Summary
Heat causes protein misfolding and aggregation, and in eukaryotic cells triggers aggregation of proteins and RNA into stress granules. We have carried out extensive proteomic studies to quantify heat-triggered aggregation and subsequent disaggregation in budding yeast, identifying more than 170 endogenous proteins aggregating within minutes of heat shock in multiple subcellular compartments. We demonstrate that these aggregated proteins are not misfolded and destined for degradation. Stable-isotope labeling reveals that even severely aggregated endogenous proteins are disaggregated without degradation during recovery from shock, contrasting with the rapid degradation observed for exogenous thermolabile proteins. Although aggregation likely inactivates many cellular proteins, in the case of a heterotrimeric aminoacyl-tRNA synthetase complex, the aggregated proteins remain active with unaltered fidelity. We propose that most heat-induced aggregation of mature proteins reflects the operation of an adaptive, autoregulatory process of functionally significant aggregate assembly and disassembly that aids cellular adaptation to thermal stress.
Introduction
Following heat shock—a rapid increase in temperature to stressful but nonlethal levels—cells accumulate protein aggregates, decelerate protein synthesis, and mount a transcriptional program called the heat shock response. Upregulated transcripts encode so-called heat-shock proteins, of which many are molecular chaperones. The standard interpretation of these events is that heat causes endogenous (species-native) proteins to misfold into aggregation-prone species whose toxicity is mitigated and reversed by chaperones (Lindquist, 1986; Mogk et al., 1999; Vabulas et al., 2010; Verghese et al., 2012). Misfolding here refers to the deleterious loss of—or failure to attain—natively folded protein structure, sometimes by adopting stable non-native conformations.
Newly synthesized proteins are particularly susceptible to heat-induced misfolding and aggregation, and appear to be the major triggers of the heat-shock response, as well as the main beneficiaries of its induction (Baler, 1992; Vabulas et al., 2010). In agreement, heat triggers rapid degradation of newly synthesized proteins, but not of bulk cellular protein (Medicherla and Goldberg, 2008).
Mature, folded proteins also aggregate in response to heat shock, forming protein/poly(A)+-RNA structures called heat shock granules (HSGs). Discovered in plants (Nover et al., 1983), HSGs form upon robust heat shock in a range of eukaryotes, including budding yeast, trypanosome, insect, and mammalian cells (Grousl et al., 2009, 2013; Cherkasov et al., 2013; Farny et al., 2009). HSGs are functionally defined by their components, notably poly(A)-binding protein and eukaryotic initiation factor 4G; some components are common to RNA/protein granules formed during other stresses (Buchan et al., 2010, 2011; Kedersha and Anderson, 2002). The mechanism(s) of HSG formation remain unclear.
Many studies demonstrate aggregation and degradation of exogenous (heterologous or other non-species-native) proteins (Cherkasov et al., 2013; Heck et al., 2010; Fredrickson et al., 2013). Colocalization of exogenous aggregated proteins and HSGs has been interpreted as signaling the presence of endogenous misfolded proteins in HSGs (Cherkasov et al., 2013). However, the identities and folding states of HSG-associated proteins is largely unknown.
When cells return to lower temperatures, HSGs dissolution is promoted by the disaggregase Hsp104 and the chaperone Hsp70 (Cherkasov et al., 2013), which also disaggregate misfolded proteins in vitro (Glover and Lindquist, 1998). It is unknown what fraction of disaggregated proteins are degraded in vivo, although evidence that stress granules are degraded by autophagy (Buchan et al., 2013) suggests that degradation might be the dominant fate of stress-induced aggregates.
Here, using the model eukaryote budding yeast (Saccharomyces cerevisiae), we report the results of experiments aimed at answering many of these fundamental questions. Which endogenous proteins aggregate during heat shock, and how do proteins differ in their propensity to aggregate? What is the relationship between protein aggregation and the formation of granules and other large subcellular foci? How does heat affect the function and fidelity of proteins determined to aggregate in response to heat shock in vivo? And what are the fates of endogenous aggregated proteins after heat shock?
Results
Aggregation profiling identifies many thermally sensitive proteins
We quantified aggregation of proteins into high-molecular-weight particles by biochemical separation into supernatant and pellet fractions using ultracentrifugation, stable-isotope labeling and liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) (Fig. 1A), and statistical estimation of the proportion of each protein in the supernatant (pSup) to control for differences in fraction mixing and inter-experiment variability (experimental procedures and Fig. S1). Here and throughout, we refer to pelletable species of proteins that are soluble before heat shock as “aggregates,” without prejudging whether the particles result from misfolding, formation of protein/RNA granules, or other homogeneous or heterogeneous oligomerization processes.
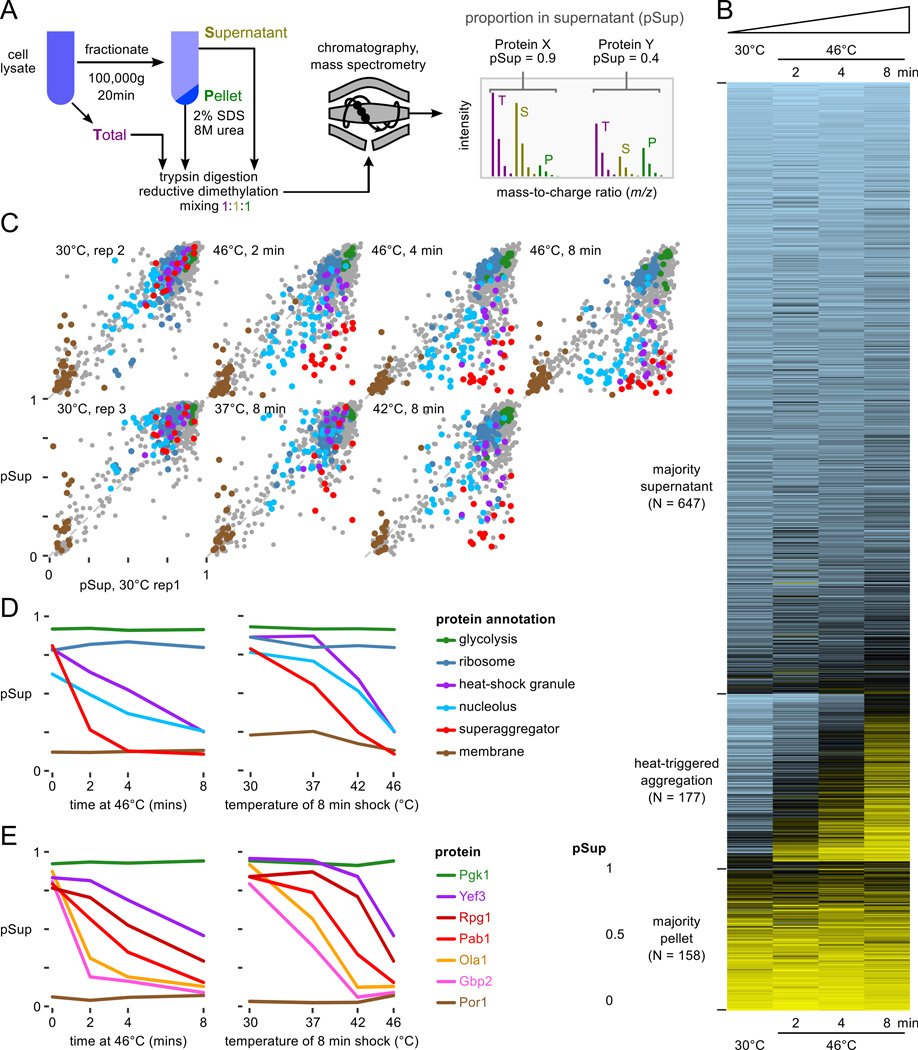
Figure 1.
