Abstract
The Gram-positive bacterium Listeria monocytogenes is a facultative intracellular pathogen that relies on the regulated secretion and activity of a variety of proteins that sustain life within diverse environments. PrsA2 has recently been identified as a secreted peptidyl-prolyl cis/trans isomerase and chaperone that is dispensable for bacterial growth in broth culture but essential for L. monocytogenes virulence. Following host infection, PrsA2 contributes to the proper folding and activity of secreted proteins that are required for bacterial replication within the host cytosol and for bacterial spread to adjacent cells. PrsA2 is one member of a family of Gram-positive secretion chaperones that appear to play important roles in bacterial physiology; however, it is not known how these proteins recognize their substrate proteins or the degree to which their function is conserved across diverse Gram-positive species. We therefore examined PrsA proteins encoded by a variety of Gram-positive bacteria for functional complementation of L. monocytogenes mutants lacking prsA2. PrsA homologues encoded by Bacillus subtilis, Streptococcus pyogenes, Streptococcus pneumoniae, Streptococcus mutans, Staphylococcus aureus, and Lactococcus lactis were examined for functional complementation of a variety of L. monocytogenes PrsA2-associated phenotypes central to L. monocytogenes pathogenesis and bacterial cell physiology. Our results indicate that while selected aspects of PrsA2 function are broadly conserved among diverse Gram-positive bacteria, PrsA2 exhibits unique specificity for L. monocytogenes target proteins required for pathogenesis. The L. monocytogenes PrsA2 chaperone thus appears evolutionarily optimized for virulence factor secretion within the host cell cytosol while still maintaining aspects of activity relevant to more general features of Gram-positive protein translocation.
INTRODUCTION
The translocation of proteins across bacterial membranes is fundamental to bacterial movement, nutrient acquisition, complex behaviors such as biofilm formation and sporulation, and survival. While the processes underlying protein secretion and folding have been well characterized in Gram-negative bacteria (1–3), less attention has generally been focused on Gram-positive bacteria. In contrast to Gram-negative bacteria, Gram-positive organisms need only target proteins across a single membrane; however, there remain a number of challenges associated with protein folding at the Gram-positive membrane-cell wall interface. The Gram-positive cell wall consists of several peptidoglycan layers containing teichoic and lipoteichoic acids that result in a high density of negative charge as well as a capacity to bind cationic molecules (4). Proteins destined for secretion are translocated across the cell membrane in an unfolded state and must fold within the space between the membrane and the cell wall; this space is not only highly charged but freely exposed to the external environment. Proteins destined for release from the bacterium must be further translocated across the thick Gram-positive peptidoglycan cell wall.
The Gram-positive bacterium Listeria monocytogenes is an environmental pathogen that is capable of life as a saprophyte within the soil while also maintaining the ability to invade and replicate within mammalian cells (5). In the United States, L. monocytogenes is a significant health threat, as it has been associated with numerous multistate foodborne outbreaks resulting in thousands of illnesses and hundreds of deaths (6, 7). L. monocytogenes infections pose serious risks to immunocompromised populations, the elderly, pregnant women, and neonates, where fatality rates range from 20 to 50% (8–11). As L. monocytogenes transitions between life in the environment to life within the cytosol of infected mammalian host cells, the bacterium requires increased expression of a number of secreted virulence factors that facilitate intracellular survival by promoting cell entry, bacterial escape from host vacuoles, replication within the cytosol, and spread to adjacent cells (12–17). Many of the secreted virulence factors required for bacterial survival within the host are regulated by a transcriptional activator known as PrfA (positive regulatory factor A) (18–20). PrfA becomes activated following bacterial entry into host cells, and this activation results in a significant increase in protein translocation across the bacterial membrane together with increased expression of factors that promote the folding and activity of secreted proteins, such as the posttranslocation secretion chaperone known as PrsA2 (15, 21, 22).
L. monocytogenes prsA2 was first identified by transcriptome analysis based on its increased expression following PrfA activation (23). Subsequent proteomic analysis of L. monocytogenes secreted proteins indicated that levels of secreted PrsA2 were increased in strains expressing a mutationally activated form of prfA, known as prfA*, that results in the constitutive expression of PrfA-regulated gene products (15). Homologues to PrsA2 appear widespread in other Gram-positive bacteria, where they are reportedly required for the folding and stability of secreted proteins (24–32). L. monocytogenes is now known to encode two PrsA proteins, PrsA1 and PrsA2, and these proteins have been characterized for their roles in L. monocytogenes protein secretion and pathogenesis (12, 15, 19, 33, 34). L. monocytogenes PrsA2 contributes to multiple facets of bacterial pathogenesis and is essential for virulence (33, 34), and it appears to be required for the proper folding and secretion of a number of proteins (12, 25, 35). L. monocytogenes mutants lacking PrsA2 exhibit decreased secreted hemolytic and phospholipase activity and are defective for cell-to-cell spread in tissue culture monolayers (33–35). Mice infected with L. monocytogenes ΔprsA2 strains have up to 100,000-fold fewer CFU recovered from livers and spleens than animals infected with the wild-type strain (33, 34). In contrast, PrsA1 shares 75% amino acid similarity and 58% identity with PrsA2 but does not contribute to L. monocytogenes pathogenesis (33). Neither prsA1 nor prsA2 is required for bacterial growth in broth culture (33).
Given what appears to be a fundamental role for PrsA family members in protein folding and secretion, we were curious to determine how broadly PrsA function is conserved among Gram-positive bacteria. We had observed that the expression of prsA1 from the prsA2 promoter did not restore L. monocytogenes virulence (12), suggesting that PrsA1 and PrsA2 do not functionally substitute for each other and therefore may recognize distinct substrates. We therefore assessed the degree to which different PrsA homologues were capable of complementing PrsA2-associated activities in L. monocytogenes. Thus far, PrsA-like proteins have been best characterized in L. monocytogenes and Bacillus subtilis (12, 33, 36–40). B. subtilis contains one prsA gene, which is essential for viability but can be deleted in the presence of high concentrations of magnesium (41), while the single prsA alleles of Streptococcus pneumoniae, Streptococcus mutans, Staphylococcus aureus, and Lactococcus lactis have been shown to be nonessential (28–30, 32). Similarly to L. monocytogenes, Streptococcus pyogenes has two prsA alleles (31), whereas its close relative S. pneumoniae has only one (30). Proteomic analyses of L. monocytogenes and B. subtilis suggest that these bacteria may have a subset of overlapping PrsA substrates with roles in cell wall metabolism, swimming motility, oligopeptide transport, and membrane bioenergetics (12, 24, 41, 42). L. monocytogenes PrsA2 has, however, been associated with the proper folding and activity of several L. monocytogenes virulence factors not expressed by B. subtilis (12, 24, 33–36). Here we provide evidence based on the complementation of L. monocytogenes ΔprsA2 strains with various prsA alleles that indicates both broad conservation of PrsA2 function and species-specific adaptations of the chaperone for L. monocytogenes pathogenesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains used in this study are listed in Table 1. L. monocytogenes 10403S is referred to as the wild-type strain, and L. monocytogenes 10403S containing an erythromycin resistance gene (erm) in place of prsA2 coding sequences is the mutant strain (L. monocytogenes ΔprsA2) (33) used for complementation with the designated heterologous Gram-positive prsA alleles. Escherichia coli One Shot TOP10 (Invitrogen) and S17 (a kind gift from N. Cianciotto, Northwestern University) were used as host strains for recombinant plasmids. Strains were grown in Luria broth (LB) or brain heart infusion (BHI) medium and supplemented with the appropriate antibiotic at the following concentrations: 50 μg/ml kanamycin and 1 mg/ml streptomycin. The integration plasmid pIMK2 (43) (a kind gift from C. Hill, Cork College) was used for providing constitutive expression of target genes for genetic complementation.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Designation | Source or reference |
