, Gilles S, Fina F, Nanni I, Ibrahimi SA, Riffi AA, et al. Mutation analysis of BRAF exon 15 and KRAS codons 12 and 13 in Moroccan patients with colorectal cancer. Int J Biol Markers. 2010;25:179–84. doi: 10.5301/JBM.2010.6091. [DOI] [PubMed] [Google Scholar]
- 27.Macedo MP, Andrade Lde B, Coudry R, Crespo R, Gomes M, Lisboa BC, et al. Multiple mutations in the Kras gene in colorectal cancer:review of the literature with two case reports. Int J Colorectal Dis. 2011;26:1241–8. doi: 10.1007/s00384-011-1238-0. [DOI] [PubMed] [Google Scholar]