Proteome-wide aggregation profiling. A, Aggregation profiling by isotope-labeling and mass spectrometry yields estimates of the proportion of each protein in the supernatant (pSup) before and after thermal stress. B, pSup values in the 46°C timecourse for all well-detected proteins show proteins consistently found in the supernatant (top), consistently found in the pellet (bottom), and transitioning from supernatant to pellet during the 8 minute heat shock (middle, see text). C, Progressive protein aggregation quantified by proportion in the supernatant fraction (pSup) during a 46°C treatment compared to unshocked replicates (top), and with increasing 8-minute shock temperature (bottom). Protein annotations in panels C and D are the same; superaggregators (see text) include 5 nucleolar proteins. D, Behavior of proteins in various categories (cf. C) as a function of temperature, for 8 minutes, and time at 46°C. E, Individual proteins aggregate at different rates in response to heat; more are shown in Fig. S4A. In panels D and E, 30°C rep 1 is shown in the timecourse plots, the same biological sample as the 46°C data; 30°C rep 3 is shown in the temperature course plots, the same biological sample as 37°C and 42°C, 8 minute, data.
We quantified protein aggregation in cells transferred from 30°C to 46°C for 2, 4, and 8 minutes, and to 37°C and 42°C for 8 minutes (Fig. 1, Table S2). During these treatments, which are shorter than those typically used to study heat shock granule (HSG) formation (Grousl et al. 2009; Cherkasov et al. 2013, and extended experimental procedures), genes upregulated in the transcriptional heat-shock response show no detectable change in protein levels (Fig. S2A). By contrast, aggregation is rapid, widespread, and increases with time and temperature (Fig. 1B,C).
Heat triggers rapid and specific protein aggregation
In the 46°C timecourse, 982 proteins are detected with least two unique peptides at all timepoints (“well-detected,” 73% of the proteome by mass, 17% of verified open reading frames (Cherry et al., 2012)), upon which we focus. Most cellular proteins remain in the supernatant throughout (Fig. 1B), and cytosolic and ribosomal proteins are the most enriched gene ontology (GO) terms describing these proteins (Fig. S3). Proteins found in the pellet in all conditions are primarily membrane-associated (Fig. S3). Heat triggers the aggregation of a large group of proteins (177 well-detected proteins), classified by consistent and substantial movement from the supernatant in unheated cells to the pellet after a shift to 46°C (Table S3; and experimental procedures). Only four proteins moved from pellet to supernatant in the same interval (Table S4).
Of 18 HSG components identified in the literature (Table S1), we detected all but one (Ngr1). Twelve of these meet our criteria for heat-triggered aggregation, including poly(A)-binding protein (Pab1), eIF4G/Tif4631, and eIF3, where our data show aggregation of all five stably complexed eIF3 subunits (Nip1/Rpg1/Prt1 reported previously (Grousl et al., 2009), Tif34/Tif35 reported here), and eukaryotic release factors eRF1/Sup45 and eRF3/Sup35 (Fig. S4A). Of the remaining five, Whi3 is not well-detected but aggregates, and three proteins (Dhh1, eIF4G2/Tif4632, and small-subunit ribosomal protein Rps30A/B) do not clearly aggregate. The behavior of Rps30A/B is consistent with the lack of aggregation in 82 other well-detected ribosomal gene products from both subunits and with in situ hybridization against ribosomal RNA (Cherkasov et al., 2013). Our experimental conditions therefore allow us to quantify biochemically the aggregation of proteins reported to form HSGs by fluorescence microscopy.
In our data, 17 proteins aggregate more than any previously reported HSG component after 2 minutes heat shock at 46°C; we dub these “superaggregators” (Table 3 and experimental procedures). For example, the nuclear protein Ett1 plunges from a supernatant proportion of 0.93 to 0.15 after two minutes at 46°C, while the mRNA-binding protein Gbp2 drops from 0.8 to 0.25. In the same interval, HSG-forming proteins such as Pab1 and eIF3 remain mostly soluble (Figs. 1C, S4). Notably, most superaggregators also show clear aggregation after 8 minutes at 37°C and 42°C. At these temperatures and times, Pab1-marked HSGs do not form (Cherkasov et al., 2013).
GO terms enriched in heat-aggregating proteins include the molecular functions RNA binding (exemplified by poly(A)-binding protein Pab1 along with Npl3, Pub1, and Gbp2) and RNA helicase activity (seven proteins including Ded1 and Dbp2/3) (Fig. S3). Enriched cellular components include cytosolic stress granules, polysomes, and notably the nucleolus (16 nucleolar proteins).
Six aminoacyl-tRNA-synthetases aggregate, including the yeast multisynthetase complex composed of methionyl-and glutamyl-tRNA synthetases Mes1 and Gus1 bound together by the aminoacylation cofactor Arc1. We return to this complex later.
Molecular chaperones, which colocalize with HSGs, largely remain soluble in our data, suggesting a biochemical distinction between aggregation and recruitment to aggregates. However, notable exceptions exist, including the ribosome-associated chaperone complex (RAC) discussed later. The small heat-shock proteins Hsp26 and Hsp42, despite poor detection in our dataset, partition into the pellet upon heat shock (Fig. S4A).
Endogenous proteins aggregate in distinct compartments
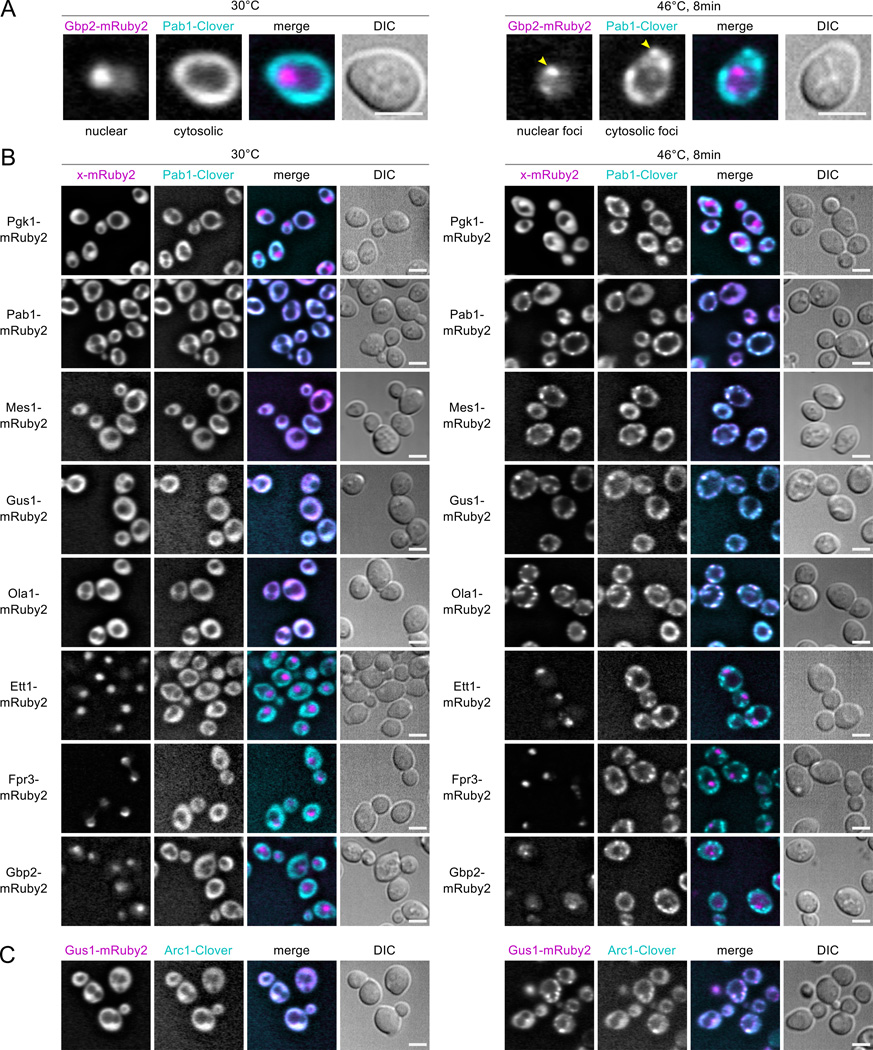
To determine the subcellular location and morphology of aggregates for MS-identified aggregators, we imaged yeast strains engineered with fluorescent C-terminally tagged proteins at their native chromosomal loci. We tagged select proteins with mRuby2, a red fluorescent protein, and tagged the HSG marker Pab1 with Clover, a green fluorescent protein (Lam et al., 2012), mating these strains to form dual-tagged diploids (Fig. 2A). Fusions of the nonaggregating glycolytic enzyme Pgk1 stay cytosolic and diffuse when heat shocked, and diploids bearing Pab1 tagged with both fluorophores (no untagged Pab1 present) form cytosolic foci containing both colored tags (Fig. 2B), indicating that these fluorophores neither cause nor prevent aggregation.