|---|---|---|---|
| Strains | |||
| TOP10 | E. coli host strain used for propagation of recombinant pIMK2 plasmids | Invitrogen | |
| S17 | E. coli host strain used for conjugation of recombinant pIMK2 plasmids | ||
| NF-L100 | L. monocytogenes 10403S parent strain | L. monocytogenes wild type | 56 |
| NF-L1651 | 10403S with ΔprsA2::erm | L. monocytogenes ΔprsA2 | 33 |
| NF-L3632 | NF-L1651 with integrated pIMK2-L. monocytogenes prsA2 | L. monocytogenes prsA2 | This work |
| NF-L3634 | NF-L1651 with integrated pIMK2-L. monocytogenes prsA1 | L. monocytogenes prsA1 | This work |
| NF-L3636 | NF-L1651 with integrated pIMK2-B. subtilis prsA | B. subtilis prsA | This work |
| NF-L3638 | NF-L1651 with integrated pIMK2-S. pyogenes prsA1 | S. pyogenes prsA1 | This work |
| NF-L3640 | NF-L1651 with integrated pIMK2-S. pyogenes prsA2 | S. pyogenes prsA2 | This work |
| NF-L3642 | NF-L1651 with integrated pIMK2-S. pneumoniae prsA | S. pneumoniae prsA | This work |
| NF-L3644 | NF-L1651 with integrated pIMK2-S. mutans prtM | S. mutans prtM | This work |
| NF-L3646 | NF-L1651 with integrated pIMK2-S. aureus prsA | S. aureus prsA | This work |
| NF-L3648 | NF-L1651 with integrated pIMK2-L. lactis pmpA | L. lactis pmpA | This work |
| NF-L3650 | NF-L1651 with integrated pIMK2-L. monocytogenes prsA2 with a C-terminal 6×His tag | L. monocytogenes prsA2-His | This work |
| NF-L3652 | NF-L1651 with integrated pIMK2-L. monocytogenes prsA1 with a C-terminal 6×His tag | L. monocytogenes prsA1-His | This work |
| NF-L3654 | NF-L1651 with integrated pIMK2-B. subtilis prsA with a C-terminal 6×His tag | B. subtilis prsA-His | This work |
| NF-L3656 | NF-L1651 with integrated pIMK2-S. pyogenes prsA1 with a C-terminal 6×His tag | S. pyogenes prsA1-His | This work |
| NF-L3658 | NF-L1651 with integrated pIMK2-S. aureus prsA with a C-terminal 6×His tag | S. aureus prsA-His | This work |
| NF-L3660 | NF-L1651 with integrated pIMK2-L. lactis pmpA with a C-terminal 6×His tag | L. lactis pmpA-His | This work |
| Plasmids | |||
| pIMK2 | ppL2-derived complementation integration vector | 43 | |
| pNF-3662 | pIMK2 with L. monocytogenes prsA2 | This work | |
| pNF-3664 | pIMK2 with L. monocytogenes prsA1 | This work | |
| pNF-3666 | pIMK2 with B. subtilis prsA | This work | |
| pNF-3668 | pIMK2 with S. pyogenes prsA1 | This work | |
| pNF-3670 | pIMK2 with S. pyogenes prsA2 | This work | |
| pNF-3672 | pIMK2 with S. pneumoniae prsA | This work | |
| pNF-3674 | pIMK2 with S. mutans prtM | This work | |
| pNF-3676 | pIMK2 with S. aureus prsA | This work | |
| pNF-3678 | pIMK2 with L. lactis pmpA | This work | |
| pNF-3680 | pIMK2 with L. monocytogenes prsA2 containing a C-terminal 6×His tag | This work | |
| pNF-3682 | pIMK2 with L. monocytogenes prsA1 containing a C-terminal 6×His tag | This work | |
| pNF-3684 | pIMK2 with B. subtilis prsA containing a C-terminal 6×His tag | This work | |
| pNF-3686 | pIMK2 with S. pyogenes prsA1 containing a C-terminal 6×His tag | This work | |
| pNF-3688 | pIMK2 with S. aureus prsA containing a C-terminal 6×His tag | This work | |
| pNF-3690 | pIMK2 with L. lactis pmpA containing a C-terminal 6×His tag | This work |
Construction of L. monocytogenes ΔprsA2 strains containing heterologous Gram-positive prsA alleles.
Genomic DNA from L. monocytogenes 10403S, B. subtilis 168, S. pyogenes MGAS5005 and S. mutans UA159 (a kind gift from M. Federle, UIC), S. pneumoniae R6 (a kind gift from D. Morrison, UIC), S. aureus USA300 (a kind gift from V. Torres, NYU), and L. lactis IL1403 (a kind gift from L. Tao, UIC) was used to amplify the respective prsA alleles using the primers specific for each sequence (see Table S1 in the supplemental material). Forward primers were designed with an NcoI restriction site, and reverse primers were designed with a ClaI restriction site, except for L. monocytogenes prsA2, for which the reverse primer had an XmaI restriction site. PCR-amplified products were cloned into the expression plasmid pIMK2, which contains a constitutive promoter for target gene expression and which integrates into a single neutral site within the L. monocytogenes chromosome (43). The use of the pIMK2 constitutive promoter for prsA gene expression provided the best current means for optimizing gene expression so as to better assess whether a specific PrsA homologue was at all capable of functional complementation. To generate constructs encoding C-terminal 6×His-tagged Gram-positive PrsA proteins, the same forward primer as specified above was used, while a new reverse primer was designed so as to insert DNA encoding a 6×His tag prior to the stop codon and a ClaI restriction site to allow for directional cloning into pIMK2 (43) (see Table S1). Plasmids were propagated in E. coli One Shot TOP10 cells, transformed into E. coli S17 cells, and subsequently introduced into the L. monocytogenes ΔprsA2 strain by conjugation. All genes and plasmids were verified by DNA sequencing prior to introduction into L. monocytogenes and subsequently amplified from L. monocytogenes transconjugants by PCR and again verified by sequencing analysis. DNA sequencing was performed at the UIC Research Resources Center Core Genomics Facility.
Isolation of bacterial cell-associated and secreted proteins for Western blot analysis.
Proteins were isolated as previously described, with minor modifications (33). Specifically, L. monocytogenes strains grown overnight were diluted 1:20 into BHI broth and then grown to an optical density at 600 nm (OD600) of 0.6. Briefly, for cell-associated proteins, the bacterial pellets from normalized culture volumes were collected by centrifugation and resuspended in 2× Laemmli sample buffer with β-mercaptoethanol (Bio-Rad). For secreted proteins, supernatants were recovered and proteins were precipitated using trichloroacetic acid (TCA) at a final concentration of 10%. The TCA-treated supernatants were incubated on ice for 30 min, and precipitated proteins were recovered by centrifugation. The supernatants were discarded, and proteins were washed with ice-cold acetone; then, protein pellets were collected by centrifugation, allowed to air dry, and resuspended in 2× Laemmli sample buffer with β-mercaptoethanol. The samples were boiled for 10 min prior to SDS-PAGE followed by Western blot analysis using an anti-His antibody.
Swimming motility assay.
Bacteria from mid-log-phase (OD600 of ∼0.8) BHI broth cultures (2 μl) were inoculated into soft BHI agar (0.3%) plates and grown at 37°C for 24 h and then at 25°C for 24 h. The motility was measured as the diameter of the spreading colony.
Growth assays at acidic and basic pH and high ionic and nonionic osmolarity.
Growth assays were performed as follows: 2 μl of an overnight culture was inoculated into 2 ml BHI liquid broth at pH 6 or pH 9 (where HCl or NaOH was used to obtain the respective pH) or into broth containing 5% NaCl (wt/vol) or 0.7 M sucrose. These cultures were grown overnight at 37°C, and growth was measured as a function of optical density (OD600).
Antibiotic MIC determination.
The MIC to prevent bacterial growth was determined following 2 μl inoculation from mid-log-phase (OD600 of ∼0.8) cultures into 2 ml BHI broth in 4 ml polypropylene test tubes containing various dilutions of bacitracin, vancomycin, penicillin G, or lysozyme. Cultures were grown at 37°C for 16 h, and the MIC was determined based on the complete inhibition of bacterial growth.
Hemolysin assays.
Hemolytic activity was measured for strains as previously described (36). Briefly, bacterial strains were grown in 3 ml LB overnight, diluted 1:10 in fresh LB, and allowed to grow for 5 h. Supernatants were collected and normalized based on OD600. Culture supernatant was serially diluted into PBS, pH 5, containing 1 mM dithiothreitol (PBS-DTT) and incubated at 37°C for 30 min. Then, 100 μl of a 1:5 dilution of PBS-DTT-washed sheep red blood cells (RBCs) was added, and the mixture was incubated for an additional 30 min at 37°C. Bacterial supernatant/RBC mixtures were pelleted by centrifugation, and the supernatant dilution resulting in 50% RBC lysis was determined by visual inspection of the pellet.
Detection of phospholipase activity.
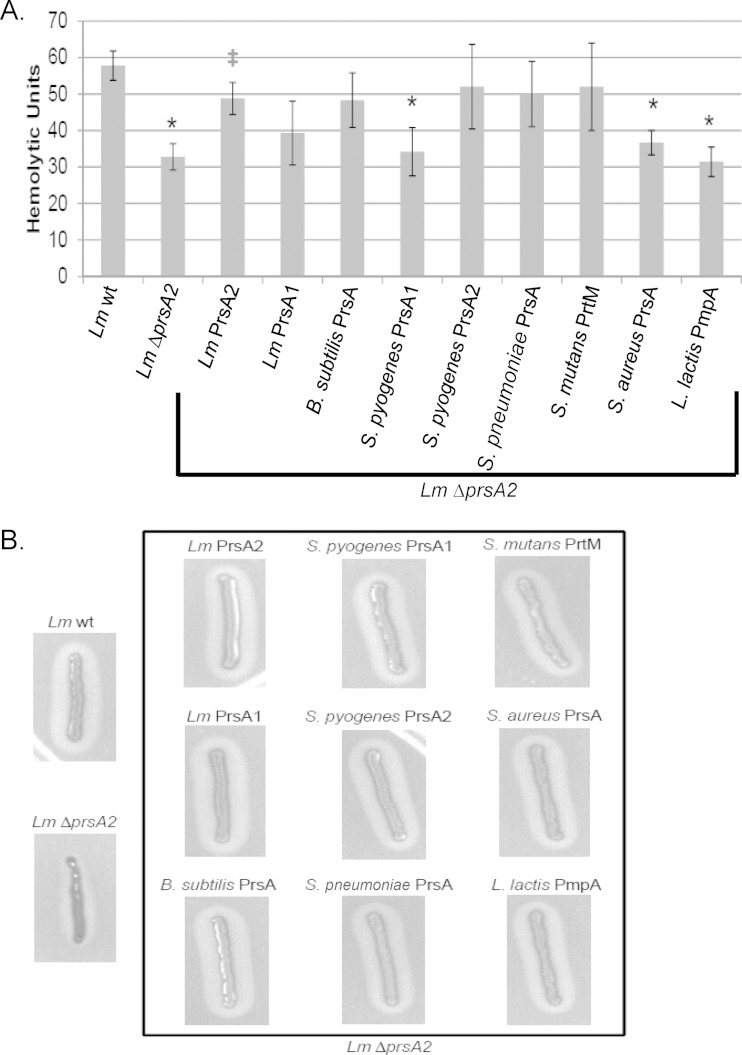
Brilliance Listeria agar with differential supplement containing lecithin (Oxoid) was used to detect phospholipase activity. L. monocytogenes phospholipase hydrolyzes lecithin in the medium, which produces an opaque halo around the streak. Colonies were streaked onto the medium and incubated for 24 h at 37°C, followed by visual inspection of the zone of opacity surrounding the bacterial streaks.
L2 plaque assays.
Plaque assays were conducted as previously described (44). Briefly, monolayers of L2 fibroblasts in 6-well culture dishes were infected at a multiplicity of infection (MOI) of 30:1 for 1 h. Then, infected monolayers were washed three times with PBS, pH 7, and a subsequent Dulbecco's modified Eagle's medium (DMEM)-agarose overlay was added, containing 10 μg/ml gentamicin to kill extracellular bacteria. Plaques were measured at 72 h with a micrometer.
Intravenous mouse infections.
Animal procedures were approved by the UIC Animal Care Committee and were conducted in the Biological Resources Laboratory. Overnight bacterial cultures were diluted 1:20 in BHI broth and grown to an OD600 of approximately 0.6. Then, bacteria were normalized to 6 × 108 CFU/ml, washed twice with PBS, pH 7, and then diluted and resuspended in PBS, pH 7, to a final concentration of 1 × 105 CFU/ml. Ten to fifteen 7- to 9-week-old female Swiss Webster mice (Charles River Laboratories) per treatment group were injected with 200 μl containing 2 × 104 CFU bacteria by tail vein injection. Livers and spleens of infected animals were collected at 24, 48, and 72 h postinfection. Organs were homogenized, and 10-fold serial dilutions were plated for total CFU.
Statistical analyses.
Two-tailed Student's t test was used for statistical analysis of data represented as bar graphs, where a P value of ≤0.05 indicates statistical significance and error bars represent the standard errors of the means. For data represented as box plots, a two-tailed Wilcoxon rank sum test was used, and a P value of ≤0.05 indicates statistical significance.
RESULTS
Construction of L. monocytogenes ΔprsA2 strains complemented with different PrsA family members.
PrsA protein family members are composed of N-terminal and C-terminal regions associated with chaperone activity that flank a central peptidyl-prolyl cis/trans isomerase (PPIase) domain (24) (Fig. 1A); however, not all PrsA-like proteins contain the complete signature PPIase motif. L. lactis PmpA, S. pneumoniae PrsA, S. pyogenes PrsA1 and PrsA2, and S. mutans PrtM all contain residues within the central region that diverge from the signature PPIase motif (24, 29, 30, 32), while L. monocytogenes PrsA1 and PrsA2, B. subtilis PrsA, and S. aureus PrsA have a centrally located conserved signature PPIase motif (29, 36, 45, 46). L. monocytogenes PrsA1 and PrsA2, B. subtilis PrsA, and S. aureus PrsA all have been confirmed to have PPIase activity in vitro (36, 40, 45), whereas S. pneumoniae PrsA and L. lactis PmpA have been demonstrated to lack PPIase activity (47, 48).
FIG 1.
Evolutionary relationship and protein conservation of PrsA homologues. (A) Domain organization of L. monocytogenes PrsA2, where the N-terminal signal peptide (SP), N terminus, PPIase domain, and C terminus are depicted and numbers designate amino acids. (B) Phylogenetic tree of PrsA homologues from L. monocytogenes and other Gram-positive bacteria, created by CLUSTAL analysis (57). The percent protein similarity of each PrsA homologue with the L. monocytogenes PrsA2 sequence is shown in parentheses. The scale bar indicates 1 substitution for every 10 amino acid residues. PrsA homologues that have a PPIase domain are indicated by asterisks, whereas more distant homologues lack a central PPIase motif.
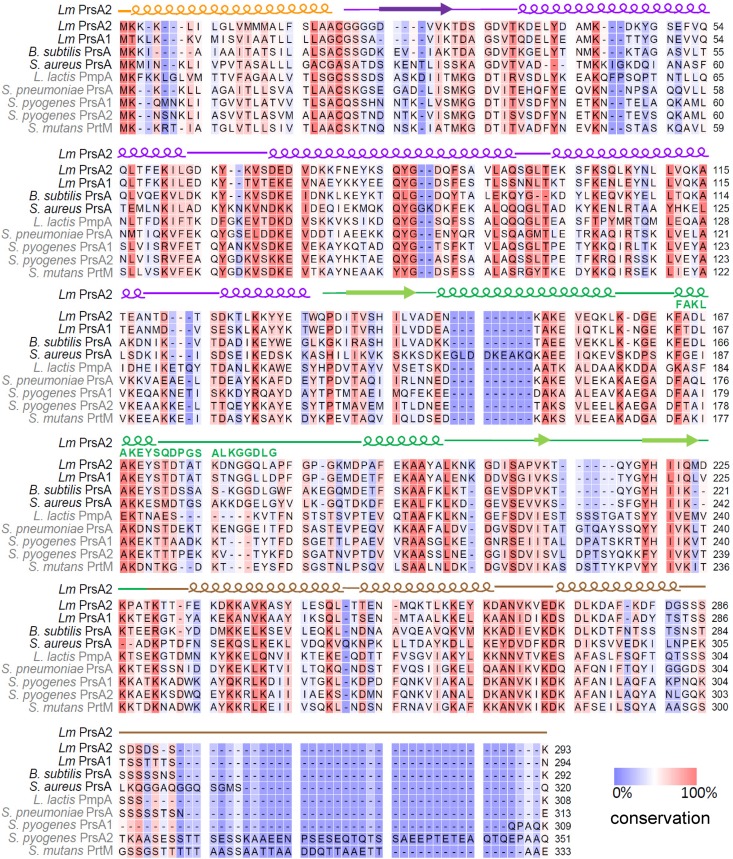
L. monocytogenes PrsA2 and PrsA1 are most closely related to the B. subtilis PrsA and most distantly related to the PrsA of streptococcal species (Fig. 1B). PrsA homologues that contain a PPIase motif—L. monocytogenes PrsA2 and PrsA1, B. subtilis PrsA, and S. aureus PrsA—are more closely related than PrsA homologues that lack this motif (Fig. 1B). Based on amino acid sequence, PrsA2 is most similar to L. monocytogenes PrsA1 (58% identity and 75% similarity), B. subtilis PrsA (46% identity and 68% similarity), and the distantly related S. pneumoniae PrsA (33% identity and 60% similarity). L. monocytogenes PrsA2 is least similar to L. lactis PmpA (31% identity and 47% similarity), S. mutans PrtM (29% identity and 48% similarity), and S. pyogenes PrsA2 (29% identity and 48% similarity). An alignment of the PrsA homologues shows regions of conservation and variation throughout the protein sequences (Fig. 2).
FIG 2.
PrsA homologue sequence conservation. Alignment of the PrsA homologues, where residues that are highly conserved are highlighted in red and less conserved in blue. The PSIPRED secondary-structure prediction of L. monocytogenes PrsA2 is also shown (58), where helices are indicated, strands are shown as arrows, and coils are represented as lines. The L. monocytogenes PrsA2 signal peptide is in orange, the N terminus is in purple, the PPIase domain is in green, and the C terminus is in brown. Homologues having a PPIase motif are shown in black, while those lacking a PPIase motif are in gray. The PPIase motif is indicated by green text.
L. monocytogenes mutants lacking the PrsA2 chaperone have been associated with a variety of defects in protein secretion, bacterial cell physiology, and virulence (12, 33–35); however, no phenotype has thus far been associated with the loss of PrsA1. We assessed the evolutionary and functional conservation of Gram-positive PrsA-like proteins by testing for their ability to complement an L. monocytogenes mutant lacking PrsA2 (L. monocytogenes ΔprsA2). L. monocytogenes prsA2 and prsA1, B. subtilis prsA, S. aureus prsA, S. pneumoniae prsA, S. pyogenes prsA1 and prsA2, S. mutans prtM, and L. lactis pmpA were placed under the control of a constitutive promoter within the integrative plasmid vector pIMK2 to optimize gene expression (43), and the plasmids were integrated into the chromosome of L. monocytogenes ΔprsA2 mutants (Fig. 3A). L. monocytogenes ΔprsA2 mutants exhibit no significant growth defects in broth culture (33), and the Gram-positive PrsA allele-complemented ΔprsA2 strains also exhibited normal patterns of growth (Fig. 3B).