Figure 2.
Live-cell microscopy identifies heat-aggregating proteins forming cytosolic or nuclear granules. Diploid strains containing the HSG component Pab1 tagged with the green fluorescent protein (FP) Clover (cyan in merged images) and test proteins tagged with the red FP mRuby2 (magenta in merge) were imaged at 30°C and after 8 minutes’ heat shock at 46°C. Scale bar is 5 µm. A, Heat-induced nuclear and cytosolic aggregation of Gbp2- mRuby2 and Pab1-Clover, respectively. B, Non-aggregating Pgk1-mRuby2 remains diffuse during heat shock, while Pab1-Clover forms HSGs. Pab1-Clover and Pab1-mRuby2 form colocalized foci during heat shock. Fusions of MS-identified heat-aggregating proteins form foci that colocalize with Pab1 during heat shock (Mes1, Gus1, Ola1), or form subnuclear foci (Ett1, Fpr3, Gbp2). C, The aminoacylation cofactor in the multisynthetase complex, Arc1, forms heat-induced foci colocalized with Gus1.
Proteins detected to aggregate by mass spectrometry after an 8-minute 46°C heat shock also form foci (Fig. 2B,C). Mes1 and Gus1, components of the multisynthetase complex, form cytosolic foci colocalized with Pab1. Arc1, the third multisynthetase component, likewise forms fluorescent foci co-localized with Gus1 (Fig. 2C). Ola1, a superaggregating cytosolic protein previously implicated in translation termination (Samanfar et al., 2014), also forms foci colocalized with Pab1. These four proteins are all thus bona fide heat shock granule components.
Some heat-aggregating proteins form nuclear foci. Gbp2, a nuclear poly(A)-RNA-binding protein involved in nuclear-cytosolic mRNA transport, forms sub-nuclear granules during heat shock (Fig. 2B). Fpr3, a nucleolar component adopting the diagnostic nucleolar crescent shape under non-shock conditions, becomes increasingly granular within the nucleolus during heat shock (Fig. 2B). Ett1, a nuclear protein and the most rapidly aggregating protein detected by mass spectrometry, forms nuclear foci during heat shock (Fig. 2B) which colocalized with Gar1, a nucleolar protein which shows no heat-triggered aggregation by MS (Fig. S5A). These results suggest that Ett1 aggregates in or near the nucleolus upon heat shock, possibly consistent with localization to the intranuclear quality-control compartment (INQ) (Miller et al., 2015a). We often observe multiple Ett1 foci per cell (Fig. S5C).
Translation inhibition impedes granule formation but does not prevent stress-triggered protein aggregation
Our data indicate clear distinctions between the heat-triggered in vivo formation of fluorescent foci and of submicroscopic, biochemically detectable aggregates. After milder shocks, several proteins producing pelletable aggregates did not form foci, such as Ett1 (at 37°C) and Pab1 (at 42°C) (Fig. S5B,C). Also, Hsp104 forms foci co-localized with Pab1 upon heat shock (Cherkasov et al., 2013) while remaining highly soluble (Fig. S4A), showing its recruitment to, but not stable association with, substrates within heat shock granules.
A series of studies have demonstrated the preferential retention of cytosolic heat-induced protein aggregates by mother cells during budding (Aguilaniu et al., 2003; Liu et al., 2010; Zhou et al., 2014); these cytosolic Hsp104 foci are heat shock granules. Zhou et al. (2014) observe that Hsp104 focus formation during heat shock is blocked by the translation elongation inhibitor cycloheximide (CHX), and conclude that heat-induced aggregation requires active translation. By contrast,Jacobson et al. (2012) observe that CHX blocks Hsp104 foci during arsenite stress, but not during heat shock. We wondered whether biochemical detection might shed useful light on the relationship between aggregation, heat-shock granules, and translation.
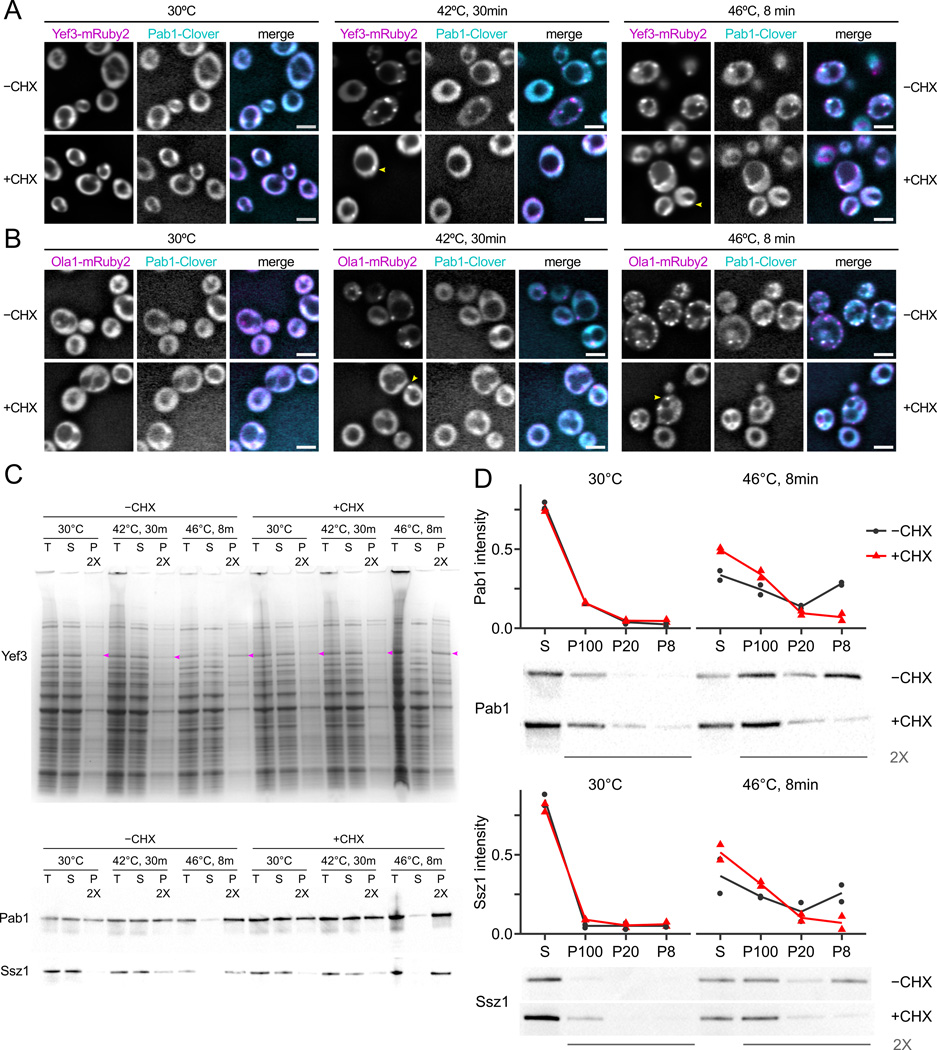
To study these phenomena, we treated cells with 100 µg/mL CHX for 5 minutes, then subjected them to either a 42°C heat shock for 30 minutes as in Zhou et al. (2014), or to a 46°C heat shock for 8 minutes. This dose of CHX attenuates formation of visible fluorescent foci by tagged Pab1 (Fig. 3A,B). However, the cytosolic heat-aggregators Yef3 and Ola1 still form some fluorescent foci in the presence of CHX (Fig. 3A,B). Thus, translation inhibition attenuates the heat-triggered formation of foci for some but not all cytosolic proteins.