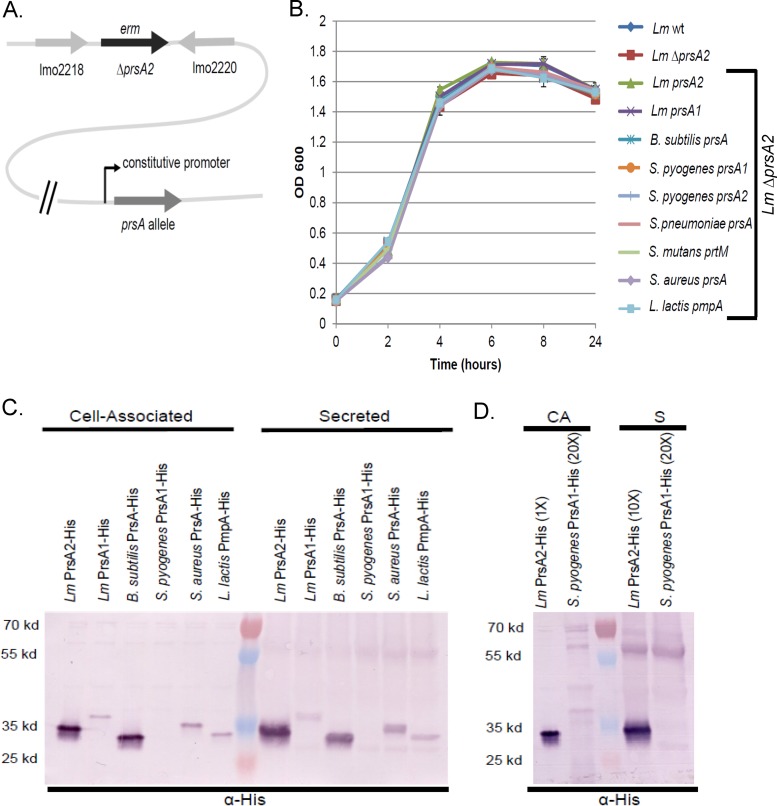
FIG 3.
Growth and protein profiles of PrsA homologue-complemented strains. (A) Schematic of the L. monocytogenes prsA2 mutant strain complemented by a prsA allele. The prsA2 gene, nucleotides 31 to 849 of 879 in total, is replaced by an erythromycin resistance gene where the prsA allele is integrated elsewhere in the chromosome and expressed by a constitutive promoter. (B) Bacterial growth for the L. monocytogenes (Lm) ΔprsA2 strains expressing the indicated Gram-positive prsA homologue in BHI broth, as determined by optical density (OD) measurements at 600 nm. The data shown are representative of results from three independent experiments. (C) Strains were constructed similarly to the procedure described for panel A, with the addition of a C-terminal 6×His tag. For secreted proteins, the bacterial supernatant from each strain was TCA precipitated; for cell-associated proteins, the pellet was isolated, resuspended in sample buffer with β-mercaptoethanol, and boiled. Samples were collected at equivalent bacterial densities, where 1 μl of surface-associated protein or 10 μl of secreted protein was loaded. Western analysis of protein fractions subjected to SDS-PAGE was done using an anti-His (α-His) antibody. (D) Western analysis of L. monocytogenes PrsA2 compared to S. pyogenes PrsA1 is shown. Cell-associated (CA) and secreted (S) samples were isolated as described for panel B, and relative amounts loaded are indicated.
Expression of the various PrsA proteins in L. monocytogenes was determined via the addition of C-terminal His tags to proteins encoded on the same pIMK2 expression plasmid, followed by Western blotting using antibodies directed against the His tag (33) (Fig. 3C and D). Expression of the B. subtilis PrsA in L. monocytogenes was found to be comparable to expression levels of L. monocytogenes PrsA2 expressed from the same constitutive promoter; however, expression of S. aureus PrsA, L. lactis PmpA, L. monocytogenes PrsA1, and S. pyogenes PrsA1 was reduced, with S. pyogenes PrsA1 in particular being barely detectable (Fig. 3C and D). Proteins migrating at the position of PrsA dimers were evident in many of the supernatant-derived fractions (Fig. 3C and D). The reasons underlying the reduced abundance of selected His-tagged PrsA proteins in L. monocytogenes are not clear, but this result was not necessarily reflective of species-dependent codon bias, as L. monocytogenes PrsA1 was produced at much lower levels than L. monocytogenes PrsA2 despite being expressed from an identical promoter (Fig. 3C). As is shown below, despite the observed differences in protein expression, sufficient protein was expressed for the assessment of complementation of a number of PrsA2-associated phenotypes, and the ability of a PrsA homologue to complement an activity did not correlate with its expression level.
PrsA proteins exhibit broad functional conservation for activities associated with swimming motility and pH tolerance but not for conditions of osmotic stress.
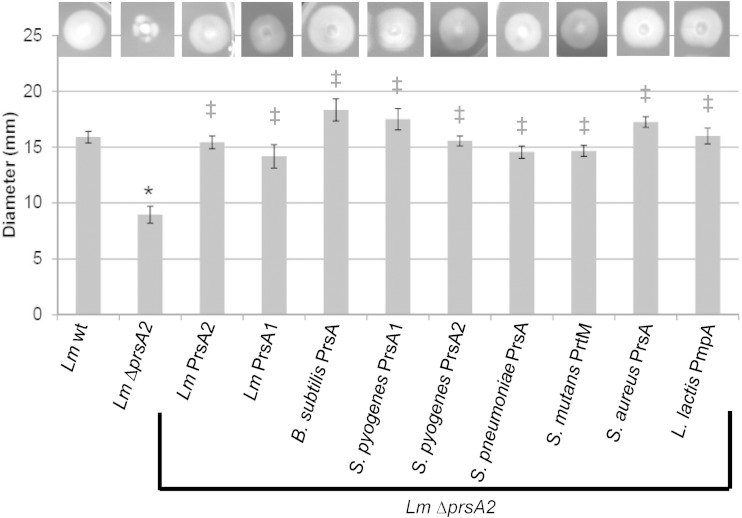
L. monocytogenes strains are flagellated and motile at low temperatures but exhibit reduced motility at higher temperatures (37°C) (49). L. monocytogenes strains lacking prsA2 exhibit reduced swimming motility at both low and high temperatures, suggesting that PrsA2 is required for a functional flagellum and/or chemotaxis (34). In addition, flagellin has been identified as a potential PrsA2/PrsA substrate in both L. monocytogenes and B. subtilis (12, 41). Interestingly, all nine prsA alleles provided full functional complementation of swimming motility to L. monocytogenes ΔprsA2, including prsA alleles derived from strains that lack flagella (streptococcal species, S. aureus, and L. lactis) (Fig. 4).
FIG 4.
Assessment of bacterial swimming motility. Motility was measured as the diameter of the swimming colony. Diameters are expressed as the averages from 4 to 9 swimming colonies from 3 independent experiments. The L. monocytogenes ΔprsA2 strain was complemented with the indicated prsA2 homologue under the control of the pIMK2 constitutive promoter (bracket). Error bars represent the standard errors of the means, whereas asterisks (*) and daggers (‡) indicate statistical significance of P ≤ 0.05, by two-tailed Student's t test, compared to L. monocytogenes wild type (wt) and L. monocytogenes ΔprsA2, respectively.
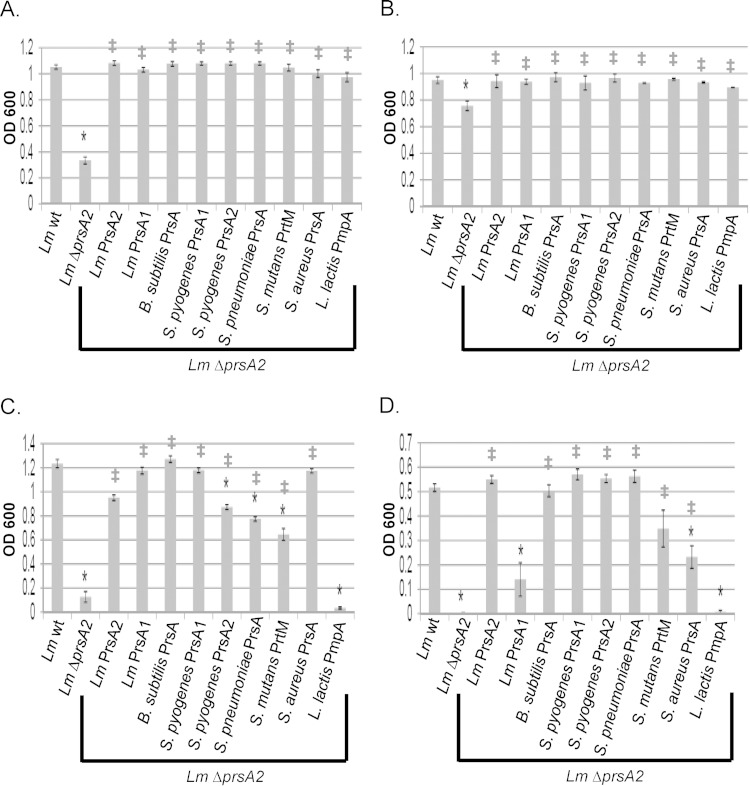
L. monocytogenes is capable of withstanding broad variations in the pH of its environment (pH 4.4 to 9.6) (50), an ability which likely contributes to its survival both inside and outside an infected host. The L. monocytogenes ΔprsA2 mutant exhibited increased sensitivity to both acidic (pH 6) and basic (pH 9) conditions, suggesting a link between chaperone function and pH tolerance (Fig. 5A and B). Similarly to what was observed for the complementation of swimming motility, all nine Gram-positive prsA alleles restored L. monocytogenes growth to wild-type levels at both pH 6 and pH 9 (Fig. 5A and B). Taken together, these results indicate a broad conservation of PrsA chaperone function as it relates to bacterial motility and pH resistance, even for homologues derived from bacterial species that lack flagella and/or exhibit only moderate levels of pH tolerance.