Figure 3.
Heat-triggered protein aggregation does not require ongoing translation. A, Cycloheximide (CHX; 100 µg/mL) blocks formation of heat-triggered cytosolic foci by fluorescently tagged Pab1 and attenuates formation of foci by Yef3. Scale bars are 5µm, arrows indicate foci. B, Ola1 forms fluorescent foci in response to heat shock in the presence of CHX. C, Pab1, Ssz1, and Yef3 (arrows on gel) are found in the 100,000 g supernatant (S) in lysate from unshocked cells, but enter the 100,000 g pellet (P) after heat shock independent of CHX treatment. Coomassie-stained protein gel and western blots against native proteins are shown. T, total protein. D, CHX inhibits Pab1 and Ssz1 entry into large aggregates, but not into small aggregates. Cell lysate was progressively fractionated at 8,000 g (pellet, P8), 20,000 g (P20), then 100,000 g (P100), and pellets and residual supernatant (S) were western blotted against Pab1 and Ssz1; intensity as proportion of total was quantified in two biological replicates (Fig. S6), and a representative blot is shown.
We also measured protein aggregation biochemically during identical heat shocks by analyzing 100,000 g pelleting particles. Pab1, Ssz1, and Yef3 all enter the 100,000 g pellet after a 46°C, 10-minute heat shock, with reduced aggregation after a 42°C, 30-minute shock. Surprisingly, biochemical aggregation was unaffected by CHX (Fig. 3C).
The data are consistent with a model of multi-stage aggregation, where initial formation of biochemical aggregates is followed by CHX-sensitive collection of these aggregates into larger bodies visible as foci. To test this model, we progressively fractionated cell lysate first at 8,000 g × 3 minutes (pellet, P8, largest aggregates), then fractionated the supernatant at 20,000 g × 5 minutes (pellet, P20, smaller aggregates), then fractionated the second supernatant at 100,000 g × 20 minutes (pellet, P100, smallest aggregates), collecting residual 100,000 g supernatant (S). Western blotting against native Pab1 showed that 75% of Pab1 remained in the supernatant from unshocked cells regardless of CHX treatment (Figs. 3D, S6). In cells heat shocked for 8 minutes at 46°C, most Pab1 entered P8 and P100; treatment with CHX blocked formation of P8 particles and increased levels of P100 particles, as predicted (Fig. 3D). Ssz1 shows the same pattern (Fig. 3D), as does Yef3 (total protein gel, Fig. S6).
These results support a CHX-blockable secondary assembly of aggregates into cytosolic foci which does not affect heat-induced formation of smaller aggregates.
Translation-related proteins aggregate in coherent groups
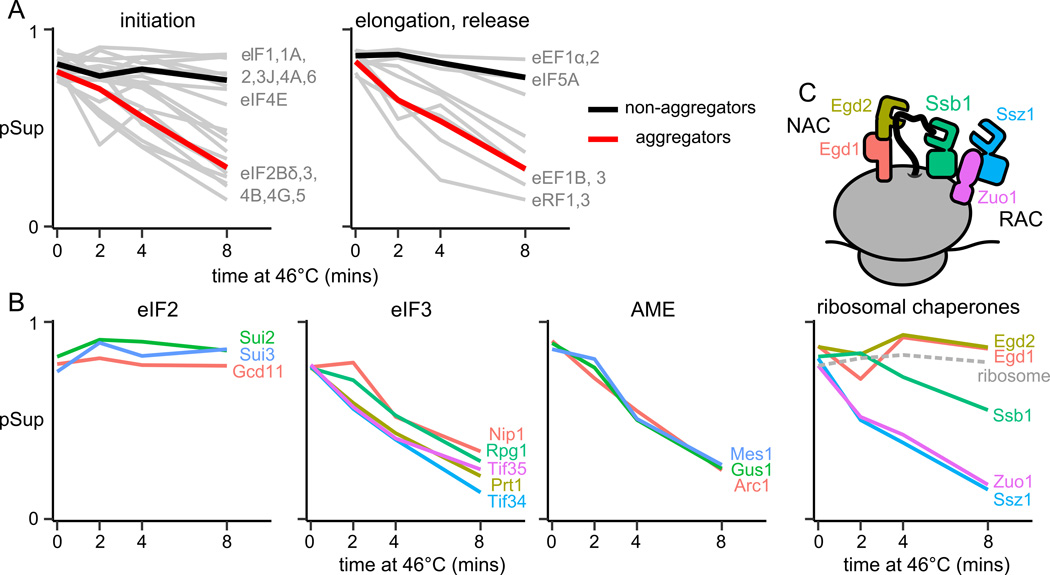
The heat-triggered aggregation of eIF3 and the multisynthetase complex prompted us to examine aggregation of other protein complexes involved in translation. Translation factors partition into heat-aggregators and non-aggregators (Fig. 4A). Assuming aggregated translation factors are inactive, the observed aggregation of eIF2B, eIF4B/G, eEF3, or eRF1 would be individually sufficient to substantially reduce net protein synthesis (Firczuk et al., 2013).
Figure 4.
Stable translation-related complexes aggregate coherently. Proportion in supernatant (pSup) is plotted against time of heat shock at 46°C; each panel shows one or more complexes and each line a protein component. A, pSup of all well-detected translation factors. Mean is shown for aggregators and non-aggregators in each plot. B, Aggregation of translation initiation factors 2 and 3, and of the multi-tRNA-synthetase complex. C, Aggregation of chaperone complexes involved in co-translational folding.
Each stable protein complex falls into a single category: of the components of eukaryotic elongation factor 1 (eEF1), all elements of the stable subcomplex eEF1B heat-aggregate, but Tef1/eEF1α does not (Fig. 4A). All components of eIF2 have similar high pSup across conditions, while all components of eIF3 heat-aggregate with similar kinetics (Fig. 4B). Aggregation of the multisynthetase complex is particularly synchronous (Fig. 4B).
Complexes with shared interaction partners show distinct aggregation patterns: for example, the nascent-polypeptide associated complex (NAC; Egd1/Egd2) and the ribosome-associated chaperone complex (RAC; Ssz1/Zuo1), along with Ssb1/2, bind the ribosome near the nascent peptide exit tunnel (Preissler and Deuerling, 2012). Both detected NAC components remain soluble across conditions, as do ribosomes; in contrast, RAC components aggregate swiftly and in lockstep (Fig. 4C). More broadly, proteins associated in annotated complexes (Pu et al., 2009) have more similar pSup trajectories than expected by chance (Fig. S7).
The yeast multisynthetase complex forms active heat-triggered aggregates in vitro
The tight correlation of the three yeast multisynthetase components during heat-triggered aggregation (Fig. 4), and their aggregation into the same subcellular location (Fig. 2), prompted us to ask how heat affects this complex and its activity in isolation. The complex, dubbed AME, is a heterotrimer formed by the aminoacylation cofactor Arc1 (A), methionyl-tRNA synthetase Mes1 (M), and glutamyl-tRNA synthetase Gus1 (E), which interact through eukaryote-specific N-terminal domains in each protein (Frechin et al., 2014).
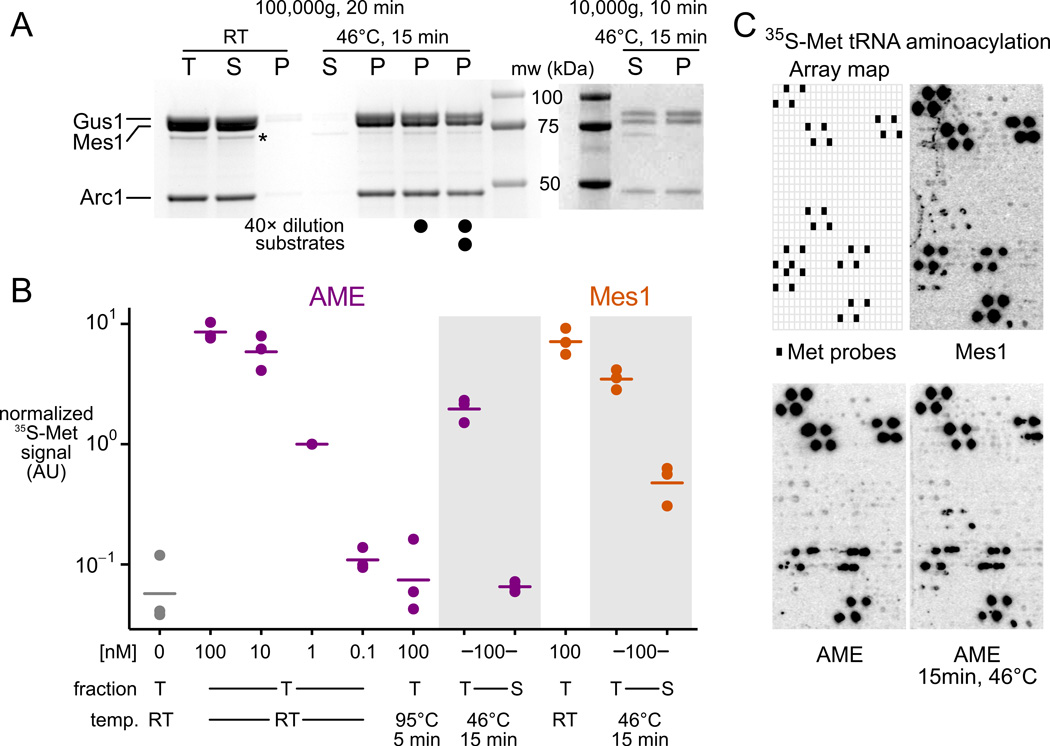
Recombinant reconstituted AME remains in the supernatant of a 100,000 g, 20 minute spin, but after a severe 46°C 15-minute treatment, aggregates completely into pelletable material and cannot be resolubilized by dilution and 1h incubation at 30°C with or without substrates (Fig. 5A).
Figure 5.
The yeast multisynthetase complex (AME) forms active aggregates in response to heat shock in vitro . A, Recombinant purified AME complex is soluble before heat treatment, and fully aggregated after (stained SDS PAGE; T, total; S, supernatant; P, pellet). A minor soluble degradation product is starred. Three heat-treated samples were incubated at room temperature (RT) for 1h (undiluted), or incubated for 1h at 30°C after 40× dilution and after addition of substrates as indicated (see Methods). Right, stoichiometry in aggregates revealed by lowerspeed centrifugation. mw, molecular weight marker. B, Activity measured by aminoacylation of total yeast tRNA with 35S-labeled methionine in a filter-binding assay. Three replicates are shown, with signal normalized by 1 nM AME RT levels. Lines indicate means. C, Fidelity measured by aminoacylation of tRNA-microarray-immobilized tRNAs with 35S-labeled methionine. Each cell contains probes complementary to a single tRNA species, with tRNA-Met probes arrayed as indicated.
Gentler centrifugation revealed that AME pellets as a stoichiometric complex (Fig. 5A), despite wide variation in the aggregation propensity of its constituents (Fig. S7C). Severely heat-shocked AME retains substantial activity, all of which resides in the aggregated fraction, as indicated by absence of activity in the supernatant after centrifugation (Fig. 5B). Similarly, the activity of heat-treated Mes1 is reduced more than 7-fold after spinning out aggregates (Fig. 5B). Gus1’s non-catalytic N-terminal domain proved necessary and sufficient for heat-induced Gus1 aggregation (Fig. S7D).
We next assessed the fidelity of tRNA-Met aminoacylation by AME before and after heat shock using tRNA microarrays. Under conditions where AME is fully aggregated (cf. Fig. 5A) it retains fidelity indistinguishable from untreated AME or Mes1 (Fig. 5C).
Bacterial inclusion bodies can contain active exogenous enzymes (Martínez-Alonso et al., 2009). Our results reveal heat-induced formation of endogenous, active, stoichiometric aggregates with normal fidelity. Here, reduced activity may indicate partial loss of function or reduced ability of large tRNA substrates to penetrate these in vitro aggregates.
Global profiling of disaggregation during recovery reveals near-complete reversibility of aggregation
Heat shock granules slowly disappear after cells are returned to non-shock temperatures (Cherkasov et al., 2013; Parsell et al., 1994), yet it has remained unclear if endogenous aggregate dispersion is due to disaggregation followed by degradation and resynthesis, or to disaggregation back into a stable soluble pool.
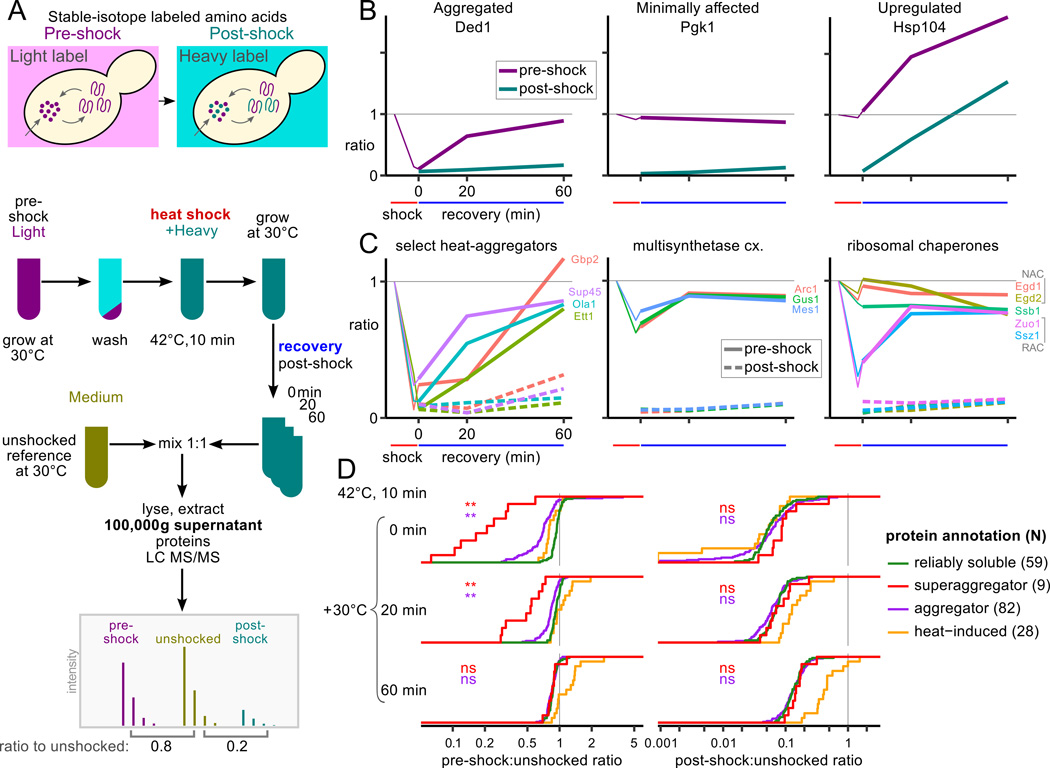
To measure disaggregation and new synthesis at the proteome scale without blocking synthesis or degradation, we performed a media-shift experiment (Fig. 6A and experimental procedures) in which cells are grown on a first set of stable-isotope-labeled amino acids, shifted to media containing a second set of labels, then heat shocked at 42°C and allowed to recover for a defined time at 30°C. Upon collection, these cells are mixed with cells from an unshocked (30°C) reference sample grown on a third label. Supernatant fractions of these mixtures measured after 0, 20, and 60 minutes of recovery allowed us to observe the depletion of aggregating proteins from the supernatant after shock followed by their recovery in both the pre- and post-shock labels, indicating new synthesis, or only in the pre-shock label, indicating disaggregation.