FIG 5.
Bacterial resistance to acidic pH, basic pH, and osmotic shock. (A) Acidic-pH sensitivity. The growth is shown as optical density (OD600) of strains inoculated from a saturated culture into liquid broth at pH 6. (B) Basic-pH sensitivity. Shown is the growth (OD600) of strains inoculated from a saturated culture into liquid broth at pH 9. (C) Ionic shock. Shown is the growth (OD600) of strains inoculated from a saturated culture into liquid broth containing 5% NaCl. (D) Nonionic shock. Growth (OD600) of strains in liquid broth containing 0.7 M sucrose inoculated from a saturated culture is shown. Error bars represent the standard errors of the means from 3 to 6 cultures, whereas asterisks (*) and daggers (‡) indicate statistical significance of P ≤ 0.05, by two-tailed Student's t test, compared to L. monocytogenes wt and L. monocytogenes ΔprsA2, respectively.
In addition to its resistance to extremes of pH, L. monocytogenes is capable of growth under conditions of high osmolarity, up to 10% (wt/vol) NaCl, where the bacterium has been shown to compensate for changes in osmolarity via the transport of compatible solutes (50). Growth of the L. monocytogenes ΔprsA2 mutant under conditions of both ionic (5% NaCl) and nonionic (0.7 M sucrose) osmotic stress revealed significantly decreased growth in comparison to that of wild-type strains; this phenotype could be fully complemented by introduction of the L. monocytogenes prsA2 allele (Fig. 5C and D). In contrast to results for swimming motility and pH resistance, the Gram-positive prsA alleles were not all capable of fully complementing resistance to conditions of ionic and nonionic osmotic stress (Fig. 5C and D). B. subtilis prsA and S. pyogenes prsA1 fully complemented L. monocytogenes ΔprsA2 for osmotic stress resistance, whereas L. lactis pmpA did not. Complementation by other prsA alleles differed for conditions of ionic versus nonionic osmotic stress (L. monocytogenes prsA1, S. pyogenes prsA2, S. pneumoniae prsA, S. mutans prtM, and S. aureus prsA), suggesting that PrsA-dependent substrates that contribute to the resistance of L. monocytogenes to ionic and nonionic shock may differ between the two conditions. Overall, all strains except those expressing L. monocytogenes prsA1 and L. lactis pmpA exhibited statistically significant improved growth in comparison to that of the L. monocytogenes ΔprsA2 mutant, indicating some level of functional PrsA conservation.
PrsA functions associated with cell wall biosynthesis are not conserved.
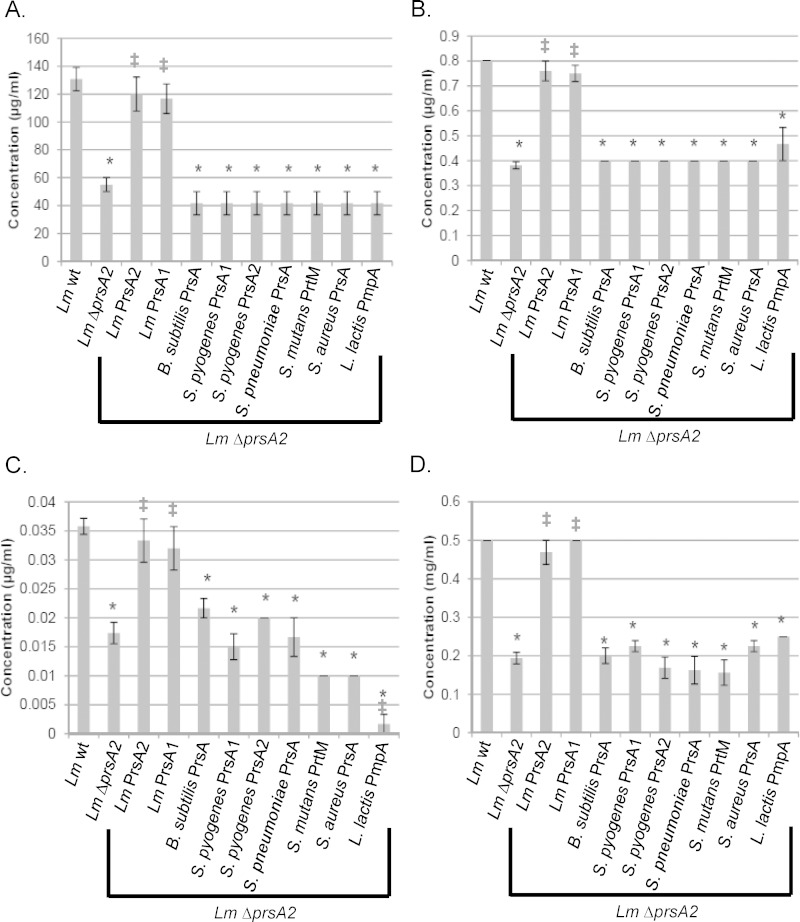
L. monocytogenes strains lacking prsA2 have reduced levels of cell wall-modifying enzymes as well as several penicillin binding proteins (PBPs) (12) and exhibit increased sensitivity to penicillin (36) and lysozyme, an enzyme that catalyzes the hydrolysis of peptidoglycan linkages (35). Given the apparent contributions of L. monocytogenes PrsA2 to cell wall biosynthesis and integrity (35, 36), we assessed the sensitivity of L. monocytogenes ΔprsA2 to additional antibiotics that target the cell wall, including those targeting lipid carrier recycling (bacitracin) and peptidoglycan cross-linking (vancomycin) (Fig. 6A to D). L. monocytogenes ΔprsA2 mutants were more susceptible to bacitracin and vancomycin than the wild type (Fig. 6A and B), and complementation with either L. monocytogenes prsA2 or prsA1 restored wild-type levels of resistance to all the cell wall-targeting antibiotics tested; however, none of the other Gram-positive prsA alleles were capable of restoring resistance (Fig. 6A to D). These data suggest that L. monocytogenes PrsA2 and L. monocytogenes PrsA1 share functional substrates that contribute to cell wall physiology and that recognition of these substrates may be specific to the L. monocytogenes chaperones.
FIG 6.
Bacterial resistance to cell wall-active antibiotics. The MIC to prevent growth of L. monocytogenes strains was determined by bacterial inoculation into broth containing dilutions of the indicated cell wall-active antibiotics. (A) MIC of bacitracin, which inhibits lipid carrier recycling. (B) MIC of vancomycin, which inhibits peptidoglycan transglycosylation and transpeptidation. (C) MIC of penicillin, which inhibits peptidoglycan transpeptidation. (D) MIC of lysozyme, which catalyzes the hydrolysis of peptidoglycan. Error bars represent the standard errors of the means from 4 to 8 cultures, whereas asterisks (*) and daggers (‡) indicate statistical significance of P ≤ 0.05, by two-tailed Student's t test, compared to L. monocytogenes wt and L. monocytogenes ΔprsA2, respectively.
Partial conservation of PrsA2 function as it relates to virulence factor secretion and bacterial spread through host cell monolayers.
The loss of L. monocytogenes prsA2 has been associated with reduced activity of proteins required for virulence (36), including listeriolysin O (LLO) and broad-range phospholipase (PC-PLC) (33, 34, 51–54). Whereas S. pyogenes prsA1, S. aureus prsA, and L. lactis pmpA were unable to complement the loss of L. monocytogenes prsA2 with respect to secreted LLO hemolytic activity (Fig. 7A), L. monocytogenes prsA1, B. subtilis prsA, S. pyogenes prsA2, S. pneumoniae prsA, and S. mutans prtM were all capable of an intermediate level of complementation of the L. monocytogenes ΔprsA2 mutant (Fig. 7A). Notably, the ability of a heterologous prsA allele to partially complement for LLO secretion was not restricted to those bacteria that produce a related cholesterol-dependent pore-forming toxin (such as streptolysin O [SLO] produced by S. pyogenes), as species (such as B. subtilis) that lack toxin production nevertheless expressed a prsA allele that restored LLO secretion and activity. All nine Gram-positive prsA alleles restored secreted PC-PLC activity, based on the assessment of lecithinase activity via growth of the bacteria on solid medium containing lecithin (Fig. 7B).
FIG 7.