Figure 6.
Heat-aggregated proteins disaggregate during recovery. A, Schematic experimental design for SILAC media-shift measurement of soluble protein dynamics during recovery from heat shock. B, SILAC ratios measure aggregation or synthesis of example proteins Ded1, Pgk1, and Hsp104. Thin purple lines show pSup after 42°C, 8 min. heat shock from Fig. 1. C, Select groups of heat-aggregating proteins during recovery after heat shock (others in Fig. S4B). D, Heat-aggregated proteins disaggregate during recovery, while many proteins change minimally and known heat-induced proteins are synthesized. Cumulative distributions of the normalized ratio in the supernatant are plotted for reliably soluble proteins, superaggregators, other heat-aggregators (annotated in the 46°C timecourse), and proteins whose ribosome occupancy increases at least 20-fold during 42°C, 20min. heat shock (Gerashchenko and Gladyshev, 2014). Wilcoxon rank sum test was used to compare the distributions of reliably soluble proteins to superaggregators and other aggregators, respectively, at each timepoint (**, p < 0.001; ns, not significant).
Heat-insensitive proteins, such as the glycolytic enzyme Pgk1, show minimal change in pre-shock ratio in the supernatant indicating no aggregation, and a slight increase in post-shock ratio indicating low levels of new synthesis during recovery (Fig. 6B). Heat-aggregating proteins have low pre-shock ratio immediately after shock, and their disaggregation is indicated by increase of the pre-shock ratio during recovery with only background-level changes in the post-shock ratio, as seen for the RNA helicase Ded1 (Fig. 6B). Proteins synthesized in response to heat shock, such as the chaperone Hsp104, show an increase in both pre- and post-shock ratios indicating new synthesis; increased signal in both channels reflects incorporation of imported post-shock amino acids and residual or recycled pre-shock amino acids (Fig. 6B). Aggregated, degraded, and resynthesized proteins would show a low pre-shock ratio after shock and an increase in post-shock ratio; we do not observe this pattern.
A biological replicate with isotopic labels permuted shows the same behavior (Fig. S2B). An additional timepoint 180 minutes post-shock, after a full cell doubling, shows that, as expected, the majority of the proteome incorporates the post-shock label (Fig. S2C).
Proteins previously identified as superaggregators by MS aggregate aggressively at 42°C and disaggregate fully after one hour of recovery (Fig. 6C, S4B). Complexes which aggregate coherently also disaggregate coherently, including the multisynthetase complex and the RAC (Fig. 6C).
To examine proteome-scale trends, we compared groups of genes identified as reliably soluble, heat-aggregators, or superaggregators in the 46°C heat-shock dataset. Immediately after heat shock, aggregators and superaggregators synthesized from pre-shock amino acids are depleted from the supernatant compared to reliable soluble proteins (Fig. 6D). After 20 minutes’ recovery at 30°C, the differences between these populations are smaller although still significant, and after 60 minutes of recovery the distributions are indistinguishable, indicating complete disaggregation (Fig. 6D). The post-shock ratios of aggregators and superaggregators are indistinguishable from those of reliably soluble proteins, indicating approximately the same level of new synthesis. At the same time, proteins whose ribosome occupancy increases at least 20-fold during heat shock (Gerashchenko and Gladyshev, 2014) show a substantial increase in new protein synthesis (Fig. 6D). New synthesis post-shock correlates well with ribosome occupancy during shock (Fig. S2D).
In summary, the data show virtually complete disaggregation of endogenous aggregated proteins during recovery without elevated levels of degradative turnover.
Discussion
The standard model of heat stress holds that heat causes protein damage and misfolding, disrupting function and causing exposure of natively buried hydrophobic residues, which triggers protein aggregation (Vabulas et al., 2010). This model was shaped by, and explains well, a wide array of observations, particularly the behavior of endogenous nascent polypeptides. A more recent regulatory interpretation holds that evolutionarily conserved heat-induced aggregation of some proteins into specific subcellular locations reflects a mechanism for attenuating translation (Grousl et al., 2009; Farny et al., 2009; Cherkasov et al., 2013) and protecting the cell during stress (Miller et al., 2015b).
Our study provides multiple lines of evidence indicating that many phenomena that correlate tightly for nascent polypeptides and exogenous unstable reporter constructs—phenomena such as heat-induced aggregation, loss of function, formation of subcellular foci, and degradation—are in many cases completely separable and thus causally unrelated for endogenous mature eukaryotic proteins under acute stress. The standard misfolding model incompletely describes the behavior of most mature proteins during heat shock.
To illustrate, consider the aggressive yet fully reversible thermally induced aggregation of nuclear proteins, exemplified by Ett1 and Gbp2, in light of recent studies on nuclear quality control. The ubiquitin ligase San1 targets nuclear misfolded proteins for degradation (Gardner et al., 2005; Fredrickson et al., 2013). GFP constructs engineered with stretches of hydrophobic residues which promote formation of fluorescent nuclear foci and pelletable aggregates undergo San1-mediated degradation detectable within one hour (Fredrickson et al., 2013). Our expectation was that endogenous nuclear proteins which aggregate should be similarly degraded. However, despite aggressive aggregation of Ett1 and Gbp2 in the nucleus, they are restored to solubility without degradation.
The organized deposition of aggregated proteins into particular subcellular sites, such as stress granules, may provide fitness benefits to organisms during stress (Miller et al., 2015b). Stress granules may facilitate preferential translation of certain mRNAs during stress (Kedersha and Anderson, 2002). By providing a view into how proteins reversibly form large assemblies during stress, without necessary restriction to granular structures or particular sites, our study reveals a separate layer of phenomena rich with exciting functional possibilities. We hypothesize that the heat-induced aggregation of mature proteins reflects the action of a vast, fast-acting regulatory system based on massive molecular assembly and disassembly. This system couples rapid protein-autonomous stress-responsive assembling elements with slower-acting disassembly machines.
Such a system invokes transient interactions beyond quaternary structure, termed quinary organization (McConkey, 1982). Molecular mechanisms and components of quinary regulation may include multivalent interactions (Li et al., 2012), low-complexity sequences (Kato et al., 2012), and phase-separation phenomena including protein and protein/RNA liquids (Weber and Brangwynne, 2012) and hydrogels (Kato et al., 2012). Our studies do not offer a mechanistic picture of aggregation or new evidence for particular physical states of quinary assemblies, but do identify targets for study.
Because heat stress necessarily involves an influx of thermal energy, it would be efficient for aggregation to result from evolved, thermally induced conformational changes which promote quinary interactions. Such processes could be all but indistinguishable from misfolding at the molecular level (Sengupta and Garrity, 2013). The fundamental distinction is in fitness: misfolding is deleterious, whereas evolved quinary regulation is beneficial, suggesting testable and opposing predictions about the fitness consequences of blocking aggregation. We also anticipate that, as in the case of the aminoacyl-tRNA synthetases, evolved quinary interactions will be domain-specific, organized, and rapidly reversible without degradation, unlike the behavior of misfolded proteins.