Assessment of L. monocytogenes secreted hemolysin and phospholipase. (A) Hemolytic activity. Dilutions of bacterial culture supernatants were assessed for their ability to lyse sheep red blood cells (RBCs) in vitro. The reciprocal of the supernatant dilution that resulted in 50% RBC lysis (hemolytic units) was determined for 4 independent experiments. Error bars represent the standard errors of the means, whereas asterisks (*) and daggers (‡) indicate statistical significance of P ≤ 0.05, by two-tailed Student's t test, compared to L. monocytogenes wt and L. monocytogenes ΔprsA2, respectively. (B) Phospholipase activity was determined by the incubation of each strain on Listeria selective agar plates containing lecithin that is hydrolyzed, producing a zone of opacity surrounding the bacterial streak.
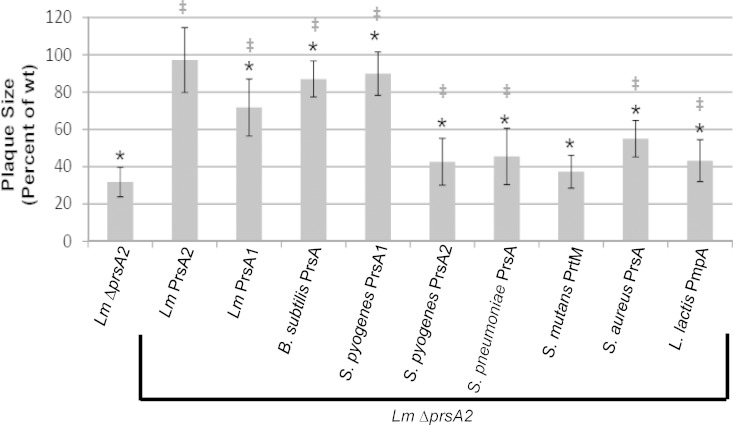
The ability of selected alleles of prsA to complement virulence factor secretion suggested that complementation may extend to activities necessary for intracellular growth and cell-to-cell spread. The L. monocytogenes ΔprsA2 mutant is defective for intracellular growth and cell-to-cell spread in monolayers of infected mouse L2 fibroblasts, forming zones of cell clearing (plaques) that are much smaller in size than those formed by wild-type L. monocytogenes (33–35) (Fig. 8). Despite intermediate complementation of secreted LLO by some prsA alleles and functional complementation of PC-PLC activity by all prsA alleles, only L. monocytogenes prsA2 was able to fully complement L. monocytogenes ΔprsA2 based on plaque formation in L2 monolayers (Fig. 8). However, most of the prsA alleles were observed to provide a partial level of complementation, with the exception of the S. mutans prtM allele (Fig. 8). These results indicate functional conservation of a subset of PrsA2-associated activities that contribute to L. monocytogenes intracellular growth and/or cell-to-cell spread but suggest that species-specific activities are required for full complementation.
FIG 8.
Bacterial intracellular growth and cell-to-cell spread as measured by plaque assays. Monolayers of mouse L2 fibroblasts were infected with the indicated L. monocytogenes strains, and plaque formation in the presence of gentamicin was determined 72 h postinfection. At least 25 plaques were measured in 3 to 5 independent experiments for all strains. Measurements represent plaque size with respect to L. monocytogenes wt (set at 100%). Error bars represent the standard errors of the means, whereas asterisks (*) and daggers (‡) indicate statistical significance of P ≤ 0.05, by two-tailed Student's t test, compared to L. monocytogenes wt and L. monocytogenes ΔprsA2, respectively.
Functional complementation of L. monocytogenes prsA2 does not extend to in vivo infection.
L. monocytogenes mutants lacking prsA2 are severely attenuated for virulence in mouse infection models, with at least 100,000-fold fewer CFU observed in mouse livers and spleens at 72 h postinfection than in mice infected with wild-type L. monocytogenes (33, 34). Given that several of the Gram-positive prsA alleles restored virulence factor secretion and provided for some restoration of intracellular growth and/or cell-to-cell spread (Fig. 9), we assessed whether any degree of functional complementation could be observed when strains were tested for virulence in mice (Fig. 9). For these experiments, we focused on those prsA alleles that supported appreciable levels of intracellular growth and/or cell-to-cell spread as indicated by plaque formation in L2 monolayers, i.e., B. subtilis prsA, S. pyogenes prsA1, and S. aureus prsA, and for contrast we included one allele with negligible complementation, L. lactis pmpA (Fig. 9). Complemented strains were examined for their ability to colonize, persist, and/or multiply in target organs of intravenously infected mice. At 24 h postinfection, all bacterial strains could be detected in livers and spleens of infected animals; however, only animals infected with the ΔprsA2 mutant complemented with L. monocytogenes prsA2 had bacterial burdens that were equivalent to those infected with wild-type L. monocytogenes (Fig. 9A). Animals infected with ΔprsA2 mutants containing the B. subtilis prsA and S. pyogenes prsA1 alleles exhibited bacterial burdens in both spleen and liver that were significantly increased in comparison to burdens after infection with the L. monocytogenes ΔprsA2 mutant without complementation (Fig. 9A). However, by 48 h postinfection, bacterial burdens continued to increase in liver and spleen for mice infected with the wild-type and L. monocytogenes prsA2-complemented strains, while the burdens associated with the remaining strains remained similar to those observed at 24 h postinfection (data not shown). By 72 h postinfection, bacterial burdens for mice infected with the wild-type and L. monocytogenes prsA2-complemented strain continued to increase, while the burdens in the remaining animals either remained similar to those observed at 24 h postinfection or decreased dramatically (Fig. 9A and B). These data demonstrate that only L. monocytogenes prsA2 is able to fully functionally complement the L. monocytogenes ΔprsA2 mutant, indicating that while some aspects of PrsA2 function may be broadly conserved, the full spectrum of activities required for virulence within infected animals appears to have been adapted specifically to L. monocytogenes PrsA2.
FIG 9.
L. monocytogenes PrsA2 is required for full virulence in mice. Mice were intravenously infected with 2 × 104 CFU of the indicated strain. At 24 h (A) and 72 h (B) postinfection, the spleens and livers were harvested, homogenized, and plated for bacterial CFU. Box plots are shown, where each point represents one mouse. A dotted line indicates the limit of detection of 500 CFU. Data were obtained from two independent experiments. Asterisks (*) and daggers (‡) indicate statistical significance of P ≤ 0.05, by two-tailed Wilcoxon rank sum test, compared to L. monocytogenes wt and L. monocytogenes ΔprsA2, respectively.
DISCUSSION
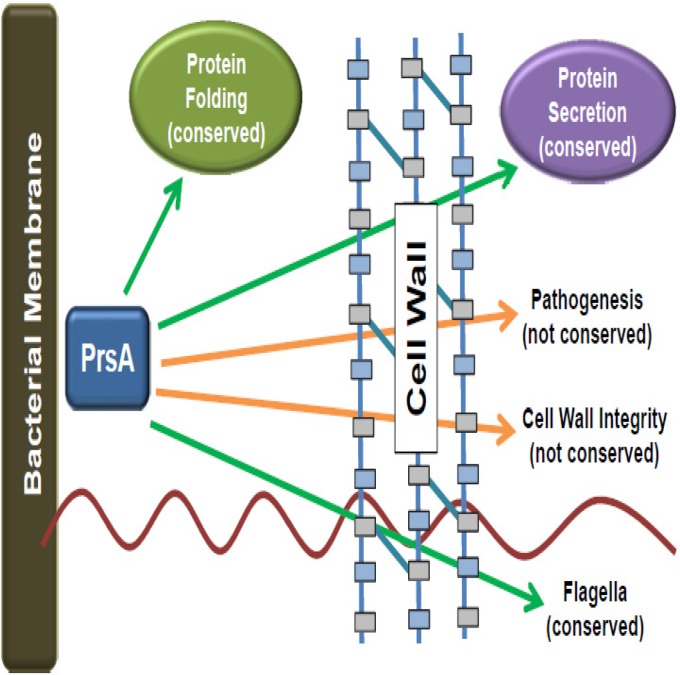
Gram-positive bacteria face challenges distinct from those faced by Gram-negative bacteria when it comes to the translocation of bacterial proteins across the cytoplasmic membrane. Membrane- and cell wall-associated proteins, as well as those secreted away from the cell, must fold in an environment that is both highly charged and freely accessible to external solutes and chemicals (4). Here we have described the functional characterization of a critical secretion chaperone, L. monocytogenes PrsA2, and its related homologues based on the ability of different prsA alleles to functionally complement L. monocytogenes strains lacking prsA2. Our data indicate a surprisingly broad degree of functional complementation for complex activities such as flagellum-mediated swimming motility and pH resistance, while other aspects of bacterial physiology, such as cell wall synthesis and bacterial replication within an infected animal, require functions specific to L. monocytogenes PrsA2. PrsA2 is thus a member of a family of chaperones that appear to maintain both broadly conserved and species-specific functions (Fig. 10).
FIG 10.
Model indicating conserved and species-specific aspects of L. monocytogenes PrsA2 activity. Shown is the L. monocytogenes bacterial membrane, extracellular space, and cell wall. The PrsA homologue is shown in the extracellular space; arrows indicate conserved functions (green) and functions that are not conserved (orange).
Chaperones often work in a stoichiometric rather than a catalytic manner, inhibiting protein aggregation while promoting folding (55); thus, the overall levels of PrsA2 protein produced are likely important for its functional activity. Each heterologous prsA allele was placed under the control of the same constitutive promoter, and yet protein levels clearly varied between L. monocytogenes strains expressing the different constructs (Fig. 3C and D). The reason underlying the variation in PrsA protein levels is not clear, as differences were observed even between L. monocytogenes prsA1 and prsA2, where, for example, potential codon bias is not anticipated to be present. Although differences in the levels of PrsA protein expressed were clearly evident, there was in general no direct correlation observed between the levels of the various PrsA proteins and the ability of each protein to functionally complement an L. monocytogenes ΔprsA2 strain. For example, L. monocytogenes PrsA1 complemented most PrsA2-associated activities, and yet its expression was severalfold lower than that of PrsA2 (Fig. 3C). B. subtilis PrsA was expressed at levels nearly equivalent to L. monocytogenes PrsA2, and it complemented a number of activities, but so did the poorly expressed S. pyogenes PrsA1, which was barely detectable. We interpret these results to indicate that, in most cases, sufficient levels of PrsA protein were present for functional complementation if complementation was possible. It should be noted, however, that increased expression of prsA1 from the pIMK2 promoter did enable complementation of secreted LLO and phospholipase activity in ΔprsA2 strains, a complementation activity that was not observed when prsA1 was expressed either from its own promoter or from the prsA2 promoter (12). The failure of even the abundantly expressed B. subtilis PrsA to complement L. monocytogenes ΔprsA2 intracellular growth and virulence in mice suggests that this version of PrsA does not recognize the full spectrum of substrates or interacting partners recognized by L. monocytogenes PrsA2.
All tested PrsA homologues were capable of restoring flagellum-mediated swimming motility and resistance to extremes of pH in L. monocytogenes ΔprsA2 strains (Fig. 4 and 5A and B). Restoration of both activities occurred regardless of whether the bacterium from which the prsA allele was derived was capable of swimming motility or of exhibiting resistance to extremes of pH. With respect to swimming motility, L. monocytogenes ΔprsA2 mutants produce visible flagella (data not shown), so it is not a complete absence of flagellar assembly that leads to the motility defect but may instead be the assembly of a nonfunctional flagellum and/or a defect in chemotactic ability. We suspect that the broad complementation of bacterial motility and pH resistance by the various prsA alleles reflects a conserved contribution of PrsA to some fundamental structural property of the bacterial surface that in turn leads to the correct assembly of membrane proteins that contribute to these two activities. Consistent with this hypothesis, there are several membrane-localized proteins that contribute to L. monocytogenes acid resistance, including the GAD and ADI transport proteins and the multicomponent FOF1 ATPase that extrudes protons, while swimming motility clearly requires the assembly of a large and complex flagellar motor.
In contrast to the broad complementation of swimming motility and pH resistance, only the L. monocytogenes PrsA homologues were capable of complementing for antibiotic resistance related to L. monocytogenes cell wall biosynthesis. Complementation with L. monocytogenes PrsA1 or PrsA2 restored resistance to antibiotics that targeted lipid carrier recycling, transglycosylation, transpeptidation, and peptidoglycan biosynthesis, while none of the other prsA alleles was capable of restoring any significant level of resistance (Fig. 6). This suggests that PrsA contributions to cell wall physiology are species specific, perhaps reflecting chaperone specificity for distinct cell wall enzymes. L. monocytogenes ΔprsA2 mutants have been shown to have reduced levels of a number of cell wall-modifying enzymes as well as several penicillin binding proteins (PBPs) (12). Similarly, PrsA has been associated with cell wall integrity and biosynthesis in several other Gram-positive organisms, including B. subtilis, S. mutans, and S. aureus (28, 32, 41).
With respect to the complementation of L. monocytogenes ΔprsA2 mutants for activities directly associated with bacterial pathogenesis, many of the heterologous prsA alleles restored secreted LLO and PC-PLC activity (Fig. 7) (33, 36). Interestingly, PrsA homologues derived from bacterial species, such as B. subtilis, that do not express a cholesterol-dependent cytolysin (CDC) restored secreted LLO activity, whereas PrsA1 derived from S. pyogenes, which expresses the CDC streptolysin O (SLO) (31), did not (Fig. 7A). S. pyogenes PrsA2 did complement for LLO secretion, and it is possible that SLO is a substrate for S. pyogenes PrsA2 but not for its PrsA1. Overall, these results indicate that PrsA substrate specificity cannot easily be predicted based on one PrsA being required for secretion of a family member of a particular type of secreted protein, such as a CDC. This observation indicates diversity between PrsA family members in substrate recognition, even between multiple alleles expressed by a single species.
With the exception of L. monocytogenes prsA2, none of the PrsA homologues was capable of restoring virulence to L. monocytogenes ΔprsA2 strains (Fig. 9). It thus appears that neither the complementation of cell wall integrity (L. monocytogenes PrsA1) nor the restoration of virulence factor secretion (B. subtilis and streptococcal homologues) suffices for restoration of L. monocytogenes ΔprsA2 virulence. L. monocytogenes PrsA1, B. subtilis PrsA, and S. pyogenes PrsA1 were all capable of partial restoration of intracellular growth and cell-to-cell spread in cell monolayers based on plaque formation; however, L. monocytogenes ΔprsA2 strains expressing these alleles remained as severely attenuated as L. monocytogenes ΔprsA2 mutants alone based on bacterial burdens recovered from the livers and spleens of infected animals (Fig. 9). These results indicate the degree to which PrsA2 has become specifically adapted for aspects of L. monocytogenes physiology necessary for animal infection.
Overall, the data presented here indicate that PrsA homologues have aspects of functional activity that are both broadly conserved (such as those activities required for swimming motility) and highly specialized (L. monocytogenes virulence in mice) (Fig. 10). It is interesting that some bacterial species maintain and express multiple PrsA homologues with at least some overlapping function (L. monocytogenes prsA1 and prsA2, S. pyogenes prsA1 and prsA2) (12, 31, 33, 36), whereas other bacteria have a single allele that may or may not be essential (B. subtilis, L. lactis) (29, 41). For L. monocytogenes, we have yet to identify a phenotype associated with prsA1 deletion strains. L. monocytogenes prsA1 is clearly expressed and functional; however, it is not clear yet as to whether it recognizes substrates that may be distinct from those recognized by PrsA2. It is possible that L. monocytogenes PrsA1 contributes to aspects of L. monocytogenes fitness that are relevant to bacterial survival in the outside environment rather than inside a mammalian host. For future studies, L. monocytogenes may prove to be an ideal organism for the molecular dissection of PrsA substrate specificity and activity, given that neither PrsA1 nor PrsA2 is required for growth of L. monocytogenes in broth culture and based on the ease with which mutations can be generated and assessed for functional consequences with respect to both bacterial physiology and virulence.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leonor Villalobos for technical assistance and members of the Freitag laboratory and the UIC Positive Thinking group for helpful discussions.
This work was supported by NIH grants R01 AI083241 and AI083241-03S1 to N.E.F.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00504-15.