Our data suggest several mechanisms for focusing translation on stress-induced transcripts (Fig. 7). Translation initiation on most yeast mRNAs depends upon initiation factors (eIFs) and auxiliary proteins (such as the RNA helicase Ded1). We find that these factors partition into two major classes, the heat-resistant factors (including eIF- 1, 1A, 2) and heat-sensitive factors (including eIF-2B, 3, 4G, 5, Ded1). Shirokikh and Spirin (2008) demonstrated assembly of a normal AUG-associated translation initiation complex on uncapped mRNA in vitro in the absence of eIF-2B/3/4A/4B/4G/4E, if a poly(A) leader sequence is present. This poly(A)-mediated cap-independent initiation mechanism may explain the cap-independent translation of heat-shock mRNAs (Rhoads and Lamphear, 1995), which often possess unstructured, A-rich 5’ UTRs (Holmgren et al., 1981) with reduced dependence on RNA unwinding (Lindquist and Petersen, 1990). We hypothesize that stress-sensitive aggregation of initiation and unwinding factors inhibits translation on most non-stress-relevant mRNAs.
Figure 7.
Mechanisms for enhancing cellular remodeling by massive assembly during heat stress.
The enzyme components of the AME complex, the aminoacyl-tRNA synthetases Mes1 and Gus1, have secondary transcriptional and translational activities in the nucleus and mitochondria, respectively, and are excluded from these compartments by complexing with Arc1 (Frechin et al., 2014). We hypothesize that autonomous heat-sensitive self-assembly of AME complexes discovered here confines active AME components to the cytosol, suppressing secondary activities in other compartments and focusing aminoacylation activity in the cytosol where it is needed during stress (Fig. 7).
Molecular chaperones may act as regulatory disassembly factors quite separate from their role in protein folding and misfolding. For example, chaperone-mediated dissolution of AME assemblies would permit return of Gus1 and Mes1 to duty in other cellular compartments. Chaperone-mediated restoration of helicases and cap-dependent initiation factors to solubility would derepress translation of most mRNAs, titrating translational activity away from stress-induced messages and thus closing a feedback loop (Fig. 7). Consistent with this, deletion of the disaggregase Hsp104 delays both heat-shock granule dissolution and reassembly of polysomes after heat shock (Cherkasov et al., 2013). Which factors (chaperones or other proteins) disassemble which assemblies, and whether and how specificity is achieved, can be addressed in large part using the methods we have introduced here.
A similar autoregulatory mechanism has been proposed as a way to link protein quality control and translation through assembly of stress granules (Cherkasov et al., 2013). Our study suggests that neither misfolding nor stressgranule formation need be involved; indeed, heat stress/aggregation/chaperones seem likely to be a special case of a broader class of signal/assembly/disassemblase regulatory systems each involving stress-specific quinary interactions. It seems likely that certain proteins will form assemblies under a wide range of stress conditions (e.g. translation initiation factors), but by stress-specific mechanisms, such as binding sites revealed by phosphorylation, pH-driven self-association, and thermally induced local unfolding. Stress-triggered formation of massive but unanchored assemblies of undamaged proteins, when reversible by stress-induced disassembly activity, allows for a fast-acting autoregulatory response.
Experimental Procedures
Full details are in extended experimental procedures.
Yeast strains and media
The yeast strains used in this study are listed in Table S6. Unless otherwise stated, S. cerevisiae were grown in SC-complete at 30°C to mid-exponential phase.
Fractionation and mass spectrometry measurement
Yeast were heat-treated, flash frozen, and lysed. Protein from total lysate, 100,000 g × 20 minutes supernatant, and pellet fractions was chloroform-methanol extracted, separately digested with trypsin using a FASP protocol (Wísniewski et al., 2009), labeled by reductive dimethylation (Boersema et al., 2009), and mixed. Mixed samples were fractionated by anion exchange and fractions were submitted for LC-MS/MS analysis on an Orbitrap Velos Pro (Thermo Fisher, San Jose, CA).
SILAC recovery assay
Yeast strains auxotrophic for arginine and lysine (RK) were grown with light (rep. 2, heavy) isotope labeled RK at 30°C to mid-exponential phase, transferred to heavy (rep. 2, light) isotope labeled RK, heat shocked for 10 minutes at 42°C, and allowed to recover at 30°C. Cells were harvested at specified times, mixed evenly with unheated cells grown in medium-isotope-labeled RK, and flash-frozen. Mixed samples were lysed and fractionated, and only the supernatant fraction was chloroform-methanol extracted, trypsin digested, and submitted for LC-MS/MS.
Data analysis for mass spectrometry
Mass spectrometry runs were analyzed with MaxQuant (Cox et al., 2011), and reported peptide intensities were further analyzed using a statistical model. In brief, three intensities per peptide detection event—light, medium, and heavy—are noisy proxies for abundance in total, supernatant, and pellet respectively. A Bayesian model, accounting for supernatant-to-total ratios, pellet-to-total ratios, variability in sample mixing, and measurement error, reports the proportion in supernatant for each detected protein.
In the SILAC recovery assay, we report median ratios of MaxQuant-estimated intensities, correcting for deviations from even mixing by fixing the median ratio to 1 for proteins reliably in the supernatant (section S1.5).
We define a protein as heat-aggregating if it is (a) well-detected, i.e. 2 or more unique peptides reported at each timepoint; (b) moves consistently from supernatant to pellet, i.e. the rank correlation of pSup with time is at least 0.8; (c) moves substantially, i.e. pSup across the timecourse declines by at least 0.3. Superaggregating proteins are defined as the subset of heat-aggregators for which pSup declines more than the most extreme HSG component at 2 minutes at 46°C (Tif4632, pSup = 0.40).
Gene Ontology (GO) enrichment analyses were performed using the topGO package (Alexa et al., 2006).
Protein gel electrophoresis and western blotting
SDS-PAGE was performed according to standard methods. Proteins were transferred to nitrocellulose membranes, detected with antibodies against Pab1 (EnCor; #MCA-1G1) or Ssz1 (Hundley et al., 2002) and visualized by chemiluminescence.
Generation of diploid yeast strains
Plasmids pJLS033 and pJLS035 were constructed for C-terminal Clover and mRuby2 labeling at the native locus. Clover/mRuby2 KanMX cassette PCR fragments generated by unique primer pairs (sequences provided upon request) were transformed into BY4741 and BY4742 according to standard lithium acetate protocol (Gietz and Schiestl, 2007) and selected. Two-color diploid yeast strains were constructed by crossing single-color labeled strains by standard methods.
Spinning-disk confocal fluorescence microscopy
Diploid yeast strains were imaged, alive, on an Olympus DSU spinning disk confocal microscope using a 100x oil immersion objective and FITC/Cy2 and DsRed filter sets for Clover and mRuby2 respectively.
Purification of multisynthetase complex
Recombinant 6xHis-tagged Arc1, Gus1, and Mes1 were overexpressed, separately, in E. coli strain BL21 (DE3), and purified.
Aminoacylation assay
Filter-based aminoacylation reactions and aminoacylation reactions for microarray analysis were performed as previously described (Netzer et al., 2009; Wiltrout et al., 2012).
Data access
Raw mass spectrometry data is available on Chorus (https://chorusproject.org, experiments 1751 and 1752); processed data and analysis scripts are available on Dryad (http://dx.doi.org/10.xxx/dryad.xxx).
Supplementary Material
Acknowledgments
We thank members of the Drummond lab for critical comments on the manuscript. This work was funded by grants from the Alfred P. Sloan Foundation to DAD and EMA, the National Institutes of Health (GM105816 via the Protein Translation Research Network, and GM096193), and the Pew Charitable Trusts. DAD is a Pew Scholar in the Biomedical Sciences.