REFERENCES
- 1.Kudva R, Denks K, Kuhn P, Vogt A, Muller M, Koch HG. 2013. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 164:505–534. doi: 10.1016/j.resmic.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 2.Jacob-Dubuisson F, Locht C, Antoine R. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol 40:306–313. doi: 10.1046/j.1365-2958.2001.02278.x. [DOI] [PubMed] [Google Scholar]
- 3.Newman CL, Stathopoulos C. 2004. Autotransporter and two-partner secretion: delivery of large-size virulence factors by gram-negative bacterial pathogens. Crit Rev Microbiol 30:275–286. doi: 10.1080/10408410490499872. [DOI] [PubMed] [Google Scholar]
- 4.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol 6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 5.Xayarath B, Freitag NE. 2012. Optimizing the balance between host and environmental survival skills: lessons learned from Listeria monocytogenes. Future Microbiol 7:839–852. doi: 10.2217/fmb.12.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mead PS, Dunne EF, Graves L, Wiedmann M, Patrick M, Hunter S, Salehi E, Mostashari F, Craig A, Mshar P, Bannerman T, Sauders BD, Hayes P, Dewitt W, Sparling P, Griffin P, Morse D, Slutsker L, Swaminathan B. 2006. Nationwide outbreak of listeriosis due to contaminated meat. Epidemiol Infect 134:744–751. doi: 10.1017/S0950268805005376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg Infect Dis 5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delgado AR. 2008. Listeriosis in pregnancy. J Midwifery Womens Health 53:255–259. doi: 10.1016/j.jmwh.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Drevets DA, Bronze MS. 2008. Listeria monocytogenes: epidemiology, human disease, and mechanisms of brain invasion. FEMS Immunol Med Microbiol 53:151–165. doi: 10.1111/j.1574-695X.2008.00404.x. [DOI] [PubMed] [Google Scholar]
- 10.Smith MA, Takeuchi K, Anderson G, Ware GO, McClure HM, Raybourne RB, Mytle N, Doyle MP. 2008. Dose-response model for Listeria monocytogenes-induced stillbirths in nonhuman primates. Infect Immun 76:726–731. doi: 10.1128/IAI.01366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swaminathan B, Gerner-Smidt P. 2007. The epidemiology of human listeriosis. Microbes Infect 9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Alonzo F III, Freitag NE. 2010. Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect Immun 78:4944–4957. doi: 10.1128/IAI.00532-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shetron-Rama LM, Mueller K, Bravo JM, Bouwer HG, Way SS, Freitag NE. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol Microbiol 48:1537–1551. doi: 10.1046/j.1365-2958.2003.03534.x. [DOI] [PubMed] [Google Scholar]
- 14.Mueller KJ, Freitag NE. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect Immun 73:1917–1926. doi: 10.1128/IAI.73.4.1917-1926.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Port GC, Freitag NE. 2007. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect Immun 75:5886–5897. doi: 10.1128/IAI.00845-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de las Heras A, Cain RJ, Bielecka MK, Vazquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr Opin Microbiol 14:118–127. doi: 10.1016/j.mib.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Mostowy S, Cossart P. 2012. Virulence factors that modulate the cell biology of Listeria infection and the host response. Adv Immunol 113:19–32. doi: 10.1016/B978-0-12-394590-7.00007-5. [DOI] [PubMed] [Google Scholar]
- 18.Freitag NE, Youngman P, Portnoy DA. 1992. Transcriptional activation of the Listeria monocytogenes hemolysin gene in Bacillus subtilis. J Bacteriol 174:1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leimeister-Wachter M, Haffner C, Domann E, Goebel W, Chakraborty T. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc Natl Acad Sci U S A 87:8336–8340. doi: 10.1073/pnas.87.21.8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengaud J, Dramsi S, Gouin E, Vazquez-Boland JA, Milon G, Cossart P. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol Microbiol 5:2273–2283. doi: 10.1111/j.1365-2958.1991.tb02158.x. [DOI] [PubMed] [Google Scholar]
- 21.Freitag NE, Port GC, Miner MD. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat Rev Microbiol 7:623–628. doi: 10.1038/nrmicro2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG, Portnoy DA. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517:170–173. doi: 10.1038/nature14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun 74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cahoon LA, Freitag NE. 2014. Listeria monocytogenes virulence factor secretion: don't leave the cell without a chaperone. Front Cell Infect Microbiol 4:13. doi: 10.3389/fcimb.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forster BM, Marquis H. 2012. Protein transport across the cell wall of monoderm Gram-positive bacteria. Mol Microbiol 84:405–413. doi: 10.1111/j.1365-2958.2012.08040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobs M, Andersen JB, Kontinen V, Sarvas M. 1993. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol Microbiol 8:957–966. doi: 10.1111/j.1365-2958.1993.tb01640.x. [DOI] [PubMed] [Google Scholar]
- 27.Kontinen VP, Saris P, Sarvas M. 1991. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol Microbiol 5:1273–1283. doi: 10.1111/j.1365-2958.1991.tb01901.x. [DOI] [PubMed] [Google Scholar]
- 28.Jousselin A, Renzoni A, Andrey DO, Monod A, Lew DP, Kelley WL. 2012. The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:3629–3640. doi: 10.1128/AAC.06264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drouault S, Anba J, Bonneau S, Bolotin A, Ehrlich SD, Renault P. 2002. The peptidyl-prolyl isomerase motif is lacking in PmpA, the PrsA-like protein involved in the secretion machinery of Lactococcus lactis. Appl Environ Microbiol 68:3932–3942. doi: 10.1128/AEM.68.8.3932-3942.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cron LE, Bootsma HJ, Noske N, Burghout P, Hammerschmidt S, Hermans PW. 2009. Surface-associated lipoprotein PpmA of Streptococcus pneumoniae is involved in colonization in a strain-specific manner. Microbiology 155:2401–2410. doi: 10.1099/mic.0.026765-0. [DOI] [PubMed] [Google Scholar]
- 31.Ma Y, Bryant AE, Salmi DB, Hayes-Schroer SM, McIndoo E, Aldape MJ, Stevens DL. 2006. Identification and characterization of bicistronic speB and prsA gene expression in the group A Streptococcus. J Bacteriol 188:7626–7634. doi: 10.1128/JB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L, Wu T, Hu W, He X, Sharma S, Webster P, Gimzewski JK, Zhou X, Lux R, Shi W. 2013. Phenotypic characterization of the foldase homologue PrsA in Streptococcus mutans. Mol Oral Microbiol 28:154–165. doi: 10.1111/omi.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonzo F III, Port GC, Cao M, Freitag NE. 2009. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect Immun 77:2612–2623. doi: 10.1128/IAI.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol 191:3950–3964. doi: 10.1128/JB.00016-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forster BM, Zemansky J, Portnoy DA, Marquis H. 2011. Posttranslocation chaperone PrsA2 regulates the maturation and secretion of Listeria monocytogenes proprotein virulence factors. J Bacteriol 193:5961–5970. doi: 10.1128/JB.05307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonzo F III, Xayarath B, Whisstock JC, Freitag NE. 2011. Functional analysis of the Listeria monocytogenes secretion chaperone PrsA2 and its multiple contributions to bacterial virulence. Mol Microbiol 80:1530–1548. doi: 10.1111/j.1365-2958.2011.07665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kontinen VP, Sarvas M. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol 8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 38.Hyyrylainen HL, Vitikainen M, Thwaite J, Wu H, Sarvas M, Harwood CR, Kontinen VP, Stephenson K. 2000. D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J Biol Chem 275:26696–26703. doi: 10.1074/jbc.M003804200. [DOI] [PubMed] [Google Scholar]
- 39.Wahlstrom E, Vitikainen M, Kontinen VP, Sarvas M. 2003. The extracytoplasmic folding factor PrsA is required for protein secretion only in the presence of the cell wall in Bacillus subtilis. Microbiology 149:569–577. doi: 10.1099/mic.0.25511-0. [DOI] [PubMed] [Google Scholar]
- 40.Vitikainen M, Lappalainen I, Seppala R, Antelmann H, Boer H, Taira S, Savilahti H, Hecker M, Vihinen M, Sarvas M, Kontinen VP. 2004. Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J Biol Chem 279:19302–19314. doi: 10.1074/jbc.M400861200. [DOI] [PubMed] [Google Scholar]
- 41.Hyyrylainen HL, Marciniak BC, Dahncke K, Pietiainen M, Courtin P, Vitikainen M, Seppala R, Otto A, Becher D, Chapot-Chartier MP, Kuipers OP, Kontinen VP. 2010. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol Microbiol 77:108–127. doi: 10.1111/j.1365-2958.2010.07188.x. [DOI] [PubMed] [Google Scholar]
- 42.Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, Vazquez-Boland JA, Kunst F, Cossart P, Buchrieser C. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 43.Monk IR, Gahan CG, Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl Environ Microbiol 74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun AN, Camilli A, Portnoy DA. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect Immun 58:3770–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heikkinen O, Seppala R, Tossavainen H, Heikkinen S, Koskela H, Permi P, Kilpelainen I. 2009. Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA—implications for the catalytic mechanism of parvulins. BMC Struct Biol 9:17. doi: 10.1186/1472-6807-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leskela S, Wahlstrom E, Kontinen VP, Sarvas M. 1999. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the Lgt gene. Mol Microbiol 31:1075–1085. doi: 10.1046/j.1365-2958.1999.01247.x. [DOI] [PubMed] [Google Scholar]
- 47.Hermans PW, Adrian PV, Albert C, Estevao S, Hoogenboezem T, Luijendijk IH, Kamphausen T, Hammerschmidt S. 2006. The streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonization. J Biol Chem 281:968–976. doi: 10.1074/jbc.M510014200. [DOI] [PubMed] [Google Scholar]
- 48.Tremillon N, Morello E, Llull D, Mazmouz R, Gratadoux JJ, Guillot A, Chapot-Chartier MP, Monlezun L, Sole V, Ginisty H, Poquet I. 2012. PpiA, a surface PPIase of the cyclophilin family in Lactococcus lactis. PLoS One 7:e33516. doi: 10.1371/journal.pone.0033516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peel M, Donachie W, Shaw A. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and western blotting. J Gen Microbiol 134:2171–2178. [DOI] [PubMed] [Google Scholar]
- 50.Chaturongakul S, Raengpradub S, Wiedmann M, Boor KJ. 2008. Modulation of stress and virulence in Listeria monocytogenes. Trends Microbiol 16:388–396. doi: 10.1016/j.tim.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cossart P, Vicente MF, Mengaud J, Baquero F, Perez-Diaz JC, Berche P. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun 57:3629–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grundling A, Gonzalez MD, Higgins DE. 2003. Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J Bacteriol 185:6295–6307. doi: 10.1128/JB.185.21.6295-6307.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeung PS, Na Y, Kreuder AJ, Marquis H. 2007. Compartmentalization of the broad-range phospholipase C activity to the spreading vacuole is critical for Listeria monocytogenes virulence. Infect Immun 75:44–51. doi: 10.1128/IAI.01001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quan S, Koldewey P, Tapley T, Kirsch N, Ruane KM, Pfizenmaier J, Shi R, Hofmann S, Foit L, Ren G, Jakob U, Xu Z, Cygler M, Bardwell JC. 2011. Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat Struct Mol Biol 18:262–269. doi: 10.1038/nsmb.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bishop DK, Hinrichs DJ. 1987. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements J Immunol 139:2005–2009. [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.