Imaging was performed at the University of Chicago Integrated Light Microscopy Facility; we thank V. Bindokas and C. Labno for their help. We thank B. Glick and K. Day for assistance with image processing, E. Craig for the generous gift of the Ssz1 antibody, Y. Gilad and B. Engelmann for use of computing resources, J. Piccirilli and B. Weissman for scintillation counting assistance, and T. Sosnick for assistance with absorbance and scattering measurements. Some computations were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental information
Supplemental information includes extended experimental procedures, 6 tables, and 7 figures, and can be found with this article online. Interactive data visualizations related to figures 1 and 6 may be found at http://drummondlab.org/endogenous-aggregates.
Author contributions
DAD, MFD, and EWJW designed the mass spectrometry experiments. EWJW and BAB prepared samples for mass spectrometry, and BAB obtained mass spectra. AMF, EMA, EWJW, and DAD designed and implemented data analysis. MFD, JLK-S, AER, and CDK constructed yeast strains. JLK-S performed in vivo microscopy. EWJW ran western blots. EVP, PRL, JAR, AER, and JLK-S purified proteins. EVP, MHS, PRL, JAR, TP, and DAD designed and performed multisynthetase experiments. DAD and EWJW wrote the manuscript with the assistance and approval of all authors.
References
- Aguilaniu H, Gustafsson L, Rigoulet M, Nyström T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Baler R. Heat shock gene regulation by nascent polypeptides and denatured proteins: hsp70 as a potential autoregulatory factor. The Journal of Cell Biology. 1992;117:1151–1159. doi: 10.1083/jcb.117.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CA, MacKenzie MM, Johnston GC, Singer RA. Efficient translation of an SSA1-derived heat-shock mRNA in yeast cells limited for cap-binding protein and eIF-4F. Mol Gen Genet. 1995;246:619–627. doi: 10.1007/BF00298969. [DOI] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJR. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Nissan T, Parker R. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Meth Enzymol. 2010;470:619–640. doi: 10.1016/S0076-6879(10)70025-2. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Yoon J-H, Parker R. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J Cell Sci. 2011;124:228–239. doi: 10.1242/jcs.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452–2462. doi: 10.1016/j.cub.2013.09.058. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA. Metazoan stress granule assembly is mediated by P-eIF2α-dependent and -independent mechanisms. RNA. 2009;15:1814–1821. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk H, Kannambath S, Pahle J, Claydon A, Beynon R, Duncan J, Westerhoff H, Mendes P, McCarthy JE. An in vivo control map for the eukaryotic mRNA translation machinery. Mol Syst Biol. 2013;9:635. doi: 10.1038/msb.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frechin M, Enkler L, Tetaud E, Laporte D, Senger B, Blancard C, Hammann P, Bader G, Clauder-Münster S, Steinmetz LM, et al. Expression of nuclear and mitochondrial genes encoding ATP synthase is synchronized by disassembly of a multisynthetase complex. Molecular cell. 2014;56:763–776. doi: 10.1016/j.molcel.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Fredrickson EK, Gallagher PS, Candadai SVC, Gardner RG. Substrate recognition in nuclear protein quality control degradation is governed by exposed hydrophobicity that correlates with aggregation and insolubility. J Biol Chem. 2013;288:6130–6139. doi: 10.1074/jbc.M112.406710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel B, Tuite MF, McCarthy JEG. The effects of 5’-capping, 3’-polyadenylation and leader composition upon the translation and stability of mRNA in a cell-free extract derived from the yeast Saccharomyces cerevisiae. Molecular Microbiology. 1992;6:2339–2348. doi: 10.1111/j.1365-2958.1992.tb01409.x. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Frydlová I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcova I, Novakova L, Janoskova D, et al. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- Grousl T, Ivanov P, Malcova I, Pompach P, Frydlova I, Slaba R, Senohrabkova L, Novakova L, Hasek J. Heat shock-induced accumulation of translation elongation and termination factors precedes assembly of stress granules in Scerevisiae. PLoS ONE. 2013;8:e57083. doi: 10.1371/journal.pone.0057083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck JW, Cheung SK, Hampton RY. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. P Natl Acad Sci Usa. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren R, Corces V, Morimoto R, Blackman R, Meselson M. Sequence homologies in the 5’ regions of four Drosophila heat-shock genes. P Natl Acad Sci Usa. 1981;78:3775–3778. doi: 10.1073/pnas.78.6.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. P Natl Acad Sci Usa. 2002;99:4203–4208. doi: 10.1073/pnas.062048399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson T, Navarrete C, Sharma SK, Sideri TC, Ibstedt S, Priya S, Grant CM, Christen P, Goloubinoff P, Tamás MJ. Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J Cell Sci. 2012;125:5073–5083. doi: 10.1242/jcs.107029. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. The heat-shock response. Annu Rev Biochem. 1986;55:1151–1191. doi: 10.1146/annurev.bi.55.070186.005443. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Petersen R. Selective translation and degradation of heat-shock messenger RNAs in Drosophila. Enzyme. 1990;44:147–166. doi: 10.1159/000468754. [DOI] [PubMed] [Google Scholar]
- Liu B, Larsson L, Caballero A, Hao X, Oling D, Grantham J, Nyström T. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Martínez-Alonso M, González-Montalbán N, García-Fruitos E, Villaverde A. Learning about protein solubility from bacterial inclusion bodies. Microb Cell Fact. 2009;8:4. doi: 10.1186/1475-2859-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey EH. Molecular evolution, intracellular organization, and the quinary structure of proteins. P Natl Acad Sci Usa. 1982;79:3236–3240. doi: 10.1073/pnas.79.10.3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Ho C-T, Winkler J, Khokhrina M, Neuner A, Mohamed M, Guilbride DL, Richter K, Lisby M, Schiebel E, et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. The EMBO journal. 2015a;34:778–797. doi: 10.15252/embj.201489524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SBM, Mogk A, Bukau B. Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J Mol Biol. 2015b;427:1564–1574. doi: 10.1016/j.jmb.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Mogk A, Tomoyasu T, Goloubinoff P, Rüdiger S, Röder D, Langen H, Bukau B. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netzer N, Goodenbour JM, David A, Dittmar KA, Jones RB, Schneider JR, Boone D, Eves EM, Rosner MR, Gibbs JS, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462:522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L, Scharf KD, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci. 2012;37:274–283. doi: 10.1016/j.tibs.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Pu S, Wong J, Turner B, Cho E, Wodak SJ. Up-to-date catalogues of yeast protein complexes. Nucleic acids research. 2009;37:825–831. doi: 10.1093/nar/gkn1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads R, Lamphear B. Cap-Independent Translation, vol. 203, of Current Topics in Microbiology and Immunology. Springer-Verlag Berlin; 1995. Cap-independent translation of heat shock messenger RNAs; pp. 131–153. [DOI] [PubMed] [Google Scholar]
- Samanfar B, Tan LH, Shostak K, Chalabian F, Wu Z, Alamgir M, Sunba N, Burnside D, Omidi K, Hooshyar M, et al. A global investigation of gene deletion strains that affect premature stop codon bypass in yeast, Saccharomyces cerevisiae. Mol Biosyst. 2014;10:916–924. doi: 10.1039/c3mb70501c. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Garrity P. Sensing temperature. Curr Biol. 2013;23:R304–R307. doi: 10.1016/j.cub.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirokikh NE, Spirin AS. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc Natl Acad Sci USA. 2008;105:10738–10743. doi: 10.1073/pnas.0804940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harbor perspectives in biology. 2010;2:a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J, Abrams J, Wang Y, Morano KA. Biology of the Heat Shock Response and Protein Chaperones: Budding Yeast (Saccharomyces cerevisiae) as a Model System. Microbiol Mol Biol Rev. 2012;76:115–158. doi: 10.1128/MMBR.05018-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Wiltrout E, Goodenbour JM, Féchin M, Pan T. Misacylation of tRNA with methionine in Saccharomyces cerevisiae. Nucleic acids research. 2012;40:10494–10506. doi: 10.1093/nar/gks805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wísniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.